A given protein sequence can be encoded by an astronomical number of alternative nucleotide sequences. Recent research has revealed that this flexibility provides evolution with multiple ways to tune the efficiency and fidelity of protein translation and folding.
Keywords: codon usage, translation accuracy, translation efficiency, tRNA
Abstract
Proper functioning of biological cells requires that the process of protein expression be carried out with high efficiency and fidelity. Given an amino-acid sequence of a protein, multiple degrees of freedom still remain that may allow evolution to tune efficiency and fidelity for each gene under various conditions and cell types. Particularly, the redundancy of the genetic code allows the choice between alternative codons for the same amino acid, which, although ‘synonymous,' may exert dramatic effects on the process of translation. Here we review modern developments in genomics and systems biology that have revolutionized our understanding of the multiple means by which translation is regulated. We suggest new means to model the process of translation in a richer framework that will incorporate information about gene sequences, the tRNA pool of the organism and the thermodynamic stability of the mRNA transcripts. A practical demonstration of a better understanding of the process would be a more accurate prediction of the proteome, given the transcriptome at a diversity of biological conditions.
Introduction
Expression of genes is one of the most central molecular processes in living cells. Organisms invest a considerable amount of their resources, including energy, raw material and information bandwidth, to carry out the process, while optimizing efficiency, responsiveness and accuracy. During evolution, organisms evolved sophisticated means to achieve all of these goals and to balance between them when needed. Efficiency of gene expression consists of the throughput of the process on one hand and of its costs on the other (Dekel and Alon, 2005). The costs of the process are numerous and they consist of investment of building blocks and energy and allocation of cellular resources, such as the ribosomes and tRNAs (Stoebel et al, 2008). Accuracy can be described as the probability that the translated protein will be error free and match the sequence prescribed by the encoding gene sequence, in addition to the likelihood that it will fold properly within the cell (Drummond and Wilke, 2008; Zhou et al, 2009). The advent of modern genomics and systems biology has revolutionized our understanding of the diversity of molecular and systems-level mechanisms that control and optimize translation efficiency and accuracy (Arava et al, 2003; Dittmar et al, 2004; Lackner et al, 2007; Hendrickson et al, 2009; Ingolia et al, 2009).
The apparent redundancy of the genetic code, in which most of the amino acids can be translated by more than one codon, offers evolution the opportunity to tune the efficiency and accuracy of protein production to various levels while maintaining the same amino-acid sequence. The various codons that correspond to the same amino acid are often considered ‘synonymous,' yet their corresponding tRNAs might differ in their amounts in cells and thus also in the speed in which they will be recognized by the ribosome (Varenne et al, 1984; Sorensen et al, 1989). Also, the alternative nucleotide sequences of the various codon choices for a protein might give rise to transcripts with different secondary structure and stability, which may affect translation (Kudla et al, 2009) and even folding (Komar et al, 1999; Kimchi-Sarfaty et al, 2007). The number of alternative nucleotide sequences that could still code for the same protein is astronomical, leaving many degrees of freedom that evolution could use for achieving control without affecting the protein sequence. While the non-random usage of synonymous codons is often correctly assumed to reflect the action of neutral drift, in an increasing number of cases it now turns out to reflect the result of natural selection, perhaps mainly for tuning efficiency and accuracy of translation (Drummond and Wilke, 2008; Cannarozzi et al, 2010; Tuller et al, 2010a). The translation process is highly regulated by diverse structural elements and sequence motifs during each of the initiation, elongation and termination steps. Recent studies have enlightened our understanding of translational regulation, for both natural and stress conditions (Loh and Song, 2010; Spriggs et al, 2010). In this review, we will focus on the dissimilar, sometimes even opposite effect of different synonymous codons on both translation efficiency and accuracy.
Quantification of translation efficiency
During evolution, cells evolved means to tune the efficiency of translation of different genes to different desired levels. Some gene products are needed in higher amounts than others, while the expression of others, such as regulatory proteins tends to be low. Perhaps more challenging are genes that need to be translated at various levels in different conditions (Takagi et al, 2005; Lu et al, 2006; Ingolia et al, 2009). A more formal treatment of the question ‘what is the optimal level of expression of a given protein' suggests that the level should be such that the benefit due to expression of the gene should exceed the costs of its production at that level (Dekel and Alon, 2005). Evolving a genome-wide translation regulation regime thus amounts to determining the efficiency of translation of various genes at different conditions, cell types and tissues.
The various genes in the genome, depending on their sequence, might be more or less efficient in consuming the cellular resources of translation, including the ribosomes, the tRNAs, the aminoacyl tRNA synthetases, amino acids, translation factors and energy. A major challenge is to model and predict translation efficiency from the sequences of genes. A sign of success in the future would be the ability to predict protein abundances genome wide in various cell types and conditions.
Traditional computations of translation elongation efficiency (see Table I) may consider the mRNA coding sequence alone and may additionally include explicit inspection of the tRNA pool. Models of the first type, which measure the codon bias of genes—i.e., the non-random assignment of codons to amino acids—revealed decades ago that a striking correlation exists between codon usage and expression levels (Grantham et al, 1981; Bennetzen and Hall, 1982; Gouy and Gautier, 1982). In these models, genes that have a codon usage pattern reminiscent of selected ‘elite' highly expressed genes are likely to be highly expressed too. The most common index of this sort is the codon adaptation index, CAI (Sharp and Li, 1987). The CAI defines the relative adaptiveness of an individual codon encoding a given amino acid as the ratio of the codon's frequency in highly expressed genes to the frequency of the most abundant codon for that amino acid. The CAI for a gene is then calculated as the geometric mean of the relative adaptiveness values of all the codons along that gene.
Table 1. Traditional measures of translation elongation efficiency.
| Index name | The model by which translation efficiency of a gene is estimated | Properties of translation elongation efficiency measure |
|||
|---|---|---|---|---|---|
| Explicitly consider the tRNAs availability | Considers the effect of amino-acid composition | Discrimination between translation efficiency of individual codons | Complexitya of implementation for many species | ||
| aThe complexity of implementation is evaluated by the nature of the required input data. Trivially, all measures weight the number of occurrence of each of the 61 codons in the gene of interest. Additionally, the tAI measure requires the identification of all tRNA genes in the genome and their classification according to their anticodons, whereas the CAI measure requires a reference set of known highly expressed genes. The implementation of the Fop and CBI measures obligates a reference set of identified ‘optimal' or ‘preferred' codons, which are dominantly used in highly expressed genes, respectively. | |||||
| bThe measure classifies codons into only two categories. | |||||
| cThe score weights different patterns of distribution of synonymous codons. Yet, the values of two hypothetical genes that differ from each other by their amino-acid composition, but use only the most abundant codons, are identical. | |||||
| dCodons that do not appear in the reference set were assigned with a fixed frequency. | |||||
| The frequency of use of optimal codons, Fop (Ikemura, 1981) | The measure quantifies the fraction of optimal codons in a gene | Yes | No | Lowb | High |
| Codon Bias Index, CBI (Bennetzen and Hall, 1982) | Measure of the fraction of codon choices, which is biased to n preferred codons (relative to random usage of synonymous codons) | Yes | No | Lowb | High |
| The codon adaptation index, CAI (Sharp and Li, 1987) | The geometric mean of the ratios of the frequency of each codon in highly expressed genes to the frequency of its most abundant synonymous codon | No | Partiallyc | Partiallyd | Moderate |
| The ‘effective number of codons', Nc (Wright, 1990) | Measures the extent to which the codon usage of a gene departs from equal usage of synonymous codons | No | No | None | Very low |
| The tRNA Adaptation Index, tAI (dos Reis et al, 2004) | The geometric mean of the availability of the tRNAs that serve each codon | Yes | Yes | High | Low |
The second type of measures explicitly considers the tRNA pool, gauging the availability of tRNA at each codon along the gene. The correspondences between tRNA concentration and translation elongation speed are based on earlier observations, indicating that translation elongation rate is positively correlated with the tRNA concentrations of the translated codons (Varenne et al, 1984). In E. coli, codons corresponding to highly abundant tRNAs are translated as much as sixfold faster than their synonymous tRNA counterparts that occur at lower concentrations (Sorensen et al, 1989). Following early works (Ikemura, 1981; Ikemura and Ozeki, 1983), the tRNA Adaptation index, tAI (dos Reis et al, 2004) was developed. The tAI follows the mathematical model of the CAI, but it estimates the translation efficiency of a given gene by assessing the availability of the tRNAs that serve each codon rather than the codon usage itself. As tRNA levels are typically not readily measured, the amount of the different tRNAs in cells is often deduced from the copy number of the tRNA-coding genes in the genome. The usage of tRNA gene copy number as a proxy of tRNA abundance is supported by several observations (Dong et al, 1996; Percudani et al, 1997; Kanaya et al, 1999; Tuller et al, 2010a). When calculating the tAI, the tRNA availability of a given codon incorporates both the approximated tRNA levels of its fully-matched tRNA, as well as contributions from tRNAs that contribute to translation through Crick's wobble rules (Crick, 1966). An obvious advantage of the tAI over the CAI is that it alleviates the need to identify a priori the ‘elite' set of highly expressed genes as a reference. Instead, it only requires the identification of all tRNA genes in the genome and their classification according to their anti-codons. The tAI measure enables a convenient implementation for many species, and yet, its assumptions regarding the relative strength of imperfect codon–anticodon pairing should be further tuned (Ran and Higgs, 2010). Nonetheless, in studies in a collection of yeast species, both measures correlated highly with mRNA levels (Pearson's correlation 0.6–0.7) in a genome-wide survey (Man and Pilpel, 2007).
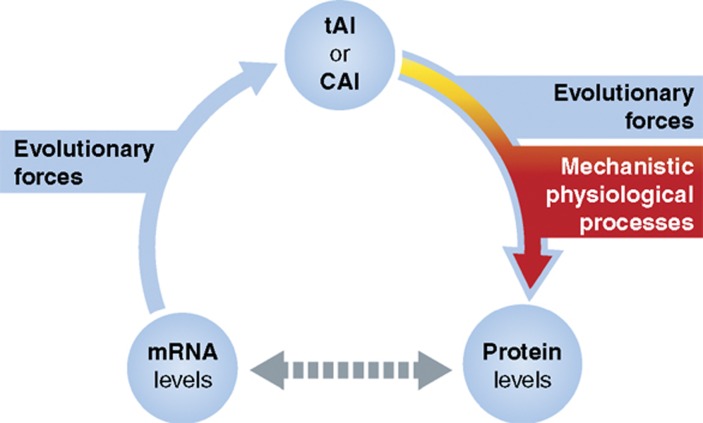
But should we expect tAI and CAI values of genes to correlate with the corresponding mRNA or protein abundances? To begin with, mRNA and protein abundances are often correlated between themselves (de Sousa Abreu et al, 2009; Vogel et al, 2010) so that any measure that correlates with one of them might show above-random levels of correlation with the other. Ideally, a measure of translation efficiency should correlate with the ratio of protein to mRNA level, and indeed the tAI has been shown to correlate with measures of this sort. In S. cerevisiae, the simple correlation between tAI and protein-to-mRNA ratio is very weak compared with the correspondence between tAI and mRNA levels, and yet it is still statistically significant (Pearson's correlation=0.123, P-value=1.47 × 10−9). The correlation between protein abundance and tAI, given the genes' mRNA levels, however, is higher (Pearson's partial correlation=0.38, P-value=8.54 × 10−81; Tuller et al, 2010b). Similarly, significant positive correlations were detected between tAI and protein levels for sets of yeast proteins having the same mRNA levels (Man and Pilpel, 2007). Furthermore, in S. cerevisiae, the contribution of codon choice to the variations in the mRNA–protein correlation remains of prime importance even where RNA decay and protein half-life are taken in consideration (Wu et al, 2008). Interestingly though, measures such as CAI and tAI have been shown (especially in unicellulars) to correlate with both mRNA and protein levels, yet probably due to completely different reasons (Figure 1). More intuitive is the correlation with protein levels—high CAI or tAI values for genes should increase translation efficiency and thus increase protein levels at a given mRNA level. Less intuitive is the correlation between mRNA levels and CAI or tAI. Non-optimal codon usage of genes can be detrimental to the cell as it will increase the sequestration of ribosomes during translation, while usage of preferred codons might optimize the allocation of ribosomes to certain genes (Andersson and Kurland, 1990; Kudla et al, 2009). The interesting point is that the weight of such effects depends on mRNA levels, so that wasteful sequestration of ribosomes on a low copy mRNA will have a minor effect on the cellular ribosomal pool. Thus, the evolutionary pressure to optimize the codons of genes should increase with their mRNA levels, thereby presumably creating the correlation between mRNA levels and measures such as CAI and tAI.
Figure 1.
mRNA levels have an evolutionary effect on translation efficiency, which in turn affects protein levels on a physiological timescale. The positive correlation between mRNAs level to measures of translation efficiency, such as CAI or tAI, might reflect an evolutionary pressure to optimize the codon usage of highly expressed mRNAs so as not to sequester too many ribosomes—the faster the elongation rate is, the shorter the time in which a ribosome is bound to any particular mRNA. The extent of evolutionary pressure to optimize a gene should thus positively correlate with its mRNA level. On the other hand, the positive correlation between translation efficiency measures and protein abundance probably acts on a much faster timescale, of mechanistic physiological processes, and it is also governed by evolutionary forces. The codon usage of proteins that are needed at high expression levels is adjusted to achieve high-translation efficiency at a given mRNA level. The significant correlation between the tAI and protein-to-mRNA ratio suggests the causal effect on protein levels.
Advanced challenges in assessing translation efficiency and accuracy
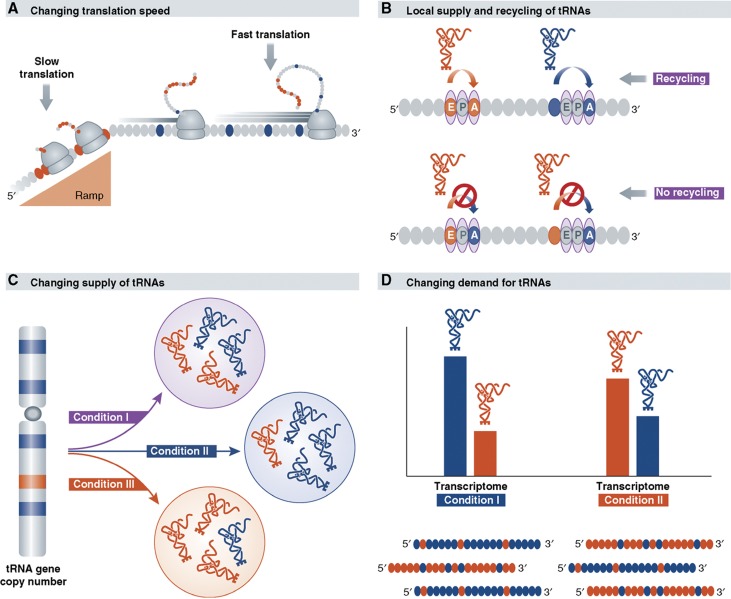
The tAI and the CAI measures predict gene expression with reasonable accuracy, yet alleviating some of the assumptions on which they are based might lead to more accurate models of translation efficiency (see Figure 2).
Figure 2.
Advanced challenges in assessing translation efficiency. New evidences challenge the common simplified assumptions in assessing translation efficiency. Shown in all sub-figures are two codon types, which may differ in their translation elongation efficiency, a ‘blue' and a ‘orange', served respectively by a ‘blue' and a ‘orange' types of tRNA. Some of the amino acids on the polypeptides are also colored blue or orange, reflecting the different efficiency of the codons that code for them. The following lines of further research into the mechanisms of translation are suggested: (A) The order of high- and low-efficiency codons (the later are colored in orange) is meaningful and can be utilized by evolution to design an optimal schedule for ribosomal flow on transcripts. In particular, the slow ‘ramp' observed in the 5′ end, especially of highly expressed genes, may avoid jamming of ribosomes once they passed it. (B) A local concentration of a tRNA molecule that was just released from the ribosome is high in the vicinity of the subsequent codons. Thus, although some tRNAs might be at low concentration over the entire cell volume, they might be present at relatively higher level in proximity of the codons they just finished translating. According to this possibility, the efficiency of translation of a codon depends also on whether that codon was used a few codons upstream on the same mRNA molecule. An indication for the mechanism might be that similar codons tend to cluster together on mRNA sequences. (C) Regulation of expression of the tRNAs could lead to dynamic changes in their availability in time or space dimensions, e.g., under various conditions, differential developmental stages, or at different tissues. (D) The efficiency of translation is a function of the ratio between the supply and the demand for each tRNA. The demand for different tRNAs, namely—the actual representation of the 61 codons at the transcriptome, might vary between different cell types, different environmental conditions and different time points along organism's life. Here, during the transition from condition I to II, the transcriptome changes from mainly consisting of genes that are rich in the blue codon to genes that more heavily biased towards the orange one; as a result the demand for the corresponding orange tRNA increases in the second condition.
First, we need to estimate the concentration of amino acid-loaded tRNAs. The life cycle of a tRNA molecule is complicated, it requires transcription, further processing including base modification and charging with amino acid. Recent measurements (Zaborske et al, 2009) are beginning to supply estimates on availability of ‘ready-to-translate' tRNAs and in general such abundance levels might deviate from the copy number of the tRNA genes, and even from just the concentration of the tRNA molecules in the cell. For example, amino-acid starvation differentially affects the charging levels of isoaccepting tRNA species, leading to wide variation in the sensitivity of the translation rate of individual codons to amino-acid deficiency (Sorensen, 2001; Elf et al, 2003).
Second, not only the global codon usage of a gene, but also the order of the high- and low-efficiency codons along the gene may affect translation efficiency. According to measures such as CAI and tAI, the order of high- and low-efficiency codons along the transcript is ignored. Recent analysis of multiple genomes revealed a trend in which the first approximately 30–50 codons in genes preferentially correspond to more rare tRNAs (Tuller et al, 2010a). Such genic sections form ‘low-efficiency ramps', which might deliberately attenuate the ribosome during early elongation. The authors showed that such a profile is particularly pronounced in highly expressed genes and, at least in yeast, it is inversely correlated with ribosomal density (experimentally measured by Ingolia et al (2009)). This correspondence with the experimentally measured ribosomal density data is an indication that the translation efficiency profile is probably a speed profile, aiming to control the rate of flow of the ribosomes by localizing an early traffic bottleneck (Figure 2A). It was proposed that such deliberate early attenuation enables a jam-free flow of ribosomes once they passed that region, thus reducing the probability of ribosome fall-off. Such a design could increase the productivity of expression while minimizing the costs of the process. This reasoning is consistent with indication of increasing selection against frameshifting errors towards the 3′ end of coding sequences (Huang et al, 2009).
Third, local pools of elevated availability of required tRNAs might promote translation elongation efficiency. An implicit assumption of traditional models such as tAI is that all codons utilize the same global tRNA pool. Surprisingly, a recent observation (Cannarozzi et al, 2010) implied that the availability of the same tRNAs might be different on different positions along the same mRNA (Figure 2B). This study showed that in subsequent occurrences of the same amino acids, genes tend to deliberately use codons that are translated by the same cognate tRNA. Similar to the ramp design, this trend was shown to be predominantly obeyed by rapidly induced genes, hinting that this is another means to boost translation efficiency. The authors hypothesized that codons at the ribosome A-site can utilize recycled tRNAs from the codons that were just translated. To further establish their hypothesis, they synthesized variants of the green fluorescent protein (GFP) gene in which the internal arrangement of synonymous codons either maximized or minimized the potential reuse of tRNAs from near-by position, and observed the expected increase or decrease in expression.
From a kinetic point of view this hypothesis is not trivial. First, it requires that the diffusion of the recycled tRNA will be slow enough compared to the rate of translation elongation. This situation may even necessitate or predict the existence of ‘local translation factories' nearby the ribosome, which will supply the re-charging services to the recycled tRNA. Studies indicating the capacity of aminoacyl–tRNA synthetases to interact with the ribosome (Kaminska et al, 2009) and reporting on colocalization of protein translation components (Barbarese et al, 1995) may serve as supported evidence.
Fourth, the tRNA pool might change dynamically rather than being constant (Figure 2C). According to the simplest models, the tRNA pool is assumed to remain constant throughout the life of a cell and in different cell types of the body. Yet measurements of the tRNA pool in different tissues and cell types showed interesting differences, suggesting that the same gene might be translated differently in each such environment (Dittmar et al, 2006). Similarly, in the transition from fermentation to respiration in yeast, the tRNA pool also seems to change (Tuller et al, 2010a). Likewise, the tRNA pool might change during development. The replacement of seven suboptimal codons by optimal ones in the ADH gene of Drosophila led to in vivo increase of its activity in third-instar larva, but in the adult flies it resulted in reduced activity of this gene (Hense et al, 2010). This result might reflect differences in tRNA pools between larvae and adult flies, though the authors consider additional possibilities.
Finally, the demand for the various tRNAs, presented by the transcriptome, might change dynamically too (Figure 2D). Presumably, the efficiency of translation is a function of the ratio between the supply and the demand for each tRNA. If a given tRNA is highly expressed, but the codons that correspond to that tRNA are highly represented in the transcriptome present at a given condition, then translation efficiency from that tRNA might be compromised in that condition. Interestingly, different codons do indeed fluctuate in their representation in the transcriptome at various conditions (H Gingold, Z Bloom, O Dahan and Y Pilpel, in preparation) emphasizing the need for parallel assessment of the representation of the codons in the transcriptome and the tRNA pool in a richer model of translation efficiency.
Challenging the above assumptions of the simple models may thus result in a more comprehensive model of translation efficiency. Such a richer model might not only improve protein level predictions, it might also explain tissue and condition variation in protein levels, the effects of mutations on translation efficiency, stochastic fluctuation in protein level and rapidity of expression response to signals and changes.
Evolutionary selection for codon—tRNA adaptation
What are the indications that genes were selected during evolution to optimize their translation efficiency? On the face of it one may ask ‘why not select for better translation efficiency even if it were to contribute only minutely to fitness?' The answer comes from population genetics that teaches us that traits are fixated in populations not only according to their fitness gain but also due to random drift caused by neutral mutations. In that respect, neutral mutations act like thermal noise in thermodynamic systems; they may prevent fixation of traits with positive, yet small fitness value. The effective population size (Hartl and Taubes, 1998) of a species determines how small the fitness value of a mutation can be while still allowing its fixation. Qualitatively, the rule is simple—the larger the species' effective population size, the higher the probability of fixation. The question of whether the genes in a genome are indeed subject to selective pressure to enhance translation efficiency is thus a priori open until rigorous criteria are met, and one would expect that while microbial species, with typically large population sizes, might manifest it, small effective population size species, such as human, might not (Bulmer, 1991; dos Reis and Wernisch, 2009).
As genomic data for coding sequences and measured levels of gene expression started accumulating, the indications of selective pressures for translational selection suggested by early evidences (Ikemura, 1985; Shields et al, 1988; Stenico et al, 1994; Moriyama and Powell, 1997) are becoming well established. A consistent trend of increased usage of codons that correspond to the most abundant tRNAs, especially in highly expressed genes, was detected in bacteria (Lithwick and Margalit, 2003). In yeast species it was found that entire gene modules, pathways and complexes might show coordinated selection for translation efficiency in some species, but not in others, depending on lifestyle needs. For instance, while genes belonging to fermentative pathways are codon-optimized in anaerobic species, respiratory genes show selection of optimal codons in aerobic yeasts (Man and Pilpel, 2007), and in related cases (Jiang et al, 2008). Selection for translation efficiency was shown also in some multicellulars such as C. elegans, D. melanogaster and Arabidopsis thaliana (Duret and Mouchiroud, 1999; Duret, 2000; Heger and Ponting, 2007; Drummond and Wilke, 2008). Yet, as expected from the above population theoretic arguments, attempts to demonstrate selection for translation efficiency in human, and to further correlate it with expression levels, yield contradictory results—reviewed in Chamary et al (2006). Some studies found no evidence for translational selection in human (Kanaya et al, 2001; dos Reis et al, 2004), suggesting that synonymous codons in human are not selected to maximize translation efficiency (Lercher et al, 2003). Conversely, other studies do indicate weak, yet significant, translational selection in human, according to estimates of codon usage adaptation to the global tRNA pool (Comeron, 2004; Lavner and Kotlar, 2005). Future related studies may further the exploration of tissue-specific expression patterns of tRNA isoaccpetors (Dittmar et al, 2006), and would ultimately be incorporated into more comprehensive measures of translation elongation efficiency.
Translational selection is also emerging in the context of adaptation between viruses and their hosts. Several studies showed codon bias in genes of bacteriophages towards their bacterial host codon bias (Sharp et al, 1984; Carbone, 2008; Lucks et al, 2008; Bahir et al, 2009), suggesting selection for efficient translation of the viral genes. Interestingly, the genomes of some viruses may contain a small selection of tRNA genes that might be added to the cellular tRNA pool and participate in translation upon infection. Why are such tRNA genes selected to be included in the typically very compact viral genome? A comprehensive analysis showed that the specific sets of viral-encoded tRNA genes were selected by the virus during evolution, presumably as they may boost translation efficiency of virus's own genes (Bailly-Bechet et al, 2007). An interesting possibility is that the viral tRNA genes might allow the virus to infect also hosts of a wide spectrum of codon usage, thus increasing the bandwidth of potential hosts, by alleviating the need to adapt precisely to the codon usage of each host separately.
Sequence-dependent determinants of translation-initiation rate
The overall speed of translation is determined by the rates of its three major steps—initiation, elongation and termination. The initiation step is regulated by a variety of structural elements and sequence motifs, some of which are uniquely associated with either prokaryotic or eukaryotic organisms (Kozak, 2005; Jackson et al, 2010). Such structural elements in eukaryotes are the 7-methylguanosine cap and the poly-(A) tail, which synergistically enhance translation-initiation efficiency (Gallie, 1991) via circularization of the mRNA, which in turn is mediated by interactions with eukaryotic-initiation factors (Tarun and Sachs, 1996; Kahvejian et al, 2005). In addition to a contribution of the 3′ end of the transcript to initiation, binding and assembly of the ribosome for a round of translation is governed by the sequence and the mRNA secondary structure in the vicinity of the start codon. In prokaryotes, ribosome binding occurs at the purine-rich Shine-Delgarno (SD) sequence (Shine and Dalgarno, 1974), located a few nucleotides upstream from the start codon, which is complementary to a sequence near the 3′ end of 16S rRNA (Steitz and Jakes, 1975; Jacob et al, 1987). In eukaryotes, translation initiation follows a scanning mechanism of the mRNA by the ribosome. The 40S ribosomal subunit enters at the 5′ end of the mRNA and migrates linearly until it encounters the first AUG codon (Kozak, 2002). The ribosome will initiate that first AUG codon if it is flanked by a short sequence motif, known as ‘Kozak sequence' (Kozak, 1986).
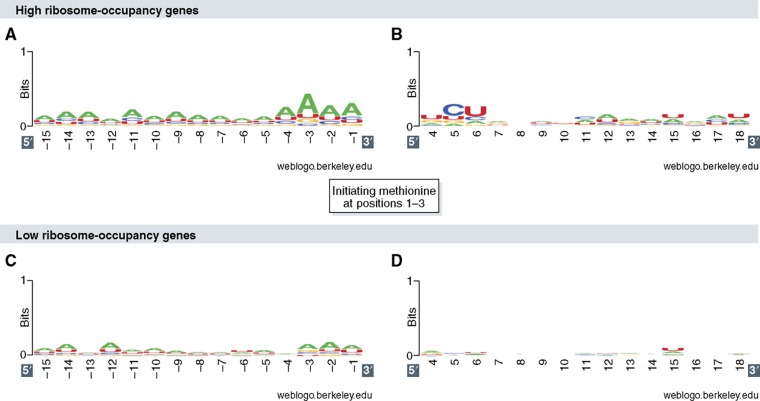
An important question is whether different variations on the sequence motif in the vicinity of the translation start site are associated with, and perhaps even determining, difference in translation-initiation efficiency. It was previously shown that the 5′ untranslated sequence of yeast mRNAs is rich in A-residues, and that highly expressed genes commonly use the Serine UCU codon as second triplet in the open-reading frame (Hamilton et al, 1987). More recently, using data on genome-wide ribosome density (Ingolia et al, 2009), Robbins-Pianka et al (2010) reported on reduced predicted secondary structure in 5′ UTRs, especially in high ribosome-density genes in yeast. Genome-wide measurements of occupancy and density of ribosomes on mRNA enable us to systematically examine how sequence in the vicinity of the initiation site may affect initiation efficiency. Figure 3 shows a sequence motif logo of the sequence flanking the AUG start codon for two sets of S. cerevisiae genes—low ribosome-occupancy genes and high ribosome-occupancy genes, based on Arava's analysis of ribosome occupancy (Arava et al, 2003). Clearly, high ribosome-occupancy genes show a motif with moderate information content, whereas the low ribosome-occupancy motif shows little or no consensus. Specifically, the analysis shows the preferred usage of the A nucleotide along the 15 positions upstream to the start codon, and in particularly at positions −4 to −1, in high ribosome-occupancy genes. This analysis suggests a hierarchy between genes in the fit of their 5′ UTR sequences to a canonical-initiation motif, which may determine the relative initiation efficiency of each gene in the genome. In addition, for high-occupancy genes, the sequence logo shows a pointed elevated usage of nucleotides C and U, in the 5th and 6th positions in the open-reading frame. Interestingly, the second codon position shows elevated tAI values on average (Tuller et al, 2010a) suggesting a selection for high-translation efficiency for efficient release and recycling of the initiator methionine tRNA. Indeed, this signal is more pronounced in genes with high ribosome occupancy compared with genes with low occupancy (H Gingold and Y Pilpel, unpublished data, 2011).
Figure 3.
Sequence motifs in the vicinity of the initiation site and ribosome occupancy. The figure displays sequence motif logos of the sequence spanning between positions −15 and +18 relative to the initiating AUG for two yeast genes sets—high ribosome-occupancy genes and low ribosome-occupancy genes (Arava et al, 2003). The sequence logos show an interesting signature of enrichment in Adenine nucleotides upstream to the initiating AUG codon in genes with high ribosome occupancy (A), accompanied with particular nucleotide preference at positions +5 and +6 (B). The 5′ UTR sequence of low ribosome-occupancy genes is also enriched with Adenine nucleotides (C), yet to a much lower extent. Genes with low ribosome occupancy show no nucleotide preference downstream to the initiating AUG codons (D). For this display, high ribosome-occupancy and low ribosome-occupancy genes (204 and 206 genes, respectively) were defined as genes at the top and at the bottom of the ribosome-occupancy distribution (occupancy>0.85, or occupancy<0.6 correspondingly). The 5′ UTR sequences of the investigated genes were derived from the study by Nagalakshmi et al (2008); the coding regions were downloaded from SGD web site. Sequence logos were created using WebLogo (Crooks et al, 2004).
Association between mRNA folding and translation rate
The mRNA molecules in the cell often assume a secondary and a tertiary structure that might be tight for some genes, and loose for others. For translation to proceed, such structure must be threaded through the ribosome. Here is thus another opportunity to regulate and induce wide variation in translation efficiency of genes—the tightness of their mRNA structure might control both the ribosome binding and the rate of its flow across them. Early evidences indicate that the stability of base pairing at the ribosome-binding site or in its vicinity is a major determinant of translation-initiation efficiency in prokaryotes (Schauder and McCarthy, 1989). In eukaryotic organisms, tight secondary structures along the 5′ UTR were shown to reduce translation efficiency, especially if they are located in proximity to the translation start site, presumably by obstructing ribosome binding (Wang and Wessler, 2001).
The effect of mRNA structure on translation was traditionally deciphered by inspecting natural genes from various genomes (Jia and Li, 2005). Now, synthetic biology may to complement this picture by allowing researchers to manipulate one property of a gene, while keeping many others constant. Recently, Kudla et al (2009) provided a good example for this modern trend by synthesizing a library of 154 GFP genes that varied randomly at synonymous sites, while encoding the same amino-acid sequence. They expressed the GFP genes in E. coli, and detected 250-fold variation in expression levels. They found that tight structure at the 5′ end of the mRNA inhibits translation, whereas loose structures promote it. These results are consistent with the notion that the initiation step is of prime importance in determining gene expression levels. In prokaryotes, ribosome binding occurs at the SD sequence (Shine and Dalgarno, 1974) located upstream from the start codon. Interestingly, it was shown before that masking of the initiation site by tight secondary structure can be offset by a stronger-than-normal SD interaction (de Smit and van Duin, 1994; Olsthoorn et al, 1995). As Kudla et al (2009) only varied the coding region of GFP, this possibility was not tested in their recent study.
The association between the stability of secondary structures in the translation-initiation region and translation efficiency is further supported by large-scale computational analysis (Gu et al, 2010), indicating a genome-wide trend of reduced mRNA stability near the start codon for both prokaryotic and eukaryotic species. Here too the trend was found to be enhanced among highly expressed genes, suggesting an effect of translation efficiency.
Determining the overall rate of translation: one key factor or a ‘combination lock'?
While it is widely accepted that mRNA folding and codon–anticodon adaptation are the key factors in the determination of initiation and elongation rates, respectively, the identity of the rate-limiting step of the overall translation efficiency remains controversial. Surprisingly, and in contradiction to many studies of natural genes, Kudla et al (2009) indicate that the variation in protein expression levels in the GFP library is not derived at all from codon bias differences (measured by the Codon Adaption Index). They proposed instead that the mRNA folding at the beginning of the transcript has the predominant role in shaping expression level of individual genes, whereas selection for codon bias aims to increase the global rate of protein synthesis by reducing the ribosomes sequestering on the mRNA. A related study inspected E. coli and S. cerevisiae and found similar trends of relatively loose secondary structure stability near 5′ ends of genes (Tuller et al, 2010b). The authors investigated the interplay between folding energy and codon bias in determining translation efficiency across all the genes of E. coli and S. cerevisiae. Unlike the results obtained by Kudla et al (2009) for synthetic genes, Tuller et al (2010b) observed a significant correlation between codon bias and protein abundance (normalized to mRNA level), but no direct correlation between folding energy and protein abundance. These authors did find, however, that the strength of association between codon bias and protein expression is modulated by folding energy. Part of the reason for this apparent discrepancy between the natural and synthetic genes was suggested to be the different distribution of folding energy values between the two gene sets (Tuller et al, 2010b).
Future studies will probably investigate the separate contribution of the diverse determinants of translation efficiency to the overall rate of translation. Such an analysis was carried out for the Desulfovibrio vulgaris bacteria, aiming to assess the contribution of sequence features associated with the initiation, elongation and termination steps to the variation in mRNA–protein correlation (Nie et al, 2006). Ideally, such studies will take into consideration in vivo estimation of mRNA decay and protein degradation as potential confounding factors. This reasoning is consistent with recent studies indicating for higher conservation of protein abundance than mRNA levels across different species, hence implying for major role of either translational or protein degradation control in maintaining proteins in desired levels (Schrimpf et al, 2009; Laurent et al, 2010).
An important challenge is to appropriately consider features in the mRNA that affect translation. For example, in addition to its prime effect on ribosome binding and initiation, the secondary structure of mRNA governs the movement of the ribosome during elongation too, suggesting a broader effect of mRNA structure on translation (Wen et al, 2008). In that respect, modern investigations broaden the scope of the classical ribosome attenuation model that was originally described as a mechanism relevant to amino-acid biosynthetic genes only (Yanofsky, 1981).
It is interesting to note the difference between the expressions of natural genes in their natural genome compared to man-made heterologous expression systems, in which one often expresses a gene from one species in another species. In both cases, the need to optimize expression of a given protein often arises, but beyond that some of the actual considerations might be very different. A native gene in its natural genome can be highly expressed but only to the extent that the benefit from the gene will not exceed the costs associated with its production. Some of the costs are direct, e.g., consumption of raw material and energy, and some are indirect, e.g., sequestration of the gene expression apparatus. Thus, even the most highly expressed genes in a natural context must be ‘considerate' of the rest of the genes in the genome. The situation could be different in artificial systems, especially in the biotechnology context in which a more ‘selfish-gene' approach could be justified. Here high expression of a gene in a host may be justified even if overall fitness of the host cell is significantly compromised, as long as the system is economically cost-effective. Another prime difference is that heterologous systems often reach very high expression levels, much beyond even highly expressed genes in their natural genomes. The design considerations of the genes' sequence and their interaction with the cellular machinery in the two cases might thus be very different. We anticipate that future studies will expand upon existing attempts to design nucleotide sequences (given amino-acid sequence constraints) that optimize either fitness of the host or productivity of a given desired protein (Kudla et al, 2009; Welch et al, 2009; Navon and Pilpel, 2011).
Codon choice may affect translation fidelity
So far we have discussed the effect of codon choice and mRNA structure on the throughput of translation, but these parameters could also govern the fidelity and accuracy of the process. In the stochastic search for the right tRNA, the ribosome might incorrectly bind a tRNA with a one base-mismatch relative to the codon, often termed ‘near-cognate tRNA' (tRNAs with more than one base-mismatch relative to the codon typically do not pass the initial screen; Rodnina and Wintermeyer, 2001). If a near-cognate tRNA binds to the A-site of the ribosome, the wrong amino acid might be incorporated, creating a ‘missense translational error'. The frequency of such translation errors in vivo was estimated to be 10−5 in yeast cells (Stansfield et al, 1998), but more recent measurements in B. subtilis showed a surprisingly high rate of 10−2 (Meyerovich et al, 2010). Missense errors can also be caused by erroneously charged tRNAs, with an overall error rate of 1 per 10 000 (Ibba and Soll, 2000). Missense errors that might disrupt protein function impose metabolic costs of wasted synthesis; if the loss of function is accompanied with improper folding, the damage might be even more pronounced. The misfolded protein may interact with other cellular components, causing protein aggregation (Bucciantini et al, 2002), disruption of membrane integrity (Stefani and Dobson, 2003) and it may ultimately result in cell dysfunction and disease—reviewed in Gregersen, 2006.
Translation can thus be thought of in terms of a competition process between the cognate and near-cognate tRNAs for a given codon, where the higher the concentration of correct tRNAs, the lower the probability of binding the wrong ones. Indeed in E. coli, the frequency of missense errors is diminished by ninefold if the same amino acid is translated by a codon that corresponds to an abundant tRNA rather than a low-abundance one (Precup and Parker, 1987).
The association between selection on synonymous site and translation accuracy was quantitatively examined for the first time by Akashi (1994). Akashi (1994) showed higher frequencies of preferred codons in evolutionarily conserved amino-acid positions among Drosophila species. Comparing only 38 orthologous genes among fly species, Akashi (1994) found that the frequency of preferred codons is significantly higher at conserved amino-acid positions compared with non-conserved ones. Akashi (1994) thus suggested that selection favors optimal codons at sites where misincorporations are most likely to disrupt protein functions. This type of pioneering analysis was later applied in the full genome era to E. coli (Stoletzki and Eyre-Walker, 2007), yeast, worm, mouse and human (Drummond and Wilke, 2008), verifying the significant association between optimal codons and evolutionary conservation, supporting Akashi's early notion that in the very same positions where evolution conserved the amino acid against DNA replication mutations it also insisted on the preferred codons that would minimize the chance for translation errors. Drummond and Wilke (2008) carried out molecular-level evolutionary simulation of the effects of misfolding due to translation errors on fitness. They concluded that selection acts on translation accuracy, but only if misfolding imposes a direct fitness cost. Their study suggested that selection for translation accuracy, although intuitively associated with production of functional proteins, might mainly be derived by the need to globally prevent the toxic consequences of misfolding errors. Selection against misfolding errors were further shown to not only associate with the usage of preferred codons but also with preference of misfolding-minimizing amino acids (Yang et al, 2010).
Selection pressure against misfolding is directly supported by studies that focus on structurally sensitive sites, where mutations are highly disruptive. Buried amino-acid residues were shown to be preferentially encoded by more optimal codons compared with solvent-exposed residues (Zhou et al, 2009). This is consistent with evidences for higher sensitivity of protein core residues, compared with surface residues, to mutations that occur during DNA replication (Tokuriki et al, 2007). The hypothesis of selection against mistranslation-induced protein misfolding is further sustained by a very different and yet complementary approach (Warnecke and Hurst, 2010). These authors demonstrated coordinated utilization of cis-acting (preferred codons) and trans-acting (molecular chaperons) elements as a strategy for misfolding prevention. They show that proteins, which attain their native structure spontaneously, or at least without the aid of the bacterial chaperonin GroEL, are enriched with preferred codons at structurally sensitive sites, compared with proteins that need the chaperonin for folding. The study thus suggests that the chaperonin alleviates the need to optimize codons as a means to prevent translation-mediated misfolding. Further, in the context of translation accuracy, selection pressures on synonymous sites also appear to act against frameshifting errors (Farabaugh and Bjork, 1999), and to reduce the cost of nonsense errors (Gilchrist et al, 2009).
But ‘errors' are sometimes beneficial, and the ability to introduce them when needed may have even been selected for. A striking recent example showed that under certain stresses, a ‘programmed translation error' may occur, which leads to increased misincorporation of methionine residues into the mammalian proteome (Netzer et al, 2009). Unlike the misincorporation errors discussed above, this phenomenon appears to feature elevation in misacylation of Met residues in non-Met tRNAs. This observation is striking because methionine has a radical oxygen-protective capacity and sure enough operates predominantly under oxidative stress.
The strategic role of the rare: advantageous usage of disadvantageous codons
In the previous sections we described the benefits associated with the usage of codons that correspond to abundant tRNAs—such codons may enhance the speed and accuracy of the translation elongation step. However, it is of interest to understand whether codons which belong to the opposite side of the scale, namely, codons that correspond to the least abundant tRNAs, are also preferred in selected cases, or whether their usage is simply the outcome of the absence of selection for abundant codons (Sharp and Li, 1986). High frequencies of rare codons in lowly expressed genes were observed in many genomes, including human (Lavner and Kotlar, 2005). Rare codons have the potential to slow down the translation elongation rate (Pedersen, 1984), due to the relatively long dwell time of the ribosome in its search for rare tRNAs. Several studies suggest that gene-wide codon bias in favor of slowly translated codons serves as a regulatory means to obtain low expression levels of protein when desired, for example, in the case of regulatory genes, or where excess of the protein appears to be detrimental or lethal to the cell (Konigsberg and Godson, 1983; Zhang et al, 1991). The level of protein secondary structure was also found to be associated with codon usage. Particularly, it was found that fast folding α-helical sequences are preferentially encoded by fast codons, whereas slower folding β-sheets strands, loops and disordered structures are enriched with rare (slow) codons (Thanaraj and Argos, 1996a).
More subtle are the cases in which only specific regions within a gene might be strategically selected to feature slow codons. For example, choice of slow codons was suggested to affect co-translational folding—reviewed in Tsai et al, 2008. A simple model suggests that the strategic usage of rare codons provides a pause during translation, during which an already translated segment of a protein may be folded in the absence of an otherwise potentially interfering segment that is not yet translated (Komar et al, 1999; Tsai et al, 2008). Supporting this notion is a study in which 16 consecutive rare codons in a gene were replaced by synonymous optimal ones in E. coli. Although the optimal codons enhanced the translation speed, they appear to have reduced folding as deduced by a 20% decrease in the encoded enzyme's specific activity (Komar et al, 1999). Such a manipulation in another gene of E. coli resulted in elevated in vivo misfolding and aggregation rates (Cortazzo et al, 2002). A small and yet significant similar effect was also obtained in yeast in a similar experiment (Crombie et al, 1992, 1994). Removal of translational attenuation sites in the bacterial SufI gene by an alternative approach, in which a global increase of the translation rate was obtained by adding a large excess of naturally rare tRNAs, also resulted in perturbed folding (Zhang et al, 2009). The hypothesis that rare codons are employed to temporally separate the synthesis of defined portions of the protein is consistent with the observation that boundaries between domains—proteins' independent folding modules—are enriched with clusters of rare codons (Thanaraj and Argos, 1996b).
In the last decade, the awareness of the fascinating biology of intrinsically unstructured proteins has grown significantly (Gsponer et al, 2008). The function of such proteins often depends on them being unstructured, and hence there have been extensive computational (Uversky et al, 2000) and experimental (Tsvetkov et al, 2008) efforts to identify such proteins genome wide. Common to such attempts is the search for signals in the protein amino-acid sequence that determine its lack of structure. A plausible hypothesis is that obtaining an unfolded structure also requires instructions from the nucleotide sequence, and in particular that coupled translation-folding determines unstructureness. Could it be that the strategic choice of certain codons, e.g., fast codons in domain boundaries, can actually serve to reverse the above-mentioned folding-promoting design, so that a protein will be unfolded? In general, is there a code of translation efficiency that is needed to create an unfolded protein? Can the effect of codon choice on folding pathways be simply referred to as either ‘beneficial' or ‘deleterious?' The answer is probably ‘no.' A naturally occurring mutation in the human MDR1 gene, involving a synonymous rare-to-frequent codon substitution, led to slight alternation in the native tertiary structure of the protein and subsequent change in its substrate specificity (Kimchi-Sarfaty et al, 2007). The wide potential impact of the co-translational folding timing is further manifested by a recent observation that codon usage might affect post-translation modification and folding, and as a consequence the stability of a protein due to a forced choice between ubiquitination and an alternative modification (Zhang et al, 2010). More generally, an interesting possibility is that proper post-translation modification of proteins, which sometimes takes place during the ‘pioneering round of translation' while the nascent chain emerges from the ribosome, may require a certain optimal tempo of translation. We may thus anticipate that some modifications, including myristylation that occur co-translationally (Wilcox et al, 1987) or others such as glycosylation, may require a certain rate of translation in their vicinity. Thus, the nucleotide sequence that codes for the protein, and not only its amino-acid sequence, may determine the modifications. In that respect it is interesting to note that highly predictive amino-acid motifs for some modifications remains elusive, and it might thus be that inclusion of nucleotide sequence information may facilitate the distinction between functional and non-functional post-translation modification sites.
Summary
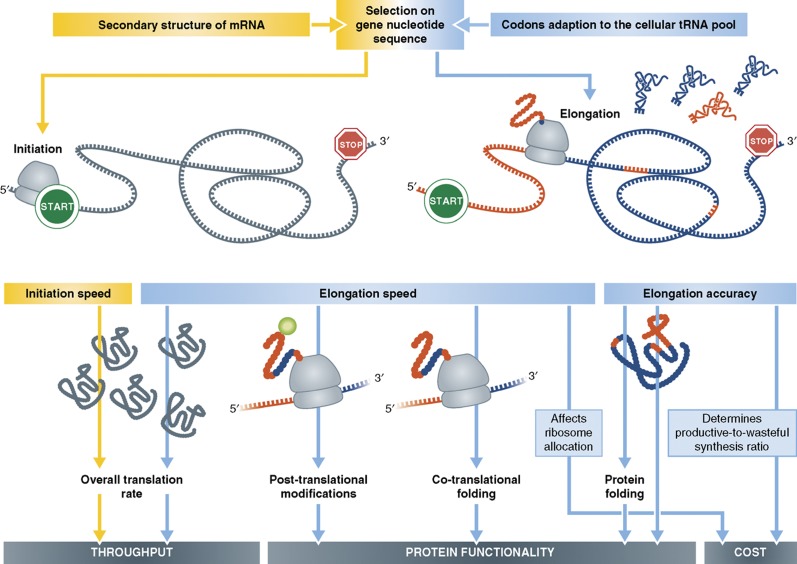
In this review, we discuss in detail the implication of selection on synonymous site to translation properties. An overall view of the effect of codon choice on gene expression is shown in Figure 4. In summary, our understanding of the process of translation has been revolutionized in the genome and systems biology era. Two important characteristics of the process, its efficiency and its fidelity, are now understood much better than just a few years ago. Still, the challenges ahead will be to integrate all of the knowledge and insight that has accumulated from these various studies, and create a consistent model of the translation process that will predict the proteome under various conditions and cell types. Such a model will greatly enhance our understanding of genomes and cellular circuits, will help to elucidate the basis of cell-to-cell variation and will shed light on the molecular basis of diseases.
Figure 4.
The predominant effect of selection on synonymous site on gene expression. Synonymous codons correspond to the same amino acid and yet might differ from each other by their adaptation to the cellular tRNA pool and also by their contribution to the secondary structure and the stability of the transcript. The effect of each attribute on translation properties and the further consequences on gene expression is marked with pale blue (for codon–anticodon adaptation) or yellow (for mRNA folding).
Current points of debate have to do with the relative role of codon choice and mRNA structure in affecting translation, the relative contribution of control at the level of translation initiation versus elongation, the relative extent of selection for efficiency versus accuracy and the role of random drift versus selection in shaping genes sequence. Even further, translation itself constitutes only one of several steps in the gene expression process, and gene expression as a whole poses only part of the constraints that genes' sequences must obey. The same nucleotide should also support other features such as nucleosome positioning, appropriate splicing (Warnecke et al, 2009) and higher order structural elements of the DNA. The apparent redundancy of the genetic code hence facilitates a choice between an astronomical number of coding possibilities of a given amino-acid sequence and may thus facilitate the coordinated satisfaction of many constraints, in addition to translation efficiency, by the same sequence.
Acknowledgments
We thank the European Research Council for an ‘ERC Ideas' grant, and the Ben May Foundation for continuous support.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akashi H (1994) Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG (1990) Codon preferences in free-living microorganisms. Microbiol Rev 54: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahir I, Fromer M, Prat Y, Linial M (2009) Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol Syst Biol 5: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M, Vergassola M, Rocha E (2007) Causes for the intriguing presence of tRNAs in phages. Genome Res 17: 1486–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH (1995) Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci 108 (Part 8): 2781–2790 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD (1982) Codon selection in yeast. J Biol Chem 257: 3026–3031 [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416: 507–511 [DOI] [PubMed] [Google Scholar]
- Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y (2010) A role for codon order in translation dynamics. Cell 141: 355–367 [DOI] [PubMed] [Google Scholar]
- Carbone A (2008) Codon bias is a major factor explaining phage evolution in translationally biased hosts. J Mol Evol 66: 210–223 [DOI] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD (2006) Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet 7: 98–108 [DOI] [PubMed] [Google Scholar]
- Comeron JM (2004) Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics 167: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazzo P, Cervenansky C, Marin M, Reiss C, Ehrlich R, Deana A (2002) Silent mutations affect in vivo protein folding in Escherichia coli. Biochem Biophys Res Commun 293: 537–541 [DOI] [PubMed] [Google Scholar]
- Crick FH (1966) Codon—anticodon pairing: the wobble hypothesis. J Mol Biol 19: 548–555 [DOI] [PubMed] [Google Scholar]
- Crombie T, Boyle JP, Coggins JR, Brown AJ (1994) The folding of the bifunctional TRP3 protein in yeast is influenced by a translational pause which lies in a region of structural divergence with Escherichia coli indoleglycerol-phosphate synthase. Eur J Biochem 226: 657–664 [DOI] [PubMed] [Google Scholar]
- Crombie T, Swaffield JC, Brown AJ (1992) Protein folding within the cell is influenced by controlled rates of polypeptide elongation. J Mol Biol 228: 7–12 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit MH, van Duin J (1994) Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J Mol Biol 235: 173–184 [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C (2009) Global signatures of protein and mRNA expression levels. Mol Biosyst 5: 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592 [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T (2006) Tissue-specific differences in human transfer RNA expression. PLoS Genet 2: e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Mobley EM, Radek AJ, Pan T (2004) Exploring the regulation of tRNA distribution on the genomic scale. J Mol Biol 337: 31–47 [DOI] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L (2004) Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res 32: 5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Wernisch L (2009) Estimating translational selection in eukaryotic genomes. Mol Biol Evol 26: 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L (2000) tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet 16: 287–289 [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D (1999) Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci USA 96: 4482–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M (2003) Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300: 1718–1722 [DOI] [PubMed] [Google Scholar]
- Farabaugh PJ, Bjork GR (1999) How translational accuracy influences reading frame maintenance. EMBO J 18: 1427–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR (1991) The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5: 2108–2116 [DOI] [PubMed] [Google Scholar]
- Gilchrist MA, Shah P, Zaretzki R (2009) Measuring and detecting molecular adaptation in codon usage against nonsense errors during protein translation. Genetics 183: 1493–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Gautier C (1982) Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res 10: 7055–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R (1981) Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res 9: r43–r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen N (2006) Protein misfolding disorders: pathogenesis and intervention. J Inherit Metab Dis 29: 456–470 [DOI] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM (2008) Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322: 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhou T, Wilke CO (2010) A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol 6: e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton R, Watanabe CK, de Boer HA (1987) Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res 15: 3581–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Taubes CH (1998) Towards a theory of evolutionary adaptation. Genetica 102–103: 525–533 [PubMed] [Google Scholar]
- Heger A, Ponting CP (2007) Variable strength of translational selection among 12 Drosophila species. Genetics 177: 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO (2009) Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense W, Anderson N, Hutter S, Stephan W, Parsch J, Carlini DB (2010) Experimentally increased codon bias in the Drosophila Adh gene leads to an increase in larval, but not adult, alcohol dehydrogenase activity. Genetics 184: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Koonin EV, Lipman DJ, Przytycka TM (2009) Selection for minimization of translational frameshifting errors as a factor in the evolution of codon usage. Nucleic Acids Res 37: 6799–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Soll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69: 617–650 [DOI] [PubMed] [Google Scholar]
- Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol 151: 389–409 [DOI] [PubMed] [Google Scholar]
- Ikemura T (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2: 13–34 [DOI] [PubMed] [Google Scholar]
- Ikemura T, Ozeki H (1983) Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol 47 (Part 2): 1087–1097 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob WF, Santer M, Dahlberg AE (1987) A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci USA 84: 4757–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M, Li Y (2005) The relationship among gene expression, folding free energy and codon usage bias in Escherichia coli. FEBS Lett 579: 5333–5337 [DOI] [PubMed] [Google Scholar]
- Jiang H, Guan W, Pinney D, Wang W, Gu Z (2008) Relaxation of yeast mitochondrial functions after whole-genome duplication. Genome Res 18: 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N (2005) Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska M, Havrylenko S, Decottignies P, Le Marechal P, Negrutskii B, Mirande M (2009) Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J Biol Chem 284: 13746–13754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T (2001) Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol 53: 290–298 [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kudo Y, Ikemura T (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238: 143–155 [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM (2007) A ‘silent' polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528 [DOI] [PubMed] [Google Scholar]
- Komar AA, Lesnik T, Reiss C (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 462: 387–391 [DOI] [PubMed] [Google Scholar]
- Konigsberg W, Godson GN (1983) Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci USA 80: 687–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292 [DOI] [PubMed] [Google Scholar]
- Kozak M (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene 299: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37 [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent JM, Vogel C, Kwon T, Craig SA, Boutz DR, Huse HK, Nozue K, Walia H, Whiteley M, Ronald PC, Marcotte EM (2010) Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics 10: 4209–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavner Y, Kotlar D (2005) Codon bias as a factor in regulating expression via translation rate in the human genome. Gene 345: 127–138 [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Pavlicek A, Hurst LD (2003) A unification of mosaic structures in the human genome. Hum Mol Genet 12: 2411–2415 [DOI] [PubMed] [Google Scholar]
- Lithwick G, Margalit H (2003) Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res 13: 2665–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PG, Song H (2010) Structural and mechanistic insights into translation termination. Curr Opin Struct Biol 20: 98–103 [DOI] [PubMed] [Google Scholar]
- Lu X, de la Pena L, Barker C, Camphausen K, Tofilon PJ (2006) Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res 66: 1052–1061 [DOI] [PubMed] [Google Scholar]
- Lucks JB, Nelson DR, Kudla GR, Plotkin JB (2008) Genome landscapes and bacteriophage codon usage. PLoS Comput Biol 4: e1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man O, Pilpel Y (2007) Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet 39: 415–421 [DOI] [PubMed] [Google Scholar]
- Meyerovich M, Mamou G, Ben-Yehuda S (2010) Visualizing high error levels during gene expression in living bacterial cells. Proc Natl Acad Sci USA 107: 11543–11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR (1997) Codon usage bias and tRNA abundance in Drosophila. J Mol Evol 45: 514–523 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon S, Pilpel Y (2011) The role of codon selection in regulation of translation efficiency deduced from synthetic libraries. Genome Biol 12: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, Embry A, Dolan B, Das S, Hickman HD, Berglund P, Bennink JR, Yewdell JW, Pan T (2009) Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462: 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W (2006) Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics 174: 2229–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn RC, Zoog S, van Duin J (1995) Coevolution of RNA helix stability and Shine-Dalgarno complementarity in a translational start region. Mol Microbiol 15: 333–339 [DOI] [PubMed] [Google Scholar]
- Pedersen S (1984) Escherichia coli ribosomes translate in vivo with variable rate. EMBO J 3: 2895–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Pavesi A, Ottonello S (1997) Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol 268: 322–330 [DOI] [PubMed] [Google Scholar]
- Precup J, Parker J (1987) Missense misreading of asparagine codons as a function of codon identity and context. J Biol Chem 262: 11351–11355 [PubMed] [Google Scholar]
- Ran W, Higgs PG (2010) The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol Biol Evol 27: 2129–2140 [DOI] [PubMed] [Google Scholar]
- Robbins-Pianka A, Rice MD, Weir MP (2010) The mRNA landscape at yeast translation initiation sites. Bioinformatics 26: 2651–2655 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Schauder B, McCarthy JE (1989) The role of bases upstream of the Shine-Dalgarno region and in the coding sequence in the control of gene expression in Escherichia coli: translation and stability of mRNAs in vivo. Gene 78: 59–72 [DOI] [PubMed] [Google Scholar]
- Schrimpf SP, Weiss M, Reiter L, Ahrens CH, Jovanovic M, Malmstrom J, Brunner E, Mohanty S, Lercher MJ, Hunziker PE, Aebersold R, von Mering C, Hengartner MO (2009) Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol 7: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH (1986) Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare' codons. Nucleic Acids Res 14: 7737–7749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH (1987) The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Rogers MS, McConnell DJ (1984) Selection pressures on codon usage in the complete genome of bacteriophage T7. J Mol Evol 21: 150–160 [DOI] [PubMed] [Google Scholar]
- Shields DC, Sharp PM, Higgins DG, Wright F (1988) ‘Silent' sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol 5: 704–716 [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L (1974) The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA 71: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MA (2001) Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(−) strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J Mol Biol 307: 785–798 [DOI] [PubMed] [Google Scholar]
- Sorensen MA, Kurland CG, Pedersen S (1989) Codon usage determines translation rate in Escherichia coli. J Mol Biol 207: 365–377 [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE (2010) Translational regulation of gene expression during conditions of cell stress. Mol Cell 40: 228–237 [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Herbert P, Lewendon A, Shaw WV, Tuite MF (1998) Missense translation errors in Saccharomyces cerevisiae. J Mol Biol 282: 13–24 [DOI] [PubMed] [Google Scholar]
- Stefani M, Dobson CM (2003) Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med 81: 678–699 [DOI] [PubMed] [Google Scholar]
- Steitz JA, Jakes K (1975) How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci USA 72: 4734–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenico M, Lloyd AT, Sharp PM (1994) Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res 22: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebel DM, Dean AM, Dykhuizen DE (2008) The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178: 1653–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A (2007) Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol 24: 374–381 [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123: 49–63 [DOI] [PubMed] [Google Scholar]
- Tarun SZ Jr, Sachs AB (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15: 7168–7177 [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P (1996a) Protein secondary structural types are differentially coded on messenger RNA. Protein Sci 5: 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P (1996b) Ribosome-mediated translational pause and protein domain organization. Protein Sci 5: 1594–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS (2007) The stability effects of protein mutations appear to be universally distributed. J Mol Biol 369: 1318–1332 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, Nussinov R (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol 383: 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov P, Asher G, Paz A, Reuven N, Sussman JL, Silman I, Shaul Y (2008) Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins 70: 1357–1366 [DOI] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y (2010a) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141: 344–354 [DOI] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E (2010b) Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA 107: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL (2000) Why are ‘natively unfolded' proteins unstructured under physiologic conditions? Proteins 41: 415–427 [DOI] [PubMed] [Google Scholar]
- Varenne S, Buc J, Lloubes R, Lazdunski C (1984) Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol 180: 549–576 [DOI] [PubMed] [Google Scholar]
- Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wessler SR (2001) Role of mRNA secondary structure in translational repression of the maize transcriptional activator Lc(1,2). Plant Physiol 125: 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Hurst LD (2010) GroEL dependency affects codon usage—support for a critical role of misfolding in gene evolution. Mol Syst Biol 6: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke T, Weber CC, Hurst LD (2009) Why there is more to protein evolution than protein function: splicing, nucleosomes and dual-coding sequence. Biochem Soc Trans 37: 756–761 [DOI] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C (2009) Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE 4: e7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I (2008) Following translation by single ribosomes one codon at a time. Nature 452: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox C, Hu JS, Olson EN (1987) Acylation of proteins with myristic acid occurs cotranslationally. Science 238: 1275–1278 [DOI] [PubMed] [Google Scholar]
- Wright F (1990) The ‘effective number of codons' used in a gene. Gene 87: 23–29 [DOI] [PubMed] [Google Scholar]
- Wu G, Nie L, Zhang W (2008) Integrative analyses of posttranscriptional regulation in the yeast Saccharomyces cerevisiae using transcriptomic and proteomic data. Curr Microbiol 57: 18–22 [DOI] [PubMed] [Google Scholar]
- Yang JR, Zhuang SM, Zhang J (2010) Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol 6: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C (1981) Attenuation in the control of expression of bacterial operons. Nature 289: 751–758 [DOI] [PubMed] [Google Scholar]
- Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC (2009) Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem 284: 25254–25267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Saha S, Shabalina SA, Kashina A (2010) Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329: 1534–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280 [DOI] [PubMed] [Google Scholar]
- Zhang SP, Zubay G, Goldman E (1991) Low-usage codons in Escherichia coli, yeast, fruit fly and primates. Gene 105: 61–72 [DOI] [PubMed] [Google Scholar]
- Zhou T, Weems M, Wilke CO (2009) Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol 26: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]