Abstract
No-go decay (NGD) and non-stop decay (NSD) are eukaryotic surveillance mechanisms that target mRNAs on which elongation complexes (ECs) are stalled by, for example, stable secondary structures (NGD) or due to the absence of a stop codon (NSD). Two interacting proteins Dom34(yeast)/Pelota(mammals) and Hbs1, which are paralogues of eRF1 and eRF3, are implicated in these processes. Dom34/Hbs1 were shown to promote dissociation of stalled ECs and release of intact peptidyl-tRNA. Using an in vitro reconstitution approach, we investigated the activities of mammalian Pelota/Hbs1 and report that Pelota/Hbs1 also induced dissociation of ECs and release of peptidyl-tRNA, but only in the presence of ABCE1. Whereas Pelota and ABCE1 were essential, Hbs1 had a stimulatory effect. Importantly, ABCE1/Pelota/Hbs1 dissociated ECs containing only a limited number of mRNA nucleotides downstream of the P-site, which suggests that ABCE1/Pelota/Hbs1 would disassemble NSD complexes stalled at the 3′-end, but not pre-cleavage NGD complexes stalled in the middle of mRNA. ABCE1/Pelota/Hbs1 also dissociated vacant 80S ribosomes, which stimulated 48S complex formation, suggesting that Pelota/Hbs1 have an additional role outside of NGD.
Keywords: ABCE1, Hbs1, no-go decay, Pelota, ribosome recycling
Introduction
Translation is a cyclical process, consisting of initiation, elongation, termination and ribosome recycling stages. Release factors eRF1 and eRF3 are involved in two successive stages of protein synthesis, termination and ribosome recycling. During termination, eRF1 recognizes the stop codon in the A-site of the 40S subunit and promotes hydrolysis of P-site peptidyl-tRNA in the peptidyl-transferase centre (PTC) of the 60S subunit (Loh and Song, 2010). eRF3 is a GTPase that is homologous to the bacterial EF-Tu and eukaryotic eEF1A elongation factors that deliver aminoacyl-tRNAs to the A-site. It comprises a non-essential N-terminal region and a C-terminal region that consists of the GTP-binding domain 1 and β-barrel domains 2 and 3 (Kong et al, 2004). eRF1 consists of three domains (Song et al, 2000): the N domain carries determinants of stop codon recognition, the M domain contains a conserved GGQ motif that is required to trigger peptide release, and the C domain interacts with domain 3 of eRF3 (Loh and Song, 2010). The eRF1/eRF3 interaction strongly increases eRF3's affinity to GTP (Pisareva et al, 2006) and stimulates eRF3's ribosome-dependent GTPase activity (Frolova et al, 1996). Binding to eRF3 also induces large conformational changes in eRF1, so that it adopts a tRNA-like structure and the eRF1•eRF3•GTP complex resembles the tRNA•EF-Tu•GTP complex (Cheng et al, 2009), which likely increases eRF1's affinity to the A-site of pre-termination complexes (pre-TCs). In the current model of termination, eRF1 binds to pre-TCs as an eRF1•eRF3•GTP complex. This induces conformational changes in pre-TCs that manifest as a 2-nt forward shift of their toe-print, but peptide release does not occur until eRF3 hydrolyses GTP (Salas-Marco and Bedwell, 2004; Alkalaeva et al, 2006). GTP hydrolysis induces further changes, likely in eRF1 (Cheng et al, 2009), that lead to accommodation of the GGQ motif in the PTC followed by rapid peptide release (Alkalaeva et al, 2006). Whereas eRF3 likely dissociates from ribosomal complexes after GTP hydrolysis, eRF1 remains bound to post-termination complexes (post-TCs).
Recycling of eRF1-associated post-TCs is mediated by ABCE1, a conserved, essential member of the ATP-binding cassette (ABC) family of proteins (Pisarev et al, 2010). ABCE1 comprises an N-terminal domain harbouring two [4Fe-4S] clusters, followed by twin nucleotide-binding domains (NBDs) arranged in a typical head-to-tail orientation, which creates two composite nucleotide-binding sites (Karcher et al, 2008). The basis for the activity of ABC proteins is their ability to undergo cyclical conformational changes depending on the nucleotide-bound state. The transition between the ‘closed’ ATP-bound and ‘open’ ADP-bound conformations results in a power stroke between NBDs (Rees et al, 2009) that could cause conformational changes in associated molecules. In a current model of recycling, ABCE1 binds to eRF1-associated post-TCs and promotes their dissociation into free 60S subunits and tRNA- and mRNA-bound 40S subunits (Pisarev et al, 2010). Subsequent release of mRNA and tRNA can be mediated by eIFs 3/1/1A (Pisarev et al, 2007a) or Ligatin, a conserved ∼65 kDa protein containing N-terminal DUF1947 and PUA domains and C-terminal SWIB/MDM2 and SUI1/eIF1 domains (Skabkin et al, 2010). The essential role of eRF1 in recycling could involve (i) induction of conformational changes in post-termination ribosomes that would create a binding site for ABCE1, (ii) enhancement of ABCE1's affinity to post-TCs due to its direct interaction with eRF1 (Khoshnevis et al, 2010; Pisarev et al, 2010) or (iii) transmission/amplification of conformational changes that occur in ABCE1 upon ATP hydrolysis.
All eukaryotes contain eRF1/eRF3 paralogues Pelota (Dom34 in yeast) and Hbs1 (Atkinson et al, 2008). Although archaea encode Pelota, they lack eRF3 and Hbs1 homologues, and their functions are performed by elongation factor aEF1α (Kobayashi et al, 2010; Saito et al, 2010). Some yeasts also encode Ski7p, a divergent variant of Hbs1 (Atkinson et al, 2008). Dom34/Pelota and Hbs1 have been implicated in the no-go decay (NGD) surveillance mechanism that targets and degrades mRNAs in stalled ribosomal elongation complexes (ECs) when stalling is caused by stable secondary structures, depurination of mRNA, rare codons or premature stop codons (Doma and Parker, 2006; Gandhi et al, 2008). mRNA degradation is initiated by endonucleolytic cleavage in the vicinity of stalled ribosomes (Doma and Parker, 2006; Passos et al, 2009), but the nuclease that mediates it is not known. Another surveillance mechanism, non-stop decay (NSD), degrades mRNAs lacking a stop codon and that have ribosomes arrested at their 3′-end (van Hoof et al, 2002). Although originally only Ski7p was implicated in NSD (van Hoof et al, 2002), recent data suggest that Dom34/Hbs1 may be involved in release of ribosomes that are stalled at the 3′-ends of non-stop mRNAs (Kobayashi et al, 2010).
The M and C domains of Dom34/Pelota are structurally related to M and C domains of eRF1, although the M domain does not contain the critical GGQ motif, but the N domain is not related to that of eRF1 and is instead similar to Sm-fold proteins (Lee et al, 2007; Graille et al, 2008). The tip of domain N contains two loops (A and B) that are important for Dom34's activity in NGD and NSD (Passos et al, 2009; Kobayashi et al, 2010), but their sequence variability suggests that they would not recognize the A-site codon in a sequence-specific manner. Similarly to eRF3, Hbs1 comprises a variable N-terminal region followed by a GTP-binding domain 1 and β-barrel domains 2 and 3 (van den Elzen et al, 2010). Like eRF1/eRF3, Dom34 and Hbs1 also bind directly (Carr-Schmid et al, 2002), and this interaction increases Hbs1's affinity to GTP (Graille et al, 2008; Chen et al, 2010). Moreover, analogously to eRF1 and eRF3 (Cheng et al, 2009), formation of the Dom34•Hbs1•GTP and aPelota•aEF1α•GTP complexes results in significant conformational changes in Dom34/aPelota and adoption by them of a tRNA-like structure (Chen et al, 2010; Kobayashi et al, 2010; van den Elzen et al, 2010), which likely increases their affinity to the A-site. Ribosomal docking suggests that these complexes would readily fit into the A-site, except for some clashes involving the Dom34 N domain and ribosomal components around the decoding centre (Chen et al, 2010), and loop A of aPelota and mRNA in the A-site (Kobayashi et al, 2010). Further conformational changes may, therefore, occur in Dom34/Pelota upon ribosomal binding.
The exact role of Dom34/Pelota and Hbs1 in NGD is only now being elucidated. The reported endonuclease activity of Dom34 and aPelota (Lee et al, 2007) could not be confirmed (Passos et al, 2009; Shoemaker et al, 2010), and endonucleolytic cleavage of mRNA near stalled ribosomes can occur even in the absence of Dom34/Hbs1 (Chen et al, 2010; Kuroha et al, 2010). However, a recent report convincingly showed that yeast Dom34/Hbs1 cooperatively promotes dissociation into subunits of stalled ECs containing P-site tRNA linked to a short peptide, which is accompanied by ‘drop-off’ of intact peptidyl-tRNA (Shoemaker et al, 2010). The process required GTP hydrolysis and occurred in an A-site codon-independent manner. The fact that unlike eRF1/eRF3, Dom34/Hbs1 did not promote peptide release is consistent with Dom34 lacking the essential GGQ motif. It has been suggested that these events may initiate NGD (Shoemaker et al, 2010).
Thus, whereas mammalian eRF1/eRF3 together with ABCE1 promote recycling of post-TCs (Pisarev et al, 2010), yeast Dom34/Hbs1 can dissociate stalled ECs in an A-site codon-independent manner (Shoemaker et al, 2010). Another situation that requires a mechanism for ribosomal dissociation occurs when vacant 80S ribosomes must be split to regenerate a pool of free ribosomal subunits capable of re-entering the translation process. Physiological processes as diverse as mitosis, hibernation and responses to stresses such as amino-acid starvation, hypoxia and serum deprivation all lead to accumulation of ribosomal subunits in a reservoir of translationally inactive vacant 80S ribosomes (Hogan and Korner, 1968; Surks and Berkowitz, 1971; Kaminskas, 1972; Frerichs et al, 1998). Importantly, these ribosomes re-enter translation following progression of the cell cycle beyond mitosis, arousal after hibernation or removal of stress (e.g. by restoration of nutrient levels, which leads to dephosphorylation and reactivation of eIF2). Although eIF3 can split 80S ribosomes in the presence of eIF2-GTP/Met-tRNAMeti or single-stranded RNA (Kolupaeva et al, 2005), it is not clear whether this mechanism can ensure dissociation of stress-accumulated 80S ribosomes at a rate sufficient for rapid resumption of translation.
Using an in vitro reconstitution approach, we investigated the activities of mammalian Pelota and Hbs1 and report that together with ABCE1, Pelota/Hbs1 promoted dissociation of stalled ECs in an A-site codon-independent manner. Like Dom34/Hbs1, Pelota/Hbs1 did not induce peptide release and dissociation was accompanied by release of intact peptidyl-tRNA, but unlike Dom34/Hbs1, Pelota/Hbs1 were unable to induce dissociation without ABCE1. Importantly, ABCE1/Pelota/Hbs1 dissociated ECs only if they contained up to 9 mRNA nucleotides downstream of the P-site, which suggests that these factors would promote disassembly of NSD, but not pre-cleavage NGD complexes. We also report that ABCE1/Pelota/Hbs1 efficiently dissociated vacant 80S ribosomes, which strongly stimulated 48S complex formation from 80S ribosomes, suggesting that Pelota/Hbs1 have an additional role outside of NGD.
Results
Influence of Pelota, Hbs1 and ABCE1 on 48S complex formation from 80S ribosomes
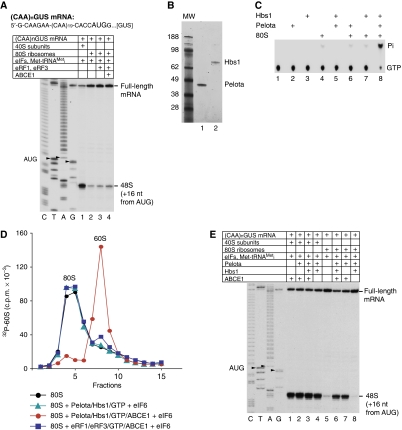
Although it has been shown in vitro that initiation factors can dissociate vacant 80S ribosomes (Kolupaeva et al, 2005), it is not known whether this activity could account for efficient dissociation of stress-accumulated 80S ribosomes in vivo. To recapitulate the process of 80S ribosome dissociation and re-entry into the translation process in vitro, we compared 48S complex formation on (CAA)nGUS mRNA (Figure 1A, upper panel) at a physiological free Mg2+ concentration of 1 mM in a reconstituted system containing eIFs and either 40S subunits or 80S ribosomes. High-yield 48S complex formation was observed only in the presence of 40S subunits (Figure 1A, compare lanes 1 and 2), which suggested that dissociation of 80S ribosomes by eIFs was not efficient. Although dissociation of post-TCs containing mRNA and P-site deacylated tRNA can be promoted by eRF1/eRF3 and ABCE1 (Pisarev et al, 2010), addition of eRF1/eRF3/ABCE1 to the reaction mixture only marginally stimulated 48S complex formation in the presence of 80S ribosomes (Figure 1A, lane 4), indicating that vacant 80S ribosomes cannot be dissociated by these factors. However, addition to the reconstituted system of small quantities of rabbit reticulocyte lysate (RRL), which alone could not promote high-yield 48S complex formation, strongly stimulated assembly of 48S complexes in the presence of 80S ribosomes (data not shown), which suggested that RRL contains a factor(s) that promotes efficient dissociation of vacant ribosomes.
Figure 1.
Pelota, Hbs1 and ABCE1 strongly stimulate 48S complex formation from 80S ribosomes by dissociating them into subunits. (A, E) Toe-printing analysis of the influence of different combinations of Pelota, Hbs1, eRF1/eRF3 and ABCE1 on 48S complex formation at 1 mM free Mg2+ on (CAA)nGUS mRNA from 40S subunits or 80S ribosomes. Lanes C, T, A and G depict cDNA sequences corresponding to (CAA)nGUS mRNA. The positions of toe-prints corresponding to ribosomal complexes are indicated. (B) Purified recombinant Pelota and Hbs1 resolved by SDS–PAGE. (C) Thin-layer chromatography analysis of the GTPase activity of Hbs1. (D) Dissociation of vacant 80S ribosomes containing [32P]60S subunits by different combinations of Pelota, Hbs1, ABCE1, eRFs and eIF6 at 1.5 mM free Mg2+, assayed by SDG centrifugation.
The fact that yeast Dom34 and Hbs1 can split stalled ECs in an A-site codon-independent manner (Shoemaker et al, 2010) prompted us to investigate whether their mammalian homologues, Pelota and Hbs1, can dissociate vacant 80S ribosomes and thereby stimulate 48S complex formation. Recombinant human Pelota and Hbs1 were purified from Escherichia coli (Figure 1B). As in the case of its yeast homologue (Shoemaker et al, 2010), the GTPase activity of mammalian Hbs1 required the presence of both 80S ribosomes and Pelota (Figure 1C). To investigate whether Pelota and Hbs1 can dissociate vacant 80S ribosomes, the latter were assembled from 40S subunits and [32P]60S subunits (Pisarev et al, 2007a). To prevent potential reassociation of subunits, reaction mixtures were supplemented with eIF6, which binds to the interface of the 60S subunit, blocking its association with the 40S subunit (Gartmann et al, 2010). In sucrose density gradient (SDG) centrifugation experiments, Pelota, Hbs1 and ABCE1 promoted near complete dissociation of vacant 80S ribosomes (Figure 1D, red circles). However, no dissociation occurred in the absence of ABCE1 (Figure 1D, green triangles). Consistently, in the presence but not in the absence of ABCE1, Pelota and Hbs1 strongly stimulated 48S complex formation from 80S ribosomes on (CAA)nGUS mRNA (Figure 1E, compare lanes 6 and 8). Only a very small proportion of 80S ribosomes dissociated in the presence of eRF1/eRF3/ABCE1 (Figure 1D, blue squares), which was in line with the inability of these factors to stimulate 48S complex formation (Figure 1A).
Dissociation of vacant 80S ribosomes by Pelota, Hbs1 and ABCE1
Pelota and ABCE1 were able to promote dissociation of vacant 80S ribosomes even in the absence of Hbs1, but efficient dissociation in the absence of Hbs1 required higher concentrations of Pelota (Figure 2A). Consistently, Pelota and ABCE1 also stimulated 48S complex formation in the absence of Hbs1 (Figure 1E, lane 7). Importantly, in the presence of GMPPNP, Hbs1 inhibited dissociation of 80S ribosomes by Pelota and ABCE1 (Figure 2B, green filled triangles). In control reactions, GMPPNP at the ratio to ATP used in these experiments did not inhibit the activity of ABCE1 during dissociation of 80S ribosomes by Pelota/ABCE1 in the absence of Hbs1 (Figure 2B, green open triangles). Pelota and Hbs1 were specific for each other, and could not be coupled with eRF3 or eRF1, respectively (data not shown).
Figure 2.
Dissociation of vacant 80S ribosomes by Pelota, Hbs1 and ABCE1. (A–D) Dissociation of vacant 80S ribosomes containing [32P]60S subunits by incubation with Pelota, Hbs1 and ABCE1 (A, B) in the presence of (A) different concentrations of Pelota and (B) different nucleotides at 1.5 mM free Mg2+, (C) at different free Mg2+ concentrations and (D) after different incubation periods at 1.5 mM free Mg2+, assayed by SDG centrifugation. Upper fractions were omitted for clarity.
Dissociation of 80S ribosomes by ABCE1/Pelota/Hbs1 occurred over a wide range of Mg2+ concentrations: it was equally efficient at 1.5–5 mM Mg2+, ∼30% less efficient at 10 mM Mg2+, and ∼85% less efficient at 20 mM Mg2+ (Figure 2C). The process was relatively rapid, and >50% of ribosomes were dissociated within 5 min of incubation (Figure 2D).
Pelota, Hbs1 and ABCE1 do not dissociate ribosomal complexes assembled on mRNA containing a long 3′-UTR
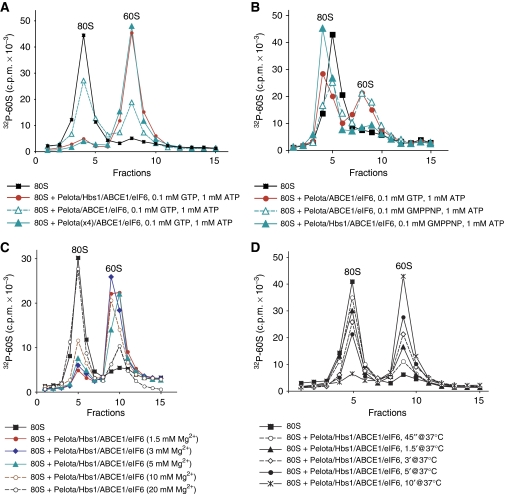
Since yeast Dom34/Hbs1 can promote dissociation of ECs in an A-site codon-independent manner (Shoemaker et al, 2010), we investigated whether Pelota and Hbs1 also possess this activity. Pre-TCs and ECs containing P-site MV-peptidyl-tRNA were assembled on MVHL-STOP mRNA encoding an MVHL tetrapeptide followed by a UAA stop codon and an ∼400-nt-long 3′-UTR (Figure 3A) from 40S subunits, [32P]60S subunits, eIFs, eEFs and appropriate aminoacylated tRNAs, and purified by SDG centrifugation. Whereas pre-TCs were efficiently dissociated by eRF1/eRF3/ABCE1 after 10 min of incubation at 1.5 mM Mg2+, no dissociation was observed in the presence of Pelota/Hbs1/ABCE1 (Figure 3B), even after 30 min of incubation at 1 mM free Mg2+ (data not shown). Pelota/Hbs1/ABCE1 were not able to dissociate ECs that contained a different A-site codon and a shorter peptide (MV) compared with pre-TCs (MVHL), and addition of puromycin did not change the situation (Figure 3C).
Figure 3.
Pelota, Hbs1 and ABCE1 cannot dissociate pre-termination and elongation complexes assembled on MVHL-STOP mRNA containing an ∼400-nt-long 3′-UTR. (A) Structure of MVHL-STOP mRNA. (B–E) Investigation by (B, C) SDG centrifugation and (D, E) toe-printing analysis of the influence of Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+ on (B, D) pre-TCs and (C, E) ECs containing MV-peptidyl-tRNA assembled on MHVL-STOP mRNA. (B, C) Pre-TCs and ECs were assembled using [32P]60S subunits. Upper fractions were omitted for clarity. (D, E) Lanes C, T, A and G depict cDNA sequence corresponding to MVHL-STOP mRNA; the positions of toe-prints corresponding to ribosomal complexes are indicated.
During eukaryotic termination/recycling processes, binding of eRF1/eRF3 to pre-TCs results in a 2-nt forward shift of their toe-prints (Figure 3D, compare lanes 1 and 2; Alkalaeva et al, 2006), which likely reflects a change in the conformation of ribosomal complexes. After peptide release, ABCE1 promotes dissociation of eRF1-bound post-TCs into 60S subunits and mRNA/tRNA-associated 40S subunits, which is accompanied by reversal of the toe-print shift (Figure 3D, lane 3; Pisarev et al, 2010). Finally, eIFs or Ligatin induce tRNA and mRNA release from recycled 40S subunits (Figure 3D, lane 4; Pisarev et al, 2007a; Skabkin et al, 2010). In contrast to eRF1/eRF3, Pelota and Hbs1 did not cause a toe-print shift in pre-TCs in the presence of GTP or GMPPNP (Figure 3D, lanes 5–8), and also did not change toe-prints of ECs containing P-site MV-peptidyl-tRNA (Figure 3E).
Treatment of ribosomal complexes with RelE renders them susceptible to dissociation by Pelota, Hbs1 and ABCE1
The fact that Pelota and Hbs1 did not induce shifts in the toe-prints of pre-TCs or ECs could theoretically indicate that these factors either do not bind to such complexes at all, or at least do not bind to them productively. Interestingly, it has recently been noted that docking of the complex of archaeal aPelota with aEF1a onto the crystal structure of the bacterial 70S ribosome results in a clash between aPelota's loop A and the A-site mRNA (Kobayashi et al, 2010). Thus, the inability of Pelota/Hbs1/ABCE1 to dissociate pre-TCs and ECs could potentially be due to the inability of Pelota to bind productively to ribosomal complexes, in which the A-site is stably occupied by mRNA.
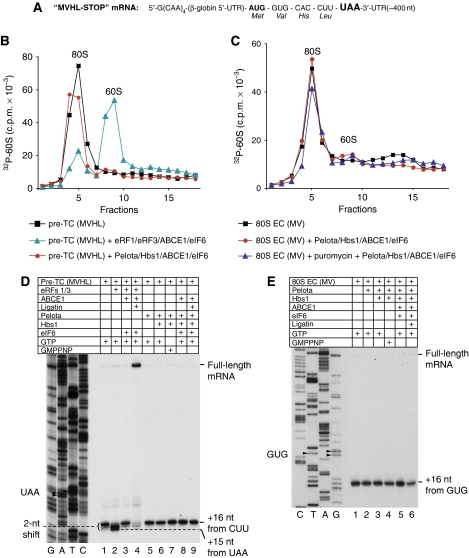
To test this hypothesis, we investigated whether Pelota/Hbs1/ABCE1 could dissociate ECs and pre-TCs assembled on MVHL-STOP mRNA after their treatment with the bacterial toxin RelE, which cleaves mRNA in the A-site (Pedersen et al, 2003; Neubauer et al, 2009). Consistent with a previous report that showed that RelE is also active on eukaryotic ribosomal complexes (Andreev et al, 2008), treatment with RelE of 80S initiation complexes and post-translocation ECs containing MV-dipeptide assembled on MVHL-STOP mRNA (Figure 4A, lanes 1 and 2) resulted in mRNA cleavage after the first and second A-site nucleotides (Figure 4A, lanes 5 and 7). As expected, RelE did not cleave mRNA in pre-translocation complexes containing MV-tRNA in the A-site that were formed in eEF2's absence (Figure 4A, lane 9). After treatment with RelE, Pelota/Hbs1/ABCE1 dissociated ∼50% of post-translocation ECs (Figure 4B, red circles). In contrast, almost no dissociation occurred after treatment with RelE of pre-translocated complexes (Figure 4C, red circles); a small amount of dissociated complexes most likely represented a mixture of 80S initiation complexes and ECs that underwent spontaneous eEF2-independent translocation. After treatment with RelE, Pelota/Hbs1/ABCE1 also dissociated ∼50% of pre-TCs (Figure 4D, lower panel, red circles). eRF1/eRF3/ABCE1, on the other hand, lost their ability to dissociate pre-TCs after their treatment with RelE (Figure 4D, compare blue triangles in upper and lower panels). These data indicate that cleavage of mRNA in the A-site by RelE renders ribosomal complexes susceptible to dissociation by Pelota/Hbs1/ABCE1.
Figure 4.
Pelota, Hbs1 and ABCE1 can dissociate pre-termination and elongation complexes assembled on MVHL-STOP mRNA after their treatment with RelE. (A) Primer-extension analysis of A-site mRNA cleavage induced by RelE in 80S initiation and in pre- and post-translocation ECs containing MV-peptidyl-tRNA that were assembled on MVHL-STOP mRNA. (B–D) Dissociation by Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+ of (B, C) post- and pre-translocation ECs containing MV-peptidyl-tRNA and (D) pre-TCs assembled on MVHL-STOP mRNA using [32P]60S subunits depending on their treatment with RelE, assayed by SDG centrifugation. Upper fractions were omitted for clarity.
Dissociation by Pelota, Hbs1 and ABCE1 of ECs assembled on non-stop mRNAs
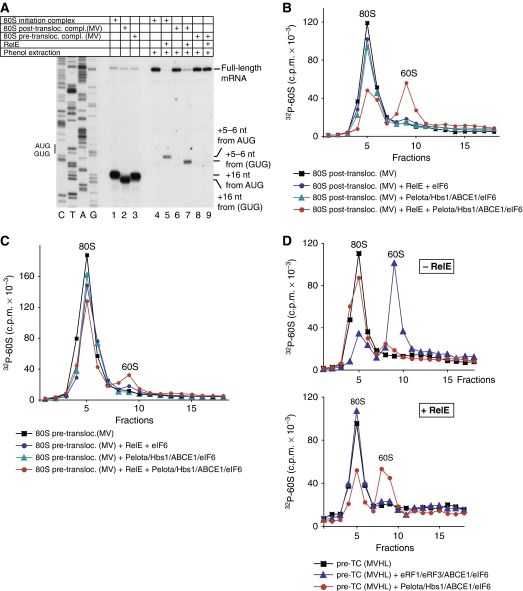
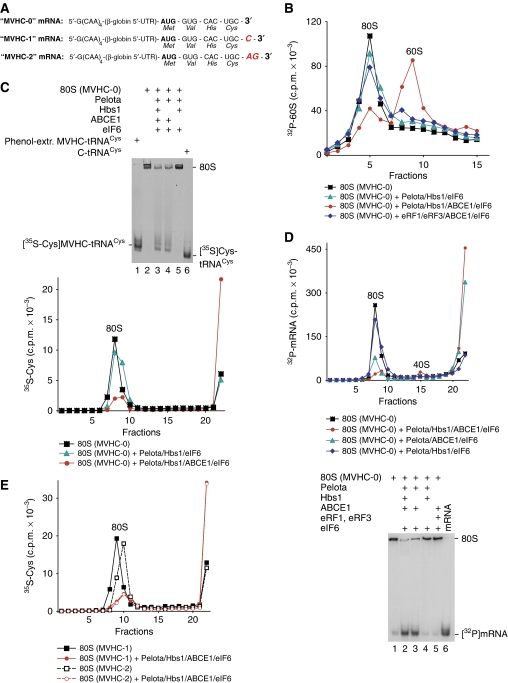
Incomplete dissociation by Pelota/Hbs1/ABCE1 of pre-TCs and ECs treated with RelE was most likely due to competition between Pelota and RelE for the ribosomal A-site. However, it could nevertheless not be excluded that only complexes, in which mRNA was cleaved after the first (but not after the second) A-site nucleotide, could undergo dissociation. To investigate this question, we assayed dissociation of ECs assembled on non-stop mRNAs encoding MVHC tetrapeptide and containing 0, 1 or 2 nts after the Cys codon (Figure 5A).
Figure 5.
Dissociation by Pelota, Hbs1 and ABCE1 of elongation complexes assembled on non-stop mRNAs. (A) Structure of non-stop mRNAs encoding MVHC tetrapeptide and containing 0, 1 or 2 nts after the last codon. (B) Dissociation into subunits of ECs assembled on MVHC-0 mRNA with [32P]60S subunits and containing MVHC-peptidyl-tRNA after their incubation with Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+, assayed by SDG centrifugation. (C) Ribosomal association of [35S]MVHC-peptidyl tRNA after incubation with Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+ of ECs assembled on MVHC-0 mRNA, assayed by (upper panel) native gel electrophoresis and (lower panel) SDG centrifugation. (C, upper panel) Lanes 1 and 6 contain [35S-Cys]MVHC-tRNACys that was phenol extracted from elongation complexes, and [35S]Cys-tRNACys, respectively. (D) Ribosomal association of [32P]MVHL-0 mRNA after incubation of ECs with Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+, assayed by (upper panel) SDG centrifugation and (lower panel) native gel electrophoresis. (E) Ribosomal association of [35S]MVHC-peptidyl tRNA after incubation with Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+ of ECs assembled on MVHC-1 and MVHC-2 mRNAs, assayed by SDG centrifugation.
Pelota/Hbs1/ABCE1 efficiently split into subunits ECs that had been assembled on MVHC-0 mRNA and did not contain mRNA downstream of the P-site (Figure 5B; red circles). However, in contrast to their yeast homologues (Shoemaker et al, 2010), Pelota/Hbs1 did not promote dissociation in the absence of ABCE1 (Figure 5B, green triangles). As expected, eRF1/eRF3/ABCE1 did not split ECs that were assembled on MVHC-0 mRNA and did not contain an A-site stop codon (Figure 5B, blue diamonds).
Dom34 and Hbs1 do not induce peptide release, but instead promote release of intact peptidyl-tRNA from ECs (Shoemaker et al, 2010). To investigate the fate of peptidyl-tRNA during dissociation of ECs by Pelota/Hbs1/ABCE1, ECs were assembled on MVHC-0 mRNA using [35S]Cys-tRNACys, incubated with Pelota/Hbs1/ABCE1 and subjected to native gel electrophoresis or SDG centrifugation (Figure 5C). Like yeast Dom34/Hbs1, Pelota and Hbs1 did not induce hydrolysis of peptidyl-tRNA, and incubation of ECs with Pelota/Hbs1/ABCE1 resulted in release of a radio-labelled product that in a native gel migrated exactly like intact [35S-Cys]MVHC-tRNA that had been phenol extracted from purified ECs (Figure 5C, upper panel, lanes 1, 3 and 4) and in SDG centrifugation experiments was found at the top of the gradient (Figure 5C, lower panel, red circles). Thus, unlike dissociation of pre-TCs by eRF1/eRF3/ABCE1 in which case deacylated tRNA remained bound to recycled 40S subunits (Pisarev et al, 2010), dissociation of ECs by Pelota/Hbs1/ABCE1 led to release of peptidyl-tRNA into solution. Dissociation of peptidyl-tRNA was efficient (Figure 5C, lower panel), could also occur in the absence of Hbs1 (Figure 5C, upper panel), but absolutely required ABCE1 (Figure 5C, both panels).
To investigate ribosomal association of mRNA after dissociation of ECs by Pelota/Hbs1/ABCE1, ECs were assembled using 32P-labelled MVHC-0 mRNA. After incubation with Pelota/Hbs1/ABCE1, only a tiny fraction of mRNA remained bound to 40S subunits, whereas the majority was also released into solution (Figure 5D, both panels).
Importantly, Pelota/Hbs1/ABCE1 also induced efficient dissociation into subunits of ECs assembled on MVHC-1 and MVHC-2 mRNAs that was accompanied by release of peptidyl-tRNA (Figure 5E; data not shown). Thus, although Pelota/Hbs1/ABCE1 could not dissociate pre-termination and ECs assembled on mRNA with an ∼400-nt-long 3′-UTR, they promoted very efficient splitting into subunits of ECs formed on non-stop mRNAs and containing 0, 1 or 2 mRNA nucleotides downstream of the P-site, which was accompanied by release of mRNA and intact peptidyl-tRNA.
The ability of Pelota/Hbs1/ABCE1 to dissociate ribosomal complexes depends on the number of mRNA nucleotides present downstream of the P-site
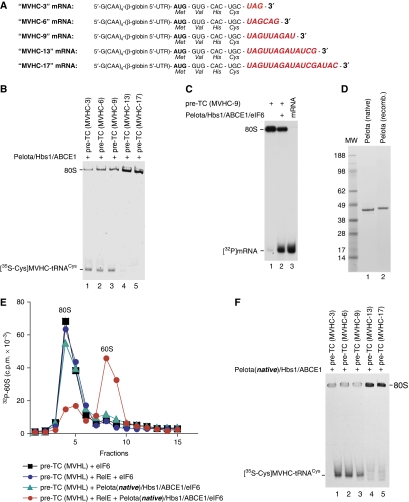
The data described above suggest that the ability of Pelota/Hbs1/ABCE1 to dissociate ribosomal complexes depends on the length of mRNA downstream of the P-site. To determine the maximum number of downstream nucleotides that are compatible with efficient dissociation, pre-TCs were assembled using [35S]Cys-tRNACys on mRNAs encoding MVHC tetrapeptide followed by 3, 6, 9, 13 or 17 nts (Figure 6A). Efficient release of [35S-Cys]peptidyl-tRNA was observed for complexes containing 3, 6 and 9, but not 13 or 17 nts downstream of the P-site (Figure 6B).
Figure 6.
Dissociation by Pelota, Hbs1 and ABCE1 of pre-termination complexes containing different numbers of mRNA nucleotides downstream of the P-site. (A) Structure of mRNAs encoding MVHC tetrapeptide followed by the UAG stop codon and containing 3, 6, 9, 13 or 17 nts after the last sense codon. (B, F) Ribosomal association of [35S-Cys]MVHC-peptidyl tRNA after incubation of pre-TCs assembled on mRNAs shown in panel (A) with (B) recombinant and (F) native Pelota, Hbs1 and ABCE1 at 1.5 mM Mg2+, assayed by native gel electrophoresis. (C) Ribosomal association of [32P]MVHC-9 mRNA after incubation of ECs with Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+, assayed by native gel electrophoresis. (D) Purified native and recombinant Pelota resolved by SDS–PAGE. (E) Dissociation by native Pelota, Hbs1 and ABCE1 at 1.5 mM free Mg2+ of pre-TCs assembled on MVHL-STOP mRNA using [32P]60S subunits depending on their treatment with RelE, assayed by SDG centrifugation. Upper fractions were omitted for clarity.
As in the case of MVHC-0 mRNA, incubation with Pelota/Hbs1/ABCE1 of pre-TCs assembled using 32P-labelled MVHC-9 mRNA resulted in release of mRNA into solution (Figure 6C), indicating that the additional 9 mRNA nucleotides were not sufficient to stabilize mRNA/40S complexes. In contrast, after treatment of yeast ECs with Dom34/Hbs1, mRNA mostly remained bound to the 40S subunits (Shoemaker et al, 2010). This difference might be due to the fact that yeast complexes contained even longer region of mRNA downstream of the P-site (24 nts).
Pelota has been reported to be phosphorylated at multiple sites (Olsen et al, 2006, 2010; Iliuk et al, 2010), which could lead to differences in the activity of its native and recombinant forms. To test whether the inability of recombinant Pelota to promote dissociation of ribosomal complexes containing >9 nts downstream of the P-site was due to the lack of post-translational modifications, native Pelota was isolated from RRL (Figure 6D). Like the recombinant protein, native Pelota promoted dissociation of pre-TCs assembled on MVHL-STOP mRNA only after their treatment with RelE (Figure 6E, compare green triangles and red circles), and could induce release of peptidyl-tRNA from ribosomal complexes containing only up to 9 nts downstream of the P-site (Figure 6F).
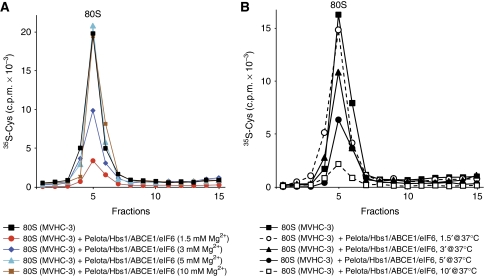
To investigate the Mg2+ dependence of dissociation of ribosomal complexes by Pelota/Hbs1/ABCE1, we assayed release of [35S-Cys]peptidyl-tRNA from pre-TCs assembled on mRNA encoding MVHC tetrapeptide followed only by a stop codon. As expected, dissociation of such pre-TCs occurred in a narrower range of Mg2+ concentrations compared with vacant 80S ribosomes. Thus, whereas peptidyl-tRNA release was very efficient at 1.5 mM Mg2+, only ∼50% of peptidyl-tRNA dissociated at 3 mM Mg2+, and no release occurred at 5 mM Mg2+ (Figure 7A). At 1.5 mM Mg2+, ∼70% of peptidyl-tRNA dissociated after 5 min incubation (Figure 7B).
Figure 7.
Kinetics and Mg2+ dependence of dissociation by Pelota, Hbs1 and ABCE1 of pre-termination complexes containing 3 mRNA nucleotides downstream of the P-site. Ribosomal association of [35S-Cys]MVHC-peptidyl tRNA after incubation of pre-TCs assembled on MVHC-3 mRNA with Pelota, Hbs1 and ABCE1 (A) at different Mg2+ concentrations, and (B) for different time periods at 1.5 mM Mg2+, assayed by SDG centrifugation. Upper fractions were omitted for clarity.
Discussion
eRF1/eRF3 not only promote hydrolysis of peptidyl-tRNA during termination, but together with ABCE1 also mediate recycling of post-termination ribosomal complexes (Pisarev et al, 2010). We now report that with the mammalian eRF1/eRF3 paralogues, Pelota and Hbs1, ABCE1 can dissociate vacant 80S ribosomes and stalled ECs in an A-site codon-independent manner. Like their yeast counterparts Dom34/Hbs1 (Shoemaker et al, 2010), Pelota and Hbs1 did not induce hydrolysis of peptidyl-tRNA, and instead promoted release of intact peptidyl-tRNA from ECs. However, in contrast to Dom34/Hbs1 (Shoemaker et al, 2010), Pelota/Hbs1 were unable to induce dissociation of ECs in ABCE1's absence.
There are substantial similarities between the mechanisms of dissociation of post-TCs by ABCE1 and eRF1/eRF3 and of vacant 80S ribosome and stalled ECs by ABCE1 and Pelota/Hbs1. Although eRF3 strongly stimulates peptide release and subsequent recycling of post-TCs in a GTP-dependent manner, the absolute requirement for it can be overcome by elevated concentrations of eRF1 (Pisarev et al, 2010). Similarly, Pelota and ABCE1 were essential for ribosomal dissociation, but Hbs1 had only a stimulatory effect, and high concentrations of Pelota could compensate for its absence. Stimulation of ribosomal disassembly by Hbs1 is consistent with the strong inhibition of release of truncated polypeptides from ribosomal complexes stalled on non-stop mRNA in yeast strains expressing Hbs1 mutants defective in GTPase activity (Kobayashi et al, 2010). In the GMPPNP-bound form, eRF3 prevents association of ABCE1 with eRF1-bound ribosomal complexes (Pisarev et al, 2010). Similarly, in the presence of GMPPNP, Hbs1 also inhibited dissociation of vacant ribosomes by Pelota and ABCE1. Thus, to allow dissociation to proceed, both Hbs1 and eRF3 would likely have to dissociate after hydrolysing GTP. The mechanism by which ABCE1 then splits eRF1- and Pelota-associated ribosomes remains unknown, but an intriguing possibility is that eRF1 and Pelota might transmit and/or amplify conformational changes that presumably occur in ABCE1 upon NTP hydrolysis. ABCE1 binds directly to eRF1 (Khoshnevis et al, 2010; Pisarev et al, 2010), but whether it also interacts with Pelota/Dom34 remains to be determined.
An important difference between eRF1/eRF3- and Pelota/Hbs1-mediated ribosomal disassembly is that the former requires prior peptide release. eRF1/eRF3-mediated splitting of post-TCs disrupts interaction of P-site tRNA with the 60S subunit, but tRNA remains based-paired with mRNA on the recycled 40S subunit (Pisarev et al, 2010). Although dissociation by Dom34/Hbs1 or by Pelota/Hbs1/ABCE1 of ECs containing P-site tRNA attached to a short peptide resulted in release of peptidyl-tRNA into solution (Shoemaker et al, 2010; this study), dissociation of ECs containing much longer nascent polypeptides (if it occurs) would likely result in disruption of the interaction of peptidyl-tRNA with the 40S subunit, but continued association with the 60S subunit, particularly if the polypeptide had already partly folded after traversing the exit tunnel. Thus, whereas eRF1/eRF3-mediated dissociation relies on disruption of the interaction of P-site tRNA with the 60S subunit and, therefore, requires prior peptide release, the potential for Pelota(Dom34)/Hbs1/ABCE1 to disrupt tRNA's interaction with the 40S subunit would account for its independence on peptide release.
The N domains of eRF1 and Pelota(Dom34) are unrelated (Kong et al, 2004; Lee et al, 2007; Graille et al, 2008), which accounts for a second major difference, namely the very strong codon dependence of eRF1/eRF3-mediated dissociation of post-TCs. The fact that eRF1/eRF3/ABCE1 did not promote dissociation of vacant 80S ribosomes suggests that eRF1 is unable to bind productively not only to complexes with an A-site sense codon, but also to ribosomes lacking mRNA, even though eRF1 stimulates eRF3's GTPase activity in the presence of vacant ribosomes (Frolova et al, 1996). Importantly, inclusion of Pelota/Hbs1 in the reconstituted system strongly stimulated 48S complex formation from 80S ribosomes. This indicates that Pelota/Hbs1/ABCE1 might be involved in active dissociation in vivo of accumulated vacant 80S ribosomes, in which case initiation factors probably have a more passive anti-association role. The involvement of ABCE1 in recycling of vacant 80S ribosomes might also account for the translation initiation defect in ABCE1-depleted cells (Dong et al, 2004). Disruption of the murine Pelota gene causes early embryonic lethality due to defects in the mitotic cell cycle and in cell proliferation (Adham et al, 2003), and genetic studies have shown that Pelota (Dom34) is required for meiotic and mitotic cell division in Drosophila and Saccharomyces cerevisiae, for example impairing progression through the G1 phase, spermatogenesis and sporulation (Eberhart and Wasserman, 1995; Davis and Engebrecht, 1998). Protein synthesis is integrated with physiological demands by reversible regulatory mechanisms (Jackson et al, 2010): inhibition of initiation during mitosis or meiosis leads to polysome disaggregation and accumulation of vacant 80S ribosomes, but subsequent resumption of translation is rapid and leads to reformation of polysomes (Johnson and Holland, 1965; Scharff and Robbins, 1966; Rinaldi and Monroy, 1969). Consequently, it is possible that the effects of mutations in or disruption of Pelota (Dom34) genes described above reflect an inability of the cell to efficiently dissociate vacant 80S ribosomes to initiate active protein synthesis when this is required, emphasizing that Pelota/Dom34 may have an additional role outside of NGD. Interestingly, genetic experiments have revealed synthetic defects between deletion of Dom34 and of some 40S subunit proteins, circumstances that reduce the level of initiation by limiting the level of active 40S subunits (Bhattacharya et al, 2010).
Another distinct feature of Pelota/Hbs1-mediated dissociation of ECs is that it depends strongly on the length of mRNA downstream of the P-site. Thus, Pelota/Hbs1/ABCE1 efficiently split ECs containing up to 9 mRNA nucleotides downstream of the P-site, but not those containing 13 nts. How could the length of mRNA influence the ability of these factors to dissociate ribosomal complexes? One possibility is that productive binding of Pelota to the A-site might require conformational changes that involve some repositioning of mRNA in the A-site, which could be impaired by stable fixation of a longer mRNA in the mRNA-binding cleft downstream of the P-site. Docking of the archaeal aPelota/aEF1a complex onto the crystal structure of the bacterial 70S ribosome results in a clash between aPelota's loop A and the A-site mRNA (Kobayashi et al, 2010), which supports the possibility that such conformational changes might occur. However, the most recent high-resolution structure of bacterial ribosomal complexes (Jenner et al, 2010) indicates that defined contacts between the 30S subunit and mRNA downstream of the A-site occur only up to position +12 relative to the first P-site mRNA nucleotide (i.e. not +15, as suggested previously; Yusupova et al, 2001), with nucleotides at positions +10–12 interacting through their phosphate backbone with basic residues of ribosomal proteins (rps) S3p, S4p and S5p. The structures of 40S and 30S subunits in the area that corresponds to the mRNA path downstream of the A-site are homologous (Ben-Shem et al, 2010) and data on UV cross-linking of mRNA in eukaryotic initiation complexes suggest that the mRNA-binding paths on 40S and 30S subunits are similar (Pisarev et al, 2008). Thus, if the mRNA path on 40S subunits extends as far downstream of the A-site as on 30S subunits, then filling it with mRNA would yield complexes containing 9 nts downstream of the P-site. However, such complexes are susceptible to dissociation by Pelota/Hbs1/ABCE1, whereas the presence of a few additional nucleotides outside the proposed mRNA-binding channel renders them resistant to dissociation. Although it cannot be excluded that in mammalian ribosomal complexes, mRNA continues to interact with rps S2 and S3 (analogues of S5p and S3p) downstream of position +12, an alternative scenario of how nucleotides downstream of this position might influence dissociation can also be envisioned. Thus, binding of eRF1/eRF3 to termination complexes causes a 2-nt forward shift of their toe-print (Alkalaeva et al, 2006), which likely reflects a change in the position of the head relative to the body of the 40S subunit that probably also extends the interaction of mRNA with the 40S subunit downstream of the P-site. If initial binding of Pelota/Hbs1 to ECs causes analogous conformational changes, a few nucleotides that would normally be outside the mRNA-binding cleft might strengthen the interaction of mRNA with the 40S subunit, interfering with accommodation of Pelota's N domain in the A-site and rendering ECs resistant to dissociation.
Interestingly, the dissociation activity of Pelota/Hbs1 appears to be noticeably lower than that of their yeast counterparts. Thus, Dom34/Hbs1 were able to promote ribosomal dissociation in the absence of ABCE1, and could also efficiently split ECs containing 24 mRNA nucleotides downstream of the P-site (Shoemaker et al, 2010). The reason for this divergence is not obvious. It might reflect the potential structural differences between yeast and mammalian ECs, but the distinct sizes of the essential loops A and B in domains N of Pelota and Dom34 (Kobayashi et al, 2010) could also influence the requirements for and outcome of their accommodation in the A-site.
An important implication of the fact that Pelota/Hbs1/ABCE1 dissociate ECs containing only a limited number of mRNA nucleotides downstream of the P-site is that they would be able to recycle mammalian ECs formed on non-stop mRNAs, but not NGD ECs that are stalled in the middle of a message. This, in turn, suggests that dissociation of the latter would require prior cleavage of mRNA to convert stable NGD complexes into dissociation-prone NSD complexes. Where should mRNA cleavage occur in this case? Specific cleavage of mRNA in the A-site would render ECs susceptible to dissociation by Pelota/Hbs1/ABCE1, since these factors could dissociate ribosomal complexes treated with RelE. However, RelE/RelB-like toxin–antitoxin systems do not occur in eukaryotes (Makarova et al, 2009), and no eukaryotic endonuclease with A-site specificity has been unambiguously identified yet. Although mRNA in the in vitro assembled ECs was stable over extended incubation periods, it cannot be excluded that under certain conditions in vivo, the A-site mRNA might be cleaved by the ribosome itself in a manner like that proposed for bacterial ribosomes during elongation pausing (Hayes and Sauer, 2003; Sunohara et al, 2004). However, if cleavage of mRNA somewhere at the 3′-side of mRNA outside the mRNA-binding cleft removed the inhibitory secondary structure and allowed elongation to resume, NGD complexes would be converted into NSD complexes, which are susceptible to dissociation. On the other hand, if the mammalian mRNA-binding cleft does indeed end at position +12, and if mRNA is cleaved directly after this position, then such complexes would also be susceptible to dissociation, even without further elongation. This hypothetical mechanism would also account for dissociation of those NGD complexes, on which mRNA cleavage would not promote further elongation (e.g. complexes stalled on rare codons). Several lines of evidence indicate that in yeast, endonucleolytic cleavage of mRNA can occur independently of Dom34/Hbs1. Thus, cleavage of mRNA containing a stable hairpin was reduced, but not abolished by deletion of Hbs1 in an ski7Δ yeast strain (Doma and Parker, 2006) and was partially restored in a dom34Δ strain by overexpression of rpS30 (Passos et al, 2009), whereas mRNAs containing clusters of rare Arg codons or encoding polylysine or polyarginine sequences underwent efficient endonucleolytic cleavage in the absence of Dom34 and/or Hbs1 (Chen et al, 2010; Kuroha et al, 2010). However, even though Dom34 and Hbs1 are not essential for endonucleolytic cleavage of mRNA in stalled complexes, the impairment of cleavage of mRNA containing a stable hairpin in the absence of Dom34 or Hbs1 (Doma and Parker, 2006; Passos et al, 2009) suggests that they might either enhance recruitment of an endonuclease to stalled ribosomes or activate a latent endonuclease that is already associated with elongating ribosomes (Harigaya and Parker, 2010). Importantly, we note that the ability of yeast Dom34/Hbs1 to dissociate ECs containing up to 24 mRNA nucleotides downstream of the P-site (Shoemaker et al, 2010) suggests that the order of events during NGD in yeast and in mammals might differ.
Materials and methods
Plasmids
Expression vectors for His6-tagged eIFs 1, 1A, 4A, 4B, 5, Ligatin, eRF1 and eRF3aC lacking the N-terminal 138 a.a., and transcription vectors for (CAA)nGUS mRNA, MVHL-STOP mRNA, tRNAMeti and tRNAVal(GUG) have been described (Pestova and Kolupaeva, 2002; Alkalaeva et al, 2006; Pisarev et al, 2007a, 2007b; Skabkin et al, 2010 and references therein). Vectors for expression of C-terminally His6-tagged human Pelota and Hbs1 were from Genecopoeia, Inc. Vectors for transcription of mRNAs encoding MVHC tetrapeptide followed by 0, 1, 2, 3, 6, 9, 13 or 17 nts (Figures 5A and 6A) were made by inserting DNA sequences flanked by an upstream T7 promoter and unique restriction sites into pJ204 (DNA2.0). mRNAs and tRNAs were transcribed using T7 RNA Polymerase. 32P-labelled MVHC-0 mRNA (5 × 106 c.p.m./μg) was transcribed in the presence of [α32P]ATP.
Purification of factors and ribosomal subunits
Rabbit 40S and 60S subunits, eIFs 2, 3, 4F, 5B and 6, eEF1H, eEF2, ABCE1 and aminoacyl-tRNA synthetases and recombinant His6-tagged eIFs 1, 1A, 4A, 4B, 5, eRF1, eRF3 and Ligatin were purified as described (Si et al, 1997; Pestova and Hellen, 2003; Alkalaeva et al, 2006; Pisarev et al, 2007a, 2010; Skabkin et al, 2010). [32P]60S subunits (2 × 107 c.p.m./pmol) were prepared using casein kinase II (Pisarev et al, 2007a).
Purification of recombinant Pelota and Hbs1
Recombinant Pelota and Hbs1 were expressed in 1 and 4 l of E. coli BL21(DE3), respectively, after induction by 0.1 mM IPTG for 16 h at 16°C. Pelota was purified by affinity chromatography on Ni-NTA-agarose followed by FPLC on a MonoQ HR5/5 column. Hbs1 was purified by affinity chromatography on Ni-NTA-agarose followed by FPLC on MonoS HR5/5 and MonoQ HR5/5 columns.
Purification of native Pelota
Pelota was purified from the 40–50% ammonium sulphate precipitation fraction of the 0.5-M KCl ribosomal salt wash that was prepared from 3 l of RRL (Green Hectares, Oregon, WI) (Pisarev et al, 2007b). This fraction was dialysed against buffer A (20 mM Tris–HCl, pH 7.5, 10% glycerol, 2 mM DTT, 0.1 mM EDTA)+100 mM KCl and applied to a DE52 column equilibrated with buffer A+100 mM KCl. Pelota was eluted by buffer A+250 mM KCl. This fraction was applied to a P11 column equilibrated with buffer A+100 mM KCl, and Pelota was eluted by buffer A+400 mM KCl. This fraction was applied to an FPLC MonoS HR 5/5 column. Fractions were collected across a 100–500 mM KCl gradient. Pelota eluted at ∼340 mM KCl. Pelota-containing fractions were applied to a hydroxyapatite column. Fractions were collected across a 20–500 mM phosphate buffer gradient. Pelota was eluted at ∼270 mM phosphate buffer. Pelota-containing fractions were concentrated and transferred into buffer A+100 mM KCl on Microcon YM30.
Aminoacylation of tRNA
In vitro transcribed tRNAMeti, tRNAVal (GUG) and total native tRNAs (Promega) were aminoacylated with Met, Val, His, Leu and Cys, as appropriate, as described (Pisarev et al, 2007a, 2007b). For peptidyl-tRNA release experiments, aminoacylation was done in the presence of [35S]Cys yielding [35S]Cys-tRNACys (spec. act. 3 × 105 c.p.m./pmol).
NTPase assay
A total of 0.5 pmol Hbs1 was incubated in a 20-μl reaction mixture containing buffer B (20 mM Tris, pH 7.5, 100 mM KCl, 0.25 mM spermidine, 2 mM DTT) supplemented with 1.5 mM MgCl2, 5 μM GTP and 0.33 μM [γ-32P]GTP in the presence/absence of 2.5 pmol 80S ribosomes and 5 pmol Pelota at 37°C for 20 min. In all, 2 μl aliquots were spotted onto polyethyleneimine cellulose plates for chromatography done using 0.8 M LiCl/0.8 M acetic acid.
Assembly and purification of vacant ribosomes and ribosomal complexes
Initiation, elongation and pre-termination ribosomal complexes were assembled on MVHL-STOP mRNA and mRNAs encoding MVHC tetrapeptide followed by 0, 1, 2, 3, 6, 9, 13 or 17 nts and purified by SDG centrifugation essentially as described (Alkalaeva et al, 2006; Pisarev et al, 2007a). For subunit dissociation experiments, elongation and pre-termination ribosomal complexes were assembled on MVHL-STOP, MVHC-0 and MVHC-3 mRNAs using [32P]60S subunits and in vitro transcribed tRNAMeti and tRNAVal or total native tRNAs aminoacylated with Met, Val, His and Leu or Cys. For mRNA release experiments, ribosomal complexes were assembled on [32P]MVHC-0 and [32P]MVHC-9 mRNAs with total native tRNAs aminoacylated with Met, Val, His and Cys. For peptidyl-tRNA release experiments, ribosomal complexes were assembled on mRNAs encoding MVHC tetrapeptide followed by 0, 1, 2, 3, 6, 9, 13 or 17 nts with total native tRNAs aminoacylated with Met, Val, His and [35S]Cys. [32P]-labelled vacant 80S ribosomes were prepared by incubating 50 pmol 40S subunits and 40 pmol [32P]60S subunits in a 200-μl reaction mixture containing buffer B supplemented with 2.5 mM MgCl2 for 10 min at 37°C and purified by SDG purification.
Dissociation of vacant ribosomes and ribosomal complexes into subunits
For dissociation of vacant ribosomes (Figures 1D and 2), 0.5 pmol [32P]80S ribosomes were incubated with different combinations of 0.75 or 3 pmol Pelota (Figure 2A and B), 5 pmol Pelota (Figures 1D, 2C and D), 5 pmol Hbs1, 5 pmol eRF1/eRF3, 2 pmol ABCE1 and 2 pmol eIF6 for 10 min (except for experiments shown in Figure 2D, in which incubation for periods ranging from 45 s to 10 min was followed by quenching by elevation of Mg2+ concentration to 20 mM) at 37°C in a 40-μl reaction mixture containing buffer B supplemented with 0.5 mM ATP and 0.5 mM GTP (Figures 1D, 2A, C and D), 1 mM ATP and 0.1 mM GTP or GMPPNP (Figure 2B), and corresponding amounts of MgCl2 to achieve 1.5 mM free Mg2+ (except for experiments shown in Figure 2C, in which free Mg2+ concentration ranged from 1.5 to 20 mM). For dissociation of ribosomal complexes (Figures 3B, C and 5B), complexes (0.5 pmol) were incubated with different combinations of 5 pmol Pelota, 5 pmol Hbs1, 5 pmol eRF1, 5 pmol eRF3, 2 pmol ABCE1, 2 pmol eIF6 and 1 mM puromycin for 10 min at 37°C in a 60-μl reaction mixture containing buffer B supplemented with 0.5 mM ATP, 0.5 mM GTP and 2.5 mM MgCl2. For dissociation of ribosomal complexes treated with RelE (Figures 4B–D and 6E), complexes (0.5 pmol) were preincubated with 24 pmol RelE for 5 min at 37°C in a 60-μl reaction mixture containing buffer B supplemented with 0.5 mM ATP, 0.5 mM GTP and 2.5 mM MgCl2, after which reaction mixture were supplemented with different combinations of 5 pmol recombinant (or native, when indicated) Pelota, 5 pmol Hbs1, 5 pmol eRF1/eRF3, 2 pmol ABCE1 and 2 pmol eIF6 and incubation continued for another 10 min. All reaction mixtures were then subjected to centrifugation through 10–30% SDG prepared in buffer B supplemented with 5 mM (Figures 1D, 2A, B, 3B, C, 4B, D, 5B and 6E) or 20 mM (Figure 2C and D) MgCl2 in a Beckman SW55 rotor at 53 000 r.p.m. for 110 min. Association of ribosomal subunits was assayed by Cerenkov counting of an aliquot of each fraction.
mRNA release
Ribosomal complexes assembled on [32P]MVHC-0 or [32P]MVHC-9 mRNAs (0.5 pmol) were incubated with different combinations of 5 pmol recombinant Pelota, 5 pmol Hbs1, 5 pmol eRF1, 5 pmol eRF3, 2 pmol ABCE1 and 2 pmol eIF6 for 10 min at 37°C in a 60-μl reaction mixture containing buffer B supplemented with 0.5 mM ATP, 0.5 mM GTP and 2.5 mM MgCl2, and subjected to centrifugation through 10–30% SDG prepared in buffer B supplemented with 5 mM MgCl2 in a Beckman SW55 rotor at 53 000 r.p.m. for 75 min, or to native gel electrophoresis (see below). Ribosomal association of mRNA was assayed by Cerenkov counting of an aliquot of each fraction after SDG centrifugation, or by autoradiography (Figures 5D and 6C).
Peptidyl-tRNA release
Ribosomal complexes assembled with [35S]Cys-tRNACys (0.5 pmol) were incubated with different combinations of 5 pmol recombinant (or native, when indicated) Pelota, 5 pmol Hbs1, 2 pmol ABCE1 and 2 pmol eIF6 for 10 min (except for the experiments shown in Figure 6D, in which incubation for periods ranging from 1.5 to 10 min was followed by quenching by elevation of Mg2+ concentration to 10 mM) at 37°C in a 60-μl reaction mixture containing buffer B supplemented with 0.5 mM ATP, 0.5 mM GTP and 2.5 mM MgCl2 (except for the experiments shown in Figure 6C, in which free Mg2+ concentration ranged from 1.5 to 10 mM) and subjected to centrifugation through 10–30% SDG prepared in buffer B supplemented with 5 mM (Figure 5C and E) or 10 mM (Figure 6C and D) MgCl2 in a Beckman SW55 rotor at 53 000 r.p.m. for 75 min, or to native gel electrophoresis (see below). Ribosomal association of peptidyl-tRNA was assayed by Cerenkov counting of an aliquot of each fraction after SDG centrifugation, or by autoradiography (Figures 5C, 6B and F).
Toe-printing analysis of ribosomal complexes
To study the influence of Pelota, Hbs1, eRF1/eRF3 and ABCE1 on 48S complex formation on (CAA)nGUS mRNA (Figure 1A and E), reaction mixtures (20 μl) containing 1 pmol mRNA, 5 pmol eIF2, 2.5 pmol eIF3, 5 pmol eIF4A, 7 pmol eIF4B, 2.5 pmol eIF4F, 10 pmol eIF1, 10 pmol eIF1A, 2.5 pmol Met-tRNAMeti, 1.5 pmol 40S subunits or preformed 80S ribosomes, and different combinations of 5 pmol eRF1, 5 pmol eRF3, 5 pmol Pelota, 5 pmol Hbs1 and 2 pmol ABCE1 were incubated for 10 min at 37°C in buffer B supplemented with 1 mM ATP, 0.5 mM GTP and 2.5 mM MgCl2. Assembled 48S complexes were analysed by primer extension using AMV-RT and [32P]-labelled primers (Pisarev et al, 2007b).
To assay the influence of Pelota, Hbs1 and ABCE1 on ribosomal ECs (Figures 3D and E), ribosomal complexes (0.2 pmol) were incubated with combinations of 2 pmol Pelota, 2 pmol Hbs1, 2 pmol ABCE1, 2 pmol Ligatin, 2 pmol eIF6, 2 pmol eRF1 and 2 pmol eRF3 in a 40-μl reaction mixture containing buffer B supplemented with 1 mM GTP or GMPPNP and 2.5 mM MgCl2 for 10 min at 37°C. After incubation, the concentration of Mg2+ was elevated to 20 mM to prevent further recycling. Ribosomal complexes were analysed by primer extension using AMV-RT and appropriate [32P]-labelled primers (Pisarev et al, 2007b).
For toe-print analysis of RelE-mediated cleavage of mRNA (Figure 4A), ribosomal complexes (0.15 pmol) were incubated with 10 pmol RelE in a 20-μl reaction mixture containing buffer B supplemented with 0.5 mM ATP, 0.5 mM GTP and 2.5 mM MgCl2 for 10 min at 37°C. After incubation, mRNA was phenol extracted and analysed by primer extension using AMV-RT and [32P]-labelled primers (Pisarev et al, 2007b).
Native gel electrophoresis
Native gel electrophoresis was performed essentially as described (Acker et al, 2007). In all, 20 μl aliquots of reaction mixtures were loaded in loading buffer (5 × =50% sucrose, 0.02% bromphenol blue, 0.02% xylene cyanol) onto 4% native polyacrylamide gel (37.5:1 acrylamide:bis-acrylamide) prepared in THEM buffer (34 mM Tris base, 57 mM HEPES, 0.1 mM EDTA, 2.5 mM MgCl2) using 22 cm × 18 cm × 0.5 mm plates. Samples were run for 1 h at 4°C at 10 W. Gels were dried and autoradiographed.
Supplementary Material
Acknowledgments
We thank Cajetan Neubauer and Venki Ramakrishnan for their gift of RelE, and L Yu Frolova for eRF1 and eRF3 expression vectors. This work was supported by NIH Grant GM80623 to TVP.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR (2007) Reconstitution of yeast translation initiation. Methods Enzymol 430: 111–145 [DOI] [PubMed] [Google Scholar]
- Adham IM, Sallam MA, Steding G, Korabiowska M, Brinck U, Hoyer-Fender S, Oh C, Engel W (2003) Disruption of the pelota gene causes early embryonic lethality and defects in cell cycle progression. Mol Cell Biol 23: 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV (2006) In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Andreev D, Hauryliuk V, Terenin I, Dmitriev S, Ehrenberg M, Shatsky I (2008) The bacterial toxin RelE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA 14: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL, Hauryliuk V (2008) Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol 8: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M (2010) Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR (2010) Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol 30: 5562–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr-Schmid A, Pfund C, Craig EA, Kinzy TG (2002) Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol Cell Biol 22: 2564–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H (2010) Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol 17: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, Nakamura Y, Svergun DI, Ito K, Song H (2009) Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev 23: 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Engebrecht J (1998) Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics 149: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Wasserman SA (1995) The pelota locus encodes a protein required for meiotic cell division: an analysis of G2/M arrest in Drosophila spermatogenesis. Development 121: 3477–3486 [DOI] [PubMed] [Google Scholar]
- Frerichs KU, Smith CB, Brenner M, DeGracia DJ, Krause GS, Marrone L, Dever TE, Hallenbeck JM (1998) Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc Natl Acad Sci USA 95: 14511–14516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341 [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Manzoor M, Hudak KA (2008) Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J Biol Chem 283: 32218–32228 [DOI] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R (2010) Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Chaillet M, van Tilbeurgh H (2008) Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. J Biol Chem 283: 7145–7154 [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Parker R (2010) No-go decay: a quality control mechanism for RNA in translation. Wiley Interdisciplinary Rev: RNA 1: 132–141 [DOI] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT (2003) Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell 12: 903–911 [DOI] [PubMed] [Google Scholar]
- Hogan BL, Korner A (1968) Ribosomal subunits of Landschütz ascites cells during changes in polysome distribution. Biochim Biophys Acta 169: 129–138 [DOI] [PubMed] [Google Scholar]
- Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA (2010) In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol Cell Proteomics 9: 2162–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M (2010) Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17: 555–560 [DOI] [PubMed] [Google Scholar]
- Johnson TC, Holland JJ (1965) Ribonucleic acid and protein synthesis in mitotic HeLa cells. J Cell Biol 27: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E (1972) Serum-mediated stimulation of protein synthesis in Ehrlich ascites tumor cells. J Biol Chem 247: 5470–5476 [PubMed] [Google Scholar]
- Karcher A, Schele A, Hopfner KP (2008) X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem 283: 7962–7971 [DOI] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H (2010) The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep 11: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, Inada T, Nureki O (2010) Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc Natl Acad Sci USA 107: 17575–17579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV (2005) Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11: 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong C, Ito K, Walsh MA, Wada M, Liu Y, Kumar S, Barford D, Nakamura Y, Song H (2004) Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol Cell 14: 233–245 [DOI] [PubMed] [Google Scholar]
- Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T (2010) Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep 11: 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Kim YS, Kim KH, Heo I, Kim SK, Kim O, Kim HK, Yoon JY, Kim HS, Kim do J, Lee SJ, Yoon HJ, Kim SJ, Lee BG, Song HK, Kim VN, Park CM, Suh SW (2007) Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol Cell 27: 938–950 [DOI] [PubMed] [Google Scholar]
- Loh PG, Song H (2010) Structural and mechanistic insights into translation termination. Curr Opin Struct Biol 20: 98–103 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV (2009) Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE (2009) The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139: 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3. [DOI] [PubMed] [Google Scholar]
- Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, Hollien J, Parker R (2009) Analysis of Dom34 and its function in no-go decay. Mol Biol Cell 20: 3025–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M (2003) The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112: 131–140 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU (2003) Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev 17: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV (2007a) Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV (2008) Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J 27: 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV (2010) The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV (2007b) Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV (2006) Kinetic analysis of interaction of kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J Biol Chem 281: 40224–40235 [DOI] [PubMed] [Google Scholar]
- Rees DC, Johnson E, Lewinson O (2009) ABC transporters: the power to change. Nat Rev Mol Cell Biol 10: 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi AM, Monroy A (1969) Polyribosome formation and RNA synthesis in the early post-fertilization stages of the sea urchin egg. Dev Biol 19: 73–86 [DOI] [PubMed] [Google Scholar]
- Saito K, Kobayashi K, Wada M, Kikuno I, Takusagawa A, Mochizuki M, Uchiumi T, Ishitani R, Nureki O, Ito K (2010) Omnipotent role of archaeal elongation factor 1 alpha (EF1α in translational elongation and termination, and quality control of protein synthesis. Proc Natl Acad Sci USA 107: 19242–19247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM (2004) GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24: 7769–7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff MD, Robbins E (1966) Polyribosome disaggregation during metaphase. Science 151: 992–995 [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R (2010) Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330: 369–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Chaudhuri J, Chevesich J, Maitra U (1997) Molecular cloning and functional expression of a human cDNA encoding translation initiation factor 6. Proc Natl Acad Sci 94: 114285–114290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV (2010) Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H (2004) Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA 10: 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surks MI, Berkowitz M (1971) Rat hepatic polysome profiles and in vitro protein synthesis during hypoxia. Am J Physiol 220: 1606–1609 [DOI] [PubMed] [Google Scholar]
- van den Elzen AM, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Séraphin B, Graille M (2010) Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat Struct Mol Biol 17: 1446–1452 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R (2002) Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF (2001) The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.