Abstract
Recent emergence of new mass spectrometry techniques (e.g. electron transfer dissociation, ETD) and improved availability of additional proteases (e.g. Lys-N) for protein digestion in high-throughput experiments raised the challenge of designing new algorithms for interpreting the resulting new types of tandem mass (MS/MS) spectra. Traditional MS/MS database search algorithms such as SEQUEST and Mascot were originally designed for collision induced dissociation (CID) of tryptic peptides and are largely based on expert knowledge about fragmentation of tryptic peptides (rather than machine learning techniques) to design CID-specific scoring functions. As a result, the performance of these algorithms is suboptimal for new mass spectrometry technologies or nontryptic peptides. We recently proposed the generating function approach (MS-GF) for CID spectra of tryptic peptides. In this study, we extend MS-GF to automatically derive scoring parameters from a set of annotated MS/MS spectra of any type (e.g. CID, ETD, etc.), and present a new database search tool MS-GFDB based on MS-GF. We show that MS-GFDB outperforms Mascot for ETD spectra or peptides digested with Lys-N. For example, in the case of ETD spectra, the number of tryptic and Lys-N peptides identified by MS-GFDB increased by a factor of 2.7 and 2.6 as compared with Mascot. Moreover, even following a decade of Mascot developments for analyzing CID spectra of tryptic peptides, MS-GFDB (that is not particularly tailored for CID spectra or tryptic peptides) resulted in 28% increase over Mascot in the number of peptide identifications. Finally, we propose a statistical framework for analyzing multiple spectra from the same precursor (e.g. CID/ETD spectral pairs) and assigning p values to peptide-spectrum-spectrum matches.
Since the introduction of electron capture dissociation (ECD)1 in 1998 (1), electron-based peptide dissociation technologies have played an important role in analyzing intact proteins and post-translational modifications (2). However, until recently, this research-grade technology was available only to a small number of laboratories because it was commercially unavailable, required experience for operation, and could be implemented only with expensive FT-ICR instruments. The discovery of electron-transfer dissociation (ETD) (3) enabled an ECD-like technology to be implemented in (relatively cheap) ion-trap instruments. Nowadays, many researchers are employing the ETD technology for tandem mass spectra generation (4–9).
Although the hardware technologies to generate ETD spectra are maturing rapidly, software technologies to analyze ETD spectra are still in infancy. There are two major approaches to analyzing tandem mass spectra: de novo sequencing and database search. Both approaches find the best-scoring peptide either among all possible peptides (de novo sequencing) or among all peptides in a protein database (database search). Although de novo sequencing is emerging as an alternative to database search, database search remains a more accurate (and thus preferred) method of spectral interpretation, so here we focus on the database search approach.
Numerous database search engines are currently available, including SEQUEST (10), Mascot (11), OMSSA (12), X!Tandem (13), and InsPecT (14). However, most of them are inadequate for the analysis of ETD spectra because they are optimized for collision induced dissociation (CID) spectra that show different fragmentation propensities than those of ETD spectra. Additionally, the existing tandem mass spectrometry (MS/MS) tools are biased toward the analysis of tryptic peptides because trypsin is usually used for CID, and thus not suitable for the analysis of nontryptic peptides that are common for ETD. Therefore, even though some database search engines support the analysis of ETD spectra (e.g. SEQUEST, Mascot, and OMSSA), their performance remains suboptimal when it comes to analyzing ETD spectra. Recently, an ETD-specific database search tool (Z-Core) was developed; however it does not significantly improve over OMSSA (15).
We present a new database search tool (MS-GFDB) that significantly outperforms existing database search engines in the analysis of ETD spectra, and performs equally well on nontryptic peptides. MS-GFDB employs the generating function approach (MS-GF) that computes rigorous p values of peptide-spectrum matches (PSMs) based on the spectrum-specific score histogram of all peptides (16).2 MS-GF p values are dependent only on the PSM (and not on the database), thus can be used as an alternative scoring function for the database search.
Computing p values requires a scoring model evaluating qualities of PSMs. MS-GF adopts a probabilistic scoring model (MS-Dictionary scoring model) described in Kim et al., 2009 (17), considering multiple features including product ion types, peak intensities and mass errors. To define the parameters of this scoring model, MS-GF only needs a set of training PSMs.3 This set of PSMs can be obtained in a variety of ways: for example, one can generate CID/ETD pairs and use peptides identified by CID to form PSMs for ETD. Alternatively, one can generate spectra from a purified protein (when PSMs can be inferred from the accurate parent mass alone) or use a previously developed (not necessary optimal) tool to generate training PSMs. From these training PSMs, MS-GF automatically derives scoring parameters without assuming any prior knowledge about the specifics of a particular peptide fragmentation method (e.g. ETD, CID, etc.) and/or proteolytic origin of the peptides. MS-GF was originally designed for the analysis of CID spectra, but now it has been extended to other types of spectra generated by various fragmentation techniques and/or various enzymes. We show that MS-GF can be successfully applied to novel types of spectra (e.g. ETD of Lys-N peptides (18, 19)) by simply retraining scoring parameters without any modification. Note that although the same scoring model is used for different types of spectra, the parameters derived to score different types of spectra are dissimilar.
We compared the performance of MS-GFDB with Mascot on a large ETD data set and found that it generated many more peptide identifications for the same false discovery rates (FDR). For example, at 1% peptide level FDR, MS-GFDB identified 9450 unique peptides from 81,864 ETD spectra of Lys-N peptides whereas Mascot only identified 3672 unique peptides, ≈160% increase in the number of peptide identifications (a similar improvement is observed for ETD spectra of tryptic peptides).4 MS-GFDB also showed a significant 28% improvement in the number of identified peptides from CID spectra of tryptic peptides (16,203 peptides as compared with 12,658 peptides identified by Mascot).
The ETD technology complements rather than replaces CID because both technologies have some advantages: CID for smaller peptides with small charges, ETD for larger and multiply charged peptides (20, 21). An alternative way to utilize ETD is to use it in conjunction with CID because CID and ETD generate complementary sequence information (20, 22, 23). ETD-enabled instruments often support generating both CID and ETD spectra (CID/ETD pairs) for the same peptide. Although the CID/ETD pairs promise a great improvement in peptide identification, the full potential of such pairs has not been fully realized yet. In the case of de novo sequencing, de novo sequencing tools utilizing CID/ETD pairs indeed result in more accurate de novo peptide sequencing than traditional CID-based algorithms (23, 24, 25). However, in the case of database search, the argument that the use of CID/ETD pairs improves peptide identifications remains poorly substantiated. A few tools are developed to use CID/ETD (or CID/ECD) pairs for the database search but they are limited to preprocessing/postprocessing of the spectral data before or following running a traditional database search tool (26, 27). Nielsen et al., 2005 (22) pioneered the combined use of CID and ECD for the database search. Given a CID/ECD pair, they generated a combined spectrum comprised only of complementary pairs of peaks, and searched it with Mascot.5 However, this approach is hard to generalize to less accurate CID/ETD pairs generated by ion-trap instruments because there is a higher chance that the identified complementary pairs of peaks are spurious. More importantly, using traditional MS/MS tools (such as Mascot) for the database search of the combined spectrum is inappropriate, because they are not optimized for analyzing such combined spectra; a better approach would be to develop a new database search tool tailored for the combined spectrum. Recently, Molina et al., 2008 (26) studied database search of CID/ETD pairs using Spectrum Mill (Agilent Technologies, Santa Clara, CA) and came to a counterintuitive conclusion that using only CID spectra identifies 12% more unique peptides than using CID/ETD pairs. We believe that it is an acknowledgment of limitations of the traditional MS/MS database search tools for the analysis of multiple spectra generated from a single peptide.
In this paper, we modify the generating function approach for interpreting CID/ETD pairs and further apply it to improve the database search with CID/ETD pairs. In contrast to previous approaches, our scoring is specially designed to interpret CID/ETD pairs and can be generalized to analyzing any type of multiple spectra generated from a single peptide. When CID/ETD pairs from trypsin digests are used, MS-GFDB identified 13% and 27% more peptides compared with the case when only CID spectra and only ETD spectra are used, respectively. The difference was even more prominent when CID/ETD pairs from Lys-N digests were used, with 41% and 33% improvement over CID only and ETD only, respectively.
Assigning a p value to a PSM greatly helped researchers to evaluate the quality of peptide identifications. We now turn to the problem of assigning a p value to a peptide-spectrum-spectrum match (PS2M) when two spectra in PS2M are generated by different fragmentation technologies (e.g. ETD and CID). We argue that assigning statistical significance to a PS2M (or even PSnM) is a prerequisite for rigorous CID/ETD analyses. To our knowledge, MS-GFDB is the first tool to generate statistically rigorous p values of PSnMs.
The MS-GFDB executable and source code is available at the website of Center for Computational Mass Spectrometry at UCSD (http://proteomics.ucsd.edu). It takes a set of spectra (CID, ETD, or CID/ETD pairs) and a protein database as an input and outputs peptide matches. If the input is a set of CID/ETD pairs, it outputs the best scoring peptide matches and their p values (1) using only CID spectra, (2) using only ETD spectra, and (3) using combined spectra of CID/ETD pairs.
EXPERIMENTAL PROCEDURES
Digestion of Cell Lysate
HEK293 cells were grown to confluence, harvested and resuspended in lysis buffer (50 ammonium bicarbonate, 8 m urea, Complete EDTA-free protease inhibitor mix (Roche Applied Science), 5 mm potassium phosphate, 1 mm potassium fluoride, and 1 mm sodium orthovanadate) and incubated for 20 min at 4 °C. An insoluble fraction was spun down at 1000 × g for 10 min at 4 °C and the protein content of the supernatant was determined using the 2DQuant Kit (GE Healthcare). Per 1 mg of lysate 45 mm dithiothreitol were used for reduction (30 min at 50 °C) and 100 mm iodoacetamide for subsequent alkylation (30 min at RT). Trypsin digests were generated by digestion of 1 mg cell lysate with 1.25 μg Lys-C for 4 h at RT followed by dilution to 2 m urea and digestion with 15 μg trypsin for 16 h at 37 °C. Lys-N digests were made by digestion of 1 mg cell lysate with 5 μg Lys-N for 4 h at RT, dilution to 2 m urea, and another digestion with 5 μg Lys-N for 16 h at 37 °C.
Peptide Prefractionation by Strong Cation Exchange (SCX)
Fractionation of peptides was performed as described earlier (28, 29). In detail, digests were acidified with formic acid and loaded onto two C18 cartridges using an Agilent 1100 high pressure liquid chromatography (HPLC) system operated at 100 μl/min with 0.05% formic acid in water. Peptides were then eluted from the C18 cartridges using 80% acetonitrile and 0.05% formic acid in water onto a PolySULFOETHYL A column (200 mm × 2.1 mm column, PolyLC). Separation of different peptide populations was performed at 200 μl/min using a nonlinear gradient as follows: 0 to 10 min 100% solvent A (5 mm KH2PO4, 30% acetonitrile, 0.05% formic acid), 10 to 15 min from 0% to 26% solvent B (350 mm KCl, 5 mm KH2PO4, 30% acetonitrile, 0.05% formic acid), 15 to 40 min from 26% to 35% solvent B and from 40 to 45 min from 35% to 60% solvent B, and from 45 to 49 min from 60% to 100% solvent B. Fractions were collected in 1 min intervals for 40 min, dried down in a vacuum centrifuge, and resuspended in 10% formic acid.
Mass Spectrometry
SCX fractions were analyzed on a reversed-phase nano-LC-coupled LTQ Orbitrap XL ETD (Thermo Fisher Scientific). An Agilent 1200 series HPLC system was equipped with a 20 mm Aqua C18 (Phenomenex) trapping column (packed in-house, 100 μm inner diameter, 5 μm particle size) and a 400 mm ReproSil-Pur C18-AQ (Dr. Maisch GmbH) analytical column (packed in-house, 50 μm inner diameter, 3 μm particle size). Trapping was performed at 5 μl/min solvent C (0.1 m acetic acid in water) for 10 min, and elution was achieved with a gradient from 10% to 30% (v/v) solvent D (0.1 m acetic acid in 1:4 acetonitrile : water) in solvent C in 110 min, followed by a gradient of 30% to 50% (v/v) solvent D in solvent C in 30 min, followed by a gradient of 50% to 100% (v/v) solvent D in solvent C in 5 min and finally 100% solvent D for 2 min. The flow rate was passively split from 0.45 ml/min to 100 nL/min. Nano-electrospray was achieved using a distally coated fused silica emitter (360 μm outer diameter, 20 μm inner diameter, 10 μm tip inner diameter, New Objective) biased to 1.7 kV. The instrument was operated in data dependent mode to automatically switch between MS and MS/MS. Survey full scan MS spectra were acquired from m/z 350 to m/z 1500 in the Orbitrap with a resolution of 60,000 at m/z 400 following accumulation to a target value of 500,000 in the linear ion trap. The two most intense ions at a threshold of above 500 were fragmented in the linear ion trap using CID at an AGC target value of 30,000 and ETD with supplemental activation at an AGC target value of 50,000. The ETD reagent AGC target value was set to 100,000 and the reaction time to 50 ms.
Data Processing
From every raw data file recorded by the mass spectrometer, representing a single SCX fraction, two different peak lists containing either CID or ETD fragmentation data were generated using Proteome Discoverer (version 1.0, Thermo Fisher Scientific) with a signal-to-noise threshold of three and the following settings for the ETD-nonfragment filter: precursor peak removal with 4 Da, charge-reduced precursor removal with 8 Da, and removal of known neutral losses from charge-reduced precursors with 8 Da within a window of 120 Da. Single-fraction peak lists of the major peptide-containing SCX fractions for trypsin-derived and Lys-N-derived peptides were then merged into four larger peak lists, denoted CID-Tryp, ETD-Tryp, CID-LysN, and ETD-LysN. The whole data set is composed of 168,960 CID/ETD pairs. Of this, 87,096 pairs (51,233 with charge 2+, 24,854 with charge 3+, and 11,009 with charges 4+ and larger) are from the trypsin digests and 81,864 (24,284 with charge 2+, 28,168 with charge 3+ and 29,412 with charges 4+ and larger) are from the Lys-N digests. Spectra with precursor charges from 2+ to 7+ were considered in the further analyses. All the spectra (Raw files and mzXML files) and database search results associated with this manuscript may be downloaded from the Tranche repository (http://proteomecommons.org/tranche/) using the following hash:
mQTEDmtWauUPq41hJMPY/tnB3+zXhc5GSMKuRm+ljChFjtJrrrnJ4WwNpkgWM0/zGE0Zy/STG0NWJwTbbqMnInXrKi8AAAAAAAB5sA==
Mascot Analysis
Mascot (version 2.3.0, Matrix Science) was used to search the peaklists against an in-house built database (74,190 entries; 31,263,418 amino acids) assembled from the IPI human database (version 3.52, http://www.ebi.ac.uk/ipi) plus common contaminants (target database). A decoy database was constructed by reversing all sequences and slightly scrambling entries using MaxQuant (version 1.0.13.8; http://www.maxquant.org) (30). The target and decoy databases were searched separately to estimate FDRs. The following parameters were used for database searching: 50 ppm precursor mass tolerance, 0.5 Da fragment ion tolerance, up to two missed cleavages allowed, carbamidomethyl cysteine as fixed modification, no variable modifications. The enzyme was specified as either trypsin or Lys-N and the instrument type either ESI-TRAP or ETD-TRAP.
Training MS-GF Scoring Parameters
MS-GF takes a set of PSMs as an input training set and outputs a scoring parameter file containing the parameters used for scoring (see Supplement 1 for details on training scoring parameters). We first generated initial scoring parameter files for the four data sets (CID-Tryp, ETD-Tryp, CID-LysN, and ETD-LysN) using PSMs with Mascot scores corresponding to peptide level FDRs less than 1% as a training set. Using these initial parameter files, we ran MS-GFDB and selected PSMs with MS-GF p values corresponding to peptide level FDRs less than 1%. These PSMs were used as a new training set to build the final scoring parameter files.
MS-GFDB Search (for CID or ETD spectra)
Because MS-GFDB automatically preprocesses spectra (see Supplement 1 for details), we converted each raw data file into an mzXML file using ReAdW 4.3.1 (31) and used the mzXML file in the MS-GFDB search (as opposed to using Proteome Discoverer for noise and (charge-reduced) precursor filtering). MS-GFDB searches were carried out against the same database with the same parameters as were used for Mascot searches.
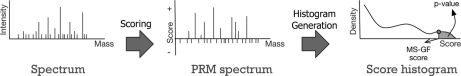
MS-GFDB uses two scores: the MS-GF score and the p value (both are computed by MS-GF). The MS-GF score is used to evaluate the quality of a PSM and the p value is used to assess the statistical significance of a PSM. To compute the MS-GF score, MS-GF first converts every spectrum into a Prefix-Residue Mass (PRM) spectrum (14, 32) using scoring parameters specific to a particular fragmentation technique and enzyme. The PRM spectrum is a scored version of a spectrum having a score at every mass up to the parent mass of the spectrum.6 As described in Dančík et al., 1999 (32), the score of a PRM spectrum at mass m represents the log likelihood ratio that the peptide from which the spectrum was derived contains a prefix of mass m.7 The MS-GF score of a peptide against a spectrum is defined as the sum of scores in the PRM spectrum corresponding to prefix masses of the peptide. To compute the p value, MS-GF generates the score histogram of all peptides using the generating function approach (see (16) for details on the generating function approach). The p value of a peptide with match score s is defined as the area under the histogram where the score value (x axis) is equal or larger than s (see Supplement 2 for details on the MS-GF scoring functions). Figure 1 illustrates the procedure to compute p values with MS-GF.
Fig. 1.
Computing p values with MS-GF for a single spectrum. Given a tandem mass spectrum, MS-GF converts the spectrum into a PRM spectrum (scored version of the tandem mass spectrum). The score of a PRM spectrum at mass m represents the log likelihood ratio that the peptide from which the spectrum was derived contains a prefix of mass m. Negative peaks in the PRM spectrum represent masses more likely to represent incorrect rather than correct prefix masses. Such negative peaks in the PRM spectrum usually correspond to low-intensity or missing peaks in the experimental spectrum. The PRM spectrum is used to compute the MS-GF score of any peptide against the spectrum. Then, MS-GF computes the histogram of the MS-GF scores of all peptides against the spectrum using the generating function approach. Finally, MS-GF computes the p value of a peptide as the area under the histogram with MS-GF scores equal or larger than the MS-GF score of the peptide.
Given a spectrum and a protein database, MS-GFDB computes MS-GF scores for all the peptides in the database (similarly to SEQUEST or Mascot), finds the peptide with the best score and reports its p value.8
MS-GFDB Search (for CID/ETD Pairs)
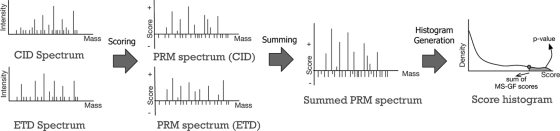
MS-GFDB combines a pair of tandem mass spectra generated from a single precursor ion (using different fragmentation techniques) and matches the combined spectrum against a database. Given a pair of spectra, it first converts each spectrum into a PRM spectrum (using fragmentation-specific parameters for each type of spectrum) and generates a summed PRM spectrum. The Summed PRM spectrum of two PRM spectra (with the same parent mass) is calculated by adding two PRM scores (log likelihood ratios) corresponding to the same mass. For example, if at mass 500, two PRM spectra have scores 7 and 3, correspondingly, the summed PRM spectrum has score 7 + 3 = 10 at mass 500. Note that summing PRM scores at mass m is equivalent to multiplying the probabilities that mass m is a prefix mass of the peptide from which each the spectrum was derived. This summed PRM model assumes that ion types are independent within the same spectrum (34) and when coming from different spectra (35), the assumption that proved to be useful in other applications. The score histogram of a CID/ETD pair is computed using the summed PRM spectrum and is used to compute p values. Figure 2 illustrates the flow of the p value computation for CID/ETD pairs. This method improves on the previous method proposed by Nielsen et al. (22) in that it merges evidence for a certain backbone cleavage (represented as a PRM score) using a probabilistic model, whereas the approach in (22) only retains a peak if it has a complementary peak or discards a peak if not. Therefore, the approach in (22) results in much stricter peak filtering, making it difficult to distinguish between correct and incorrect peptide identifications. For example, given a CID/ETD pair with a poor-quality CID spectrum and a high-quality ETD spectrum, the method in (22) is unlikely to interpret the pair, because the CID spectrum does not help to identify “complementary pairs of peaks” and the resulting spectrum contains only a few peaks identified from the ETD spectrum itself. In contrast, the summed PRM scores retain most of the sequence information in the ETD spectrum contributing to successful peptide identifications.
Fig. 2.
Computing p values with MS-GF for CID/ETD pairs. Given a CID/ETD pair, MS-GFDB converts each spectrum into a PRM spectrum and merges two PRM spectra by summing scores of peaks sharing the same mass. This “summed” PRM spectrum is used to generate the score histogram of all peptides and p values are computed using the histogram.
Note that this method can be generalized to the case of analyzing more than two tandem mass spectra generated from a single precursor ion (e.g. by adding a high energy collisional dissociation beam-type CID spectrum).
RESULTS
Analysis of Individual Spectra
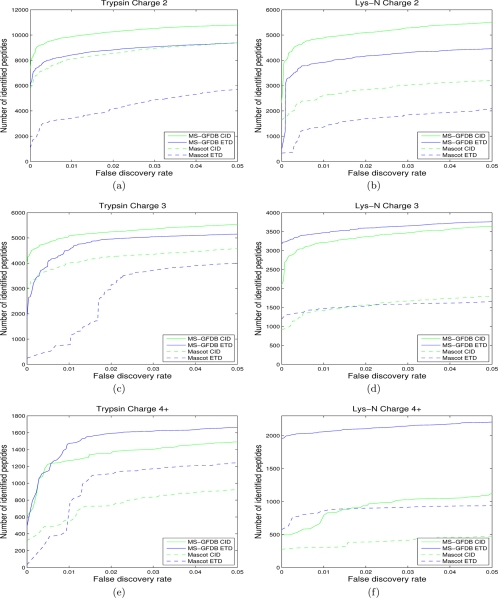
For each of the CID-Tryp, ETD-Tryp, CID-LysN, and ETD-LysN data sets, we compared the performance of MS-GFDB with Mascot by counting the number of identified peptides for each FDR (peptide-level FDR) using the separate target-decoy search approach (36). For all the four data sets, MS-GFDB outperformed Mascot (Fig. 3). For example, at 1% FDR, MS-GFDB identified 14,409 peptides in ETD-Tryp whereas Mascot identified 5310 peptides. The difference is more notable for ETD spectra than CID spectra and for Lys-N digests than trypsin digests. This indicates that Mascot is poorly optimized for the analysis of new data types whereas MS-GFDB automatically adapts to novel types of data. Even in the case of the CID-Tryp data set where Mascot has been subjected to a decade-long development, MS-GFDB identified ≈30% more peptides across entire FDR range. Similar results were obtained using the spectrum-level FDR (see Supplement 3).
Fig. 3.
Number of identified peptides with Mascot and MS-GFDB from (a) charge 2 spectra in CID-Tryp and ETD-Tryp, (b) charge 2 spectra in CID-LysN and ETD-LysN, (c) charge 3 spectra in CID-Tryp and ETD-Tryp, (d) charge 3 spectra in CID-LysN and ETD-LysN, (e) spectra with charges 4 and larger in CID-Tryp and ETD-Tryp, and (f) spectra with charges 4 and larger in CID-LysN and ETD-LysN. The number of peptide identifications is plotted against the corresponding peptide level FDR. Solid curves represent MS-GFDB and dashed curves represent Mascot. Green curves represent CID and blue curves represent ETD. Mascot ion scores and MS-GFDB p values were used for computing FDRs. FDRs were separately computed for spectra of precursor charge 2, precursor charge 3, and precursor charge 4 and larger. For all the cases considered, MS-GFDB outperformed Mascot.
MS-GFDB also outperformed SEQUEST and OMSSA (see Supplement 4). To boost the performance of existing MS/MS database search tools, PeptideProphet (37), iProphet, and Percolator (38, 39) rescore their PSMs, resulting in a significant increase in the number of peptide identifications.9 However, MS-GFDB outperformed even PeptideProphet, iProphet, and Percolator, which take advantage of extra information unavailable to MS-GF such as the score distribution of all PSMs and the retention time information (Supplements 4 and 5).
In this experiment, we used the same data for both training and testing of the performance, thus raising a valid concern about over-fitting. This was done because we observed that MS-GF parameters characterize a particular protocol (e.g. ETD for a particular enzyme) and are rather stable with respect to specific data sets, i.e. variable data sets with the same protocol result in similar MS-GF parameters. To address this concern, we demonstrated that if we derive MS-GF scoring parameters from a training data set A and apply it to a test data set B, the results hardly change as compared with deriving MS-GF scoring parameters from the data set B and apply it to the same data set B (see Supplement 6).
For further analyses below, PSMs with FDRs below 1% were selected from the four data sets using MS-GFDB; if multiple spectra of the same charge are matched to the same peptide, only that with the best score was chosen. From CID-Tryp/ETD-Tryp/CID-LysN/ETD-LysN data set, 16,203/14,409/8893/9450 PSMs were selected and denoted by CID-Tryp-Confident/ETD-Tryp-Confident/CID-LysN-Confident/ETD-LysN-Confident.
Comparison of Ion Fragmentation Statistics Across Different Spectral Data Sets
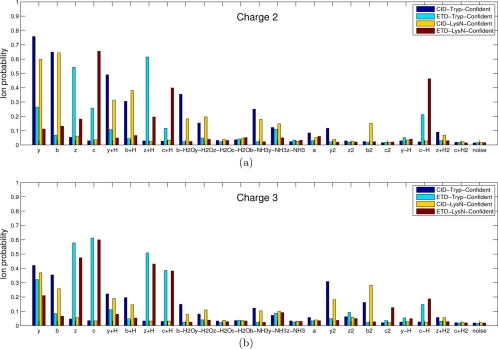
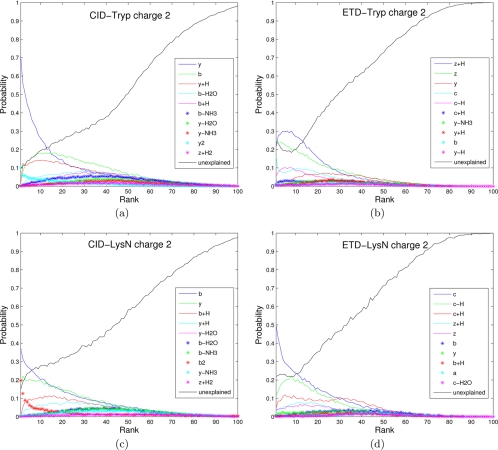
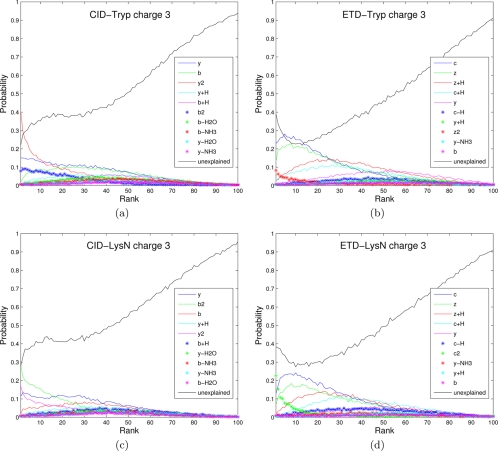
The spectra of the same peptide are different depending on the fragmentation methods and precursor ion charges. Moreover, spectra of peptides produced by one enzyme (e.g. tryptic peptides ending with Lys or Arg) do have different fragmentation propensities than spectra of peptides produced by other enzyme (e.g. Lys-N peptides starting from Lys) (28, 40). The common knowledge that ETD spectra are mainly comprised of c and z· ions (and their neutral losses) whereas CID spectra are of b and y ions (and their neutral losses) is insufficient for designing a good scoring function because one has to know the propensities (likelihood) of these ions and many other neutral losses (41). To analyze such propensities for different types of spectra, we measured the probability of a certain ion type being observed (Fig. 4) and plotted the distribution of a peak of a given rank being a certain ion type (Figs. 5 and 6) as presented in (17, 32).10 Note the high abundance of c ions with high intensities in Figure 5d, confirming the previously published result (18). Features shown in Figures 4, 5, and 6 were automatically derived by MS-GF scoring functions and contributed to the improved performance of MS-GFDB over other tools.
Fig. 4.
Probabilities of various ion types for the four types of (a) charge 2 spectra and (b) charge 3 spectra (see (32) for similar analysis). Spectra in CID-Tryp-Confident, ETD-Tryp-Confident, CID-LysN-Confident, and ETD-LysN-Confident were used. All the spectra were filtered to remove noisy peaks as follows: given a peak at mass M, we retained the peak if it is among the top six peaks within a window of size 100 Da around M. Precursor ions (or charge-reduced precursor ions) and their derivatives were also filtered out. A colored bar represents the probability (y axis) of a certain type of ion (x axis) being present in a filtered spectrum. Each data set is color coded. For example, a charge 2 spectrum in CID-Tryp-Confident generated from a length 10 peptide is expected to have 10–1 (number of potential cleavage sites) × 0.76 (probability of y ion) = 6.8 y ions, whereas a charge 2 spectrum in ETD-Tryp-Confident is expected to have only 9 × 0.26 = 2.3 y ions. In MS-GFDB, all ion types with probabilities exceeding 0.15 are used for scoring (see Supplement 1 for details).
Fig. 5.
Rank distributions of different ion types for different data sets: a, CID-Tryp-Confident; b, CID-LysN-Confident; c, ETD-Tryp-Confident; and d, ETD-LysN-Confident. Only charge two spectra were considered and all spectra were filtered to remove precursor ions (or charge-reduced precursor ions) and their derivatives. For each data set, 10 different ion types with highest probabilities were selected and the probability of a peak of a given rank (x axis) being a certain ion type (color-coded) is plotted for peaks with rank 1 to 100. The black curve (labeled as unexplained) represents the peaks that are not explained by any of the 10 selected ion types. For example, for CID-Tryp-Confident charge 2, the highest ranked peak represents a singly charged y ion with probability 0.7, a doubly charged y ion (y2) with probability 0.1, a singly charged b ion with probability 0.04, etc. It remains unexplained with probability 0.1.
Fig. 6.
Analog of Fig. 5 for charge 3 spectra.
Pitfalls of “Intersection” and “Union“ Approaches to Identifying CID/ETD Pairs
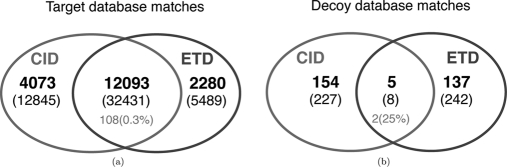
It is believed that utilizing CID/ETD pairs is helpful to improve confidence of peptide identifications because the identification from one method cross-validates the other. However, there is no consensus on how to utilize CID/ETD pairs for the database search. The common practice is to run database search for CID spectra and ETD spectra separately as if the pairing is not even known, identify confident PSMs using a predefined threshold (e.g. peptide level FDR 1% or a predefined score threshold) and take the intersection of CID PSMs and ETD PSMs (intersection approach). For example, in CID-Tryp and ETD-Tryp there are 50,765 spectral pairs where either CID or ETD spectra (or both) are confidently identified with MS-GFDB within the peptide level FDR 1%. In 32,431 spectral pairs (representing 12,093 distinct peptides), the CID identification and ETD identification were the same, indicating that these identifications are reliable (Fig. 7a). To measure the FDR of these “intersection” spectral pairs, we repeated the same procedure with the identifications to the decoy database and obtained eight pairs (representing five peptides) where CID and ETD identifications agree (Fig. 7b); hence, the peptide level FDR corresponds to 5/12,093 = 4.1·10−4. Although taking the intersection improved the confidence of the resulting peptide identifications (12,093 peptides at FDR close to 0), at the same confidence level, MS-GFDB identified 7% more peptides using only CID spectra (not shown in Fig. 7)!11 This indicates that this approach is inefficient considering that half of the instrument time was wasted generating ETD spectra that did not help to improve the number of peptide identifications.
Fig. 7.
Venn diagrams of (a) spectral pairs identified against the IPI-Human database within peptide level FDR 1% and (b) spectral pairs identified against the decoy database with p values corresponding to peptide level FDR 1% or less. The number of peptides (the number of spectral pairs in parentheses) are shown. The grey numbers correspond to the number (percentage in parentheses) of spectral pairs where CID and ETD identifications disagree.
The poor performance of the intersection approach can be explained by the dependences in scores of CID and ETD spectra from the same pair. Examination of hits in the decoy database revealed that a high scoring PSM for CID spectra often corresponds to a high scoring PSM for ETD spectra from the same pair. As a result, contrary to the common belief, the intersection approach has limited ability to remove incorrect PSMs. On the other hand, many hits in the target database have high scores for CID spectra and low scores for ETD spectra (or vice versa), thus reducing the number of correct PSMs returned by the intersection approach.
Similarly, it is possible to take the “union“ of identified peptides (all significant CID identifications plus all significant ETD identifications) to get more peptide identifications. For instance, from the above 50,765 spectral pairs, one may take the 4073 + 12,093 + 2280 = 18,446 peptides, corresponding to FDR (154 + 5 + 137)/18,446 = 1.6%.12 At the same FDR level, MS-GFDB identified 16,636 peptides only from CID spectra, thus this union approach resulted in 11% increase in the number of peptides. Although this improvement in the number of peptides (with a larger FDR) is meaningful, our proposed approach results in a comparable number of identified peptides at a stricter level of confidence (1% FDR instead of 1.6%).
Identifications from Combined CID/ETD Spectra
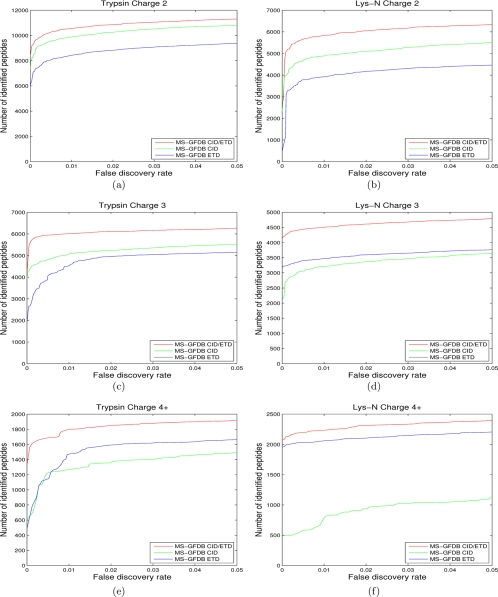
Given a CID/ETD pair, one can generate a “combined spectrum” and search a database with the combined spectrum. We used the summed PRM spectra as described above (denoted by MS-GFDB CID/ETD) and compared its performance with MS-GFDB using only CID spectra (MS-GFDB CID) or ETD spectra (MS-GFDB ETD). MS-GFDB CID/ETD identified more peptides across entire FDR range compared with MS-GFDB CID or MS-GFDB ETD for both trypsin digests and Lys-N digests (Fig. 8). For example, at 1% FDR, MS-GFDB CID/ETD identified 18,342 peptides from CID/ETD pairs of trypsin digests and 12,561 peptides from LysN digests, corresponding to 13%, 27%, 41%, and 33% improvement over when CID-Tryp, ETD-Tryp, CID-LysN and ETD-LysN data sets are separately used, respectively. If we consider spectra of charge 3 and larger (where ETD has advantages over CID), the improvement becomes even more significant: 23%, 30%, 68%, and 21%.
Fig. 8.
Number of identified peptides with MS-GFDB CID/ETD from (a) charge 2 spectral pairs in CID-Tryp and ETD-Tryp, (b) charge 2 spectral pairs in CID-LysN and ETD-LysN, (c) charge 3 spectral pairs in CID-Tryp and ETD-Tryp, (d) charge 3 spectral pairs in CID-LysN and ETD-LysN, (e) spectral pairs of charges 4 and larger in CID-Tryp and ETD-Tryp, and (f) spectral pairs of charges 4 and larger in CID-LysN and ETD-LysN. Number of identified peptides with MS-GFDB are also shown for reference. The number of peptide identifications is plotted against the corresponding peptide level FDR. FDRs were separately computed for spectra of precursor charge 2, precursor charge 3, and precursor charge 4 and larger. Red curves represent MS-GFDB CID/ETD, green curves represent MS-GFDB CID and blue curves represent MS-GFDB ETD. For all the cases considered, MS-GFDB outperformed both MS-GFDB CID and MS-GFDB ETD.
The improved performance of MS-GFDB CID/ETD is because of the probabilistic model for constructing combined spectra. We remark that a brute-force approach to constructing combined spectra actually reduces the number of peptide identifications (Supplement 7).
DISCUSSION
We demonstrated that the generating function approach is easily adaptable to the analysis of novel types of spectra. For all types of spectral data sets we have tested, MS-GFDB outperformed state-of-the-art MS/MS database search tools. We further demonstrated how to utilize the combined CID/ETD spectra generated from CID/ETD pairs using MS-GFDB.
We emphasize that MS-GFDB analyzes all different data sets in exactly the same way using different scoring parameters that are automatically derived by the same training procedure. Although it may seem counterintuitive that the MS-GF scoring function (defined as a simple dot-product of vectors) improves on more complex scoring functions used in traditional MS/MS tools, it was made possible by deriving rigorous MS-GF p values using the generating function approach. We are not claiming that MS-GF scores are “better“ than Mascot scores, but we do show that p values derived from MS-GF scores greatly improve on Mascot scores. This observation emphasizes the importance of rigorous p values that remain unavailable for popular tools such as Mascot and SEQUEST.
The problem of analyzing spectral pairs from the same precursor is related to the problem of combining database search scores of MS2 and MS3 spectra from the same peptide addressed by Olsen and Mann, 2004 (42), Bandeira et al., 2008 (35), and Ulintz et al., 2008 (43). Olsen and Mann, 2004 and Bandeira et al., 2008 developed a probabilistic scoring model for MS2 spectra and used it to adjust the MS3 score by summing the MS2 and MS3 scores. Although this approach is similar to our approach in that both use the sum of (log-likelihood) scores as the score of a pair, it did not provide a rigorous framework to compute the p value of the pair. On the other hand, Ulintz et al., 2008 developed an approach searching the database separately for MS2 and MS3 spectra and adjusting the probabilities of both spectra if the top scoring sequences match (similar to the intersection approach described above). In contrast, our approach considers all possible cases (e.g. including peptides having poor scores against CID spectrum and good scores against ETD spectrum) and uses them to compute p values, something that was missing in previous studies.
ETD has certain advantages over CID in the analysis of peptides with post-translational modifications (PTMs) (18, 44–46). MS-GFDB can be used to identify modified peptides. When PTMs are selected in advance (restrictive search for PTMs), MS-GFDB only needs to add the masses of amino acids with PTMs to the standard 20 amino acid set. In the analysis of a sample of phosphorylated peptides, MS-GFDB identified about 30%-40% more peptides from CID spectra and about 60%-90% more peptides from ETD spectra than Mascot (Supplement 8). The gain from MS-GFDB over Mascot in this data set was smaller than in the other data sets described above. This is because we used the parameters trained from unmodified spectra to score spectra of phosphorylated peptides. It is well known that some post-translational modifications (PTMs) such as phosphorylation change the fragmentation propensity of the spectrum, especially in the case of CID spectra (47). Therefore, to efficiently analyze such PTMs, one needs to develop a scoring function that is specific to the target PTM (48). Designing a PTM-specific scoring function and the generating function for modified peptides is beyond the scope of this paper.
Supplementary Material
Footnotes
* This work was supported by National Institutes of Health Grant 1-P41-RR024851 from the National Center for Research Resources.
 This article contains supplemental material 1–8.
This article contains supplemental material 1–8.
2 The term “p-value” here and the term “spectral probability” used in Kim et al., 2008 (16) are synonymous. Throughout the paper, we use “p-value,” because it is more generally used.
3 A thousand PSMs of unique peptides is usually sufficient.
4 The peptide level FDR is defined as the number of unique peptides in the decoy database over the number of unique peptides in the target database at a certain threshold. At 1% spectrum level FDR, MS-GFDB identified 22,003 spectra, whereas Mascot identified 9027 spectra, a 140% increase in the number of identified spectra for ETD spectra of Lys-N peptides.
5 The combined spectrum is a pseudo-spectrum generated from the set of pairs of peaks supporting the same backbone cleavage. The pair may come from the same spectrum (e.g. two peaks with the sum of masses equals to the parent mass) or different spectra (e.g. a peak from CID spectrum and a peak from ECD spectrum with the mass difference 16.02 Da, representing a possible pair of y and z fragment ions).
6 One can define the granularity of a mass depending on the resolution of the mass spectrum. Throughout this paper, the granularity is set as 1 Da (equivalent to the fragment ion tolerance 0.5 Da). Although this paper focuses on MS/MS spectra with inaccurate fragment masses, MS-GFDB can be adapted to analyze spectra with accurate fragment masses by changing the granularity.
7 Every peptide of length n defines n-1 prefix masses representing masses of the first i amino acids (for 1<i<n).
8 MS-GFDB search takes only ≈0.1 second per spectrum against a database containing 31 million amino acids for a computer with Core i7 2.7Ghz CPU with 12GB memory. We have recently published a study to further speed up MS-GFDB using gapped peptides (MS-GappedDictionary, Jeong et al., 2010 (33)), an approach that is similar to using peptide sequence tags in Inspect (14). MS-GappedDictionary uses MS-GF scores to generate gapped peptides that are used for fast database scan such as peptide sequence tags. Combining MS-GappedDictionary and MS-GFDB enables orders of magnitudes speed-up.
9 Shteynberg, D., et al., Postprocessing and validation of tandem mass spectrometry data sets improved by iProphet. (In preparation.)
10 Rank of a peak is defined as the number of peaks (in the same spectrum) with intensities higher than or equal to intensity of the peak (17).
11 For Lys-N digests, we identified 5788 peptides using the intersection approach with a corresponding to 3.5·10−4 FDR; at the same FDR, MS-GFDB identified a similar number of peptides using only CID spectra.
12 Spectral pairs where CID and ETD identifications disagree (red numbers in Fig. 7) were discarded.
1 The abbreviations used are:
- ECD
- electron capture dissociation
- ETD
- electron transfer dissociation
- MS/MS
- tandem mass spectrometry
- CID
- collision induced dissociation
- FDR
- false discovery rate
- PSM
- peptide-spectrum match
- PS2M
- peptide-spectrum-spectrum match
- SCX
- strong cation exchange
- PRM
- prefix-residue mass
- PTM
- post-translational modification
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1. Zubarev R., Kelleher N., McLafferty F. (1998) Electron capture dissociation of multiply charged protein cations. a nonergodic process. J. Am. Chem. Soc. 120, 3265–3266 [Google Scholar]
- 2. Cooper H. J., Håkansson K., Marshall A. G. (2005) The role of electron capture dissociation in biomolecular analysis. Mass Spectrom. Rev. 24, 201–222 [DOI] [PubMed] [Google Scholar]
- 3. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taverna S. D., Ueberheide B. M., Liu Y., Tackett A. J., Diaz R. L., Shabanowitz J., Chait B. T., Hunt D. F., Allis C. D. (2007) Long-distance combinatorial linkage between methylation and acetylation on histone h3 n termini. Proc. Natl. Acad. Sci. U.S.A. 104, 2086–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., Hsieh-Wilson L. C. (2007) Probing the dynamics of o-glcnac glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 [DOI] [PubMed] [Google Scholar]
- 6. Appella E., Anderson C. W. (2007) New prospects for proteomics–electron-capture (ecd) and electron-transfer dissociation (etd) fragmentation techniques and combined fractional diagonal chromatography (cofradic). FEBS J. 274, 6255. [DOI] [PubMed] [Google Scholar]
- 7. Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A. (2007) Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Altelaar A. F., Mohammed S., Brans M. A., Adan R. A., Heck A. J. (2009) Improved identification of endogenous peptides from murine nervous tissue by multiplexed peptide extraction methods and multiplexed mass spectrometric analysis. J. Proteome Res. 8, 870–876 [DOI] [PubMed] [Google Scholar]
- 9. Mohammed S., Lorenzen K., Kerkhoven R., van Breukelen B., Vannini A., Cramer P., Heck A. J. (2008) Multiplexed proteomics mapping of yeast rna polymerase ii and iii allows near-complete sequence coverage and reveals several novel phosphorylation sites. Anal. Chem. 80, 3584–3592 [DOI] [PubMed] [Google Scholar]
- 10. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 11. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 12. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 13. Craig R., Beavis R. C. (2004) Tandem: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 14. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) Inspect: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 15. Sadygov R. G., Good D. M., Swaney D. L., Coon J. J. (2009) A new probabilistic database search algorithm for etd spectra. J. Proteome Res. 8, 3198–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S., Gupta N., Pevzner P. A. (2008) Spectral probabilities and generating functions of tandem mass spectra: A strike against decoy databases. J. Proteome Res. 7, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S., Gupta N., Bandeira N., Pevzner P. A. (2009) Spectral dictionaries: Integrating de novo peptide sequencing with database search of tandem mass spectra. Mol. Cell. Proteomics 8, 53–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taouatas N., Drugan M. M., Heck A. J., Mohammed S. (2008) Straightforward ladder sequencing of peptides using a lys-n metalloendopeptidase. Nat. Methods 5, 405–407 [DOI] [PubMed] [Google Scholar]
- 19. Eppstein D. (2008) Targeted scx based peptide fractionation for optimal sequencing by collision induced, and electron transfer dissociation. J. Proteomics Bioinform. 1, 379–388 [Google Scholar]
- 20. Zubarev R. A., Zubarev A. R., Savitski M. M. (2008) Electron capture/transfer versus collisionally activated/induced dissociations: solo or duet? J. Am. Soc. Mass Spectrom. 19, 753–761 [DOI] [PubMed] [Google Scholar]
- 21. Swaney D. L., McAlister G. C., Coon J. J. (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nielsen M. L., Savitski M. M., Zubarev R. A. (2005) Improving protein identification using complementary fragmentation techniques in fourier transform mass spectrometry. Mol. Cell. Proteomics 4, 835–845 [DOI] [PubMed] [Google Scholar]
- 23. Savitski M. M., Nielsen M. L., Kjeldsen F., Zubarev R. A. (2005) Proteomics-grade de novo sequencing approach. J. Proteome Res. 4, 2348–2354 [DOI] [PubMed] [Google Scholar]
- 24. Datta R., Bern M. (2009) Spectrum fusion: Using multiple mass spectra for de novo peptide sequencing. J. Comput. Biol. 16, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 25. Bertsch A., Leinenbach A., Pervukhin A., Lubeck M., Hartmer R., Baessmann C., Elnakady Y. A., Müller R., Böcker S., Huber C. G., Kohlbacher O. (2009) De novo peptide sequencing by tandem ms using complementary cid and electron transfer dissociation. Electrophoresis 30, 3736–3747 [DOI] [PubMed] [Google Scholar]
- 26. Molina H., Matthiesen R., Kandasamy K., Pandey A. (2008) Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal. Chem. 80, 4825–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Good D. M., Wenger C. D., McAlister G. C., Bai D. L., Hunt D. F., Coon J. J. (2009) Post-acquisition etd spectral processing for increased peptide identifications. J. Am. Soc. Mass Spectrom. 20, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taouatas N., Altelaar A. F., Drugan M. M., Helbig A. O., Mohammed S., Heck A. J. (2009) Strong cation exchange-based fractionation of lys-n-generated peptides facilitates the targeted analysis of post-translational modifications. Mol. Cell. Proteomics 8, 190–200 [DOI] [PubMed] [Google Scholar]
- 29. Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. (2009) Lys-n and trypsin cover complementary parts of the phosphoproteome in a refined scx-based approach. Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 30. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 31. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics ms/ms analysis platform utilizing open xml file formats. Mol Syst Biol 1, 2005.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dančík V., Addona T. A., Clauser K. R., Vath J. E., Pevzner P. A. (1999) De novo peptide sequencing via tandem mass spectrometry. J. Comput. Biol. 6, 327–342 [DOI] [PubMed] [Google Scholar]
- 33. Jeong K., Kim S., Bandeira N., Pevzner P. (2010) Gapped spectral dictionaries and their applications for database searches of tandem mass spectra. Lecture Notes Computer Sci. 1, 208–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pevzner P. A., Dančík V., Tang C. L. (2000) Mutation-tolerant protein identification by mass spectrometry. J. Comput. Biol. 7, 777–787 [DOI] [PubMed] [Google Scholar]
- 35. Bandeira N., Olsen J. V., Mann J. V., Mann M., Pevzner P. A. (2008) Multi-spectra peptide sequencing and its applications to multistage mass spectrometry. Bioinformatics 24, i416–i423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 37. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by ms/ms and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 38. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 39. Brosch M., Yu L., Hubbard T., Choudhary J. (2009) Accurate and sensitive peptide identification with mascot percolator. J. Proteome Res. 8, 3176–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boersema P. J., Taouatas N., Atelaar A. F., Gouw J. W., Ross P. L., Pappin D. J., Heck A. J., Mohammed S. (2009) Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol. Cell. Proteomics 8, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coon J. J. (2009) Collisions or electrons? protein sequence analysis in the 21st century. Anal. Chem. 81, 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olsen J. V., Mann M. (2004) Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. U.S.A. 101, 13417–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ulintz P. J., Bodenmiller B., Andrews P. C., Aebersold R., Nesvizhskii A. I. (2008) Investigating ms2/ms3 matching statistics: a model for coupling consecutive stage mass spectrometry data for increased peptide identification confidence. Mol. Cell. Proteomics 7, 71–87 [DOI] [PubMed] [Google Scholar]
- 44. Domon B., Bodenmiller B., Carapito C., Hao Z., Huehmer A., Aebersold R. (2009) Electron transfer dissociation in conjunction with collision activation to investigate the drosophila melanogaster phosphoproteome. J Proteome Res 8, 2633–2639 [DOI] [PubMed] [Google Scholar]
- 45. Swaney D. L., Wenger C. D., Thomson J. A., Coon J. J. (2009) Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. L. (2009) Identification of protein o-glcnacylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boersema P. J., Mohammed S., Heck A. J. R. (2009) Phosphopeptide fragmentation and analysis by mass spectrometry. J Mass Spectrom. 44, 861–878 [DOI] [PubMed] [Google Scholar]
- 48. Payne S. H., Yau M., Smolka M. B., Tanner S., Zhou H., Bafna V. (2008) Phosphorylation-specific ms/ms scoring for rapid and accurate phosphoproteome analysis. J. Proteome Res. 7, 3373–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.