Abstract
The paracaspase mucosa-associated lymphoid tissue 1 (MALT1) is central to lymphocyte activation and lymphomagenesis. MALT1 mediates antigen receptor signalling to NF-κB by acting as a scaffold protein. Furthermore, MALT1 has proteolytic activity that contributes to optimal NF-κB activation by cleaving the NF-κB inhibitor A20. Whether MALT1 protease activity is involved in other signalling pathways, and the identity of the relevant substrates, is unknown. Here, we show that T-cell receptors (TCR) activation, as well as overexpression of the oncogenic API2–MALT1 fusion protein, results in proteolytic inactivation of CYLD by MALT1, which is specifically required for c-jun N-terminal kinase (JNK) activation and the inducible expression of a subset of genes. These results indicate a novel role for MALT1 proteolytic activity in TCR-induced JNK activation and reveal CYLD cleavage as the underlying mechanism.
Keywords: AP-1, cytokine, MAP kinase, signal transduction, ubiquitination
Introduction
Lymphocyte activation has a critical role in immunity. Dysregulation of lymphocyte activation can cause autoimmune diseases, immunodeficiency, or leukaemia and lymphoma. Stimulation of T-cell receptors (TCR) or B-cell receptors (BCR) on the surface of T or B lymphocytes, respectively, induces a series of signal transduction cascades leading to the activation of multiple transcription factors including NF-κB, NF-AT, and AP-1. The paracaspase mucosa-associated lymphoid tissue 1 (MALT1) has emerged as a critical mediator of TCR and BCR signal transduction (Ruefli-Brasse et al, 2003; Ruland et al, 2003; Staal et al, 2011). MALT1 constitutively associates with the CARD-containing protein BCL10. TCR or BCR stimulation triggers the phosphorylation of the CARD-containing adaptor protein CARMA1 (also known as CARD11) by, respectively, protein kinase C (PKC) θ or β (Thome, 2004). This induces a conformational change of CARMA1 and allows BCL10/MALT1 to bind. The assembled CARMA1/BCL10/MALT1 (CBM) complex subsequently recruits TRAF6 and NEMO (also known as IKKγ), resulting in IKKβ activation and NF-κB-dependent gene expression. In parallel, IKKβ is activated by the kinase TAK1 (also known as MAP3K7) (Wang et al, 2001), which is recruited to the CBM complex in an ADAP-dependent manner (Srivastava et al, 2010). TAK1 can also activate AP-1 by stimulating c-jun N-terminal kinase (JNK) mitogen-activated protein kinase (MAPK) phosphorylation via MAPK kinase (MKK)4 and MKK7 (Davis, 2000; Blonska et al, 2007), and enhances NF-AT activation through phosphorylation of RCAN1 (Liu et al, 2009). NF-κB, AP-1, and NF-AT transcription factors regulate the expression of interleukin-2 (IL-2) (Jain et al, 1995), which promotes proliferation and survival of the activated T cells. The above-mentioned signalling pathways can be cell-context dependent. For example, in B cells, MALT1 is specifically important for activation of the NF-κB subunit c-Rel (Ferch et al, 2007), and in primary mouse effector T cells, TAK1 is dispensable for NF-κB but not JNK activation in response to TCR triggering (Wan et al, 2006). According to the work of Ruefli-Brasse et al (2003), MALT1-deficient primary T cells can still activate JNK in response to TCR stimulation. In contrast, Ruland et al (2003) showed that MALT1-deficient T cells are unable to activate JNK. The reason for these differences is still unclear, but might reflect differences in mouse background, gene targeting strategy, or experimental settings. Furthermore, MALT1 might regulate specific JNK isoforms, as suggested by the finding that different JNK isoforms are regulated by CARMA1-Bcl10-dependent and -independent mechanisms (Blonska et al, 2007).
In addition to its role in TCR and BCR signalling, MALT1 also mediates NF-κB activation in response to a number of other receptors in immune and non-immune cells (Dong et al, 2006; Gross et al, 2006, 2008; Klemm et al, 2006, 2007; McAllister-Lucas et al, 2007; Martin et al, 2009; Rehman and Wang, 2009; Tusche et al, 2009). Finally, oncogenic mutations or other genetic events affecting CARMA1, BCL10, or MALT1 in specific B-cell lymphomas such as ABC-type diffuse large B-cell lymphoma and MALT lymphoma enforce NF-κB activation, which is recognized as a key factor in lymphomagenesis.
Besides functioning as a scaffold protein for other signalling molecules, we and others recently showed that MALT1 has proteolytic activity. Although the precise role of MALT1 proteolytic activity is not completely understood, we showed that TCR stimulation results in the MALT1-mediated cleavage of the NF-κB inhibitor A20 (also known as TNFAIP3), which disrupts its NF-κB inhibitory function and results in increased IL-2 production (Coornaert et al, 2008). In addition, the group of Margot Thome described MALT1-mediated cleavage of its binding partner BCL10, which does not modulate NF-κB activity, but is thought to affect integrin-mediated T-cell adhesion (Rebeaud et al, 2008). MALT1 protease activity is necessary for optimal NF-κB activation in antigen receptor-stimulated T cells and has a growth promoting role in ABC-DLBCL (Ferch et al, 2009; Hailfinger et al, 2009). Whether MALT1 protease activity has a functional role in other signalling pathways, and the identity of the relevant substrate, is currently unknown. Here, we demonstrate a role for MALT1 proteolytic activity in TCR-induced JNK activation and describe MALT1-mediated CYLD cleavage as a molecular mechanism.
Results
CYLD is cleaved after TCR and BCR stimulation
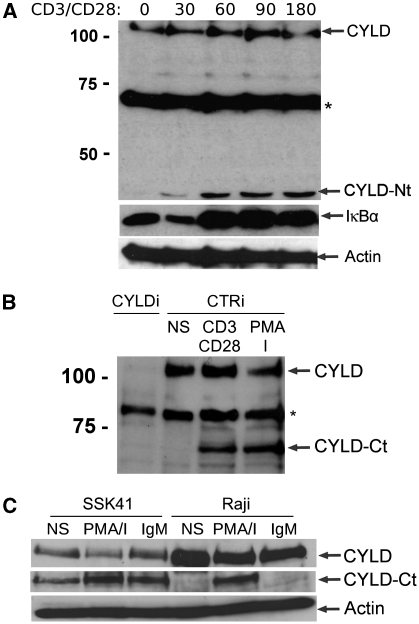
While exploring the role of MALT1 proteolytic activity, we discovered that the deubiquitinating protein CYLD is cleaved in response to TCR stimulation. Upon triggering of the TCR complex of Jurkat cells with anti-CD3 and anti-CD28 for different time points, followed by western blot analysis of complete cell extracts, we noticed the rapid appearance of a band of ∼40 kilodalton (kDa) that was specifically recognized by an antibody raised against CYLD (Figure 1A). The appearance of this band coincided in time with the degradation of the NF-κB inhibitor IκBα that is indicative of NF-κB activation. As the used CYLD antibody was raised against the N-terminus of CYLD, the 40 kDa band represents an N-terminal protein fragment of CYLD. In line with these observations, an antibody raised against the C-terminus of CYLD resulted in the detection of a band of ∼70 kDa, representing the other C-terminal fragment of CYLD (Figure 1B). This band could no longer be detected upon siRNA-mediated silencing of CYLD (Figure 1B), demonstrating CYLD specificity. The above data indicate that CYLD is proteolytically cleaved after TCR stimulation. Similar results were obtained when Jurkat cells were stimulated with phorbol myristate acetate (PMA) plus ionomycine, which mimics TCR signalling by activating PKCθ (Figure 1B). Both generated CYLD fragments were stable over time and their levels were not affected by treatment of the cells with the proteasome inhibitor MG-132 (data not shown). In addition, stimulation of the neoplastic B-cell lines Raji (human Burkitt's lymphoma cell line) and SSK41 (MALT-type B-cell lymphoma that overexpresses MALT1) with PMA plus ionomycin or anti-IgM led to CYLD cleavage (Figure 1C). Of note, the MALT1 overexpressing SSK41 cells already show spontaneous CYLD cleavage.
Figure 1.
CYLD is cleaved upon T-cell and B-cell activation. (A) Jurkat cells were stimulated for the indicated times (minutes) with anti-CD3 plus anti-CD28. CYLD, IκBα (as a read-out for NF-κB activation), and actin (loading control) were analysed by immunoblotting. The CYLD antibody that was used for detection recognizes an N-terminal epitope and detects an N-terminal fragment of CYLD (CYLD-Nt). *Non-specific band. (B) Jurkat cells were stimulated overnight with anti-CD3 plus anti-CD28 or PMA plus ionomycin (PMA/I) and analysed for CYLD expression by immunoblotting (NS, non-stimulated cells). The CYLD antibody that was used for detection recognizes a C-terminal epitope and detects a C-terminal fragment of CYLD (CYLD-Ct). To illustrate the specificity of the signal, Jurkat cells were transfected with CYLD specific (CYLDi) and control (CTRi) siRNA 2 and 3 days prior to stimulation. *Non-specific band. (C) The B-cell lines SSK41 and Raji were stimulated for 1 h with anti-IgM or PMA plus ionomycin and analysed by immunoblotting with a CYLD antibody raised against C-terminal fragment of CYLD. Data are representative of three independent experiments.
CYLD cleavage requires MALT1 proteolytic activity
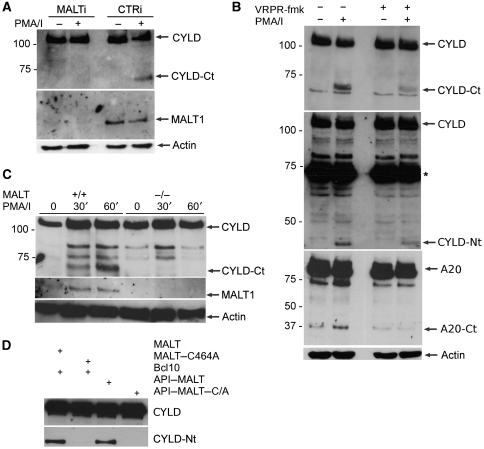
Signalling pathways initiated by TCR and BCR triggering both result in the assembly of the CBM complex and activation of MALT1 proteolytic activity. So far, A20 and BCL10 are the only known substrates for MALT1. Our present results suggest a potential role for MALT1 in CYLD cleavage. To further analyse the function of MALT1 in TCR-induced CYLD cleavage, we transfected Jurkat cells with siRNA directed against MALT1, and stimulated the cells with PMA plus ionomycin. Immunoblot analysis showed that siRNA treatment efficiently downregulated MALT1 expression (Figure 2A). In contrast to cells transfected with control siRNA, cells treated with MALT1-specific siRNA did not have detectable CYLD cleavage after TCR stimulation (Figure 2A), indicating that MALT1 expression is essential for CYLD cleavage. To establish whether CYLD cleavage was dependent on MALT1 proteolytic activity, Jurkat cells were treated with the MALT1 tetrapeptide protease inhibitor z-VRPR-fmk (Rebeaud et al, 2008) prior to PMA plus ionomycin stimulation. z-VRPR-fmk significantly decreased the formation of both CYLD cleavage fragments (Figure 2B), demonstrating that CYLD cleavage is dependent on MALT1 proteolytic activity. Similarly, z-VRPR.fmk inhibited A20 cleavage, which is also mediated by MALT1 (Coornaert et al, 2008), in the same cells (Figure 2B, lower panel). The only partial inhibition of CYLD cleavage may be explained by the fact that z-VRPR.fmk is a rather weak MALT1 inhibitor that is only active in the μM range (Rebeaud et al, 2008). The more pronounced inhibition of A20 cleavage most likely reflects our observation that MALT1 cleaves A20 less efficiently than CYLD. We also compared PMA plus ionomycin-induced CYLD cleavage in primary T cells isolated from, respectively, wild-type and MALT1-deficient mice, and could only detect CYLD cleavage in stimulated wild-type cells (Figure 2C). Finally, we tested the ability of MALT1 and its catalytically inactive C464A mutant to induce the cleavage of CYLD carrying an N-terminal E-tag epitope after overexpression in HEK293T cells. BCL10 was cotransfected to activate MALT1. Immunoblotting with anti-E-tag revealed a CYLD protein fragment of ∼40 kDa after overexpression of MALT1 plus BCL10, but not MALT1–C464A plus BCL10 (Figure 2D), further indicating that the cleavage of CYLD depends on the proteolytic activity of MALT1. Consistent with these findings, overexpression of the constitutively active MALT lymphoma-associated fusion protein API2–MALT1, which lacks the N-terminal death domain and two immunoglobulin-like domains of MALT1, resulted in the generation of the same CYLD fragment, whereas the catalytically inactive API2–MALT1–C464A mutant failed to do so (Figure 2D). These data collectively indicate involvement of MALT1 proteolytic activity in the cleavage of CYLD after TCR stimulation and API2–MALT1 overexpression.
Figure 2.
CYLD cleavage requires MALT1 catalytic activity. (A) Effect of MALT1 silencing. (Top) Immunoblot analysis of CYLD cleavage in Jurkat cells transfected with MALT1-specific siRNA or control siRNA twice, 2 and 3 days prior to stimulation for 1 h with PMA plus ionomycin. CYLD was detected with an antibody raised against the C-terminus and recognizes the C-terminal fragment (CYLD-Ct). (Middle) Immunoblotting for MALT1 to demonstrate MALT1 silencing efficiency. (Bottom) Immunoblot analysis of β-actin (loading control). (B) Effect of an MALT1 peptide inhibitor. Immunoblot analysis of CYLD and A20 cleavage in Jurkat cells stimulated for 1 h with PMA plus ionomycine in the presence or absence of 100 μM of the MALT1 inhibitor z-VRPR-fmk, which was administered to the cells 1 h prior to stimulation. CYLD was detected with an antibody raised against the C-terminus or N-terminus. A20 was detected with an antibody raised against the C-terminus. (Bottom) Immunoblot analysis of β-actin (loading control). (C) Effect in primary T cells derived from MALT1-deficient mice. Primary CD4+ T cells derived from spleens of MALT1 wild-type (+/+) and MALT1-deficient (−/−) mice by MACS separation were stimulated with PMA plus ionomycin for 30 or 60 min and analysed with an antibody raised against the C-terminal fragment of CYLD (top). (Middle) MALT1 deficiency was confirmed by immunoblotting with anti-MALT1 (middle). (Bottom) Immunoblot analysis of β-actin (loading control). (D) Effect of overexpression of MALT1 in HEK293T cells. Cells were transiently transfected with N-terminally E-tagged CYLD and either wild-type MALT1 or catalytically inactive MALT1–C464A plus Bcl10 as indicated (lanes 1 and 2). Similarly, cells were transfected with API2–MALT1 or the catalytically inactive mutant API2–MALT1–C464A (lanes 3 and 4). CYLD cleavage was detected by immunoblotting with anti-E-tag. Data are representative of at least two independent experiments.
MALT1 directly cleaves CYLD at a single conserved site between its second and third Cap-Gly domains
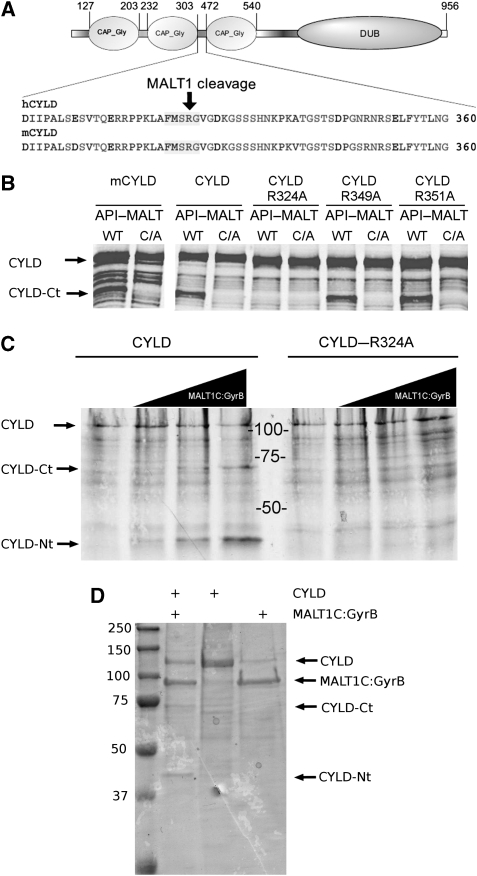
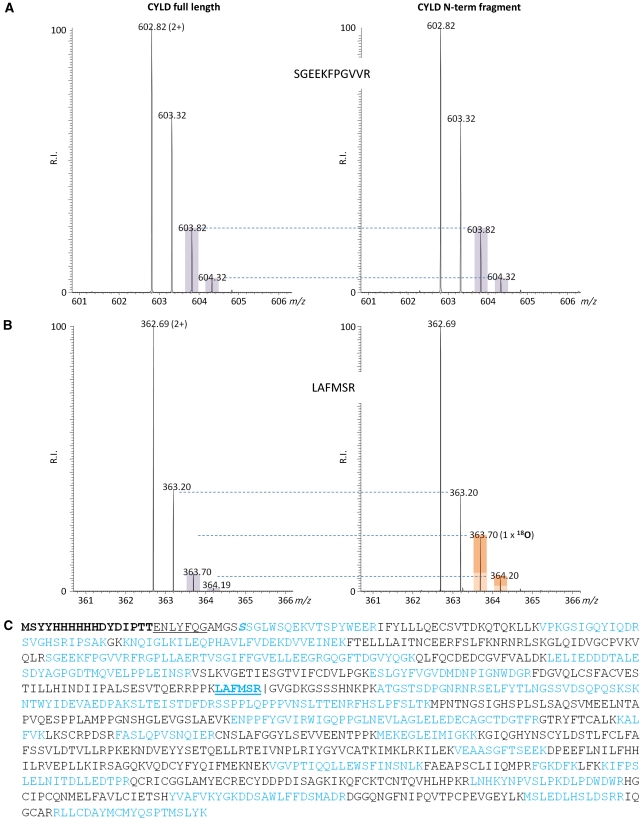
CYLD is a protein of ∼110 kDa with three N-terminal cytoskeletal-associated protein-glycine-conserved (CAP-Gly) domains and a C-terminal catalytic ubiquitin-specific protease (USP) domain that is able to hydrolyse lysine 63-linked ubiquitin chains (Komander et al, 2009). Our observation that TCR stimulation generates an N-terminal CYLD fragment of 40 kDa and a C-terminal fragment of 70 kDa led us predict that CYLD was cleaved at a single site between the second and third CAP-Gly domains (Figure 3A). MALT1 was previously shown to cleave its substrates A20 and BCL10 at the C-terminal side of arginine (P1 site). Three potential cleavage sites are present in the linker region between the second and third CAP-Gly domain. To evaluate the potential cleavage of CYLD at one of these sites, we generated three distinct mutants with substitution of alanine for arginine at position 324 (CYLD–R324A), 349 (CYLD–R349A), and 351 (CYLD–R351A), respectively. MALT1-mediated processing of wild-type CYLD and its mutants was tested upon coexpression with API2–MALT1 in HEK293T cells. Proteolytically inactive API2–MALT1–C464A was used as a negative control. In contrast to wild-type CYLD, CYLD–R349, and CYLD–R351A, we did not detect any cleavage of CYLD–R324A (Figure 3B), indicating that MALT1-mediated cleavage of CYLD occurs at arginine 324, thereby physically separating the two first N-terminal CAP-Gly domains from the C-terminal CAP-Gly and USP domains. The human CYLD cleavage site is conserved in mouse CYLD (Figure 3A) and MALT1-mediated CYLD cleavage generates in both species fragments with a similar size (Figures 2C and 3B). Although we did not determine the exact cleavage site in mouse CYLD, these data indicate that human and mouse CYLD are cleaved at the same site. To confirm that MALT1 directly cleaves CYLD at R324, [35S]methionine-labelled in vitro translated wild-type CYLD and CYLD–R324A was incubated in vitro with increasing concentrations of a recombinant fusion protein of the C-terminal MALT1 catalytic domain (residues 334–824) and an oligomerization domain of bacterial GyraseB (MALT1C–GyraseB), which was previously shown to activate MALT1 catalytic activity (Figure 3C). Incubation of wild-type CYLD with MALT1C–GyraseB generated two fragments of, respectively, 40 and 70 kDa, corresponding to the size of the N-terminal and C-terminal CYLD fragments observed in stimulated T cells or in HEK293 cells transfected with API2–MALT1. In contrast, no fragments were detectable with CYLD–R324A. Similar results were obtained using recombinant CYLD expressed in bacteria (Figure 3D). Finally, the cleavage site was confirmed by mass spectrometry. Therefore, in vitro cleavage was performed in the presence of H218O and gel-pieces of the full length and resulting proteolytic fragments were analysed by liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS) and isotope peak distribution was calculated (Julka et al, 2008). Incorporation of 18O at the C-terminal carboxyl group of the N-terminal CYLD fragment, and its corresponding tryptic peptide, confirms that MALT1 directly cleaves CYLD at arginine 324 (Figure 4).
Figure 3.
CYLD is directly cleaved by MALT1 at R324. (A) Cartoon of the domain structure of CYLD showing the three N-terminal CAP-Gly domains and the C-terminal DUB domain, as well as the amino-acid sequence of the region between the second and third CAP-Gly domains of human and mouse CYLD, respectively. The MALT1 cleavage site is indicated. (B) Mapping of the CYLD cleavage site. (Right) HEK293T cells were transfected with Flag-tagged human wild-type CYLD or the indicated human CYLD mutants together with API2–MALT1 (WT) or its inactive mutant API2–MALT1–C464A (C/A). Immunoblot analysis was performed with the CYLD-specific antibody raised against the C-terminus of CYLD. (C) In vitro cleavage of [S35]methionine-labelled CYLD (left) or CYLD–R324A (right) by increasing concentrations (0, 250, 500, and 1000 nM; wedges) of recombinant MALT1C–GyraseB, incubated for 1.5 h at 37°C, and analysed by SDS–PAGE and autoradiography. Data are representative of two independent experiments. (D) In vitro cleavage of recombinant CYLD by recombinant MALT1C–GyraseB, incubated for 1.5 h at 37°C, and analysed by SDS–PAGE and coomassie staining.
Figure 4.
Determination of the CYLD cleavage site by LC–MS/MS. Recombinant CYLD was treated with recombinant MALT1C–GyraseB (5 μg) in the presence of 18O-labelled water (20% v/v of 93.7% H218O in reaction buffer), incubated for 1.5 h at 37°C, and analysed by SDS–PAGE. Gel-pieces of the full length and resulting proteolytic fragments were cut out and processed as described in the Materials and methods section. (A) MS spectra of representative peptide ions (doubly charged) contained in the N-terminal proteolytic CYLD fragment. The isotopic envelope distributions of the peptides derived from full-length CYLD or the N-terminal proteolytic CYLD fragment, here identified as 137-SGEEKFPGVVR-147, are indistinguishable, hinting that these peptides were not generated upon MALT1C–GyraseB-mediated CYLD proteolysis. (B) The peptide identified as 319-LAFMSR-324 is representative of the MALT1C–GyraseB generated neo-C-terminus in CYLD, as the difference in isotopic envelope distributions are indicative for the stable isotopic incorporation of 18O at the newly exposed carboxyl function of arginine 324 generated upon proteolysis. The extent of LAFMSR(18O), as determined by MS-isotope pattern calculator (http://prospector.ucsf.edu), was calculated 16%. (C) Obtained peptide coverage information of CYLD is highlighted in blue. The N-terminal His-tag is indicated in bold, the thrombin protease cleavage site (ENLYFQ▾G) underlined and the tryptic reporter-peptide indicative for MALT-1-specific cleavage at arginine 324 is in blue and underlined.
TCR-induced JNK activation requires proteolytic inactivation of CYLD
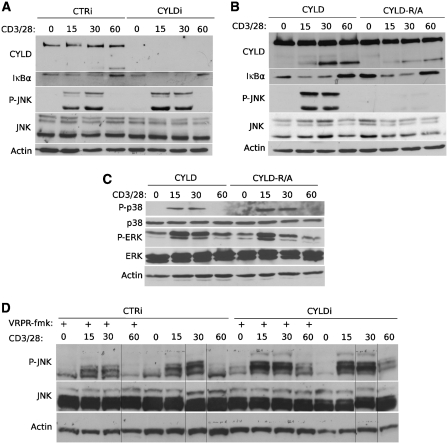
CYLD has been established as a negative regulator of inflammation by inhibiting NF-κB and JNK signalling in response to multiple immune receptors such as Toll-like receptors, TNF receptors, and antigen receptors (Kovalenko et al, 2003; Trompouki et al, 2003; Sun, 2010). However, several studies with CYLD-deficient mice suggest that the regulatory function of CYLD is complex and varies among different cell types and stimulatory conditions (Massoumi et al, 2006; Reiley et al, 2006, 2007; Hövelmeyer et al, 2007). As CYLD also has a pivotal role in regulating T-cell activation (Reiley et al, 2006, 2007), we tested the effect of CYLD silencing on TCR-induced NF-κB and JNK activation. TCR stimulation with anti-CD3 and anti-CD28 rapidly activated NF-κB and JNK, as illustrated by decreased IκBα expression and increased JNK phosphorylation, respectively (Figure 5A). Consistent with the function of CYLD as a negative regulator of TCR-induced NF-κB and JNK signalling, CYLD silencing increased IκBα degradation and JNK phosphorylation (Figure 5A and D). It must be noted, however, that the effect of CYLD silencing was rather weak and did not cause a more sustained IκBα degradation or JNK phosphorylation, suggesting a role for additional regulatory mechanisms. To further investigate the functional relevance of the observed CYLD cleavage, we generated Jurkat cells that stably express comparable levels of either wild-type CYLD or non-cleavable CYLD–R324A. Expression of transfected CYLD in these cells was 4–6 times higher compared with endogenous CYLD (Supplementary Figure S1). Remarkably, TCR-induced activation of JNK was completely and specifically inhibited in cells expressing CYLD–R324A, but not in cells expressing wild-type CYLD (Figure 5B). Interestingly, the inhibitory effect of non-cleavable CYLD expression was specific for JNK, as TCR-induced activation of NF-κB, p38 and ERK MAPK were not affected (Figure 5B and C; the small change in ERK activity that can be noted upon R324A CYLD expression at 30 and 60 min is not reproducible). It should be noted that endogenous CYLD is still present in the above used cell system, which is also reflected by the presence of its cleavage fragment in CYLD–R324A expressing cells (Figure 5B). We therefore also silenced endogenous CYLD in the transfected cells using a morpholino that is specific for the CYLD 5′UTR. Again, CYLD silenced cells complemented with wild-type CYLD do activate JNK in response to TCR stimulation, whereas cells complemented with non-cleavable CYLD do not respond (Supplementary Figure S2).
Figure 5.
MALT1 proteolytic activity and CYLD cleavage are required for JNK signalling. (A) JNK and NF-κB signalling in CYLD silenced Jurkat cells. Immunoblot analysis of CYLD, IκBα, phospho-JNK (P-JNK), JNK, and β-actin from cells that are treated for the indicated times (minutes) with anti-CD3 plus anti-CD28. CYLD was silenced by transfection of CYLD-specific siRNA (CYLDi) or control siRNA (CTRi) 2 and 3 days prior to stimulation. (B) JNK and NF-κB signalling in Jurkat cells transfected with non-cleavable CYLD–R324A. Immunoblot analysis of CYLD, IκBα, phospho-JNK (P-JNK), JNK, and β-actin from anti-CD3 plus anti-CD28-stimulated cells that stably express wild-type CYLD or non-cleavable CYLD–R324A. The cleavage fragment in CYLD–R324A expressing cells results from endogenous CYLD expression. Identical results were obtained with several independent clones showing comparable expression of wild-type and CYLD–R324A. (C) p38 and ERK signalling in Jurkat cells transfected with non-cleavable CYLD–R324A. Immunoblot analysis of phospho-p38 (P-p38), p38, phospho-ERK (P-ERK), or β-actin from Jurkat cells that stably express wild-type CYLD or uncleavable CYLD–R324A. (D) JNK signalling in Jurkat cells in the presence of a MALT1 protease inhibitor. Immunoblot analysis of phospho-JNK (P-JNK), JNK, and β-actin from cells treated for the indicated times (minutes) with anti-CD3 plus anti-CD28. CYLD was silenced by transfection of CYLD-specific siRNA (CYLDi) or control siRNA (CTRi) 2 and 3 days prior to stimulation. Where indicated, cells received 100 μM of the MALT1 inhibitor z-VRPR-fmk 1 h prior to stimulation. Samples were loaded on a single gel. Data are representative of two (C, D) or three (A, B) independent experiments.
The above results indicate that TCR-induced JNK activation requires the proteolytic inactivation of CYLD by MALT1. Consistent with this, the MALT1 inhibitor z-VRPR.fmk significantly reduced TCR-induced JNK activation (Figure 5D, left part). The only partial inhibition of TCR-induced JNK activation most likely reflects our previous observation that this inhibitor also only partially prevents CYLD cleavage (Figure 2B). As z-VRPR.fmk did no longer inhibit JNK activation in CYLD silenced cells (Figure 5D, right part), we can conclude that inhibition of JNK activation by z-VRPR.fmk reflects the inhibition of MALT1-mediated CYLD cleavage and not any other effect. Altogether, these results indicate a novel role for MALT1 proteolytic activity in TCR-induced JNK activation and reveal proteolytic inactivation of CYLD as the underlying mechanism.
CYLD cleavage is required for the inducible expression of specific genes
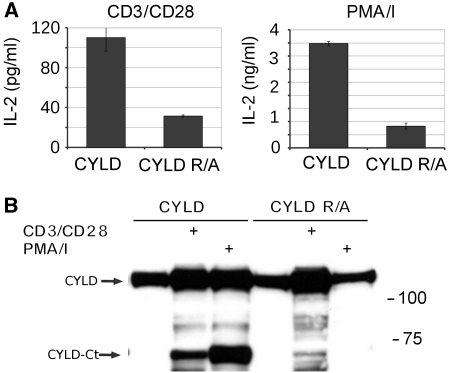
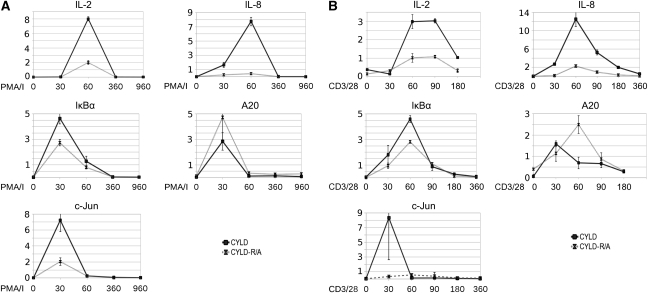
JNK activation has a key role in the activation of multiple transcription factors including AP-1, which together with NF-κB and NF-AT regulates the TCR-inducible expression of IL-2 in T cells. We, therefore, compared anti-CD3 plus anti-CD28-induced IL-2 production in Jurkat cells expressing either wild-type or CYLD–R324A, which is resistant to MALT1 cleavage. Consistent with the absence of JNK activation in CYLD–R324A expressing cells, the amount of IL-2 protein and mRNA produced by these cells was much lower compared with cells expressing wild-type CYLD (Figure 6). Similar results were obtained when TCR-induced signalling was mimicked by stimulation of the cells with PMA plus ionomycin. Also, inducible expression of IL-8 and c-jun, which requires AP-1 (Angel et al, 1988; Khalaf et al, 2010), was much lower in cells expressing CYLD–R324A compared with wild-type CYLD (Figure 7). IκBα and A20 are two genes whose expression is NF-κB dependent, but AP-1 independent (Krikos et al, 1992; Zhang et al, 2005). In agreement with our findings that CYLD cleavage does not affect TCR-induced NF-κB activation, TCR-inducible expression of IκBα was only marginally decreased, and A20 expression was even increased in cells expressing non-cleavable CYLD. Altogether, these data demonstrate that MALT1-mediated CYLD cleavage determines the specific gene expression profile of stimulated T cells.
Figure 6.
CYLD cleavage is required for TCR-induced IL-2 production in Jurkat cells. (A) IL-2 secretion. Jurkat cells stably expressing either wild-type CYLD or uncleavable CYLD–R324A were stimulated with anti-CD3 plus anti-CD28 (left) or PMA plus ionomycine (right) for 16 h, and the amount of IL-2 in the supernatant was analysed by ELISA. Endogenous CYLD was silenced with CYLD-specific morpholinos that were electroporated in the cells 2 and 3 days prior to stimulation. ELISA was performed in duplicate. Values and s.d. represent results from two independent duplicate experiments. (B) CYLD cleavage. Immunoblotting for CYLD of the cells used in (A). Data are representative of two independent experiments.
Figure 7.
CYLD cleavage is required for TCR-induced expression of a subset of genes in Jurkat cells. (A) Jurkat cells stably expressing either wild-type CYLD (black line with ▪ symbol) or uncleavable CYLD–R324A (grey line with x symbol) were stimulated with PMA plus ionomycine (A) or anti-CD3 plus anti-CD28 (B) for the times indicated (minutes), and IL-2, IL-8, A20, IκBα, and c-Jun expression were determined by Q-PCR and normalized against the two house-keeping genes HPRT1 and PPIA. Data are representative of three independent experiments.
Discussion
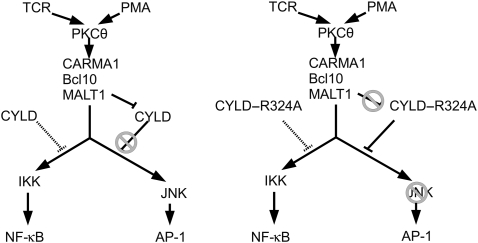
The deubiquitinating protein CYLD regulates diverse physiological functions ranging from immunity and inflammation, to endocytosis, mitosis, spermatogenesis, and osteoclastogenesis, and has a predominant role in the regulation of NF-κB and MAPK signalling pathways that can contribute in several ways to its physiological functions (Sun, 2010). The mechanism by which CYLD interferes with these pathways is quite complex and only partially understood. The removal of K63-linked polyubiquitin chains from several specific substrates, including NEMO, TRAF2, TRAF6, TAK1, and RIP1, has been suggested to mediate its biological activities (Sun, 2010). In most cells, CYLD is constitutively expressed, suggesting an important role for posttranslational regulatory mechanisms. It should be noted that increased CYLD expression upon TCR stimulation can be observed in several of our experiments. As this is the case for endogenous (Figures 1A and 2C) as well as transfected CYLD (Figure 6B), and occurs as early as 15 min after stimulation (Supplementary Figure S2A), increased CYLD expression most likely reflects stabilization of CYLD mRNA or protein. Here, we show that CYLD activity can also be regulated by proteolytic processing. More specifically, we show that TCR and BCR stimulation results in the rapid cleavage of CYLD by the paracaspase MALT1, which is a prerequisite for TCR-induced JNK activation (Figure 8). In contrast, TCR-induced activation of p38 and ERK MAPK, as well as NF-κB activity, occurs independent of CYLD cleavage. CYLD has also been shown to control NF-AT activation (Koga et al, 2008) and the shuttling of BCL3 between the nucleus and the cytoplasm (Massoumi et al, 2006; Hövelmeyer et al, 2007), but also these activities seem to be independent of CYLD cleavage (data not shown). Consistent with the observed requirement of CYLD cleavage for TCR-induced JNK signalling, we found that inhibition of CYLD cleavage specifically prevents the expression of a subset of genes, most likely reflecting their dependency on AP-1, which is regulated by JNK. Differential regulation of, respectively, JNK and NF-κB signalling by CYLD has previously also been observed in TNF-stimulated cells (Reiley et al, 2004). However, it is unlikely that differences in TNF signalling result from MALT1-mediated CYLD cleavage, as TNF signalling does not involve MALT1. Presumably, other mechanisms control the regulatory function of CYLD on JNK signalling in response to TNF. In this context, IKK-mediated phosphorylation of CYLD has already been shown to prevent the inhibitory activity of CYLD on TNF-induced JNK activation (Hutti et al, 2009). We were not able to observe JNK activation in TNF-treated Jurkat cells (Supplementary Figure S3A), and consistent with this, we also did not observe AP-1-dependent gene expression (c-jun, IL-2) in response to TNF (data not shown). In contrast, TNF activates NF-κB and increases IκBα expression (which is largely AP-1 independent, but NF-κB dependent) in Jurkat cells, but this TNF effect was comparable in non-transfected cells or cells transfected with wild-type CYLD or CYLD–R324A (Supplementary Figure S3B).
Figure 8.
Scheme illustrating our model for the role of MALT1-mediated CYLD cleavage in TCR-induced JNK activation. TCR stimulation or direct activation of PKC by the phorbol ester TPA results in the activation of the CARMA1/BCL10/MALT1 complex, leading to IKK- and JNK-mediated activation of NF-κB and AP-1 transcription factors, respectively. CYLD is negatively regulating both signalling pathways. (Left panel) MALT1 rapidly cleaves CYLD (as part of a signalling complex that specifically controls JNK activation), preventing it from blocking JNK activation. (Right panel) MALT1 cannot cleave the CYLD–R324A mutant, which can now fully inhibit JNK activation.
CYLD has been reported to regulate NF-κB and JNK signalling in a cell type and stimulus-dependent manner (Reiley et al, 2007). In the present study, CYLD silencing in Jurkat cells weakly increased TCR-induced JNK and NF-κB activation. However, cells reconstituted with a non-cleavable CYLD mutant were completely defective in TCR-induced JNK activation, indicating that CYLD does have a key role in the negative regulation of JNK signalling, but is rapidly inactivated by MALT1-mediated cleavage upon TCR stimulation. The observation that NF-κB activation is not affected by CYLD cleavage might be explained by functional redundancy with other NF-κB regulatory proteins such as A20 or Cbl-b (Qiao et al, 2008; Coornaert et al, 2009). Alternatively, CYLD could be part of an NF-κB signalling complex that is not accessible for MALT1-mediated cleavage. It is worth mentioning that only a subfraction of CYLD is cleaved upon TCR stimulation, consistent with the existence of specific MALT1/CYLD/JNK signalling complexes. Such complexes might exist only transiently as we were not able to coimmunoprecipitate MALT1 and CYLD (data not shown).
Signalling to JNK and NF-κB bifurcate downstream of the MAPK kinase kinase (MAP3K) TAK1, which phosphorylates and activates IKKβ, MKK6, and MKK7, leading to the activation of NF-κB, p38, and JNK kinase pathways, respectively (Wang et al, 2001; Liu et al, 2006; Wan et al, 2006). In addition to TAK1, other MAP3Ks might also be involved in the regulation of JNK activation. MAPK specificity is obtained by the binding of specific scaffold proteins to components of MAPK signalling modules. For example, JNK-interacting protein (JIP) 1 forms a complex with MEKK3, MKK7, and JNK to specifically activate JNK (Engström et al, 2010). Of note, BCL10 has been described as a JIP-like scaffold to assemble JNK2, MKK7, and TAK1 in the TCR signalling pathway (Blonska et al, 2007). TCR-induced NF-κB and JNK signalling has also been reported to be differentially regulated by the death domain containing protein Siva, which forms a ternary complex with TAK1 and XIAP (Resch et al, 2009). CYLD might regulate the formation of these protein complexes by affecting the ubiquitination of specific components. MALT1-mediated cleavage of CYLD after arginine 324 results in the generation of an N-terminal fragment containing the first two CAP-Gly domains and a C-terminal fragment containing the third CAP-Gly domain and the USP domain. It has previously been shown that deletion of the CAP-Gly domains does not affect the deubiquitinating activity of CYLD in vitro (Kovalenko et al, 2003). CYLD cleavage can, therefore, be expected not to affect CYLD catalytic activity as such, but might affect its function in other ways. The C-terminal fragment contains the binding sites for NEMO (amino acids 472–540) (Kovalenko et al, 2003), TRAF2 (amino acids 453–457) (Kovalenko et al, 2003), and BCL3 (amino acids 470–957; exact binding site has not been defined) (Massoumi et al, 2006). The first CAP-Gly motif in the N-terminal fragment mediates the binding of CYLD to tubulin, which might be implicated in the regulation of microtubule assembly and cell migration, and possibly also cell cycle progression, by CYLD (Gao et al, 2008). CYLD cleavage by MALT1 removes the first two CAP-Gly domains and can, therefore, be expected to release the catalytically active USP domain from microtubules. This could in turn affect the ubiquitination of tubulin or microtubule-associated proteins. Whether such proteins are involved in JNK activation is, however, still unknown. It is worth mentioning that naturally occurring shorter CYLD variants, with a different activity than their full-length cousin, have been described before in other cells. For example, a shorter splice variant of CYLD that is devoid of the TRAF2 and NEMO-binding domains is a positive regulator of NF-κB activity and induces a hyperactive phenotype in dendritic cells (Srokowski et al, 2009). CYLD gene mutations that are associated with different skin appendage tumours also lead to truncated versions of the protein, some of which only retain the first two CAP-Gly domains (Courtois, 2008), corresponding to the N-terminal CYLD fragment that is generated by MALT1. It cannot be excluded that some of these truncated proteins or fragments acquired novel biological functions. It should be noted, however, that the absence of JNK activation in CYLD–R324A expressing cells, despite the presence of the endogenous CYLD fragment (Figure 5B), excludes a gain of function effect of MALT1-mediated CYLD cleavage in our system.
In summary, our findings show an important role for MALT1 proteolytic activity in TCR-induced JNK activation and the fine tuning of gene expression, and reveal MALT1-mediated CYLD cleavage as the underlying mechanism. It remains to be investigated if and how CYLD cleavage affects its physiological function in the regulation of immune responses in vivo. As CYLD is also cleaved upon overexpression of the oncogenic fusion protein API2–MALT1, it will also be of interest to determine if CYLD cleavage is involved in the pathogenesis of MALT lymphoma or other B-cell lymphomas that are characterized by hyperactivation of the MALT1 pathway. It is intriguing that MALT1 cleaves and inactivates two deubiquitinating proteins, CYLD and A20, that both have a key role in the negative regulation of inflammation and immunity. Clearly, MALT1 serves as a master regulator and represents an interesting drug target for immuno-suppression or lymphoma treatment.
Materials and methods
Plasmids
Details of plasmids and cloning procedures are presented along with detailed sequence maps at the BCCM-LMBP plasmid databank http://bccm.belspo.be/index.php. Mouse CYLD (LMBP 6356 and 6692), human CYLD (LMBP 5579 and 6613), human CYLD–R324A (LMBP 6114 and 6645), human CYLD–R349A (LMBP 6099), human CYLD–R351A (LMBP 6100), Bcl-10 (LMBP 5535), MALT1 (LMBP 5536), MALT1–C464A (LMBP 5541), API2–MALT1 (LMBP 5537), and API2–MALT1–C464A (LMBP 5538).
Cell culture and transfections
HEK293T, Jurkat, Raji, and SSK41 cells were cultured as described before. Silencing of CYLD in Jurkat cells was obtained by electroporating (300 V and 1050 μF; Genepulser Bio-Rad) the cells twice, 2 and 3 days prior to stimulation, with 400 nM siRNA (L-004609-00-005, Dharmacon) in 0.5 ml serum-free medium per 5 × 106 cells. For stable transfection, wild-type CYLD and CYLD–R324A plasmids were linearized with NruI and electroporated into Jurkat cells (250 V and 900 μF). Stably transfected cells were selected on 1 mg/ml geneticin for 6 weeks prior to single clone selection. Single clones were isolated by limiting dilution in 96-well format using conditioned RPMI medium and geneticin selection with an average of 1 cell/3 wells. Single clones were analysed for CYLD expression following immunoprecipitation with anti-Flag and immunoblot detection with anti-CYLD. Two independent comparison pairs of equal expression level (wild-type versus CYLD–R324A; Supplementary Figure S1) were used for further experiments. For reconstitution experiments, endogenous CYLD was silenced in the stably CYLD expressing lines using a CYLD 5′ UTR-specific (GGGAAACTACTAATAACTTTGTCAT) morpholino (Gene Tools LLC). As silencing control, standard non-target morpholino (CCTCTTACCTCAgTTACAATTTATA) was used. Morpholinos (15 nmol per 5 × 106 cells) were electroporated into Jurkats twice, 2 and 3 days prior to stimulation (300 V and 1050 μF).
MALT1-deficient mice were used as a source of primary T cells. Spleens were collected in ice cold PBS-0.5% BSA and cells were washed out. This suspension was passed through a 70-μm cell strainer. CD4+ T cells were isolated by MACS separation according to the manufacturer's protocol (Miltenyi Biotec, Germany).
Cells were treated with 10 μg/ml anti-CD3 (UCHT-1; Pharmingen) plus 10 μg/ml anti-CD28 (28.2; Pharmingen), or 200 ng/ml PMA (SIGMA-Aldrich) plus 1 μM ionomycin (Calbiochem). Raji and SSK41 cells were stimulated for 1 h with 20 μg/ml anti-IgM.
Immunoblot analysis
Cells were lysed in 50 mM Hepes, pH 7.6, 250 mM NaCl, 5 mM EDTA, and 0.5% (vol/vol) NP-40, plus phosphatase and protease inhibitors, and proteins were separated by SDS–PAGE and analysed by semi-dry immunoblotting and detection via enhanced chemiluminescence (Perkin-Elmer Life Sciences). The antibodies that were used are anti-CYLD recognizing the N-terminal fragment (gift from Dr SC Sun) or the C-terminal fragment (E10, Santa Cruz Biotechnology Inc.), anti-MALT1 (H-300, Santa Cruz Biotechnology Inc.), anti-P-JNK (Cell Signaling, #9251 and 44682G, Invitrogen), anti-JNK (sc-571, Santa Cruz Biotechnology Inc.), anti-IκBα (sc-371, Santa Cruz Biotechnology Inc.), anti-P-p38 (#9215, Cell Signaling), anti-p38 (#9212, Cell Signaling), anti-P-ERK (#9101, Cell Signaling), anti-ERK (#9102, Cell Signaling), anti-actin (MP 6472J, MP Biomedicals), HRP-coupled anti-Flag (A-8592, Sigma), anti-E-tag (Ab66152-200, Abcam), and anti-A20 (clone 59A426, eBioscience).
ELISA and quantitative real-time PCR
IL-2 in the supernatant of cells was detected with an ELISA (BD OptEIA hIL-2 ELISA kit, Pharmingen) according to the manufacturer's instructions. For quantitative real-time PCR, total RNA was isolated from Jurkat cells using the Aurum Total RNA Mini Kit (Bio-Rad), prior to cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. A total of 10 ng of cDNA was used for quantitative PCR in a total volume of 10 μl with LightCycler 480 SYBR Green I Master Mix (Roche) and specific primers on a LightCycler 480 (Roche). Real-time PCR reactions were performed in triplicates and normalized based on two house-keeping genes (HPRT1 and PPIA). The following primers were used: A20 forward, AGAAGAGCAACTGAGATCGAG; A20 reverse, GCCATACATCTGCTTGAACTG; c-Jun forward, GCCGGTCTACGCAAACCTC; c-Jun reverse, GGACTCCATGTCGATGGGG; HPRT1 forward, TGACACTGGCAAAACAATGCA; HPRT1 reverse, GGTCCTTTTCACCAGCAAGCT; IκBα forward, GCTGATGTCAACAGAGTTACC; IκBα reverse, CTCTCCTCATCCTCACTCTC; IL-2 forward, AACTCACCAAGGATGCTCACATTT; IL-2 reverse, TTAGCACTTCCTCCAGAGGTTTG; IL-8 forward, ACTGAGAGTGATTGAGAGAGTGGAC; IL-8 reverse, AACCCTCTGCACCCAGTTTTC; PPIA forward, TCCTGGCATCTTGTCCATG; PPIA reverse, CCATCCAACCACTCAGTCTTG.
In vitro cleavage of CYLD
pCDNA3 expression vectors of CYLD and CYLD–R324A under the control of the T7 promoter were used for in vitro coupled transcription translation of [S35]methionine-labelled CYLD using wheat germ and reticulocyte extracts according to the manufacturer's instructions (Promega Biosystems Inc.) and served as a source of CYLD. Alternatively, we used 4 μg of recombinant CYLD (#64-0010-050, Ubiquigent) that was expressed and purified from bacteria. In vitro cleavage with recombinant MALT1C–GyraseB was performed as described previously (Coornaert et al, 2008) in a buffer consisting of sodium citrate (1 M), NaCl (150 mM), glycerol (10%), MES (50 mM), and 10 mM DTT, pH=6.8. CYLD was incubated with 5 μg recombinant MALT1C–GyraseB and ∼20% H218O for mass spectrometry confirmation of the cleavage site.
Protein identification by LC–MS/MS analysis
SDS–PAGE-separated and coomassie-stained gel bands were excised and washed for 15 min with water, twice with water/acetonitrile (1/1), and finally with acetonitrile. Proteins were in-gel digested overnight at 37°C with 0.1 μg of sequencing-grade modified trypsin (Promega Corporation, Madison). The resulting peptide mixtures were acidified (0.1% formic acid) and analysed by LC–MS/MS analysis. LC–MS/MS analysis was done using an Ultimate 3000 HPLC system (Dionex) in-line connected to a LTQ Orbitrap XL mass spectrometer (Thermo Electron) as described before (Arnesen et al, 2010). Peak lists of the generated MS/MS spectra were then searched with Mascot using the Mascot Daemon interface (version 2.2.0, Matrix Science). Spectra were searched against the human section of the SwissProt database. Variable modifications were set to methionine oxidation and pyro-glutamate formation of N-terminal glutamine. Mass tolerance on the precursor ion was set to 10 p.p.m., and 0.5 Da mass tolerance was used for peptide fragment ions. The peptide charge was set to 1+, 2+, 3+ and 1 missed cleavage by trypsin, without restriction to RP- and RK-cleavage, was allowed. Peptide identifications were considered significant when their Mascot ion score was above the identity threshold set at a 95% confidence interval and when they were top ranked.
The extent of 18O-incorparation in the MALT1C–GyraseB-mediated neo-C-terminus of CYLD (i.e. the C-terminus of the N-terminal CYLD fragment) was calculated from the peptide ion signals observed in the MS spectra. Here, the modified peptide sequences were used to calculate the theoretical isotope peak distribution using the MS-isotope pattern calculator (http://prospector.ucsf.edu). The predicted intensity of the third isotope in the isotopic pattern of unincorporated peptide was subtracted from the measured intensity of the monoisotopic peak of the isotope cluster of the 18O-incorparated peptide to correct for the overlapping isotopic envelopes.
Statistical analysis and data processing
Statistical analysis was performed in OpenOffice.org Calc on openSUSE. Images were prepared using GIMP (http://www.gimp.org).
Supplementary Material
Acknowledgments
We thank Zhijian Chen (Department of Molecular Biology, Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas) for the generous gift of recombinant MALT1 protein, Sun Shao-Cong and Xuhong Cheng (MD Anderson Cancer Center, University of Texas, Huston) for CYLD antibodies, and Tak Mak (Ontario Cancer Institute, Toronto) for MALT1-deficient mice. SSK41 cells were generously provided by Mathijs Baens (University of Leuven, Belgium). JS, DM and PVD are postdoctoral researchers, and TB is a PhD fellow with the ‘Fund for Scientific Research of Flanders’ (FWO). AD is a PhD fellow with the ‘Agency for Innovation by Science and Technology in Flanders’ (IWT). Work in the authors’ lab is further supported by research grants from the ‘Interuniversity Attraction Poles program’ (IAP6/18), the FWO (G.0089.10), the ‘Belgian Foundation against Cancer’, the ‘Strategic Basis Research program’ of the IWT, the ‘Centrum voor Gezwelziekten’, and the ‘Concerted Research Actions’ (GOA grant 01G06B6) and ‘Group-ID MRP’ initiative of the Ghent University. JS, YD, TB, AD, DM, KG and PVD performed the experiments; JS and RB wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Angel P, Hattori K, Smeal T, Karin M (1988) The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 55: 875–885 [DOI] [PubMed] [Google Scholar]
- Arnesen T, Starheim K, Van Damme P, Evjenth R, Betts M, Ryningen A, Vandekerckhove J, Gevaert K, Anderson D (2010) The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol Cell Biol 30: 1898–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonska M, Pappu BP, Matsumoto R, Li H, Su B, Wang D, Lin X (2007) The CARMA1-Bcl10 signaling complex selectively regulates JNK2 kinase in the T cell receptor-signaling pathway. Immunity 26: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R (2008) T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat Immunol 9: 263–271 [DOI] [PubMed] [Google Scholar]
- Coornaert B, Carpentier I, Beyaert R (2009) A20: central gatekeeper in inflammation and immunity. J Biol Chem 284: 8217–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G (2008) Tumor suppressor CYLD: negative regulation of NF-κB signaling and more. Cell Mol Life Sci 65: 1123–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103: 239–252 [DOI] [PubMed] [Google Scholar]
- Dong W, Liu Y, Peng J, Chen L, Zou T, Xiao H, Liu Z, Li W, Bu Y, Qi Y (2006) The IRAK-1-BCL10-MALT1-TRAF6-TAK1 cascade mediates signaling to NF-κB from Toll-like receptor 4. J Biol Chem 281: 26029–26040 [DOI] [PubMed] [Google Scholar]
- Engström W, Ward A, Moorwood K (2010) The role of scaffold proteins in JNK signalling. Cell Prolif 43: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferch U, Buschenfelde CMZ, Gewies A, Wegener E, Rauser S, Peschel C, Krappmann D, Ruland J (2007) MALT1 directs B cell receptor-induced canonical nuclear factor-[kappa]B signaling selectively to the c-Rel subunit. Nat Immunol 8: 984–991 [DOI] [PubMed] [Google Scholar]
- Ferch U, Kloo B, Gewies A, Pfänder V, Düwel M, Peschel C, Krappmann D, Ruland J (2009) Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med 206: 2313–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Huo L, Sun X, Liu M, Li D, Dong J, Zhou J (2008) The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem 283: 8802–8809 [DOI] [PubMed] [Google Scholar]
- Gross O, Gewies A, Finger K, Schäfer M, Sparwasser T, Peschel C, Förster I, Ruland J (2006) Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 442: 651–656 [DOI] [PubMed] [Google Scholar]
- Gross O, Grupp C, Steinberg C, Zimmermann S, Strasser D, Hannesschlager N, Reindl W, Jonsson H, Huo H, Littman DR, Peschel C, Yokoyama WM, Krug A, Ruland J (2008) Multiple ITAM-coupled NK cell receptors engage the Bcl10/Malt1 complex via Carma1 for NF-κB and MAPK activation to selectively control cytokine production. Blood 112: 2421–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, Thome M (2009) Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 106: 19946–19951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hövelmeyer N, Wunderlich FT, Massoumi R, Jakobsen CG, Song J, Worns MA, Merkwirth C, Kovalenko A, Aumailley M, Strand D, Bruning JC, Galle PR, Wallach D, Fassler R, Waisman A (2007) Regulation of B cell homeostasis and activation by the tumor suppressor gene CYLD. J Exp Med 204: 2615–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, Hahn WC, Cantley LC (2009) Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKε promotes cell transformation. Mol Cell 34: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J, Loh C, Rao A (1995) Transcriptional regulation of the IL-2 gene. Curr Opin Immunol 7: 333–342 [DOI] [PubMed] [Google Scholar]
- Julka S, Dielman D, Young SA (2008) Detection of C-terminal peptide of proteins using isotope coding strategies. J Chromatogr B Analyt Technol Biomed Life Sci 874: 101–110 [DOI] [PubMed] [Google Scholar]
- Khalaf H, Jass J, Olsson P (2010) Differential cytokine regulation by NF-κB and AP-1 in Jurkat T-cells. BMC Immunol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm S, Gutermuth J, Hültner L, Sparwasser T, Behrendt H, Peschel C, Mak TW, Jakob T, Ruland J (2006) The Bcl10-Malt1 complex segregates Fc epsilon RI-mediated nuclear factor kappa B activation and cytokine production from mast cell degranulation. J Exp Med 203: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm S, Zimmermann S, Peschel C, Mak TW, Ruland J (2007) Bcl10 and Malt1 control lysophosphatidic acid-induced NF-κB activation and cytokine production. Proc Natl Acad Sci USA 104: 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Lim JH, Jono H, Ha UH, Xu H, Ishinaga H, Morino S, Xu X, Yan C, Kai H, Li J (2008) Tumor suppressor cylindromatosis acts as a negative regulator for Streptococcus pneumoniae-induced NFAT signaling. J Biol Chem 283: 12546–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D (2009) Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G (2003) The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature 424: 801–805 [DOI] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM (1992) Transcriptional activation of the tumor necrosis factor α-inducible zinc finger protein, A20, is mediated by κB elements. J Biol Chem 267: 17971–17976 [PubMed] [Google Scholar]
- Liu H, Xie M, Schneider MD, Chen ZJ (2006) Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci USA 103: 11677–11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Busby JC, Molkentin JD (2009) Interaction between TAK1-TAB1-TAB2 and RCAN1-calcineurin defines a signalling nodal control point. Nat Cell Biol 11: 154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Galisteo R, Gutkind JS (2009) CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFκB through the CBM (Carma3/Bcl10/Malt1) complex. J Biol Chem 284: 6038–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fässler R (2006) Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125: 665–677 [DOI] [PubMed] [Google Scholar]
- McAllister-Lucas LM, Ruland J, Siu K, Jin X, Gu S, Kim DSL, Kuffa P, Kohrt D, Mak TW, Nune˜z G, Lucas PC (2007) CARMA3/Bcl10/MALT1-dependent NF-κB activation mediates angiotensin II-responsive inflammatory signaling in nonimmune cells. Proc Natl Acad Sci USA 104: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao G, Li Z, Molinero L, Alegre M, Ying H, Sun Z, Penninger JM, Zhang J (2008) T-cell receptor-induced NF-κB activation is negatively regulated by E3 ubiquitin ligase Cbl-b. Mol Cell Biol 28: 2470–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, Fasel N, Thome M (2008) The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol 9: 272–281 [DOI] [PubMed] [Google Scholar]
- Rehman AO, Wang C (2009) CXCL12/SDF-1 alpha activates NF-κB and promotes oral cancer invasion through the Carma3/Bcl10/Malt1 complex. Int J Oral Sci 1: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley W, Zhang M, Sun S (2004) Negative regulation of JNK signaling by the tumor suppressor CYLD. J Biol Chem 279: 55161–55167 [DOI] [PubMed] [Google Scholar]
- Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, Leonard TO, Norbury CC, Fitzpatrick L, Zhang M, Sun S (2007) Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med 204: 1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, Sun S (2006) Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol 7: 411–417 [DOI] [PubMed] [Google Scholar]
- Resch U, Schichl YM, Winsauer G, Gudi R, Prasad K, de Martin R (2009) Siva1 is a XIAP-interacting protein that balances NF-κB and JNK signalling to promote apoptosis. J Cell Sci 122: 2651–2661 [DOI] [PubMed] [Google Scholar]
- Ruefli-Brasse AA, French DM, Dixit VM (2003) Regulation of NF-κB-dependent lymphocyte activation and development by paracaspase. Science 302: 1581–1584 [DOI] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Wakeham A, Mak TW (2003) Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 19: 749–758 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Burbach BJ, Shimizu Y (2010) NF-κB activation in T cells requires discrete control of IKK-α/β phosphorylation and IKKγ ubiquitination by the ADAP adapter protein. J Biol Chem 285: 11100–11105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srokowski CC, Masri J, Hövelmeyer N, Krembel AK, Tertilt C, Strand D, Mahnke K, Massoumi R, Waisman A, Schild H (2009) Naturally occurring short splice variant of CYLD positively regulates dendritic cell function. Blood 113: 5891–5895 [DOI] [PubMed] [Google Scholar]
- Staal J, Bekaert T, Beyaert R (2011) Regulation of NF-κB signaling by caspases and MALT1 paracaspase. Cell Res 21: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S (2010) CYLD: a tumor suppressor deubiquitinase regulating NF-κB activation and diverse biological processes. Cell Death Differ 17: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M (2004) CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol 4: 348–359 [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 424: 793–796 [DOI] [PubMed] [Google Scholar]
- Tusche MW, Ward LA, Vu F, McCarthy D, Quintela-Fandino M, Ruland J, Gommerman JL, Mak TW (2009) Differential requirement of MALT1 for BAFF-induced outcomes in B cell subsets. J Exp Med 206: 2671–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YY, Chi H, Xie M, Schneider MD, Flavell RA (2006) The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat Immunol 7: 851–858 [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- Zhang N, Ahsan MH, Zhu L, Sambucetti LC, Purchio AF, West DB (2005) Regulation of IκBα expression involves both NF-κB and the MAP kinase signaling pathways. J Inflamm (Lond) 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.