Abstract
A key question in nuclear RNA surveillance is how target RNAs are recognized. To address this, we identified in vivo binding sites for nuclear RNA surveillance factors, Nrd1, Nab3 and the Trf4/5–Air1/2–Mtr4 polyadenylation (TRAMP) complex poly(A) polymerase Trf4, by UV crosslinking. Hit clusters were reproducibly found over known binding sites on small nucleolar RNAs (snoRNAs), pre-mRNAs and cryptic, unstable non-protein-coding RNAs (ncRNAs) (‘CUTs’), along with ∼642 predicted long anti-sense ncRNAs (asRNAs), ∼178 intergenic ncRNAs and, surprisingly, ∼1384 mRNAs. Five putative asRNAs tested were confirmed to exist and were stabilized by loss of Nrd1, Nab3 or Trf4. Mapping of micro-deletions and substitutions allowed clear definition of preferred, in vivo Nab3 and Nrd1 binding sites. Nrd1 and Nab3 were believed to be Pol II specific but, unexpectedly, bound many oligoadenylated Pol III transcripts, predominately pre-tRNAs. Depletion of Nrd1 or Nab3 stabilized tested Pol III transcripts and their oligoadenylation was dependent on Nrd1–Nab3 and TRAMP. Surveillance targets were enriched for non-encoded A-rich tails. These were generally very short (1–5 nt), potentially explaining why adenylation destabilizes these RNAs while stabilizing mRNAs with long poly(A) tails.
Keywords: RNA degradation; RNA processing; RNA surveillance; yeast, RNP structure
Introduction
Quality control of RNA processing and nuclear surveillance of aberrant RNAs are integral features of eukaryotic gene expression. The exosome is a conserved complex with endonuclease and 3′ exonuclease activities, which functions together with a set of cofactors to degrade many types of defective transcript as well as processing the 3′ ends of stable RNAs (reviewed by Houseley and Tollervey, 2009). Characterized nuclear cofactors include the Trf4/5–Air1/2–Mtr4 polyadenylation (TRAMP) complexes and the Nrd1–Nab3 RNA-binding heterodimer. The TRAMP complexes contain three proteins; a poly(A) polymerase (either Trf4 or Trf5), the DExH-box helicase Mtr4 and a zinc-knuckle protein (either Air1 or Air2) (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). Analyses of strains lacking Trf4 or Trf5 show that they have partially overlapping functions, but Trf4 appears to have the major role in nuclear surveillance (reviewed by Houseley and Tollervey, 2009). Adenylation by the TRAMP complexes promote exosome-mediated degradation, in contrast to the role of poly(A) addition in promoting mRNA stability and translation. This distinction was proposed to derive from the lower processivity observed in vitro for Trf4/5, compared with the mRNA poly(A) polymerase (LaCava et al, 2005) but the actual lengths of tails added by TRAMP in vivo was unclear.
The Nrd1–Nab3 complex participates in transcription termination on RNA polymerase II transcribed small nucleolar RNAs (snoRNAs), cryptic unstable intergenic transcripts (CUTs) and some mRNAs (Steinmetz et al, 2001; Thiebaut et al, 2006; Arigo et al, 2006a, 2006b; Carroll et al, 2007; Rondon et al, 2009; Kim et al, 2010). Nrd1–Nab3 termination is also proposed to be available to all mRNAs transcripts as an alternative pathway (Rondon et al, 2009). Nrd1 binds directly to the Ser5/Ser7 phosphorylated C-terminal domain (CTD) of the large subunit of Pol II (Gudipati et al, 2008; Vasiljeva et al, 2008; Kim et al, 2010). This suggested that their direct role in termination is restricted to short transcripts, since phosphorylation at Ser5/Ser7 is generally replaced by Ser2 during elongation (Komarnitsky et al, 2000; Schroeder et al, 2000; Egloff et al, 2007). In addition, Nrd1–Nab3 act as exosome cofactors, promoting degradation of CUTs and 3′ processing of precursors to snoRNAs (Arigo et al, 2006b; Thiebaut et al, 2006; Vasiljeva and Buratowski, 2006; Grzechnik and Kufel, 2008).
Pol II ChIP and microarray data have identified many non-protein-coding RNAs (ncRNAs), including CUTs and stable unannotated transcripts (SUTs) (David et al, 2006; Steinmetz et al, 2006). Moreover, the promoter regions of protein-coding genes generate short, bidirectional, promoter-associated RNAs (PARs) (Neil et al, 2009; Xu et al, 2009; Churchman and Weissman, 2011). Genome-wide tiling arrays and nucleosome position analyses indicate that Pol II can initiate wherever DNA is accessible. Accumulation of the resulting transcripts is, however, greatly limited by the surveillance machinery, which acts as the gatekeeper of the transcriptome.
RNA Pol III transcribes tRNAs, 5S rRNA and many other small stable RNAs. Surveillance of the processing and modification of at least some Pol III transcribed RNAs involves TRAMP and the exosome (Kadaba et al, 2004, 2006; Vanacova et al, 2005; Huang et al, 2006; Schneider et al, 2007; Wang et al, 2008), which are also key players in surveillance of Pol I transcribed pre-rRNAs (Allmang et al, 2000; Dez et al, 2006; Wery et al, 2009). In contrast, the known functions and associations of Nrd1–Nab3 suggested that they were specific for Pol II transcripts.
Numerous studies have revealed specific targets for the nuclear RNA surveillance machinery. However, general recognition mechanisms that discriminate between functional RNAs and aberrant transcripts remain elusive. In vivo UV crosslinking is a powerful approach for the identification of sites of RNA–protein interactions (Ule et al, 2003, 2005; Granneman et al, 2009; Hafner et al, 2010). Here, we use a UV crosslinking approach to identify RNAs bound by nuclear RNA surveillance factors. High-throughput sequencing analysis of crosslinked RNAs revealed that the TRAMP complex marks its targets with a short oligo(A) tail and that Nrd1–Nab3 functions upstream of TRAMP and exosome in surveillance of a wide range of previously unrecognized targets.
Results
Identification and validation of targets for surveillance factors
In order to identify novel targets for the nuclear RNA surveillance machinery, we applied an in vivo RNA–protein crosslinking approach (CRAC) (Granneman et al, 2009) to the RNA-binding proteins Nrd1, Nab3 and the polyA polymerase Trf4. Yeast strains were constructed expressing genomically encoded, C-terminal tagged Nrd1–HTP, Nab3–HTP and Trf4–HTP, respectively, expressed under the control of the endogenous promoter. All three strains showed wild-type (WT) growth rates, indicating that the fusion proteins were functional. The trf4Δ strain is cs-lethal at 18°C (Sadoff et al, 1995), and impairs growth at all temperatures. The cs-lethal phenotype was fully complemented by the tagged construct (Supplementary Figure S1A). To assess the expression levels of the tagged proteins, lysates of strains expressing Nab3–HTP and Trf4–HTP was treated with TEV protease and western blotted using anti-Nab3 and anti-Trf4 antibodies, respectively. Nab3–HTP was mildly overexpressed (∼1.5-fold) relative to the endogenous proteins, whereas Trf4 was underexpressed (∼3-fold). Antibodies raised against Nrd1 failed to give a clear result, but northern analyses revealed that the mRNA was close to the WT levels (data not shown). HTP-tagged versions of the Air1 and Air2 proteins, which associate with Trf4 in the TRAMP complex, were also generated but failed to give usable crosslinking efficiencies (data not shown).
Crosslinking was performed in these strains and a non-tagged control strain in three biological replicates. RNA–protein complexes were purified by two-step purification and recovered RNAs were reverse transcribed. cDNA libraries were initially analysed by cloning and Sanger sequencing. Two independent data sets from each strain were further analysed by Solexa sequencing (Supplementary Figure S1). Solexa data sets were analysed separately and results shown represent averages over both experiments, unless stated otherwise. Graphs over single genomic locations are shown for one representative experiment only. Notably, in each case the low- and high-throughput data sets were similar in distribution of targets and presence of oligo(A) tails. The sequences recovered were assigned to genomic locations as described (Granneman et al, 2009) and grouped into functional categories (Figure 1A–E; Supplementary Figure S2). For each of the tagged strains, 0.4–4.5 M sequence reads were obtained and mapped to the genome. In contrast, only 23 K reads could be mapped for the non-tagged control (see also Supplementary data), largely corresponding to 25S rRNA fragments that are common contaminants in CRAC analyses (Supplementary Figure S1) (Granneman et al, 2009). The ribosome synthesis factor Nop58 was used as positive control; 71% of recovered sequences represented snoRNAs and 22% rRNA, consistent with its known functions (Granneman et al, 2009). The full data sets are available from the authors.
Figure 1.
Nuclear RNA surveillance factors are crosslinked to many RNA targets. (A–D) High-throughput sequencing of cDNA libraries generated from crosslinked RNAs associated with purified Nrd1–HTP (based on 3.1 M mapped reads), Nab3–HTP (5.8 M mapped reads), Trf4–HTP (6.5 M mapped reads) and a no-tag control (23 K mapped reads). Sequencing data were mapped to the yeast genome using NOVOALIGN. Pie charts illustrate proportion of mapped reads corresponding to classes of RNAs in the indicated IP. The results from two independent, high-throughput analyses are combined. (E) Single track distribution of CRAC hits of the indicated IP along chromosome IV. (F) Bar diagram representing percentage of all mapped sequences carrying at least three non-encoded terminal A residues in the indicated purification from the combined data sets. (G) Length distribution of non-encoded tails on RNAs associated with the indicated proteins using the combined data sets and allowing up to 20% non-A residues. See also Supplementary Figure S1F and G.
In all data sets, the three factors tested were associated with classes of RNA corresponding to known targets (Figure 1A–D). For Trf4–HTP, 50% of all sequences mapped to the Pol I transcribed rDNA, consistent with the role of Trf4 in pre-rRNA surveillance (Dez et al, 2006) and the degradation of truncated fragments generated by transcriptional pausing and R-loop formation in the 18S rRNA 5′ region (El Hage et al, 2010). In contrast, pre-rRNAs were largely absent from the Nrd1 and Nab3 data sets, despite their very high abundance in total RNA. Other stable RNAs, snRNAs and snoRNAs, were found in all data sets (Figure 1A–D), consistent with reported roles for Nrd1–Nab3 in termination of their transcription (Steinmetz et al, 2001) and surveillance by TRAMP (Houalla et al, 2006; Grzechnik and Kufel, 2008). Many non-coding, cryptic unstable transcripts (CUTs) were also recovered, as anticipated. Unexpectedly, all low- and high-throughput data sets contained numerous tRNAs, which were not believed to be targets for surveillance by Nab3 or Nrd1, with similar distributions in the low-throughput analyses (data not shown).
Nab3 consistently showed higher crosslinking efficiency and more hit clusters than Nrd1 (Supplementary Figures S1 and S2). Together with the better representation of Nab3 consensus sequences in the recovered fragments (below), this suggests that Nab3 might be the primary RNA-binding protein at many, but not all, target sites.
Surveillance targets carry A-rich, non-encoded tails
Oligoadenylation of RNAs by the TRAMP complex is an important signal for degradation mediated by the exosome (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). cDNAs associated with Nrd1, Nab3 and Trf4 were compared with the genomic sequence and analysed for the presence of non-encoded, 3′ residues. Non-encoded tails were initially defined as any sequence located between the genome-mapped and 3′-linker-mapped fragments of reads (see Supplementary data). As a control, we analysed a data set obtained with the snoRNP protein Nop58 (Granneman et al, 2009).
A large majority of non-encoded tails identified were oligo(A). In all Nab3, Nrd1 and Trf4 experiments, between 50 and 80% of non-encoded tails contained only A residues. Manual analysis of the sequence data revealed that tails with large numbers of non-A nucleotides could usually be explained by incorrect mapping of the reads. To improve mapping quality, in the subsequent analyses we focused on non-encoded tails with a maximum of 20% non-A residues.
The number of adenylated reads recovered will underestimate the true association with oligo(A)+ RNA, since the small fragments sequenced include the tail only when the protein-binding site is located in very close vicinity. Despite this, between 6 and 18% of Nrd1, Nab3 and Trf4 reads carried three or more non-encoded, 3′-terminal adenosine residues. In contrast, Nop58–HTP recovered <1% of oligoadenylated reads. This provides strong support for the recovery of bona fide targets for the surveillance machinery (Figure 1F).
Analysis of non-encoded tails can provide an estimate of the in vivo nucleotide specificity of the poly(A) polymerases involved in RNA surveillance. The per-nucleotide frequencies of non-A residues in non-encoded tails in Nab3, Nrd1 and Trf4 experiments, were 2.5, 3.1 and 5.1%, respectively. Among the non-A residues, between 54 and 81% were G, 15–36% were U and 4–14% were C, consistent with in vitro measurements of the nucleotide specificity of Trf4 (LaCava et al, 2005).
Analysis of the distribution of oligoA tail lengths allowing up to 20% non-A nucleotides in the A-rich tails gave the distribution of tail lengths shown in (Figure 1G; Supplementary Figure S1F and G). The median tail length is between 3 and 5 nt. This was not simply due to the short read lengths in the deep sequencing data, since oligoadenylated sequences with a similar length distribution were also identified by Sanger sequencing (data not shown). Moreover, the RNA was fragmented with RNases A+T1, which do not cut adjacent to As, so oligo(A) tails will remain intact. The relatively small population of long-tailed RNAs presumably give rise to the fractions previously identified as poly(A)+ by oligo(dT) selection. Notably, this result implies that the RNAs identified here as surveillance substrates would predominately be overlooked in microarray analyses that involve oligo(dT) selection or priming for cDNA synthesis.
An outstanding question was how A tail addition by TRAMP could target aberrant RNAs for degradation, while mRNAs were stabilized by polyadenylation? These data indicate that oligo(A) tails added by TRAMP are predominately too short to bind the canonical poly(A)-binding protein Pab1, which stabilizes mRNAs and stimulates translation but requires around A12 to bind (Sachs et al, 1987). Therefore, the oligo(A) and A-rich tails are left unprotected and provide an entry side for exonuclease degradation of the marked RNAs.
Identification of preferred binding motifs
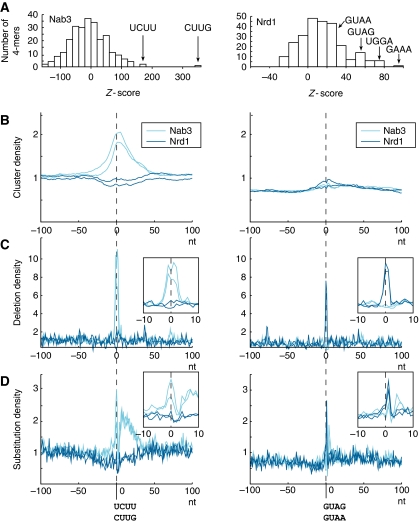
During studies on snoRNA transcription, consensus-binding motifs for Nrd1 and Nab3 were characterized in vitro and in vivo (Carroll et al, 2004, 2007). We assessed the extent to which these and other motifs are enriched among all RNAs recovered with each protein. All motif analyses were performed on reads with a 100% match in the genome. Figure 2A shows the statistical overrepresentation scores for all possible 4 mers in the actual data set compared with a simulated control data set (see Materials and methods). In the case of Trf4, this analysis failed to identify any clear consensus-binding sites. Recovered sequences appeared to contain fewer C nucleotides than expected (data not shown), suggesting that overall nucleotide composition might contribute to the affinity of Trf4 binding. Alternatively, multiple protein cofactors might have the primary role in Trf4 recruitment to its targets. For both Nab3 Solexa data sets, the previously identified UCUU motif is the second most overrepresented 4 mer in the data set. Only a variant of this motif, CUUG, scored higher, and 65% of reads contained one of these motifs. Alignment of the top scoring k mers revealed that UCUUG forms the core of preferred binding motifs for Nab3 (Supplementary Figure S1H). For Nrd1 the reported binding motifs, GUAA and GUAG, were significantly overrepresented in the sequencing data (Figure 2A), but other purine-rich motifs such as UGGA and GAAA had higher scores. No single 4 mer was present in >30% of reads, indicating that the presence of GUAA/G is not strictly required to recruit Nrd1 in vivo. Similar findings were obtained from low-throughput analyses, as around 60% of the Nab3 sequences contained either UCUU or CUUG and for Nrd1 around 30% of all reads contained GUAA/G or UGGA.
Figure 2.
Nrd1 and Nab3 bind consensus motifs. (A) Statistical overrepresentation scores of 4 mer sequences in Nab3 and Nrd1. (B–D) Density of clusters (B), deletions (C) and substitutions (D) around Nab3-binding motifs UCUU/CUUG or Nrd1-binding motifs GUAG/GUAA. See also Supplementary Figure S1H.
Nucleotide substitutions and deletions can identify precise crosslinking sites (Ule et al, 2005; Granneman et al, 2009). We therefore plotted the distribution of clusters of hits, putative crosslinking-induced deletions and putative crosslinking-induced substitutions in a 200-nt window around all TCTT, CTTG, GTAA and GTAG motifs found in the genome (Figure 2B–D). As expected, Nab3 hit clusters were enriched in a relatively broad, 50 nt region around the TCTT and CTTG sequences (Figure 2B). Strikingly, crosslinking-induced deletions in Nab3 data were enriched 10-fold in a very narrow region of 5–6 nt around the Nab3-binding motif (Figure 2C). Crosslinking-induced substitutions were also strongly enriched around the Nab3 motif, although the main peak was accompanied by a broad shoulder towards the 3′ end of the reads (Figure 2D). In contrast, the Nrd1 data showed a mild decrease in signal over the Nab3 consensus sites. This is consistent with binding as a heterodimer, in which Nab3 contacts the UCUU/CUUG motif. No peak of deletions or substitutions was seen for Nrd1, indicating that the deletions are genuinely caused by Nab3 crosslinking, rather than by an increase of sequencing error rate or background crosslinking efficiency near UCUU or CUUG motifs.
An analysis of genomic GTAA and GTAG sequences, previously identified as Nrd1-binding motifs, revealed at most a weak enrichment of Nrd1 hit clusters (Figure 2B). This is consistent with our finding that GUAA and GUAG are not among the most strongly enriched 4 mers in the Nrd1 data set. Despite this, deletions and substitutions in Nrd1 data were strongly enriched in a 3-nt window around the Nrd1-binding motifs (Figure 2C and D). No such enrichment could be seen in the Nab3 data. This indicates that the analysis of crosslinking-induced deletions and substitutions can greatly aid the determination of in vivo specificity of RNA-binding proteins and identify sites of direct protein–RNA interaction.
Characterization of known snoRNA and mRNA targets
Nrd1–Nab3 are involved in transcription termination and coupled 3′ processing of snoRNAs, snRNAs and some mRNAs, whereas Nrd1–Nab3 plus TRAMP terminate and degrade cryptic, unstable ncRNAs and the truncated Nrd1 mRNA (Steinmetz et al, 2001; Arigo et al, 2006a, 2006b; Thiebaut et al, 2006; Ciais et al, 2008; Grzechnik and Kufel, 2008; Rondon et al, 2009). snoRNAs comprised ∼10% of Nrd1–Nab3 targets and 2% of Trf4 hits (Figure 1A–D) in the Sanger and both Solexa sequencing data sets.
Previous analyses of SNR13 identified two terminator elements downstream of the 3′ end of the snoRNA (Steinmetz et al, 2001; Carroll et al, 2004). These include consensus Nrd1 and Nab3 binding sequences and their mutation leads to transcriptional read-through (Carroll et al, 2004). Pre-snR13 was bound by Nrd1 and Nab3, but also interacted with Trf4. The majority of reads were mapped to the downstream terminator elements, rather than the mature snoRNA (Figures 1E and 3A; Supplementary Figure S3). Nop58 crosslinked to snR13 and many other boxC/D snoRNAs, but preferentially associated with the internal boxD′ element, which is different from the preferred surveillance factor binding sites (Figure 1E). Terminator I was bound by both Nrd1 and Nab3, with the reads covering the consensus-binding sequences (Figure 3A; Supplementary Figure S3).
Figure 3.
Crosslinking experiments recover known targets for the nuclear RNA surveillance machinery. Densities in hits per million of high-throughput sequencing reads of the indicated IP mapped to SNR13 (A), SNR3 (B), NRD1 (C), CTH2 (D), IGS1-R CUT (E) and SRG1-SER3 (F). Open reading frames are represented by arrows, ncRNAs by boxes. UTRs and terminator elements are indicated where appropriate. (E) 5S and 25S transcripts are represented by bold arrows and cryptic ncRNAs by slim arrows. See also Supplementary Figure S3.
Analyses of micro-deletions revealed that Nab3 directly binds the UCUU consensus motifs positioned 40 and 85 nt downstream of snR13. For Nrd1 nucleotide substitutions in and around a GUAG motif 50 nt downstream of snR13 (Supplementary Figure S3) also indicated sequence-specific recognition.
Transcription termination on SNR3 is impaired in nrd1 mutants (Steinmetz et al, 2001) and Nrd1 and Nab3 crosslinked to the 3′ end of this snoRNA and in the flanking region (Figure 3B). Multiple consensus Nab3 binding sites and fewer Nrd1 sites are located in several short regions up to 300 nt downstream of the mature 3′ end. These regions were recovered with the respective proteins and presumably contain the signals for Nrd1–Nab3-dependent snR3 termination/processing. Specific association with consensus Nrd1 and Nab3 binding sites was also observed for other snoRNA genes (data not shown), demonstrating the specificity of in vivo crosslinking and providing a detailed view of Nrd1–Nab3-dependent snoRNA terminator elements.
The 5′ UTR and 5′ coding region of NRD1 contain consensus-binding sites for Nrd1 and Nab3, which autoregulate NRD1 mRNA levels via premature transcription termination (Steinmetz et al, 2001; Arigo et al, 2006a). Nrd1 and Nab3 recovered sequences from the 5′ UTR and the 5′ end of the NRD1 ORF, including the consensus motifs (Figure 3C).
Previous analyses indicated that mRNAs from the CTH2 gene are generated by post-transcriptional processing from a precursor that is 3′ extended by ∼1.6 kb (Ciais et al, 2008). Maturation involves recognition of the pre-mRNA by Nrd1–Nab3 and subsequent 3′ processing by TRAMP and the exosome. Sequences associated with Nrd1, Nab3 and Trf4 were consistent with binding to the 3′ extended pre-CTH2 RNA (Figure 3D). Nrd1 and Nab3 bound a cluster of previously predicted binding sites located at +900 relative to the 3′ end of the ORF, supporting both the conclusions concerning the processing pathway and the reliability of the CRAC technique.
Identification of novel mRNA targets
In all data sets, a surprisingly large number of sequences were mapped to mRNAs (19–31%). A few nuclear mRNA transcripts were previously shown to be targets for Nrd1–Nab3 binding and all known targets were recovered (Figure 3).
For ∼1384 mRNAs, the sense strands were identified by hit clusters in both Solexa data sets (Supplementary Figure S2). In all data sets, hit clusters were distributed across the entire coding sequence, (Supplementary Figure S3 and data not shown) and are therefore unlikely to correspond to the short, unstable, promoter-associated transcripts (PARs) previously detected in strains lacking TRAMP and exosome activities (Wyers et al, 2005; Davis and Ares, 2006) or to reflect the roles of Nrd1–Nab3 in failsafe transcription termination (Rondon et al, 2009). The oligo(A) tails recovered were generally located at sites within the ORF, rather than at the expected mRNA polyadenylation site (Supplementary Figure S3), indicative of crosslinking to degradation intermediates. The recovery of oligoadenylated fragments demonstrates that hits recovered do not reflect non-specific binding to intact mRNAs. Many well-expressed housekeeping genes were not recovered in these analyses, including the components of the exosome itself. Notably, the oligo(A) tail length data indicate that RNAs identified here would not have been included in most previous analyses. We conclude that nuclear turnover of mRNA precursors is substantially more active than previously believed.
Intergenic ncRNA targets
A large number of reads from each data set were mapped to intergenic regions (unannotated and not overlapping with any feature annotated in SGD) or were antisense to protein-coding genes; 26% of all reads for Nrd1, 30% for Nab3 and 13% for Trf4 (Figure 1A–D). A number of CUTs were previously shown to be stabilized by loss of Trf4, Nrd1 or Nab3 (Wyers et al, 2005; Arigo et al, 2006b) and all CUTs previously characterized as surveillance targets contained clusters of hits in the CRAC analyses. In addition, 178 other intergenic regions were reproducibly identified by hit clusters. The identification of these ncRNA confirms that they are actively transcribed in WT cells, and are not only induced by mutation of the surveillance machinery.
Characterized CUTs include the IGS1-R ncRNA, derived from the intergenic spacer region of the rDNA repeat (Kobayashi and Ganley, 2005; Houseley et al, 2007). IGS1-R was frequently recovered with Nrd1, Nab3 and Trf4 (Figure 3E; >1000 hits per million).
SRG1 is a ncRNA transcript that partially overlaps with the promoter of the downstream gene SER3, which it represses by nucleosome positioning (Martens et al, 2004, 2005; Hainer et al, 2011). SRG1 is oligoadenylated and degraded in the nucleus in a Nrd1–Nab3/TRAMP/exosome-dependent manner or in the cytoplasm by the 5′ exonuclease Xrn1 after decapping (Arigo et al, 2006b; Thiebaut et al, 2006; Thompson and Parker, 2007). We found Nrd1, Nab3 and Trf4 associated with SRG1 ncRNA but not significantly with the downstream SER3 mRNA (Figure 3F).
Numerous long antisense RNAs are targets for Nrd1–Nab3, TRAMP and the exosome
A small number of asRNAs have been functionally analysed and shown to participate in regulating the expression of cognate sense mRNA (Camblong et al, 2007). In the GAL cluster, a 4.0-kb long ncRNA (GAL10as) is expressed when transcription of the GAL10 mRNA is repressed by glucose (Houseley et al, 2008; Pinskaya et al, 2009). GAL10as is subject to TRAMP-dependent degradation and present at only 0.07 copies per cell (i.e. about one cell in 13 has a copy of the RNA at steady state) (Houseley et al, 2008). Despite this low abundance, we were readily able to detect the association of GAL10as with Trf4, Nrd1 and Nab3 in all Solexa data sets (Supplementary Figure S4). Depletion of Nrd1 or Nab3 increased the level of the GAL10as (data not shown), confirming that it is indeed a target. We conclude that the CRAC technique can identify even low abundance targets of the surveillance machinery. Around 642 other putative transcripts that lie antisense to protein-coding genes (asRNAs) were reproducibly identified by hit clusters (Supplementary Figure S2).
To determine whether these putative novel asRNAs were actually present and subject to surveillance, selected candidates were examined in detail. Transcription start sites (TSSs) were mapped for the asRNAs CAF17as and HPF1as by 5′ RACE in trf4Δ and WT strains (Figure 4A and B; Supplementary Figure S4). For CAF17as two TSSs were identified, one lying 10 nt upstream and one 10 nt downstream of the stop codon of the corresponding mRNA. The TSS for HPF1as is located 265 nt downstream of the mRNA stop codon. Strong asRNA accumulation was seen in strains depleted for Nrd1 or Nab3, lacking the nuclear exosome component Rrp6 or lacking Trf4, but not in strains lacking the homologous poly(A) polymerase Trf5 (Figure 4C and D). The same strains accumulated three other asRNAs tested; DBP2as, MAL32as and PCH2as (data not shown). All asRNAs detected were long (0.5–8 kb) but notably heterogeneous in size, with multiple bands being visible by northern hybridization. Heterogeneity was also observed for previously analysed asRNAs and intergenic RNAs (Arigo et al, 2006b; Thiebaut et al, 2006) and may be a common feature of yeast ncRNAs.
Figure 4.
Long non-coding asRNAs are targets for the nuclear RNA surveillance machinery. Densities of reads mapped to HPF1as RNA (A) and CAF17as RNA (B). Arrows represent major asRNA species, read-through products as detected in nrd1 and nab3 mutants (C, D) are illustrated as dashed arrows. (C, D) Northern analyses of total RNA from BY4741, GAL∷nrd1, GAL∷nab3, GAL∷nrd1 GAL∷nab3, trf4Δ, trf5Δ and rrp6Δ strains. Riboprobes were directed against indicated RNA species; sizes of detected RNAs are given. Ethidium bromide stain of 18S rRNA serves as loading control. (*) Marks an unknown contaminant RNA. See also Supplementary Figure S4.
The major northern bands for CAF1as and HPF1as observed in trf4Δ and rrp6Δ strains and weakly in the WT were shorter than in strains depleted for Nrd1 or Nab3. This would be consistent with a role for Nrd1 and Nab3 in transcription termination on these asRNAs, with the longer RNAs representing read-through products.
Analyses using genome-wide tilling arrays (Xu et al, 2009) grouped ncRNAs into CUTs (both intergenic and antisense), which accumulate in strains lacking the non-essential exosome component Rrp6 and SUTs that are unaffected by loss of Rrp6. Comparison of the CRAC data sets with the CUTs and SUTs revealed ∼266 CUTs and ∼150 SUTs that were reproducibly identified by CRAC hit clusters (Supplementary Figure S2). Notably, the averaged density of Nrd1 and Nab3 hits over all annotated CUTs was substantially higher than over annotated SUTs, ORFs or intergenic regions in each of the high-throughput data sets (Figure 5).
Figure 5.
CUTs are preferentially targeted by Nrd1 and Nab3. Density of high-throughput sequencing reads mapped to CUTs, SUTs, mRNAs and intergenic regions, according to Xu et al (2009).
Nrd1 and Nab3 participate in surveillance of Pol III transcripts
The most unexpected feature of the CRAC data was the apparent association of Nrd1 and Nab3 with RNA Pol III transcripts, which comprised 31% of Nrd1 and 17% of Nab3 hits over all low- and high-throughput data sets. Interactions of TRAMP and the exosome with RNA Pol III transcripts were previously shown for 3′ truncated 5S rRNA (5S*) and undermethylated tRNAiMet (Kadaba et al, 2004, 2006; Vanacova et al, 2005; Schneider et al, 2007). In contrast, other defective tRNAs tested were predominately 5′ degraded by Rat1 (Chernyakov et al, 2008) and the roles of Nrd1–Nab3 were suspected to be restricted to Pol II due to the interactions between Nrd1 and the CTD region (Vasiljeva et al, 2008).
Nrd1, Nab3 and Trf4 were each most frequently associated with 5S sequences that terminated between nucleotides 50 and 100. These often but not exclusively carried oligo(A) tails, indicating that they represent degradation intermediates (Supplementary Figure S5). In addition, oligoadenylated sequences were found at, and downstream of, the mature 3′ end of 5S (Supplementary Figure S5), probably representing precursors to the truncated species. 5S rRNA contains several consensus Nrd1-binding motifs and sequencing data revealed nucleotide substitution in the second, and deletions in the fourth GUAA/G motif (deleted nucleotides underlined), indicating direct Nrd1 binding at these positions. Nab3 and Trf4 also bound this region of 5S, with crosslinking to the fourth GUAG motif (Nab3; deleted nucleotide underlined) and the nucleotides downstream of the motif (Trf4; Supplementary Figure S5). In vivo analysis did not reveal clear stabilization of any distinct, truncated 5S species following depletion of Nrd1, Nab3 or both (data not shown). This may reflect the redundancy observed in many yeast RNA surveillance pathways (Houseley and Tollervey, 2009).
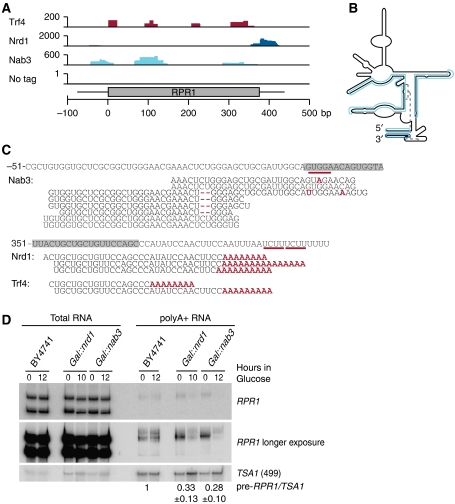
RPR1 encodes the RNA component of RNase P and is transcribed by Pol III as a precursor containing a 5′ leader and 3′ trailer, removal of which requires RNP assembly (see Srisawat et al (2002) and references therein). CRAC revealed association of Nab3 and Trf4 with the 5′ leader and Nrd1 and Trf4 with the 3′ trailer of pre-RPR1 (Figure 6A–C). Sequences recovered with Nrd1 and Trf4 did not contain the full 3′ trailer up to the transcription stop site but carried extensions with non-encoded oligo(A) tails (Figure 6C). Trf4 and Nab3 bound an overlapping set of sites within the mature RNA, which are brought into proximity in the predicted secondary structure (Figure 6B).
Figure 6.
Pre-Rrp1 is polyadenylated and targeted by the surveillance machinery. (A) Densities of high-throughput sequencing reads mapped to pre-RPR1. (B) Two-dimensional structure of RPR1 according to Srisawat et al (2002). High-throughput sequencing reads mapped to RPR1 are highlighted. Trf4 crosslinks overlapped with Nab3 hits and have been omitted for clarity. (C) Alignment of high-throughput sequencing reads of RNAs in the indicated IP to pre-RPR1. Grey boxes mark the mature RPR1 sequence and numbering indicates the nucleotide position with respect to nucleotide +1 in the RPR1 gene. Mismatches and deletions in sequencing reads are displayed in red. Numbers in brackets indicate the frequency with which each specific sequence was recovered in reads per million mapped sequences. (D) Northern analyses of total and poly(A)+ RNA from GAL∷nrd1, GAL∷nab3 and BY4741 strains. Quantification of expression levels of polyadenylated pre-RPR1 relative to TSA1 mRNA is displayed. Ratio of expression after 12 h compared with 0 h is set to 1 for WT and given as average of three biological replicates with s.d.
We predicted that defects during RNase P assembly in WT cells lead to recognition of pre-RPR1 RNA by Nrd1–Nab3, oligoadenylation by Trf4 and exosome degradation. To test this hypothesis, poly(A)+ RNA from trf4Δ and strains depleted of Nrd1 or Nab3 was analysed by northern hybridization. Pre-RPR1 was detectably polyadenylated in WT cells (Figure 6D), presumably reflecting normal surveillance activity. Polyadenylation was lost when Trf4 was absent (data not shown) and following depletion of Nrd1 or Nab3 (Figure 6D). The GAL∷nrd1 and GAL∷nab3 strains showed reduced levels of pre-RPR1 in the poly(A)+ fraction, while RPR1 and pre-RPR1 RNA levels remained constant in the total RNA of all tested strains, demonstrating that the primary defect is not in RPR1 processing. We conclude that Nrd1 and Nab3 are required to recognize defective pre-RPR1 and to recruit Trf4 to add an oligo(A) tail.
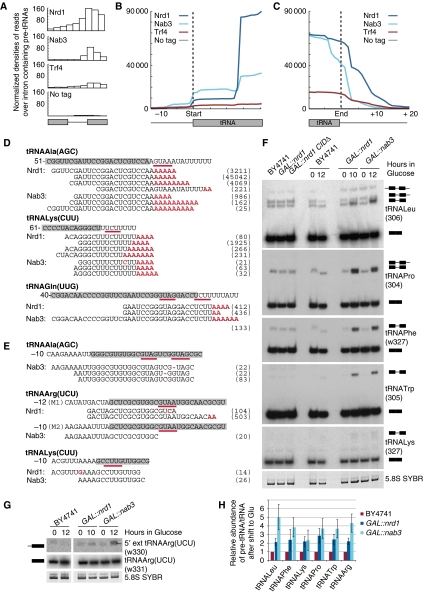
Pre-tRNAs are transcribed with a 5′ leader and 3′ trailer and in some cases also contain introns. These are removed during tRNA maturation, while the 3′ CCA tail is added and many base modifications are introduced. Nrd1, Nab3 and Trf4 were associated with many pre-tRNA fragments, which generally contained introns (Figure 7A), 5′ leaders (Figure 7B) or 3′ extensions (Figure 7C). Almost no tRNA recovered carried the 3′ CCA tail, whereas many retained the 3′ oligo(U) Pol III termination signal, followed by a non-encoded oligo(A) tail, or just an oligo(A) tail following the coding region (Figure 7D and data not shown). The recovery of many oligoadenylated RNAs that extend to the Pol III terminator indicated that tRNA-like species detected in CRAC analyses are derived from bona fide pre-tRNAs. In the case of tRNAIle(AAU), which is reported to be edited (Auxilien et al, 1996), the Nrd1 CRAC recovered only RNAs that had failed to undergo editing of the anticodon loop (data not shown). We conclude that the RNAs recovered were predominately derived from pre-tRNA species rather than mature tRNAs. Pre-tRNAs recovered with Nrd1–Nab3 generally carried consensus-binding motifs and mutations were frequently found within and around the GUAA/G and UCUU/CUUG sequences (underlined in Figure 7D and E), indicating direct protein binding.
Figure 7.
Pre-tRNAs are targets for the nuclear RNA surveillance machinery. (A) Average densities of reads mapped to intron-containing tRNAs. tRNA exons and introns have various lengths; all exons and introns were divided into three bins and density of reads in each bin is displayed. (B, C) Average densities of reads of all tRNAs associated with the indicated proteins are plotted with respect to start (B) and end of the tRNA (C). (D, E) Alignments of high-throughput sequencing reads of RNAs associated with the indicated proteins to 3′ extended pre-tRNAs (D) and 5′ extended pre-tRNAs (E). Grey boxes mark mature tRNA sequences and numbering indicates nucleotide positions with respect to nucleotide +1 of the tRNA. Mismatches and deletions in sequencing reads are displayed in red. Nrd1 and Nab3 consensus-binding motifs are underlined. Numbers in brackets indicate the frequency with which each specific sequence was recovered in reads per million mapped sequences. (F, G) Northern analyses of total RNA from BY4741, GAL∷nrd1CIDΔ, GAL∷nrd1 and GAL∷nab3 strains. Oligonucleotide probes are given with the probe number in brackets. A schematic representation of the identified species is shown. (H) Quantification of pre-tRNA relative to mature tRNA is shown; expression at 12 versus 0 h is set to 1 for the WT. Average of three biological replicates is shown with s.d. See also Supplementary Figure S6.
CRAC analyses of Nrd1 and Nab3 recovered many hits on pre-tRNAArg(ACG)J, frequently with oligo(A) tails (Supplementary Figure S6). Notably, tRNAArg(ACG) is encoded by a multigene family but tRNAArg(ACG)J has two single-nucleotide substitutions (A43G and A56G) relative to the five other genes. Folding predictions indicate that these mutations are likely to interfere with formation of the normal tRNA structure (Supplementary Figure S6). The strongly preferential recovery of tRNAArg(ACG)J relative to other isoforms indicates that the misfolded pre-tRNA was targeted by Nrd1–Nab3 binding. Primer extension analysis revealed that the level of pre-tRNAArg(ACG)J is similar to that of the other isoforms but the corresponding mature tRNA could not be detected, supporting the selective surveillance of this species prior to pre-tRNA processing (Supplementary Figure S6).
To confirm the participation of Nrd1–Nab3 in pre-tRNA surveillance, depleted strains were analysed by northern hybridization for pre-tRNAs identified in CRAC experiments (Figure 7F and G). Accumulation of unspliced forms of five pre-tRNAs tested was seen in strains depleted of Nrd1 and Nab3, but no decrease in mature tRNA levels was observed (Figure 7F). In contrast, metabolic depletion of the tRNA splicing endouclease Sen34 lead to a stronger accumulation of pre-tRNAs and loss of mature tRNA (Supplementary Figure S6). To further test the possibility that the tRNAs identified by CRAC represent spurious Pol II transcripts, we analysed a nrd1 mutant, that is unable to interact with the CTD (nrd1CIDΔ; Vasiljeva et al, 2008). In this mutant, levels of mature and pre-tRNAs were unchanged (Figure 7F), indicating that the observed effects of Nrd1 and Nab3 on tRNAs are independent of their association with Pol II. The CRAC analyses also identified 5′ extended pre-tRNAs (Figure 7B and E) and northern hybridization revealed that depletion of Nab3 and, to a lesser extent Nrd1, increased the level of 5′ extended tRNAArg(UCU) (Figure 7G and H). These phenotypes were not accompanied by loss of the mature tRNA, indicating that the pre-tRNA accumulation does not reflect a processing defect.
We conclude that pre-tRNAs with defects in folding or maturation are bound by Nrd1–Nab3 and targeted to the TRAMP–exosome degradation pathway.
Discussion
Our results provide a genome-wide view of the RNA population that is targeted by the nuclear RNA surveillance system in WT cells. Known substrates of the Nrd1–Nab3 and TRAMP complexes were recovered including cryptic ncRNA transcripts as well as defective RNAs generated by Pol I and Pol II. The identification of the ncRNAs confirms that these are actively transcribed in WT cells, and not solely produced in response to deficient surveillance activities. These ncRNAs included the GAL10as RNA, which is present at around one molecule per 13 cells, supporting the sensitivity and reliability of the technique. The unexpected recovery of Pol III targets demonstrates that the roles of Nrd1 and Nab3 in surveillance are not obligatorily dependent on association with the CTD of RNA Pol II.
Nab3 showed a clear preference for targets that contained the consensus-binding site previously identified (UCUU), or closely related sequences (CUUG). Mapping of crosslinking-induced micro-deletions and substitutions onto genomic sequences showed that their analysis can greatly enhance the identification of direct, in vivo RNA-binding motifs. In the case of Nrd1, enrichment of the previously identified binding site (GUAA/G) in the recovered target sequences was modest, but analysis of deletions clearly pin-pointed this motif as a preferred in vivo binding site.
Many RNAs recovered carried non-templated oligo(A) or A-rich tails that are characteristic of RNAs recognized and targeted for degradation by TRAMP. A long-standing question was how adenylation can stabilize mRNAs and promote translation while inducing degradation of surveillance targets? This was resolved by the observation that surveillance targets generally carried A3–A5. Poly(A) tails on mRNAs associate with the poly(A)-binding protein (Pab1 in yeast), which both stabilizes the mRNA and stimulates translation. However, Pab1 requires a minimum binding site of ∼A12 (Sachs et al, 1987) and is therefore not expected to bind most surveillance targets detected here. We cannot formally exclude the possibility that surveillance substrates initially have longer tails that are truncated to the observed lengths. However, Jankowski and colleagues (personal communication) have observed that TRAMP preferentially adds A4 tails when assayed in vitro, indicating that this is an intrinsic property of the surveillance system. The frequency of non-A residues in the non-encoded tails (∼2.5–5%) is not consistent with their addition by the canonical poly(A) polymerase, but is broadly similar to other systems from Escherichia coli to human cells (Deutscher, 2006; Slomovic et al, 2006; West et al, 2006). The preference for inclusion G>U>C is also consistent with the in vitro activity of Trf4 (LaCava et al, 2005).
Recent analyses have revealed pervasive transcription. A key question is, how the large numbers of ncRNAs can be systematically distinguished from the complex mRNA population? The finding that the averaged density of Nrd1 and Nab3 hits over all annotated CUTs (Xu et al, 2009) was substantially higher than over annotated SUTs, ORFs or intergenic regions in each of the high-throughput data sets, strongly supports the idea that Nrd1–Nab3 binding constitutes one general feature that targets ncRNAs to the exosome.
The tilling array and CRAC data sets each contained transcripts that were not identified with the other approach (Supplementary Figure S2). Predicted CUTs not recovered in the CRAC analyses may be targeted for degradation by other nuclear surveillance factors, several of which are known (reviewed in Houseley and Tollervey, 2009). Conversely, Figure 1G shows that most of the oligo(A) tails present on RNA surveillance substrates are too short to be recovered by oligo(dT) selection or to act as primers for oligo(dT) primed cDNA synthesis. Since these are important steps in previous microarray analyses, such RNAs will be poorly identified.
Transcription termination mediated by Nrd1–Nab3 on CUTs and small stable RNAs was thought to be limited to short transcripts due to the association of the complex with Ser5/Ser7 phosphorylated CTD (Gudipati et al, 2008; Vasiljeva et al, 2008; Kim et al, 2010). However, depletion of either Nrd1 or Nab3 caused a termination defect on the 5-kb HPF1as-RNA, suggesting that Nrd1–Nab3 termination activity is not fully dependent on Ser5 phosphorylation of the CTD.
CTD-independent activity of Nrd1–Nab3 in surveillance was further shown by the recovery of oligoadenylated pre-tRNAs and other Pol III transcripts. One concern was that the tRNA-like RNAs might not arise from Pol III transcription, but from spurious Pol II transcription through the region. However, a nrd1 mutant, defective in CTD association (nrd1CIDΔ) did not exhibit an accumulation of pre-tRNAs, supporting the conclusion that the effect on Pol III transcripts is Pol II independent. In addition, many oligoadenylated clones stopped at the Pol III terminator. Moreover, tRNAArg(ACG) is encoded by six genes; five of these have identical tRNA sequences but one carries two single-nucleotide substitutions. The CRAC analyses of Nrd1 and Nab3 preferentially recovered the altered pre-tRNA, frequently with oligo(A) tails (Supplementary Figure S6). RNA-folding algorithms predict that this RNA is unlikely to fold into the correct tRNAArg structure, providing a clear rationale for its targeting by the surveillance system. These differences in folding would not, however, have been predicted to alter the fate of a spurious RNA Pol II transcript.
Depletion of Nrd1 or Nab3 resulted in accumulation of unspliced pre-tRNAs, but was not associated with loss of mature tRNAs, indicating that it reflects pre-tRNA stabilization rather than the inhibition of pre-tRNA splicing. Mutations in NRD1 and NAB3 potentially cause transcription read-through into SEN2, which encodes a pre-tRNA splicing factor (Steinmetz et al, 2001). However, depletion of the tRNA splicing endonuclease Sen34 leads to a different phenotype, with strong pre-tRNA accumulation and loss of mature tRNAs (Supplementary Figure S6), making it unlikely that Sen2 depletion underlies the nrd1/nab3 phenotypes. The precursor to the RNA component of RNase P, pre-RPR1, was also bound by Nrd1, Nab3 and Trf4. Analysis of poly(A)+ RNA revealed that pre-RPR1 is polyadenylated by Trf4 and that this polyadenylation is strongly reduced in the absence of Nab3 or Nrd1. This supports the model that Nrd1–Nab3 acts upstream of TRAMP in recognizing defective Pol III transcripts and targeting them for degradation by the exosome. RNA Pol III transcribes very structured RNAs and we predict that during normal transcription and folding, binding sites for Nrd1–Nab3 are not exposed. Misfolding of the RNA, for whatever reason, would make these sites available leading to targeting for degradation. The recognition of tRNAArg carrying point mutations predicted to alter its structure supports this model for structure-dependent targeting.
Together, our findings suggest a revised model of nuclear RNA surveillance (Figure 8). Nrd1–Nab3 can bind the Pol II CTD and nascent transcripts cotranscriptionally but also act post-transcriptionally on Pol III RNAs. The TRAMP complex is recruited to the defective RNA by the Nrd1–Nab3 complex, which remains associated with the RNA through the process of polyadenylation, until the exosome degrades the aberrant transcript. Hit clusters and oligoadenylated fragments were recovered at multiple sites on many transcripts, suggesting that repeated rounds of surveillance factor binding and oligo(A) addition may be needed for complete substrate degradation. Budding yeast lacks the miRNA systems present in most other eukaryotes analysed. The miRNAs are believed to modestly reduce the expression of large numbers of genes, with some stronger, more specific effects. A surprisingly large number of mRNAs were targeted by the surveillance factors. We speculate that nuclear surveillance similarly acts to modulate the expression of many genes, in addition to specifically targeting defective RNAs.
Figure 8.
Model for transcription termination and surveillance mediated by the Nrd1–Nab3, TRAMP and exosome complexes. (A) Nrd1–Nab3 interact with the CBC and CTD of Pol II to initiate transcription termination through recognition of consensus-binding motifs of sn(o)RNAs, CUTs and long ncRNAs. They subsequently recruit TRAMP and exosome complexes for oligoadenylation and degradation/processing. (B) Nrd1–Nab3 interact post-transcriptionally with aberrant Pol III transcripts, recognizing consensus-binding motifs or structural abnormalities in the RNA. They subsequently recruit TRAMP and exosome complexes for oligoadenylation and degradation.
Materials and methods
Yeast strains and depletion experiments
Strains were constructed by standard methods (Gietz et al, 1992) and are listed in Supplementary Table S1. For crosslinking experiments, colonies from HTP-tagged strains were pre-grown overnight in YPD (2% glucose), diluted to OD600 0.05 and grown to OD600 0.5 at 30°C. Plasmids used are listed in Supplementary Table S2. For RNA analysis, cells were grown at 25°C to OD600 0.2–0.6 in YPD. GAL strains for depletion experiments were grown overnight to OD600 0.2–0.5 in YPGalSuc (2% galactose, 1% sucrose), diluted to OD600 0.2 in YPD and proteins were depleted for the indicated times.
Crosslinking and analysis of Solexa data
The CRAC method was performed as previously described (Granneman et al, 2009). Solexa sequencing data were aligned to the yeast genome using NOVOALIGN (http://www.novocraft.com). A detailed description of the bioinformatics analysis can be found in the Supplementary data.
RNA preparation and northern hybridization
Yeast RNA extraction and northern blotting were performed as described in Tollervey (1987). Details for generation and hybridization of riboprobes can be found in the Supplementary data. Poly(A)+ RNA was prepared using PolyA tract mRNA isolation System IV (Promega) as amended by LaCava et al (2005). Northern blots contained 10 μg total RNA (on 2% BPTE agarose gels and 8% PAA, TBE, 8.3 M urea gels), or 2 μg total RNA and 60 μg polyA+ RNA with respect to the input for polyA+ analyses. Hybridization probes are listed in Supplementary Tables S3 and S4.
Supplementary Material
Acknowledgments
We thank Phil Mitchell for providing plasmids, Claudia Schneider and Jonathan Houseley for helpful discussions and Alastair Kerr for bioinformatic support. This work was supported by the Wellcome Trust (DT), a Darwin Trust Studentship (WW), European Commission 7th Framework Programme (UNICELLSYS, Grant No. 201142) (WW), long-term EMBO Fellowships (GK and SG) and a Marie Curie EIF Fellowship (SG).
Footnotes
The authors declare that they have no conflict of interest.
References
- Allmang C, Mitchell P, Petfalski E, Tollervey D (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 28: 1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL (2006a) Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651 [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL (2006b) Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851 [DOI] [PubMed] [Google Scholar]
- Auxilien S, Crain PF, Trewyn RW, Grosjean H (1996) Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J Mol Biol 262: 437–458 [DOI] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F (2007) Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, Corden JL (2007) Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL (2004) Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol 24: 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM (2008) Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev 22: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS (2011) Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciais D, Bohnsack MT, Tollervey D (2008) The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res 36: 3075–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM (2006) A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares MJ (2006) Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP (2006) Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res 34: 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D (2006) Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S (2007) Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D (2010) Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D (2009) Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high throughput analysis of cDNAs. Proc Natl Acad Sci USA 106: 9613–9818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P, Kufel J (2008) Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell 32: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D (2008) Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol 15: 786–794 [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA (2011) Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev 25: 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D (2006) Microarray detection of novel nuclear RNA substrates for the exosome. Yeast 23: 439–454 [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D (2007) Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M (2008) A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D (2009) The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Huang Y, Bayfield MA, Intine RV, Maraia RJ (2006) Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol 13: 611–618 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT (2006) Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL (2010) Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR (2005) Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 21: 713–724 [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martens JA, Wu PY, Winston F (2005) Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev 19: 2695–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A (2009) H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J 28: 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ (2009) Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell 36: 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW, Kornberg RD (1987) A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol 7: 3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff BU, Heath-Pagliuso S, Castano IB, Zhu Y, Kieff FS, Christman MF (1995) Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 141: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D (2007) The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell 27: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D (2000) Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G (2006) Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res 34: 2966–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Houser-Scott F, Bertrand E, Xiao S, Singer RH, Engelke DR (2002) An active precursor in assembly of yeast nuclear ribonuclease P. RNA 8: 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA (2006) Genome-wide distribution of yeast RNA polymerase II and Its control by Sen1 Helicase. Mol Cell 24: 735–746 [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D (2006) Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the Nrd1-Nab3 pathway in genome surveillance. Mol Cell 23: 853–864 [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R (2007) Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol 27: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D (1987) High level of complexity of small nuclear RNAs from fungi and plants. J Mol Biol 196: 355–361 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB (2005) CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37: 376–386 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB (2003) CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S (2006) Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A (2008) The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jia H, Jankowsky E, Anderson JT (2008) Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA 14: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery M, Ruidant S, Schillewaert S, Lepore N, Lafontaine DL (2009) The nuclear poly(A) polymerase and exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 15: 406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Norbury CJ, Proudfoot NJ (2006) Adenylation and exosome-mediated degradation of co-transcriptionally cleaved pre-messenger RNA in human cells. Mol Cell 17: 437–443 [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, Libri D, Jacquier A (2005) Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.