Abstract
The filamentous fungus Penicillium chrysogenum is well-known by its ability to synthesize β-lactam antibiotics as well as other secondary metabolites. Like other filamentous fungi, this microorganism is an excellent host for secretion of extracellular proteins because of the high capacity of its protein secretion machinery. In this work, we have characterized the extracellular proteome reference map of P. chrysogenum Wisconsin 54–1255 by two-dimensional gel electrophoresis. This method allowed the correct identification of 279 spots by peptide mass fingerprinting and tandem MS. These 279 spots included 328 correctly identified proteins, which corresponded to 131 different proteins and their isoforms. One hundred and two proteins out of 131 were predicted to contain either classical or nonclassical secretion signal peptide sequences, providing evidence of the authentic extracellular location of these proteins. Proteins with higher representation in the extracellular proteome were those involved in plant cell wall degradation (polygalacturonase, pectate lyase, and glucan 1,3-β-glucosidase), utilization of nutrients (extracellular acid phosphatases and 6-hydroxy-d-nicotine oxidase), and stress response (catalase R). This filamentous fungus also secretes enzymes specially relevant for food industry, such as sulfydryl oxidase, dihydroxy-acid dehydratase, or glucoamylase. The identification of several antigens in the extracellular proteome also highlights the importance of this microorganism as one of the main indoor allergens. Comparison of the extracellular proteome among three strains of P. chrysogenum, the wild-type NRRL 1951, the Wis 54–1255 (an improved, moderate penicillin producer), and the AS-P-78 (a penicillin high-producer), provided important insights to consider improved strains of this filamentous fungus as versatile cell-factories of interest, beyond antibiotic production, for other aspects of white biotechnology.
Filamentous fungi have an extraordinary ability to secrete proteins, secondary metabolites, and organic acids to the culture medium. The secreted proteins play important roles in nutrition, substrate colonization, or pathogenicity (1). This high secretory capacity has made filamentous fungi attractive for the commercial production of extracellular proteins (2), especially for the food and beverage industries (3), which have been using the compounds secreted by filamentous fungi for decades. Examples are provided by Aspergillus oryzae, which has been important for the production of traditional fermented foods and beverages in Japan and is used in modern biotechnology because of its ability to secrete large amounts of proteins (4) or Aspergillus niger, which has been widely used in biotechnology for the production of organic acids, food ingredients, and industrial enzymes (5). Because A. oryzae, A. niger, and Penicillium chrysogenum belong to the same fungal family, the latter microorganism might be considered of interest for the secretion of extracellular proteins.
Although the understanding of the molecular basis of the secretion process in filamentous fungi is still limited (1), it is generally accepted that the secretion pathway in these microorganisms does not differ greatly from that present in yeasts and higher eukaryotes and protein secretion is believed to occur mainly at hyphal tips (6). The classical secretory pathway of proteins is driven by a canonical N-terminal signal peptide. These proteins enter the endoplasmic reticulum, where they are properly folded and modified (glycosylation, phosphorylation, etc.) and subsequently reach the Golgi compartment packed in transport vesicles. In this compartment, proteins can undergo further additional modifications such as glycosylation and peptide processing. Following this step, proteins are packed in secretory vesicles directed to the plasma membrane for secretion, or targeted to the vacuole either to become resident proteins or to undergo proteolytic degradation (7). In addition to the classical endoplasmic reticulum-Golgi pathway, it has been suggested that various kinds of mechanistically distinct nonclassical export routes may exist (8, 9). Cytoplasmic, nuclear and signal-peptide-containing proteins have been shown to reach the cell surface by nonconventional transport pathways (10). In yeasts, other mechanisms of secretion, which drive proteins lacking the signal peptide outside the plasma membrane, have also been described (11).
P. chrysogenum is a filamentous fungus well-known by its ability to synthesize β-lactam antibiotics such as benzylpenicillin and isopenicillin N (12). Because the isolation of the wild-type strain NRRL 1951 from an infected cantaloupe in Peoria, Illinois in 1943 (13), this microorganism has undergone artificial selection by mutagenesis during industrial strain improvement programs, which gave rise to the improved-producing Wisconsin 54–1255 strain (hereafter named Wis 54-1255) (14). This strain became a laboratory model strain and was used for the genome sequencing project (15) and the intracellular proteome reference map (16). P. chrysogenum Wis 54–1255 was the ancestor of penicillin high-producing mutants, such as the AS-P-78 strain developed by Antibióticos S.A (León, Spain). The mutagenesis processes undergone by the P. chrysogenum strains during the industrial selection have introduced several important modifications in their metabolic networks (16).
The recent advances in the Proteomics tools and the availability of genome sequences, has allowed an analysis of the secretomes of a few filamentous fungi, but the available information is still scarce (17–19). However, because of the availability of several fungal genomes and diverse prediction programs for secretory proteins, an integrated platform for annotation of fungal secretomes (Fungal Secretome Database) has been established and implemented in a web-based database (20). This database has been proposed as an integrated environment for the study of secretory proteins in the fungal kingdom.
In order to fully characterize P. chrysogenum and to establish how the modifications acquired during the industrial strain improvement programs affected the wild type plant pathogenicity, analysis of the secreted proteins present in the culture broths was carried out. Using two-dimensional gel electrophoresis (2-DE)1 gels coupled to peptide mass fingerprint (PMF) and tandem MS we describe here for the first time the extracellular proteome of P. chrysogenum and the differences found in secreted protein among the wild type and two improved strains of this microorganism. Results reveal the nutritional versatility of this filamentous fungus and its potential interest for other biotechnological purposes different from antibiotic production, because nonpenicillin producer strains have been previously developed (21) that lack the penicillin biosynthesis genes (22) and can be used for other biotechnological uses.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Three strains of P. chrysogenum were used in this work: the wild-type NRRL 1951, the Wis 54–1255 (reference strain for the genome sequencing project and intracellular proteome reference map), and the high-producer AS-P-78 (kindly provided to us by Antibioticos S.A., León, Spain). They were grown for 7 days at 28 °C on solid Power medium as described before (16). Conidia from one Petri dish were collected, inoculated into a flask with 100 ml of PMMY defined medium containing 1 g of potassium phenylacetate, and incubated as previously described (16). All experiments were carried out using three individual cultures for each strain (biological replicates).
Protein Extraction from the Culture Medium
Samples were taken from cultures and passed through a Nytal membrane. The medium containing the extracellular proteins was immediately centrifuged at 4 °C and 3900 × g for 5 min to discard any residual mycelia. The supernatant was then filtered on ice using the 0.45 μm membrane filters (Millipore) and a vacuum pump. The filtered supernatant was mixed with an equal volume of prechilled 20% (w/v) 2,2,2-trichloroacetic acid/acetone containing 0.14% (w/v) 1,4-dithiothreitol. The mixture was incubated overnight at −20 °C. Precipitated proteins were separated from supernatant by centrifuging at 3900 × g for 5 min and 4 °C. The pellet was washed twice with prechilled acetone containing 0.07% (w/v) 1,4-dithiothreitol and once with 80% (v/v) acetone/MilliQ water. Protein concentration was determined according to the Bradford method, which showed a high reproducibility for this protein extraction protocol. The final pellet was resuspended in sample buffer: 8 m urea, 2% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 0.5% (v/v) ampholytes, 25 mm 1,4-dithiothreitol and 0.002% bromphenol blue) and stored at −80 °C.
Two-dimensional Gel Electrophoresis
A solution containing 450 μg of soluble proteins in the sample buffer (see above), was loaded onto 18-cm IPG strips (GE Healthcare), with nonlinear pH 4–7 gradient. Focusing of proteins and equilibration of the focused IPG strips were achieved as described before, as well as the second dimension, which was run by SDS-PAGE in 12.5% polyacrylamide in an Ettan Dalt Six apparatus (GE Healthcare) (16). Gels were dyed with Colloidal Coomassie following the “Blue Silver” staining method (23), which provides high reproducibility, as indicated before (16).
Analysis of Differential Protein Expression
Scanned two-dimensional gels were analyzed using an ImageScanner II (GE Healthcare) as described before (16). Three biological replicates were used for each condition and each strain. Variability in the number of protein spots detected among biological replicates was less than 10%, which may be because of experimental variability. Spot normalization, as an internal calibration to make the data independent from experimental variations among gels, was made using relative volumes to quantify and compare the gel spots. Relative volume corresponds to the volume of each spot divided by the total volume of all the spots in the gel. Differentially expressed proteins between two strains were considered when the ratio of the relative volume average for one specific spot (present in the three biological replicates) was higher than 1.5 and the p value was <0.05.
Protein Identification by MALDI-TOF MS and MS/MS
The protein spots of interest were manually excised from Colloidal Commassie-stained gels by biopsy punches, placed in an Eppendorf tube, and washed twice with double distilled water. The proteins were digested following the method of Havlis and coworkers (24) and processed for further analysis as indicated before (16). The samples were analyzed with a 4800 Proteomics Analyzer matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF/TOF) mass spectrometer (Applied Biosystems, Foster City, CA). A 4700 proteomics analyzer calibration mixture (Cal Mix 5; Applied Biosystems) was used as external calibration. All MS spectra were internally calibrated using peptides from the trypsin digestion. The analysis by MALDI-TOF/TOF mass spectrometry produced peptide mass fingerprints, and the peptides observed (up to 65 peptides per spot) were collected and represented as a list of monoisotopic molecular weights with a signal to noise (S/N) ratio greater than 20 using the 4000 Series Explorer v3.5.3 software (Applied Biosystems). All known contaminant ions (trypsin- and keratin-derived peptides) were excluded for later MS/MS analysis. Hence, from each MS spectra, the six most intensive precursors with a S/N greater than 20 were selected for MS/MS analyses with CID (atmospheric gas was used) in 2-kV ion reflector mode and precursor mass windows of ±7 Da. The default calibration was optimized for the MS/MS spectra.
For protein identification, Mascot Generic Files combining MS and MS/MS spectra were automatically created and used to interrogate a nonredundant protein database using a local license of Mascot v 2.2 from Matrix Science through the Global Protein Server v 3.6 (Applied Biosystems). The search parameters for peptide mass fingerprints and tandem MS spectra obtained were set as follows: (i) NCBInr (2009.11.03) sequence databases were used; (ii) taxonomy: All entries (9993394 sequences, 3409286210 residues); (iii) fixed and variable modifications were considered (Cys as S carbamidomethyl derivative and Met as oxidized methionine); (iv) one missed cleavage site was allowed; (v) precursor tolerance was 100 parts per million and MS/MS fragment tolerance was 0.3 Da; (vi) peptide charge: 1+; and (vii) the algorithm was set to use trypsin as the enzyme. Protein candidates produced by this combined peptide mass fingerprinting/tandem MS search were considered valid when the global Mascot score was greater than 83 with a significance level of p < 0.05. Additional criteria for confident identification were that the protein match should have at least 15% sequence coverage; for lower coverages, only those proteins with a Mascot ions score above 54 and at least two peptides identified in the tandem MS analysis (with a significance level of p < 0.05), were considered valid.
Prediction of the Secretion Mechanism
Prediction of the presence of secretion signal motifs was achieved using SignalP (for classical secretion signal motifs) and SecretomeP (for nonclassical signal motifs) softwares (25, 26).
RESULTS
Growth Phases and Extracellular Protein Concentration
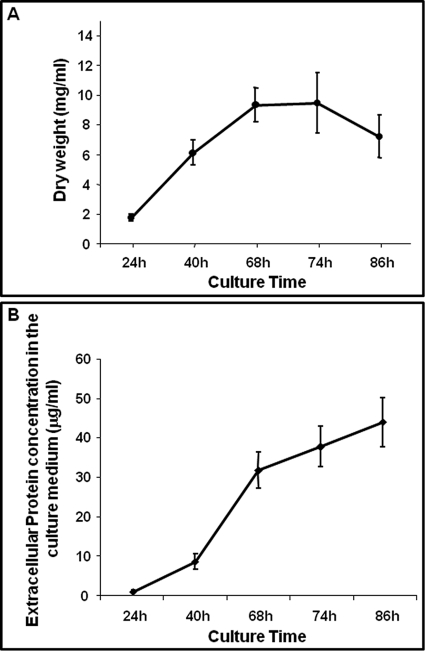
The presence of extracellular proteins in the culture medium is correlated to the growth phase (1). Therefore, first we determined the optimal conditions for sample preparations. Growth of P. chrysogenum in submerged cultures in defined DP medium was analyzed at different time points. Cells were collected at 24 h, 40 h, 68 h, 74 h, and 86 h to quantify the dry weight. As shown in Fig. 1A, P. chrysogenum hyphal growth reached the end of the linear phase at 68 h, where it entered the stationary phase until 74 h. A small decrease in the dry weight was observed from this time point, indicating that hyphal lysis and fragmentation coupled with a progressive loss of biomass was taking place. Hence, in order to evaluate the possibility of contamination with intracellular proteins resulting from the cell lysis, we determined the extracellular protein concentration by the Bradford method at 24 h, 40 h, 68 h, 74 h, and 86 h. The amount of proteins present in the culture medium (Fig. 1B) was very low at 24h (0.86 μg/ml), reaching the concentration of 8.5 μg/ml at 40 h. This concentration rapidly increased from this time point until 68 h, reaching 31.7 μg/ml. Protein concentration increased until the end of the culture, reaching a maximum concentration of 43.8 μg/ml at 86 h. However, because dry weight remains constant following 68 h and decreases following 72 h, the increase in the protein concentration following 68 h likely corresponds to cell lysis events. According to these results, we selected 68 h for the characterization of the P. chrysogenum extracellular proteome reference map.
Fig. 1.
Growth phase and extracellular protein concentration. A, Dry weight obtained from samples taken from P. chrysogenum Wis 54–1255 liquid cultures at different time points. Data correspond to three biological replicates performed in duplicate. Results for the NRRL 1951 and AS-P-78 strains were similar to those obtained for the Wis 54–1255 strain (data not shown). B, Extracellular protein concentration plotted versus the culture time. Data correspond to three biological replicates performed in duplicate.
Extracellular Proteome Reference Map
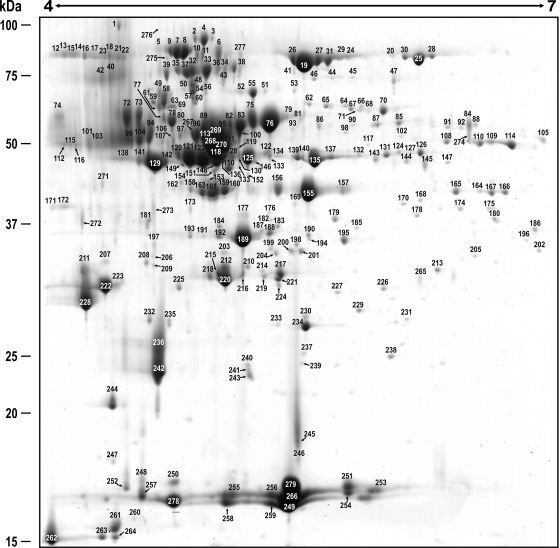
The extracellular proteome reference map was made using 68 h cultures of P. chrysogenum Wis 54-1255, which was the reference strain for the genome sequencing project (15) and the cytoplasmatic proteome reference map (16). Initially, wide range IPG strips (pH 3–10, nonlinear) were used, but the majority of spots appeared distributed in the acidic side of the gel (data not shown). Therefore, to get a better resolution of the spots, IPG strips with a pH range from 4 to 7 were used. Visualization of 317 spots by “Blue Silver” staining was obtained under this condition and allowed the correct identification by peptide mass fingerprinting and tandem MS of the proteins present in 279 spots (Fig. 2 and supplemental Table 1). The 279 spots represented 328 correctly identified proteins (with an average error of 7 parts per million), because some spots contained more than one protein. The extracellular proteome reference map with a direct link to the identified proteins can be accessed through the URL: http://isa.uniovi.es/P_chrysogenum_secretome/.
Fig. 2.
Reference map of the P. chrysogenum extracellular proteome. A total of 317 spots were visualized in a 4–7 pH range and 279 spots (numbered in the figure) were identified by PMF and tandem mass spectrometry (see supplemental Table 1). Molecular weights are shown on the left. Assigned numbers over each spot correlate with those shown in supplemental Table I.
All the significant MASCOT searches provided an exact match with proteins inferred from the P. chrysogenum Wis 54–1255 genome (except spots 185 and 257, see below), which indicates high accuracy of the genome annotation process. From the 328 proteins identified in the reference map, 62 proteins showed a total of 259 isoforms and 69 proteins had no isoforms (supplemental Table 2). The 10 spots showing more relative volume represented 31.31% of the total spot volume. Spot 279, which includes a hypothetical cell wall protein BinB (Pc22g00190) provided the highest relative volume (5.22%).
The final number of different proteins that were found in the extracellular proteome of P. chrysogenum was 131 (62 different proteins having isoforms + 69 individual proteins without isoforms). Seventy three proteins out of 131 were predicted by the SignalP software (25) to have classical signal peptide sequences (supplemental Table 2). Fifty-eight of the different proteins identified lacked a predicted N-terminal signal peptide sequence, indicating that these proteins might be secreted through a nonclassical secretory mechanism, such as that reported for yeasts (11). In order to assess whether the 58 proteins lacking the classical N-terminal signal peptide sequence include nonclassical secretion signals, the SecretomeP prediction software (26) was used. This tool predicted 29 proteins out of 58 having a nonclassical secretory targeting signal (supplemental Table 2), pointing to the existence of nonclassical secretory pathways in P. chrysogenum (see Discussion).
Functional Classification of the Extracellular Proteins of P. chrysogenum
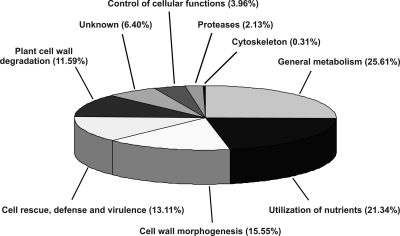
The 328 proteins identified (131 different proteins and isoforms) were functionally classified according to their biological roles. As shown in Fig. 3 and supplemental Table 1, 84 proteins were implicated in general metabolism (25.61%); 70 proteins were involved in utilization of nutrients (21.34%); and 7 additional proteins had protease activity that are relevant for nutrient utilization (2.13%); 51 proteins were in charge of cell wall morphogenesis (15.55%); 43 proteins were involved in cell rescue, defense, and virulence (13.11%); 38 proteins were in charge of plant cell wall degradation (11.59%); 21 proteins had an unknown function (6.40%); 13 proteins were involved in the control of cellular functions (3.96%); and one protein belonged to the cytoskeleton (0.31%). The main findings for each functional category are summarized below.
Fig. 3.
Functional classification of the proteins identified in the extracellular reference map. The 328 correctly identified proteins (131 different proteins and isoforms) were classified according to their biological function.
General Metabolism
Some proteins identified in the culture medium of P. chrysogenum are involved in metabolic reactions of special interest for food industry (see Discussion).
The finding of a probable glyceraldehyde-3-phosphate dehydrogenase (Pc21g14560, spot 202), which is predicted to be a nonclassical secreted protein (supplemental Table 2), in the extracellular proteome is highly interesting, because it appears to be located on several subcellular locations in eukaryotes (see Discussion). Other examples of enzymes predicted to be secreted that might play a dual role according to their location are listed in supplemental Table 2.
Utilization of Nutrients
Several proteins identified by mass spectrometry were grouped according to their ability to utilize phospate, carbon, or nitrogen sources from the culture medium. Phosphatases (specially acid phosphatases) were highly represented in the extracellular proteome both in several isoforms and relative volumes, such as the AFPhoA (Pc18g02580; spots 6, 10, 11, 34, 38, 43, and 277). This protein from Aspergillus ficuum has 64% homology to a phosphate-repressible acid phosphatase from P. chrysogenum and has been reported to be secreted (27). Probable alkaline phosphatases were also identified in spots 2–4 (Pc12g15470; 85% homology and 73% identity to an alkaline phosphatase from Aspergillus clavatus) and 5, 7, 8, and 9 (Pc22g06490). In addition, spot 74 contained a protein (Pc22g12670) that shared 72% homology (60% identity) with a phytase from Ajellomyces dermatitidis. This protein can break down undigestible phytates, which are the principal storage form of phosphorus in cereals and leguminous plants (28), thus releasing digestible phosphorus, calcium, and other nutrients.
Enzymes for the utilization of several carbon sources were also identified. Spots 119 and 136 include the glucan 1,4-alpha-glucosidase or glucoamylase (Pc22g01850), which catalyzes the release of glucose from the nonreducing ends of starch and related polysaccharides. This activity is of industrial importance in the production of sugar from starch (29). Spots 164–166 include a protein (Pc12g04310, encoded by a probable acetate-inducible gene aciA) with strong similarity to a NAD-dependent formate dehydrogenase AciA/Fdh from Neosartorya fischeri (90% homology, 84% identity). This enzyme catalyzes the oxidation of formate, which serves as an auxiliary energy substrate for yeasts and fungi (30). Protein Pc16g04220 (probable trehalose metabolism factor Pmu1) was identified in spots 84 and 274. This protein is required in addition to the MAL4 gene-encoded protein, for both constitutive maltase synthesis and maltose utilization in Saccharomyces cerevisiae (31). Probable sorbitol (a sugar alcohol found in stone fruits) utilization proteins Sou2 (Pc12g00830; spots 230, 233, 234, and 238) and sorbitol dehydrogenases (Pc16g08460; spots 173 and 175), also known as glucitol-, polyol-, or l-iditol-dehydrogenases, were present in the extracellular fraction. Although these enzymes were not predicted to be secreted by the classical system (supplemental Table 2), extracellular sorbitol dehydrogenase activity (oxidation of sorbitol to fructose) has been detected extracellularly in fungal species of the genus Fusarium (32), thus supporting our results.
Another interesting finding is the presence of several isoforms of two probable 6-hydroxy-d-nicotine oxidases (Pc18g05530 in spots 107, 127, 129, 138, 141, 142, 145, and 197; Pc21g12590 in spots 89, 95, 96, 100, 111, 113, 118, 148, and 266–269). All these isoforms are also highly represented and implies that nicotine (or similar compounds) degradation is an important source of carbon and nitrogen in this fungus. This is well known for some soil bacteria, which use nicotine as a suitable carbon and nitrogen source and degrade it through a single pathway involving this enzyme (33).
Cell Wall Morphogenesis
Enzymes relevant for fungal cell wall biosynthesis and morphogenesis are β-1,3-glucanosyltransferases (see Discussion). Probable 1,3-β-glucanosyltransferases Bgt1 and Gel1 (Pc13g08730 with six isoforms, Pc15g01030 with six isoforms, and Pc21g22000 with one isoform) are identified in the extracellular proteome of P. chrysogenum (spots 61, 95, 96, 148, 150, 151, 153, 203, 212, 215, 216, 218, and 220). In addition, a probable glycosylphosphatidylinositol-anchored β(1–3)glucanosyltransferase gel3 (Pc22g09380) is present in spots 32, 33, 35, 37, 48, 50, 54, and 57. Chitin (and its partially or fully deacetylated derivative chitosan) are major components of most fungal cell walls. Probable chitinases are identified in spots 138 (Pc13g09520) and 106 (Pc20g02250). Related to this protein is the identification of a probable chitosanase (Pc12g07820, spot 194), which has been purified from the culture medium of the koji mold A. oryzae (34), and 11 isoforms of a cell wall protein Crh1 (Pc21g23210, spots 22, 40, 74, 99, 101, 103, 112, 115, 116, 232, and 235). The latter transfers chitin to 1,6-βglucan and has been located at the cell surface, particularly in chitin-rich areas at the incipient bud site, around the septum area in later stages of budding, and in ascospore envelopes, suggesting that it might exert a common role in cell wall organization of S. cerevisiae (35).
Spots 51, 52, 55, and 56 include a β-N-acetylhexosaminidase (Pc20g10360), which is widely distributed in nature including fungi (36, 37). Several roles have been given to fungal β-N-acetylhexosaminidases, β-N-acetylglucosaminidases, and chitobiases, such as the invasion of host organisms, the digestion of chitinous nutrients, the apical growth and branching of hyphae, the autolysis of mycelia and fruiting bodies in chitinous fungi, and in the recycling and mobilization of intracellular polysaccharides and glycoproteins especially during starvation (36), although it has been reported that chitin is not utilized by P. chrysogenum even in the state of carbon starvation (38).
Cell Rescue, Defense, and Virulence
One of the proteins showing a high representativity in the extracellular proteome both in number and intensity of isoforms (18, 6.03%) is a probable catalase R (Pc16g11860, spots 12–24, 26, 27, 29, 31, and 275). Correlated with this finding is the presence of a probable glucose oxidase (Pc20g09560) in spot 41. This enzyme, initially known as notatin, has been isolated from the culture filtrate of Penicillium notatum Westling and possesses antibacterial properties (39). This protein catalyzes the oxidation of β-d-glucose into d-glucono-1,5-lactone, which then is hydrolyzed to gluconic acid, which is involved in virulence (see Discussion).
Other proteins important for virulence are phospholipases (see Discussion). Proteins with a putative phospholipase activity are found in spot 1 (Pc18g02900; lysophospholipase phospholipase B), spots 69, 77, 78, and 80 (Pc14g00170; 81% homology and 68% identity to a phosphatidylglycerol specific phospholipase from Aspergillus clavatus), spots 81–83 and 100 (Pc15g01880; probable nonhemolytic phospholipase C). Another virulence protein is found in spots 181, 270, and 272 (Pc16g06700). This protein shares 82% homology and 75% identity with a muraminidase (lysozime) from A. flavus, which functions as an antibacterial agent. This protein has been reported to be secreted also in Agaricus bisporus (40). Spots 85, 87, and 90 include a protein (Pc18g00980) with 96% similarity (90% identity) to an UDP-galactopyranose mutase from A. fumigatus. This mutase enzyme is essential for the viability of mycobacteria (41). Galactofuranose containing molecules have been described at the cell surface of several eukaryotes and shown to contribute to the virulence of A. fumigatus (42).
Spot 242 includes a protein (Pc12g13600) with strong similarity to hypothetical necrosis and ethylene inducing protein BH0395. A similar protein, which induces ethylene and necrosis in leaves of Erythroxylum coca, has been purified from culture filtrates of Fusarium oxysporum (43). These proteins are elicitors of defense responses in tobacco and tomato and induce ethylene biosynthesis, production of phytoalexins, and pathogenesis-related proteins and cause cell death resembling the hypersensitive response reaction (44, 45).
Other proteins identified in the extracellular fraction were involved in cell rescue or defense. Spots 211 and 228 contain a probable ribonuclease T2 precursor (Pc06g00430). RNase T2 was first reported by Sato and Egami (46) as a ribonuclease in Takadiastase produced from a culture extract of A. oryzae. This enzyme might be a defense mechanism against viral pathogens (47). Oxidative stress response proteins were also found. Spot 250 contains a type 2 peroxiredoxin (Pc22g23760), a potent thiol-specific antioxidant protein with alkyl hydroperoxidase activity involved in osmotic stress resistance and detoxification of the cell (48). Spot 47 includes a probable l-galactonolactone oxidase Alo1p (Pc16g04640), which participates in the biosynthesis of vitamin C (l-ascorbic acid) in plants. Some fungi, such as S. cerevisiae do not normally synthesize ascorbate, but Alo1p, which is embedded in the mitochondrial membrane, can convert several related substrates to either dehydro-d-arabino-1,4-lactone or ascorbate, two antioxidants (49).
The finding of a probable cephalosporin esterease (Pc12g13400) in spot 53 is also remarkable. This enzyme, like other cephalosporin acetylhydrolases, is present in many organisms and may be in charge of cephalosporin deacetylation, thus generating deacetylcephalosporin. The presence of cephalosporin acetylhydrolases in the culture broths of Acremonium chrysogenum has been described since 1975 (50) and a extracellular cephalosporin C acetylhydrolase has been purified from this microorganism (51). Although P. chrysogenum does not produce cephalosporin C, cephalosporin esterases cleave a variety of acetylated substrates.
Several extracellular fungal enzymes, such as polygalacturonase, pectate lyase, xylanase, and proteases, have been shown or postulated to be required for virulence in at least one host/pathogen interaction and certain polygalacturonases and xylanases are elicitors of plant defense responses (18, 52). These proteins have been included in subsequent categories (see below).
Plant Cell Wall Degradation
Cellulolytic enzymes are represented by endoglucanases (Pc22g19230, spots 22, 23, 40, and 42; Pc20g07340, spot 271; Pc13g13110, spot 273) and β-glucosidases (Pc15g00210, spots 270 and 272; Pc18g01940, spot 276). For hemicellulose degradation, P. chrysogenum secretes an endo 1,4-β-xylanase, which was found to be the major protein in the culture filtrate of this fungus when grown on 1% xylan (53). It is well-known that P. crysogenum is able to grow on xylan as a sole carbon source (54). The precursor of this endo-1,4-beta-xylanase (Pc20g07020) is present in spot 193.
Pectins form another group of heteropolysaccharides from the plant cell wall. It is noteworthy that 11 isoforms (3.56% of the total intensity) of a probable pectate lyase (Pc22g24890) have been identified in spots 184, 187–194, 206, and 265. In addition, a putative pectate lyase is also found in spot 222 (Pc22g05880, 80% homology and 65% identity to a pectate lyase D from Pyrenophora tritici-repentis). Other pectinolytic enzymes found in the extracellular fraction are rhamnogalacturonase B (Pc14g00370, spot 88), which has a positive effect in the apple hot-mash liquefaction process (55) and polygalacturonases (Pc20g13360, spot 149; Pc22g20290, spots 154–157). The finding of a probable ferulic acid esterease (Pc20g07010) in spots 206 and 208 is also remarkable (see Discussion).
Other accessory enzymes involved in the degradation of plant cell wall polysaccharides described in the genus Aspergillus (56) including endoarabinases, arabinoxylan arabinofuranohydrolase, or acetyl xylan esterase are also found in the P. chrysogenum secretome.
Proteases
Proteolytic enzymes secreted by fungi are important enzymes for fungal pathogenicity and some of them are important in the food industry. Examples of probable carboxypeptidases are found in spots 94 (Pc12g14680), 56 (Pc20g10240), and 22 (Pc21g06670), the latter two being probable serine carboxypeptidases. Extracellular serine carboxypeptidases are found in the vacuoles of fungi and higher plants and in the lysosomes of animal cells and additionally, many fungi secrete serine carboxypeptidases (57). Other probable secreted serine protease (Pc20g15140) with putative antigenic properties (see below) is found in spot 264. The group of aspartyl proteases is represented by homologues of candidapepsin (Pc13g09680, spot 181) and aspergillopepsin (Pc21g02370, spot 271), the latter being one of the major proteins secreted by A. fumigatus (58) and Aspergillus awamori (59). Some of these proteins may contribute to pathogenicity as has been reported for other fungi (52).
It is also important to indicate that spot 45 contains an aminopeptidase-like protein (Pc22g15910). This protein shares 92% homology and 84% identity with a putative aminopeptidase P from A. flavus. This enzyme, also known as proline aminopeptidase, and aminoacylproline aminopeptidase catalyzes the release of any N-terminal amino acid, including proline itself, that is linked with proline. Prolyl aminopeptidase activity is important during the ripening of the Camembert cheese by decreasing the amount of bitter tasting peptides (60).
Allergenic Proteins
In Penicillium spp., many allergens show higher IgE-binding activity in culture filtrate extracts than in cellular extracts (61). Orthologs of human IgE-reactive proteins from Penicillium citrinum are identified in the filtrate extracts of P. chrysogenum, such as pectate lyase (Pc22g24890) with 11 isoforms (spots 184, 187–194, 206, and 265), catalase R (Pc16g11860) with 18 isoforms (spots 12–24, 26, 27, 29, 31, and 275) or 1,3-β-glucanosyltransferase (Pc15g01030) with 6 isoforms (spots 95, 96, 148, 150, 151, and 153). In addition, two isoforms of a probable IgE binding protein (Pc22g22290) are present in spots 236 and 242. Spot 257 includes a 16-kDa allergen specific of P. chrysogenum and spots 51, 52, 55, and 56 contain the 68-kDa allergen N-acetylhexosaminidase (Pc20g10360) specific for P. chrysogenum (62). This protein is one of the highly represented proteins in P. chrysogenum, increasing along the fermentation process (37). Orthologs of proteins that have been shown to possess antigenic properties in the filamentous fungus P. citrinum are also present in the P. chrysogenum secretome. Spot 264 contains a protein (Pc20g15140) with strong similarity to a secreted serine protease (19 kDa CS antigen) from Coccidioides immitis. In addition, an enolase (Pc14g01740, spots 53–59) and an elongation factor 1-β(Pc13g08810, spot 255) are present in the culture medium of P. chrysogenum, although these proteins are not predicted to be secreted as shown in supplemental Table 2 (see Discussion). These findings highlight the importance of P. chrysogenum as an indoor allergen.
Differences in the Extracellular Proteome of the Wild-type P. chrysogenum NRRL 1951 and Wis 54–1255 at 68 h: How a Fruit-rotting Fungus was Converted to an Industrial Penicillin Producer
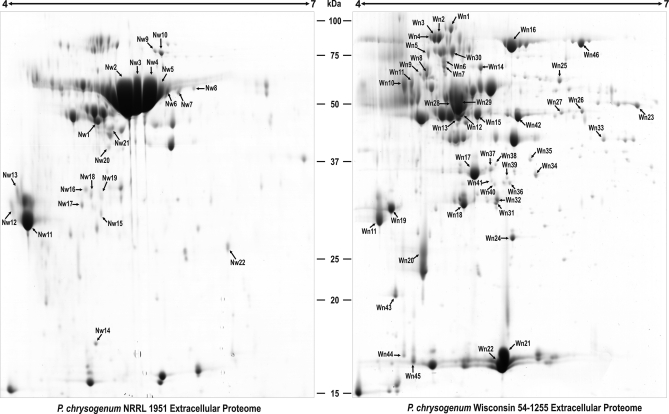
In order to find the extracellular protein variations introduced during the early strain improvement program, the wild-type strain NRRL 1951 was compared with the improved, but still low producer Wis 54–1255. Analysis of the 2-DE gels (Fig. 4) showed 22 spots that were overrepresented in the NRRL 1951 strain (totalling 31 proteins with isoforms), whereas in the Wis 54–1255, 46 spots (51 proteins and isoforms) had higher representation compared with the wild-type strain. Supplemental Tables 3 and 4 display the findings summarized below.
Fig. 4.
Comparison of the extracellular proteomes of P. chrysogenum NRRL 1951 and Wis 54–1255 at 68 h. Proteins were separated by 2-DE. The word “Nw” is used for those spots over-represented or present only in the wild-type strain, whereas “Wn” is used for those spots over-represented or present only in the Wis 54–1255 strain. The spots differentially represented in each strain are numbered and correspond to those proteins listed in supplemental Table 3 (for Nw) and supplemental Table 4 (for Wn).
Cell Rescue, Defense, and Virulence
It is striking that 18 out of the 22 spots overrepresented or only present in the wild-type strain NRRL 1951 include proteins related to fungal virulence. Isoforms of a probable nonhemolytic phospholipase C are highly abundant (15 isoforms, spots Nw1-Nw3, Nw5, Nw6, Nw8, Nw12, and Nw15-Nw22). In addition, two isoforms of a glusose oxidase GOX are only detected in this strain (spots Nw9 and Nw10). All these proteins showed lower concentrations in the secretome of the improved penicillin producer Wis 54–1255 strain. However, in the Wis 54–1255 strain, proteins involved in stress defense mechanisms are predominant, such as a probable catalase R (spot Wn16), a probable glyoxalase I (spot Wn35), which detoxifies methylglyoxal, a typical 2-oxoaldehyde in living cells that inhibits the growth of various kinds of cells from microorganisms to mammals by condensation with glutathione, and the hypothetical protein present in spot Wn29. The latter shares 82% similarity and 75% identity with a muramidase (lysozime) from Aspergillus flavus NRRL3357. This antibacterial enzyme catalyzes the hydrolysis of 1,4-β-linkages between N-acetylmuramic acid and N-acetyl-d-glucosamine residues in peptidoglycan and between N-acetyl-d-glucosamine residues in chitodextrin (see Discussion).
Utilization of Nutrients
Acid phosphatases are predominant in the wild-type strain. Seven isoforms of PhoA (spots Nw3-Nw8 and Nw20) and one isoform of AphA (spot Nw19) are overrepresented in the NRRL 1951 strain, whereas alkaline phosphatases are overrepresented in the Wis 54–1255 strain (spots Wn1-Wn4). Glucan 1,4-α-glucosidase (glucoamylase) is only detected in the Wis 54–1255 strain (spot Wn12). Because it catalyzes the release of glucose from the nonreducing ends of starch and related polysaccharides, the overproduction of this enzyme may have represented one important milestone during the industrial improvement program, allowing P. chrysogenum to utilize carbon sources more efficiently.
General Metabolism
One of the proteins that are overrepresented in the Wis 54–1255 strain is a probable isoamyl alcohol oxidase, which is included in spots Wn8 and Wn9. This protein is of relevant interest for the production of alcoholic beverages (see Discussion). Strikingly some proteins overrepresented or only detected in the Wis 54–1255 strain lack predicted secretion signal sequences. These proteins might play different roles according to their location (see Discussion). Examples are provided by a probable d-arabinose dehydrogenase Ara1 (spot Wn34), an enolase (spot Wn42), a probable mitochondrial aconitate hydratase Aco1 (spot Wn46), and a probable transaldolase Tal1 (spots Wn36, Wn40, and Wn41).
Plant Cell Wall Degradation
One interesting finding is the overrepresentation of several enzymes involved in degradation of complex nutrients in the Wis 54–1255 strain probably as the result of selection of mutants showing good growth in complex plant derived nutrients. These enzymes are a probable pectate lyase PlyA (spots Wn19 and Wn37), glucan 1,3-β-glucosidase Bgl2 (spot Wn29), and endo 1,5-α arabinanase AbnA (sopts Wn31 and Wn32).
Specific Allergens
The Wis 54–1255 strain also overproduces some specific antigens, such as one ortholog of the IgE-binding protein from A. fumigatus (spot Wn20) and a 16 kDa allergen (spot Wn45).
Other Proteins
There are two proteins that were not identified in the extracellular proteome reference map of Wis 54–1255 and therefore, seem to be specific of the wild-type strain NRRL 1951 under the conditions used. These proteins are a hypothetical protein of unknown function (spots Nw14) and a protein with strong similarity to a hypothetical protein smik_17056 (spots Nw12 and Nw13). The latter shows a 58% similarity and a 51% identity with the Ddr48p protein of S. cerevisiae. This corresponds to a DNA damage-responsive protein, whose expression is increased in response to heat-shock stress or treatments that produce DNA lesions (63).
Extracellular Proteome Differences between the Wis 54–1255 and the High Producer Strain AS-P-78 (Industrial stage of P. chrysogenum) at 68 h
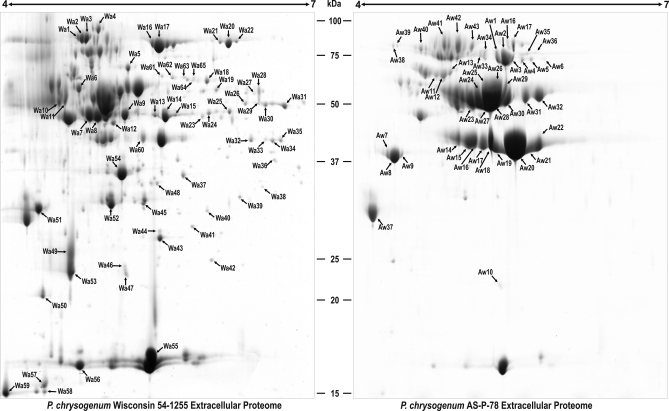
To assess some of the protein variations that occurred during the development of the high producer AS-P-78 from Wis 54–1255, extracellular proteins from these two strains were compared with each other as indicated above. Analysis of the 2-DE gels (Fig. 5) identified 65 spots (68 proteins and isoforms) that were overrepresented in the Wis 54–1255 strain, whereas 44 spots (49 proteins and isoforms) had higher representation in the AS-P-78 strain. The findings summarized below are detailed in supplemental Tables 5 and 6.
Fig. 5.
Comparison of the extracellular proteomes of P. chrysogenum Wis 54–1255 and AS-P-78 at 68 h. Proteins were separated by 2-DE. The word “Wa” is used for those spots over-represented or present only in the Wis 54–1255 strain, whereas “Aw” is used for those spots over-represented or present only in the AS-P-78 strain. The spots differentially represented in each strain are numbered and correspond to those proteins listed in supplemental Table 5 (for Wa) and supplemental Table 6 (for Aw).
General Metabolism
One of the proteins that are overrepresented in the Wis 54–1255 strain when compared with the high producer AS-P-78 is a probable sulfydryl oxidase Sox (spots Wa7 and Wa8), an important enzyme for food industry (see Discussion). Another enzyme overrepresented in the Wis 54–1255 that is related to food industrial processes is a dihydroxy-acid dehydratase Ilv3 (Wa62 and Wa63), which is involved in the biosynthesis of branched-chain amino acids (see Discussion). As it was observed during the comparison of the the wild-type strain versus the Wis 54–1255 strain, some proteins overrepresented or only detected in the latter strain lack predicted secretion signal sequences. Examples are provided by an enolase (spots Wa13-Wa15), a probable mitochondrial (homo) aconitate hydratase Aco1 (spots Wa20-Wa22), a probable glucose-6-phosphate 1-dehydrogenase (spot Wa27), a probable ribose 5-phosphate isomerase RPI (spot Wa40), a probable transaldolase Tal1 (spots Wa48), and the glutamate dehydrogenase GDHA (spots Wa26, Wa30), the latter reported to be involved in regulation of β-lactam production (64). Several of these enzymes play roles related to the mitochondrial formation of the penicillin precursor α-aminoadipic acid and may be released by some type of vesicles. As it was indicated before, these proteins might play different roles according to their location (see Discussion).
In the AS-P-78 strain, a probable isoamyl alcohol oxidase is overrepresented (spot Aw11). In addition, a hypothetical amidase is overrepresented in this strain (spots Aw4-Aw6).
Utilization of Nutrients
Several proteins involved in the utilization of alternative carbon sources as auxiliary energy substrates (see before) are overrepresented in the Wis 54–1255 strain, such as three isoforms of a protein encoded by an acetate-inducible gene aciA (spots Wa32, Wa33, and Wa35), two isoforms of a probable trehalose metabolism factor Pmu1 (spots Wa28 and Wa29), and two isoforms of a probable sorbitol utilization protein Sou2 (spots Wa42-Wa44).
One of the most remarkable differences found between AS-P-78 strain and the Wis 54–1255 strain is represented by phosphatases. Ten isoforms of the acid phosphatase PhoA (spots Aw23–Aw32) and three isoforms of another probable acid phosphatase AFPhoA (spots Aw41–Aw43) are overexpressed in the high producer strain. On the contrary, the Wis 54-1255 strain overexpresses three isoforms of a probable alkaline phosphatase (spots Wa1–Wa3).
Cell Rescue, Defense, Virulence, and Plant Cell Wall Degradation
Another interesting feature shown by the AS-P-78 strain is the overrepresentation of seven isoforms of a probable glucose oxidase GOX (spots Aw1–Aw3 and Aw33–Aw36) and two isoforms of a probable nonhemolytic phospholipase C (spots Aw23 and Aw24).
Although both strains secrete plant cell wall-degrading enzymes, the overrepresentation of 9 isoforms of a probable polygalacturonase in the AS-P-78 strain is remarkable (spots Aw14–Aw22). The high content of a polygalacturonase is likely because of the selection of mutant colonies showing better growth on plant polymeric materials during the selection procedure to compensate for the slow growth rate of the high penicillin producing strains (65). This pectinolytic enzyme is of relevant importance for beverage industries (see Discussion).
Proteases
The high producer strain AS-P-78 showed several proteases overrepresented, such as a probable aspergillopepsin (spots Aw7–Aw9) or carboxypeptidases (spots Aw11–Aw13 and Aw38–Aw40). Strikingly, the carbosypeptidase protein included in spot Aw40 is not present in the reference map (strain Wis54–1255), which indicates that it is specific of the AS-P-78 strain under the conditions used.
Specific Allergens and Other Proteins
The Wis 54–1255 strain contained two overrepresented isoforms of the IgE-binding protein ortholog from A. fumigatus (spots Wa49 and Wa53). As it was indicated before, some of the proteins overrerpresented in the Wis54–1255 strain lacked predicted signal sequences. This is the case of a probable cyclophilin CypD, which is found in spots Wa46 and Wa47.
DISCUSSION
In this work, we present for the first time, the extracellular proteome reference map of P. chrysogenum. Of the total number of different proteins in the extracellular proteome of P. chrysogenum, 102 out of 131 were predicted to contain either classical or nonclassical signal sequences using the SignalP and SecretomeP programs. Prediction of secretory proteins has been improved by the implementation of the Fungal Secretome Database (20), which is based on nine prediction programs (including SignalP and SecretomeP) and has been used to identify putative secretory proteins in 158 fungal/oomycete genomes. The presence of proteins with nonpredicted signal sequences in the secreted fraction has also been described in the filamentous fungus Botrytis cinerea (19). These authors suggested that these proteins could be truly secreted multifunctional proteins with different activities according to their intracellular or extracellular location, a fact that has been confirmed in other organisms (66). Therefore, a dual role for those 29 proteins identified in P. chrysogenum cannot be excluded, specially when major cell lysis events were not observed at 68h (Fig. 1A). Interestingly, Kiel and coworkers (67) also identified some of these extracellular proteins in the microbody (peroxisomes) matrix of P. chrysogenum, even those presumed to be located on the cytosol, mitochondria, or vacuoles (supplemental Table 7). An alternative explanation might be given by the selective autophagic degradation of peroxisomes (pexophagy). It is known that in late stage cultures, peroxisomes are integrated into vacuoles by pexophagy (68). Integration of peroxisomes into vacuoles may lead to secretion of the proteins located in the peroxisomal matrix by exocytosis, a mechanism that has been discussed as an alternative route for the release of penicillin from peroxisomes to the culture medium (12).
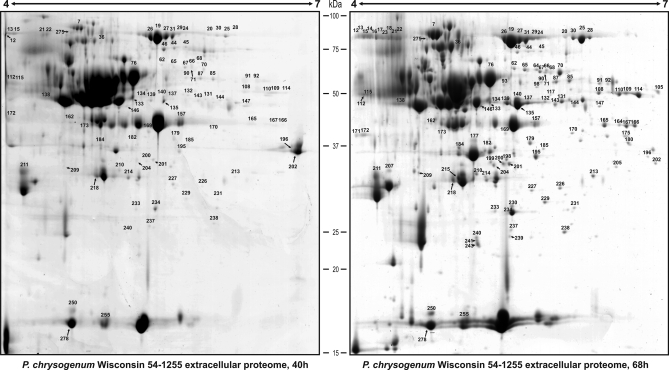
Cell lysis is very unlikely because proteins that have been reported to be the most abundant in the intracellular proteome of P. chrysogenum, such as a flavohemoglobin, a hypothetical ECM33 homolog SPCC1223.12c, a manganese superoxide dismutase, or a methyltransferase (16) are not detected in the extracellular proteome. Only traces of a probable glyceraldehyde-3-phosphate dehydrogenase (Pc21g14560), which is also relatively abundant in the intracellular proteome of P. chrysogenum (16), are found in the secretome, suggesting that contribution of proteins from cell lysis to the extracellular proteome is minimal. The case of the latter protein is especially relevant, because it appears to be a multifunctional protein found in several subcellular locations in eukaryotes, displaying functions unrelated to glycolysis, such as membrane transport and fusion, as well as nuclear RNA transport (69, 70). In some pathogenic fungi, this enzyme has been found on the cell wall, where it may have a role in host-pathogen interactions (71–73). A similar extracellular role for this enzyme is likely in P. chrysogenum. As an additional control of cell lysis events that might contaminate the secretome at 68 h, we analyzed the extracellular proteome at 40 h (Fig. 6) and searched for those proteins detected at 68 h that lacked a classical secretion signal peptide. Results (supplemental Table 8) indicated that 32 out of the 118 spots were not present at 40 h. This may indicate that 32 out of the 279 spots analyzed in this work (11.47%) may be contaminating the extracellular reference map at 68 h. Although some spots are not detected at 40 h, other spots including isoforms of the same protein are detected at this early time point (e.g. Pc16g11860), which indicates that the presence of such protein at 40 and 68 h is not because of cell lysis. In addition, 15 out of the 32 spots not detected at 40 h, included isoforms of 10 different proteins predicted to be secreted through a nonclassical pathway. Therefore, it seems that only 17 out of the 279 spots analyzed (6.09%) might correspond to contamination from intracellular proteins. Because of the low representativity of those proteins and taking into account our previous observations, cell lysis seems not to be an important event at 68 h.
Fig. 6.
Comparison of the extracellular proteomes of P. chrysogenum Wis 54–1255 at 40 h and 68 h. Proteins were separated by 2-DE. Those spots present at each time point were numbered according to the numbers given in Fig. 2 and supplemental Table 1. The differences between these time-points are listed in supplemental Table 8.
Many of the Observed Extracellular Enzymes are Clearly Related to the Saprophytic Growth of the Fungus on Plant Material
The major structural component of the fungal wall is β-1,3-glucan, which serves as an anchor for other polysaccharides, such as chitin and galactomannan (74). Following the biosynthtesis of β-1,3-glucan, this compound is subjected to downstream processing leading to extensive branching and crosslinking to other cell wall components (75). The β-1,3-glucanosyltransferases are key enzymes involved in this processing and play active roles in fungal cell wall biosynthesis and morphogenesis. These enzymes have also being related to virulence, because one of the key factors underlying fungal attack is the capacity to grow as hyphae (76), a process that requires the polarized biogenesis of the cell wall at the apical tip (77). The β-1,3-glucanosyltransferases are present in the extracellular proteome of P. chrysogenum together with chitinases and chitosanases. Similarly, Coccidiodes immitis secretes in both the saprobic and parasitic phases a 48-kDa antigen acting as a chitinase (78). It was suggested that the active chitinase associates with the autolysis of the parasitic cell wall during early asexual reproductive cycle endosporulation (79).
Glucose oxidases are enzymes involved in virulence, because gluconic acid and glucose oxidase have been associated with the pathogenicity of Penicillium expansum in apples (80). An extracellular localization of glucose oxidase activity implies an extracellular production of hydrogen peroxide. The sequestration of this toxic compound will prevent damage to the cell and extracellular catalases might be involved in this process. Extracellular catalases are detected in the culture medium of P. chrysogenum as it has been reported for other fungi, such as Claviceps purpurea and Blumeria graminis. These microorganisms secrete a catalase during infection of rye (81) and barley (82), respectively. The production of extracellular phospholipases has also been shown to be important in the pathogenesis, because they cause damage to host cell membranes and allow tissue invasion (83). Phospholipases are also produced by certain fungi, including Candida albicans and P. notatum (84, 85) and could represent a virulence determinant in Aspergillus fumigatus (86).
Relevance for the Food Industries
Some proteins identified in the culture medium of P. chrysogenum are specially relevant because of their interest for food industry. This is the case of a probable isoamyl alcohol oxidase, an extracellular enzyme in A. oryzae catalyzing the formation of isovaleraldehyde—the main component of mureka that gives sake an off-flavor (87) —or sulfydryl oxidase. The treatment with sulfhydryl oxidase has been reported of value in the removal of a burnt flavor from Ultra-High Temperature sterilized milk (88). Therefore, this enzyme may be used for treatment of bakery products or for removal of off-flavor from milk or beer. Another relevant enzyme for its application in food industry is a probable dihydroxy-acid dehydratase Ilv3, which is related to the biosynthesis of branched-chain amino acids. This enzyme is important to remove vicinal diketones formed by nonenzymatic reactions from intermediaries of the biosynthesis of valine and isoleucine. When vicinal diketones are present above certain threshold, they give foods the flavor of diacetyl (butter flavor or sweaty flavor in beer, “tsuwari-ka” or nauseating flavor in sake), a representative off-flavor in brewed alcoholic beverages such as beer, sake, and wine. Amplification of the ilv3 gene encoding the dihydroxyacid dehydratase of S. cerevisiae has been patented to produce alcoholic beverages with superior flavor (89). Related to this finding is the identification of a probable ketol-acid reductoisomerase Ilv2, which is also involved in the biosynthesis of branched-chain amino acids isoleucine and valine. Also related to these two previous enzymes is the presence of a probable branched-chain-amino acid aminotransferase Bat2p. During alcoholic fermentation, S. cerevisiae produces higher alcohols that can influence the flavor of the end-product, such as the taste of wines (90). The higher alcohols produced by yeast can originate from the degradation of imported branched-chain amino acids or from endogenous biosynthesis (91). The P. chrysogenum Bat2p ortholog is predicted to possess a signal peptide (supplemental Table 2) and it has been found in the extracellular proteome. Other proteins that also have an important application for beverage industries are pectinolytic enzymes (see below).
A full arsenal of proteins involved in plant cell wall degradation is identified in the extracellular proteome of P. chrysogenum. Among these enzymes, pectinolytic enzymes represented by polygalacturonases or pectate lyases are important for beverage industries and preparation of juices. Other important enzymes are ferulic acid esterases. They hydrolyze the ester linkages of ferulic and diferulic acids present in plant cell walls releasing β-glucan and pentosans from the cell walls, which increases the susceptibility to further enzymatic attack (92).
Extracellular Proteins as Allergens
Penicillium species are important indoor allergens (93) and a limited number of allergens have been reported for these microorganisms (94–99). Among the Penicillium allergens, alkaline or vacuolar serine proteases have been described as the major allergen of P. citrinum, Penicillium brevicompactum, P. chrysogenum, and Penicillium oxalicum (100). Two proteins that have been shown to possess antigenic properties in the filamentous fungus P. citrinum are allergen Pen c 22, which is an enolase with similar immunological characteristics to Asp f22 from A. fumigatus (101), and allergen Pen c 24, which shows sequence homology with elongation factor 1-b from A. fumigatus (99). Orthologs of all these proteins are found in the extracellular proteome of P. chrysogenum.
Modifications of the Extracellular Proteome During the Industrial Strain Improvement Program
Comparison of the extracellular proteins of P. chrysogenum strains NRRL 1951, Wis 54–1255, and AS-P-78, which represent three different stages of the industrial strain improvement program, provides a further step in the characterization of the protein modifications that occurred during this process (16). In general, it seems that enzymes related to plant pathogenesis, tissue invasion, and infectivity, were diminished during the improvement of the wild-type strain, result that correlates well with the fact that the wild-type strain was isolated from an infected cantaloupe. It is also interesting that proteins involved in stress defense mechanisms are predominant in the Wis 54–1255 strain. Antibiotic production can be considered as a defense mechanism against stress caused by competing microorganisms. In fact, it was observed in our previous work on the intracellular proteome, that one the main changes observed in the high producer strains were increases of stress response proteins (16). The amount of pectinolytic enzymes seems to have been increased during the improvement program, specially polygalacturonases, as shown by the amount and intensity of the isoforms found in the AS-P-78 strain. Other proteins following this pattern are proteases, which are predominant in the culture broths of the high producer AS-P-78.
P. chrysogenum as a Cell Factory for White Biotechnology
Fungi have been described as nature's toolbox for white biotechnology (102), although there are other systems for the production of useful proteins, such as bacteria, insects, mammalian cell culture, transgenic animal, plant cell cultures, and transgenic plants. Each system offers some unique advantages and features regarding yield and purity of the proteins overproduced (103). In general, overproduction of proteins in bacteria is a scalable low-cost process, which reaches high yields and purity. The main drawback of this system is protein solubility, together with the absence post-translational modifications that may be necessary for the correct expression and biological activity of functional eukaryotic proteins. Eukaryotic systems, such as insect cells, yeasts, or mammalian host organisms, are capable of a variety of post-translational modifications, but are expensive to operate and have a longer growth period. On the other hand, large amounts of recombinant proteins can be produced at low cost in plants using genetically engineered chloroplasts (104).
Filamentous fungi are also able to produce and secrete high amounts of homologous and heterologous proteins (g/L), although the levels obtained for heterologous proteins were always lower than those for native fungal proteins (2). Secreted proteases by fungal host strains (partially solved by improved protease-deficient host strains) and low secretion levels of heterologous proteins (improved by the gene-fusion approach or fusion to a carrier secreted fungal protein, are the major limitations for this system. The availability of fungal genomes has allowed a detailed analysis of several fungal secretomes, highlighting the potential of these microorganisms for white biotechnology (102). The versatility of P. chrysogenum, its plasticity, its ability to grow on different media and conditions and the availability of several mutants and tools for its genetic manipulation, may confer some advantages on P. chrysogenum over other fungi.
In conclusion, the data provided here show that despite the rounds of classical mutagenesis and the artificial selection undergone by this microrganism just exclusively to increase the production of β-lactam antibiotics, the versatility of this filamentous fungus is high. This is supported by the secretion of a variety of enzymes of relevant interest for white biotechnology. The importance of this filamentous fungus in the immunological response as indoor allergen is confirmed following the identification of several proteins with antigenic properties in the culture medium.
Supplementary Material
Acknowledgments
The technical support of M. D. Gutiérrez (Universidad Complutense de Madrid, Spain), E. Calvo, and L. E. Camafeita (Fundacion Centro Nacional de Investigaciones Cardiovasculares (CNIC), Madrid, Spain), and the excellent technical assistance of B. Martín, J. Merino, A. Casenave, and A. Mulero (INBIOTEC) is acknowledged.
Footnotes
* This research was supported by grants of the Eupean Union (Eurofung QLRT-1999-00729 and Eurofungbase), of the Agencia de Inversiones y Servicios de Castilla y León (Proyecto Genérico de Desarrollo Tecnológico 2008) and the Ministry of Industry of Spain (PROFIT: FIT-010000-2007-74). C. Barreiro is supported by the Program ‘Personal Técnico de Apoyo’ of the Ministry of Science and Innovation of Spain and the European Social Fund (ESF) (PTA-2003-01-00689).
 This article contains supplemental Tables 1–8.
This article contains supplemental Tables 1–8.
1 The abbreviations used are:
- 2-DE
- two dimensional gel electrophoresis
- ACN
- acetonitrile
- DTT
- 1,4-dithiothreitol
- PMF
- peptide mass fingerprinting.
REFERENCES
- 1. Peberdy J. F. (1994) Protein secretion in filamentous fungi—trying to understand a highly productive black box. Trends Biotechnol. 12, 50–57 [DOI] [PubMed] [Google Scholar]
- 2. Punt P. J., van Biezen N., Conesa A., Albers A., Mangnus J., van den Hondel C. (2002) Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20, 200–206 [DOI] [PubMed] [Google Scholar]
- 3. Cardoza R. E., Gutiérrez S., Ortega N., Colina A., Casqueiro J., Martín J. F. (2003) Expression of a synthetic copy of the bovine chymosin gene in Aspergillus awamori from constitutive and pH-regulated promoters and secretion using two different pre-pro sequences. Biotechnol. Bioeng. 83, 249–259 [DOI] [PubMed] [Google Scholar]
- 4. Machida M., Asai K., Sano M., Tanaka T., Kumagai T., Terai G., Kusumoto K., Arima T., Akita O., Kashiwagi Y., Abe K., Gomi K., Horiuchi H., Kitamoto K., Kobayashi T., Takeuchi M., Denning D. W., Galagan J. E., Nierman W. C., Yu J., Archer D. B., Bennett J. W., Bhatnagar D., Cleveland T. E., Fedorova N. D., Gotoh O., Horikawa H., Hosoyama A., Ichinomiya M., Igarashi R., Iwashita K., Juvvadi P. R., Kato M., Kato Y., Kin T., Kokubun A., Maeda H., Maeyama N., Maruyama J., Nagasaki H., Nakajima T., Oda K., Okada K., Paulsen I., Sakamoto K., Sawano T., Takahashi M., Takase K., Terabayashi Y., Wortman J. R., Yamada O., Yamagata Y., Anazawa H., Hata Y., Koide Y., Komori T., Koyama Y., Minetoki T., Suharnan S., Tanaka A., Isono K., Kuhara S., Ogasawara N., Kikuchi H. (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 5. Pel H. J., de Winde J. H., Archer D. B., Dyer P. S., Hofmann G., Schaap P. J., Turner G., de Vries R. P., Albang R., Albermann K., Andersen M. R., Bendtsen J. D., Benen J. A., van den Berg M., Breestraat S., Caddick M. X., Contreras R., Cornell M., Coutinho P. M., Danchin E. G., Debets A. J., Dekker P., van Dijck P. W., van Dijk A., Dijkhuizen L., Driessen A. J., d'Enfert C., Geysens S., Goosen C., Groot G. S., de Groot P. W., Guillemette T., Henrissat B., Herweijer M., van den Hombergh J. P., van den Hondel C. A., van der Heijden R. T., van der Kaaij R. M., Klis F. M., Kools H. J., Kubicek C. P., van Kuyk P. A., Lauber J., Lu X., van der Maarel M. J., Meulenberg R., Menke H., Mortimer M. A., Nielsen J., Oliver S. G., Olsthoorn M., Pal K., van Peij N. N., Ram A. F., Rinas U., Roubos J. A., Sagt C. M., Schmoll M., Sun J., Ussery D., Varga J., Vervecken W., van de Vondervoort P. J., Wedler H., Wösten H. A., Zeng A. P., van Ooyen A. J., Visser J., Stam H. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25, 221–231 [DOI] [PubMed] [Google Scholar]
- 6. Wösten H. A., Moukha S. M., Sietsma J. H., Wessels J. G. (1991) Localization of growth and secretion of proteins in Aspergillus niger. J. Gen. Microbiol. 137, 2017–2023 [DOI] [PubMed] [Google Scholar]
- 7. Conesa A., Punt P. J., van Luijk N., van den Hondel C. A. (2001) The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33, 155–171 [DOI] [PubMed] [Google Scholar]
- 8. Nickel W. (2003) The mystery of nonclassical protein secretion - A current view on cargo proteins and potential export routes. Eur. J. Biochem. 270, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 9. Nickel W. (2005) Unconventional secretory routes: Direct protein export across the plasma membrane of mammalian cells. Traffic. 6, 607–614 [DOI] [PubMed] [Google Scholar]
- 10. Nickel W., Rabouille C. (2009) Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10, 148–155 [DOI] [PubMed] [Google Scholar]
- 11. Nombela C., Gil C., Chaffin W. L. (2006) Non-conventional protein secretion in yeast. Trends Microbiol. 14, 15–21 [DOI] [PubMed] [Google Scholar]
- 12. Martín J. F., Ullán R. V., García-Estrada C. (2010) Regulation and compartmentalization of β-lactam biosynthesis. Microb. Biotechnol.. 3, 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raper K. B., Alexander D. F., Coghill R. D. (1944) Penicillin II. Natural variation and penicillin production in Penicillium notatum and allied species. J. Bacteriol. 48, 639–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demain A. L., Elander R. P. (1999) The β-lactam antibiotics: past, present and future. Antonie Van Leeuwenhoek. 75, 5–19 [DOI] [PubMed] [Google Scholar]
- 15. van den Berg M. A., Albang R., Albermann K., Badger J. H., Daran J. M., Driessen A. J., Garcia-Estrada C., Fedorova N. D., Harris D. M., Heijne W. H., Joardar V., Kiel J. A., Kovalchuk A., Martín J. F., Nierman W. C., Nijland J. G., Pronk J. T., Roubos J. A., van der Klei I. J., van Peij N. N., Veenhuis M., von Döhren H., Wagner C., Wortman J., Bovenberg R. A. L. (2008) Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 26, 1161–1168 [DOI] [PubMed] [Google Scholar]
- 16. Jami M. S., Barreiro C., García-Estrada C., Martín J. F. (2010) Proteome analysis of the penicillin producer Penicillium chrysogenum: Characterization of protein changes during the industrial strain improvement. Mol. Cell. Proteomics 9, 1182–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim Y., Nandakumar M. P., Marten M. R. (2007) Proteomics of filamentous fungi. Trends Biotechnol. 25, 395–400 [DOI] [PubMed] [Google Scholar]
- 18. Paper J. M., Scott-Craig J. S., Adhikari N. D., Cuomo C. A., Walton J. D. (2007) Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics. 7, 3171–3183 [DOI] [PubMed] [Google Scholar]
- 19. Shah P., Atwood J. A., Orlando R., El Mubarek H., Podila G. K., Davis M. R. (2009) Comparative proteomic analysis of Botrytis cinerea secretome. J. Proteome Res. 8, 1123–1130 [DOI] [PubMed] [Google Scholar]
- 20. Choi J., Park J., Kim D., Jung K., Kang S., Lee Y. H. (2010) Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics. 11, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cantoral J. M., Gutiérrez S., Fierro F., Gil-Espinosa S., van Liempt H., Martín J. F. (1993) Biochemical characterization and molecular genetics of nine mutants of Penicillium chrysogenum impaired in penicillin biosynthesis. J. Biol. Chem. 268, 737–744 [PubMed] [Google Scholar]
- 22. Fierro F., Montenegro E., Gutiérrez S., Martín J. F. (1996) Mutants blocked in penicillin biosynthesis show a deletion of the entire penicillin gene cluster at a specific site within a conserved hexanucleotide sequence. Appl. Microbiol. Biotechnol. 44, 597–604 [DOI] [PubMed] [Google Scholar]
- 23. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 24. Havlis J., Thomas H., Sebela M., Shevchenko A. (2003) Fast-response proteomics by accelerated in-gel digestion of proteins. Anal. Chem. 75, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 25. Bendtsen J. D., Nielsen H., von Heijne G., Brunakm S. (2004) Improved prediction of signal peptides: Signal P 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 26. Bendtsen J. D., Jensen L. J., Blom N., von Heijne G., Brunak S. (2004) Feature based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 27. Ehrlich K. C., Montalbano B. G., Mullaney E. J., Dischinger H. C., Jr., Ullah A. H. (1994) An acid phosphatase from Aspergillus ficuum has homology to Penicillium chrysogenum PhoA. Biochem. Biophys. Res. Commun. 204, 63–68 [DOI] [PubMed] [Google Scholar]
- 28. Reddy N. R., Sathe S. K., Salunkhe D. K. (1982) Phytates in legumes and cereals. Adv. Food. Res. 28, 1–92 [DOI] [PubMed] [Google Scholar]
- 29. Kumar P., Satyanarayana T. (2009) Microbial glucoamylases: characteristics and applications. Crit. Rev. Biotechnol. 29, 225–255 [DOI] [PubMed] [Google Scholar]
- 30. Harris D. M., van der Krogt Z. A., van Gulik W. M., van Dijken J. P., Pronk J. T. (2007) Formate as an auxiliary substrate for glucose-limited cultivation of Penicillium chrysogenum: impact on penicillin G production and biomass yield. Appl. Environ. Microbiol. 73, 5020–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khan N. A. (1982) Suppression of maltose-negative phenotype by a specific nuclear gene (PMU1) in the petite cells of the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 186, 40–43 [DOI] [PubMed] [Google Scholar]
- 32. Bilai V. I., Efimov A. S., Sokolova E. V., Ellanskaia I. A., Obrosova I. G. (1986) Extracellular sorbitol dehydrogenase activity of fungal species in the genus Fusarium. Mikrobiol. Zh. 48, 33–37 [PubMed] [Google Scholar]
- 33. Hochstein L. I., Rittenberg S. C. (1959) The bacterial oxidation of nicotine I. Nicotine oxidation by cell-free preparations. J. Biol. Chem. 234, 151–155 [PubMed] [Google Scholar]
- 34. Zhang X. Y., Dai A. L., Kuroiwa K., Kodaira R., Nogawa M., Shimosaka M., Okazaki M. (2001) Cloning and characterization of a chitosanase gene from the koji mold Aspergillus oryzae strain IAM 2660. Biosci. Biotechnol. Biochem. 65, 977–981 [DOI] [PubMed] [Google Scholar]
- 35. Rodríguez-Peña J. M., Cid V. J., Arroyo J., Nombela C. (2000) A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20, 3245–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horsch M., Mayer C., Sennhauser U., Rast D. M. (1997) Beta-N-acetylhexosaminidase: a target for the design of antifungal agents. Pharmacol. Ther. 76, 187–218 [DOI] [PubMed] [Google Scholar]
- 37. Díez B., Rodríguez-Sáiz M., de la Fuente J. L., Moreno M. A., Barredo J. L. (2005) The nagA gene of Penicillium chrysogenum encoding beta-N-acetylglucosaminidase. FEMS Microbiol. Lett. 242, 257–264 [DOI] [PubMed] [Google Scholar]
- 38. Pusztahelyi T., Pócsi I., Szentirmai A. (1997) Aging of Penicillium chrysogenum cultures under carbon starvation: II protease and N-acetyl-b-D-hexosaminidase production. Biotechnol. Appl. Biochem. 25, 87–93 [Google Scholar]
- 39. Coulthard C. E., Michaelis R., Short W. F., Sykes G. (1945) Notatin: an anti-bacterial glucose-aerodehydrogenase from Penicillium notatum Westling and Penicillium resticulosum sp. nov. Biochem. J. 39, 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lincoln S. P., Fermor T. R., Wood D. A. (1997) Production and detection of muramidase and acetylglucosaminidase from. Agaricus bisporus. Lett. Appl. Microbiol. 25, 24–29 [DOI] [PubMed] [Google Scholar]
- 41. Pan F., Jackson M., Ma Y., McNeil M. (2001) Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183, 3991–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmalhorst P. S., Krappmann S., Vervecken W., Rohde M., Müller M., Braus G. H., Contreras R., Braun A., Bakker H., Routier F. H. (2008) Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell. 7, 1268–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bailey B. A., Jennings J. C., Anderson J. D. (1997) The 24-kDa protein from Fusarium oxysporum f. sp. erythroxyli: occurrence in related fungi and the effect of growth medium on its production. Can. J. Microbiol. 43, 45–55 [DOI] [PubMed] [Google Scholar]
- 44. Bailey B. A., Korcak R. F., Anderson J. D. (1992) Alteration in Nicotiana tabacum L. cv Xanthi cell membrane integrity following treatment with ethylene biosynthesis-inducing endoxylanase. Plant Physiol. 100, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yano A., Suzuki K., Uchimiya H., Shinshi H. (1998) Induction of hypersensitive cell death by a fungal protein in cultures of tobacco cells. Mol. Plant-Microbe Interact. 11, 115–123 [Google Scholar]
- 46. Sato K., Egami F. (1957) The specificity of T1 ribonuclease. C. R. Seances Soc. Biol. Fil. 151, 1792–1796 [PubMed] [Google Scholar]
- 47. Irie M. (1999) Structure-function relationships of acid ribonucleases: Lysosomal, vacuolar, and periplasmic enzymes. Pharmacol. Ther. 81, 77–89 [DOI] [PubMed] [Google Scholar]
- 48. Jeong J. S., Kwon S. J., Kang S. W., Rhee S. G., Kim K. (1999) Purification and characterization of a second type thioredoxin peroxidase (type II TPx) from Saccharomyces cerevisiae. Biochemistry 38, 776–783 [DOI] [PubMed] [Google Scholar]
- 49. Hancock R. D., Galpin J. R., Viola R. (2000) Biosynthesis of L-ascorbic acid (vitamin C) by Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186, 245–250 [DOI] [PubMed] [Google Scholar]
- 50. Fujisawa Y., Shirafuji H., Kanzaki T. (1975) Deacetylcephalosporin C formation by cephalosporin C acetylhydrolase induced in a Cephalosporium acremonium mutant. Agric. Biol. Chem. 39, 1303–1309 [Google Scholar]
- 51. Velasco J., Gutiérrez S., Casqueiro J., Fierro F., Campoy S., Martín J. F. (2001) Cloning and characterization of the gene cahB encoding a cephalosporin C acetylhydrolase from. Acremonium chrysogenum. Appl. Microbiol. Biotechnol. 57, 350–356 [DOI] [PubMed] [Google Scholar]
- 52. Monod M., Borg-von Z. M. (2002) Secreted aspartic proteases as virulence factors of Candida species. Biol. Chem. 383, 1087–1093 [DOI] [PubMed] [Google Scholar]
- 53. Haas H., Herfurth E., Stöffler G., Redl B. (1992) Purification, characterization and partial amino acid sequences of a xylanase produced by Penicillium chrysogenum. Biochim. Biophys. Acta. 1117, 279–286 [DOI] [PubMed] [Google Scholar]
- 54. Haas H., Friedlin E., Stöffler G., Redl B. (1993) Cloning and structural organization of a xylanase-encoding gene from Penicillium chrysogenum. Gene. 126, 237–242 [DOI] [PubMed] [Google Scholar]
- 55. Suykerbuyk M. E., Schaap P. J., Stam H., Musters W., Visser J. (1995) Cloning, sequence and expression of the gene coding for rhamnogalacturonase of Aspergillus aculeatus; a novel pectinolytic enzyme. Appl. Microbiol. Biotechnol. 43, 861–870 [DOI] [PubMed] [Google Scholar]
- 56. de Vries R. P., Visser J. (2001) Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65, 497–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Breddam K. (1986) Serine carboxypeptidases. A review. Carlsberg Res. Commun. 51, 83–128 [Google Scholar]
- 58. Schwienbacher M., Weig M., Thies S., Regula J. T., Heesemann J., Ebel F. (2005) Analysis of the major proteins secreted by the human opportunistic pathogen Aspergillus fumigatus under in vitro conditions. Med. Mycol. 43, 623–630 [DOI] [PubMed] [Google Scholar]
- 59. Moralejo F. J., Cardoza R. E., Gutiérrez S., Sisniega H., Faus I., Martín J. F. (2000) Overexpression and lack of degradation of thaumatin in an aspergillopepsin-A defective mutant of Aspergillus awamori containing an insertion in the pepA gene. Appl. Microbiol. Biotechnol. 54, 772–777 [DOI] [PubMed] [Google Scholar]
- 60. Fuke Y., Kaminogawa S., Matsuoka H., Yamauchi K. (1988) Purification and properties of aminopeptidase I from Penicillium caseicolum. J. Dairy Sci. 71, 1423–1431 [Google Scholar]
- 61. Chiu L. L., Lee K. L., Lin Y. F., Chu C. Y., Su S. N., Chow L. P. (2008) Secretome analysis of novel Ig E-binding proteins from Penicillium citrinum. Proteomics Clin. Appl. 2, 33–45 [DOI] [PubMed] [Google Scholar]
- 62. Shen H. D., Liaw S. F., Lin W. L., Ro L. H., Yang H. L., Han S. H. (1995) Molecular cloning of cDNA coding for the 68 kDa allergen of Penicillium notatum using MoAbs. Clin. Exp. Allergy 25, 350–356 [DOI] [PubMed] [Google Scholar]
- 63. Sheng S., Schuster S. M. (1993) Purification and characterization of Saccharomyces cerevisiae DNA damage-responsive protein 48 (DDRP 48). J. Biol. Chem. 268, 4752–4758 [PubMed] [Google Scholar]
- 64. Thykaer J., Rueksomtawin K., Noorman H., Nielsen J. (2008) NADPH-dependent glutamate dehydrogenase in Penicillium chrysogenum is involved in regulation of β-lactam production. Microbiology 154, 1242–1250 [DOI] [PubMed] [Google Scholar]
- 65. Lein J. (1986) The Panlabs Penicillium strain improvement program, in Overproduction of Microbial Metabolites (Vanek Z., Hostalek Z., eds), pp. 105–140, Butterworths, Stoneham, MA [Google Scholar]
- 66. Radisky D. C., Stallings-Mann M., Hirai Y., Bissell M. J. (2009) Single proteins might have dual but related functions in intracellular and extracellular microenvironments. Nat. Rev. Mol. Cell Biol. 10, 228–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kiel J. A., van den Berg M. A., Fusetti F., Poolman B., Bovenberg R. A., Veenhuis M., van der Klei I. J. (2009) Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct. Integr. Genomics. 9, 167–184 [DOI] [PubMed] [Google Scholar]
- 68. Sakai Y., Oku M., van der Klei I. J., Kiel J. A. (2006) Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta. 1763, 1767–1775 [DOI] [PubMed] [Google Scholar]
- 69. Singh R., Green M. R. (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 259, 365–368 [DOI] [PubMed] [Google Scholar]
- 70. Sirover M. A. (1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta. 1432, 159–184 [DOI] [PubMed] [Google Scholar]
- 71. Gil-Navarro I., Gil M. L., Casanova M., O'Connor J. E., Martínez J. P., Gozalbo D. (1997) The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 179, 4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gozalbo D., Gil-Navarro I., Azorín I., Renau-Piqueras J., Martínez J. P., Gil M. L. (1998) The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 66, 2052–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Barbosa M. S., Báo S. N., Andreotti P. F., de Faria F. P., Felipe M. S., dos Santos Feitosa L., Mendes-Giannini M. J., Soares C. M. (2006) Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74, 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fontaine T., Mouyna I., Hartland R. P., Paris S., Latgé J. P. (1997) From the surface to the inner layer of the fungal cell wall. Biochem. Soc. Trans. 25, 194–199 [DOI] [PubMed] [Google Scholar]
- 75. Klis F. M., Mol P., Hellingwerf K., Brul S. (2002) Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26, 239–256 [DOI] [PubMed] [Google Scholar]
- 76. Madhani H. D., Fink G. R. (1998) The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 8, 348–353 [DOI] [PubMed] [Google Scholar]
- 77. Harris S. D., Momany M. (2004) Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41, 391–400 [DOI] [PubMed] [Google Scholar]
- 78. Johnson S. M., Pappagianis D. (1992) The coccidioidal complement fixation and immunodiffusion-complement fixation antigen is a chitinase. Infect. Immun. 60, 2588–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cole G. T., Kruse D., Seshan K. R., Pan S., Szaniszlo P. J., Richardson J., Bian B. (1993) Factors regulating morphogenesis in Coccidioides immitis, in Dimorphic fungi in biology and medicine (Vanden Bossche H., Odds F. C., Kerridge D., eds.), pp. 191–212, Plenum Press, New York, NY [Google Scholar]
- 80. Hadas Y., Goldberg I., Pines O., Prusky D. (2007) Involvement of gluconic acid and glucose oxidase in the pathogenicity of Penicillium expansum in apples. Phytopathology. 97, 384–390 [DOI] [PubMed] [Google Scholar]
- 81. Garre V., Tenberge K. B., Eising R. (1998) Secretion of a fungal extracellular catalase by Claviceps purpurea during infection of rye: putative role in pathogenicity and suppression of host defense. Phytopathology. 88, 744–753 [DOI] [PubMed] [Google Scholar]
- 82. Zhang Z., Henderson C., Gurr S. J. (2004) Blumeria graminis secretes an extracellular catalase during infection of barley: potential role in suppression of host defence. Mol. Plant Pathol. 5, 537–547 [DOI] [PubMed] [Google Scholar]
- 83. Meyers D. J., Palmer K. C., Bale L. A., Kernacki K., Preston M., Brown T., Berk R. S. (1992) In vivo and in vitro toxicity of phospholipase C from Pseudomonas aeruginosa. Toxicon. 30, 161–169 [DOI] [PubMed] [Google Scholar]
- 84. Saito K., Sugatani J., Okumura T. (1991) Phospholipase B from Penicillium notatum. Methods Enzymol. 197, 446–456 [DOI] [PubMed] [Google Scholar]
- 85. Ibrahim A. S., Mirbod F., Filler S. G., Banno Y., Cole G. T., Kitajima Y., Edwards J. E., Jr., Nozawa Y., Ghannoum M. A. (1995) Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect Immun. 63, 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Birch M., Robson G., Law D., Denning D. W. (1996) Evidence of multiple extracellular phospholipase activities of Aspergillus fumigatus. Infect. Immun. 64, 751–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yamashita N., Motoyoshi T., Nishimura A. (1999) Purification and characterization of isoamyl alcohol oxidase (“Mureka”-forming enzyme). Biosci. Biotechnol. Biochem. 63, 1216–1222 [DOI] [PubMed] [Google Scholar]
- 88. Swaisgood H. E., Janolino V. G., Skudder P. J. (1987) Continuous treatment of ultrahigh temperature sterilized milk using immobilized sulfhydryl oxidase. Methods Enzymol. 136, 423–431 [Google Scholar]
- 89. Kodama Y., Nakao Y., Shimonaga T. (2009) USPTO Patent Application 20090148555 [Google Scholar]
- 90. Rankine B. C. (1967) Formation of higher alcohols by wine yeasts, and relationship to taste thresholds. J. Sci. Food Agriculture 18, 583–589 [Google Scholar]
- 91. Didion T., Grauslund M., Kielland-Brandt M. C., Andersen H. A. (1996) Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J. Bacteriol. 178, 2025–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bartolome B., Faulds C. B., Tuohy M., Hazlewood G. P., Gilbert H. J., Williamson G. (1995) Influence of different xylanases on the activity of ferulic acid esterase on wheat bran. Biotechnol. Appl. Biochem. 22, 65–73 [Google Scholar]
- 93. Kurup V. P., Shen H. D., Vijay H. (2002) Immunobiology of fungal allergens. Int. Arch. Allergy Immunol. 129, 181–188 [DOI] [PubMed] [Google Scholar]
- 94. Su N. Y., Yu C. J., Shen H. D., Pan F. M., Chow L. P. (1999) Pen c 1, a novel enzymic allergen protein from Penicillium citrinum. Purification, characterization, cloning and expression. Eur. J. Biochem. 261, 115–123 [DOI] [PubMed] [Google Scholar]
- 95. Chow L. P., Su N. Y., Yu C. J., Chiang B. L., Shen H. D. (1999) Identification and expression of Pen c 2, a novel allergen from Penicillium citrinum. Biochem. J. 341, 51–59 [PMC free article] [PubMed] [Google Scholar]
- 96. Shen H. D., Au L. C., Lin W. L., Liaw S. F., Tsai J. J., Han S. H. (1997) Molecular cloning and expression of a Penicillium citrinum allergen with sequence homology and antigenic crossreactivity to a hsp 70 human heat shock protein. Clin. Exp. Allergy 27, 682–690 [PubMed] [Google Scholar]
- 97. Shen H. D., Wang C. W., Chou H., Lin W. L., Tam M. F., Huang M. H., Kuo M. L., Wang S. R., Han S. H. (2000) Complementary DNA cloning and immunologic characterization of a new Penicillium citrinum allergen (Pen c 3). J. Allergy Clin. Immunol. 105, 827–833 [DOI] [PubMed] [Google Scholar]
- 98. Chou H., Lai H. Y., Tam M. F., Chou M. Y., Wang S. R., Han S. H., Shen H. D. (2002) cDNA cloning, biological and immunological characterization of the alkaline serine protease major allergen from Penicillium chrysogenum. Int. Arch. Allergy Immunol. 127, 15–26 [DOI] [PubMed] [Google Scholar]
- 99. Tang R. B., Chen Y. S., Chou H., Lee S. S., Tai H. Y., Shen H. D. (2005) cDNA cloning and immunologic characterization of a novel EF-1beta allergen from Penicillium citrinum. Allergy 60, 366–371 [DOI] [PubMed] [Google Scholar]
- 100. Shen H. D., Tam M. F., Tang R. B., Chou H. (2007) Aspergillus and Penicillium allergens: focus on proteases. Curr. Allergy Asthma Rep. 7, 351–356 [DOI] [PubMed] [Google Scholar]
- 101. Lai H. Y., Tam M. F., Tang R. B., Chou H., Chang C. Y., Tsai J. J., Shen H. D. (2002) cDNA cloning and immunological characterization of a newly identified enolase allergen from Penicillium citrinum and Aspergillus fumigatus. Int. Arch. Allergy Immunol. 127, 181–190 [DOI] [PubMed] [Google Scholar]
- 102. Bouws H., Wattenberg A., Zorn H. (2008) Fungal secretomes-nature's toolbox for white biotechnology. Appl. Microbiol. Biotechnol. 80, 381–388 [DOI] [PubMed] [Google Scholar]
- 103. Desai P. N., Shrivastava N., Padh H. (2010) Production of heterologous proteins in plants: strategies for optimal expression. Biotechnol. Adv. 28, 427–435 [DOI] [PubMed] [Google Scholar]
- 104. Bally J., Nadai M., Vitel M., Rolland A., Dumain R., Dubald M. (2009) Plant physiological adaptations to the massive foreign protein synthesis occurring in recombinant chloroplasts. Plant Physiol. 150, 1474–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.