How do molecular networks at the single-cell level define collective cell behavior? This Review discusses recent studies on synthetic and natural microbial communities that dissect the molecular and dynamical mechanisms underlying microbial social evolution.
Keywords: cooperation, quorum sensing, social evolution, social interaction motif, synthetic biology
Abstract
Social interaction among cells is essential for multicellular complexity. But how do molecular networks within individual cells confer the ability to interact? And how do those same networks evolve from the evolutionary conflict between individual- and population-level interests? Recent studies have dissected social interaction at the molecular level by analyzing both synthetic and natural microbial populations. These studies shed new light on the role of population structure for the evolution of cooperative interactions and revealed novel molecular mechanisms that stabilize cooperation among cells. New understanding of populations is changing our view of microbial processes, such as pathogenesis and antibiotic resistance, and suggests new ways to fight infection by exploiting social interaction. The study of social interaction is also challenging established paradigms in cancer evolution and immune system dynamics. Finding similar patterns in such diverse systems suggests that the same ‘social interaction motifs' may be general to many cell populations.
Introduction
For more than a decade, the field of molecular systems biology has advanced our knowledge of how networks of molecular processes enable cells to perceive their environment and trigger phenotypic changes in response. This view centers on the single cell as a computational unit. However, many biological processes are multicellular in nature and are the product of cell–cell interaction within populations. How do molecular networks at the single-cell level ultimately define collective cell behaviors via social interaction? Understanding how interaction among cells enables the spread of information and leads to dynamic population behaviors is a fundamental problem in biology. A closely related question is how adaptive social interactions evolved in spite of the conflicting selection pressures at the individual and the population levels.
A range of recent studies takes advantage of microbes as model organisms to probe social interaction at the molecular level. These studies fall mostly into two distinct categories. First, synthetic social evolution experiments apply genetic engineering to construct artificial social interactions. Such well-defined systems are powerful models to test predictions of social evolution theory in the laboratory by allowing independent manipulation of parameters. A second set of studies investigates interaction in natural microbial populations. In contrast to synthetic systems, the main goal of these studies aims mostly at understanding how altruistic cooperation evolves in spite of strong selective pressures for selfish growth.
The studies of social interaction in synthetic and natural microbial communities have produced important complementary insights into the social biology of microbes and cell populations in general. In this article I review key work in the emerging field of microbial social evolution and discuss how it contributes to our growing understanding of cell–cell interactions in all multicellular systems.
Cooperation and conflict in microbial populations
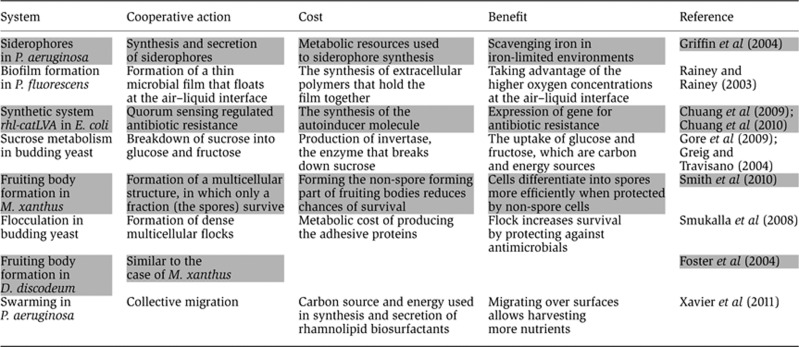
Often viewed as solitary organisms, most microbes in fact live in populations and rely on population-level traits for their survival. Bacteria achieve strength in numbers by collectively secreting virulence factors required for pathogenesis (Rumbaugh et al, 2009) or when producing the extracellular polymeric matrices to make biofilms (Costerton et al, 1999). The yeast Saccharomyces cerevisae secretes sucrose-digesting enzymes and thus carries out part of its sucrose metabolism as a population (Greig and Travisano, 2004). The amoeba Dictyostelium discoideum forms multicellular assemblies such as slugs and fruiting bodies that help communities survive under stressful environments (Weijer, 2004). However, although microbial social interaction spans over a wide range of sophistication (Crespi, 2001; Table I; Glossary), even the simplest cooperative interaction can be difficult to explain when it brings population benefits but comes at the expense of individuals (West et al, 2006). How do adaptive social interactions evolve in spite of the conflict that exists between individual- and population-level?
Table 1. Examples of microbial cooperative actions, and the costs and benefits associated.
 |
Cells can rely on molecular mechanisms of decision-making to tune costly gene expression for optimal self-benefit (Dekel and Alon, 2005). In some cases, cells can even trigger phenotypic changes in anticipation to environmental changes (Tagkopoulos et al, 2008). Genes for individual-level traits are simply favored if the benefits to the individual outweigh any fitness costs of carrying the gene (Perkins and Swain, 2009). However, cells can also have traits that increase group performance but are costly to individual cells (Figure 1). A good illustration of this conflict is the trade-off between slow growth rates with a high yield versus fast but wasteful growth. The trade-off can be a consequence of irreversible thermodynamics on heterotrophic cell metabolism and has important consequences for populations (Pfeiffer, 2001). Higher yields make a more economic use of limited resources, and therefore can be beneficial to the entire population (Pfeiffer and Bonhoeffer, 2003). The population benefit comes at the expense of individual-level restraint, as cells could grow faster with lower yields. Another example is the persister phenotype, which has a role in bacterial antibiotic resistance (Lewis, 2008). Persisters are cells in a dormant state that typically compose a small fraction of all cells in a population (Balaban et al, 2004). As many antibiotics act on growing cells, dormant cells can resist short treatments and afterwards revert back to active growth to restore the population. The persister phenotype is therefore a bet-hedging strategy (Perkins and Swain, 2009) that confers antibiotic resistance, but does so at the expense of the growth of the individuals that slow down their own growth by entering the dormant state (Gardner, 2007).
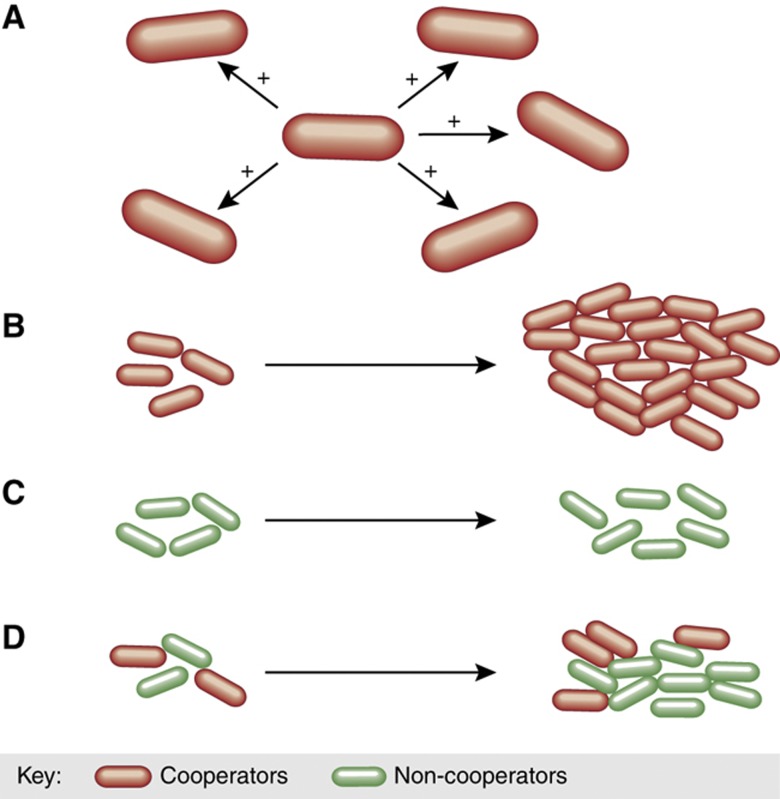
Figure 1.
Cooperative social interactions that provide a population-level benefit often come at a cost to individual's cells. (A) A cooperative interaction provides a fitness benefit to recipients. (B) A population of cooperators has a higher productivity than (C) a population of non-cooperators. (D) Non-cooperators can exploit cooperators in mixed populations by benefiting from cooperation without contributing.
Both high-yield metabolism and persistence are examples of altruistic social interaction (West et al, 2006). The evolutionary advantage of such traits is easy to understand if populations are monoclonal and the fitness cost to individual cells is outweighed by the benefit to the population (Figure 1B and C). However, in many realistic situations microbial populations are not monoclonal but rather heterogeneous populations where cooperators and non-cooperators interact (Figure 1D). In natural settings, bacteria are often mixed with other strains that may differ in the level of cooperation, which makes social interaction open to exploitation. For example, Pseudomonas aeruginosa, a opportunistic pathogenic bacterium, must secrete siderophores that scavenge host-bound iron in order to grow in the iron-limited environment of the host (Ratledge and Dover, 2000). Siderophore synthesis is costly to individual cells but provides a group benefit. However, siderophore production is open to exploitation by non-producers (West and Buckling, 2003). Competition experiments showed that a siderophore-producing strain of P. aeruginosa is fitter than a non-producing one when both were cultivated as monoclonal populations in iron-limited media. However, when both strains were mixed, the non-producers gained the advantage. Once secreted, siderophores become a ‘public good'—they benefit all cells irrespective of who produces them (Griffin et al, 2004). This phenomenon is relevant to the pathogenesis of P. aeruginosa, as it can explain why siderophore-negative mutants often evolve in the lungs of cystic fibrosis patients (De Vos et al, 2001). This effect is widespread as it occurs in P. aeruginosa strains with a range of siderophore production (Jiricny et al, 2010).
The emergence of non-cooperators through mutation is a major challenge to cooperative phenotypes. Rainey and Rainey (2003) observed this effect in Pseudomonas fluorescens biofilms that formed spontaneously in unshaken flasks. The biofilms are examples of cooperation evolving de novo from within a population. While wild-type P. fluorescens is incapable of forming such biofilms, spontaneous mutants that overproduce an adhesive polymer always arise and form a biofilm allowing them to gain better access to oxygen. However, the polymer production is costly to individual cells and, therefore, is a trait open to exploitation. Every time the experiment was performed, the experimentalists also observed the de novo evolution of non-producers within the film around day 5. These new mutants gained advantage among the biofilm population by not paying the cost of polymer production. Eventually, non-producers increased in proportion enough to cause the film to collapse and sink into the broth (Rainey and Rainey, 2003).
Cell–cell communication is also open to exploitation. Many bacteria use a process called quorum sensing to synchronize group behaviors (Bassler and Losick, 2006). Quorum sensing cells secrete small signaling molecules called autoinducers that accumulate in the medium in a density-dependent way. The same cells also detect the extracellular concentration of autoinducer through a cognate receptor. Autoinducer signal concentration becomes a proxy for cell density and allows synchronization of population gene expression. Quorum sensing regulates many traits including bioluminescence (Eberhard et al, 1981), expression of virulence factors (Miller et al, 2002), biofilm formation (Davies et al, 1998) and collective cell migration (Kearns, 2010). However, when autoinducer production is costly, the signal becomes a ‘public good' and mutants lacking signal production can have an advantage in mixed populations (Diggle et al, 2007). When the traits under quorum sensing regulation are themselves costly, quorum sensing bacteria can also be exploited by mutants lacking signal detection (‘signal-blind' mutants) that do not embark in costly cooperative behaviors once the quorum is reached (Diggle et al, 2007; Sandoz et al, 2007). This example is also relevant to pathogenesis as it explains why signal-blind mutants often arise spontaneously in cystic fibrosis lung infections (D'Argenio et al, 2007).
Microbial populations therefore face a major challenge to their collective functioning—the evolution of exploitation from within. To understand how interaction among cells can evolve in the face of selection at the individual level we recur to social evolution theory.
Social evolution theory
Social evolution theory has devoted considerable attention to the fundamental problem of the evolution of cooperative behavior (Pennisi, 2005). In 1964, Hamilton formalized his famous explanation that cooperation can evolve if cooperators preferentially favor other, related individuals (Hamilton, 1964). A cooperative trait is favored if
This inequality, known as Hamilton's rule, highlights three factors central to social interaction. c is the fitness cost to the actor, b is the fitness benefit to the recipient and r is the correlation between the genotypes of actors and recipients, also called relatedness. Hamilton's rule is appealingly simple. However, the simplifying assumptions on which it is based (weak selection, linear and additive interactions) can limit its applicability.
How does Hamilton's rule apply to concrete systems? In the case of the siderophores of P. aeruginosa (Griffin et al, 2004), c is the fitness cost that a siderophore-producing cell pays for production. The siderophores released can boost the fitness of individuals in the same population by a factor b. However, if the population is heterogeneous, then the siderophores can also benefit non-producers, which would be disadvantageous. Hamilton's rule takes this into account using the relatedness coefficient r to quantify the proportion of related cooperators within those receiving the benefit. Hamilton's rule predicts that siderophore production is favored if r>c/b (West and Buckling, 2003). This example illustrates the key feature of Hamilton's analysis, which is the concept of inclusive fitness. To determine the evolutionary success of a cooperative action, one must take into account not only its direct effect on the actor (the direct fitness) but also its effect on related individuals that share the same gene (the inclusive fitness).
Another key concept is the existence of multiple levels of selection, which is the fact that a focal sub-population will compete with other sub-populations within a global gene pool. Sub-populations with a high proportion of siderophore producers will have a higher productivity, and therefore will contribute to a larger fraction of the global gene pool (Griffin et al, 2004). A cooperative genotype can, therefore, increase in frequency within the global gene pool even if its frequency decreases locally in every sub-population. Chuang et al (2009) demonstrated this counterintuitive prediction with a series of elegant experiments with synthetic cooperation in Escherichia coli. They engineered a ‘public-good' system, in which a quorum sensing signal activated the expression of antibiotic resistance genes. They also created non-cooperator cells that lacked signal production, but could still receive it. When grown as monocultures in antibiotic-containing media, producers fared better than non-producers as expected. When cultivated together, however, producers lost the competition to non-producers because of ‘public-good' exploitation.
This system was then used to make a subtler point. Several sub-populations were inoculated separately with different proportions of producers. At the end of a round of competition, the proportions of producers in each sub-population (the local proportions) were quantified. The global proportion of producers was also determined by adding up all sub-populations. What they observed was that the proportion of producers had decreased in every sub-population yet the global proportion had increased. The explanation for this apparent disparity—that the proportion of producers decreased locally but increased globally—was as follows. When a sub-population has a higher proportion of producers, its overall productivity is higher. Therefore, those sub-populations contribute more to the global gene pool (Figure 2). This is explained by Simpson's paradox, a statistical phenomenon in which a trend observed in different groups is reversed when the groups are combined (Blyth, 1972). Importantly, the effect was dissipated when the initial proportion of producers varied less among sub-populations (Chuang et al, 2009).
Figure 2.
Multilevel selection is essential for the evolution of cooperation among microbes. Cooperative strategies can have an evolutionary advantage in the global population, even though the same strategy is disadvantageous in local sub-populations. This happens because groups with a higher proportion of cooperators are more productive and therefore contribute more to the global gene pool.
These experiments with synthetic cooperators followed the qualitative predictions of Hamilton's rule well. Decreasing the variability among inocula decreased relatedness and that made cooperation less favorable. The result encouraged the same authors to test Hamilton's rule further by independently manipulating the cost and benefit at the molecular level (Chuang et al, 2010). The system was first extended by introducing an arginine auxotrophic mutation (Arg−) into producers, which implemented an increased fitness cost to production. As expected by Hamilton's rule, adding arginine to the medium (decreasing the cost) favored producers. In a second extension, they increased the number of the autoinducer receptors in both producers and non-producers. This modification was intended to increase the benefit of cooperation by making cells more sensitive to the ‘public good'. However, the results obtained were non-intuitive as they went against what was expected from Hamilton's rule. Rather than favoring producers, the alteration increased the advantage of non-producers. The explanation found for this was that the benefit was not a constant parameter but rather a nonlinear function of the receptor copy number. The relative benefit decreased as the copy number increased, which produced conditions that were less favorable to cooperators than previously expected (Chuang et al, 2010).
Extending Hamilton's rule
The experiments by Chuang et al (2010) highlighted a central problem faced by all those who try to bridge molecular mechanism and social evolution. Even in well-controlled, engineered systems the factors governing social interaction are often non-trivial. Rather than being constant parameters, costs and benefits depend on many factors, including the proportion at which cooperators and non-cooperators are mixed, i.e., the relatedness. This is well known to evolutionary biologists, who recognize that Hamilton's rule achieves its generality by bundling complicated details in its components (Gardner et al, 2007). Nonlinear and non-additive effects limit the practical utility of Hamilton's rule beyond the specific conditions at which costs and benefits are measured.
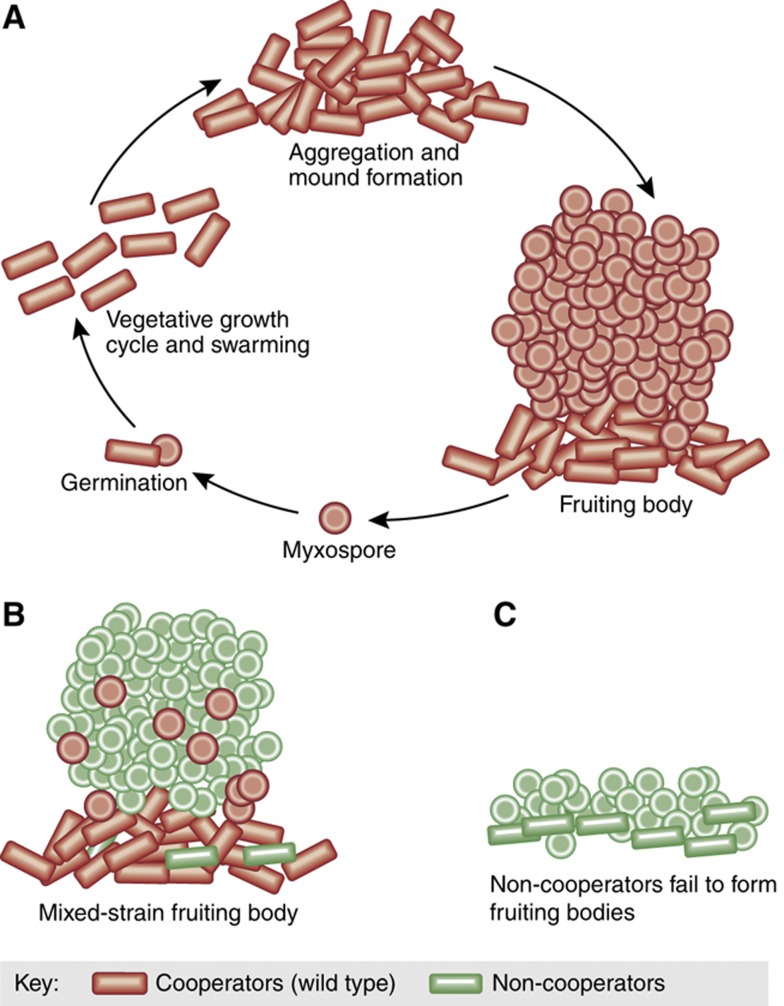
Smith et al (2010) showed that it is nevertheless possible to overcome this limitation with a simple generalization. Their extension acknowledged that br−c>0 is a simplified representation, the same way that a strongly nonlinear function can be crudely approximated by the first-order term of its Taylor series. Under this perspective, relatedness corresponds to a first-order moment of population structure, and the rule can be extended simply by including higher order terms. Smith et al (2010) demonstrated the applicability of the generalized Hamilton's rule to experiments in fruiting body formation in Myxococcus xanthus bacteria (Figure 3A). Unlike the examples above where social interaction is based on the secretion of a ‘public good', fruiting body formation is a complex developmental process. When starved, M. Xanthus cells come together and form multicellular assemblies, where some cells differentiate into highly resistant spores (Kaiser and Welch, 2004). The remaining cells that support the fruiting body eventually die. Non-cooperator cells can sporulate at higher efficiency when mixed with others (Figure 3B), yet sporulate less when cultured in isolation (Figure 3C). As only the spores survive, the fitness of a colony is defined by its sporulation efficiency. The fitness of fruiting bodies was a monotonically increasing function of the initial proportion of cooperators. However, the function was strongly nonlinear and cooperators and non-cooperators benefited asymmetrically from the cooperation. These two features challenged the applicability of Hamilton's rule. Conventional relatedness, i.e., the first-order moment of population structure, had limited predictive power. However, taking into account higher-order terms extended the predictive range of Hamilton's rule and more accurately described the system (Smith et al, 2010).
Figure 3.
Cooperation and conflict in fruiting body formation. (A) The life cycle of M. xanthus (Zusman et al, 2007). (B) Non-cooperators preferentially differentiate into spores and have an advantage when mixed with wild-type cooperators. (C) Non-cooperators fail to form proper fruiting bodies when alone.
The implications of the generalization proposed by Smith et al (2010) go well beyond M. Xanthus. Generalizing Hamilton's rule can extend its applicability to systems where the original assumptions of weak selection and non-additive effects do not hold. As the experiments of Chuang et al (2010) illustrates, nonlinearity is likely to be widespread in molecular experiments of social evolution. Therefore, in order to determine the evolutionary outcomes, one must characterize the functions governing costs and benefits of a social interaction, including the influence of population structure and environmental factors.
Synthetic biology models of ecosystems
Hamilton's rule describes the evolution of cooperative behaviors by isolating the effect of a single gene encoding a cooperative behavior. Yet, the molecular networks that govern cell behavior evolve from different social interactions, both cooperative and competitive, occurring simultaneously (Foster, 2011). Recent studies in synthetic biology set out to study the inverse problem—how molecular networks lead to complex social interactions? Mutualisms are social interactions that benefit both the actor and the recipient. A minimal mutualism system can be engineered using two auxotrophic yeast strains, each producing a metabolite essential to the other strain (Shou et al, 2007). Experiments conducted with each auxotroph in isolation suggested that the mutualism would not work because each strain delayed synthesizes until near death. However, when both auxotrophs were mixed together the mutualism worked consistently well, and even showed a resilience to reductions in cell density (Shou et al, 2007). Mutualistic systems were also extended to higher orders by using a set of E. coli auxotrophic mutants (Wintermute and Silver, 2010). Pairs of auxotrophs were combined in a high-throughput fashion to identify which combinations produced synergistic interactions. This study revealed that the metabolites with less cost to the producing strain are those most readily exchanged (Wintermute and Silver, 2010).
Auxotrophy was also used to manipulate the parameters governing cooperation in experiments with Saccharomyces cerevisiae (Gore et al, 2009). Yeast lacking the sucrose invertase gene exploited the metabolic conversion carried out by others (Greig and Travisano, 2004). Invertase is necessary to convert sucrose into glucose and fructose, which allow faster growth. Invertase producers are cooperators as 99% of converted sugars are lost to neighboring cells (Gore et al, 2009). By engineering producers that were also histidine auxotrophic, their fitness cost could be tuned by controlling histidine concentration in the media. Gore et al (2009) observed that cooperators and non-cooperators (cells lacking invertase production but harboring histidine genes) were capable of mutual reinvasion because both cell types were fitter when rare. The feature that rare strategies outperform common ones, they pointed out, is a characteristic of the snowdrift game, a theoretical framework that explains coexistence of cooperative strategies in nature (Gore et al, 2009). These results may explain the natural diversity in the sucrose metabolism of yeast (Naumov et al, 1996).
Balagaddé et al (2008) implemented a more complex dynamic by genetically engineering two strains of E. coli to play the role of ‘predator' and ‘prey'. The interactions between predator and prey were mediated by two quorum sensing autoinducers. In agreement with theoretical predator–prey models, the synthetic ecosystem produced a range of outcomes including predator extinction, coexistence of prey and predator and even oscillations between the two types. The same system was also used to investigate the role of spatial structure in maintaining biodiversity (Song et al, 2009). The new experiments revealed a key role for the length scale of diffusion of the autodinducers mediating the social interaction in the maintenance of biodiversity.
Spatial interactions
Experiments that manipulate relatedness by varying the proportion of cooperators in a mixed group are common in the study of microbial social interaction. However, this assumes that all individuals within the same group interact equally with each other. Nevertheless, as shown by the experiments with the predator–prey system, spatial structure can strongly affect social interaction. In nature, many species of bacteria form biofilms, where interaction occurs preferentially among cells in close proximity (Kreft, 2004; Xavier and Foster, 2007; Nadell et al, 2008, 2009; Xavier et al, 2009). Secreted substances such as digestive enzymes or quorum sensing signals commonly diffuse away from producers, and therefore the benefits are much lower far away from the producer. Under these conditions, the central parameter governing the success of a cooperative trait is genetic relatedness evaluated within the length scale of ‘public-good' dispersal.
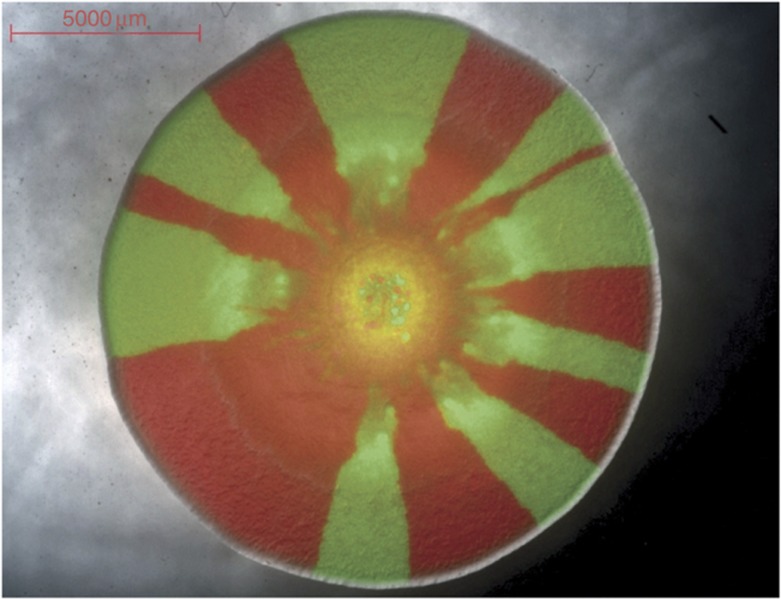
The emergence of genetic structure in expanding microbial colonies was investigated empirically by Hallatschek et al (2007). They constructed two variants of E. coli that were phenotypically identical in everything, except for a neutral fluorescent label that enabled them to be distinguished from each other. When the experimentalists cultivated mixed colonies of the two neutral cell types, they observed large mono-colored sectors of spontaneous genetic segregation (Figure 4). The segregation occurred in the absence of any fitness difference between the two genotypes and was the result of genetic drift as the colony expanded, an effect that is well captured by population dynamics models of neutral evolution (Korolev et al, 2010). Could this spontaneous spatial segregation be an important factor for the evolution of cooperation in microbial populations?
Figure 4.
Genetic drift in an expanding bacterial colony. The colony shown here was prepared by mixing two strains of P. aeruginosa labeled with green and red fluorescent markers, respectively. Besides the fluorescent label, the two strains have identical phenotypes and therefore are neutral. Nevertheless, they segregate into sectors as the colony expands. This method was first used in E. coli colonies (Hallatschek et al, 2007).
Computer simulations of colony growth revealed that genetic segregation can result from simple diffusion limitation of growth (Nadell et al, 2010). When penetration of nutrient into the colony is limited, only a thin layer of cells is actively growing, whereas cells further inside the colony are starved. The limited nutrient penetration reduces the effective population size, causing a continuous population bottleneck as the colony expands. The same simulations showed that the evolutionary success of ‘public-good' producers is favored when spatial segregation is high. Therefore, the same conditions of growth limitation that induce spontaneous segregation of neutral genotypes can also favor the evolution of cooperation by making cooperators stay close to each other (Nadell et al, 2010).
Molecular mechanisms that stabilize social interaction
Discrimination is an important mechanism governing the evolution of social behavior, and microbial interaction is no exception. For example, quorum sensing can be a form of molecular discrimination as it cannot be ‘eavesdropped' by cells lacking a cognate receptor. Another example is the formation of protective flocks in S. cerevisae. Flocculation requires the costly expression of a membrane-bound protein, FLO1, which induces cells to adhere to each other (Smukalla et al, 2008). FLO1+ cells avoid exploitation by adhering preferentially to other FLO1+ cells, regardless of genetic similarity across the remaining genome. This molecular discrimination makes FLO1 a gene that directs cooperation toward other carriers of the same gene, also known as a ‘green beard gene' (Dawkins, 1976). Discrimination can also have an important role in competitive interactions. Many species of bacteria engage in intraspecies warfare through the secretion of toxins called bacteriocins. Bacteriocins are very diverse in terms of function and their secretion is under strong selection for specificity (Majeed et al, 2010). Bacteriocin producers carry genes that encode for resistance against the self-produced toxin, and therefore affect only unrelated strains. Interestingly, strains that carry bacteriocin resistance genes, but lack bacteriocin production, can have a competitive advantage against bacteriocin producers but are outcompeted by sensitive cells. A system composed by three strains—a sensitive strain, a resistant strain and a bacteriocin producer—can exhibit complex dynamics like an evolutionary game of rock-paper-scissors (Kerr et al, 2002).
Another molecular mechanism that stabilizes cooperation is pleiotropy. The social amoeba Dictyostlium discodeum aggregates upon starvation and assembles into a multicellular structure composed of cells that differentiate into stalk and spores. Cells in the stalk eventually die, and therefore only the spore formers can pass on their genes. The gene dimA is required to receive the signal for differentiation into prestalk cells (Thompson et al, 2004). dimA− are signal-blind, and therefore should act as cheaters. However, when mixed with the wild-type cells, cells lacking dimA were excluded from spores (Foster et al, 2004). This pleiotropic linkage of stalk and spore formation prevented exploitation in D. discoideum, because defecting on prestalk cell production reduced spore formation. Another case of pleiotropic constraints is the siderophore production in P. aeruginosa. Strains producing lower siderophore levels are also affected in their ability to form biofilms (Harrison and Buckling, 2009), a trait that is difficult to exploit (Xavier and Foster, 2007). This observation links two cooperative phenotypes, siderophore secretion and biofilm production, to the same gene and effectively raises the cost of exploitation (Harrison and Buckling, 2009).
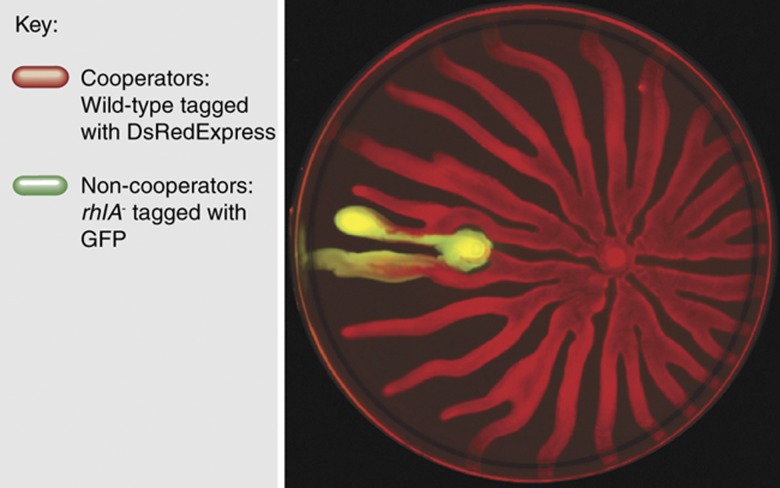
In cooperation, discrimination increases the likelihood that the benefits are properly directed to other cooperators, and therefore increases relatedness. Pleiotropy, on the other hand, attributes a fitness penalty to non-cooperators, thereby reducing the net benefit attributed to cheaters. Recently, a new molecular mechanism that stabilizes cooperation by reducing the cost was found in swarming in P. aeruginosa (Xavier et al, 2011). Swarming relies on the synthesis and secretion of large quantities of biosurfactants called rhamnolipids (Deziel et al, 2003; Caiazza et al, 2005). Cells lacking a rhamnolipid synthesis gene are incapable of swarming on their own, but can swarm by using the surfactants secreted by wild-type bacteria (Figure 5). Counter to naive expectations, competition experiments showed that non-producers did not increase in proportion when mixed with wild-type producers. The unexpected observation suggested the absence of a fitness cost to rhamnolipid synthesis, which was in striking contrast to the massive biosurfactant secretions amounting to 20% of the producer's biomass. Further experiments then showed that P. aeruginosa regulates the expression of rhamnolipid synthesis genes to ensure that synthesis occurred only when growth was limited by the nitrogen source. With this strategy, production occurred only when carbon was in excess of that needed for growth. By regulating gene expression, P. aeruginosa delays investment into cooperation to times when its impact on to fitness becomes negligible (Xavier et al, 2011).
Figure 5.
Social exploitation in P. aeruginosa swarming. Colonies of P. aeruginosa swarm over soft agar, but this collective trait requires that individual cells synthesize and secrete massive amounts of rhamnolipid biosurfactants. Cells lacking the synthesis gene rhlA are capable of swarming using the biosurfactants produced by others. Wild-type cells restrict rhlA expression to times when carbon source is in excess of that needed for growth, which lowers the cost that biosurfactant synthesis has on their fitness. This mechanism, called metabolic prudence, prevents exploitation by non-cooperator rhlA− mutants (Xavier et al, 2011).
Clinical implications
Understanding social traits in microbes can assist in the rational development of novel therapies that specifically target interaction in pathogenic populations (Foster, 2005). Drugs that target individual level traits of bacteria, such as antibiotics and other antimicrobials, create a strong selective pressure for resistance. In a heterogeneous population where only a small proportion of cells are resistant, these drugs benefit the resistant cells by harming its drug-sensitive competitors. On the other hand, our growing understanding of social interaction opens the way to novel therapies that do not select for resistance (Griffin et al, 2004; Foster, 2005). For example, non-cooperator strains that can exploit wild-type bacteria can potentially be introduced to outcompete more virulent conspecifics, or even variants of non-producers could be engineered in the laboratory to carry medically beneficial traits or anticompetitor phenotypes (Brown et al, 2009). Importantly, however, the exact nature of the social interaction being targeted must be taken into careful consideration. In a recent study aimed at testing a quorum sensing inhibitor against P. aeruginosa infections, the drug actually benefited the virulent quorum sensing capable strains by reducing the selective advantage of its cheating competitors (Kohler et al, 2010).
The study of microbial populations is also leading to important new revelations on the nature of antibiotic resistance as a population-level trait. Lee et al (2010) studied the emergence of antibiotic resistance in continuous cultures. They consistently observed that individual cells within a population of resistant bacteria varied enormously in their level of resistance and that a vast majority of isolates was less resistant than the overall population level. The explanation found was that a small fraction of highly resistant mutants improved the survival of less resistant cells within the same population. The highly resistant clones secreted indole, a signal that induces all cells to turn on protective mechanisms and upregulates the expression of drug efflux pumps. Indole was, therefore, a ‘public good' that increased the survival of the entire population. The mechanism, which the authors termed ‘bacterial charity work', allowed the population to resist antibiotics beyond the average resistance level of its composing individuals (Lee et al, 2010).
Beyond microbes—social interaction in cancer evolution and immune system dynamics
Cooperation has vast implications in biology as it is intimately related to evolutionary transitions. The transition to eukaryotic life resulted from an initially facultative interaction between prokaryotes that became obligatory and led to the primordial eukaryotic cells (Lane and Martin, 2010). The transition from eukaryotic unicellular organisms to multicellular organisms required the evolution of obligatory cooperation among individual cells (Bonner, 1999). Multicellular organisms, such as in our own species, can be composed of trillions of cells (Dobzhansky, 1971), of which only the small fraction of cells in the germ line can directly pass its genes on to future generations. In contrast, somatic cells are in an evolutionary dead end. The somatic cells compose a vehicle that assists the germ line in passing on the individual's genes (Dawkins, 1999).
Cancer is a departure from the developmental process of a multicellular organism. It is a disease of evolution within the body (Nowell, 1976) where neoplasms evolve from its cell of origin to accumulate increasingly aggressive features, such as uncontrolled growth, invasiveness and metastasis (Hanahan and Weinberg, 2000). In the course of a cancer's progression, tumor cells interact among each other and with cells within the microenvironment (Joyce and Pollard, 2009), competing for resources but also cooperating by sharing secreted substances (Axelrod et al, 2006). These aggressive features may be recovered ancient functions that once helped our metazoan ancestors to thrive as loosely associated colonies of single-celled organisms (Davies and Lineweaver, 2011). The immune system and anticancer therapies exert additional selective pressures. All this occurs in a spatially structured, dynamic environment. Cancer is, therefore, an example of social exploitation. Cancer cells outcompete the normal cells of its host, but like the other exploitations analyzed here, the advantage gained is short lived. Cancer proliferation eventually harms the host organism and often leads to its death. The features that make cancer cells thrive within the body do not enable the cancer to pass on its genes to future generations. Beyond rare but notable exceptions, such as the transmissible cancer in Tasmanian Devils (Murchison et al, 2010) and tumor cell lines artificially maintained for the purpose of scientific research (Skloot, 2010), cancer cells do not survive outside the body where they originated. Discovering the social interactions underlying cancer evolution can greatly improve our understanding of how cancers progress and how they react to treatment. Similar to the case of antibiotic resistance in bacteria, cancer therapies themselves create a selective pressure for resistance. Exploiting cancer social interaction may help us better develop new therapies that do not select for resistance (Merlo et al, 2006).
Social interaction among cells is also important in the self/non-self discrimination of the immune system. When challenged by a foreign antigen, effector T cells start producing and secreting the cytokine interleukin-2 (IL-2). Secreted IL-2 can be taken up by the same effector T cells to induce their proliferation and set off an immune response. However, there is the constant danger that effector T cells generate an autoimmune response by upregulating IL-2 production when weakly activated by a native peptide. Such autoimmune responses are prevented, in part, by the presence of cell type, the regulatory T cells (Tregs; Sakaguchi, 2005; Pandiyan et al, 2007). Tregs take up IL-2, but do not produce it (Pandiyan et al, 2007; Scheffold et al, 2007). Treg depletion of IL-2 is a discriminatory negative feedback on the effector T-cell population that helps prevent weakly activated but not strongly activated cells from proliferating (Busse et al, 2010; Feinerman et al, 2010).
Conclusions and outlook
Populations of interacting cells face a major challenge to their functioning: the emergence of conflict from within. All cell populations are prone to exploitative individuals that may not contribute to shared resources, yet still benefit from them (Perkins and Swain, 2009). This is true in microbial populations where the conflict between individual and population level of selection can lead to the emergence of ‘cheaters' (West et al, 2006). But it is also true in multicellular organisms, in which the same conflict leads to the evolution of cancer (Michod and Roze, 2001) or autoimmune disorders (Feinerman et al, 2010).
All adaptive population traits must have evolved with features to eliminate potential conflicts. Understanding what these features are and how they confer robustness to social interaction is a challenge for Systems Biologists that holds much potential. We can now easily manipulate microbes at the molecular level leading to unprecedented control of social interaction. This has already led to the identification of molecular mechanisms that govern social interaction by reducing the costs of interaction to prevent exploitation, such as pleiotropy (Foster et al, 2004) and metabolic prudence (Xavier et al, 2011).
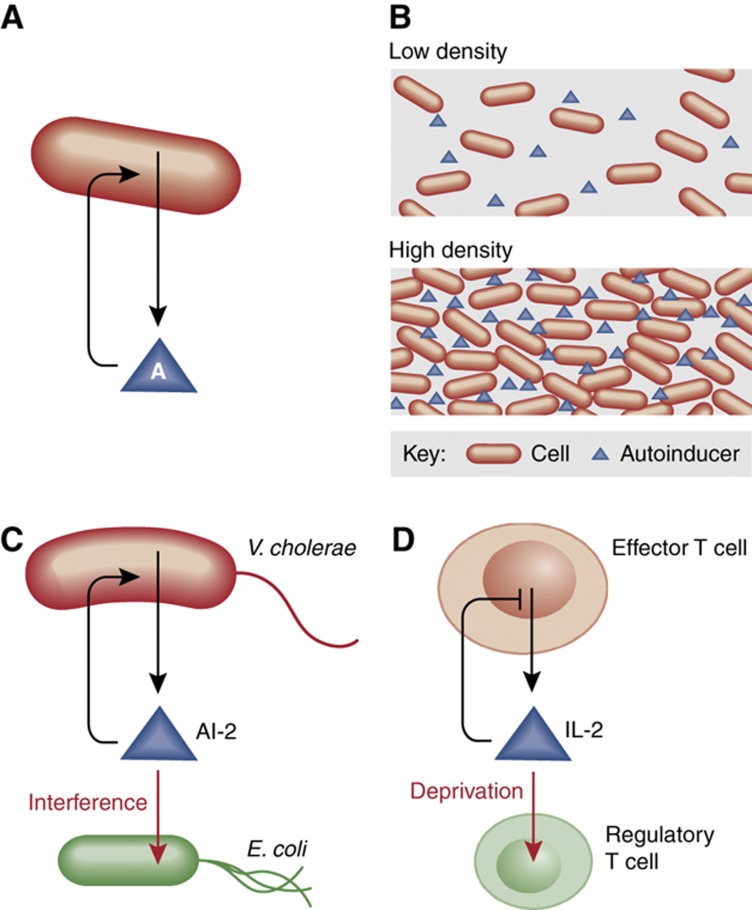
Social interaction is key to understanding the functions of cell populations. Just as we can interpret circuit motifs in biochemical networks as ‘design principles' (Alon, 2007), we may find motifs in networks of social interactions irrespective of the specific molecular players involved. Quorum sensing is a clear example of such a motif. The ability to secrete small molecules and to sense their extracellular concentration enables cells to sense population density or other features of their environment (Goryachev, 2009). Quorum sensing is repeatedly found in many organisms, even in diverse across taxonomic kingdoms, in which the specific molecules may be quite different but the diagram of influences for the social interaction is similar (Figure 6). It is found in a vast number of Gram-positive and Gram-negative bacteria (Pai and You, 2009), in yeast (Chen and Fink, 2006) and in the adaptive immune system (Burroughs et al, 2006; Feinerman et al, 2010). Density-dependent effects in cancer metastasis suggest that quorum sensing also plays a role in cancer (Hickson et al, 2009). Finding how quorum sensing and other social interaction motifs govern cell populations is a challenge at the frontier of molecular and systems biology. The search has already begun and there are many opportunities in this emerging field.
Figure 6.
Quorum sensing as a social interaction motif. (A) The ability of a cell to produce a signaling molecule (an autoinducer) and sense its extracellular concentration can enable the cell to sense changes in population density (B). Quorum sensing can be found in diverse systems such as (C) pathogenic bacteria (Vibrio cholerae) and (D) the adaptive immune system of mammals with common principles but different molecular players. Interestingly, quorum sensing in both V. cholerae and effector T cells is perturbed by competitor cells that sequester the signaling molecule. The enteric E. coli interferes with V. cholerae by taking up the autoinducer AI-2 (Xavier and Bassler, 2005). The immune response is mediated by IL-2 quorum sensing in effector T cells, but IL-2 deprivation by regulatory T cells is important to prevent autoimmune responses. An important difference between the T-cell system and others is that the feedback on signal production is negative (Feinerman et al, 2010).
Finally, there is a pressing need to understand social interaction within complex microbial communities. The study of communities such as the intestinal microbial flora is advancing rapidly, thanks to metagenomic culture-independent methods (Eckburg et al, 2005) and novel sequencing technologies (Wooley et al, 2010). The current focus is in the census of the bacteria that make up the communities, but more biologically significant is to understand how these species interact in a mechanistic way (Blaser, 2010). There is a flood of potential applications of microbiome ecology to the environmental and medical fields (e.g., Ubeda et al, 2010), but we must first seek to understand the ecological processes governing microbiome composition dynamics and function thoroughly. The next challenge is therefore to take the study of social interaction beyond pairwise interactions in minimal models and into networks of social interactions in natural microbial ecosystems.
Glossary
Autoinducers—Small diffusible molecules used in quorum sensing signaling. Homoserine lactones are well-known examples of quorum sensing in the bacterium P. aeruginosa.
Auxotrophic strain—Strain incapable of synthesizing a compound required for its own growth.
Cheaters—An individual that benefits from cooperation of others, but does not cooperate itself.
Cooperator—An individual that carries out an action that benefits other individuals.
Non-producers—Cheaters in a scenario of public-good cooperation.
Public good—a resource that is available to all individuals in a population irrespective of which individual produces it.
Quorum sensing—A mechanism for density-dependent regulation of gene expression that works by cells producing and detecting autoinducers.
Rhamnolipids—Biosurfactants secreted by P. aeruginosa and used in cooperative behaviors such as swarming motility.
Siderophores—Iron scavenging molecules secreted by bacteria in iron-limited environments, such as a host tissue. Siderophores are examples of public goods.
Swarming—Collective form of surface motility displayed by many bacteria.
Acknowledgments
I thank Grégoire Altan-Bonnet, Kevin Foster, Jeff Smith, Dave van Ditmarsch, Vanni Bucci and Debra Bemis for comments on the manuscript. This work was supported by a National Institute of Cancer Center for Integrated Cancer Biology grant (1U54CA148967-01) and a seed grant from the Lucille Castori Center for Microbes, Inflammation and Cancer.
Footnotes
The author declares that he has no conflict of interest.
References
- Alon U (2007) An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton: Taylor & Francis [Google Scholar]
- Axelrod R, Axelrod DE, Pienta KJ (2006) Evolution of cooperation among tumor cells. Proc Natl Acad Sci USA 103: 13474–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQJ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305: 1622–1625 [DOI] [PubMed] [Google Scholar]
- Balagaddé FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L (2008) A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol 4: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B, Losick R (2006) Bacterially Speaking. Cell 125: 237–246 [DOI] [PubMed] [Google Scholar]
- Blaser MJ (2010) Harnessing the power of the human microbiome. Proc Natl Acad Sci USA 107: 6125–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth CR (1972) On Simpson's paradox and the sure-thing principle. J Am Stat Assoc 67: 364–366 [Google Scholar]
- Bonner JT (1999) The origins of multicellularity. Integr Biol 1: 27–36 [Google Scholar]
- Brown SP, West SA, Diggle SP, Griffin AS (2009) Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. Philos T R Soc B 364: 3157–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs NJ, Miguel Paz Mendes de Oliveira B, Adrego Pinto A (2006) Regulatory T cell adjustment of quorum growth thresholds and the control of local immune responses. J Theor Biol 241: 134–141 [DOI] [PubMed] [Google Scholar]
- Busse D, de la Rosa M, Hobiger K, Thurley K, Flossdorf M, Scheffold A, Höfer T (2010) Competing feedback loops shape IL-2 signaling between helper and regulatory T lymphocytes in cellular microenvironments. Proc Natl Acad Sci 107: 3058–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Shanks RMQ, O'Toole GA (2005) Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187: 7351–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fink GR (2006) Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Develop 20: 1150–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Rivoire O, Leibler S (2009) Simpson's paradox in a synthetic microbial system. Science 323: 272–275 [DOI] [PubMed] [Google Scholar]
- Chuang JS, Rivoire O, Leibler S (2010) Cooperation and Hamilton's rule in a simple synthetic microbial system. Mol Syst Biol 6: 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322 [DOI] [PubMed] [Google Scholar]
- Crespi BJ (2001) The evolution of social behavior in microorganisms. Trends Ecol Evol 16: 178–183 [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Wu MH, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, Nguyen H, Ernst RK, Freeman TJL, Spencer DH, Brittnacher M, Hayden HS, Selgrade S, Klausen M, Goodlett DR, Burns JL, Ramsey BW, Miller SI (2007) Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64: 512–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298 [DOI] [PubMed] [Google Scholar]
- Davies PCW, Lineweaver CH (2011) Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Phys Biol 8: 015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R (1976) The Selfish Gene. Oxford, UK: Oxford University Press [Google Scholar]
- Dawkins R (1999) The Extended Phenotype: The Long Reach of the Gene. Oxford, UK: Oxford University Press [Google Scholar]
- De Vos D, De Chial M, Cochez C, Jansen S, Tümmler B, Meyer J-M, Cornelis P (2001) Study of pyoverdine type and production by<SMALL>Pseudomonas aeruginosa</SMALL>isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch Microbiol 175: 384–388 [DOI] [PubMed] [Google Scholar]
- Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592 [DOI] [PubMed] [Google Scholar]
- Deziel E, Lepine F, Milot S, Villemur R (2003) rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiol-SGM 149: 2005–2013 [DOI] [PubMed] [Google Scholar]
- Diggle S, Griffin A, Campbell G, West S (2007) Cooperation and conflict in quorum-sensing bacterial populations. Nature 450: 411–414 [DOI] [PubMed] [Google Scholar]
- Dobzhansky T (1971) Genetics of the Evolutionary Proces, Vol. 1, New York: Columbia University Press [Google Scholar]
- Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ (1981) Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20: 2444–2449 [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinerman O, Jentsch G, Tkach KE, Coward JW, Hathorn MM, Sneddon MW, Emonet T, Smith KA, Altan-Bonnet G (2010) Single-cell quantification of IL-2 response by effector and regulatory T cells reveals critical plasticity in immune response. Mol Syst Biol 6: 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K, Shaulsky G, Strassmann J, Queller D, Thompson C (2004) Pleiotropy as a mechanism to stabilize cooperation. Nature 431: 693–696 [DOI] [PubMed] [Google Scholar]
- Foster KR (2005) Biomedicine. Hamiltonian medicine: why the social lives of pathogens matter. Science 308: 1269–1270 [DOI] [PubMed] [Google Scholar]
- Foster KR (2011) The sociobiology of molecular systems. Nat Rev Genet 12: 193–203 [DOI] [PubMed] [Google Scholar]
- Gardner A (2007) Is bacterial persistence a social trait? Plos One 2: e752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A, West SA, Barton NH (2007) The relation between multilocus population genetics and social evolution theory. Am Nat 169: 207–226 [DOI] [PubMed] [Google Scholar]
- Gore J, Youk H, van Oudenaarden A (2009) Snowdrift game dynamics and facultative cheating in yeast. Nature 459: 253–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryachev AB (2009) Design principles of the bacterial quorum sensing gene networks. Wires Syst Biol Med 1: 45–60 [DOI] [PubMed] [Google Scholar]
- Greig D, Travisano M (2004) The Prisoner's Dilemma and polymorphism in yeast SUC genes. P Roy Soc Lond B Bio 271: S25–S26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin AS, West SA, Buckling A (2004) Cooperation and competition in pathogenic bacteria. Nature 430: 1024–1027 [DOI] [PubMed] [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR (2007) Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA 104: 19926–19930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD (1964) The genetical evolution of social behaviour I & II. J Theor Biol 7: 1–52 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Harrison F, Buckling A (2009) Siderophore production and biofilm formation as linked social traits. ISME J 3: 632–634 [DOI] [PubMed] [Google Scholar]
- Hickson J, Diane Yamada S, Berger J, Alverdy J, O'Keefe J, Bassler B, Rinker-Schaeffer C (2009) Societal interactions in ovarian cancer metastasis: a quorum-sensing hypothesis. Clin Exper Metastasis 26: 67–76 [DOI] [PubMed] [Google Scholar]
- Jiricny N, Diggle SP, West SA, Evans BA, Ballantyne G, Ross-Gillespie A, Griffin AS (2010) Fitness correlates with the extent of cheating in a bacterium. J Evol Biol 23: 738–747 [DOI] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW (2009) Microenvironmental regulation of metastasis. Nat Rev Cancer 9: 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D, Welch R (2004) Dynamics of fruiting body morphogenesis. J Bacteriol 186: 919–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB (2010) A field guide to bacterial swarming motility. Nat Rev Microbiol 8: 634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJM (2002) Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418: 171–174 [DOI] [PubMed] [Google Scholar]
- Kohler T, Perron GG, Buckling A, van Delden C (2010) Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. Plos Pathog 6: e1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev KS, Avlund M, Hallatschek O, Nelson DR (2010) Genetic demixing and evolution in linear stepping stone models. Rev Modern Phys 82: 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft JU (2004) Biofilms promote altruism. Microbiol-SGM 150: 2751–2760 [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W (2010) The energetics of genome complexity. Nature 467: 929–934 [DOI] [PubMed] [Google Scholar]
- Lee HH, Molla MN, Cantor CR, Collins JJ (2010) Bacterial charity work leads to population-wide resistance. Nature 467: 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K (2008) Multidrug tolerance of biofilms and persister cells. In Bacterial Biofilms. Romeo T (ed), Vol. 322, pp 107–131. Springer: Berlin Heidelberg [DOI] [PubMed] [Google Scholar]
- Majeed H, Gillor O, Kerr B, Riley MA (2010) Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J 5: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LMF, Pepper JW, Reid BJ, Maley CC (2006) Cancer as an evolutionary and ecological process. Nat Rev Cancer 6: 924–935 [DOI] [PubMed] [Google Scholar]
- Michod RE, Roze D (2001) Cooperation and conflict in the evolution of multicellularity. Heredity 86: 1–7 [DOI] [PubMed] [Google Scholar]
- Miller M, Skorupski K, Lenz D, Taylor R, Bassler B (2002) Parallel quorum sensing systems converge to regulate virulence in vibrio cholerae. Cell 110: 303–314 [DOI] [PubMed] [Google Scholar]
- Murchison EP, Tovar C, Hsu A, Bender HS, Kheradpour P, Rebbeck CA, Obendorf D, Conlan C, Bahlo M, Blizzard CA, Pyecroft S, Kreiss A, Kellis M, Stark A, Harkins TT, Graves JAM, Woods GM, Hannon GJ, Papenfuss AT (2010) The Tasmanian devil transcriptome reveals schwann cell origins of a clonally transmissible cancer. Science 327: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Foster KR, Xavier JB (2010) Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol 6: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Foster KR (2009) The sociobiology of biofilms. FEMS Microbiol Rev 33: 206–224 [DOI] [PubMed] [Google Scholar]
- Nadell CD, Xavier JB, Levin SA, Foster KR (2008) The evolution of quorum sensing in bacterial biofilms. PLoS Biol 6: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov GI, Naumova ES, Sancho ED, Korhola MP (1996) Polymeric SUC genes in natural populations of Saccharomyces cerevisiae. Fems Microbiol Lett 135: 31–35 [DOI] [PubMed] [Google Scholar]
- Nowell P (1976) The clonal evolution of tumor cell populations. Science 194: 23–28 [DOI] [PubMed] [Google Scholar]
- Pai A, You LC (2009) Optimal tuning of bacterial sensing potential. Mol Syst Biol 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ (2007) CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat Immunol 8: 1353–1362 [DOI] [PubMed] [Google Scholar]
- Pennisi E (2005) How did cooperative behavior evolve. Science 309: 93. [DOI] [PubMed] [Google Scholar]
- Perkins TJ, Swain PS (2009) Strategies for cellular decision-making. Mol Syst Biol 5: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T (2001) Cooperation and competition in the evolution of ATP-producing pathways. Science 292: 504–507 [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Bonhoeffer S (2003) An evolutionary scenario for the transition to undifferentiated multicellularity. Proc Natl Acad Sci USA 100: 1095–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey P, Rainey K (2003) Evolution of cooperation and conflict in experimental bacterial populations. Nature 425: 72–74 [DOI] [PubMed] [Google Scholar]
- Ratledge C, Dover LG (2000) Iron metabolism in pathogenic bacteria. Ann Rev Microbiol 54: 881–941 [DOI] [PubMed] [Google Scholar]
- Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA (2009) Quorum sensing and the social evolution of bacterial virulence. Curr Biol: CB 19: 341–345 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S (2005) Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol 6: 345–352 [DOI] [PubMed] [Google Scholar]
- Sandoz KM, Mitzimberg SM, Schuster M (2007) Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA 104: 15876–15881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffold A, Murphy KM, Hofer T (2007) Competition for cytokines: Treg cells take all. Nat Immunol 8: 1285–1287 [DOI] [PubMed] [Google Scholar]
- Shou WY, Ram S, Vilar JMG (2007) Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA 104: 1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skloot R (2010) The Immortal Life of Henrietta Lacks. NY, USA: Random House [Google Scholar]
- Smith J, Van Dyken JD, Zee PC (2010) A generalization of Hamilton's rule for the evolution of microbial cooperation. Science 328: 1700–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé J-P, Fink GR, Foster KR, Verstrepen KJ (2008) FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135: 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Payne S, Gray M, You L (2009) Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem. Nat Chem Biol 5: 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagkopoulos I, Liu YC, Tavazoie S (2008) Predictive behavior within microbial genetic networks. Science 320: 1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CRL, Fu Q, Buhay C, Kay RR, Shaulsky G (2004) A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development 131: 513–523 [DOI] [PubMed] [Google Scholar]
- Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, Pamer EG (2010) Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120: 4332–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijer CJ (2004) Dictyostelium morphogenesis. Curr Opin Genet Devel 14: 392–398 [DOI] [PubMed] [Google Scholar]
- West SA, Buckling A (2003) Cooperation, virulence and siderophore production in bacterial parasites. Proc R Soc Lond B Biol Sci 270: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP (2006) Social evolution theory for microorganisms. Nat Rev Micro 4: 597–607 [DOI] [PubMed] [Google Scholar]
- Wintermute EH, Silver PA (2010) Emergent cooperation in microbial metabolism. Mol Syst Biol 6: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley JC, Godzik A, Friedberg I (2010) A primer on metagenomics. PLoS Comput Biol 6: e1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier JB, Foster KR (2007) Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA 104: 876–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier JB, Kim W, Foster KR (2011) A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol 79: 166–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier JB, Martinez-Garcia E, Foster KR (2009) Social evolution of spatial patterns in bacterial biofilms: when conflict drives disorder. Am Nat 174: 1–12 [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL (2005) Interference with Al-2-mediated bacterial cell-cell communication. Nature 437: 750–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR (2007) Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol 5: 862–872 [DOI] [PubMed] [Google Scholar]