Abstract
Induced pluripotent stem (iPS) cell reprogramming is a gradual epigenetic process that reactivates the pluripotent transcriptional network by erasing and establishing repressive epigenetic marks. In contrast to loci-specific epigenetic changes, heterochromatin domains undergo epigenetic resetting during the reprogramming process, but the effect on the heterochromatin ultrastructure is not known. Here, we characterize the physical structure of heterochromatin domains in full and partial mouse iPS cells by correlative electron spectroscopic imaging. In somatic and partial iPS cells, constitutive heterochromatin marked by H3K9me3 is highly compartmentalized into chromocentre structures of densely packed chromatin fibres. In contrast, chromocentre boundaries are poorly defined in pluripotent embryonic stem and full iPS cells, and are characterized by unusually dispersed 10 nm heterochromatin fibres in high Nanog-expressing cells, including pluripotent cells of the mouse blastocyst before differentiation. This heterochromatin reorganization accompanies retroviral silencing during conversion of partial iPS cells by MEK/GSK3 2i inhibitor treatment. Thus, constitutive heterochromatin is compacted in partial iPS cells but reorganizes into dispersed 10 nm chromatin fibres as the fully reprogrammed iPS cell state is acquired.
Keywords: electron spectroscopic imaging (ESI), heterochromatin reorganization, induced pluripotent stem (iPS) cells, Nanog, retrovirus silencing
Introduction
The cascade of events in somatic cell reprogramming to a pluripotent state involves large-scale epigenetic remodelling to establish repressive epigenetic marks on tissue-specific genes, and to erase these marks on key members of the pluripotency network (Takahashi and Yamanaka, 2006; Maherali et al, 2007; Wernig et al, 2007; Barrero et al, 2010). Reprogramming proceeds through at least three stages: intermediate, partial and full induced pluripotent stem (iPS) cell states (Jaenisch and Young, 2008; Yamanaka, 2009). Partial mouse iPS cells attain some aspects of pluripotency including embryonic stem (ES) cell-like colony morphology, teratoma-forming ability and partial activation of pluripotency genes accompanied by downregulation of differentiation-specific genes (Okita et al, 2007; Mikkelsen et al, 2008; Sridharan et al, 2009). Partial iPS cells can be converted into full iPS cells using cell signalling or epigenetic inhibitors (Mikkelsen et al, 2008; Silva et al, 2008). As they complete reprogramming, full mouse iPS cells acquire epigenetic marks of pluripotency including X chromosome reactivation and genome-wide establishment of ES cell-like histone H3K27 and H3K4 trimethylation patterns (Maherali et al, 2007).
ES cells epigenetically inactivate exogenous retroviruses, similarly full iPS cells robustly activate endogenous pluripotency genes during the reprogramming process while silencing the retroviral reprogramming factors (Okita et al, 2007; Silva et al, 2008; Stadtfeld et al, 2008). In contrast, partial iPS cells maintain retroviral gene expression indicating that an ES cell-specific transcriptional network is required for successful silencing of retroviruses in pluripotent cells. Clearly, expression of Oct4, Sox2 and Klf4 in primary mouse embryonic fibroblasts (MEFs) is not sufficient for retrovirus silencing, nor is their persistent expression in partial iPS cells, suggesting that additional endogenous pluripotency factors are required for retroviral silencing in full iPS cells. For example, Nanog, which controls the pluripotency ground state (Silva et al, 2009), is reactivated in full mouse iPS cells (Okita et al, 2007), and is capable of silencing gene expression (Liang et al, 2008). Epigenetic mechanisms normally employed in establishing heterochromatin have prominent functional roles in silencing retrovirus vectors in ES cells. Viral DNA sequences are recognized by a ZFP809/TRIM28 complex that establishes H3K9me2 marks bound by HP1 (Wolf and Goff, 2007, 2009). In addition, H3 is deacetylated (Pannell et al, 2000; Lorincz et al, 2001) and H3K9me3 is deposited by Eset/Setdb1 (Matsui et al, 2010). Additional epigenetic involvement includes recruitment of Swi/Snf components (Golding et al, 2010) and participation of de novo DNA methylases (Cherry et al, 2000; Dodge et al, 2002). These enzymes also reorganize chromatin structure on a global scale. As retroviral silencing occurs at late time points in the reprogramming process, it can be a useful marker to identify molecular changes that take place in the fully reprogrammed iPS cell state.
ES cells are known to have unique heterochromatin domain organization with hyperdynamic binding of histone and associated heterochromatin structure proteins (Meshorer et al, 2006). In contrast to loci-specific epigenetic changes compatible with altered gene expression, changes to the physical structure of heterochromatin domain organization during reprogramming remain unexplored. Mouse cells are unusual in that pericentric constitutive heterochromatin, comprising major and minor satellite repeat sequences, cluster into structures known as chromocentres (Wong and Rattner, 1988; Joseph et al, 1989; Guenatri et al, 2004). These chromocentres are easily identified in mouse nuclei by their 4′,6-diamidino-2-phenylindole (DAPI)-rich staining, and are specifically marked by H3K9me3 and H4K20me3 (Peters et al, 2001). This clustering makes mouse heterochromatin an attractive model system for studying chromatin domain organization. Human cells, on the other hand, contain repetitive sequences that are distributed more evenly across the genome and in most contexts do not cluster to the same degree as in mouse cells. Chromocentre organization has typically been investigated by measuring the amount of clustering or changes in the number of observable chromocentre foci, within differentiating nuclei (Meyer-Ficca et al, 1998; Brero et al, 2005; Tessadori et al, 2007). Notably, the number of these H3K9me3 foci increases when Chd1 is knocked down in ES cells and pluripotency is simultaneously lost (Gaspar-Maia et al, 2009). Thus, like retrovirus silencing, heterochromatin organization in ES cells also correlates with the pluripotent state. However, the timing of its reorganization during iPS cell reprogramming and the specific structures of the heterochromatin fibres remain to be identified.
As an alternative to using molecular biology approaches or visible light imaging, chromatin organization has been studied using conventional transmission electron microscopy (CTEM). Somatic cell nuclei imaging by CTEM reveals condensed ‘closed’ chromatin domains along the nuclear envelope and at the nucleolus periphery (Belmont et al, 1989; Kireev et al, 2008). These silenced compartments, including chromocentres, are widely accepted to be comprising 30 nm and higher-order chromatin fibre assemblies (Rego et al, 2008). Currently, evidence for 30 nm fibres in vivo is largely restricted to non-mammalian cell types (van Holde and Zlatanova, 1995; Tremethick, 2007; Maeshima et al, 2010). When visualized by electron spectroscopic imaging (ESI) (Ahmed et al, 2009), the only technique that provides high contrast of unstained chromatin at high-molecular resolution (Bazett-Jones and Ottensmeyer, 1981; Dehghani et al, 2005), the predominant chromatin configuration in ES cells is a mesh of ‘open’ dispersed chromatin fibres, and displays a paucity of the blocks of condensed ‘closed’ chromatin observed in somatic cells (Efroni et al, 2008). Upon differentiation of ES cells to neural progenitor cells, however, some of the dispersed chromatin becomes organized into compact heterochromatin domains, particularly along the nuclear envelope (Hiratani et al, 2009). In addition to cell culture models, we have shown that pluripotent pre-implantation embryos also have globally decondensed chromatin, most strikingly after the 8-cell stage (Ahmed et al, 2010). These observations led us to ask whether reprogramming is accompanied by the loss of compact heterochromatin domains at chromocentres to the more dispersed open structures that is typical of pluripotent ES cells.
The significant difference in global nuclear architecture between somatic and ES cells implies that large-scale reorganization events may be involved in iPS cell reprogramming. Our primary goal was to identify the timing and structural features of heterochromatin reorganization during reprogramming. We compare H3K9me3-marked constitutive heterochromatin organization in full and partial iPS cells with that of the parental MEFs and the J1 ES cell line. We demonstrate that chromocentres in full iPS cells with high Nanog levels are characterized by dispersed open domains comprised entirely of 10 nm chromatin fibres, like those of ES cells and high Nanog-expressing cells of the inner cell mass (ICM) of the mouse blastocyst before differentiation. Failure to disrupt the heterochromatin domains seen in MEFs by retaining tightly packed closed chromatin domains is a characteristic of partial iPS cells. Conversion of partial iPS cells with the mitogen-activated protein/Erk kinase and glycogen synthase kinase-3 (MEK/GSK3) inhibitor (2i) cocktail (Silva et al, 2008) shows that heterochromatin reorganization is a novel signature of the fully reprogrammed state that accompanies retrovirus silencing.
Results
Chromocentre compartment differences in partial and full iPS cell lines
To study the relationship between global chromatin organization and cellular reprogramming events, we compared MEFs, iPS and J1 ES cells. We previously reported the derivation of three mouse iPS cell lines, EOS3F-24, EOS3F-28 and EOS3F-29, using pMX-based retroviral delivery of three ‘Yamanaka’ factors, Klf4, Sox2 and Oct4. All three iPS cell lines were shown to maintain expression of the EOS-EGFP pluripotency reporter vector and have a pluripotent capacity to differentiate in vitro and form teratomas in vivo (Hotta et al, 2009). Interestingly, only the EOS3F-24 cell line maintained its teratoma-forming capacity after embryoid body-mediated differentiation, indicative of the inefficient differentiation capacity of partial iPS cells (Okita et al, 2007; Jaenisch and Young, 2008). In contrast, differentiation of both EOS3F-28 and EOS3F-29 is accompanied by EOS-EGFP inactivation and subsequent loss of their teratoma-forming capacity (Hotta et al, 2009).
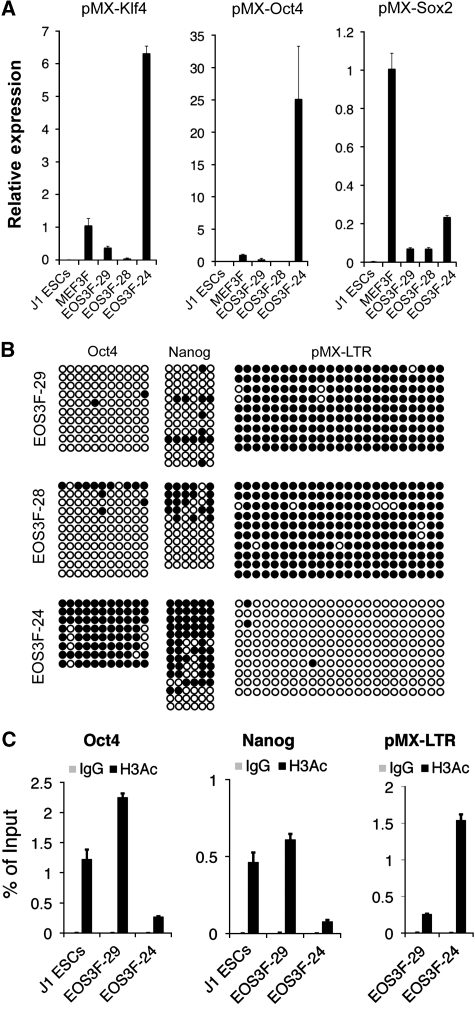
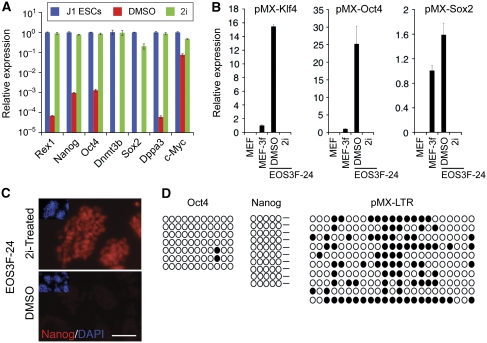
A reliable molecular marker for full reprogramming is retrovirus silencing, and partially reprogrammed cell lines fail to silence the retroviral transgenes. Indeed, EOS3F-24 iPS cells maintain high level of expression of all three exogenous reprogramming factor transgenes compared with the retrovirus silencing observed in EOS3F-28 and EOS3F-29 cells (Figure 1A). Accordingly, bisulphite sequencing reveals that the LTR promoters of the reprogramming vectors are hypomethylated in EOS3F-24 but hypermethylated in EOS3F-29 and EOS3F-28 iPS cells (Figure 1B). Furthermore, the endogenous Oct4 and Nanog promoters remain hypermethylated at CpG sites in EOS3F-24 but not in EOS3F-28 and EOS3F-29 iPS cells (Figure 1B). Chromatin immunoprecipitation (ChIP) analysis with an anti-H3Ac antibody reveals that Nanog and Oct4 promoters are hypoacetylated in EOS3F-24 compared with J1 ES and EOS3F-29 iPS cells while the viral transgenes are hyperacetylated compared with EOS3F-29 iPS cells (Figure 1C). These data demonstrate that, in contrast to EOS3F-28 and EOS3F-29, the epigenetic state of the viral transgenes and endogenous pluripotency genes have not been reprogrammed in EOS3F-24 iPS cells.
Figure 1.
Characterization of induced pluripotent stem cells. (A) qRT–PCR was performed on RNA from infected MEFs (MEF3F) and uninfected controls (J1 ES cells), EOS3F-29, EOS3F-28 and EOS3F-24 iPS cell lines using primers specific for pMX-Klf4, pMX-Oct4 and pMX-Sox2. (B) Bisulphite sequencing of CpG sites of endogenous Oct4 and Nanog promoters and pMX-LTRs. Open and closed circles indicate unmethylated and methylated CpG sites, respectively. (C) ChIP analysis of histone H3 acetylation of the Oct4 and Nanog promoters and the pMX vector of EOS3F-29 and EOS3F-24 cells. J1 cells are included as control for ChIP analyses of endogenous promoters. Error bars represent ±s.e.m. of triplicate reactions.
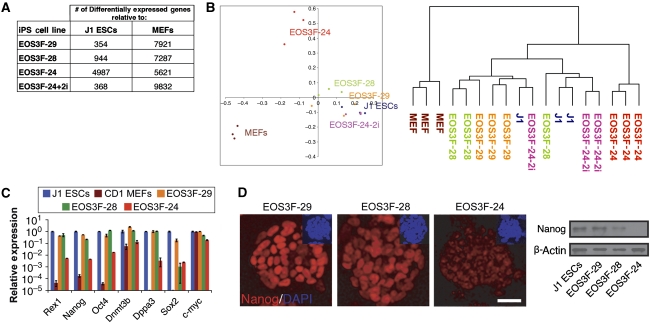
To directly examine expression of endogenous pluripotency genes in the iPS cell lines in comparison with reference pluripotent J1 cells and MEFs, global gene expression analyses with microarray chips were performed. These results show that the EOS3F-29 expression profile is similar to J1 cells (354 genes differentially expressed, see Materials and methods). In contrast, EOS3F-24 cells are distinct from both J1 ES cells (4987 differentially expressed genes) and MEFs (5621 differentially expressed genes) (Figure 2A). These distance relationships are further illustrated in the hierarchical clustering (Figure 2B) of the arrays. Importantly, pluripotency-related genes are expressed at significantly reduced levels compared with J1 ES cells only in EOS3F-24 iPS cells as validated by qRT–PCR (Figure 2C). Nanog is a strong marker of full reprogramming in the mouse system and is less easily detected in EOS3F-24 as analysed by immunocytochemistry and western blot analysis compared with EOS3F-28 and EOS3F-29 cells (Figure 2D). Furthermore, EOS3F-24 is lacking any detectable levels of the SSEA1 cell surface marker measured by flow cytometry (data not shown). Collectively, these data show that EOS3F-28 and EOS3F-29 have the molecular marks of full iPS cells whereas EOS3F-24 has molecular features of partial iPS cells.
Figure 2.
Microarray and pluripotent gene expression analysis of partial and full iPS cells. (A) Microarray data summary of differentially expressed genes in iPS cell lines shows that EOS3F-29, EOS3F-28 and 2i-treated and not EOS3F-24 iPS cells have expression profiles that are very close to J1 cells. Table indicates the number of genes over- or under-represented in different iPS cells compared with J1 ES cells and MEF controls. (B) Principal component analysis was performed to compare the microarray expression profiles of genes in MEFs, iPS (EOS3F-24, EOS3F-24+2i, EOS3F-28 and EOS3F-29) and J1 ES cells. The left panel represents a scatter plot of the microarray expression profiles on the planes spanned by the first and second principal components. The right panel represents a dendrogram of the hierarchical clustering of expression profiles. (C) Pluripotency-related gene expression in iPS cells compared with J1 ES cells as measured by qRT–PCR analysis. Error bars represent ±s.e.m. of triplicate reactions. (D) Nanog expression measured by immunofluorescence microscopy and western blot analysis of EOS3F-29, EOS3F-28 and EOS3F-24 iPS cell lines. Scale bar, 20 μm.
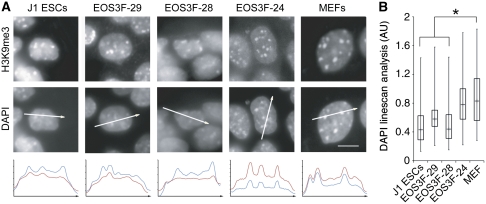
To determine whether the pluripotent versus differentiated state was reflected in global chromatin organization we compared parental MEFs, partial and full iPS cells and J1 ES cells. These cells all have physically confined heterochromatin regions enriched in H3K9me3 as seen by fluorescence microscopy (Figure 3A). These domains were identified as chromocentres in all analysed cell lines on the basis of H4K20me3 enrichment, DAPI density and proximity to centromeres, visualized with CREST antisera (Supplementary Figure S1). Analysis of optical z-stack images and two-channel linescans confirmed that DAPI enrichment is always associated with H3K9me3 enrichment in these cells (Figure 3A; Supplementary Movies 1 and 2). These data are consistent with previously published results indicating that both pluripotent and differentiated mouse cells have compartmentalized heterochromatin, which form chromocentres (reviewed in Ahmed et al (2009); Gaspar-Maia et al, 2009; Martin et al, 2006). Although H3K9me3 enrichment is useful for identifying chromocentres in all analysed cell types, it cannot be used to measure chromatin fibre density. On the other hand, though DAPI binds AT-rich DNA sequences preferentially, its signal intensity is a more accurate measure of chromatin compaction than detection of specific histone modifications. Indeed, close inspection of DAPI-stained nuclei revealed significant differences between differentiated and pluripotent cell types. A description of the advantages and caveats of these optical methods to measure chromatin density is provided in Supplementary data. DAPI linescan analysis demonstrate that MEF and partial iPS cell chromocentres appear as bright foci while those of J1 ES and full iPS cells contain lower DAPI signal relative to nucleoplasmic background (Figure 3B). These linescan analyses are supported by whole nucleus variance analyses which show a nine-fold increase in the signal variance between MEF feeder and J1 ES cells within the same image field. Thus, based on DAPI distribution, chromocentres are poorly compartmentalized in ES and full iPS cells.
Figure 3.
Constitutive heterochromatin organization differs between partial and full iPS cells. (A) H3K9me3 immunofluorescence and DAPI counterstain in iPS, J1 ES and MEF cells. White arrows through DAPI images indicate intensity linescan plot and direction. DAPI (red) and H3K9me3 (blue) linescans are shown below image panels. Scale bar, 5 μm. (B) Box plot analysis of 100 chromocentres from at least 30 different cells and three independent trials of the background to chromocentre DAPI ratio, asterisks indicate significance to P<0.001. These DAPI linescan analyses were entirely consistent with a variance analysis. The average variance of feeder cells (MEF) was measured to be 48 477±19 115 compared with the average variance of J1 ES cells, which was 5964±4005. Sixteen cells of each cell type were measured to calculate the variance.
Chromocentres in full iPS cells are composed of 10 nm chromatin fibres
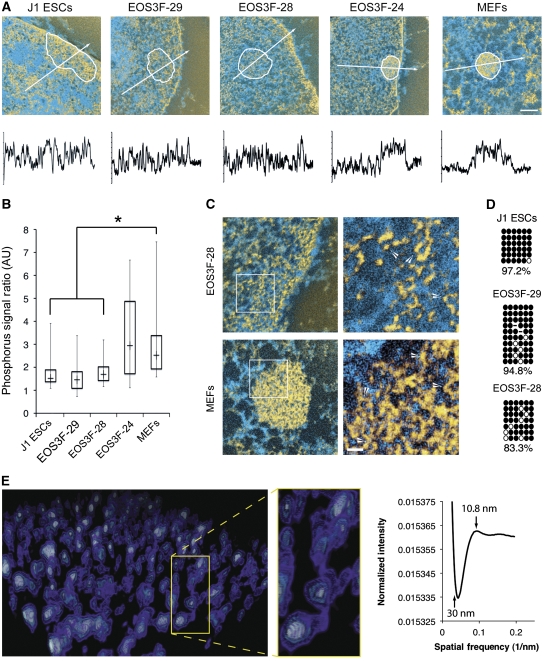
DAPI is not a good indicator of chromatin fibre density per se because it is also affected by AT richness and nucleosome-repeat length. Therefore, to interpret differences in heterochromatin organization observed by DAPI counterstain we employed ESI. Indirect labelling of H3K9me3 and H4K20me3 (Supplementary Figure S2) was used to demark the constitutive heterochromatin domains for imaging the underlying chromatin fibre organization by correlative light microscopy (LM)/ESI (Figure 4A; Supplementary Figure S2) (Dellaire et al, 2004). Integrative phosphorus density analysis demonstrates that the chromocentres of ES and full iPS cells are difficult to delineate from the surrounding chromatin (Figure 4B). Therefore, the presence of H3K9me3 is compatible with dispersed chromatin packing density. In contrast, chromocentres in partial iPS cells and MEFs were densely packed, displaying a significantly higher fibre density than the surrounding chromatin.
Figure 4.
Chromocentres in full iPS cells are composed of 10 nm heterochromatin fibres. Correlative light microscopy/Electron Spectroscopic Imaging with indirectly labelled H3K9me3 delineates chromocentres. (A) White circles delineate H3K9me3 enrichment. Quantitative phosphorus and nitrogen ratio images were segmented to show chromatin in yellow and protein-based structures in blue. White arrows indicate length and direction of phosphorus linescan analysis shown below image panels. Scale bar, 0.5 μm. (B) Box plot analysis of the phosphorus density shows the distribution of chromatin compaction within these cells, 50 chromocentres were measured from at least 30 different nuclei from three independent trials, asterisks indicate significant differences in fibre density, P<0.001. (C) The 10 nm fibres within constitutive heterochromatin in a pluripotent cell and a differentiated cell, chromatin fibres are shown in yellow with zoomed panels to the right. Zoomed regions are shown in non-enhanced image in white boxes. White arrows in the zoomed images indicate 10 nm fibres. Scale bar, 30 nm. (D) Bisulphite sequencing of CpG sites in major satellite repeats, open and closed circles represent unmethylated and methylated CpG sites, respectively. (E) Perspective image of Chimera generated model of tomographic reconstruction of H3K9me3-enriched region and surrounding chromatin of a J1 ES cell pseudo-coloured by electron density. Connected nucleosomes and intervening linker sequence of these 10 nm fibres clearly visualized in the zoomed panel to the right. Representative Fourier transform analysis of the average chromatin fibre size at 10.84 nm is shown.
To investigate whether this disruption of constitutive heterochromatin was a general feature of iPS cell reprogramming we analysed previously characterized full (Ng-20D17) and partial (Fb-20A10) iPS cell lines generated independently (Okita et al, 2007). When analysed by DAPI linescan analysis and integrated ESI-phosphorus analysis, the results from these cell lines were in full agreement with those of the EOS-derived iPS cell lines (Supplementary Figure S3). The fully reprogrammed iPS cell line had disrupted heterochromatin regions whereas the partially reprogrammed cell line had well-delineated closed heterochromatin domains with open dispersed surrounding regions.
At higher resolution, we observed an abundance of dispersed chromatin within chromocentre domains of ES and full EOS-28 iPS cells that is difficult and sometimes impossible to distinguish from the surrounding chromatin (Figure 4C). Surprisingly, we also observed a prevalence of 10 nm fibres within the compact heterochromatin domains of MEFs, where only 30 nm or higher-level fibre organization might be expected. Although unexpected, this result is consistent with cryo-EM studies of metaphase chromatin in situ, where only 10 nm chromatin fibres were detected using phase contrast algorithms (Eltsov et al, 2008). These observations can be quantitatively confirmed since the phosphorus signal is a direct measure of chromatin density (Bazett-Jones and Ottensmeyer, 1981; Woodcock et al, 1990; Bazett-Jones et al, 1999). In addition, all of the chromatin in a physical section can be detected by ESI (Bazett-Jones et al, 1999). We further demonstrate this (Supplementary data) by using the integrated phosphorus intensities of individual nucleosomes to predict total chromatin content of the image field and ultimately of an entire nucleus. This analysis correctly estimates the real chromatin content of a diploid mouse cell (Table S2). Previous low angle X-ray scattering (Langmore and Paulson, 1983) and EM with conventional heavy atom contrast agents or of isolated chromatin experiments have indicated that the genome is comprised entirely of 30 nm and higher-order chromatin fibres (Gilbert et al, 2004; Sinclair, 2010). Our observation of 10 nm chromatin fibres in compact H3K9me3-marked heterochromatin structures challenges current models in which chromatin condensation is based on a transition between 10 and 30 nm chromatin fibres. Bisulphite sequencing shows that major satellite DNA repeats located in chromocentres are still highly methylated in the full iPS cells (Figure 4D), indicating that DNA methylation is also compatible with dispersed chromocentre formation.
To confirm that these dispersed fibres were in fact 10 nm we combined ESI with tomography to generate a three-dimensional representation of the phosphorus content in ES cells (Figure 4E; Supplementary Figure S5; Supplementary Movies 3 and 4). This analysis clearly shows the 10-nm chromatin fibre comprising both nucleosomes and intervening linker sequences. Both the H3K9me3-defined heterochromatin regions and the surrounding euchromatin were comprised exclusively of 10 nm chromatin fibres. Although we detect no 30 nm fibres in these dispersed chromatin regions, 30 nm fibres can be detected by ESI. For example, 30 nm chromatin fibres have been imaged by ESI in starfish sperm (Supplementary Figure S4) (Bazett-Jones, 1992). Fourier transformed analysis of five tomograms containing both H3K9me3-enriched regions and surrounding chromatin demonstrates an average particle size of 10.8 nm with a complete absence of higher-order chromatin fibres of greater diameters (Figure 4E). The 3D analysis clearly demonstrates that 10 nm chromatin fibres exclusively populate both the heterochromatin regions and surrounding chromatin domains in pluripotent J1 ES cells.
Taken together, these data indicate that dispersion of blocks of constitutive heterochromatin correlates with complete reprogramming, whereas partial iPS cells maintain compact constitutive heterochromatin domains resembling those in MEFs. This is the first demonstration of 10 nm chromatin fibres within constitutive heterochromatin domains.
Heterochromatin reorganizes during 2i conversion of partial iPS cells
To address whether constitutive heterochromatin reorganization is indeed a feature of the acquisition of the fully reprogrammed state, we induced complete reprogramming of EOS3F-24 iPS cell line using an MEK/GSK3 2i inhibitor cocktail (Silva et al, 2008). Preliminary readouts of successful conversion after 1-week chemical treatments were evaluated by SSEA1 and EOS-EGFP activation in EOS3F-24 iPS cells. Three concentrations of the 2i cocktail ([1 × ], [2 × ] and [4 × ], see Materials and methods section) resulted in dosage-dependent emergence of a high EOS-EGFP expressing EOS3F-24 subpopulation of cells that was also SSEA1 positive (Supplementary Figure S6). To investigate whether molecular changes at the DNA/RNA level were taking place in the newly derived SSEA1+ EOS3F-24 cells, we sorted for the high EGFP expressing cells and cultured them for an additional week in the presence of 2i. Following 2i treatment, the converted EOS3F-24 cells robustly activated pluripotency-related loci (Figure 5A) and silenced the reprogramming retroviral transgenes (Figure 5B). Accordingly, Nanog protein is abundant in the 2i-converted EOS3F-24 colonies (Figure 5C). Bisulphite sequencing demonstrates appropriate changes in DNA methylation at the LTRs and promoter regions of Oct4 and Nanog following the 2i treatment (Figure 5D). These expression patterns were reproduced in independently converted [4 × ] 2i-treated EOS3F-24 cells, and microarray analysis on these samples strongly indicates that 2i-treated cells cluster with ES cells and are as similar to them (368 differentially expressed genes) as the fully reprogrammed EOS3F-29 iPS cells (Figure 2A and B).
Figure 5.
Characterization of 2i conversion of partial EOS3F-24 iPS cells into full iPS cells. (A) qRT–PCR of upregulated endogenous pluripotency-associated genes during 2i conversion of EOS3F-24 iPS cells. (B) qRT–PCR of pMX-Klf4, pMX-Oct4 and pMX-Sox2 transgenes. Error bars represent +/− s.e.m. of triplicate reactions. (C) Upregulation of Nanog after 2i conversion of EOS3F-24 iPS cells demonstrated by indirect immunofluorescence microscopy, Nanog shown in red and DAPI inset in blue. Scale bar, 40 μm. (D) Bisulphite sequencing of CpG sites of endogenous and transgene promoters in 2i-treated EOS3F-24 cells. Open and closed circles represent unmethylated and methylated CpG sites, respectively.
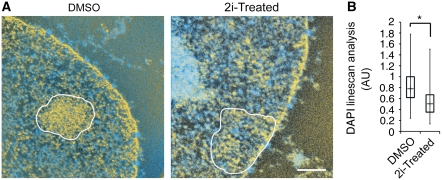
To determine whether conversion of partial iPS cells was accompanied by global heterochromatin changes we performed LM/ESI. H3K9me3-enriched regions were significantly more disrupted in the 2i-treated cells but not in DMSO-treated controls (Figure 6A). These trends were consistent with our LM observations, where the chromocentre DNA density relative to nucleoplasmic background, as measured by DAPI intensity, was significantly different between the control and 2i-treated cells (Figure 6B). Thus, DAPI linescan analysis supports the LM/ESI data and is a simple method to identify dispersed heterochromatin in converted and fully reprogrammed iPS cells. The 2i treatment caused dispersion of densely packed heterochromatin fibres during the conversion of partial iPS cells. We speculate that this transition of heterochromatin domain reorganization occurs at a late stage in reprogramming as only the cells which have silenced the transgenes, a previously described late-stage event in iPS reprogramming, have disrupted heterochromatin.
Figure 6.
Heterochromatin reorganizes during 2i-mediated conversion of partial iPS cells. (A) Correlative LM/ESI analysis of constitutive heterochromatin in DMSO control and 2i-treated cells, H3K9me3 enriched regions outlined in white. Scale bar, 0.5 μm. (B) DAPI linescan analysis of 2i versus DMSO control cells indicates a significant reduction in chromocentre partitioning, asterisk indicates significance of P<0.001.
Heterochromatin reorganizes in Nanog high pluripotent cells
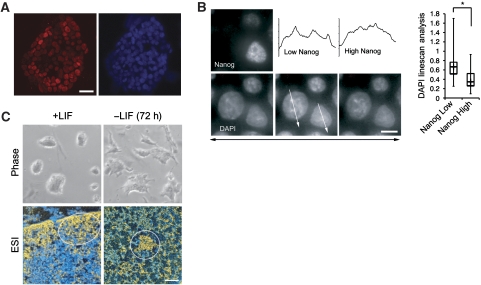
Disruption of constitutive heterochromatin in the 2i-treated EOS3F-24 cells was accompanied by reactivation of endogenous Nanog expression. To determine whether Nanog levels correlate with heterochromatin reorganization, we analysed ES cell colonies exhibiting Nanog variegation (Chambers et al, 2007) (Figure 7A). To assess chromatin compaction, we measured DAPI intensity of chromocentres relative to nucleoplasmic intensity in relatively high and low Nanog-expressing J1 ES cell nuclei. We observed a correlation of Nanog expression with heterochromatin compaction, where high Nanog-expressing cells exhibited the most disrupted and least compartmentalized chromocentres (Figure 7B). The lowest Nanog-expressing ES cells had significantly more compacted chromocentres, suggesting that heterochromatin reorganization initiates when Nanog is downregulated as, or even before, ES cells begin to differentiate. It should be noted, however, that Nanog-low ES cells still have more disrupted chromocentres than differentiated cells. Retrovirus silencing also occurs in the presence of Nanog in 2i-converted iPS cells but not Nanog-negative partial iPS cells. This is consistent with other reports showing that the retroviral reprogramming factors are specifically silenced in Nanog–GFP-positive iPS cell colonies (Nakagawa et al, 2008). These data show that high Nanog expression in pluripotent cells in vitro correlates with heterochromatin reorganization. Furthermore, withdrawal of LIF from J1 ES cells for 72 h results in rapid reappearance of fully compact chromocentres detected by LM/ESI (Figure 7C). These results indicate that dispersed heterochromatin in pluripotent cells is reorganized into the compact structures seen in somatic cells as Nanog expression is lost at the onset of differentiation.
Figure 7.
Nanog expression level impacts chromatin organization in J1 ES cells. (A) Low magnification immunofluorescence of variegated Nanog expression in a J1 ES cell colony with DAPI shown to the right; scale bar, 20 μm. (B) High magnification and resolution representative image of a J1 ES cell field with high and low Nanog-expressing cells (top left panel) and DAPI counterstain of a 0.3-μm z-stack series of these same nuclei. DAPI linescan analysis quantification is adjacent, 30 nuclei of each expression level were analysed and asterisk represents significance of P<0.001. White arrows indicate direction of linescan (shown above) through optimal DAPI z-stack. Scale bar, 5 μm. (C) Withdrawal of LIF from J1 ES cell culture results in the loss of typical smooth colony morphology of the ES cells (phase) and formation of compact chromocentres (ESI) within 72 h. Scale bar, 0.5 μm.
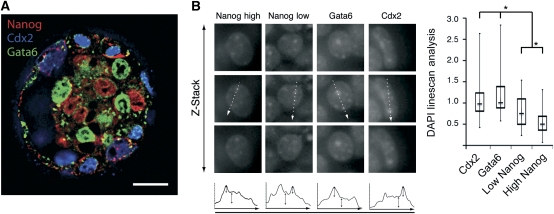
To assess the relationship between Nanog expression and heterochromatin organization in vivo we took advantage of Nanog variegation that briefly occurs in the ICM of E3.75 blastocysts (Figure 8; Supplementary Figure S7) (Yamanaka et al, 2010). Cells were binned according to Nanog expression levels, excluding lineage-committed Gata6 and Cdx2-positive cells, which are no longer pluripotent (Figure 8A). Using a DAPI linescan analysis, we found that Nanog levels correlated directly with heterochromatin compartmentalization and significant differences were observed between high and low Nanog-expressing nuclei. Chromatin in both high and low Nanog-positive nuclei was significantly less compartmentalized than the lineage-committed Cdx2 or Gata6-positive cells (Figure 8B). Together, these data indicate that in the absence of Nanog heterochromatin in lineage-committed cells during early development is organized into a compact structure. In contrast, high Nanog protein levels in pluripotent cells of the blastocyst directly correlate with their dispersed heterochromatin organization. These findings are consistent with the global analysis of chromatin structures associated with embryonic development and with the striking chromatin compaction of the ICM nuclei in the Oct4-null embryos (Ahmed et al, 2010). The implications of these data combined are two-fold: (1) the pluripotent transcription factor network regulates chromatin structure and dictates unique chromatin architecture of the pluripotent state and (2) chromatin reorganization precedes differentiation both in vitro and in vivo.
Figure 8.
Heterochromatin reorganizes in Nanog high pluripotent cells in vivo in the early mouse blastocyst. (A) Immunofluorescence of variegated Nanog (red), GATA-6 (green) and Cdx2 (blue) expression in E3.75 day early mouse blastocyst. Scale bar, 20 μm. (B) DAPI images of three optical sections above and below the optimal image plane, representative chromocentres in these cells are adjacent to IF image, z=2.1 μm. White arrows indicate direction of DAPI linescans, shown below and quantified. Double-headed black arrows represent chromocentre DAPI peak height relative to nucleoplasmic background. Asterisk represents minimum significance to P<0.005, from a minimum of 40 nuclei per category.
Discussion
We used LM/ESI to identify novel heterochromatin structure reorganization during iPS cell reprogramming. We demonstrate that constitutive heterochromatin in chromocentres transitions from a very compact closed chromatin fibre domain in MEFs and partial iPS cells to a more open domain of loosely packed chromatin fibres in ES and full iPS cells. Although generally dispersed chromatin was previously observed in ES cells (Efroni et al, 2008), we show here that heterochromatin specifically enriched in H3K9me3 is composed entirely of 10 nm fibres in ES and full iPS cells. This is compatible with the general concept that pluripotent stem cells have more open chromatin structure to make the cells more responsive to differentiation cues that they receive. We were surprised to observe 10 nm chromatin fibres in the very densely packed chromocentres of MEFs and partial iPS cells although we cannot exclude the possibility that 30 nm fibres can also be found in these structures. However, prevalence of 10 nm fibres in both compact and disrupted heterochromatin domains indicates the transition between open and closed chromatin domains involves, at least in part, transitions between closely packed and highly folded 10 nm chromatin fibres. This challenges the absolute requirement for transitions between 10 and 30 nm chromatin fibres in defining heterochromatin domains.
To directly demonstrate that heterochromatin reorganization specifically occurs in fully reprogrammed cells, we converted partial iPS cells into full iPS cells via 2i treatment. We thus confirm that 2i conversion is an effective means to complete reprogramming, and that during conversion heterochromatin reorganization coincides with other late events in reprogramming including retrovirus silencing and Nanog gene activation. These events are specific for ES and full iPS cells, and therefore are not induced by the combined expression of Oct4, Sox2 and Klf4 in the primary transduced MEFs or the partial iPS cells. This implies that heterochromatin reorganization is dependent on establishment of the endogenous pluripotency transcriptional network. Since endogenous Nanog expression is a reliable marker of mouse full iPS cells, we took advantage of its natural variegation in ES cells and blastocysts. These analyses show that high Nanog ES cells have the most dispersed heterochromatin, whereas low Nanog cells have less dispersed heterochromatin. These findings are confirmed in vivo during normal development, where heterochromatin is dispersed most in Nanog high cells and is more compartmentalized in Nanog-negative differentiated cells. Thus, expression of Nanog, or its targets in the pluripotency network, correlates with late heterochromatin reorganization events in reprogramming. This finding is consistent with the known ability of Nanog both to activate pluripotent gene expression and to silence genes normally expressed in somatic cells. Candidate epigenetic pathways that participate in heterochromatin reorganization may in future be identified by comparing the gene expression differences between the partial EOS3F-24 iPS cells and the 2i-treated cells. Experimental manipulation of such pathways in partial iPS cells may direct chromocentre reorganization and establish the exact timing of heterochromatin disruption during the acquisition of the fully reprogrammed state.
Materials and methods
Cell culture
J1 ES cells and iPS cell clones were grown on feeders with recombinant LIF. EOS-generated iPS cell lines were maintained under 1 μg/ml puromycin selection. The 2i conversion was performed using MEK inhibitor (PD0325901, 0.5 μM) and GSK3 inhibitor (CHIR99021, 3 μM) (StemGent), represented as [1 × ] concentration in the text, in LIF supplemented ES media, in absence of puromycin selection. Blastocysts were collected from uteri of ICR outbreed mice at embryonic day 3.75 and fixed in 4% formaldehyde solution in PBS for 15 min at RT.
Quantitative RT–PCR analysis
In all, 1 μg of RNA, collected using TRIZOL (Invitrogen), was reverse transcribed using SSII RT kit with random hexamer primers (Invitrogen) using manufacturer's instructions. In all, 50 ng of cDNA was used for qRT–PCR using SYBR green master mix (ABI) in triplicate. The primer sequences are in Table S1.
Bisulphite sequencing
Bisulphite sequencing was performed on DNA isolated from iPS lines at passage 10. DNA was bisulphite converted and purified using EZ DNA methylation Gold Kit (Zymo Research). In all, 50 ng of converted DNA was subjected to PCR (see Supplementary Table S1 for primer sequences). PCRs were subcloned into TOPO-TA vectors (Invitrogen) and sequenced.
Chromatin immunoprecipitation
ChIP was performed using the Upstate chromatin immunoprecipitation kit. Briefly, 1.0 × 106 cells were crosslinked in 1% paraformaldehyde, cells were washed, scraped into a conical tube and sonicated for 25 min in the Bioruptor™ XL at the high setting with cycles of 30 s ON, 30 s OFF. ChIPs were performed using EZ-ChIP kit (Millipore) under manufacturer's instructions with 4 μg of anti-acetyl histone H3 (Upstate), DNA was purified using phenol/chloroform extraction and DNA was resuspended in 50 μl of H2O. In all, 1 μl of DNA was subjected to qPCR using SYBR green.
Microarray analysis
Total RNA isolated from different clones was labelled and hybridized to Affymetrix GeneChIP Mouse Gene 1.0 ST gene expression microarrays using standard conditions. Intensity values from biological triplicates were processed by variance stabilization and normalization (Huber et al, 2002) and summarized by RMA (Irizarry et al, 2003), iPS expression profiles were contrasted both to MEFs and to J1 ES cell profiles. Differentially expressed transcript in each contrast with an adjusted P-value <0.05 were determined using limma (Smyth, 2004). The raw and processed data with additional information are available at the GEO database website under accession number GSE21595.
Western blot analysis
Protein was isolated from different clones with RIPA lysis buffer and run on an 8% SDS–PAGE gel. Proteins were transferred onto a nitrocellulose membrane, blocked with 5% blocking buffer and probed with anti-Nanog (Cosmobio) and β-actin (Sigma) antibody at a 1:500 and 1:2000 dilution, respectively. Secondary HRP-conjugated antibodies were used with SuperSignal West Pico detection reagents (Thermo Scientific).
Immunofluorescence microscopy and immunostaining
Cells were prepared for immunolabelling as described (Ahmed et al, 2009), except for blastocysts which were washed in PBS-Tween and permeabilized with 0.2% Triton X-100. Primary antibodies used were rabbit anti-H3K9me3, rabbit anti-H4K20me3, human anti-CREST (Immunovision), rabbit anti-Nanog (Cosmobio), Cdx2 (BioGenex), Gata6 (R&D), and SSEA1 (Developmental Studies Hybridoma Bank). Secondary antibodies used were donkey anti-rabbit or mouse Cy2, Cy3 and Cy5 (Jackson Laboratories) and PE-Cy5 (eBiosciences), Alexa488 and Alexa633 (Molecular Probes). Cells used for DAPI linescan analyses were mounted in 1 mg/ml paraphenlenediamine and 1 μg/ml DAPI or soaked in 35 ng/ml DAPI. Images were collected on a Leica Microsystems DMRA2 microscope equipped with a Hamamatsu ORCA-ER camera or an Olympus IX81 inverted microscope equipped with a Cascade II CCD (Photometrics) camera with or without the spinning disc. OpenLab 3.5.1 (Improvision) software was used to collect images. Fluorescence images of embryos were processed with Image-Pro Plus 6.2 3D-blind deconvolution software. All other images were processed with Photoshop 7.0 (Adobe).
Correlative LM/ESI microscopy
Sample preparation and detailed ESI procedure are described fully in Ahmed et al (2009). Following immunolabelling H3K9me3 or H4K20me3 cells were post fixed in 1% glutaraldehyde, dehydrated and embedded in Quetol (Electron Microscopy Sciences). Samples were sectioned by an ultramicrotome (Leica) 70 nm. Energy filtered electron micrographs were taken on a transmission electron microscope (Tecnai 20, FEI) operated at 200 kV collected using a GATAN post column imaging filter at 120 and 155, and 385 and 415 eV to generate the phosphorus and nitrogen images, respectively. Pre- and post-edge images were recorded on a CCD camera. Digital micrograph software was used to collect images. Nitrogen images were subtracted from phosphorus images, so that the net nitrogen signal in chromatin structures was normalized to zero. These phosphorus subtracted nitrogen images are coloured blue. The phosphorus images are pseudo-coloured yellow and overlaid onto the phosphorus subtracted nitrogen image. Images were processed with Digital micrograph and Photoshop 7.0 (Adobe).
Image analysis
DAPI linescan analyses were performed using ImageJ on optical sections where DAPI foci were at optimal focal planes. Fifty pixel (approximately half the diameter of the average chromocentre analysed) histograms were generated through the foci and the background (outside the nucleus) was subtracted from the nucleoplasmic and chromocentre signals. Variations in these data were calculated as a ratio of chromocentre peak height to nucleoplasmic signal.
Integrative phosphorus analyses of ESI images were preformed on unprocessed phosphorus jump ratio maps in ImageJ. The integrated average phosphorus intensities within H3K9me3-enriched regions were compared with the integrated average phosphorus intensity outside the H3K9me3-enriched region within the same image field. Phosphorus intensities were background subtracted using a small region within the field devoid of nucleic acid or phosphorus signal.
Electron tomography
Phosphorus ESI jump ratio tilt series of H3K9me3-enriched regions and surrounding chromatin of sectioned J1 ES cells were acquired at 120 and 155 eV using SerialEM (Mastronarde, 2005) with 2° increment over a tilt range of ±60°. The images in the series were aligned and processed into jump ratios using a combination of IMOD (Kremer et al, 1996), SPARX (Hohn et al, 2007) and ImageJ. Three-dimensional phosphorus maps were reconstructed using the IMOD implementation of SIRT from the aligned jump ratio tilt series. Rendering of the volumes was done in UCSF Chimera (Pettersen et al, 2004).
Fourier analysis
Power spectra of overlapping subareas of a tomographic slice from each tomogram were added and rotationally averaged to yield a 1-dimensional trace in reciprocal units using SPARX. These traces were normalized by integral area and averaged to give a single representative profile of reciprocal distance.
Flow cytometry
Flow cytometry was performed as previously described (Hotta et al, 2009). Briefly, trypsinized cells were suspended in PBS with 5% FBS and were analysed by FACScan using CellQuest software (Becton Dickinson) and data were analysed with FlowJo (Tree Star).
Supplementary Material
Acknowledgments
We thank Dr Prim Singh for antibodies. We thank Dr Keisuke Okita and Dr Shinya Yamanaka for providing the Fb-20A10 and Ng-20D17 iPS cell lines. Dr Ren Li, Reagan Ching, Kashif Ahmed and Anna Toulina for intellectual and/or technical contributions and Doug Holmyard and Robert Temkin at the Advanced Bioimaging facility for technical advice. This work was funded by Canadian Institutes of Health Research operating grants to DPB-J (MOP-14311), JE (MOP-81129) and JD (RMF-92090).
Author contributions: EF, UD, AH, JE and DPB-J conceived the project; EF, UD, JE and DPB-J designed the experiments; AH derived the iPS cell lines, UD characterized the iPS cell lines by qRT–PCR, bisulphite sequencing and ChIP analysis and performed the 2i conversion experiments. EF, MS and DPB-J performed the ultrastructural analyses; MS analysed and generated the ESI-tomographic reconstructions; JD and CP-I preformed the microarray and analysis; FL performed the embryo experiments; EF, UD, DPB-J and JE wrote the manuscript. DPB-J holds the Canada Research Chair in Molecular and Cellular Imaging.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP (2010) Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE 5: e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, Li R, Bazett-Jones DP (2009) Electron spectroscopic imaging of the nuclear landscape. Methods Mol Biol 464: 415–423 [DOI] [PubMed] [Google Scholar]
- Barrero MJ, Boue S, Izpisua Belmonte JC (2010) Epigenetic mechanisms that regulate cell identity. Cell Stem Cell 7: 565–570 [DOI] [PubMed] [Google Scholar]
- Bazett-Jones DP (1992) Electron spectroscopic imaging of chromatin and other nucleoprotein complexes. Electron Microsc Rev 5: 37–58 [DOI] [PubMed] [Google Scholar]
- Bazett-Jones DP, Hendzel MJ, Kruhlak MJ (1999) Stoichiometric analysis of protein- and nucleic acid-based structures in the cell nucleus. Micron 30: 151–157 [DOI] [PubMed] [Google Scholar]
- Bazett-Jones DP, Ottensmeyer FP (1981) Phosphorus distribution in the nucleosome. Science 211: 169–170 [DOI] [PubMed] [Google Scholar]
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA (1989) Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma 98: 129–143 [DOI] [PubMed] [Google Scholar]
- Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC (2005) Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol 169: 733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234 [DOI] [PubMed] [Google Scholar]
- Cherry S, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R (2000) Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol 20: 7419–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani H, Dellaire G, Bazett-Jones DP (2005) Organization of chromatin in the interphase mammalian cell. Micron 36: 95–108 [DOI] [PubMed] [Google Scholar]
- Dellaire G, Nisman R, Bazett-Jones DP (2004) Correlative light and electron spectroscopic imaging of chromatin in situ. Methods Enzymol 375: 456–478 [DOI] [PubMed] [Google Scholar]
- Dodge J, Ramsahoye B, Wo Z, Okano M, Li E (2002) De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene 289: 41–48 [DOI] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E (2008) Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J (2008) Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA 105: 19732–19737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M (2009) Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA (2004) Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118: 555–566 [DOI] [PubMed] [Google Scholar]
- Golding MC, Zhang L, Mann MR (2010) Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell 6: 457–467 [DOI] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G (2004) Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 166: 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, Rathjen PD, Gilbert DM (2009) Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ (2007) SPARX, a new environment for Cryo-EM image processing. J Struct Biol 157: 47–55 [DOI] [PubMed] [Google Scholar]
- Hotta A, Cheung A, Farra N, Vijayaragavan K, Séguin C, Draper J, Pasceri P, Maksakova I, Mager D, Rossant J, Bhatia M, Ellis J (2009) Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods 6: 370–376 [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 (Suppl 1): S96–104 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph A, Mitchell AR, Miller OJ (1989) The organization of the mouse satellite DNA at centromeres. Exp Cell Res 183: 494–500 [DOI] [PubMed] [Google Scholar]
- Kireev I, Lakonishok M, Liu W, Joshi VN, Powell R, Belmont AS (2008) In vivo immunogold labeling confirms large-scale chromatin folding motifs. Nat Methods 5: 311–313 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Langmore JP, Paulson JR (1983) Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol 96: 1120–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung S, Qin J, Wong J, Cooney A, Liu D, Songyang Z (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10: 731–739 [DOI] [PubMed] [Google Scholar]
- Lorincz M, Schübeler D, Groudine M (2001) Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol Cell Biol 21: 7913–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Hihara S, Eltsov M (2010) Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol 22: 291–297 [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K (2007) Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70 [DOI] [PubMed] [Google Scholar]
- Martin C, Beaujean N, Brochard V, Audouard C, Zink D, Debey P (2006) Genome restructuring in mouse embryos during reprogramming and early development. Dev Biol 292: 317–332 [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36–51 [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler P, Brown D, Misteli T (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca M, Muller-Navia J, Scherthan H (1998) Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci 111 (Part 10): 1363–1370 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein B, Jaenisch R, Lander E, Meissner A (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106 [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448: 313–317 [DOI] [PubMed] [Google Scholar]
- Pannell D, Osborne C, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, Okano M, Li E, Lipshitz H, Ellis J (2000) Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J 19: 5884–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS (2008) The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci 121 (Part 7): 1119–1127 [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen T, Smith A (2008) Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 6: e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen T, Guo G, van Oosten A, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A (2009) Nanog is the gateway to the pluripotent ground state. Cell 138: 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J (2010) Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim Biophys Acta 1799: 286–295 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason M, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K (2009) Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault D, Hochedlinger K (2008) Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tessadori F, Chupeau MC, Chupeau Y, Knip M, Germann S, van Driel R, Fransz P, Gaudin V (2007) Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J Cell Sci 120 (Part 7): 1200–1208 [DOI] [PubMed] [Google Scholar]
- Tremethick DJ (2007) Higher-order structures of chromatin: the elusive 30 nm fiber. Cell 128: 651–654 [DOI] [PubMed] [Google Scholar]
- van Holde K, Zlatanova J (1995) Chromatin higher order structure: chasing a mirage? J Biol Chem 270: 8373–8376 [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein B, Jaenisch R (2007) In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448: 318–324 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff S (2007) TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP (2009) Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Rattner JB (1988) Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res 16: 11645–11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Horowitz RA, Bazett-Jones DP, Olins AL (1990) Localization of DNA in chromatin using electron spectroscopic imaging and osmium staining. In Proc of XIIth ICEM, pp 116–117 [Google Scholar]
- Yamanaka S (2009) Elite and stochastic models for induced pluripotent stem cell generation. Nature 460: 49–52 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J (2010) FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137: 715–724 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.