Abstract
Human African trypanosomiasis, or sleeping sickness, is a parasitic disease endemic in sub-Saharan Africa, transmitted to humans through the bite of a tsetse fly. The first or hemolymphatic stage of the disease is associated with presence of parasites in the bloodstream, lymphatic system, and body tissues. If patients are left untreated, parasites cross the blood-brain barrier and invade the cerebrospinal fluid and the brain parenchyma, giving rise to the second or meningoencephalitic stage. Stage determination is a crucial step in guiding the choice of treatment, as drugs used for S2 are potentially dangerous. Current staging methods, based on counting white blood cells and demonstrating trypanosomes in cerebrospinal fluid, lack specificity and/or sensitivity. In the present study, we used several proteomic strategies to discover new markers with potential for staging human African trypanosomiasis. Cerebrospinal fluid (CSF) samples were collected from patients infected with Trypanosoma brucei gambiense in the Democratic Republic of Congo. The stage was determined following the guidelines of the national control program. The proteome of the samples was analyzed by two-dimensional gel electrophoresis (n = 9), and by sixplex tandem mass tag (TMT) isobaric labeling (n = 6) quantitative mass spectrometry. Overall, 73 proteins were overexpressed in patients presenting the second stage of the disease. Two of these, osteopontin and β-2-microglobulin, were confirmed to be potential markers for staging human African trypanosomiasis (HAT) by Western blot and ELISA. The two proteins significantly discriminated between S1 and S2 patients with high sensitivity (68% and 78%, respectively) for 100% specificity, and a combination of both improved the sensitivity to 91%. The levels of osteopontin and β-2-microglobulin in CSF of S2 patients (μg/ml range), as well as the fold increased concentration in S2 compared with S1 (3.8 and 5.5 respectively) make the two markers good candidates for the development of a test for staging HAT patients.
Human African trypanosomiasis (HAT), or sleeping sickness, is caused by an extracellular protozoan parasite of the genus Trypanosoma, which is transmitted through the bite of a tsetse fly (genus Glossina). Two morphologically identical subspecies of the parasite, are responsible for the two geographically and clinically different forms of HAT: a chronic form, widespread in West and Central Africa, caused by T. b. gambiense, and an acute form, endemic in eastern Africa, caused by T. b. rhodesiense (1). In both forms of the disease, parasites are initially localized in the blood stream, lymph, and peripheral tissues; this is the first or hemolymphatic stage (S1). During this stage, patients present generic clinical features that are common to other infectious diseases such as human immunodeficiency virus (HIV), malaria, and tuberculosis (TB), which can coexist with HAT, thus making its early diagnosis difficult (2). If treatment is not carried out, the disease progresses to the second or meningoencephalitic stage (S2) after trypanosomes cross the blood-brain barrier (BBB) and invade the central nervous system (CNS). This phase is characterized by a broad range of neurological signs that are indicative of CNS involvement (1). Diagnosis of HAT is based on parasitological demonstration of parasites in blood or lymph-node aspirate (3). All positive or suspect patients have to undergo a lumbar puncture and cerebrospinal fluid (CSF)1 examination, to determine whether they have second stage disease (4). According to the World Health Organization (WHO) guidelines, the meningoencephalitic stage is defined by the presence of parasites in CSF and/or a white blood cell (WBC) count of more than 5 cells per μl (5). Other parameters, such as intrathecal IgM production could also provide additional information to determine whether the CNS is involved (6, 7).
Treatment of HAT patients varies depending on the infecting parasite and the stage of disease (5, 8). S2 drugs in current use, including melarsoprol, eflornithine, and a combination of nifurtimox and eflornithine have several limitations, such as a high rate of toxicity (melarsoprol causes death to 5% of treated patients) (9), complex logistics, and mode of administration (6, 10). Consequently, staging is a vital step in the diagnosis and treatment of HAT. However, the poor specificity or sensitivity of WBC counting and of parasitological techniques for demonstration of parasites in CSF, highlight the need for discovery of better tools for staging the disease.
Several attempts have been made during the last decade to identify potential biomarkers able to discriminate between the two stages of sleeping sickness. Most of the efforts focused on cytokines and chemokines, because the patient's immune system plays a crucial role in the brain pathology (11–14).
Proteomic approaches are increasingly being applied in biomedical research and clinical medicine to investigate body fluids as a source of biomarkers (15), including the diagnosis of neurological disorders such as Alzheimer's disease (16), Parkinson's disease (17), and multiple sclerosis (18, 19). The protein composition of CSF is strictly regulated and can reflect the physiological or pathological state of the CNS (15). Thus in the present study, we addressed the challenge of staging HAT by analyzing CSF from T. b. gambiense patients using two complementary proteomic strategies: a classical approach based on two-dimensional gel electrophoresis (2-DE), and quantitative mass spectrometry (MS) using isobaric tandem mass tag (TMT) technology (sixplex TMT® MS/MS) (20).
EXPERIMENTAL PROCEDURES
Samples
The CSF samples used in the present study were collected at Dipumba hospital in Mbuji-Mayi (East Kasai province, Democratic Republic of Congo) as part of a longitudinal study monitoring the outcome of treated HAT patients, whose results are described elsewhere (21). The patients were enrolled prospectively using the following inclusion criteria: presence of trypanosomes in lymph node aspirate, blood, or CSF, age ≥12 years, and living within a 100-km radius around Mbuji-Mayi. The exclusion criteria were pregnancy, follow-up not guaranteed, moribund condition, hemorrhagic CSF, and serious concurrent illness such as tuberculosis and bacterial or cryptococcal meningitis. No information on their HIV status was available at the moment of inclusion, but HIV prevalence was retrospectively found to be 3.1% (21). No systematic testing for malaria was done, but because the prevalence of the disease in the region is high, antimalarial drugs were administered to all patients prior to treatment for HAT. A lumbar puncture was performed on each patient and the CSF examined within 30 min to determine the stage of disease before treatment (22). This was done counting CSF WBC in disposable counting chambers (Uriglass, Menarini, Vienna, Austria) under a microscope. When the number of WBC was less than 20/μl, a second count was carried out. The modified single centrifugation method (23) was used to determine whether parasites were present in the CSF. The stage of disease was established in accordance with the guidelines of the national sleeping sickness control program, Programme National de Lutte contre la Trypanosomiase Humaine Africaine. Patients with a WBC count of ≤5/μl and no trypanosomes were classified as S1, and those with >5 WBC/μl and/or trypanosomes in the CSF as S2. The patients were further classified based on three categories of neurological signs as described by Hainard et al. (14): absent (no neurological signs), moderate (at least one major neurological sign but no generalized tremors), or severe (at least two major neurological signs, including generalized tremors).
The CSF used in the present study was taken from the supernatant following the modified single centrifugation, frozen in liquid nitrogen, shipped in dry ice, and stored at −80 °C until use. The samples were handled at room temperature (30–35 °C) for not more than 30 min between collection and freezing. They were then thawed and aliquoted, such that a different aliquot was used for each subsequent test.
The ethical committees of The Ministry of Health, Democratic Republic of Congo and of the University of Antwerp, Belgium, approved the study. Patients, or their relatives, were informed about the objectives and modalities of the study, and provided written consent prior to inclusion.
Analytical 2-DE Gels
Five S1 and 4 S2 CSF samples (without significant difference in age and sex) were analyzed by 2-DE. For each sample, 250 μl were precipitated with cold acetone prior to protein separation. Two-DE experiments were performed as described by Sanchez et al. (24), except for the second dimension separation, which was performed on 12.5% polyacrylamide gels.
Preparative 2-DE Gels
Duplicate preparative 2-DE gels were obtained by separating a pool of S2 CSF samples (n = 5, final volume 250 μl), in order to generate protein spots for identification by MS. The protocol was similar to that used for the analytical gels, apart from the staining procedure. In order to enable subsequent identification, preparative gels were stained following the protocol for MS-compatible silver staining (25), and the protein spots cut from the gels manually.
Image Analysis
Gel images were analyzed by ImageMaster™ 2D Platinum 6.0 software (GE Healthcare). Selection of differentially expressed protein spots was performed by carrying out interclass statistical analysis using the Kolmogorov-Smirnov test comparing the percentage volume of all matched spots. All protein spots whose percent volume was significantly different between the two groups (p value < 0.05) were considered. Additionally, S2/S1 ratios were calculated on the basis of the corresponding mean spot percent volume, and finally only spots with a ratio higher than two were selected for identification by MS.
In-gel Tryptic Protein Digestion
Excised protein spots were in-gel digested as described by Burgess et al. (26) for identification by matrix-assisted laser desorption ionization time-of-flight (MALDI TOF)-TOF MS (37 spots) and linear trap quadrupole-orbitrap (LTQ-OT) MS (52 spots). Following peptide extraction, samples were completely dried under speed vacuum.
MALDI TOF-TOF MS
Samples were desalted and then spotted in duplicate onto a 384-well MALDI plate. Matrix (α-cyano-4-hydroxycinnamic acid in H2O/acetonitrile 50:50, 10 mm NH4H2PO4) was then added and mass spectra were acquired with a MALDI TOF-TOF 4800 analyzer (Applied Biosystems, Foster City, CA) using the positive ionization mode and an m/z scan window of 800–4000 Th. The 20 most intense precursors were then subjected to MS/MS analysis. Argon was used as collision gas with the medium collision energy mode.
LTQ-OT MS
Electrospray ionization (ESI) LTQ-OT MS was performed on a LTQ Orbitrap XL from Thermo Electron (San Jose, CA) equipped with a NanoAcquity system from Waters (Milford, MA). Separation was run on a home-made analytical column using a gradient of H2O and CH3CN. Mass spectra were acquired in the positive mode with an m/z window of 400–2000 Th. A maximum of four precursors were selected for collision-induced dissociation with analysis in the LTQ (isolation width of 2 m/z). The normalized collision energy was set to 35%.
Protein Identification
Peak lists were generated using either the 4000 Series Explorer software from Applied Biosystem (MALDI TOF/TOF) or the embedded software (extract MSN.exe) from Thermo Electron (LTQ-OT). This was followed by searching for peak lists individually against UniProt-Swiss-Prot database (57.4 of June 16 2009, 565,634 protein entries) using Phenyx 2.6 (GeneBio, Geneva, Switzerland). Homo sapiens taxonomy (40,335 protein entries) was specified for database searching. Variable amino acid modification was oxidized methionine, whereas carbamidomethylation of cysteines was set as fixed modification. Trypsin was selected as the enzyme. The peptide p value was 1 E-6. Protein accession number and peptide scores were set up at 7.0 for both instruments. The minimum peptide length was six amino acids. The parent ion tolerance was 1.0 Da for MALDI TOF-TOF and 10 ppm for LTQ-OT. The scores were set to have a false discovery rate below 1%. For all subsequent analyses, only proteins identified with two different peptide sequences were kept.
Quantitative Mass Spectrometry with Sixplex TMT
Eighteen age and sex matched CSF samples, comprising nine S1 and nine S2, were analysed by quantitative MS. Samples were pooled in groups of three to obtain six differents pools (i.e. three S1 and three S2 pools).
Depletion by Immunoaffinity
Each pool was spiked with 1 μg of bovine β-lactoglobulin (Sigma, St Louis, MO) and then subjected to depletion of 14 abundant proteins using MARS Hu-14 column (Agilent Technologies, Wilmington, DE). Following collection of the flow-through fractions (containing unbound proteins), buffer was exchanged with H2O using AMICON ultra-15 centrifugal filter units (Millipore, Billerica, MA) and samples dried completely under speed vacuum.
Reduction, Alkylation, Digestion, and TMT Labeling
Reduction, alkylation, digestion, and TMT labeling were mainly performed as described by Dayon et al. (20). Briefly, reduction was carried out for 1 h at 60 °C following addition of tris-(2-carboxyethyl) phosphine hydrochloride, 50 mm. Alkylation was performed with iodo-acetamide, 400 mm (30 min in the dark) and overnight digestion was performed at 37 °C with freshly prepared trypsin (0.2 μg/μl). Each sample was then labeled with one of the 6 TMT reagents (Proteome Sciences, Frankfurt, Germany) according to manufacturer's instructions. The three pools corresponding to S1 patients were labeled with TMTs 126.1, 128.1, and 130.1 respectively. Pools corresponding to S2 patients were labeled with the three other TMTs (i.e. 127.1, 129.1 and 131.1). All the samples were finally pooled and evaporated under speed-vacuum.
Off-gel Electrophoresis
Off-gel electrophoresis was performed according to manufacturer's instructions (Agilent). Briefly, desalted and dehydrated samples were reconstituted in OFFGEL solution. Focusing was done on an IPG dry strip (13 cm, pH 3–10, linear; GE Healthcare) set up with a 12-well frame, for 20 kVh with a maximum current of 50 μA and power of 200 mW. The collected fractions were desalted, evaporated under speed-vacuum, and stored at −20 °C.
Liquid Chromatography MALDI TOF-TOF MS
Liquid chromatography-MS/MS was performed as described by Dayon et al. (20). Each sample was subjected to reverse-phase chromatography using an Alliance LC system (Waters) and deposited directly on a MALDI plate using a home-made spotter. Following matrix addition, mass spectra were acquired with a MALDI TOF-TOF 4800 analyzer as described previously.
Liquid Chromatography ESI LTQ-OT MS
ESI LTQ-OT MS was performed as described elsewhere (27). Mass spectra were acquired in the positive mode with an m/z window of 400–2000 Th. A maximum of three precursors were selected for high-energy C-trap dissociation with analysis in the OT. The normalized collision energy was set to 40% for high-energy C-trap dissociation.
Protein Identification
Generation of peak lists was done in the same way as for 2-DE identifications, using either the 4000 Series Explorer software from Applied Biosystems (MALDI TOF/TOF) or the embedded software (extract MSN.exe) from Thermo Electron (LTQ-OT). The peak lists generated from the 12 off-gel fractions were analyzed as described for 2-DE, with the following modifications. TMT-sixplex amino terminus and TMT-sixplex lysine (+229.1629 Da) were additionally set as fixed modifications. The AC and peptide scores were set up at 7.7 for the analysis with MALDI TOF-TOF and 12.5 for the analysis with LTQ-OT with a false peptide discovery rate evaluated at 0.95% and 0.99% respectively. The parent ion tolerance was set to 1.1 Da for MALDI TOF-TOF and to 6 ppm for LTQ-OT. Bos taurus taxonomy (8168 entries) was separately specified to search for the spiked β-lactoglobulin. For all analyses, only proteins identified with two different peptide sequences were selected. To search for parasite proteins, a database restricted to Homo sapiens and Kinetoplastida taxonomies (92,136 protein entries) was created using the FASTA files available from Uniprot (www.uniprot.org) and the same parameters described earlier were applied.
Protein Quantification
Quantification of proteins was mainly performed following the procedure described by Dayon et al. (20), who demonstrated the accuracy of relative quantitation with TMT method using both protein mixture model and CSF samples and estimated a technical variation between 7% and 20%. The following procedure was used in the present study. On the basis of the data obtained with Phenyx, TMT reporter peak intensities (or area under peak for MALDI TOF-TOF results) of each identified peptide were extracted (Supplemental MS information). An isotopic purity correction was performed for each reporter on the basis of the isotopic distribution of sixplex-TMT provided by the manufacturer. Isotopic purity equations were calculated using Maple software (Maple 11, Maple Inc., Waterloo, Canada). Peptides with missing reporter intensities were removed from the quantification. Spiked β-lactoglobulin was used to minimize experimental biases, and a normalization of the reporter intensities by the sum of all the reporter intensities was performed. A S2/S1 ratio was calculated for each peptide as the sum of S2 channels (127.1, 129.1, and 131.1) divided by the sum of S1 channels (126.1, 128.1, and 130.1). Peptide ratios were further subjected to outlier removal using the Outlier software (http://www.sediment.uni-goettingen.de/staff/dunkl/software/outlier.html). This web-free software computes four different outlier tests: the Grubbs test, the Dixon test, the IQR test, and the Gauss g-test. A peptide was considered an outlier and the protein removed from the quantification if one of these tests was positive. The protein ratio S2/S1 was calculated as the geometric mean of its corresponding peptide ratios. For each protein ratio, the geometric standard deviation (S.D.) was calculated as described by Tan et al. (28) and the coefficient of variation (CV) determined as the S.D. divided by the protein ratio, and multiplied by 100. To determine the cutoff threshold for considering a protein differentially expressed between the two stages, a method based on random ratios was applied. For each identified peptide, the normalized intensities of S1 reporters were duplicated in order to mimic a sixplex experiment (126.1, 128.1, 130.1, 126.1*, 128.1*, and 130.1*). All these intensities were then randomly mixed and ratios calculated as described earlier: sum (126.1*, 128.1*, 130.1*)/sum (126.1, 128.1, 130.1). The S.D. was evaluated among the random ratios obtained. In an ideal case, all the ratios calculated should be equal to one. Thus, the significant cutoff threshold to consider a protein up-regulated in S2 patients was determined as 1 + 2 S.D (28). The corresponding cutoff for down-regulated proteins was calculated as the reciprocal value. The results of these calculations are presented in Supplemental MS information. Quantified proteins presenting a CV > 50% were removed. Proteins quantified with both instruments were only excluded if in both cases the CV was higher than 50%.
Western Blot
The expression of complement factor H (CFH), osteopontin (OPN), and β-2-microglobulin (B2MG) was evaluated by Western blot in four S1 and four S2 CSF samples. Goat anti-complement factor H polyclonal antibody (Calbiochem - Merck, Darmstadt, Germany) was used at a concentration of 8.4 μg/ml. Mouse anti-osteopontin and mouse anti-β-2-microglobulin monoclonal antibodies (Abcam, Cambridge, UK) were used at a final concentration of 1 μg/ml and 5 μg/ml respectively. All horseradish peroxidase-conjugated secondary antibodies were purchased from Dako (Glostrup, Denmark) and applied at 1:1000 (anti-mouse secondary antibody) and 1:2000 (anti-goat secondary antibody) dilutions. The images obtained were analyzed with ImageQuant™ TL 7.0 (GE Healthcare) and band volume data analyzed with GraphPad Prism software (version 4.03, GraphPad software Inc., San Diego, CA) to determine significant differences.
ELISA
The concentrations of CFH, OPN, and B2MG were measured in CSF of HAT patients using commercially available sandwich ELISA kits (B2MG, Calbiotech, Spring Valley, CA; OPN, R&D Systems, Minneapolis, MN; CFH, Hycult Biotech, The Netherlands), following manufacturer's instructions. Detailed description of patients whose CSF was analyzed are reported in Table I. CSF samples were diluted 1:50 for CFH and 1:101 for OPN and B2MG. Following color development, absorbance was read on a Vmax Kinetic microplate reader (Molecular Devices Corporation, Sunnyvale, CA) at a wavelength of 450 nm. The concentration of the three proteins in the CSF samples was back-calculated using either four-parameter logistic or five-parameter logistic curves (SoftMax Pro software, Molecular Devices, CA) based on the measured respective standard values.
Table I. Characteristics of the population assessed with ELISA for verification experiments.
| Stage 1 (S1) | Stage 2 (S2) | ||
|---|---|---|---|
| Population | n | 21 | 37 |
| Gender | Male | 8 | 24 |
| Female | 13 | 13 | |
| Age | Median (range) | 32 (14–60) | 35 (16–65) |
| WBC/μl | Median (range) | 2 (0–5) | 91 (6–2064) |
| Patients with parasites in CSF | n | 0 | 25 |
| Neurological signs* | Absence | 11 | 10 |
| Moderate | 10 | 21 | |
| Severe | 0 | 5 |
* Neurological signs were not reported for one patient.
Data and statistical analysis
Descriptive statistics were performed using GraphPad Prism 4.03 software. As none of the proteins presented a normal distribution (Kolmogorov-Smirnov test), differences between groups were tested with the nonparametric Mann-Whitney U test (comparison between two groups) and Kruskal-Wallis test followed by Dunn's post-hoc test (comparison between three groups). Statistical significance for the tests was set at 0.05 (two-tailed test). The concentrations of the different molecules were considered as independent variables. Bivariate nonparametric correlations using the Spearman rho coefficient were carried out with statistical significance set at 0.01 (two-tailed test). To calculate sensitivity and specificity of individual predictors with respect to staging, the specific receiver operating characteristic curve of each analyte was determined. The cutoff value was selected as the threshold predicting stage two patients with 100% specificity. Aabel software (version 2.4.2, Gigawiz Ltd. Co., Tulsa, OK) was used for box plots.
Protein combination and panel selection
To evaluate the possibility of improving the potential of the three molecules in staging HAT patients, they were combined in a panel as described by Hainard et al. (14). Briefly, the optimized cut-off values were obtained by modified iterative permutation-response calculations (rule-induction-like) using the 3 analytes. Each cutoff value was changed iteratively by quantiles of 2% increment and sensitivity determined following each iteration, until a maximum sensitivity for 100% specificity was achieved.
RESULTS
Two-Dimensional Gel Electrophoresis
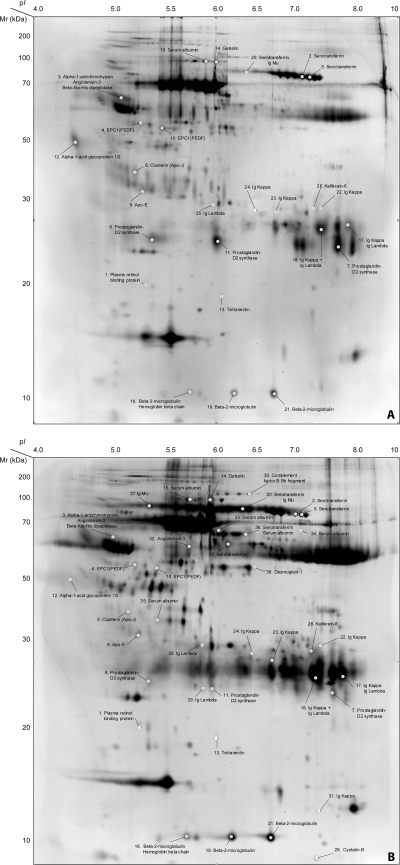
The protein expression patterns of the nine CSF samples separated by 2-DE, just before image analysis, showed macroscopic differences between the two stages of disease, principally in expression of immunoglobulin. IgM and IgG heavy and light chains were particularly increased in samples from second stage compared with first stage patients, as previously shown by nephelometry (7). The data on percent spot volume provided by the software were used to evaluate protein spot expression. This resulted in 754 spots matched between S1 and S2 master gels, of which 59 had a p value < 0.05. The 59 comprised 25 spots expressed in S2 gels only, 13 overexpressed in S2 gels compared with S1 with an average percent volume ratio S2/S1 larger than 2, and 21 overexpressed in S1 compared with S2 (average percent volume ratio S1/S2 > 2). Among the 59 spots, 52 were visualized and excised from the preparative S2 gels and subsequently analyzed by LTQ-OT MS and/or with MALDI TOF-TOF MS. Out of the 52 spots, 38 (73.1%) were successfully identified as corresponding to 25 different proteins, as shown in Table IIa–c and in Fig. 1.
Table IIA. Protein spots overexpressed in stage 1 CSF samples.
| Spot # | MW | pI | DB entry | Description | S1/S2Ratio |
|---|---|---|---|---|---|
| 1 | 23′010.01 | 6.15 | P02753 | Plasma retinol-binding protein (1–176) | 3 |
| 69′366.70 | 5.98 | P02768 | Serum Albumin | ||
| 2 | 75′181.46 | 6.84 | P02787 | Serotransferrin (Transferrin) | 2.29 |
| 49′306.61 | 6.55 | P01871 | Ig mu chain C region | ||
| 3 | 45′265.84 | 5.40 | P01011 | Alpha-1-antichymotrypsin His-Pro-less | 8.48 |
| 53′154.22 | 5.96 | P01019 | Angiotensin-3 (Ang III) | ||
| 53′863.55 | 5.13 | Q96KN2 | Beta-Ala-His dipeptidase | ||
| 4 | 46′342.31 | 6.12 | P36955 | Pigment epithelium-derived factor (PEDF) | 2.43 |
| 5 | 77′049.89 | 6.97 | P02787 | Serotransferrin (Transferrin) | 2.25 |
| 6 | 18′698.03 | 8.68 | P41222 | Prostaglandin-H2 d-isomerase | 3.19 |
| 7 | 18′698.03 | 8.68 | P41222 | Prostaglandin-H2 d-isomerase | 2.89 |
| 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | ||
| 11.236.69 | 5.56 | P01842 | Ig lambda chain C region | ||
| 8 | 50′062.57 | 5.97 | P10909 | Clusterin alpha chain (Apo J) | 3.38 |
| 9 | 34′236.69 | 5.56 | P02649 | Apolipoprotein E (Apo E) | 3.03 |
| 10 | 46′342.31 | 31 | P36955 | Pigment epithelium-derived factor (PEDF) | 3.58 |
| 11 | 18′698.03 | 8.68 | P41222 | Prostaglandin-H2 d-isomerase | 4.07 |
| 11′236.52 | 7.90 | P01842 | Ig lambda chain C region | ||
| 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | ||
| 12 | 23′511.57 | 5.02 | P02763 | Alpha-1-acid glycoprotein 1 (AGP 1) (OMD 1) | 51.99 |
| 23′602.63 | 5.13 | P19652 | Alpha-1-acid glycoprotein 2 (AGP 2) (OMD 2) | ||
| 13 | 20′168.93 | 6.22 | P05452 | Tetranectin (TN) | 5.99 |
| 14 | 69′366.70 | 5.98 | P02768 | Serum Albumin | 11.16 |
| 80′640.64 | 5.64 | P06396 | Gelsolin (ADF) [ISOFORM 2] | ||
| 49′306.61 | 6.55 | P01871 | Ig mu chain C region | ||
| 15 | 69′366.70 | 5.98 | P02768 | Serum Albumin | 3.24 |
Table IIB. Protein spots overexpressed in stage 2 CSF samples.
| Spot # | MW | pI | DB entry | Description | S2/S1Ratio |
|---|---|---|---|---|---|
| 16 | 15′998.41 | 7.13 | P68871 | LVV-hemorphin-7 | 2.64 |
| 11′731.17 | 6.46 | P61769 | Beta-2-microglobulin form pI 5.3 | ||
| 69′366.70 | 5.98 | P02768 | Serum Albumin | ||
| 17 | 11′608.65 | 6.04 | P01834 | Ig kappa chain C region | 2.00 |
| 11′236.52 | 7.90 | P01842 | Ig lambda chain C region | ||
| 18 | 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | 4.68 |
| 11′236.52 | 7.90 | P01842 | Ig lambda chain C region | ||
| 69′366.70 | 5.98 | P02768 | Serum Albumin | ||
| 19 | 11′731.17 | 6.46 | P61769 | Beta-2-microglobulin form pI 5.3 | 2.02 |
| 20 | 75′181.46 | 6.84 | P02787 | Serotransferrin (Trnasferrin) | 5.25 |
| 51′790.42 | P01871 | Ig mu chain C region [ISOFORM 2] | |||
| 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | ||
| 21 | 13′714.57 | 6.41 | P61769 | Beta-2-microglobulin form pI 5.3 | 1.77 |
| 22 | 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | 6.17 |
| 23 | 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | 5.02 |
| 24 | 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | 12.06 |
| 69′366.70 | 5.98 | P02768 | Serum Albumin | ||
| 25 | 11′236.52 | 7.90 | P01842 | Ig lambda chain C regions | 9.43 |
| 26 | 26′855.73 | 7.57 | Q92876 | Kallikrein-6 | 9.07 |
| 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | ||
| 31′105.92 | 8.60 | P01857 | Ig gamma-1 chain C region |
Table IIC. Protein spots expressed only in stage 2 CSF samples.
| Spot # | MW | pI | DB entry | Description |
|---|---|---|---|---|
| 27 | 49′306.61 | 6.55 | P01871 | Ig mu chain C region [ISOFORM 2] |
| 28 | 11′139.59 | 7.91 | P04080 | Cystatin-B |
| 69′366.70 | 5.98 | P02768 | Serum Albumin | |
| 29 | 11′236.52 | 7.90 | P01842 | Ig lambda chain C region |
| 30 | 85′532.90 | 6.81 | P00751 | Complement factor B Bb fragment |
| 49′306.61 | 6.55 | P01871 | Ig mu chain C region | |
| 31 | 11′608.85 | 6.04 | P01834 | Ig kappa chain C region |
| 32 | 49′761.12 | 5.68 | P01019 | Angiotensin-3 (Ang III) |
| 33 | 69′366.70 | 5.98 | P02768 | Serum Albumin |
| 34 | 69′366.70 | 5.98 | P02768 | Serum Albumin |
| 36′105.92 | 8.60 | P01857 | Ig gamma-1 chain C region | |
| 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | |
| 35 | 69′366.70 | 5.98 | P02768 | Serum Albumin |
| 36 | 75′181.46 | 6.84 | P02787 | Serotransferrin (Transferrin) |
| 69′366.70 | 5.98 | P02768 | Serum Albumin | |
| 37′654.65 | 6.26 | P01876 | Ig alpha-1 chain C region | |
| 11′608.85 | 6.04 | P01834 | Ig kappa chain C region | |
| 37 | 75′181.46 | 6.84 | P02787 | Serotansferrin (Transferrin) |
| 69′366.70 | 5.98 | P02768 | Serum Albumin | |
| 38 | 113′715.57 | 4.92 | Q02413 | Desmoglein-1 (DG1) (DGI) |
Fig. 1.
Representative 2-DE image of cerebrospinal fluid from stage 1 (A) and stage 2 (B) HAT patients. Master gels obtained separating 250 μl of CSF of a S1 (A) and a S2 patient (B). For each gel, proteins were separated on 18 cm pH 3–10 NL IPG-strips (GE Healthcare). The second dimension was performed on 12.5% polyacrylamide gels and proteins were finally visualized with silver staining. The 38 identified protein spots are reported on the gels. Spots 1–15: spots overexpressed in stage 1 patients (S1/S2 > 2.0, p value < 0.05); spots 16–26: spots overexpressed in stage 2 patients (S2/S1 > 2.0, p value < 0.05); spots 27–38: spots expressed only in stage 2 patients. The approximate pI and MW (kDa) have been assigned according to the CSF SWISS-2DPAGE map available on the ExPASy website (http://www.expasy.org/ch2d/).
Quantitative Mass Spectrometry with Sixplex TMT
The six TMT-labeled pools were subjected to protein identification and relative quantitation using both MALDI TOF-TOF MS and LTQ-OT MS. With MALDI TOF-TOF MS, 128 proteins were identified from a total of 916 tryptic peptides. In the same way, LTQ-OT MS allowed the identification of 141 proteins from a total of 3334 tryptic peptides (Supplemental MS information). In all, 172 proteins were identified, each with at least two unique peptides. Among these proteins, 97 were identified with both instruments, 44 with LTQ-OT only, and 31 with MALDI TOF-TOF only. Interestingly, no parasite protein was identified following the simultaneous search against Homo sapiens and Kinetoplastida databases.
Following removal of the unquantifiable and outlier peptides, the ratio and corresponding S.D. were calculated for each protein (Supplemental MS information). The significant cutoff thresholds were then evaluated for each set of data as described previously. With MALDI-TOF-TOF MS, the S.D. for random S1 ratio was 0.323, thus 1.65 (1 + 2 S.D.) was selected as the up-regulation significant threshold ratio (S2/S1) and reciprocally, 0.61 was identified as the down-regulation significant threshold. With ESI LTQ-OT MS, calculated S.D. was 0.370 and up-regulation and down-regulation thresholds were 1.74 and 0.57 respectively (Supplemental MS information). According to these thresholds, one protein (i.e. C-reactive protein) was down-regulated whereas 59 were significantly up-regulated in second stage patients. Out of the 59 proteins, 29 were quantified with both MS instruments, 8 with the LTQ-OT only, and 22 with the MALDI TOF-TOF only (Supplemental TableS1 A–C).
The two proteomic approaches used in the present study identified 85 proteins differentially expressed between S1 and S2 HAT. Among these, 73 were overexpressed in S2 CSF samples. The two discovery techniques were highly complementary as, among all proteins overexpressed in S2 patients, only immunoglobulin chains, β-2-microglobulin and complement factor B were identified with the two approaches. Three overexpressed proteins, including complement factor H, osteopontin and β-2-microglobulin were chosen for verification by immunoassay methods on a larger number of patients.
Verification by Western Blot
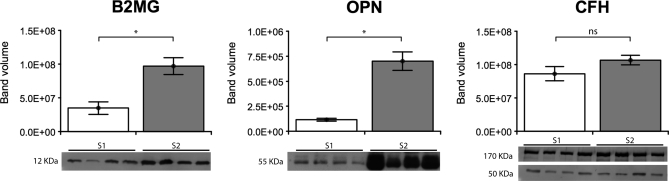
CFH was identified as overexpressed using the TMT approach, with S2/S1 ratios of 1.93 and 1.97 for LTQ-OT and MALDI TOF-TOF instruments, respectively. The detection of CFH with a polyclonal anti-CFH antibody on Western blot resulted in the visualization of two intense bands (50 and 170 kDa) among other weaker ones (data not shown). According to the molecular weight, the higher band most likely corresponded to the complete form of CFH, whereas the lower one might correspond to the factor H-like protein 1 (FHL-1), obtained from alternative splicing (29). However, when considered together or separately, the volumes of the two bands were not significantly different between the S1 and S2 groups (Table III and Fig. 2).
Table III. Staging performance of the three verified proteins according to the different techniques.
| Protein name | Quantitative MS (n = 18) |
Western blot (n = 8) |
ELISA (n = 52 - 58) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Ratioa | S.D.a | Ratio | p valueb | Ratio | pa valueb | % AUC | % Sensitivity | % Specificity | |
| Beta-2-microglobulin (B2MG) | 6.33/4.90 | 1.78/1.98 | 2.37 | <0.05 | 5.51 | <0.0001 | 92 | 78 | 100 |
| Osteopontin (OPN) | 3.64 | 0.28 | 6.00 | <0.05 | 3.83 | <0.0001 | 85 | 68 | 100 |
| Complement factor H (CFH) | 1.93/1.97 | 0.22/0.30 | 1.29 | ns | 1.51 | <0.05 | 73 | 31 | 100 |
a When available, data from both MS experiments are given in the following way: LTQ-OT MS/MALDI TOF-TOF MS.
b Mann-Whitney U test.
Fig. 2.
Western blot results for B2MG, OPN, and CFH. Expression of B2MG, OPN, and CFH in cerebrospinal fluid of early and late stage HAT patients (n = 8). The same volume for each sample (10 μl for B2MG, 20 μl for OPN and CFH) was analyzed on a 12.5% (B2MG) or 10% (OPN and CFH) polyacrylamide gel. Bars represent the mean quantified band volume with the respective standard error. * corresponds to significant p value < 0.05; ns corresponds to nonsignificant p value (Mann-Whitney U test). Images of the corresponding analyzed bands are presented below each graph.
Western blot results confirmed the overexpression of OPN in the CSF of S2 patients, which was previously detected with the TMT approach (MALDI TOF/TOF MS ratio S2/S1 = 3.64). The acidity and characteristic behavior of the protein during gel separation (30) is likely to hinder its visualization on 2-DE gels. Although the reported molecular weight is ∼35 kDa, the observed 55 kDa band should correspond to the principal form of OPN as the protein undergoes extensive posttranslational modifications, which make its molecular weight higher than the theoretical one (30). In Western blot the quantified volume of this 55 kDa band was significantly increased in S2 compared with S1 patients (p value < 0.05, Mann-Whitney U test; median band volume S2/S1 ratio = 6.00) (Table III and Fig. 2).
The third protein, B2MG, was identified with both discovery techniques. Three spots were identified as B2MG on 2-DE gels and the geometric mean of the percentage volume ratio S2/S1 of the three spots was of 2.11. With quantitative MS B2MG was identified with S2/S1 ratios of 6.33 (LTQ-OT MS) and 4.90 (MALDI TOF-TOF MS) (Table III). These results were confirmed by Western blot, where a single 12-kDa band was visualized in all HAT samples, with a statistically significant increase in band volume in S2 samples (p value < 0.05, Mann-Whitney U test) (Fig. 2) and a calculated ratio on the median band volume S2/S1 of 2.37 (Table III).
Verification by ELISA
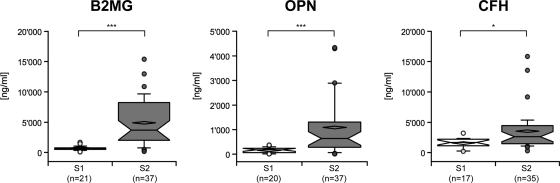
All CSF samples analyzed by ELISA were classified in two groups on the basis of their stage (21 S1 and 37 S2). The concentrations of B2MG and OPN were significantly increased in the CSF of S2 patients (p < 0.0001, Mann-Whitney U test). CFH showed a significant, but less marked, increased concentration in the same group of patients (p < 0.05, Mann-Whitney U test) (Fig. 3). In order to assess the sensitivity and specificity of the three molecules, Receiver operating characteristic curves were built (Supplemental Fig. S1). The staging efficiency of the three molecules was evaluated using the area under the curve (AUC) and the sensitivity for 100% specificity. Indeed, this configuration (maximum of specificity) avoided false negatives, such that a patient positive with the test was truly in the second stage. CFH had the lowest AUC (0.723) and a sensitivity of 31% (95% CI; 17%–49%) for 100% specificity. OPN and B2MG showed higher performances with AUC of 0.848 and 0.915, and sensitivity of 68% (95% CI; 50%–82%) and 78% (95% CI; 62%–90%) respectively. The S2/S1 ratio was calculated for each molecule, on the basis of the median value of each group of patients. This ratio corresponded to 1.51 for CFH, which was the lowest, and 3.83 and 5.51 for OPN and B2MG respectively (Table III). To evaluate the staging potential of the three molecules further, we assessed the correlation between their concentration in CSF and the number of WBC, the reference staging method. B2MG and OPN showed a high correlation with the number of WBC, with Spearman rho coefficients at 0.725 and 0.723 respectively. There was also a significant correlation between CFH and the number of WBC but with a lower Spearman rho coefficient (0.562) compared with the others. Detailed results for the three molecules are reported in Supplemental Table S2.
Fig. 3.
Box-plot of B2MG, OPN, and CFH concentrations according to HAT staging. ELISA results representing the measured concentrations (ng/ml) in S1 and S2 CSF samples of B2MG (n = 58), OPN (n = 57), and CFH (n = 52). Median and mean are represented as a solid line in the box and a diamond, respectively. Whisks are defined as 5th–95th percentile with outliers. Half-width of the notch was calculated automatically by the software. *** and * correspond to a significant difference between the two groups, respectively < 0.0001 and < 0.05 (Mann-Whitney U test). S1, stage 1 samples; S2, stage 2 samples.
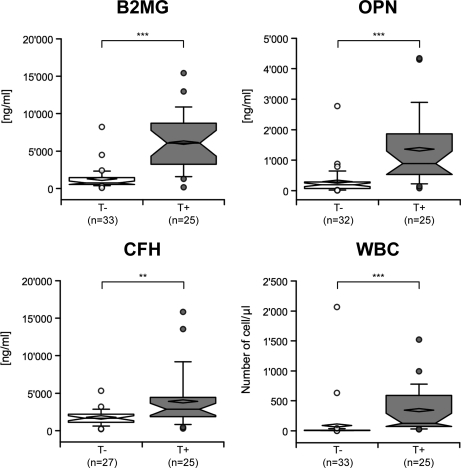
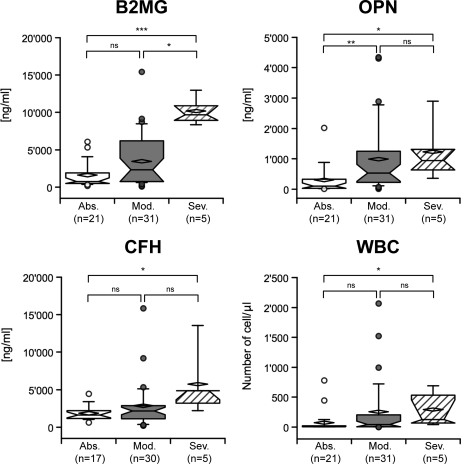
The concentration of the three molecules was then evaluated in relation to the presence or absence of parasites in CSF, and the neurological signs reported before treatment. The three proteins were significantly increased in the CSF of patients with parasites in CSF (Fig. 4), with p values < 0.0001 for B2MG and OPN, and < 0.01 for CFH. When the severity of neurological signs was considered, CFH could only discriminate between the absence and the presence of severe neurological signs with a p value < 0.05 (Kruskal-Wallis test) (Fig. 5). OPN showed a lower p value (p < 0.001, Kruskal-Wallis) and significantly discriminated between all the groups, except between the moderate and severe ones. B2MG had the lowest p value (p < 0.0001, Kruskal-Wallis) and significantly discriminated between absent and severe neurological signs, as well as between the moderate and severe ones (Fig. 5).
Fig. 4.
Box-plot of B2MG, OPN, CFH concentrations, and WBC number, classified according to the presence of the parasite in CSF. Median and mean are represented as a solid line in the box and a diamond, respectively. Whisks are defined as 5th–95th percentile with outliers. Half-width of the notch was calculated automatically by the software. *** and ** correspond to a significant difference between the two groups, respectively < 0.0001 and < 0.01 (Mann-Whitney U test). T-, patients without parasite in CSF; T+, patients having parasites in CSF.
Fig. 5.
Box-plot of B2MG, OPN, CFH concentrations, and WBC number, classified according to the severity of neurological signs. Median and mean are represented as a solid line in the box and a diamond, respectively. Whisks are defined as 5th–95th percentile with outliers. Half-width of the notch was calculated automatically by the software. *** corresponds to a significant difference between the two groups < 0.0001; ** corresponds to a significant difference < 0.01 and * < 0.05; ns indicates a nonsignificant difference (Dunn's post-hoc test). Abs, absence of neurological signs; Mod, moderate neurological signs; Sev, severe neurological signs.
When the three molecules were combined using the rule-induction-like process, a panel comprising B2MG (cutoff: 1802.5 ng/ml) and OPN (cutoff: 408.8 ng/ml) that discriminated between first and second stage disease with a sensitivity of 91% for 100% specificity was identified (Supplemental Fig. S1). This panel gave a positive test response (i.e. S2 patient identification) any time the concentration of one molecule was above its cutoff value.
DISCUSSION
In this study, we analyzed CSF samples from T. b. gambiense infected patients with a combination of proteomic strategies to identify new biomarkers that could complement or replace current methods of staging HAT. A total of 73 host proteins whose expression was increased in patients presenting the second stage of disease were identified. No parasite proteins were identified with the applied approaches, probably because CSF samples were first centrifuged for the parasitological examination (23) leading to removal of all parasites and cells.
The application of two different proteomic strategies was particularly useful in obtaining complementary information, such that only few proteins were commonly found. This can be explained if the different workflows as well as the intrinsic limitations associated to both techniques are taken into account. All samples analyzed by TMT mass spectrometry were first depleted of the 14 most abundant proteins whereas whole CSF samples were separated by 2-DE. Further, 2-DE is based on protein separation with potential identification of specific protein isoforms but also loss of hydrophobic proteins, whereas TMT quantitative mass spectrometry is based on peptide separation. Many steps in the workflow of the latter approach could lead to peptide loss, such as the use of TCEP for protein reduction (31) (32), peptide-gel interaction during the off-gel electrophoresis separation, and the peptide tagging. In addition, proteins identified with one unique peptide were excluded from subsequent analysis. The use of more optimized protocols, especially for the TMT MS approach, could therefore lead to the discovery of less abundant proteins or specific trypanosome antigens. These antigens are known to be present in host's CSF (33), but probably they are not concentrated enough to be detected with the techniques applied in the present study.
A preliminary analysis of the functions of the proteins identified indicates that they are involved in the immune response, both cell-mediated and humoral, as well as in cell-cell adhesion and transport. The three proteins chosen for further verification, CFH, OPN, and B2MG, were differentially expressed between the two stages (i.e. S2/S1 ratios higher than 2 for 2-DE, and higher than 1.65 or 1.74 for TMT MS), and to our knowledge, they have never been described in HAT patients. Furthermore, on the basis of their known functions, these proteins could potentially be involved in disease progression.
It is well established that many pathogens can find mechanisms to escape the host immune response and one of the main targets of these evasion mechanisms is the complement cascade (34). Trypanosomes are able to activate the alternative complement pathway in blood (35), whereas decreased complement activation was reported during the 1980s in infective cultures of T. cruzi, responsible for Chagas' disease (36). Complement factor H is the principal inhibitor of the alternative pathway, and is also involved in the protection of epithelial, endothelial, and some cancer cells against complement action (37). In our population of T. b. gambiense patients, overexpression of CFH during the second stage of disease was only confirmed with ELISA. Despite the high similarity between the results obtained by TMT and ELISA, a well established quantitative method widely applied in clinical research (38), CFH did not come out as a promising marker for staging HAT, because the AUC was only 0.73, which was below the 0.8 arbitrary limit that we established for considering a test as having staging potential. Furthermore, the ratio calculated from the TMT results was close to the cutoff for considering a protein overexpressed in S2 patients.
The data for OPN and B2MG were very promising, with both discriminating S1 and S2 patients with high accuracy, as indicated by the elevated AUC values. OPN, also known as early T-lymphocyte activation 1 (Eta-1), is expressed by a variety of immune and nonimmune cells, including brain cells, macrophages, and activated Th1 cells (30) (39), and is believed to act as a pro-inflammatory cytokine (40). The protein is a ligand for two classes of adhesion molecules: CD44, expressed on activated and memory T cells, and different types of integrins, including αVβ3 and α4β1, expressed by T lymphocytes. It also binds VCAM-1, which, in turn, is expressed on cytokine-activated endothelial cells (41). At the same time, OPN induces the production of interferon-γ and IL-12 by macrophages, and inhibits the production of IL-10 (42), participating in the polarization of the cellular immune response toward the Th1 type involved in phagocytosis and killing of microbes. Expression of OPN is highly increased during chronic inflammatory diseases or tissue injury, especially in proximity of activated T cells and monocytes/macrophages (43). The protein has been extensively studied in multiple sclerosis, where it appears to be involved, through the α4β1 integrin, in the entry of effector T cells in the brain (41), and through CD44 receptor, in the permanence of T cells at the site of inflammation. Recent information has suggested that the levels of OPN in CSF are not disease specific, but can point to involvement of the CNS or damage to the blood-brain barrier (BBB) (44, 45). The levels of OPN in the CSF of S2 HAT patients analyzed in the present study were significantly higher than in S1 patients, suggesting an association between CSF OPN and disease progression.
B2MG, found differentially expressed with both discovery approaches, was revealed to have the highest staging potential. This 11.8-kDa protein, expressed on the surface of all nucleated cells, is noncovalently associated to the MHC class I molecules and therefore involved in cellular immune response against invading pathogens mediated by cytotoxic T lymphocytes CD8+ (46). Several studies have reported that the levels of free B2MG in body fluids are increased in many malignant conditions (47) and they can be an indicator of a high cellular turnover (48). It has been suggested that the levels of B2MG could correlate with both the degree of CNS involvement and neuronal damage in children with symptomatic congenital CMV infection (48). Furthermore, proteomic and nonproteomic approaches have associated B2MG with many neurological disorders, including Alzheimer's disease (16, 19) and cancers (47). In the present study, CSF B2MG was significantly elevated in second stage patients, enabling the molecule to distinguish S1 and S2 patients with the best sensitivity and specificity.
The potential of B2MG and OPN as new markers for staging HAT patients was further supported by a highly significant correlation between their levels and the number of WBC in CSF. Furthermore, when compared with the number of WBC, CSF B2MG and OPN were better indices of both the presence of parasites in patients' CSF and the severity of neurological signs. Finally, when the concentrations of both proteins were considered together as a panel, they identified S2 patients with a sensitivity of 91%.
The CSF concentration of OPN and B2MG in both S1 and S2 HAT patients was relatively high (μg/ml range). This finding could be particularly relevant in the development of an antibody-based field test such as a lateral-flow assay, resulting in major improvement in accuracy of staging and reduction in costs, although still limited by the necessity of a lumbar puncture. The lumbar puncture could be eliminated during staging of HAT patients only if markers could be found in patients' blood. However, preliminary tests for B2MG on 30 plasma samples (15 S1 and 15 S2) did not reveal any significant differences between S1 and S2 (data not shown), probably as a consequence of the presence of parasites in the blood of both groups of patients.
In order to further validate the potential of CSF OPN and B2MG in staging HAT, a larger multicentric cohort, including T. b. gambiense and T. b. rhodesiense patients, as well as control CSF from patients with other infectious diseases (e.g. TB, and HIV), will be carried out.
In conclusion, the present study has revealed β-2-microglobulin and osteopontin as good markers that could potentially replace WBC count in staging HAT patients. Treatment of HAT patients is hampared by lack of safe drugs effective for both stages of the disease. Erroneous determination of the stage could, in fact, have serious consequences on the safety and health of patients, with S1 patients being unnecessarily exposed to the toxicity of stage 2 drugs, and S2 patients not getting cured with stage 1 drugs, and thus exposed to the risk of relapses and death. The present discovery of biomarkers that increase the accuracy of staging HAT represents an important improvement for guiding treatment decision.
Supplementary Material
Acknowledgments
The authors thank Noémie Roze-Fumeaux, Nadia Walter and Catherine Fouda for technical and scientific assistance, and the Foundation for Innovative New Diagnostics (FIND) for scientific and financial support. We also thank Proteome Science plc. for providing TMT reagents.
Footnotes
* The THARSAT study and D. M. N. received financial support from the Belgian Directorate General for International Cooperation.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CSF
- cerebrospinal fluid
- HAT
- human African trypanosomiasis
- S1
- stage 1
- BBB
- blood-brain barrier
- S2
- stage 2
- WBC
- white blood cell
- TMT
- tandem mass tag
- HIV
- human immunodeficiency virus
- TB
- tuberculosis
- CNS
- central nervous system
- MS/MS
- tandem mass spectrometry
- LTQ-OT
- linear trap quadrupole-orbitrap
- LTQ
- linear trap quadrupole
- SD
- standard deviation
- CV
- coefficient of variation
- CFH
- complement factor H
- OPN
- osteopontin
- B2MG
- β-2-microglobulin
- AUC
- area under the curve
- CI
- confidence interval
- 2-DE
- two-dimensional gel electrophoresis.
REFERENCES
- 1. Kennedy P. G. (2004) Human African trypanosomiasis of the CNS: current issues and challenges. J. Clin. Invest. 113, 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chappuis F., Loutan L., Simarro P., Lejon V., Büscher P. (2005) Options for field diagnosis of human african trypanosomiasis. Clin. Microbiol. Rev. 18, 133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kennedy P. G. (2008) The continuing problem of human African trypanosomiasis (sleeping sickness). Ann. Neurol. 64, 116–126 [DOI] [PubMed] [Google Scholar]
- 4. Kennedy P. G. (2008) Diagnosing central nervous system trypanosomiasis: two stage or not to stage? Trans. R. Soc. Trop. Med. Hyg. 102, 306–307 [DOI] [PubMed] [Google Scholar]
- 5. WHO (1998) Control and surveillance of African trypanosomiasis. Report of a WHO Expert Committee. World Health Organ Tech. Rep. Ser. 881, I–VI, 1–114 [PubMed] [Google Scholar]
- 6. Bisser S., Lejon V., Preux P. M., Bouteille B., Stanghellini A., Jauberteau M. O., Büscher P., Dumas M. (2002) Blood-cerebrospinal fluid barrier and intrathecal immunoglobulins compared to field diagnosis of central nervous system involvement in sleeping sickness. J. Neurol. Sci. 193, 127–135 [DOI] [PubMed] [Google Scholar]
- 7. Lejon V., Reiber H., Legros D., Djé N., Magnus E., Wouters I., Sindic C. J., Büscher P. (2003) Intrathecal immune response pattern for improved diagnosis of central nervous system involvement in trypanosomiasis. J. Infect. Dis. 187, 1475–1483 [DOI] [PubMed] [Google Scholar]
- 8. Fairlamb A. H. (2003) Chemotherapy of human African trypanosomiasis: current and future prospects. Trends Parasitol. 19, 488–494 [DOI] [PubMed] [Google Scholar]
- 9. Blum J., Nkunku S., Burri C. (2001) Clinical description of encephalopathic syndromes and risk factors for their occurrence and outcome during melarsoprol treatment of human African trypanosomiasis. Trop. Med. Int. Health. 6, 390–400 [DOI] [PubMed] [Google Scholar]
- 10. Priotto G., Kasparian S., Mutombo W., Ngouama D., Ghorashian S., Arnold U., Ghabri S., Baudin E., Buard V., Kazadi-Kyanza S., Ilunga M., Mutangala W., Pohlig G., Schmid C., Karunakara U., Torreele E., Kande V. (2009) Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet. 374, 56–64 [DOI] [PubMed] [Google Scholar]
- 11. Lejon V., Lardon J., Kenis G., Pinoges L., Legros D., Bisser S., N'Siesi X., Bosmans E., Büscher P. (2002) Interleukin (IL)-6, IL-8 and IL-10 in serum and CSF of Trypanosoma brucei gambiense sleeping sickness patients before and after treatment. Trans. R. Soc. Trop. Med. Hyg. 96, 329–333 [DOI] [PubMed] [Google Scholar]
- 12. Maclean L., Odiit M., Macleod A., Morrison L., Sweeney L., Cooper A., Kennedy P. G., Sternberg J. M. (2007) Spatially and genetically distinct African Trypanosome virulence variants defined by host interferon-gamma response. J. Infect. Dis. 196, 1620–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Courtioux B., Boda C., Vatunga G., Pervieux L., Josenando T., M'Eyi P. M., Bouteille B., Jauberteau-Marchan M. O., Bisser S. (2006) A link between chemokine levels and disease severity in human African trypanosomiasis. Int. J. Parasitol. 36, 1057–1065 [DOI] [PubMed] [Google Scholar]
- 14. Hainard A., Tiberti N., Robin X., Lejon V., Ngoyi D. M., Matovu E., Enyaru J. C., Fouda C., Ndung'u J. M., Lisacek F., Müller M., Turck N., Sanchez J. C. (2009) A Combined CXCL10, CXCL8 and H-FABP Panel for the Staging of Human African Trypanosomiasis Patients. PLoS Negl. Trop. Dis. 3, e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu S., Loo J. A., Wong D. T. (2006) Human body fluid proteome analysis. Proteomics. 6, 6326–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Puchades M., Hansson S. F., Nilsson C. L., Andreasen N., Blennow K., Davidsson P. (2003) Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer's disease. Brain Res. Mol. Brain Res. 118, 140–146 [DOI] [PubMed] [Google Scholar]
- 17. Pienaar I. S., Daniels W. M., Götz J. (2008) Neuroproteomics as a promising tool in Parkinson's disease research. J. Neural. Transm. 115, 1413–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rithidech K. N., Honikel L., Milazzo M., Madigan D., Troxell R., Krupp L. B. (2009) Protein expression profiles in pediatric multiple sclerosis: potential biomarkers. Mult. Scler. 15, 455–464 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J., Keene C. D., Pan C., Montine K. S., Montine T. J. (2008) Proteomics of human neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 67, 923–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dayon L., Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D. F., Burkhard P. R., Sanchez J. C. (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal. Chem. 80, 2921–2931 [DOI] [PubMed] [Google Scholar]
- 21. Mumba Ngoyi D., Lejon V., Pyana P., Boelaert M., Ilunga M., Menten J., Mulunda J. P., Van Nieuwenhove S., Muyembe Tamfum J. J., Büscher P. (2010) How to shorten patient follow-up after treatment for Trypanosoma brucei gambiense sleeping sickness. J. Infect. Dis. 201, 453–463 [DOI] [PubMed] [Google Scholar]
- 22. WHO (2000) Guidelines for the collection of clinical specimens during field investigation of outbreaks. WHO/CDS/CSR/EDC/2000.4 Available: http://www.who.int/csr/resources/publications/surveillance/whocdscsredc2004.pdf [Google Scholar]
- 23. Miézan T. W., Meda H. A., Doua F., Djè N. N., Lejon V., Büscher P. (2000) Single centrifugation of cerebrospinal fluid in a sealed pasteur pipette for simple, rapid and sensitive detection of trypanosomes. Trans. R. Soc. Trop. Med. Hyg. 94, 293. [DOI] [PubMed] [Google Scholar]
- 24. Sanchez J. C., Converset V., Nolan A., Schmid G., Wang S., Heller M., Sennitt M. V., Hochstrasser D. F., Cawthorne M. A. (2002) Effect of rosiglitazone on the differential expression of diabetes-associated proteins in pancreatic islets of C57Bl/6 lep/lep mice. Mol. Cell. Proteomics. 1, 509–516 [DOI] [PubMed] [Google Scholar]
- 25. Mortz E., Krogh T. N., Vorum H., Görg A. (2001) Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 1, 1359–1363 [DOI] [PubMed] [Google Scholar]
- 26. Burgess J. A., Lescuyer P., Hainard A., Burkhard P. R., Turck N., Michel P., Rossier J. S., Reymond F., Hochstrasser D. F., Sanchez J. C. (2006) Identification of brain cell death associated proteins in human post-mortem cerebrospinal fluid. J. Proteome Res. 5, 1674–1681 [DOI] [PubMed] [Google Scholar]
- 27. Dayon L., Pasquarello C., Hoogland C., Sanchez J. C., Scherl A. (2010) Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J. Proteomics. 73, 769–777 [DOI] [PubMed] [Google Scholar]
- 28. Tan H. T., Tan S., Lin Q., Lim T. K., Hew C. L., Chung M. C. (2008) Quantitative and temporal proteome analysis of butyrate-treated colorectal cancer cells. Mol. Cell Proteomics. 7, 1174–1185 [DOI] [PubMed] [Google Scholar]
- 29. Cheng Z. Z., Corey M. J., Pärepalo M., Majno S., Hellwage J., Zipfel P. F., Kinders R. J., Raitanen M., Meri S., Jokiranta T. S. (2005) Complement factor H as a marker for detection of bladder cancer. Clin. Chem. 51, 856–863 [DOI] [PubMed] [Google Scholar]
- 30. Sodek J., Ganss B., McKee M. D. (2000) Osteopontin. Crit. Rev. Oral Biol. Med. 11, 279–303 [DOI] [PubMed] [Google Scholar]
- 31. Wang Z., Rejtar T., Zhou Z. S., Karger B. L. (2010) Desulfurization of cysteine-containing peptides resulting from sample preparation for protein characterization by mass spectrometry. Rapid Commun. Mass Spectrom. 24, 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu P., O'Mara B. W., Warrack B. M., Wu W., Huang Y., Zhang Y., Zhao R., Lin M., Ackerman M. S., Hocknell P. K., Chen G., Tao L., Rieble S., Wang J., Wang-Iverson D. B., Tymiak A. A., Grace M. J., Russell R. J. (2010) A tris (2-carboxyethyl) phosphine (TCEP) related cleavage on cysteine-containing proteins. J. Am. Soc. Mass Spectrom. 21, 837–844 [DOI] [PubMed] [Google Scholar]
- 33. Lejon V., Büscher P. (2005) Review Article: cerebrospinal fluid in human African trypanosomiasis: a key to diagnosis, therapeutic decision and post-treatment follow-up. Trop. Med. Int. Health. 10, 395–403 [DOI] [PubMed] [Google Scholar]
- 34. Lambris J. D., Ricklin D., Geisbrecht B. V. (2008) Complement evasion by human pathogens. Nat. Rev. Microbiol. 6, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Donelson J. E., Hill K. L., El-Sayed N. M. (1998) Multiple mechanisms of immune evasion by African trypanosomes. Mol. Biochem. Parasitol. 91, 51–66 [DOI] [PubMed] [Google Scholar]
- 36. Joiner K., Sher A., Gaither T., Hammer C. (1986) Evasion of alternative complement pathway by Trypanosoma cruzi results from inefficient binding of factor B. Proc. Natl. Acad. Sci. U.S.A. 83, 6593–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Junnikkala S., Jokiranta T. S., Friese M. A., Jarva H., Zipfel P. F., Meri S. (2000) Exceptional resistance of human H2 glioblastoma cells to complement-mediated killing by expression and utilization of factor H and factor H-like protein 1. J. Immunol. 164, 6075–6081 [DOI] [PubMed] [Google Scholar]
- 38. Lequin R. M. (2005) Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 51, 2415–2418 [DOI] [PubMed] [Google Scholar]
- 39. Cantor H., Shinohara M. L. (2009) Regulation of T-helper-cell lineage development by osteopontin: the inside story. Nat. Rev. Immunol. 9, 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ashkar S., Weber G. F., Panoutsakopoulou V., Sanchirico M. E., Jansson M., Zawaideh S., Rittling S. R., Denhardt D. T., Glimcher M. J., Cantor H. (2000) Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 287, 860–864 [DOI] [PubMed] [Google Scholar]
- 41. Steinman L. (2009) A molecular trio in relapse and remission in multiple sclerosis. Nat. Rev. Immunol. 9, 440–447 [DOI] [PubMed] [Google Scholar]
- 42. Chabas D., Baranzini S. E., Mitchell D., Bernard C. C., Rittling S. R., Denhardt D. T., Sobel R. A., Lock C., Karpuj M., Pedotti R., Heller R., Oksenberg J. R., Steinman L. (2001) The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 294, 1731–1735 [DOI] [PubMed] [Google Scholar]
- 43. Denhardt D. T., Giachelli C. M., Rittling S. R. (2001) Role of osteopontin in cellular signaling and toxicant injury. Annu. Rev. Pharmacol. Toxicol. 41, 723–749 [DOI] [PubMed] [Google Scholar]
- 44. Chowdhury S. A., Lin J., Sadiq S. A. (2008) Specificity and correlation with disease activity of cerebrospinal fluid osteopontin levels in patients with multiple sclerosis. Arch. Neurol. 65, 232–235 [DOI] [PubMed] [Google Scholar]
- 45. Iwanaga Y., Ueno M., Ueki M., Huang C. L., Tomita S., Okamoto Y., Ogawa T., Ueda N., Maekawa N., Sakamoto H. (2008) The expression of osteopontin is increased in vessels with blood-brain barrier impairment. Neuropathol. Appl. Neurobiol. 34, 145–154 [DOI] [PubMed] [Google Scholar]
- 46. Cresswell P., Bangia N., Dick T., Diedrich G. (1999) The nature of the MHC class I peptide loading complex. Immunol. Rev. 172, 21–28 [DOI] [PubMed] [Google Scholar]
- 47. Shi C., Zhu Y., Su Y., Chung L. W., Cheng T. (2009) Beta2-microglobulin: emerging as a promising cancer therapeutic target. Drug Discov. Today. 14, 25–30 [DOI] [PubMed] [Google Scholar]
- 48. Alarcon A., Garcia-Alix A., Cabañas F., Hernanz A., Pascual-Salcedo D., Martin-Ancel A., Cabrera M., Tagarro A., Quero J. (2006) Beta2-microglobulin concentrations in cerebrospinal fluid correlate with neuroimaging findings in newborns with symptomatic congenital cytomegalovirus infection. Eur. J. Pediatr. 165, 636–645 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.