Abstract
The first step in ergosterol biosynthesis in Saccharomyces cerevisiae consists of the condensation of two acetyl coenzyme A (acetyl-CoA) moieties by acetoacetyl-CoA thiolase, encoded by ERG10. The inhibition of the sterol pathway results in feedback activation of ERG10 transcription. A cell-based reporter assay, in which increased ERG10 transcription results in elevated specific β-galactosidase activity, was used to find novel inhibitors of ergosterol biosynthesis that could serve as chemical starting points for the development of novel antifungal agents. A class of pyridines and pyrimidines identified in this way had no detectable activity against the major fungal pathogen Candida albicans (MICs > 64 μg · ml−1). However, a strain of C. albicans lacking the Cdr1p and Cdr2p efflux pumps was sensitive to the compounds (with MICs ranging from 2 to 64 μg · ml−1), suggesting that they are efficiently removed from wild-type cells. Quantitative analysis of sterol intermediates that accumulated during growth inhibition revealed the accumulation of lanosterol at the expense of ergosterol. Furthermore, a clear correlation was found between the 50% inhibitory concentration at which the sterol profile was altered and the antifungal activity, measured as the MIC. This finding strongly suggests that the inhibition of growth was caused by a reduction in ergosterol synthesis. The compounds described here are a novel class of antifungal pyridines and pyrimidines and the first pyri(mi)dines to be shown to putatively mediate their antifungal activity against C. albicans via lanosterol demethylase.
The sterol biosynthesis pathway, which is taken here to include the mevalonate pathway, converts acetyl coenzyme A (acetyl-CoA) into farnesyl-diphosphate, which subsequently leads to the synthesis of ergosterol. This metabolic pathway has many putative targets that vary in their degrees of genetic conservation relative to fungal and human orthologs. Furthermore, the exploitation of many of these targets has led to therapeutics for the treatment of human disease, and these targets are therefore considered proper objects of drugs. The therapeutics include drugs used for the treatment of fungal infection (azoles, allylamines, thiocarbamates, and morpholines, which all act against fungal targets that have human homologs [21]) and also for the treatment of osteoporosis (2) and hypercholesterolemia (e.g., reference 19).
Dimster-Denk and Rine (5) and Dixon et al. (7) developed virtually identical gene reporter assays for Saccharomyces cerevisiae for the identification of fungal sterol biosynthesis inhibitors that could serve as chemical starting points for new drug discovery programs. The attractiveness of this assay resides in the fact that it can in principle identify inhibitors of any of the essential steps in the pathway. Furthermore, since this is a cell-based assay, all of these inhibitors are expected to have at least some degree of antifungal activity. The use of this assay has led to the identification of a new class of antifungal pyridines and pyrimidines that is also distinct from the most closely related class of antifungal pyrimidines, exemplified by triarimol. Furthermore, whereas triarimol-like pyrimidines have been described as inhibitors of lanosterol demethylase in fungal plant pathogens (20), the compounds described here are the first examples of pyridines and pyrimidines inhibiting lanosterol demethylase (Erg11p) of Candida albicans.
MATERIALS AND METHODS
Strains and media.
Strains used in this study were S. cerevisiae FSB1 (MATα leu2-3,112 ura3-52-pERG10-Escherichia coli lacZ-URA3 rme1 trp1 his4), referred to as MEY133::pACoAT by Dixon et al. (7); C. albicans CAF 2-1 (SC5314 URA3/ura3::λimm434) (9); and C. albicans DSY654 (SC5314 ura3::λimm434/ura3::λimm434 cdr1::hisG/cdr1::hisG cdr2::hisG-URA3-hisG/cdr2::hisG) (17).
S. cerevisiae FSB1 was grown in uracil-deficient yeast minimal broth. One-liter volumes of broth were prepared by adding yeast nitrogen base without amino acids (6.7 g; Difco), adenine sulfate (40 mg), l-arginine-HCl (20 mg), l-methionine (20 mg), l-tyrosine (30 mg), l-isoleucine (30 mg), l-lysine-HCl (30 mg), l-phenylalanine (50 mg), l-glutamic acid (100 mg), l-aspartic acid (100 mg), l-valine (150 mg), l-threonine (200 mg), and l-serine (400 mg). This broth was brought to a pH value of 5.4 and a final volume of 900 ml and autoclaved. Before the broth was used, 100 ml of filter-sterilized 20% glucose was added along with 200 μl of filter-sterilized stock solutions (10 g · liter−1) of each histidine, tryptophan, and leucine. YPD consisted of yeast extract (10 g · liter−1), Bacto Peptone (20 g · liter−1), and glucose (20 g · liter−1).
Susceptibility testing.
The susceptibility of the isolates was determined according to the NCCLS M-27A broth microdilution method (13).
Control inhibitors.
The following control inhibitors were purchased from commercial sources (in parentheses): alendronate (Calbiochem), amphotericin B (Sigma), chlorhexidine (Sigma), cycloheximide (Calbiochem), fluconazole (ICN Biomedicals), flucytosine (Aldrich), 5-fluoro-orotic acid (Acros Organics), lovastatin (Sigma), terbinafine (TCI), and zaragozic acid (Sigma). Lovastatin was activated by heating a 6-mg · ml−1 stock solution in 50% (vol/vol) ethanol-0.2 N NaOH for 40 min at 65°C, after which 1 volume of 1 M Tris HCl (pH 8.0) was added; this stock solution was stored at −20°C (6). A mock solution that did not contain lovastatin but was treated identically was generated. This “lovastatin control” did not contain antifungal activity and did not induce β-galactosidase activity.
Reporter assay.
S. cerevisiae FSB1 was grown overnight in 25 ml of yeast minimal broth in 125-ml flasks at 30°C at 120 rpm in a shaking incubator (ISF-4-W; Kühner, Birsfelden, Switzerland). The culture, typically with an optical density at 600 nm (OD600) of 1 to 3, was diluted to a final OD600 of 0.03 in 50-μl volumes of yeast minimal broth containing a final concentration of 2% (vol/vol) dimethyl sulfoxide (DMSO) and 10 subsequent twofold dilutions of the compound of interest, starting at a maximum concentration of 64 μg · ml−1 (except for control compounds amphotericin B and 5-fluoroorotic acid, for which the starting concentrations were 18 and 512 μg · ml−1, respectively). Samples were prepared in quadruplicate and were distributed into 384-well plates (Costar) and incubated for 24 h at 30°C without shaking (model 5025 incubator; VWR Scientific). Subsequently, well contents were mixed and cell densities were determined with a plate reader (Spectra Fluor Plus; Tecan). To this end, the densities of one set of duplicate wells were measured without dilution; for the other duplicate set, 25 μl of each well was diluted twofold into medium containing the compound, if any, at an identical concentration. This process resulted in the establishment of a reference OD600 for the undiluted wells; wells with OD600s higher than 1.5 or lower than 0.1 were rejected because the cultures in these wells were too dense or too dilute, respectively, for reliable quantitative measurement. To each well, 50 μl of reaction buffer (0.4 M Na2HPO4, 0.2 M NaH2PO4, 0.05 M KCl, 0.01 M MgSO4, 0.135% [vol/vol] β-mercaptoethanol, 0.1% sodium dodecyl sulfate, 2.5 mM chlorophenol red-β-d-galactopyranoside [pH 7.0]) was added as described by Dixon et al. (7), and plates were incubated at 30°C (model 5025 incubator; VWR Scientific). Color development was monitored hourly by mixing well contents, followed by measurement of the A570. The highest level of specific activity was typically found at the highest compound concentration that allowed fungal growth.
Acetate incorporation and analysis of sterol intermediates.
The method for acetate incorporation and the analysis of sterol intermediates was based on that of Ryder et al. (16). Strains of C. albicans were grown overnight in 25 ml of YPD in 125-ml flasks at 30°C with shaking at 130 rpm (ISF-4-W; Kühner), which typically resulted in an OD600 of 5 to 10. Cultures were centrifuged at 4°C at 3,500 rpm for 10 min (Allegra 6R centrifuge; Beckman) and subsequently resuspended in 25 mM NaH2PO4-1% glucose (pH 6.5). Centrifugation was repeated, and the cells were resuspended at a final OD600 of 5.
One-milliliter volumes of resuspended cells were incubated in six-well plates at 30°C with shaking at 150 rpm (Environ shaker; Labline). To each well, 20 μl of DMSO containing various amounts of test compound was added. After 30 min, 10 μl of 100 mM acetic acid was added along with 30 μl of [2-14C]acetate (6 μCi [60 mCi · mmol−1]; Amersham). Labeled acetate was allowed to incorporate for 90 min, after which the contents of each well were added to 2 ml of freshly prepared 15% (wt/vol) ethanolic KOH in 15-ml Falcon tubes. Mixtures were transferred to an 80°C water bath and kept there for 90 min, after which they were cooled to room temperature.
One 3-ml volume of distilled water was added to each tube, followed by the addition of 3 ml of petroleum ether (40 to 60°C, vol/vol). Samples were mixed, and the petroleum ether layer was removed and retained. Another volume of petroleum ether (40:60, vol/vol) was then added, mixed, and recovered. The two aliquots were combined and dried by rotary evaporation (Universal Vacuum System Plus, model UVS800DDA; Savant). The dry preparations were resuspended in 50 μl of hexanes. For thin-layer chromatographic analysis, samples with equal amounts of radioactivity (10,000 to 50,000 dpm) were loaded on Silica Gel 60 F254 plates (Merck, Darmstadt, Germany) and were developed in chloroform. Plates were exposed to a low-energy phosphor screen (Molecular Dynamics) for 1 to 2 days and analyzed with a PhosphorImager (Molecular Dynamics).
In order to confirm the chromatographic properties of various metabolites as they were reported by Ryder et al. (16), 10- to 50-nmol samples of purchased, unlabeled intermediates were loaded and run on Silica Gel 60 plates and visualized by spraying the plates with 40% (wt/vol) sulfuric acid in ethanol, followed by baking for 10 min at 110°C. Rf values determined for intermediates were 0.08 to 0.13 for ergosterol (Sigma), 0.13 to 0.18 for farnesol (ICN), 0.17 to 0.22 for lanosterol (ICN), 0.44 to 0.53 for oxidosqualene (Echelon Biosciences), and 0.72 to 0.75 for squalene (Sigma).
Untreated cells were found to accumulate lanosterol (11%), 4-α-methylated sterols (13%), and ergosterol (76%), whereas the maximum inhibition of lanosterol demethylase resulted in 76% lanosterol, 15% 4-α-methylated sterols, and 9% 4-ergosterol. The IC50, lanosterol was therefore defined as the compound concentration causing half the effect of maximum inhibition, i.e., resulting in a lanosterol content of 45%, and the inhibition of lanosterol demethylase was quantitated by determining the IC50, lanosterols of various compounds.
Chemical synthesis of pyridine and pyrimidine derivatives.
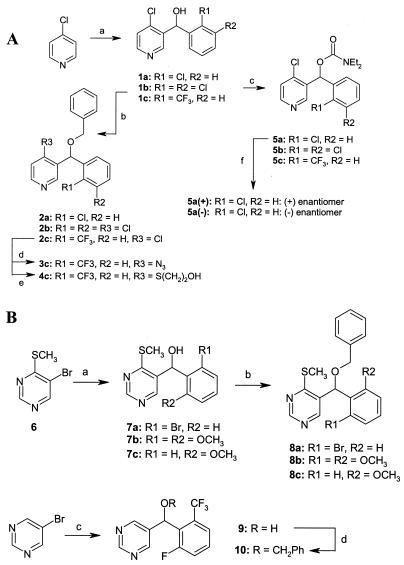
Figure 1 outlines the synthetic schemes and experimental conditions used to prepare the compounds under study. Pyridine derivatives (Fig. 1A) were prepared from 4-chloropyridine. Treatment with a strong base (lithium diisopropylamide [1 eq]-tetrahydrofuran [THF] at −70°C for 1.5 h) produced the 3-lithio species that was reacted in situ with the required substituted benzaldehydes (R1R2PhCHO [1.1 eq] for 1.5 h at −70°C), affording the alcohol intermediates 1a through 1c (30 to 40% yield). Alcohols 1a through 1c were reacted with benzyl bromide (sodium hydride [2.5 eq]-nBu4NI [cat]-PhCH2Br [2 eq]-dimethylformamide [DMF] at 0°C for 2 h; 80 to 90% yield) or diethyl carbamoylchloride (sodium hydride [3 eq]-diethyl carbamoylchloride [2 eq]-THF at 0 to 25°C for 1.5 h; 80 to 90% yield) to give ethers 2a and 2b or carbamates 5a through 5c, respectively. The chloride group of compound 2a was displaced with azide ion (sodium azide [5 eq]-18-crown-6 [cat]-DMF-water [5:1, vol/vol] at 95°C for 18 h; 70% yield) or 2-mercaptoethanol (2-mercaptoethanol [2 eq]-potassium carbonate [6 eq]-DMF at 80°C for 45 min; 80% yield) to yield the derivatives 3c and 4c, respectively. Carbamate 5a was resolved into its component enantiomers via chiral preparative high-pressure liquid chromatography (Chiracel AD, 50 by 500 mm; isocratic elution, 5% isopropanol-95% hexane at 100 ml · min−1; UV detection at 254 nm). The absolute configuration of the enantiomerically pure materials was not determined.
FIG. 1.
Preparation of pyridine (A) and pyrimidine (B) compounds (numbers in boldface refer to the various compounds). (A) Reaction a, lithium diisopropylamide, THF, and (R1R2)PhCHO; reaction b, NaH, PhCH2Br and DMF; reaction c, NaH, diethyl carbamoylchloride, and THF; reaction d, NaN3, DMF, and water; reaction e, 2-mercaptoethanol, K2CO3, and DMF; reaction f, chiral-phase preparative high-pressure liquid chromatography. (B) Reaction a, nBuLi, THF, diethylether, and R1R2PhCHO; reaction b, NaH, PhCH2Br, and DMF; reaction c, nBuLi, THF, diethylether; and 2-fluoro-6-trifluoromethylbenzaldehyde; reaction d, NaH, PhCH2Br, and THF.
Pyrimidine derivatives (Fig. 1B) were prepared from 4-(thiomethyl)-5-bromopyrimidine 6 (4) (nBuLi [1.25 eq]-THF-diethyl ether [2:1, vol/vol] at −70°C for 5 min; then R1R2PhCHO [1.3 eq] for 45 min at −70°C; 35 to 50% yield) or 5-bromopyrimidine (nBuLi [1.4 eq]-THF-diethyl ether [1:1, vol/vol] at −100°C for 30 min; then 2-fluoro-6-trifluoromethylbenzaldehyde [1.4 eq] at −100 to 25°C for 16 h; 65% yield). Alcohols 7a through 7c and 9 were subsequently prepared and converted to the corresponding benzyl ethers, 8a through 8c (sodium hydride [2.5 eq]-nBu4NI [cat]-PhCH2Br [2 eq]-DMF at 0°C for 2 h; 80 to 90% yield) and 10 (sodium hydride [3.5 eq]-THF, with reflux for 5 min; then PhCH2Br [1.4 eq]-NaI [cat] was added, with reflux for 30 min; then 25°C for 16 h; 35% yield), respectively.
RESULTS
A number of years ago, Dimster-Denk and Rine (5) and Dixon et al. (7) developed in parallel a cell-based reporter assay for S. cerevisiae. This assay allows the detection of elevated β-galactosidase activities resulting from the increased transcription of ERG10. In principle, this assay identifies inhibitors of enzymes that make up the biosynthetic pathway that leads to the synthesis of ergosterol from acetyl-CoA, which was the purpose of our study. In this assay, compounds were typically tested at a maximum concentration of 64 μg · ml−1 and at nine subsequent twofold dilutions to a lowest concentration of 0.125 μg · ml−1. As a first step, the assay was validated by using positive and negative control inhibitors. Treatment of cells with lovastatin, zaragozic acid, terbinafine, and fluconazole, which each inhibit a different enzyme in the ergosterol biosynthetic pathway (HMG-CoA reductase [8], squalene synthase [14], squalene epoxidase [15], and lanosterol demethylase [11], respectively), resulted in increased specific β-galactosidase activities compared to that of the DMSO-only control (Table 1). The exposure of cells to the negative-control inhibitors amphotericin B (3), cycloheximide (18), and chlorhexidine (12), which do not inhibit the synthesis of ergosterol but instead disrupt membrane integrity or inhibit translation, did not lead to increased specific β-galactosidase activities. Unexpectedly, however, the negative-control inhibitor flucytosine, which inhibits DNA and RNA biosynthesis (21), also increased β-galactosidase activity (see Discussion).
TABLE 1.
Effects of control inhibitors and pyri(mi)dines on the induction of specific β-galactosidase activity in S. cerevisiae FSB1a
| Compound | β-Galactosidase activity (mA570 · min−1 · OD600−1) |
|---|---|
| Terbinafine | 48 (64) |
| 5-Fluorocytosine | 44 (32) |
| Zaragozic acid | 16 (32) |
| Fluconazole | 12 (64) |
| Lovastatin | 12 (32) |
| Amphotericin B | 2 (9) |
| Cycloheximide | 0 (0.063) |
| Chlorhexidine | −2 (4) |
| DMSO | 1 (NA) |
| 2b | 3 (32) |
| 3c | 12 (64) |
| 4c | 2 (64) |
| 5a(+) | 14 (64) |
| 5a(−) | 4 (64) |
| 5b | 9 (64) |
| 5c | 8 (64) |
| 8a | 10 (32) |
| 8b | 5 (64) |
| 8c | 3 (32) |
| 10 | 3 (64) |
The actual compound concentration (in micrograms per milliliter) at which the maximum activity was obtained is indicated in parentheses. NA, not applicable.
The reporter assay of Dixon et al. (7) was used to screen a corporate compound collection to identify those compounds that possibly mediated their antifungal activities via one of the enzymes required for ergosterol biosynthesis. This assay resulted in a hit rate of 0.3%, and one of the compounds identified in this way was compound 5b (Fig. 1). Compounds related to 5b were synthesized, and the structures and antimicrobial activities of a representative subset of these compounds are shown in Fig. 1 and Table 2, respectively. The reporter assay identified about half of the compounds as putative inhibitors of sterol biosynthesis in S. cerevisiae, as could be detected with maximum compound concentrations of 64 μg · ml−1 (Table 1).
TABLE 2.
Activities of pyridines and pyrimidines against C. albicans CAF2-1 and its efflux-negative derivative C. albicans DSY654 and their potency in inducing lanosterol accumulation in C. albicans DSY654
| Compounda | MIC (μg · ml−1) for:
|
IC50, lanosterol (μg · ml−1) for C. albicans DSY654 | |
|---|---|---|---|
| C. albicans CAF2-1 | C. albicans DSY654 | ||
| 2b | >64 | 16 | 1 |
| 3c | >64 | 2 | 0.125 |
| 4c | >64 | 32 | 8 |
| 5a(+) | >64 | 16 | 16 |
| 5a(−) | >64 | 64 | 32 |
| 5b | >64 | 4 | 0.75 |
| 5c | >64 | 64 | 64 |
| 8a | >64 | 2 | 0.25 |
| 8b | >64 | 4 | 0.125 |
| 8c | >64 | 4 | 0.125 |
| 10 | >64 | 8 | 0.125 |
For compound 5a, because the absolute configuration of the enantiomerically pure materials was not determined, they are referred to here as the (+) and (−) isomers.
None of the compounds were active against the actual pathogen C. albicans, here exemplified by wild-type C. albicans CAF2-1 (9), at concentrations lower than or equal to 64 μg · ml−1. However, when we tested C. albicans DSY654, a strain isogenic with C. albicans CAF2-1 except for the removal of CDR1 (both copies) and CDR2, both of which encode efflux pumps (17), the compounds were found to display antifungal activities in the concentration range tested (Table 2). This finding suggests that this class of compounds is efficiently removed from the wild-type fungal cell by these transporters. This detectable activity against strain DSY654 allowed the antifungal mode of action of these compounds in this strain to be determined.
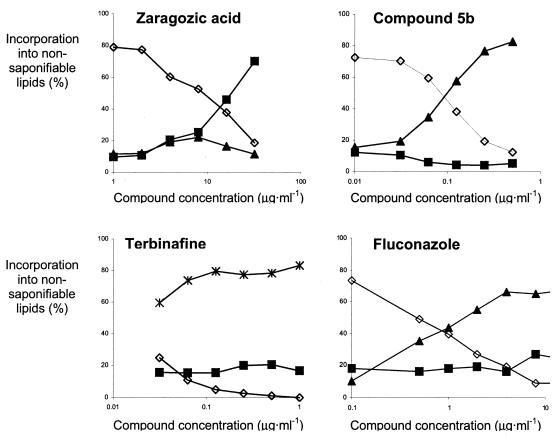
In order to determine which step in fungal ergosterol biosynthesis was inhibited, the fate of incorporated 14C-labeled acetate was monitored as described by Ryder et al. (16). To validate this assay, cells were treated with characterized sterol biosynthesis inhibitors (Fig. 2). In the absence of inhibition, three bands, corresponding to lanosterol, 4-α-methylated sterols, and ergosterol, were observed (16). Treatment with the lanosterol demethylase inhibitor fluconazole led to the accumulation of lanosterol, whereas the squalene epoxidase inhibitor terbinafine caused the accumulation of squalene. As expected, the inhibition of squalene synthase by zaragozic acid did not result in the accumulation of squalene but did lead to the appearance of a previously described (16), uncharacterized intermediate migrating with 4-α-methylated sterol, which we believe to be farnesol (see below). Since lovastatin acts in the mevalonate pathway, the accumulation of sterol-like intermediates was not expected. At a lovastatin concentration of 64 μg · ml−1, the cellular levels of all intermediates were reduced fivefold, but that left the relative amounts of each sterol (95% ergosterol and 5% 4-α-methylated sterols) virtually unchanged relative to those in untreated cells. Finally, since flucytosine resulted in high β-galactosidase activities, the effect of this compound at a concentration of 64 μg · ml−1 on sterol biosynthesis was evaluated, but no effect was found (82% ergosterol, 8% 4-α-methylated sterols, and 8% lanosterol).
FIG. 2.
Incorporation of 14C-labeled acetate into nonsaponifiable lipids of C. albicans DSY654 upon treatment with various concentrations of ergosterol biosynthesis inhibitors. Concentrations of ergosterol (diamonds), lanosterol (triangles), squalene (asterisks), and 4α-methylsterols/farnesol (squares) are shown.
Since the uncharacterized band accumulated when cells were treated with zaragozic acid (Fig. 2) but not when they were treated with either the farnesyl-diphosphate synthase inhibitor alendronate (75% ergosterol, 10% 4-α-methylated sterols, and 14% lanosterol) or lovastatin, it is likely that the metabolite was related to farnesyl-diphosphate. Commercially available 3H-farnesyl-diphosphate was subjected to the same treatment as cell extracts were, i.e., incubation in hot ethanolic KOH and subsequent extraction with petroleum ether. Whereas the nontreated control did not migrate upon thin-layer chromatography in chloroform, the treated control preparation contained a metabolite migrating to a position similar to those of both the uncharacterized band and the commercially available farnesol (data not shown). This finding suggests that the presence of farnesol among nonsaponifiable lipids obtained from zaragozic acid-treated cells of C. albicans is the result of farnesol formation by the cells (10) and the conversion of farnesyl-diphosphate into farnesol during saponification.
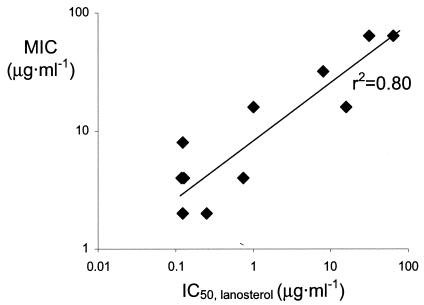
Treatment of cells with the first-discovered compound, 5b, and a number of similar compounds of that class all resulted in the accumulation of lanosterol and thus a pattern identical to that caused by fluconazole. In order to quantitate this inhibition, cells were treated with various concentrations of compound, and as a result, IC50, lanosterols could be determined. Untreated cells were found to accumulate lanosterol (11%), 4-α-methylated sterols (13%), and ergosterol (76%), whereas the maximum inhibition of lanosterol demethylase inhibition resulted in 76% lanosterol, 15% 4-α-methylated sterols, and 9% 4-ergosterol. The IC50, lanosterol was therefore defined as the compound concentration causing half the effect of maximum inhibition, i.e., resulting in a lanosterol content of 45%. Compounds were selected from a range of antifungal activities, and the effects of their presence on ergosterol biosynthesis were determined qualitatively and quantitatively. All compounds caused the accumulation of lanosterol, and their IC50, lanosterols were typically below the MICs of the compounds (Table 2). In addition, a correlation between the IC50, lanosterol and the MIC was obtained (Fig. 3). This finding suggests that the inhibition of lanosterol demethylase is not merely a secondary effect but that the inhibition of lanosterol metabolism is the antifungal mode of action of these pyri(mi)dines.
FIG. 3.
Antifungal activities of pyridines and pyrimidines correlate with their potency in reducing ergosterol biosynthesis in C. albicans DSY654, as indicated by their IC50, lanosterols.
DISCUSSION
With the reporter assay developed by Dimster-Denk and Rine (5) and Dixon et al. (7), a positive signal can be obtained with four antifungal compounds, each inhibiting a different step in the biosynthesis of ergosterol (Table 1). However, it is intriguing that the sizes of the signals varied considerably. The rationale of the assay is that the inhibition of ergosterol biosynthesis leads to lowered levels of this sterol, thereby activating compensatory feedback mechanisms, one of which is the increased transcription of ERG10. This mechanism is common for all four inhibitors, and therefore they would be expected to result in similar signals, albeit possibly at different compound concentrations. The fact that they did not suggests that the feedback mechanism that leads to the transcriptional activation of ERG10 is more complex and depends quantitatively upon where in the pathway inhibition occurs.
The most obvious factor differentiating the control inhibitors is the possible accumulation of intermediates and their incorporation into the cell membrane. Treatment with zaragozic acid or lovastatin led to very similar activities (12 to 16 mA570 · min−1 · OD600−1) (Table 1). Since under these conditions no accumulated intermediates are incorporated into the cell membrane, this finding suggests that this signal was solely the result of diminished ergosterol levels. Treatment with fluconazole also led to β-galactosidase activities of 12 mA570 · min−1 · OD600−1 (Table 1), showing that the accumulation of lanosterol is of no consequence to the signal. Treatment with terbinafine, however, resulted in a three- to fourfold-higher signal (48 mA570 · min−1 · OD600−1) (Table 1), which indicates that the accumulation of squalene has a profound effect on the feedback mechanism that leads to the transcription of ERG10. The implication is that although this assay may in principle detect inhibitors of each step in the pathway, screening results will be biased towards inhibition of more sensitive steps, such as the squalene epoxidase step.
A further complication of the assay is that flucytosine was a strong inducer of the reporter, whereas its antifungal activity is mediated by the inhibition of RNA and DNA biosynthesis (21). In accordance with its mechanism, no alteration in sterol biosynthesis was observed (data not shown). 5-Fluoroorotic acid, which is converted in 5-fluorouracil and has a mode of action similar to that of flucytosine, induced the reporter enzyme as well (9.4 mA570 · min−1 · OD600−1). Since both compounds interfere with the availability of uridine metabolites for RNA biosynthesis, it can be expected that genes involved in uridine biosynthesis will be activated upon exposure to these compounds. One of these genes, URA3, was used to integrate the reporter construct (7), but it is not clear whether terminator sequences that would prevent read-through from URA3 into ERG10-lacZ are present. One explanation for our results is that such transcriptional read-through, leading to β-galactosidase activity, might have occurred. It should be stated, though, that recently published microarray analyses do not provide evidence that an upregulation of URA3 upon the exposure of S. cerevisiae to flucytosine does indeed occur (1, 22).
The induction of the reporter enzyme in the presence of pyridines or pyrimidines suggested that these compounds inhibited sterol biosynthesis in S. cerevisiae. Hence, the effects of this class of compounds on sterol biosynthesis in C. albicans were evaluated in order to rule out interference with uridine metabolism and assess the mode of action in this pathogen. The mutant DSY654 was used because antifungal activity could be observed only in the absence of efflux. Analysis of sterol metabolites showed that lanosterol accumulated but that ergosterol levels decreased. Quantification of this effect led to the determination of IC50, lanosterols, which were typically well below the MICs. This finding implies that when this strain of C. albicans is incubated with compound concentrations at approximately the MIC, sterol metabolism is severely compromised. Furthermore, the antifungal potency of compounds in this series ranks with the potency of inhibition of lanosterol demethylase, as assessed by lanosterol accumulation (Fig. 3). This finding strongly suggests that the inhibition of this step in sterol biosynthesis is the antifungal mode of action of these compounds.
Acknowledgments
We thank Shanta Bist and Julie Demeritt for the synthesis of various compounds and Linda Otterson for antifungal susceptibility testing.
REFERENCES
- 1.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. [DOI] [PubMed] [Google Scholar]
- 2.Bergstrom, J. D., R. G. Bostedor, P. J. Masarachia, A. A. Reszka, and G. Rodan. 2000. Alendronate is a specific, nanomolar inhibitor of farnesyl diphosphate synthase. Arch. Biochem. Biophys. 373:231-241. [DOI] [PubMed] [Google Scholar]
- 3.Bolard, J. 1991. Mechanism of action of an anti-Candida drug: amphotericin B and its derivatives, p. 214-238. In R. Prasad (ed.), Candida albicans. Cellular and molecular biology. Springer-Verlag, Berlin, Germany.
- 4.Dietsche, T. J., D. B. Gorman, J. A. Orvik, G. A. Roth, and W. R. Shiang. 2000. Process chemistry related to the experimental rice herbicide 2,2-dimethyl-1-(4-methylthio-5-pyrimidinyl)indane. Org. Process Res. Dev. 4:275-285. [Google Scholar]
- 5.Dimster-Denk, D., and J. Rine. 1996. Transcriptional regulation of a sterol-biosynthetic enzyme by sterol levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimster-Denk, D., M. K. Thorsness, and J. Rine. 1994. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol. Biol. Cell 5:655-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon, G., D. Scanlon, S. Cooper, and P. Broad. 1997. A reporter gene assay for fungal sterol biosynthesis inhibitors. J. Steroid Biochem. Mol. Biol. 62:165-171. [DOI] [PubMed] [Google Scholar]
- 8.Endo, A. 1988. Chemistry, biochemistry, and pharmacology of HMG-CoA reductase inhibitors. Klin. Wochenschr. 66:421-427. [DOI] [PubMed] [Google Scholar]
- 9.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornby, J. M., B. W. Kebaara, and K. W. Nickerson. 2003. Farnesol biosynthesis in Candida albicans: cellular response to sterol inhibition by zaragozic acid B. Antimicrob. Agents Chemother. 47:2366-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly, S. L., D. C. Lamb, A. J. Corran, B. C. Baldwin, and D. E. Kelly. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem. Biophys. Res. Commun. 207:910-915. [DOI] [PubMed] [Google Scholar]
- 12.Maris, P. 1995. Modes of action of disinfectants. Rev. Sci. Tech. Off. Int. Epizoot. 14:47-55. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth microdilution antifungal susceptibility testing of yeasts: approved standard. Document M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Ponpipom, M. M., N. N. Girotra, R. L. Bugianesi, C. D. Roberts, G. D. Berger, R. M. Burk, R. W. Marquis, W. H. Parsons, K. F. Bartizal, J. D. Bergstrom, M. M. Kurtz, J. C. Onishi, and D. J. Rew. 1994. Structure-activity relationships of C1 and C6 side chains of zaragozic acid A derivatives. J. Med. Chem. 37:4031-4051. [DOI] [PubMed] [Google Scholar]
- 15.Ryder, N. S. 1985. Specific inhibition of fungal sterol biosynthesis by SF 86-327, a new allylamine antimycotic agent. Antimicrob. Agents Chemother. 27:252-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryder, N. S., G. Seidl, and P. F. Troke. 1984. Effect of the antimycotic drug naftifine on growth of and sterol biosynthesis in Candida albicans. Antimicrob. Agents Chemother. 25:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungals: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 18.Stöcklein, W., and W. Piepersberg. 1980. Binding of cycloheximide to ribosomes from wild-type and mutant strains of Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 18:863-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teo, K. K., and J. R. Burton. 2002. Who should receive HMG CoA reductase inhibitors? Drugs 62:1707-1715. [DOI] [PubMed] [Google Scholar]
- 20.Vanden Bossche, H. 1988. Mode of action of pyridine, pyrimidine and azole antifungals, p. 79-119. In D. Berg and M. Plempel (ed.), Sterol biosynthesis inhibitors: pharmaceutical and agrochemical aspects. Ellis Horwood, New York, N.Y.
- 21.Vanden Bossche, H., P. Marichal, and F. C. Odds. 1994. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 2:393-400. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, L., Y. Zhang, Y. Zhou, Y. Zhao, Y. Zhou, and J. Cheng. 2002. Expression profiling of the response of Saccharomyces cerevisiae to 5-fluorocytosine using a DNA microarray. Int. J. Antimicrob. Agents 20:444-450. [DOI] [PubMed] [Google Scholar]