Abstract
Gastric cardia cancer (GCC), which occurs at the gastric-esophageal boundary, is one of the most malignant tumors. Despite its high mortality and morbidity, the molecular mechanism of initiation and progression of this disease is largely unknown. In this study, using proteomics and metabolomics approaches, we found that the level of several enzymes and their related metabolic intermediates involved in glucose metabolism were deregulated in GCC. Among these enzymes, two subunits controlling pyruvic acid efflux, lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase B (PDHB), were further analyzed in vitro. Either down-regulation of LDH subunit LDHA or overexpression of PDH subunit PDHB could force pyruvic acid into the Krebs cycle rather than the glycolysis process in AGS gastric cancer cells, which inhibited cell growth and cell migration. Our results reflect an important glucose metabolic signature, especially the dysregulation of pyruvic acid efflux in the development of GCC. Forced transition from glycolysis to the Krebs cycle had an inhibitory effect on GCC progression, providing potential therapeutic targets for this disease.
Gastric cardia cancer (GCC),1 which occurs at the gastric-esophageal boundary, is one of the most malignant tumors. Despite the steadily falling incidence of gastric non-cardia cancer in the past two decades (1), the rate of GCC has risen rapidly, establishing gastric cancer as the second major cause of cancer-related deaths throughout the world (2). GCC has become a significant cause of mortality and morbidity both in the west (3–5) and in Asia (6, 7), especially in China (8). Although this cancer has become an important health problem worldwide, the its pathogenesis has not been well characterized (1). Most patients are diagnosed at an advanced stage, contributing to the high mortality rate of the disease.
Systematic proteomics analysis has proved to be a powerful approach in a variety of human cancer research, including lung (9), esophagus (10), gastric (11), liver (12), breast (13), and brain cancer (14). Metabolomics, another new bio-omics technology recently introduced into cancer research (15), is the global analysis of the small metabolites produced by normal or pathologic cellular processes. Some metabolic intermediates have been identified as new cancer biomarkers (16).
Using proteomics and metabolomics methods in this study, we found that a series of proteins and metabolic intermediates, mainly involved in glucose metabolism, were altered during the development of GCC. The high activity of anaerobic glycolysis and the impairment of aerobic respiration occurring in these cells recapitulated the Warburg effect (17). Further studies using a gastric cancer cell line demonstrated that the predominant anaerobic glycolysis was essential for tumor cells to sustain rapid proliferation, whereas forced transition from anaerobic glycolysis to aerobic respiration inhibited the growth of tumor cells. In conclusion, our study revealed the major metabolic alterations essential for the development of GCC and discovered a biomarker signature of GCC. Such a finding has the potential to improve early diagnosis and prognosis and helps to identify new therapeutic targets.
MATERIALS AND METHODS
Patients
All GCC tissues and corresponding non-cancerous gastric cardia tissues were obtained from patients treated at the First Affiliated Hospital of Zhengzhou University (Henan, China) with written consent from the patients. Our study was approved by the Ethics Committee Board of Zhengzhou University. TNM classification, according to the criteria of the American Joint Committee on Cancer and the International Union against Cancer, was used by pathologists to stage the tumors (18). The corresponding non-cancerous tissues were resected 5 cm away from the cancer tissues from the same patient. All the samples were quickly frozen by liquid nitrogen and then kept at −80 °C. The basic histopathologic variables are summarized in Table I.
Table I. Histopathological characteristics of 65 samples selected for analysis (proteomics and metabolomics analysis, RNA, and protein validation).
| Characteristic | No. of patients |
|---|---|
| Gender | |
| Female | 7 |
| Male | 58 |
| Age | |
| <60 | 33 |
| ≥60 | 32 |
| Tumor size (cm3) | |
| <5 | 10 |
| ≥5 | 55 |
| Tumor differentiation | |
| Moderate | 42 |
| Poor | 23 |
| Tumor infiltration depth | |
| ≤T3 | 21 |
| T4 | 44 |
| Lymph node metastasis | |
| N0 | 11 |
| N1 | 54 |
| TNM stage | |
| I | 3 |
| IIa | 16 |
| IIb | 6 |
| III | 40 |
Proteomics Analysis
Five pairs of matched samples, randomly selected and mixed according to their differentiation level or lymph node metastasis status, were crushed and dissolved in lysis buffer (7 m urea, 2 m thiourea, 4% CHAPS, 0.5% IPG buffer, 3–10 NL, 0.5% Triton) containing protease inhibitor mixture. The process of two-dimensional gel separation and protein identification was carried out as described previously (10). The peptides were generated by trypsin. Keratin and trypsin peptide peaks were excluded from analysis. Both ESI-TOF and MS/MS spectra were processed by Analysis QS 1.1 Build 9865 (Applied Biosystems) and were searched using Mascot 1.6b13 (Matrix Science). All spectra were searched against the mass spectrometry protein sequence database (MSDB) from the National Center for Biotechnology Information (release date, September 8, 2006; including 3,239,079 sequences). The following parameters were used for the searches: one missed and/or nonspecific cleavage permitted; mass tolerance for precursor ions, ±0.2 Da; mass tolerance for fragment ions, ±0.5 Da; MS/MS filtering: reject spectra if less than 10 peaks or precursor <50 or precursor >10,000. Probability-based Mowse score was used as the tool to determine whether the hit was a random event.
Metabolite Profiling
50 mg of tissue samples were pulverized after being frozen in liquid nitrogen with the addition of 250 μl of mixed solvent (chloroform:methanol:water, 1:2:1, v/v/v). The lysate was ultrasonicated for 1 min and centrifuged at 12,000 rpm for 10 min. A total of 150 μl of aqueous supernatant was transferred to a GC vial containing two internal standards, l-2-chlorophenylalanine (10 μl, 0.3 mg/ml) and heptadecanoic acid (10 μl, 1.0 mg/ml). The deposit was rehomogenized with a T10 basic homogenizer (IKA, Staufen, Germany) for 30 s at 0 °C after adding 250 μl of methanol. After a second centrifugation, another 150-μl aliquot of supernatant was added to the mixture in the GC vial and vacuum-dried. Following the derivatization process, GC-TOF MS spectral acquisition and data analysis were performed according to the procedure described previously (19) with some modifications. Details of the method are provided in the supplemental material.
RNA Isolation and Real Time PCR
Total RNA was extracted from GCC specimens and their matched normal counterparts using TRIzol according to the standard protocol. 2 μg of RNA were processed directly to cDNA using a reverse transcription kit (Promega, Madison, WI) according to the manufacturer's instructions. Amplification reactions were performed in a 15-μl volume with 0.2 μl of SYBR Green. All of the reactions were performed in triplicate using an iCycler iQ System (Bio-Rad). To confirm the specificity of amplification, PCR products from each primer pair were subjected to a melting curve analysis and subjected to electrophoresis using a 2% agarose gel. POLR2A was used as an internal control for normalization. The relative gene expression level of each pair of GCC samples was calculated as described previously (20). The primers used in the experiments are listed in Table II.
Table II. Primers used for real time PCR.
| Gene | Primer sequences |
|---|---|
| ALDOA | CCCCAAGTTATCAAATCC, GAAGTCAGCTCCGTCCT |
| GAPDH | ATGTTCGTCATGGGTGTGAA, GTCTTCTGGGTGGCAGTGAT |
| ENO1 | CAGGTCTGGGAAGTATGA, GCGATCCTCTTTGGGT |
| Pyruvate kinase isozymes M2 isoform 1 (PKM2a) | TGGAGAAACAGCCAAAG, GGCGGAGTTCCTCAAAT |
| Pyruvate kinase isozymes M2 isoform 2 (PKM2b) | GTGCGAGCCTCAAGTCA, ACGTGGGCGGTATCTGG |
| LDHA | TGAATGTTGCTGGTGTC, GTAGCCTTTGAGTTTGAT |
| LDHB | CTAGATTTCGCTACCTTAT, TCATTGTCAGTTCCCATT |
| PDHB | GTGATAAATATGCGTACCATTAGACC, CAGCACCAGTGACACGAACAGC |
| ACO2 | CCACCACTTCCGTGTTCC, CACCTTGCCCACTTCTGC |
| FH | GTGAACTAAAGGTGCCAAAT, CATACCACGAGAGGAAAATG |
| MDH | GCTCGAAAACTATCCAGTG, TAGGGAGACCTTCAACAAA |
| UQCRC1 | GGGAGATGCAGGAGAATGAT, TCTGGGCGAGGTCTAACAGT |
| IDH1 | GAAGAAGGTGGTGGTGTTGCC, CTGGGACTTGTACTGCTTGTCATAT |
| Polymerase (RNA) II (DNA-directed) polypeptide A (POLR2A) | GCAGGACGTAATAGAGG, CGGACACGACCATAGA |
Western Blot
Proteins concentrations of samples extracted from tissue specimens and cell lines using radioimmune precipitation assay lysis buffer were determined using Bradford reagent (Sigma) according to the manufacturer's instructions. Western blot analysis was carried out by standard protocol. The following antibodies were used: murine anti-α-enolase (ENO1) (1:2000) from Abnova Corp., rabbit anti-lactate dehydrogenase A (LDHA) (1:1000) and -LDHB (1:1000) from Epitomics, and rabbit anti-aconitate hydratase (ACO2) (1:1000) from Proteintech Group Inc.
Cell Culture
Gastric cancer cell line AGS was obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai Institute of Cell Biology, Chinese Academy of Sciences) and cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum and 10 units/ml penicillin-G at 37 °C in a humidified atmosphere containing 5% CO2.
Establishment of AGS/Pyruvate Dehydrogenase B (PDHB) Stable Cell Line
The pcDNA3.1-PDHB and pcDNA3.1 plasmids were transfected into AGS cells using FuGENE reagent. The transfected cells were selected in the presence of G418 (800 μg/ml), and after 2 weeks of selection, single clones were isolated for further analysis.
RNA Interference of LDHA
Target sequences of small interfering RNA (siRNA) for LDHA as well as a randomly aligned sequence used as negative control are listed in supplemental Table S1. FG12 lentiviral vector was used to produce double-stranded siRNA to inhibit LDHA expression in AGS cells; the information and usage of this vector system have been described previously (21).
High Performance Liquid Chromatography Analysis of Lactic Acid
Supernatants and cell lysis, respectively, were collected after the cells were grown for 2 days. Three volumes of methanol were added to remove protein from the samples above. After centrifugation and filtration through 0.22-μm PVDF membrane, a 20-μl aliquot of supernatant was injected into an Agilent 1100 instrument. Separation was achieved on an Agilent Zorbax Extend C18 column (4.6 × 250 mm, 5 μm) with phosphate buffer (100 mm NaH2PO4 in 0.01 m H3PO4, pH 2.5) containing 5% acetonitrile as the mobile phase at a constant flow rate of 0.8 ml/min at 25 °C. The total time of separation was 7 min, and lactic acid was detected by a diode array detector at 210 nm at about 3.7 min. The relative level of lactic acid was normalized to the corresponding control samples according to their peak area.
MTT and Soft Agar Assay
For cell growth analysis, equal numbers of cells were seeded in 48-well plates. Cell numbers were measured by MTT assay according to the manufacturer's protocol (Roche Applied Science) every 2 days. For the clonogenic assay, cells were plated into 24-well, flat bottom plates using a two-layer soft agar system with 2000 cells/well in a volume of 400 μl/well as described previously (22). After 7 days of incubation, the colonies were counted and measured.
Statistical Analysis
SPSS 13.0 for Windows was used for statistical analysis, and data are expressed as mean ± S.E. Statistically significant differences were determined by Student's t test, one-way analysis of variance, and bivariate correlation analysis where appropriate and defined as p < 0.05 (*) and p < 0.01 (**).
RESULTS
Proteomics Analysis of GCC
To identify proteins involved in either initiation or progression of GCC, protein expression profiles were compared between pooled cancer and corresponding normal tissues, which were mixed according to the status of either tumor differentiation or lymph node metastasis. After two-dimensional gel separation and subsequent profile analysis, we identified a series of proteins with changes in their expression level (Fig. 1, A–D). ENO1 and LDHB were two representative spots on the gels indicating different intensity between cancer tissues and their corresponding normal counterparts (Fig. 1E and supplemental Fig. S1). Our results identified 44 protein spots produced by 38 genes whose expression showed an association with the degree of tumor differentiation. Furthermore, 29 protein products of 25 genes were differentially expressed during the process of tumor metastasis (Table III and supplemental Table S2). The expression of 17 proteins from 15 genes was closely related to the status of both tumor dedifferentiation and tumor metastasis, indicating that these proteins play important roles in tumor progression and providing new candidates for early disease detection and prediction. These differentially expressed proteins participate in a variety of cellular functions, which could be classified into seven categories: cytoskeletal organization, cell proliferation, apoptosis, signal transduction, protein chaperone, metabolic enzymes, and proteins with other functions.
Fig. 1.
An overview of two-dimensional gel images between GCC samples and corresponding normal samples. A–D, representative images of a two-dimensional gel for normal (left) versus cancer (right) tissues with poor differentiation (top) or with lymph node metastasis (bottom), respectively. Numbers indicated on the map highlight several of the protein spots that had differential expression. E, detailed alteration patterns of two glycolytic enzymes, ENO1 and LDHB, in different pooled samples.
Table III. Differentially expressed proteins involved in glucose metabolism in gastric cardia cancer compared with normal tissues.
NA, not applicable; ID, identity; M, Modest; P, Poor.
| Spot no. | Protein ID | Accession no. | Theoretical molecular mass (kDa)/pI | Experimental mass (kDa)/pI | Peptides matched | Sequence coverage | Total score | Differential ratio |
Function | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor differentiation |
Lymph node metastasis |
|||||||||||
| M | P | N0 | N1 | |||||||||
| % | ||||||||||||
| G1 | Fructose-bisphosphate aldolase (EC 4.1.2.13) | AAA51697 | 39.4/8.2 | 32/8.2 | 1 | 3.8 | 61 | 3 | 2.1 | 6.6 | 4.4 | Glycolysis |
| G2 | Glyceraldehyde-3- phosphate dehydrogenase (EC 1.2.1.12) | AAA52496 | 36.05/8.73 | 33/8.4 | 1 | 4.2 | 97 | 2.1 | 5.8 | 3.8 | 4.8 | Glycolysis |
| G3 | 2-phospho-d-glycerate hydrolyase (EC 4.2.1.11) | AAA52387 | 47.2/7.46 | 42/6.5 | 2 | 7.1 | 130 | 3.9 | 2.9 | 1.1 | 3.4 | Glycolysis |
| G4 | Pyruvate kinase isozymes M1/M2 (EC 2.7.1.40) | AAA36672 | 58/7.7 | 58/8.5 | 2 | 4.5 | 126 | −2.6 | −3 | −11.8 | −5.9 | Glycolysis |
| G5 | Pyruvate kinase isozymes M1/M2 (EC 2.7.1.40) | AAA36672 | 58/7.7 | 36/6.0 | 3 | 7.7 | 224 | 4 | 1.8 | NA | NA | Glycolysis |
| G6 | l-Lactate dehydrogenase B (EC 1.1.1.27) | NP_002291 | 36.6/5.9 | 37/5.5 | 4 | 15.8 | 247 | −3 | −2 | −1.4 | −2.1 | Anaerobic respiration |
| G7 | Pyruvate dehydrogenase (lipoamide) (EC 1.2.4.1) | NP_000916 | 39.2/6.6 | 38/5.0 | 2 | 8.9 | 116 | −0.9 | −3 | NA | NA | Krebs cycle |
| G8 | Aconitate hydratase (EC 4.2.1.3) | CAI17931 | 85.4/7.6 | 85/8.5 | 5 | 7.1 | 289 | NA | NA | −9.7 | −16.9 | Krebs cycle |
| G9 | Aconitate hydratase (EC 4.2.1.3) | CAI17931 | 85.4/7.6 | 85/8.2 | 7 | 11.5 | 470 | NA | NA | −6.4 | −6.8 | Krebs cycle |
| G10 | Isocitrate dehydrogenase (EC 1.1.1.42) | NP_005887 | 46.6/7.0 | 46/7.0 | 5 | 11.8 | 219 | 1 | −1.5 | NA | NA | Krebs cycle |
| G11 | Fumarate hydratase (EC 4.2.1.2) | CAI13908 | 54.6/9.2 | 50/9.0 | 3 | 7.8 | 239 | NA | NA | −2.8 | −2.8 | Krebs cycle |
| G12 | Malate dehydrogenase (EC 1.1.1.37) | CAG33686 | 36.4/7.4 | 36/7.2 | 3 | 8.7 | 106 | −10.4 | −3.9 | NA | NA | Krebs cycle |
| G13 | Ubiquinol-cytochrome-c reductase (EC 1.10.2.2) core protein I | EAW64898 | 52.6/6.4 | 52.0/6.0 | 3 | 6 | 144 | −2.5 | −2.3 | NA | NA | Oxidative phosphorylation |
Metabolite Profiling of GCC
Alterations of metabolic enzymes were the prominently detected major changes in the proteomics analysis. These changes spanned from lipids to amino acids, especially glucose metabolism, suggesting that the tumor cells had acquired totally different metabolite profiles compared with their normal cellular counterparts. The metabolite profiles of 40 paired GCC samples underwent further analysis to identify the metabolic intermediates showing changes consistent with the related enzymes.
Typical GC-TOF MS methods were used to analyze the samples from both GCC and normal tissues (Fig. 2A), and in total, 223 variables were introduced to the subsequent analysis. The OPLS-DA model provided optimal modeling and predictive abilities with one predictive component and two orthogonal components (R2Ycum > 0.875, Q2cum > 0.7), achieving a distinct separation between the metabolite profiles of the two groups (Fig. 2B and supplemental Fig. S2). Significant differences were confirmed in a total of 44 metabolites involved in the metabolism of glucose, lipid, amino acids, nucleic acids, and vitamins. As shown in supplemental Table S3, 30 metabolites were up-regulated in cancer tissues, whereas 14 were down-regulated compared with their matched normal tissues.
Fig. 2.
Results from GC-TOF MS analysis. A, typical total ion current chromatograms of tissue samples obtained from a GCC specimen and its matched normal counterparts. The total ion current chromatogram for the normal tissues is shown below the GCC samples. B, OPLS-DA score plot discriminating the tissue samples from the GCC specimen and its matched normal counterpart using GC-TOF MS analysis.
Unique Glucose Metabolism in GCC
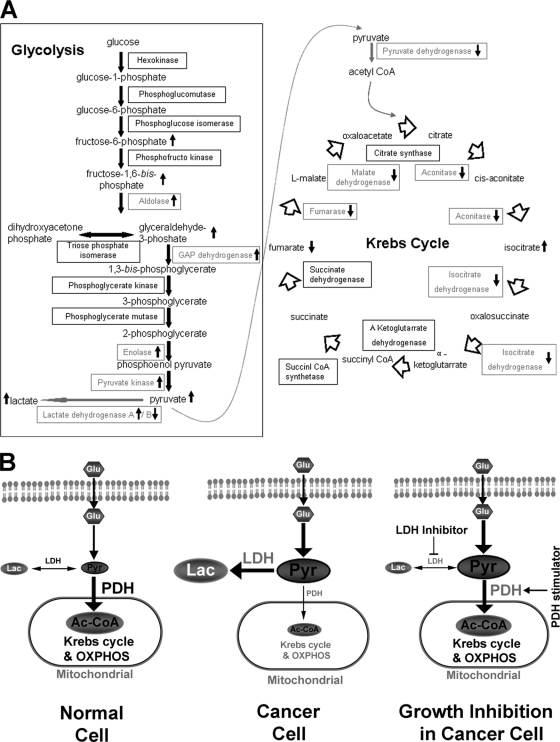
As revealed by proteomics analysis, 11 glucose metabolic enzymes identified from 13 spots showed different expression levels between cancer and normal tissues (Table III). These important enzymes in glycolysis, including fructose-1,6-diphosphate aldolase A (ALDOA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and ENO1, exhibited an elevated expression level in the cancer cells. In contrast, five enzymes involved in the Krebs cycle and oxidative phosphorylation were remarkably down-regulated in cancer tissues, including PDHB, ACO2, fumarate hydratase (FH), malate dehydrogenase (MDH), and ubiquinol-cytochrome-c reductase core protein I (UQCRC1). Another enzyme in the Krebs cycle, isocitrate dehydrogenase (IDH1), also showed a slightly decreased expression level in the cancer cells. Additionally, the enzyme LDHB was down-regulated in GCC. This enzyme normally performs a major function in aerobic pathways transforming lactate to pyruvic acid (23).
Further metabolomics study discovered that six intermediates in the glucose metabolism were changed during tumorigenesis (Table IV). As expected, fructose (fructose 6-phosphate and fructose 1,6-diphosphate), glyceraldehyde (glyceraldehyde 3-phosphate), and pyruvic acid produced during glycolysis were elevated in the tumor samples, and the end product of anaerobic pathway, lactic acid, was also increased. Fumaric acid produced by the Krebs cycle was decreased, which was consistent with down-regulation of fumarate hydratase. However, another product in the Krebs cycle, isocitric acid, was greatly enhanced in the tumor samples. The higher isocitric acid level was presumably due to the difference in metabolic rate from citrate to α-ketoglutarate involving citrate synthase. The conversion of isocitric acid to ketoglutarate and then to succinyl-CoA is relatively slow because it is the critical step to build up NADH, causing the accumulation of isocitric acid. In general, the integrated results from proteomics and metabolite profiling clearly revealed that, unlike normal cells, glycolysis followed by anaerobic respiration was utilized as the major pathway to metabolize glucose in GCC cells, whereas the Krebs cycle and oxidative phosphorylation were impaired.
Table IV. Differential metabolites associated with glycometabolism derived from OPLS-DA mode of GC-TOF MS analysis with t test.
| No. | Retention (min) | Metabolitea | -Fold changeb,c | pc | VIPd |
|---|---|---|---|---|---|
| 1 | 5.22 | Pyruvic acid | 1.20 | 0.12 | |
| 2 | 5.37 | Lactic acid | 1.14 | 2.01 × 10−3 | 1.13 |
| 3 | 7.47 | Glyceraldehyde | 2.21 | 3.22 × 10−3 | 1.12 |
| 4 | 9.54 | Fumaric acid | 0.86 | 0.03 | |
| 5 | 15.55 | Isocitric acid | 1.40 | 1.64 × 10−5 | 1.13 |
| 6 | 16.25 | d-Fructose | 1.93 | 1.69 × 10−4 | 1.37 |
a Metabolites were identified by comparing the exact mass and retention time of our reference metabolites in our laboratory.
b -Fold change with a value >1 indicates a relatively higher concentration present in GCC patients, whereas a value <1means a relatively lower concentration as compared with the healthy controls.
c -Fold change and p value were calculated from a non-parametric t test.
d Variable importance in the projection was obtained from OPLS-DA with a threshold of 1.0.
Validation of Glucose Metabolic Characteristics in Additional GCC Patients: RNA and Protein Levels of Key Pathways
To extend our initial observations of changes in the glucose metabolism of GCC, RNA and protein levels of the corresponding genes were examined by real time PCR and Western blot of additional GCC patients. Because the isoenzymes of PKM2 (M1 and M2; labeled as PKM2a and PKM2b in the following) and the subunits of LDH (LDHA and LDHB) may play different roles in cancer, the mRNA levels of both PKM2 isoenzymes and LDHA were measured. When compared with matched normal gastric cardia samples, six of the seven glycolytic enzymes (combined with LDHA and LDHB for ease of analysis) were significantly overexpressed with the exception of LDHB, which was obviously down-regulated (Fig. 3A). However, most enzymes in the Krebs cycle (combined with UQCRC1 for the same reason) were greatly down-regulated in GCC with the exception of IDH1 and FH, which were only slightly decreased (Fig. 3B). Moreover, a significant correlation was found between the change in levels of relative mRNA expression and the expression ratio of the same protein identified in the two-dimensional gel (Fig. 3C), adding to our knowledge that these enzyme levels were truly altered in GCC. Nevertheless, no significant relationship was detected between the mRNA expression of these target genes and the clinical features of GCC (supplemental Table S4), which may indicate that such metabolic conversions might happen in the early stage of tumorigenesis.
Fig. 3.
Validation of glucose metabolic characteristics in GCC from RNA to protein level. A and B, relative mRNA expression of seven glycolysis- and anaerobic respiration-related genes and six Krebs cycle- and oxidative phosphorylation-related genes in 33 pairs of matched human GCC and corresponding normal tissues by quantitative real time PCR. The mRNA expression level was normalized to that of RP2; and each box plot shows the distribution of the relative expression level of the corresponding gene individually. Error bars, maximum and minimum values except for the outlier represented as an open circle in the figure. C, scatter plots demonstrating the concordance between the protein expression level from proteomics analysis and the RNA expression level given by real time PCR. D, protein expression level of ACO2, ENO1, LDHA, and LDHB in randomly selected eight pairs of matched human GCC and corresponding normal tissues using Western blot. Ponceau S staining was used as a loading control. 2D, two-dimensional gel; T, tumor; N, normal.
On the other hand, the protein expression levels of ENO1, LDHA, LDHB, and ACO2 were analyzed by Western blot (Fig. 3D). ACO2 showed decreased expression in seven of eight cancer samples, whereas the level of ENO1 was increased in six of eight cancer samples compared with their normal counterparts. Due to the high homology of LDHA and LDHB, the antibodies could not distinguish the two bands clearly on the gel. Thus, we differentiated the two bands using specific siRNA to knock down either LDHA or LDHB, which facilitated the identification of each band successfully. As shown in supplemental Fig. S3, the upper band was LDHB, whereas the lower band was LDHA. Seven of eight cancer samples showed LDHA overexpression, and six of eight tumor tissues had decreased expression of LDHB. Notably, none of the GCC samples had a normal glucose metabolic pathway; each had an abnormality in one or several steps in this process, which might facilitate the early detection of GCC.
Fate of Pyruvic Acid Regulated by LDH and PDH Is Intrinsically Intertwined with Survival of Cancer Cells
One metabolic characteristic of GCC discovered from our results was that more pyruvic acid was transformed into lactic acid, rather than acetyl-CoA, following the Krebs cycle. We speculated that the ratio of the two enzyme complexes, LDH and PDH, determined the fate of pyruvic acid, which changed the glucose metabolism pathway and influenced the cell transformation process. When LDHA was knocked down by siRNA (Fig. 4A), the activity of LDH to catalyze pyruvic acid (supplemental Fig. S4A) and produce lactic acid (Fig. 4B) dramatically decreased, which further confirmed the effect of siRNA in these function assays. Meanwhile, a zymogram of the LDH isoenzyme pattern showed that in LDHA knockdown cells the intensities of the five bands obviously shifted from LDH5 (A4) to LDH1 (B4) (supplemental Fig. S4B), which was found to be consistent with diminished enzymatic activity. The MTT assay revealed that these LDHA knockdown cells grew much slower than the control cells (Fig. 4C). Moreover, LDHA knockdown cells formed fewer colonies in soft agar (Fig. 4D) and also had a relatively impaired ability to migrate in a Boyden chamber (supplemental Fig. S5). We concluded that an overexpression of LDHA and high activity of LDH complex were necessary for tumor cells to sustain rapid proliferation and migration. In contrast, in vitro stable overexpression of PDHB (Fig. 4E) (a subunit of PDH complex that was down-regulated in GCC as mentioned above) diverted more pyruvic acids to the Krebs cycle and caused an impairment in the production of lactic acid (Fig. 4F), resulting in cell growth inhibition (Fig. 4G) and a decrease in the ability to form colonies in soft agar (Fig. 4H). Interestingly, overexpression of PDHB had almost no effect on cell migration (supplemental Fig. S5), which was slightly different from knockdown of LDHA, although both of them resulted in the increasing conversion from acetyl-CoA to pyruvic acids. In conclusion, transforming more pyruvic acids to lactic acids by GCC cells was necessary to sustain their rapid proliferation and was controlled by the levels of LDH and PDH.
Fig. 4.
LDH and PDH influence cancer cell growth by controlling the fate of pyruvic acid. A, Western blot analysis showed RNAi-mediated knockdown of LDHA in AGS. Equal loading was ascertained using tubulin as an internal control. B, HPLC analysis indicated the decrease of lactic acid in both supernatant and cytoplasm of cells with LDHA knockdown. C, MTT assay showed the effect of LDHA gene silencing on cell growth of AGS. D, soft agar assay showed the effect of LDHA gene silencing on colony formation of AGS in vitro. E, Western blot analysis showed overexpression of PDHB in AGS. F, HPLC analysis indicated the decrease of lactic acid in both supernatant and cytoplasm of cells with PDHB overexpression. G, MTT assay showed the effect of PDHB overexpression on cell growth of AGS. H, soft agar assay showed the effect of PDHB overexpression on colony formation of AGS in vitro. All the experiments were performed in triplicate and expressed as mean ± S.E. *, p < 0.05; **, p < 0.01.
DISCUSSION
In the last two decades, the incidence of GCC, or proximal gastric cancer, has increased almost all over the world (24–26), including China (27). This cancer is often diagnosed late in its course and is often fatal. Little progress has been achieved in the prevention and treatment of this disease because of the lack of an effective and sensitive diagnostic biomarker as well as knowledge of its etiology (28). Therefore, systematic research of GCC is required to better understand its molecular pathogenesis.
In this study, we first discovered the comprehensive differences of proteins and metabolic intermediates between paired normal and GCC samples, especially those involved in glucose metabolism. Glucose is the major source for energy production and macromolecule biosynthesis to maintain continuous rapid proliferation of tumor cells (29). Many groups have demonstrated that a variety of solid tumors, including renal (30), colon, stomach (31), and breast (32), prefer anaerobic glycolysis to the Krebs cycle for energy production even under normoxia, or the Warburg effect (17). In our study, we also found a predominance of anaerobic glycolysis, rather than the Krebs cycle, in GCC at different levels, spanning from RNA to protein to the corresponding metabolic intermediates, thus validating that the Warburg effect also exists in GCC (Fig. 5A). Several enzymes involved in the glycolysis are considered ubiquitously overexpressed in a variety of cancers, such as ENO1, GAPDH, and PKM (33), which is consistent with our observation in GCC. A high level of glycolysis endows cancer cells with a growth advantage, which enhances unconstrained proliferation and invasion. As for the enzymes involved in the Krebs cycle, the situation is more complicated, depending on the cancer type. Succinate dehydrogenase (SDH) and FH are often dysregulated in renal cancer, both of which are associated with hypoxia-inducible factor (HIF) activation (34). Genes encoding enzymes, such as IDH1, have been found frequently mutated, demonstrating early genetic alterations in glioma (35). The activity of ACO2 is usually elevated in prostate cancer, promoting the transformation of normal citrate-producing epithelial cells to malignant citrate-oxidizing cells (36). However, ACO2 is often deleted in colorectal cancer (37). In our GCC samples, IDH1, ACO2, FH, and MDH were all down-regulated, which indicated an impaired Krebs cycle. Highly active glycolysis and an impaired Krebs cycle guarantee enough metabolic intermediates by avoiding thorough oxidation of glucose, which is essential for the synthesis of macromolecules, such as lipid, protein, and nuclear acid, during cell division (38–40). Moreover, the hypoxic environment in tumor tissues causes increased reactive oxygen species production (41, 42), which leads to severe oxidative damage and cell apoptosis, so that tumor cells often adopt impaired oxidative phosphorylation to evade such reactive oxygen species-mediated cell death (43). Our work revealed that the Warburg effect is also prevalent in GCC, spanning from genes to metabolites, suggesting that such metabolic transition is important for the development of GCC, and the glycolysis signature provides potential biomarker identification for clinical diagnosis of this intractable cancer.
Fig. 5.
Unique glucose metabolic characteristics in GCC may have potential as targets for cancer therapy. A, the fingerprint map of the glucose metabolic network in GCC highlighting those enzymes and metabolic intermediates that are dysregulated. B, normal cells preferred the Krebs cycle and oxidative phosphorylation (OXPHOS) compared with anaerobic respiration. This favored pathway is controlled by the high activity of PDH and relatively low activity of LDH (left). GCC cells not only favor glycolysis by up-regulating the enzymes involved but also strongly prefer anaerobic respiration with production of prominent lactic acids rather than the Krebs cycle and oxidative phosphorylation because of activating LDH and suppressing PDH (middle). This divergence between GCC and normal tissues will give us new therapeutic opportunities (right). GAP, glyceraldehyde 3-phosphate; Lac, lactic acid; Pyr, pyruvate.
The balance between aerobic glycolysis and the Krebs cycle in cancer cells ensures the maximal and rapid production of both energy and metabolic intermediates for cell proliferation. Two enzyme complexes, LDH and PDH, control the metabolism of pyruvic acids, transforming them to either lactic acids or acetyl-CoA, then entering the Krebs cycle. The tetrameric LDH contains five isoenzymes formed by random association of two subunits, LDHA and LDHB, interconverting pyruvate and lactate with concomitant interconversion of NADH and NAD+ (44). The change of LDHB expression suggested by our two-dimensional gel analysis encouraged us to further examine the expression of LDHA in GCC. In contrast to the down-regulation of LDHB, LDHA was found to be overexpressed in GCC. Overexpression of LDHA has been previously reported to favor the formation of cathodic LDH (isoenzymes 4 and 5) and generation of lactate, which is often observed in solid tumors and correlated with advanced tumor grade, poor prognosis, VEGF up-regulation, and hypoxia (45–47). In our study, we also observed the influence of deregulated LDHA on the efflux of pyruvic acid. A knockdown of LDHA by siRNA resulted in a shift of LDH isoenzymes from Stages 3/4/5 to 1/2 with a concomitant significant decrease in catalytic activity as well as lactic acid production. Furthermore, a knockdown of LDHA could obviously hamper both anchorage-dependent and -independent growth of tumor cells, which is consistent with the previous study reporting that LDHA was essential for the transformation process (48, 49).
The other pyruvic acid efflux controller, PDH multienzyme complex, catalyzes the first reaction of an oxidative decarboxylation sequence and converts pyruvate to acetyl-CoA and CO2. It has been reported that in various cancer types, such as lung (50) as well as head and neck squamous cancer (51), pyruvate dehydrogenase kinase induced by overexpressed HIF-1 phosphorylates and inactivates the PDH complex, inhibits the conversion of pyruvate to acetyl-CoA, and thereby attenuates mitochondrial function and respiration (52). In our study, we found that one of the components in PDH multienzyme complex, PDHB, was down-regulated in GCC, leading to decreased enzymatic activity and an impaired Krebs cycle. Consistent with our observation, overexpression of PDHB reduced lactate acid production and decreased the proliferation of tumor cells in both an anchorage-dependent and -independent manner. Thus, our study clearly demonstrated that LDHA and PDHB can determine the fate of pyruvic acid, anaerobic glycolysis or aerobic respiration, via the regulation of LDH and PDH complex, respectively. These studies have improved our understanding of the Warburg effect and its influence on GCC development and progression, and the metabolic pathway can serve as a potential target for GCC therapy (Fig. 5B).
Although either knockdown of LDHA or overexpression of PDHB promoted the conversion from acetyl-CoA to pyruvic acids, leading to the retardation of tumor growth, only LDHA but not PDHB affected cell migration, indicating that the corresponding molecules, such as lactic acid, pyruvic acid, and acetyl-CoA, might not influence cell migration directly, whereas other concomitant changes such as the ratio of NAD+/NADH might influence the cell migration. Of course, we cannot exclude that LDHA has its own special role in cancer cell migration except for its usual role in cell metabolism.
This study highlighted several other proteins that expand our understanding of the molecular pathology of GCC. For example, selenium-binding protein 1 (SELENBP1) was significantly down-regulated in these cancers, consistent with the findings in colorectal (53), ovarian (54), and lung (55) cancers. Some investigators have suggested that selenium may have an anticarcinogenic property toward GCC (56). In addition, elevated levels of many metabolic intermediates of the nucleotide metabolism were noted that were also observed in previous reports (19, 57), indicating active nucleic acid turnover in tumor tissues. Also of interest, fatty acids were all down-regulated in the tumor samples examined, which was consistent with the dramatically decreased expression of lipase, gastric (LIPF). Down-regulation of lipase in GCC resulted in impaired triglyceride and phospholipid hydrolysis, leading to the lack of dissociative fatty acids. This was further confirmed by the decrease of another two intermediates (glycerol phosphate and myoinositol). As expected, a number of amino acids and their polyamine derivatives were increased in cancer tissues to meet the requirement of protein synthesis. A low expression level of the enzyme aspartate aminotransferase (AspT) was noted in our proteomics analysis of the GCC cells. This enzyme normally helps aspartate to enter the Krebs cycle through transamination. Large amounts of aspartic acid and asparagine in cancer tissues might result from impairment of this enzyme, diverting more aspartic acids to participate in the synthesis of purines as well as proteins rather than entering the impaired Krebs cycle.
In summary, with the help of proteomics and metabolomics analysis, we discovered that a series of proteins and related metabolites in glucose metabolism showed an altered expression level in GCC tissues. These findings will not only benefit early diagnosis of this cancer at the molecular level but also improve our understanding of the initiation and development of GCC. A highly activated anaerobic glycolysis and an impaired aerobic respiration are important for maintaining the rapid proliferation of this cancer, which is achieved by highly active LDH concomitant with relatively inactivated PDH to divert more pyruvic acids into the anaerobic glycolysis pathway. Our fin dings suggest that an antagonist to LDH as well as an agonist of PDH may be a novel therapy for GCC.
Supplementary Material
Footnotes
* This work was supported in part by Ministry of Science and Technology Key Program Grant 2008ZX10002-017, National Basic Research Program of China Grant 2010CB91200, 863 Program Grant 2007AA02Z4, 973 Program Grant 2007CB914704, National Natural Science Funds for Distinguished Young Scholars Grant 30725010, National Natural Science Foundation of China Grants 30930023 and 90813023, Chinese Academy of Sciences Grants KSCX2-YW-R-152 and KSCX-YW-R-73, and Science and Technology Commission of Shanghai Municipality Grant 08140902300.
 This article contains supplemental Figs. S1–S5 and Tables S1–S4.
This article contains supplemental Figs. S1–S5 and Tables S1–S4.
The abbreviations used are:
- GCC
- gastric cardia cancer
- LDH
- lactate dehydrogenase
- PDH
- pyruvate dehydrogenase
- TNM
- Tumor, Node and Metastasis
- OPLS-DA
- orthogonal partial least squares discriminant analysis
- ENO1
- α-enolase
- ACO2
- aconitate hydratase
- UQCRC1
- ubiquinol-cytochrome-c reductase core protein I
- IDH1
- isocitrate dehydrogenase
- PKM
- pyruvate kinase, muscle
- FH
- fumarate hydratase
- MDH
- malate dehydrogenase
- HIF
- hypoxia-inducible factor
- ALDOA
- fructose-1,6-diphosphate aldolase A
- MTT
- Methyl Thiazolyl Tetrazolium.
REFERENCES
- 1. Wang L. D., Zheng S., Zheng Z. Y., Casson A. G. (2003) Primary adenocarcinomas of lower esophagus, esophagogastric junction and gastric cardia: in special reference to China. World J. Gastroenterol. 9, 1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. He Q. Y., Cheung Y. H., Leung S. Y., Yuen S. T., Chu K. M., Chiu J. F. (2004) Diverse proteomic alterations in gastric adenocarcinoma. Proteomics 4, 3276–3287 [DOI] [PubMed] [Google Scholar]
- 3. Botterweck A. A., Schouten L. J., Volovics A., Dorant E., van Den Brandt P. A. (2000) Trends in incidence of adenocarcinoma of the oesophagus and gastric cardia in ten European countries. Int. J. Epidemiol. 29, 645–654 [DOI] [PubMed] [Google Scholar]
- 4. Bareiss D., Stabenow R., Müller R., Eisinger B., Stegmaier C., Däubler P., Zeitz M., Scherübl H. (2002) Current epidemiology of carcinoma of the esophagus and cardia in Germany. Dtsch. Med. Wochenschr. 127, 1367–1374 [DOI] [PubMed] [Google Scholar]
- 5. Wijnhoven B. P., Louwman M. W., Tilanus H. W., Coebergh J. W. (2002) Increased incidence of adenocarcinomas at the gastrooesophageal junction in Dutch males since the 1990s. Eur. J. Gastroenterol. Hepatol. 14, 115–122 [DOI] [PubMed] [Google Scholar]
- 6. Blaser M. J., Saito D. (2002) Trends in reported adenocarcinomas of the oesophagus and gastric cardia in Japan. Eur. J. Gastroenterol. Hepatol. 14, 107–113 [DOI] [PubMed] [Google Scholar]
- 7. Derakhshan M. H., Yazdanbod A., Sadjadi A. R., Shokoohi B., McColl K. E., Malekzadeh R. (2004) High incidence of adenocarcinoma arising from the right side of the gastric cardia in NW Iran. Gut 53, 1262–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He Y. T., Hou J., Chen Z. F., Qiao C. Y., Song G. H., Meng F. S., Jin H. X., Chen C. (2008) Trends in incidence of esophageal and gastric cardia cancer in high-risk areas in China. Eur. J. Cancer Prev. 17, 71–76 [DOI] [PubMed] [Google Scholar]
- 9. Au J. S., Cho W. C., Yip T. T., Law S. C. (2008) Proteomic approach to biomarker discovery in cancer tissue from lung adenocarcinoma among nonsmoking Chinese women in Hong Kong. Cancer Invest. 26, 128–135 [DOI] [PubMed] [Google Scholar]
- 10. Liu D. P., Qi R. Z., Wang Y., Chen P. P., Koeffler H. P., Xie D. (2007) Discovery of stage-related proteins in esophageal squamous cell carcinoma using proteomic analysis. Proteomics Clin. Appl. 1, 312–320 [DOI] [PubMed] [Google Scholar]
- 11. Liu R., Li Z., Bai S., Zhang H., Tang M., Lei Y., Chen L., Liang S., Zhao Y. L., Wei Y., Huang C. (2009) Mechanism of cancer cell adaptation to metabolic stress: proteomics identification of a novel thyroid hormone-mediated gastric carcinogenic signaling pathway. Mol. Cell. Proteomics 8, 70–85 [DOI] [PubMed] [Google Scholar]
- 12. Chaerkady R., Harsha H. C., Nalli A., Gucek M., Vivekanandan P., Akhtar J., Cole R. N., Simmers J., Schulick R. D., Singh S., Torbenson M., Pandey A., Thuluvath P. J. (2008) A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J. Proteome Res. 7, 4289–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stemke-Hale K., Gonzalez-Angulo A. M., Lluch A., Neve R. M., Kuo W. L., Davies M., Carey M, Hu Z., Guan Y., Sahin A., Symmans W. F., Pusztai L., Nolden L. K., Horlings H., Berns K., Hung M. C., van de Vijver M. J., Valero V., Gray J. W., Bernards R., Mills G. B., Hennessy B. T. (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 68, 6084–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khwaja F. W., Nolen J. D., Mendrinos S. E., Lewis M. M., Olson J. J., Pohl J., Van Meir E. G., Ritchie J. C., Brat D. J. (2006) Proteomic analysis of cerebrospinal fluid discriminates malignant and nonmalignant disease of the central nervous system and identifies specific protein markers. Proteomics 6, 6277–6287 [DOI] [PubMed] [Google Scholar]
- 15. Tainsky M. A. (2009) Genomic and proteomic biomarkers for cancer: a multitude of opportunities. Biochim. Biophys. Acta 1796, 176–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 17. Elton E. (2005) Esophageal cancer. Dis. Mon. 51, 664–684 [DOI] [PubMed] [Google Scholar]
- 18. Qiu Y., Cai G., Su M., Chen T., Zheng X., Xu Y., Ni Y., Zhao A., Xu L. X., Cai S., Jia W. (2009) Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 8, 4844–4850 [DOI] [PubMed] [Google Scholar]
- 19. Chen P. P., Li W. J., Wang Y., Zhao S., Li D. Y., Feng L. Y., Shi X. L., Koeffler H. P., Tong X. J., Xie D. (2007) Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS One 2, e534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qin X. F., An D. S., Chen I. S., Baltimore D. (2003) Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. U.S.A. 100, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie D., Miller C. W., O'Kelly J., Nakachi K., Sakashita A., Said J. W., Gornbein J., Koeffler H. P. (2001) Breast cancer. Cyr61 is overexpressed, estrogen-inducible, and associated with more advanced disease. J. Biol. Chem. 276, 14187–14194 [DOI] [PubMed] [Google Scholar]
- 22. Chen Y., Zhang H., Xu A., Li N., Liu J., Liu C., Lv D., Wu S., Huang L., Yang S., He D., Xiao X. (2006) Elevation of serum l-lactate dehydrogenase B correlated with the clinical stage of lung cancer. Lung Cancer 54, 95–102 [DOI] [PubMed] [Google Scholar]
- 23. Wilkinson N. W., Howe J., Gay G., Patel-Parekh L., Scott-Conner C., Donohue J. (2008) Differences in the pattern of presentation and treatment of proximal and distal gastric cancer: results of the 2001 gastric patient care evaluation. Ann. Surg. Oncol. 15, 1644–1650 [DOI] [PubMed] [Google Scholar]
- 24. Falk J., Carstens H., Lundell L., Albertsson M. (2007) Incidence of carcinoma of the oesophagus and gastric cardia. Changes over time and geographical differences. Acta Oncol. 46, 1070–1074 [DOI] [PubMed] [Google Scholar]
- 25. Blot W. J., Devesa S. S., Kneller R. W., Fraumeni J. F., Jr. (1991) Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA 265, 1287–1289 [PubMed] [Google Scholar]
- 26. Fan Y. J., Song X., Li J. L., Li X. M., Liu B., Wang R., Fan Z. M., Wang L. D. (2008) Esophageal and gastric cardia cancers on 4238 Chinese patients residing in municipal and rural regions: a histopathological comparison during 24-year period. World J. Surg. 32, 1980–1988 [DOI] [PubMed] [Google Scholar]
- 27. Stein H. J., Feith M., Siewert J. R. (2000) Cancer of the esophagogastric junction. Surg. Oncol. 9, 35–41 [DOI] [PubMed] [Google Scholar]
- 28. Ortega A. D., Sánchez-Aragó M., Giner-Sánchez D., Sánchez-Cenizo L., Willers I., Cuezva J. M. (2009) Glucose avidity of carcinomas. Cancer Lett. 276, 125–135 [DOI] [PubMed] [Google Scholar]
- 29. Semenza G. L. (2007) HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J. Bioenerg. Biomembr. 39, 231–234 [DOI] [PubMed] [Google Scholar]
- 30. Hirayama A., Kami K., Sugimoto M., Sugawara M., Toki N., Onozuka H., Kinoshita T., Saito N., Ochiai A., Tomita M., Esumi H., Soga T. (2009) Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 69, 4918–4925 [DOI] [PubMed] [Google Scholar]
- 31. Robey I. F., Stephen R. M., Brown K. S., Baggett B. K., Gatenby R. A., Gillies R. J. (2008) Regulation of the Warburg effect in early-passage breast cancer cells. Neoplasia 10, 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altenberg B., Greulich K. O. (2004) Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84, 1014–1020 [DOI] [PubMed] [Google Scholar]
- 33. King A., Selak M. A., Gottlieb E. (2006) Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 25, 4675–4682 [DOI] [PubMed] [Google Scholar]
- 34. Yan H., Bigner D. D., Velculescu V., Parsons D. W. (2009) Mutant metabolic enzymes are at the origin of gliomas. Cancer Res. 69, 9157–9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh K. K., Desouki M. M., Franklin R. B., Costello L. C. (2006) Mitochondrial aconitase and citrate metabolism in malignant and nonmalignant human prostate tissues. Mol. Cancer 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laiho P., Hienonen T., Mecklin J. P., Järvinen H., Karhu A., Launonen V., Aaltonen L. A. (2003) Mutation and LOH analysis of ACO2 in colorectal cancer: no evidence of biallelic genetic inactivation. J. Med. Genet 40, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gatenby R. A., Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 38. Deberardinis R. J., Sayed N., Ditsworth D., Thompson C. B. (2008) Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18, 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tong X., Zhao F., Thompson C. B. (2009) The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 19, 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hervouet E., Simonnet H., Godinot C. (2007) Mitochondria and reactive oxygen species in renal cancer. Biochimie 89, 1080–1088 [DOI] [PubMed] [Google Scholar]
- 41. Pelicano H., Carney D., Huang P. (2004) ROS stress in cancer cells and therapeutic implications. Drug Resist. Updat. 7, 97–110 [DOI] [PubMed] [Google Scholar]
- 42. Santamaría G., Martínez-Diez M., Fabregat I., Cuezva J. M. (2006) Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis 27, 925–935 [DOI] [PubMed] [Google Scholar]
- 43. Read J. A., Winter V. J., Eszes C. M., Sessions R. B., Brady R. L. (2001) Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins 43, 175–185 [DOI] [PubMed] [Google Scholar]
- 44. Thorn C. C., Freeman T. C., Scott N., Guillou P. J., Jayne D. G. (2009) Laser microdissection expression profiling of marginal edges of colorectal tumours reveals evidence of increased lactate metabolism in the aggressive phenotype. Gut 58, 404–412 [DOI] [PubMed] [Google Scholar]
- 45. Giatromanolaki A., Sivridis E., Gatter K. C., Turley H., Harris A. L., Koukourakis M. I. (2006) Lactate dehydrogenase 5 (LDH-5) expression in endometrial cancer relates to the activated VEGF/VEGFR2(KDR) pathway and prognosis. Gynecol. Oncol. 103, 912–918 [DOI] [PubMed] [Google Scholar]
- 46. Koukourakis M. I., Giatromanolaki A., Sivridis E., Gatter K. C., Harris A. L. (2006) Lactate dehydrogenase 5 expression in operable colorectal cancer: strong association with survival and activated vascular endothelial growth factor pathway–a report of the Tumour Angiogenesis Research Group. J. Clin. Oncol. 24, 4301–4308 [DOI] [PubMed] [Google Scholar]
- 47. Shim H., Dolde C., Lewis B. C., Wu C. S., Dang G., Jungmann R. A., Dalla-Favera R., Dang C. V. (1997) c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. U.S.A. 94, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fantin V. R., St-Pierre J., Leder P. (2006) Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434 [DOI] [PubMed] [Google Scholar]
- 49. Koukourakis M. I., Giatromanolaki A., Sivridis E., Gatter K. C., Harris A. L. (2005) Pyruvate dehydrogenase and pyruvate dehydrogenase kinase expression in non small cell lung cancer and tumor-associated stroma. Neoplasia 7, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wigfield S. M., Winter S. C., Giatromanolaki A., Taylor J., Koukourakis M. L., Harris A. L. (2008) PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br. J. Cancer 98, 1975–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim J. W., Dang C. V. (2006) Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66, 8927–8930 [DOI] [PubMed] [Google Scholar]
- 52. Kim H., Kang H. J., You K. T., Kim S. H., Lee K. Y., Kim T. I., Kim C., Song S. Y., Kim H. J., Lee C., Kim H. (2006) Suppression of human selenium-binding protein 1 is a late event in colorectal carcinogenesis and is associated with poor survival. Proteomics 6, 3466–3476 [DOI] [PubMed] [Google Scholar]
- 53. Huang K. C., Park D. C., Ng S. K., Lee J. Y., Ni X., Ng W. C., Bandera C. A., Welch W. R., Berkowitz R. S., Mok S. C., Ng S. W. (2006) Selenium binding protein 1 in ovarian cancer. Int. J. Cancer 118, 2433–2440 [DOI] [PubMed] [Google Scholar]
- 54. Chen G., Wang H., Miller C. T., Thomas D. G., Gharib T. G., Misek D. E., Giordano T. J., Orringer M. B., Hanash S. M., Beer D. G. (2004) Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. J. Pathol. 202, 321–329 [DOI] [PubMed] [Google Scholar]
- 55. Rayman M. P. (2005) Selenium in cancer prevention: a review of the evidence and mechanism of action. Proc. Nutr. Soc. 64, 527–542 [DOI] [PubMed] [Google Scholar]
- 56. Denkert C., Budczies J., Weichert W., Wohlgemuth G., Scholz M., Kind T., Niesporek S., Noske A., Buckendahl A., Dietel M., Fiehn O. (2008) Metabolite profiling of human colon carcinoma—deregulation of TCA cycle and amino acid turnover. Mol. Cancer 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.