RNA is an essential component of telomerase. It plays roles in the polymerization cycle that replicates telomeric DNA repeats and recruits proteins involved in telomere regulation.
SUMMARY
Telomerase is a eukaryotic ribonucleoprotein (RNP) whose specialized reverse transcriptase action performs de novo synthesis of one strand of telomeric DNA. The resulting telomerase-mediated elongation of telomeres, which are the protective end-caps for eukaryotic chromosomes, counterbalances the inevitable attrition from incomplete DNA replication and nuclease action. The telomerase strategy to maintain telomeres is deeply conserved among eukaryotes, yet the RNA component of telomerase, which carries the built-in template for telomeric DNA repeat synthesis, has evolutionarily diverse size and sequence. Telomerase shows a distribution of labor between RNA and protein in aspects of the polymerization reaction. This article first describes the underlying conservation of a core set of structural features of telomerase RNAs important for the fundamental polymerase activity of telomerase. These include a pseudoknot-plus-template domain and at least one other RNA structural motif separate from the template-containing domain. The principles driving the diversity of telomerase RNAs are then explored. Much of the diversification of telomerase RNAs has come from apparent gain-of-function elaborations, through inferred evolutionary acquisitions of various RNA motifs used for telomerase RNP biogenesis, cellular trafficking of enzyme components, and regulation of telomerase action at telomeres. Telomerase offers broadly applicable insights into the interplay of protein and RNA functions in the context of an RNP enzyme.
1. AN ENDOGENOUS REVERSE TRANSCRIPTASE FOR TELOMERIC REPEAT SYNTHESIS
DNA and protein assemblies at the ends of eukaryotic chromosomes constitute the end-portions, or “telomeres,” essential for genomic stability. Jeopardy is inherent in the terminal location of these chromosome domains: Terminal DNA sequences are under-replicated and even actively eroded in each cell cycle, because of incomplete DNA-templated DNA replication and the nuclease processing required to generate a 3′ overhang for end-capping proteins. The widespread eukaryotic strategy to counter-balance telomeric repeat erosion involves telomerase, a specialized reverse transcriptase capable of de novo DNA synthesis. Telomerase action coupled with additional DNA replication and end-repair activities allows telomere length homeostasis in single-celled organisms and generational maintenance of telomere length in the germline of multicellular organisms (Gilson and Geli 2007; Verdun and Karlseder 2007). Somatic tissues of long-lived organisms repress telomerase, adding a hurdle to tumor progression but limiting tissue renewal (Collins and Mitchell 2002; Garcia et al. 2007). Cancer cells typically activate telomerase and telomere maintenance, with clinical implications described in detail elsewhere (Harley 2008).
The telomerase mechanism is deeply conserved throughout eukaryotes, from flagellated protozoans including Giardia and trypanosomes to higher plants and metazoans. Even the genome of a virus that infects chickens has been discovered to encode a telomerase RNA component, likely acquired by lateral transfer from its metazoan host. Eukaryotic species lacking telomerase-dependent telomere maintenance are the exception rather than the rule, with indications that alternative strategies can be independently derived. Soon after its discovery in the pond ciliate Tetrahymena thermophila (Greider and Blackburn 1985), the enzymatic activity of telomerase was shown to be sensitive to ribonuclease and protease treatment, implicating an RNA as well as protein contribution to enzymatic activity (Greider and Blackburn, 1987). A region within the integral RNA subunit of the enzyme provides the template for telomeric-repeat DNA synthesis (Greider and Blackburn 1989), defining telomerase as a reverse transcriptase (RT). Many telomerases can reiteratively copy the internal template (Fig. 1) without completely releasing product DNA, achieving both nucleotide addition processivity across the template and repeat addition processivity in the synthesis of a product ladder.
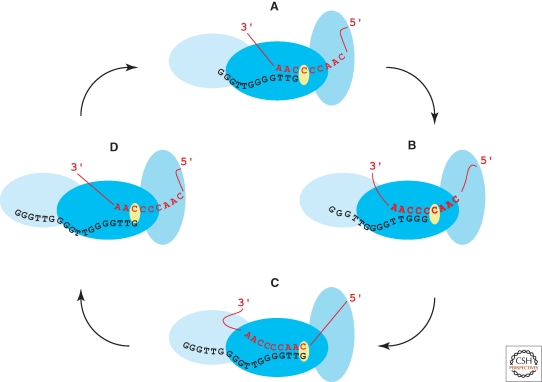
Figure 1.
The telomerase catalytic cycle. Stages in the catalytic cycle are illustrated using T. thermophila sequences of the TER template (red) and telomeric-repeat DNA (black). The active site (yellow) is within the TERT RT domain (filled with the darkest shade of blue). Some TERT-TER contacts that influence template use are stable across all states of the catalytic cycle (shown for T. thermophila as the template 5′-flanking region bound to the TERT TRBD domain in an intermediate shade of blue). Other TERT-TER and TERT-DNA contacts may be specific to particular configurations of template and product relative to the active site (shown here as changing contact between the template 3′-flanking region and the TERT TEN domain in the lightest shade of blue, which also contacts single-stranded DNA). Copying across the template with nucleotide addition processivity is accompanied by changes in the length of hybrid between template RNA and product DNA, depicted in states (A–C). Product released from the template can be held in association with the active enzyme by other interactions, as depicted in state (D). Realignment of the product 3′ end at the template 3′ end, as depicted in the transition from state (D) to state (A), allows for repeat addition processivity.
2. INTERPLAY OF RNA AND PROTEIN FUNCTION IN A MIXED-SUBSTANCE POLYMERASE
The demonstration that an RNA moiety provides the template for polymerization of telomeric DNA repeats established a new paradigm for RNA function in a cellular RNP. But how do the various catalytic steps required of a DNA polymerase—binding of primer and nucleotide substrates, conformational change(s) required for substrate discrimination and alignment, catalysis itself, product and template translocation, and eventual product release—distribute as a division of labor between the RNA and protein components of the telomerase RNP enzyme? Together the telomerase RNA (TER) and telomerase reverse transcriptase protein (TERT) can reconstitute telomeric repeat synthesis in vitro, without the additional proteins that coassemble in vivo to form telomerase holoenzymes (Autexier and Lue 2006; Collins 2006). TERT active site motifs shared with other protein-only RTs play an essential role (Lingner et al. 1997), consistent with their expected function in binding the magnesium ions that activate the deoxynucleoside triphosphate for incorporation. In addition to this general polymerase chemistry, there must be enzyme determinants that adapt telomerase for its unique specificity of template and primer use. This article aims to highlight how the catalytic specialization of telomerase occurred in large part through gains of RNA motif function. Coevolution of coordinated roles for protein and RNA subunits of telomerase presents a challenge to deciphering the earliest stages of its evolution (discussed in section 6.1).
In addition to the gains of function necessary for its biochemical and biological specialization, telomerase is notably lacking in some features typically associated with protein RTs. For example, to reuse its internal template, telomerase must eschew the RT-associated RNase H activity important for degradation of the RNA strand of a retroviral RT RNA-cDNA duplex. Although specializations unique to telomerase are described later, we note that the human LINE-1 retroelement RT lacks RNase H activity and like telomerase shows strong preference for copying an associated RNA (Kulpa and Moran 2006; Piskareva and Schmatchenko 2006).
3. GENERAL FEATURES OF TELOMERASE RNAS
3.1. Uncovering the Core Beneath Sequence Divergence and Evolutionary Innovation
Despite broad evolutionary conservation of the telomerase mechanism, the size and sequence of TERs show great diversity. The TERs of smallest size are the group cloned from ciliated protozoa, only 147–209 nt in length (Ye and Romero 2002). Many TERs have been cloned from vertebrate species as well, with lengths ranging from 312 to 559 nt (Xie et al. 2008). On the long side of the spectrum are the sequenced group of budding yeast TERs, with lengths of 779–1817 nt (Gunisova et al. 2009). The current length record is held by the malarial parasite Plasmodium falciparum, which has a TER of 2.2 kb (Chakrabarti et al. 2007).
What drives this evolutionary diversification, when TERT length and sequence have remained generally more consistent (Fig. 2)? TER phylogenetic diversity could be merely a reflection of freedom to drift. Alternatively, RNA may offer more opportunities than protein for the occurrence, selection, or fixation of functional gains. RNA sequence expansion, gain of interaction, and functional adaptation may be sampled readily over evolution because of inherent properties of RNA structure, leading to advantageous RNP enzyme properties.
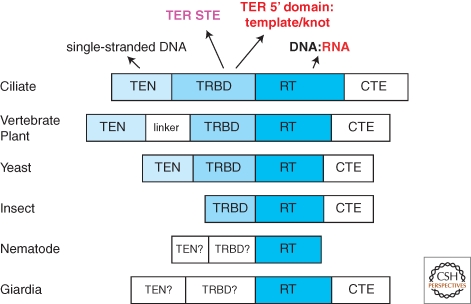
Figure 2.
Conserved TERT domains. TERT domain evolutionary conservation and protein-nucleic acid interactions are summarized, with the same color scheme of TERT and TER domains used in other figures.
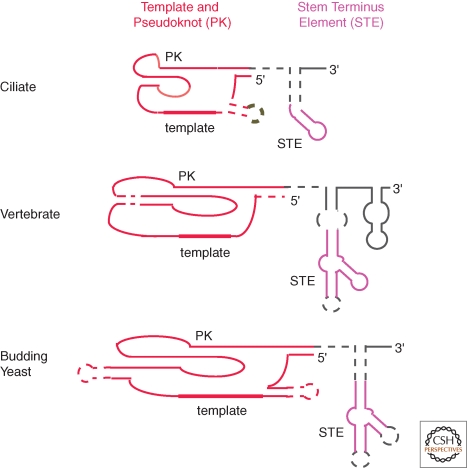
It is difficult to chart the evolutionary steps of TER divergence between the phylogenetic groups of ciliates, vertebrates, and budding yeasts using only modern-day TER secondary structure models. Despite this limitation, we will draw within-group and in some cases between-group comparisons of TER structure and function in the sections to follow, beginning in this section with a description of three TER motifs arguably shared by all known TERs with significance for the catalytic cycle: the template, the pseudoknot, and a stem-loop or bulged stem-junction with conserved paired and unpaired nucleotides (Fig. 3).
Figure 3.
Conserved TER motifs within phylogenetically divergent secondary structures. The thickest region of line represents the template, whereas dashed lines represent sites of sequence variability within the phylogenetic group. PK indicates the pseudoknot. The color scheme of TER motifs matches that in the other figures.
3.2. The Template
The template region in all TERs includes a 5′ portion that is frequently copied and a 3′ portion that is typically used for the alignment of a primer (a synthetic DNA oligonucleotide in vitro or a single-stranded chromosome 3′ end in vivo). Hybridization-based primer alignment allows the synthesis of a precise repeat sequence. Also, if product released from the end of the template is retained by other enzyme associations, the alignment region facilitates repeat addition processivity (Fig. 1). The template-product hybrid must reach a length of up to four to five base-pairs to uniquely match a primer 3′ end with its proper template position. However, at least for the telomerases assayed to date, the maximal possible length of template-product hybrid does not form (Wang et al. 1998; Förstemann and Lingner 2005). Instead, as polymerization progresses along the template, unpairing occurs at the template 3′ end, even if this requires partial dissociation of formerly annealed primer (Fig. 1). Polymerization along the template halts at a typically, but not always, fixed template 5′ boundary. Copying generally results in synthesis of the template complement, but again this is not always true: there are a few examples of deviant nucleotide selection that are biologically used in the normal course of synthesis of mixed or degenerate telomeric repeats, as discussed in detail elsewhere (Collins 2009). Also, some telomerases can extend a primer without hybridization, using a specific template position as the default for initiation (Yu and Blackburn 1991; Wang and Blackburn 1997).
Mutational analyses show that the template is not just a passive carrier of sequence information. Substitutions of one or a few template bases can have large, mutation-specific impacts on rates of dNTP misincorporation, propensity for template slippage or misalignment, or premature product dissociation (Gilley and Blackburn 1996; Lin et al. 2004). Substrate dGTP also affects enzyme function independently of its incorporation, for example altering repeat addition processivity through a presumed allosteric influence on the active site (Hammond and Cech 1997; Hardy et al. 2001). Also, single-stranded DNA can interact with active RNP beyond 3′ base-pairing with the template, affecting several aspects of the polymerization cycle (Collins and Greider 1993; Lee and Blackburn 1993). These and additional results point to a well-tuned codependence of template, nucleotide, and product sequence identity in determining the fidelity and overall activity of internal template use.
3.3. A Shared Pseudoknot
The likelihood of strongly conserved nontemplate TER structures emerged from comparison of the phylogenetically derived TER secondary structures of ciliates, vertebrates, and budding yeasts (Legassie and Jarstfer 2006; Theimer and Feigon 2006). All contain a similarly folded pseudoknot. The template and pseudoknot together are enclosed within a TER domain formed by long-distance pairing of the 5′ end of TER (Fig. 3). Ciliate TERs show thermodynamically weak pseudoknot folding within their compact overall structure, whereas yeast TERs tolerate long stem-loop insertions in the template/pseudoknot domain. Curiously, rodent TERs have lost the long-distance 5′ end pairing.
One set of open questions concerns the function(s) of the pseudoknot and pseudoknot structural dynamics. The pseudoknot is one of the last elements of ciliate TERs to adopt a stable fold during assembly of the minimal RNP in vitro, which it does only if TERT contacts are established with flanking TER motifs (Sperger and Cech 2001). On the other hand, the vertebrate TER pseudoknot folds independently into a rigid structure with highly determined triple-helix U-A-U base-pairing architecture (Theimer et al. 2005). Modeling and mutagenesis indicate that triple-helix structure is conserved and essential in budding yeasts as well (Shefer et al. 2007). The thermodynamic stability of human TER pseudoknot folding would seem to argue against catalytic cycle dynamics. However, tertiary structure dynamics could be coupled to distortions of RNA and protein conformation that occur during a cycle of repeat synthesis, as the template transits the active site.
RNA motif transplantation assays have suggested that a particular TER pseudoknot can be replaced by another structure with retention of TER function. Also, circular permutation and trans complementation assays of ciliate and vertebrate TERs indicate that the pseudoknot can stimulate the activity of recombinant TERT even if it is disrupted in backbone continuity or covalently unlinked from adjacent sequence. However, these chimeric, fragmented, and mutated pseudoknot success stories may not have detected important roles of the pseudoknot in the catalytic cycle. In vivo expression of some but not all T. thermophila TER pseudoknot-disruption variants reduce cell viability and enzyme activity (Gilley and Blackburn 1999; Cunningham and Collins 2005). Some Kluyveromyces lactis pseudoknot mutations that do not abrogate enzyme activity reduce the extent of template copying, leading to variably truncated repeat synthesis in vivo (Tzfati et al. 2003). Also, in vitro assays with a minimized Saccharomyces cerevisiae TER suggest that pseudoknot 2′-OH–mediated interactions contribute to primer positioning in the active site (Qiao and Cech 2008).
3.4. A Stem Terminus Element
Beyond the template and pseudoknot, TERs have at least one additional motif that may have been present in the ancestral enzyme. In all groups of TERs characterized to date, a stem-loop or bulged stem-junction is important for activity, with unpaired loop or bulge residues adjacent to a stem terminus being particularly critical. In the context of modern-day TER secondary structures (Fig. 3), this stem terminus element (STE) can occur as a terminal stem-loop (loop IV of ciliate TER), a stem-loop emerging from a stem junction with bulged nucleotides (P6.1 of vertebrate TER), or a stem junction with bulged nucleotides (the three-way junction of budding yeast TER). It is reasonable to surmise that the function of the STE may be conserved among ciliate, vertebrate, and yeast telomerases, but unrelated functions are also possible. The ciliate and vertebrate STE motifs both bind TERT in a manner separable from TERT interaction with the template/pseudoknot domain (discussed in section 4.2) and greatly stimulate telomerase catalytic activity. Robust, highly processive activity can be reconstituted using ciliate or vertebrate TER fragments separately carrying the template/pseudoknot domain and STE, indicating that STE function does not require covalent linkage with the template (Mitchell and Collins 2000; Lai et al. 2003; Mason et al. 2003). Curiously, although K. lactis telomerase holoenzyme activity is severely inhibited by STE mutations, the STE is missing from a minimal TER that reconstitutes core S. cerevisiae telomerase enzyme activity in vitro (Zappulla et al. 2005; Brown et al. 2007). In contrast, whereas STE substitutions cripple the activity of the T. thermophila minimal active RNP reconstituted in vitro, a small fraction of STE-mutant holoenzyme reconstituted in vivo that is stable to purification has normal catalytic activity (Robart et al. 2009).
One plausible model for STE function is in the allosteric modulation of interactions among independently folded domains of TERT (Fig. 2). STE interaction with the TERT high-affinity RNA binding domain (TRBD) could order the adjacent RT domain for a correct disposition of template docking, perhaps holding off active site closure by the TERT amino-terminal domain (TEN) and carboxy-terminal extension (CTE) until the template has been properly placed. The STE could also play a more direct role in the catalytic cycle. Sequence substitutions of certain residues of the STE cripple catalytic activity without severely impacting TERT binding. These residues could map an RNA surface involved in TERT conformational rearrangement, or they could instead map a surface that is oriented by TERT to face the active site. The STE lies far from the template in TER secondary structure, but purified human TER crosslinking and single-molecule FRET studies of ciliate telomerase RNP assembly suggest a tertiary structure proximity of the STE and template (Ueda and Roberts 2004; Stone et al. 2007). Possibly like the pseudoknot, the STE could have a role in shaping the active site that is obscured in the modern-day RNP enzyme by subsequent gains of function.
4. REFINEMENT OF THE CATALYTIC CYCLE, INCLUDING ROLES FOR PHYLOGENETICALLY DIVERSE TER MOTIFS
4.1. Fine-Tuning Repeat Synthesis Activity
Beyond the potentially universal TER motifs described earlier, TER motifs specific to individual phylogenetic groups also contribute to the functional elaboration of telomerase enzymes. Indeed, TERs appear to have finessed the art of telomerase evolutionary adaptation, exploiting it both to fine-tune the catalytic core (discussed in this section) and to recruit holoenzyme proteins important for RNP biogenesis and regulation (discussed in section 5.1).
Importantly, stable association of TERT and TER is not mediated by the RT domain and template. TERT by itself cannot recruit an RNA oligonucleotide to the active site, even an RNA comprised of the native template and its flanking sequence, and even if this RNA is pre-annealed to a telomeric DNA primer (Miller and Collins 2002). If telomerase evolved from a protein-only RT (discussed in section 6.1), a loss-of-function in active site binding to template may have been evolutionarily advantageous as a mechanism for limiting cDNA insertion into the genome. By this model, telomerase would have had to acquire gain-of function mechanisms that productively position the TER internal template and a chromosome 3′ end substrate in the TERT active site.
4.2. RNA Motifs for Binding TERT
One obvious mechanism for telomerase gain-of-function was the physical partnership of protein and RNA, creating the first telomerase RNP. A TERT domain adjacent to the RT motifs, the TRBD (Fig. 2), is necessary and sufficient for the high biological specificity of TERT-TER interaction in ciliate and vertebrate TERTs assayed to date (Lai et al. 2001; O’Connor et al. 2005). Curiously, the TRBD binds to both the template/pseudoknot domain and the STE (Figs. 2 and 3). The logic of two separate TERT-TER interactions appears not to be to increase specificity per se, as one or other TER motif still allows TERT-specific interaction. Rather, these TRBD-TER contacts may be part of a much more complex set of RNP folding cues required to create an active site around an internal template and accommodate the changes in RNA topology that occur during a cycle of repeat synthesis.
4.3. TER Roles in Enforcing the Template 5′ Boundary
TER also functions in template 5′ boundary demarcation. An accurate halt in synthesis at the template 5′ end is an inherent requirement for precise repeat synthesis. In budding and fission yeasts, a stem just 5′ of the template limits template copying (Tzfati et al. 2000; Box et al. 2008). TER substitutions that alter base-pairing of this stem suggest that it acts as a simple steric block. In ciliates, the terminal base pair of a template-adjacent helix and its surrounding single-stranded residues bind TERT to establish the 5′ limit of the template. TER or TERT substitutions that weaken the interaction also weaken the boundary (Lai et al. 2002). Human telomerase template boundary definition also requires a template-flanking helix, but it is separated from the template 5′ end by intervening single-stranded residues (Chen and Greider 2003). Albeit set in different RNP structural contexts, all known template 5′ boundary determination mechanisms apparently share the feature of steric block or strain against progression of template through the active site. In rodent species in which the TER 5′ end is just a few nucleotides beyond the template 5′ boundary, the biochemical mechanism of template boundary definition remains unknown. The theme of steric occlusion could still hold true, potentially involving the vertebrate TER 5′ trimethylguanosine cap (Chen and Greider 2003; Jády et al. 2004).
4.4. Exchange of Protein-Nucleic Acid Associations across the Catalytic Cycle
Instead of a constant grip on the duplex of RNA template and DNA product, the telomerase active site must recognize a variable length of duplex and also allow for its complete dissociation (Fig. 1). TERT and TER both play important roles in this template handling dynamic, as described in detail elsewhere (Collins 2009). Activity defects imposed by TERT mutagenesis suggest that template residues within and nearby the active site are positioned in part by amino-acid side-chains shared among all RTs (Miller et al. 2000). On the other hand, protein-only RTs would not experience single-stranded RNA at the 3′ end of the template, such as the template residues liberated from the hybrid (Fig. 1). Thus, instead of RT domain interactions, the 3′ end of the template and its adjacent flanking region are likely to make dynamic contact with a TERT-specific protein domain. Transient TER-TERT contacts that fine-tune template use in a dynamic manner over the catalytic cycle appear distinct from stable TER-TRBD interactions that maintain RNP integrity. In addition to several types of TER interactions, TERT also mediates interactions with single-stranded DNA involving the TEN domain (Jacobs et al. 2006; Romi et al. 2007).
5. BELLS AND WHISTLES: TER MOTIFS FOR HOLOENZYME PROTEIN INTERACTIONS
5.1. TER Motifs for Holoenzyme Biogenesis and Regulation
Every phylogenetic group of TERs characterized to date contains group-specific motifs dispensable for catalytic activity reconstitution in vitro yet critical for holoenzyme assembly and function in vivo (Legassie and Jarstfer 2006; Theimer and Feigon 2006). Even the compact ciliate TERs include a ciliate-specific motif for binding of a holoenzyme protein essential for RNP biogenesis. Vertebrate TER elaboration likewise provides for stable RNP biogenesis and gives the RNP its own nuclear address code, perhaps made necessary by nuclear envelope breakdown with each cell cycle. Fungal TER elaborations are the most extensive, with variable arms extending from the core that offer binding sites for the Sm protein complex (required for RNP biogenesis), the telomerase holoenzyme protein Est1 (required for telomere elongation), and in some species the DNA end-binding heterodimer Ku (required for efficient RNP nuclear localization and recruitment to chromosome ends). The geometry of these binding sites has a degree of positional flexibility for TER function in vivo, leading to the notion of protein beads on an RNA string (Zappulla and Cech 2006). It remains uncertain whether modules are actually flexible relative to one another or rigid but distance-independent in function and whether they are alternately, independently, or cooperatively engaged in protein-RNA interactions.
5.2. Vertebrate TER: Nuclear Assembly and Addressing
The vertebrate-specific motifs of TER create a hairpin-Hinge-hairpin-ACA (H/ACA) motif shared by a large family of RNPs (snoRNPs) that catalyze the site-specific isomerization of uridine to pseudouridine. Each H/ACA-motif stem assembles in highly chaperoned fashion with a set of four proteins: the pseudouridine synthase Cbf5p/NAP57/dyskerin, NHP2, NOP10, and GAR1 (Collins 2008). Bulged regions in each snoRNA stem occur at fixed spacing relative to the Hinge or ACA; unpaired residues of these stem “pockets” hybridize to a target ribosomal or small nuclear RNA. The pocket sequences of vertebrate TERs are not conserved, suggesting that TER does not function as a guide for pseudouridine modification. Why then do vertebrate TERs harbor an H/ACA motif? This motif directs RNP assembly and precursor RNA processing, providing a mechanism for nonpolyadenylated RNA 3′ end formation. In addition, the H/ACA motif and its associated proteins provide vertebrate TERs with nuclear address codes.
Cajal bodies are nuclear domains of concentrated RNP biogenesis and recycling. Subsets of H/ACA and other RNPs, including the human telomerase RNP, are enriched in Cajal bodies. A short RNA element called the CAB box is found in both loops of the small Cajal body H/ACA RNAs that modify small nuclear rather than ribosomal RNA targets and is also found as a single copy in the 3′ loop of many but not all vertebrate TERs (Jády et al. 2004; Xie et al. 2008). CAB box mutation shifts the predominant concentration of human TER from Cajal bodies to nucleoli and slows telomerase-mediated telomere over-elongation in transformed cells but not telomere elongation in primary fibroblasts. The CAB box and H/ACA protein dyskerin bind to a conserved, Cajal body-concentrated WD40-domain protein, WDR79/TCAB1 (Tycowski et al. 2009; Venteicher et al. 2009). Long-term WDR79 depletion in transformed cells shortens telomeres, suggesting that in these cells either TER RNP concentration in Cajal bodies or its lack of concentration in nucleoli improves telomerase function. Cajal bodies are not detectable in all cell types and are disrupted by some forms of cell stress, raising the question of whether TER distribution and its access to telomeres also vary with the state of the cell.
5.3. Yeast TER: Finding and Elongating the Telomere
Beyond base-pairing of the template with single-stranded chromosome-terminal telomeric repeats, TER can play other roles in bringing telomerase to telomeres. One of the S. cerevisiae TER “beads on a string” modules binds to Ku80, the larger subunit of a Ku heterodimer that binds double-stranded DNA ends. Telomere maintenance by S. cerevisiae telomerase is partially dependent on Ku-TER interaction (Stellwagen et al. 2003). Also, nuclear localization of S. cerevisiae telomerase RNP is enhanced by TER association with Ku (Gallardo and Chartrand 2008). At least some budding yeast TERs also harbor a transplantable motif for interaction with the telomerase holoenzyme protein Est1 (Zappulla and Cech 2004). Est1 contributes to telomerase-telomere association in part by bridging TER and Cdc13, a single-stranded telomeric DNA binding protein and component of the Cdc13-Stn1-Ten1 telomeric 3′ overhang capping complex (Pennock et al 2001; Gao et al. 2007). The S. cerevisiae Ku-TER and Cdc13-Est1-TER mechanisms of telomerase RNP recruitment to the telomere are distinguished by differential timing: Ku-mediated recruitment of telomerase to telomeres is evident over most of the cell cycle, whereas Cdc13-mediated recruitment of telomerase to telomeres is largely restricted to the DNA replication phase (Chan et al. 2008).
6. TELOMERASE AS A WINDOW TO AN EVOLUTIONARY RNP RENAISSANCE
6.1. Evolutionary Origins of TERT and TER
Even deeply branching eukaryotes harbor a TERT gene with active-site motifs common not just to TERTs but also mobile Group II intron RTs, non-LTR retroelement RTs, and retroviral RTs (Nakamura and Cech 1998). Did the earliest version of a chromosome-end extending telomerase activity begin with the active site of a protein RT that subsequently gained an RNA cofactor, or did this activity begin as a catalytic RNA that later transferred metal-binding active-site duty to a protein? Telomerase likely arose in the last common ancestor of all eukaryotes after the occurrence of fragmented chromosomes, which would have set the stage for RT active-site exaption from a mobile element (as required by the protein-first model). On the other hand, the presence of conserved TER motifs such as the pseudoknot raises the prospect that much of the ancestral telomerase enzyme function could have been RNA-mediated (as required by the RNA-first model). The reshapings of TER over relatively short evolutionary time frames obscure its early origin, but also open a window of opportunity to appreciate the evolving interplay of protein and RNA function in RNP enzyme context.
6.2. RNA Motif Gain-Of-Function in RNP Context
The high thermodynamic stability of even short segments of random RNA sequence creates a barrier to homogeneous folding of large RNAs (Herschlag 1995). Transition from the RNA World to an RNP World of better RNA catalysts exploited peptide chaperones and stably RNA-bound cofactors of hierarchical RNP biogenesis, which gave specificity to the assembly of ribosomes and spliceosomes (described in other articles of this collection). This same machinery has been exploited by telomerase: ciliate, yeast, and vertebrate TERs all require RNP biogenesis chaperones and/or holoenzyme protein cofactors to direct successful maturation and folding of TER in vivo (Collins 2006; Collins 2008; Gallardo and Chartrand 2008). This suggests that the same principles of RNP biogenesis allowed expansion of RNA motif function whether the RNP harbors a catalytic RNA or protein active site. The example of telomerase, among various recently evolving RNPs, provides evidence of an increase in noncoding RNA complexity occurring in RNP context (Hogg and Collins 2008). Future studies of TER motifs and their roles will uncover new insights about telomerase mechanism and also illuminate how an RNP enzyme can accomplish gain-of-function through RNA, providing a window to the evolutionary RNP Renaissance
REFERENCES
- Autexier C, Lue NF 2006. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem 75: 493–517 [DOI] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P 2008. A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem 283: 24224–24233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y 2007. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res 35: 6280–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti K, Pearson M, Grate L, Sterne-Weiler T, Deans J, Donohue JP, Ares MJ 2007. Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA 13: 1923–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A, Boulé JB, Zakian VA 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet 4: e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Greider CW 2003. Template boundary definition in mammalian telomerase. Genes Dev 17: 2747–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K 2006. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol 7: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K 2008. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev 129: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K 2009. Forms and functions of telomerase RNA. In Non-protein coding RNAs (ed. Walter NG, Woodson SA, Batey RT), pp. 285–301 Springer-Verlag, Berlin [Google Scholar]
- Collins K, Greider CW 1993. Nucleolytic cleavage and non-processive elongation catalyzed by Tetrahymena telomerase. Genes Dev 7: 1364–1376 [DOI] [PubMed] [Google Scholar]
- Collins K, Mitchell JR 2002. Telomerase in the human organism. Oncogene 21: 564–579 [DOI] [PubMed] [Google Scholar]
- Cunningham DD, Collins K 2005. Biological and biochemical functions of RNA in the Tetrahymena telomerase holoenzyme. Mol Cell Biol 25: 4442–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Lingner J 2005. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep 6: 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Chartrand P 2008. Telomerase biogenesis: The long road before getting to the end. RNA Biol 5: 212–215 [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V 2007. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14:208–214 [DOI] [PubMed] [Google Scholar]
- Garcia CK, Wright WE, Shay JW 2007. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res 35: 7406–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH 1996. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol 16: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH 1999. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Nat Acad Sci 96: 6621–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V 2007. How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51: 887–898 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L 2009. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15: 546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond PW, Cech TR 1997. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res 25: 3698–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CD, Schultz CS, Collins K 2001. Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J Biol Chem 276: 4863–4871 [DOI] [PubMed] [Google Scholar]
- Harley CB 2008. Telomerase and cancer therapeutics. Nat Rev Cancer 8: 167–179 [DOI] [PubMed] [Google Scholar]
- Herschlag D 1995. RNA chaperones and the RNA folding problem. J Biol Chem 270: 20871–20874 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Collins K 2008. Structured non-coding RNAs and the RNP Renaissance. Curr Opin Chem Biol 12: 684–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol 13: 218–225 [DOI] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV 2006. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol 13: 655–660 [DOI] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K 2002. Template boundary definition in Tetrahymena telomerase. Genes Dev 16: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell 11: 1673–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K 2001. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol 21: 990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Blackburn EH 1993. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol 13: 6586–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legassie JD, Jarstfer MB 2006. The unmasking of telomerase. Structure 14: 1603–1609 [DOI] [PubMed] [Google Scholar]
- Lin J, Smith DL, Blackburn EH 2004. Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol Biol Cell 15: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Mason DX, Goneska E, Greider CW 2003. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol 23: 5606–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Collins K 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci 99: 6585–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Liu JK, Collins K 2000. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J 19: 4412–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Collins K 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol Cell 6: 361–371 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR 1998. Reversing time: Origin of telomerase. Cell 92: 587–590 [DOI] [PubMed] [Google Scholar]
- O’Connor CM, Lai CK, Collins K 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem 280: 17533–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387–396 [DOI] [PubMed] [Google Scholar]
- Piskareva O, Schmatchenko V 2006. DNA polymerization by the reverse transcriptase of the human L1 retrotransposon on its own template in vitro. FEBS Lett 580: 661–668 [DOI] [PubMed] [Google Scholar]
- Qiao F, Cech TR 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, O’Connor CM, Collins K 2010. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA 16: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romi E, Baran N, Gantman M, Shmoish M, Min B, Collins K, Manor H 2007. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc Natl Acad Sci 104: 8791–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y 2007. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol 27: 2130–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger JM, Cech TR 2001. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 40: 7005–7016 [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MS, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X 2007. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Feigon J 2006. Structure and function of telomerase RNA. Curr Opin Struct Biol 16: 307–318 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Blois CA, Feigon J 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell 17: 671–682 [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288: 863–867 [DOI] [PubMed] [Google Scholar]
- Tzfati Y, Knight Z, Roy J, Blackburn EH 2003. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev 17: 1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda CT, Roberts RW 2004. Analysis of a long-range interaction between conserved domains of human telomerase RNA. RNA 10: 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J 2007. Replication and protection of telomeres. Nature 447: 924–931 [DOI] [PubMed] [Google Scholar]
- Wang H, Blackburn EH 1997. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J 16: 866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gilley D, Blackburn EH 1998. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J 17: 1152–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen JJ 2008. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem 283: 2049–2059 [DOI] [PubMed] [Google Scholar]
- Ye AJ, Romero DP 2002. Phylogenetic relationships amongst tetrahymenine ciliates inferred by a comparison of telomerase RNAs. Int J Syst Evol Microbiol 52: 2297–2302 [DOI] [PubMed] [Google Scholar]
- Yu G, Blackburn EH 1991. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell 67: 823–832 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2004. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc Natl Acad Sci 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2006. RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol 71: 217–224 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12: 1072–1077 [DOI] [PubMed] [Google Scholar]