Abstract
The evolutionarily conserved ATP-dependent nucleosome remodelling factor ISWI can space nucleosomes affecting a variety of nuclear processes. In Drosophila, loss of ISWI leads to global transcriptional defects and to dramatic alterations in higher-order chromatin structure, especially on the male X chromosome. In order to understand if chromatin condensation and gene expression defects, observed in ISWI mutants, are directly correlated with ISWI nucleosome spacing activity, we conducted a genome-wide survey of ISWI binding and nucleosome positioning in wild-type and ISWI mutant chromatin. Our analysis revealed that ISWI binds both genic and intergenic regions. Remarkably, we found that ISWI binds genes near their promoters causing specific alterations in nucleosome positioning at the level of the Transcription Start Site, providing an important insights in understanding ISWI role in higher eukaryote transcriptional regulation. Interestingly, differences in nucleosome spacing, between wild-type and ISWI mutant chromatin, tend to accumulate on the X chromosome for all ISWI-bound genes analysed. Our study shows how in higher eukaryotes the activity of the evolutionarily conserved nucleosome remodelling factor ISWI regulates gene expression and chromosome organization genome-wide.
Keywords: chromatin remodelling, D. melanogaster , ISWI, nucleosome spacing
Introduction
The eukaryotic cell has evolved regulatory mechanisms to change chromatin structure and to adapt to specific transcriptional programmes in response to a variety of environmental and cellular stimuli. Chromatin covalent modifiers catalyse specific post-translational modifications of the histones amino- and carboxy-terminal tails (Kouzarides, 2007), while chromatin remodelling complexes use the energy of ATP hydrolysis to change nucleosome positions or to incorporate histone variants into chromatin (Eberharter and Becker, 2004; Saha et al, 2006). These modifications set different chromatin functional states that contribute to local chromatin structure and gene expression profiles (Imhof, 2006; Martin and Zhang, 2007).
ISWI is the catalytic subunit of several ATP-dependent chromatin remodelling complexes, conserved in composition and function across species (Corona et al, 2004; Dirscherl and Krebs, 2004). In higher eukaryotes, ISWI is an abundant and ubiquitously expressed protein that is essential for cell viability (Deuring et al, 2000; Stopka and Skoultchi, 2003; Arancio et al, 2010). In vitro, ISWI uses the energy of ATP hydrolysis to catalyse nucleosome spacing and sliding reactions (Corona et al, 1999). Loss of ISWI function in Drosophila results in dramatic chromosome condensation defects and in the reduction of chromatin-bound histone H1 levels, suggesting that ISWI has a general role in chromosome condensation in vivo by promoting the loading of the linker histone H1 on chromatin (Deuring et al, 2000; Lusser et al, 2005; Corona et al, 2007; Siriaco et al, 2009).
Genetic and biochemical studies have also supported a role for ISWI in promoting transcription (Corona and Tamkun, 2004; Dirscherl and Krebs, 2004). However, the preferential association of ISWI with transcriptionally silent chromatin and microarray gene expression studies have suggested that ISWI also has an important role in transcriptional repression (Deuring et al, 2000; Corona et al, 2007). Indeed, works conducted in several model organisms have shown that ISWI family complexes appear to both activate and repress transcription (Corona and Tamkun, 2004). However, to date, how ISWI chromatin binding and its associated nucleosome remodelling activity can influence chromatin condensation and gene expression in higher eukaryotes is poorly understood.
In order to determine if changes in nucleosome spacing could account for chromosome condensation and transcription defects observed in ISWI mutants, we conducted a genome-wide identification of ISWI chromatin-binding sites and nucleosome spacing activity in the higher eukaryote Drosophila melanogaster. Our analysis revealed that ISWI has ∼1200 high-affinity chromatin-binding sites, in both genic and intergenic regions. In particular, we identified a number of ISWI-binding elements (IBE) corresponding to specific ISWI–DNA-binding consensus motifs. Remarkably, we found that ISWI tends to bind genes near their transcription start site (TSS) causing alterations in nucleosome positioning after the TSS, providing an important insight in understanding ISWI role in higher eukaryote transcriptional regulation. Surprisingly, global changes in nucleosome spacing caused by loss of ISWI activity does not seem to be directly correlated with the X chromosome condensation defects observed in ISWI mutants (Deuring et al, 2000). However, when looking at differences in nucleosome spacing only on all ISWI-bound genes, they tend to accumulate on the X chromosome. Indeed, we found a significant enrichment of ISWI binding on genes mapping the X chromosome. In particular, we found that ISWI is active on dosage compensated genes in order to keep their TSS nucleosome free. Our study reveals how higher eukaryote transcription and chromosome organization is regulated genome-wide by the activity of the chromatin remodelling factor ISWI.
Results
ISWI binds chromatin near the TSS
Virtually, all we know about ISWI function in vivo derives from studies conducted in Drosophila larval tissues (Corona and Tamkun, 2004). We reasoned that the genome-wide identification of ISWI chromatin-binding sites in larvae could provide some important insights in order to understand the transcription and chromosome condensation defects observed in ISWI mutants. Therefore, we sought to identify ISWI chromatin target sites across the D. melanogaster genome by chromatin immunoprecipitation (ChIP) in larvae. Larval chromatin was immunoprecipitated (ChIPISWI) using the highly specific affinity purified anti-ISWI antibody and hybridized against the input chromatin on a set of tiled arrays covering the entire Drosophila genome with an average probe spacing of 100 bp.
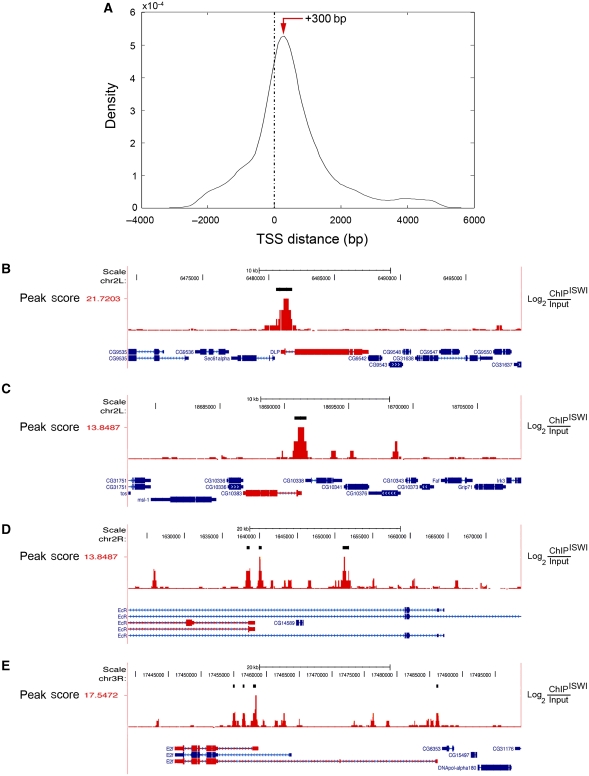
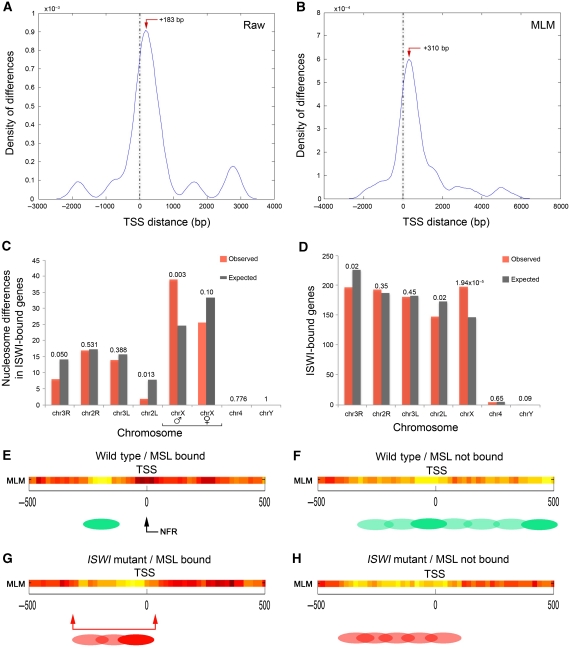
We quantified ISWI enrichment over the fly genome using a ‘peak score’ function that takes into account the length and intensity of the normalized raw log2(ChIPISWI/input) signals. Our analysis identified 1176 distinct ISWI-enriched chromatin regions corresponding to discrete ISWI peaks, mapping 925 unique gene loci and 141 intergenic regions with an average peak length of ∼500 bp (Supplementary Table S1A; Supplementary Figure S1). Since, ISWI has an important role in both transcriptional activation and repression (Corona and Tamkun, 2004; Dirscherl and Krebs, 2004), we mapped ISWI binding relative to the TSS of genes present in the proximity of ISWI peaks. Remarkably, we found that ISWI preferentially binds genes near their regulatory regions with an average peak at about ∼300 bp after the TSS (Figure 1A). Although the majority of ISWI peaks maps in the proximity of the TSS (for specific examples, see Figure 1B–E), we also found the presence of many ISWI peaks at the level of exons, introns and at the 3′-end of several genes (Supplementary Figure S2).
Figure 1.
ISWI chromatin binding is enriched after the transcription start site (TSS). (A) The density of ISWI-enriched genomic sequences is plot relative to the distance from the TSS of nearby genes. ISWI on average tends to bind with a peak (red arrow) at about ∼300 bp after the TSS. (B) Representative example of a gene bound by ISWI at the level of the promoter–exon region. As expected, ISWI also binds genes whose expression is (C) increased or (D) decreased in ISWI mutant larval tissues relative to wild-type levels (for a complete list, see Supplementary Table S1B and C) (Corona et al, 2007). (E) ISWI binds near the TSS of genes involved in nurse cell apoptosis. ISWI enrichment is proportional to the normalized raw log2(ChIPISWI/input) red signals reported to the right y axis. The thick black bars highlight ISWI peaks. The peak score values of ISWI peaks are reported to the left y axis. Exons are represented by filled-in boxes, introns by a continuous line, while the 5′- and 3′-UTRs by thick lines. The transcription direction for each gene can be deduced by the repeated small blue arrows present in the intron segments. Genes bound by ISWI are shown in red.
ChIP-on-chip data on total larval chromatin were validated on salivary gland chromatin immunoprecipitated with the affinity purified anti-ISWI antibody (Deuring et al, 2000), and analysed by semi-quantitative RT–PCR using specific primers for the representative ISWI-enriched regions identified in total larval chromatin (compare Figure 1B–E with Supplementary Figure S3). Next, we checked whether ISWI-binding sites with higher peak scores values correlated with a specific TSS distance or gene functional element (i.e. promoter, promoter–exon boundary, exon, exon–intron boundary, intron and intron–exon boundary). Although we could not find a direct correlation between peak score values and TSS distance for each specific gene functional element analysed (Supplementary Figure S4A–G), we found that ISWI binding at the promoter–exon boundary regions tend to correlate with higher peak scores (Supplementary Figure S4H). Collectively, our data suggest that in higher eukaryotes, ISWI could regulate gene expression by remodelling chromatin near gene regulatory regions in the proximity of the TSS.
Indeed, when we intersected genes whose expression changes at least × 1.5-fold in ISWI mutant larvae, obtained from previous array expression data (Corona et al, 2007), with ISWI-bound genes, we found ∼50% of overlap (Supplementary Table S1B; for specific examples, see Figure 1C and D). However, this level of overlap dropped to ∼10% when we looked at genes that change their expression in ISWI mutant larvae at least × 4-fold (Supplementary Table S1C; Figure 1C and D) (Corona et al, 2007). While ChIP data are the result of bona fide chromatin protein binding, array expression data underlie direct and indirect transcriptional effects caused by the loss of the chromatin protein studied. Therefore, the two biological processes (binding versus activity) do not have to necessarily overlap. Indeed, data coming from ISW2, the yeast homologue of ISWI, have shown that ISW2 chromatin recruitment is poorly correlated with genes whose expression is dependent on ISW2 activity (Gelbart et al, 2005). In yeast, ISW2 chromatin binding was shown to be transient and, as a result in certain genes, ISW2 binding could not be detected above background in ChIP experiments (Gelbart et al, 2005). To circumvent these problems, a catalytically inactive ISW2 was used to block chromatin remodelling, leading to persistence of a normally transient ISW2 recruitment (Whitehouse et al, 2007).
A K159R substitution in the putative ATP-binding pocket of Drosophila ISWI reduces ATPase activity to background levels without affecting ISWI complex formation (Deuring et al, 2000). However, when we compared fly stocks expressing wild-type ISWI or the catalytically inactive ISWIK159R, we did not appreciate significant differences in the distribution and intensity of the wild type and the catalytically inactive ISWI on interphase polytene chromosomes (Supplementary Figure S5). Therefore, we reasoned that the partial overlap existing between ISWI-bound genes and genes whose expression change greatly in ISWI mutant larvae are likely due to changes in gene expression that are indirectly caused by ISWI loss (Corona et al, 2007). In alternative, we cannot exclude that for some genes ISWI could locally regulate their expression not based on an in cis association, but in trans through tridimensional chromatin interactions, thus explaining the low correlation we observe between ISWI binding and its transcriptional activity.
ISWI binds several DNA consensus motifs on chromatin
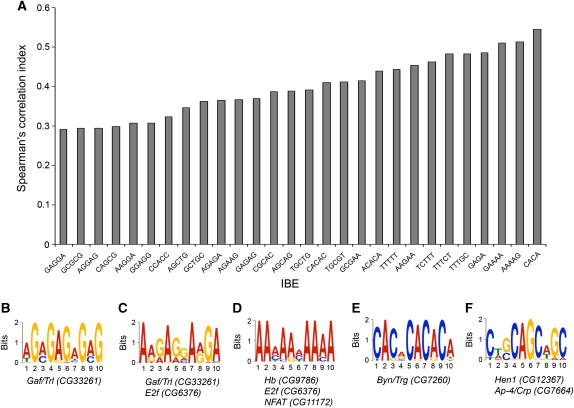
Our data show that ISWI binding near gene regulatory regions tend to correlate with higher peak score values. Given the high ‘peak score’ values obtained for ISWI-binding sites near the TSS, we also reasoned that ISWI could preferentially bind specific DNA consensus motifs at the level of gene regulatory regions. Therefore, we used motif discovery scan (MDscan) (Liu et al, 2002), a robust motif discovery tool, to search for potential consensus ISWI–DNA interaction modules. Our analysis resulted in the identification of a variety of IBE, corresponding to DNA sequence motifs associated directly or indirectly with ISWI in the context of chromatin. In particular, we found that ISWI binds several 4–5 letter IBE that are positively correlated with the ‘peak score’ function, strongly indicating that the identified motifs are likely associated with ISWI binding (Figure 2A).
Figure 2.
Identification of ISWI-binding elements (IBE). (A) DNA consensus motif corresponding to IBE identified with MDscan where correlated with the ‘peak score’ function using the Spearman's correlation index. The Spearman's rank assesses how well the relationship between two variables can be described using a monotonic function. The positive Spearman's correlation index values we obtained indicate that the number of occurrences for every analysed IBE within each ISWI-bound DNA sequence increases as a function of the ‘peak score’, strongly indicating that the identified motifs are likely associated with ISWI binding. (B–F) The top five motifs identified with multiple EM for motif elicitation (MEME) along with their putative D. melanogaster DNA-binding factors are shown. These logos are a graphical representation of DNA multiple sequence alignment. Each logo consists of a series of stacks of the four DNA deoxynucleotide (A, T, G, C), one stack for each position in the sequence. The overall height of the stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each nucleic acid at that position.
Using multiple EM for motif elicitation (MEME), another motif searching algorithm (Bailey et al, 2006), we identified similar IBE that contained a subset of the motifs found with MDscan (Figure 2B–F). Interestingly, one of the top motifs identified with both algorithm corresponds to the GAGA factor (encoded by the Gaf/Trl gene) consensus sequence GAGAGA (Figure 2B). Noteworthy, the ISWI-containing multisubunit complex NURF was isolated based on its ability to recruit the GAGA factor to its target sites on chromatin in vitro (Tsukiyama and Wu, 1995). However, double label immunofluorescence microscopy has also shown that both GAGA factor and ISWI are associated with hundreds of sites in the euchromatin of polytene chromosomes, but the distribution of the two proteins only partially overlaps (nearly 30% of overlap) (Deuring et al, 2000). In line with the immunostaining data, when we intersected the ChIP-on-chip data available for the GAGA factor (Lee et al, 2008) with ISWI-binding sites, we found that nearly 1/3 of the ISWI-bound genes we identified were also bound by the GAGA factor (Supplementary Figure S6A) with roughly an equal percentage of induced versus repressed ISWI-regulated genes (Supplementary Figure S6B). However, it is important to point out that this analysis does not address whether the binding of GAGA factor and ISWI occurs simultaneously or are mutually exclusive. In conclusion, our motif discovery analysis on ISWI-bound DNA sequences did not find a single consensus sequence, but instead it allowed us to identify a collection of IBEs that probably reflects the variety of factors that may recruits ISWI (Figure 2B–F), that are associated with it or that can be targeted to specific chromatin loci by the remodelling activity of ISWI.
ISWI remodels nucleosomes after the TSS
Our ChIP-on-chip data revealed that ISWI preferentially binds near the TSS; however, this analysis did not tell us whether ISWI-enriched chromatin regions corresponded to active sites of chromatin remodelling. To answer this question and to determine if the transcription and chromosome condensation defects observed in ISWI mutant larvae could be directly correlated with ISWI chromatin remodelling alterations, we conducted a genome-wide identification of nucleosome spacing changes between wild-type and ISWI mutant salivary gland chromatin. Drosophila salivary glands are actively transcribing interphase cells. These cells undergo repeated rounds of DNA replication without cell division forming giant polytene chromosomes containing ∼1000 copy of chromatinized genomic DNA (GenDNA) per cell (Johansen et al, 1999; Stephens et al, 2004). This chromatin organization reflects the interphase chromosome physiology occurring in other diploid tissues (Johansen et al, 1999; Stephens et al, 2004). Therefore, we reasoned that salivary gland cells could be a good source of Drosophila chromatin coming from an homogenous population of cells in interphase. Moreover, having a very high chromatin/cell ratio, salivary gland cells are an abundant source of nucleosomal DNA (NucDNA) suitable for classic microarray-based identification of nucleosome positions (Yuan et al, 2005).
Thus, we isolated NucDNA from wild-type and ISWI mutant male and female salivary gland cells, and the purified NucDNA was fluorescently labelled and competitively hybridized against total GenDNA on a custom-tiled array with a resolution of 20 bp (Supplementary Figure S7). Due to the restraint in the number of sequences that we could print on a single chip, the array was designed to have some of the top ISWI-enriched sequences we found by ISWI-ChIP. We also printed an equal number of DNA sequences corresponding to genes whose expression is altered in ISWI mutants (Corona et al, 2007). Finally, we printed contiguous DNA sequences, equally distributed on the different Drosophila chromosomes and covering each nearly 1 Mbp, encoding for genes not linked with ISWI functions (ISWI-binding or ISWI-dependent expression) and representing >90% of genes present in the tiled array (Supplementary Figure S8; Supplementary Table S2). The rational of such design was to have an array not biased on genomic sequences exclusively linked to ISWI function.
Based on this approach, the normalized raw log2(NucDNA/GenDNA) ratio signal for each spot along the overlapping tiled DNA sequences highlights nucleosomes as peaks surrounded by lower ratio values corresponding to linker regions (Supplementary Figure S7B) (Yuan et al, 2005). However, the complexity of higher eukaryote chromatin usually generates more complex and structured noise that usually limits the identification of NucDNA (Kharchenko et al, 2008). To overcome this limitations, we have recently developed a new algorithm called multi-layer model (MLM) that belongs to a class of methods successfully used in the analysis of very noisy data because of their ability to recover statistical properties of a signal, starting from several views of the input data set (Di Gesu et al, 2009). Based on simulated data, one of the predicted features of the MLM is its ability to identify nucleosome positions on chromatin regions with highly mobile or dense nucleosomes, which is typical of higher eukaryote chromatin (Di Gesu et al, 2009).
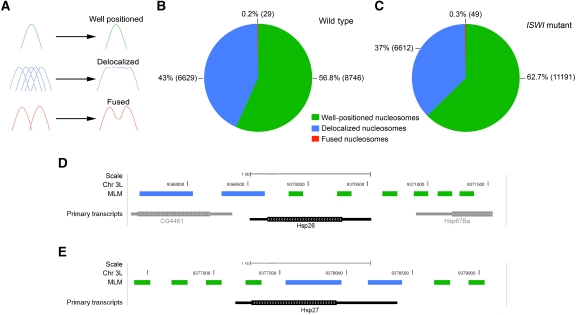
One of the properties linked to the pattern/shape recognition feature integrated into the MLM approach is the ability to recognize nucleosomes with different mobility ‘features’. In particular, the MLM can identify ‘well-positioned’ nucleosomes corresponding to nucleosome characterized by well-defined peaks (bell-shaped curve) covering about 150 bp, ‘delocalized’ nucleosomes representing single nucleosomes or arrays of nucleosomes with high mobility, and finally ‘fused’ nucleosomes defining a single nucleosome that occupies two distinct close positions (Figure 3A) (Di Gesu et al, 2009). Distinct nucleosome mobility ‘features’ can underlie important regulatory functions, highlighting the impact that the MLM can have on genome-wide nucleosome identification studies in higher eukaryotes. Indeed, when we applied the MLM approach on wild-type chromatin, we successfully identified the typical nucleosome architecture of the well-studied hsp26 and hsp27 gene promoters (Thomas and Elgin, 1988; Quivy and Becker, 1996), highlighting the reliability and power of the MLM approach in identifying nucleosome positions in higher eukaryotes (Figure 3D and E).
Figure 3.
Multi-layer model (MLM) nucleosome classification. (A) ‘Well-positioned’ nucleosomes (in green) are shown as peaks of a bell-shaped curve, ‘delocalized’ nucleosomes (in blue) represent single nucleosomes or arrays of nucleosomes with high mobility, while ‘fused’ nucleosomes (in red) reflect a single nucleosome that occupies two distinct close positions. On the left of the arrows are shown examples of nucleosome configuration that may contribute to the log2(Nuc-DNA/GenDNA) ratio signal shape. The MLM detected all three classes of nucleosomes with similar relative abundances between wild type (B) and ISWI mutant (C) chromatin. The MLM identifies with high reliability well-positioned and delocalized nucleosomes around the well-characterized nucleosome promoter architecture of the (D) hsp26 and (E) hsp27 genes (Thomas and Elgin, 1988; Quivy and Becker, 1996). Delocalized nucleosomes are depicted as blue lines, while well-positioned nucleosomes as green lines.
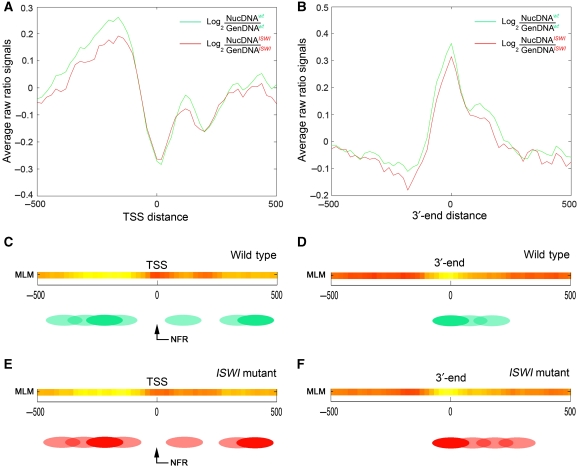
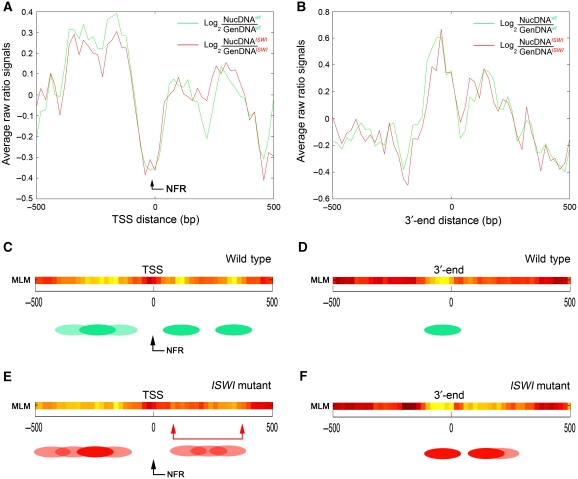
When we applied the MLM to our tiled array data, we did not find significant differences in the global relative percentage of nucleosome mobility ‘features’ existing between wild-type and ISWI mutant salivary gland chromatin (Figure 3B and C). Thus, we next looked at potential differences in nucleosome positioning near the TSS of all the genes present in the tiled array. As previously reported in other eukaryotes including flies, we found a well-defined nucleosome-free region (NFR) mapping the TSS on all genes analysed (Figure 4A and C). Interestingly, we also found one delocalized nucleosome preceding and two delocalized nucleosomes following the TSS in wild-type chromatin (Figure 4A and C). When we looked at the 3′-end of the genes present in the tiled array, we also found one delocalized nucleosome mapping the end of the transcripts (Figure 4B and D). To our surprise, when we extended our analysis to ISWI mutant chromatin, we did not find any significant difference in nucleosome positioning when compared with wild-type chromatin (Figure 4A, B, E and F).
Figure 4.
Differences in nucleosome positioning around the TSS and 3′-end between wild-type and ISWI male chromatin. The average log2(NucDNA/GenDNA) raw ratio of all the signals coming from wild-type (green) and ISWI mutant (red) male salivary gland chromatin of all genes present in the nucleosome-tiled array is plot relative to the (A) TSS distance or the (B) 3′-end distance. Nucleosome positions were calculated with the MLM for (C) wild-type and (E) ISWI mutant chromatin near the TSS, and for (D) wild-type and (F) ISWI mutant chromatin around the 3′-end. The MLM analysis shows well-positioned nucleosomes as shadings of yellow, linker regions in red, while delocalized nucleosomes in orange. The green and red ovals represent an interpretation of the MLM nucleosome positioning analysis in wild-type and ISWI mutant chromatin, respectively. Filled ovals indicate well-positioned nucleosomes, while shaded ovals show delocalized nucleosomes.
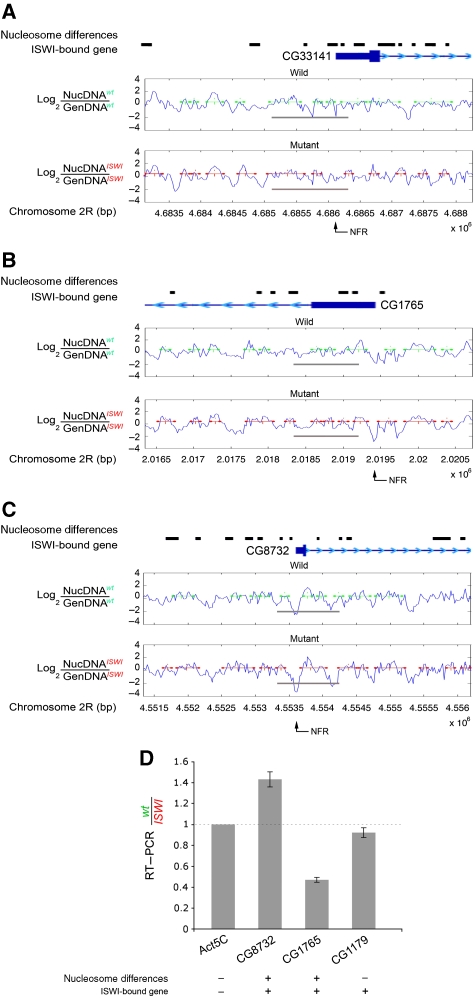
However, when we focused our attention exclusively on ISWI-bound genes present in the tiled array, besides the NFR mapping at the TSS, and the delocalized nucleosome preceding the TSS, we found two well-positioned nucleosomes following the TSS in wild-type chromatin (Figure 5A and C). Remarkably, these two well-positioned nucleosomes present in ISWI-bound genes after the TSS become delocalized in ISWI mutant chromatin (Figure 5A and E). Similarly, the nucleosome mapping at the 3′-end also showed a tendency to nucleosome delocalization in ISWI mutant when compared with wild-type chromatin (Figure 5B, D and F). The presence of delocalized nucleosomes at the level of the TSS and 3′-end of ISWI-bound genes was also validated by classic clustered representation of raw nucleosomal data signals (Supplementary Figure S9).
Figure 5.
Loss of ISWI causes nucleosome spacing defects after the TSS on ISWI-bound genes. The average log2(NucDNA/GenDNA) raw ratio of the signals coming from wild-type (green) and ISWI mutant (red) male salivary gland chromatin of all ISWI-bound genes present in the nucleosome positioning array is plot relative to the (A) TSS distance or the (B) 3′-end distance. The nucleosome-free region (NFR) mapping the TSS is indicated by the black arrow. Nucleosome positions were calculated with the MLM for (C) wild-type and (E) ISWI mutant chromatin relative to the TSS, and for (D) wild-type and (F) ISWI mutant chromatin around the 3′-end. The MLM analysis shows well-positioned nucleosomes as shadings of yellow, linker regions in red, while delocalized nucleosomes in orange. The green and red ovals represent an interpretation of the MLM nucleosome positioning analysis in wild-type and ISWI mutant chromatin, respectively. Filled ovals indicate well-positioned nucleosomes, while shaded ovals show delocalized nucleosomes. The double-arrowed red bar indicates the region after the TSS where we observe higher nucleosome mobility in the absence of ISWI.
Interestingly, similar defects in nucleosome positioning after the TSS were also found in genes whose expression is repressed (Supplementary Figure S10), and to a lesser extent in the ones that are de-repressed in the ISWI mutant (Supplementary Figure S11) (Corona et al, 2007). These effects are specific for ISWI because when we mapped nucleosomes at the level of the TSS on genes that change their expression in ISWI mutants, but with low frequency of ISWI association (from now on referred as not bound), we did not find significant differences in nucleosome positioning between wild-type and ISWI mutant chromatin (Supplementary Figure S12).
The presence of an average delocalized nucleosome after the TSS in the ISWI mutant chromatin (Figure 5E), may reflect the presence of an heterogeneous population of well-positioned nucleosomes with different phasing (see green and red nucleosome tracks in Figure 6A) or highly mobile nucleosomes in different ISWI-bound genes (Figure 6B and C). Collectively, our data support a role for ISWI in organizing nucleosome positions after the TSS of ISWI-bound genes. However, the changes in nucleosome positions we observe can be due to the indirect effects of changes in transcription in the ISWI mutant. Therefore, we looked at a possible correlation existing between changes in transcription levels and nucleosomes positions in the ISWI mutant, based on published genome-wide transcriptome array data (Corona et al, 2007) as well as on new RT–PCR data. Among ISWI-bound genes with changes in nucleosome positioning, we found without distinction both an increase or a decrease in transcription (Figure 6D; data not shown). In conclusion, our analysis did not find any positive or negative correlation between changes in transcription levels and nucleosomes positions in ISWI-bound or not-bound genes, when comparing wild-type and ISWI mutants. Nevertheless, we cannot exclude that the changes in nucleosome positions we observe could be in part indirectly caused by changes in transcription resulting from loss of ISWI function.
Figure 6.
Analysis of nucleosome positioning at specific gene loci bound by ISWI. The normalized log2(NucDNA/GenDNA) ratio (blue signal) was used to calculate the MLM nucleosome positions in wild-type (green segments) and ISWI mutant (red segments) chromatin in ISWI-bound genes (A) at the level of the promoter–exon region, and on genes (B) negatively regulated or (C) positively regulated by ISWI (for a complete list, see Supplementary Table S1B and C) (Corona et al, 2007). Exons are represented by filled-in boxes, introns by a continuous line, while the 5′ and 3′-UTRs by thick lines. The transcription direction for each gene can be deduced by the repeated small blue arrows present in the intron segments. The grey bars highlight ISWI-bound region. The thick black bars represent regions of chromatin with nucleosome positioning differences between wild-type and ISWI mutant chromatin. The black arrows point at the nucleosome-free regions (NFR) present at the level of the TSS. (D) Changes in transcription levels, between wild-type and ISWI mutants, are measured by semi-quantitative RT–PCR on some representative examples of ISWI-bound (+) and not-bound (−) genes, where nucleosomes position differences (+) or no-differences (−) are detected. The data were normalized for the house-keeping gene Act5C that is not bound by ISWI and where no nucleosome differences were observed. While genes bound by ISWI with no nucleosome position changes (i.e. CG1179) do not show changes in transcription, genes bound by ISWI (i.e. CG8732 & CG1765; the same analysed in panels (B, C)) showing nucleosome position changes also show a change in the level of their transcripts when comparing wild type with ISWI mutants.
ISWI modulates nucleosome positions in gene regulatory regions of dosage compensated genes in both males and females
Apart from its role in transcription regulation, ISWI also has an important global function in the regulation of higher-order chromatin structure in vivo. Loss of ISWI in larval salivary gland cells leads to the dramatic decondensation of the male X chromosome (Deuring et al, 2000). In order to determine if global differences in nucleosome spacing could account for the male X chromosome condensation defects observed in ISWI mutants, we systematically identified differences in nucleosomal positioning occurring in genic and intergenic regions between wild-type and ISWI mutant male and female salivary gland chromatin (Supplementary Figure S13).
Using the normalized raw log2(NucDNA/GenDNA) ratio signals or the MLM-calculated nucleosome positions (on ISWI-bound and not-bound chromatin regions), the comparison between wild-type and ISWI mutant chromatin revealed many sites of altered nucleosome positioning (Supplementary Figure S14A and B). Interestingly, these nucleosome differences also tend to globally accumulate near the TSS, though with peak values that do not match the gene intervals where we observe the major changes in nucleosome positions in ISWI-bound genes (see the red arrows on Supplementary Figure S14A and B and compare it with the red double-arrowed bar on Figure 5E). Surprisingly, when we mapped these genic and intergenic differences on each Drosophila chromosomes, we could not detect a significant enrichment in the relative abundance of nucleosome spacing differences for the X chromosome or any particular chromosome arm (Supplementary Figure S14C and D). Our data show that ISWI chromosome condensation defects are not correlated with global ISWI-dependent nucleosome spacing changes.
Since the majority of sequences present in our nucleosome positioning array are represented by genes with low frequency of ISWI association (>90%; see Supplementary Table S2; for simplicity from now on referred as not bound), we reasoned that some of the global nucleosome spacing differences we analysed and mapped were not enriched in ISWI-bound sequences. Therefore, we decided to focus our attention only on nucleosome position changes existing in ISWI-bound genes between wild-type and ISWI mutant chromatin. Using both the raw data or the MLM-calculated nucleosome positions, we found that nucleosome position differences in ISWI-bound genes map after the TSS, where we observed the major changes in nucleosome spacing in ISWI-bound genes (see red arrows in Figure 7A and B and compare it with the red double-arrowed bar in Figure 5E). Interestingly, differences in nucleosome positioning, between wild-type and ISWI mutant chromatin, tend to significantly accumulate on the male X chromosome with respect to all ISWI-bound genes analysed as well as the female X chromosome (Figure 7C). One easy way to explain the male X chromosome tendency to accumulate differences in nucleosome positions is that ISWI is enriched on genes mapping the X chromosomes. Indeed, we observed that ISWI binds genes on the X with a statistically significant higher frequency (Figure 7D).
Figure 7.
ISWI binds dosage compensated genes to make their TSS nucleosome free. The density of nucleosome positioning changes occurring between wild-type and ISWI mutant chromatin was calculated using the (A) normalized raw log2(NucDNA/GenDNA) ratio signal or (B) the MLM, and was plot against the distance from the transcription start site (TSS) of ISWI-bound genes. The red arrow indicates where the nucleosome difference density peaks relative to the TSS. (C) Nucleosome position differences between wild-type and ISWI mutant chromatin for all ISWI-bound genes were calculated for each chromosome arm and plot against their expected values, based on the relative representation of the different chromosome sequences in the nucleosome-tiled array. (D) The number of ISWI-bound genes for each chromosome arm was calculated and plot against their expected values, based on the known number of genes mapping each chromosome. The P-values calculated comparing the expected with the observed values are shown on top of each histogram pair. Nucleosome positions were calculated with the MLM for (E) wild-type and (G) ISWI mutant chromatin relative to the TSS of ISWI- and MSL-bound genes, and for (F) wild-type and (H) ISWI mutant chromatin relative to the TSS of genes not bound neither by ISWI nor MSL. The MLM analysis shows well-positioned nucleosomes as shadings of yellow, linker regions in red, while delocalized nucleosomes in orange. The green and red ovals represent an interpretation of the MLM nucleosome positioning analysis in wild-type and ISWI mutant chromatin, respectively. Filled ovals indicate well-positioned nucleosomes, while shaded ovals show delocalized nucleosomes. The black arrow indicates the nucleosome-free region at the TSS of wild-type ISWI- and MSL-bound genes. The red double-arrowed bar indicates the region at the TSS where we observe nucleosome occupancy in the absence of ISWI in ISWI- and MSL-bound genes.
Sex chromosomes in flies, like in many eukayotes, undergo the process of dosage compensation. In Drosophila, dosage compensation is dependent on a multisubunit complex that is specifically targeted to the male X chromosome, leading to the acetylation of dosage compensated genes in lysine 16 of histone H4 (H4K16Ac) by the MOF histone acetyltransferase (Park and Kuroda, 2001). This histone modification is thought to increase by two-fold the transcription of dosage compensated genes, thus compensating for the presence of one X chromosome in males (Smith et al, 2001). Remarkably, when we compared the binding of ISWI to genes strongly bound with the ones weakly bound by the dosage compensation complex on the X chromosome (Larschan et al, 2007), we found a strong enrichment of ISWI binding on or near dosage compensated genes (Supplementary Table S3; Supplementary Figure S15A). Therefore, we decided to analyse differences in nucleosome positions existing in genes strongly and weakly bound by the dosage compensation complex on the X chromosome, present in our nucleosome position array, and the effect of ISWI loss on those genes.
Our analysis revealed that ISWI-bound genes strongly bound by the dosage compensation complex present on average a well-positioned nucleosome before the nucleosome-free TSS (Figure 7E). On the other hand, several delocalized nucleosomes occupy the TSS of ISWI-bound genes weakly bound by the dosage compensation complex on the X chromosome (Figure 7F). Similarly, when we analysed nucleosome positions in wild-type female chromatin, we found nucleosome positions around the TSS very different from those found in all ISWI-bound genes (compare Supplementary Figure S16 with Supplementary Figure S5C). Our analysis shows that non-dosage compensated genes mapping the X chromosome show a very peculiar nucleosome organization at the level of their TSS (absence of NFR) in both males and females. Probably, in the absence of dosage compensation on both males and females, this nucleosome organization is functional to a specific transcriptional regulation of those genes.
Remarkably, while the loss of ISWI has little effect on genes weakly bound by the dosage compensation complex (compare Figure 7F and H), the TSS of genes strongly bound by the dosage compensation complex becomes occupied by a delocalized nucleosome in the absence of ISWI (compare Figure 7E and G). However, when we looked at nucleosome position changes resulting from ISWI loss at the 3′-end of dosage compensated genes and in genes with low frequency of association with the dosage compensation complex, we found that loss of ISWI had little effect (data not shown). Moreover, when we looked at female chromatin, we found that changes in nucleosome positions around the TSS of MSL-bound genes were very different from those found in males. Remarkably, those changes did not affect the nucleosome-free TSS (compare Figure 7E and G with Supplementary Figure S16A and C). Collectively, our data indicate that ISWI binding on the X-linked genes causes specific nucleosome changes in both male and female chromatin.
Discussion
The unusual sensitivity of the male X chromosome to the loss of ISWI function has suggested in the past that changes in chromatin structure that accompany dosage compensation might regulate the ability of ISWI to remodel chromatin in vivo. Indeed, dosage compensation is necessary and sufficient for the chromosome defects observed in ISWI mutant larvae (Corona et al, 2002). In particular, genetic studies have shown that in the absence of ISWI the decondensed male X chromosome is dosage compensated and that in vitro H4K16Ac counteracts ISWI nucleosome spacing activity by reducing ISWI binding to chromatin (Corona et al, 2002; Shogren-Knaak et al, 2006). However, our data show that ISWI binds dosage compensated genes affecting nucleosome positioning near the TSS. Interestingly, it has been recently shown that the dosage compensation complex preferentially binds its target genes towards their 3′-end (Larschan et al, 2007). When we mapped ISWI binding on MSL-bound genes, we found that ISWI tend to bind the 5′-end on those genes (see Supplementary Figure S15B). Therefore, ISWI and the MSL occupy complementary gene functional elements along dosage compensated genes, making our nucleosome changes observation consistent with published data.
In conclusion, we found that the loss of ISWI globally causes subtle changes in nucleosome positioning. However, these changes are highly localized and mainly concentrated at level of the TSS, including the ones mapping the X chromosome that are particularly enriched in dosage compensated genes, probably explaining the specific sensitivity of the male X for ISWI loss (Deuring et al, 2000; Corona et al, 2002). Although our data suggest that in higher eukaryotes ISWI could regulate gene expression by remodelling chromatin near gene regulatory regions, additional studies will be necessary to understand the advantage of having ISWI-sensitive-nucleosome-free TSS on dosage compensated genes. Furthermore, our genome-wide analysis of ISWI binding and nucleosome spacing activity in Drosophila also revealed that ISWI-dependent alterations in nucleosome positioning at the level of the TSS are unlikely to be correlated with global chromatin structural alterations observed when ISWI activity is lost. Collectively, our data provide new insights on the role played by the evolutionarily conserved chromatin remodelling factor ISWI in transcriptional regulation and chromosome organization in higher eukaryotes.
Materials and methods
Drosophila stocks and genetic crosses
Flies were raised at 25 or 18°C on K12 medium (USBiological). Unless otherwise stated, strains were obtained from Bloomington Stock Center and are described in FlyBase (http://www.flybase.org). For nucleosomal and GenDNA preparation, ISWI1/ISWI2 male larvae were obtained as previously described (Burgio et al, 2008).
ISWI ChIP
ChIPs were conducted starting from larval chromatin according to previously published protocols (Larschan et al, 2007) with some minor variations (see Supplementary data). All raw data files relative to the ISWI-ChIP experiments are available at http://math.unipa.it/pinello/iswi/iswi_raw.zip.
ISWI ChIP-on-chip and data analysis
Microarray hybridizations were performed according to NimbleGen protocols on a three-array set covering the entire fly genome. Microarray scanning and data extraction were conducted using NimbleScan software (Kim et al, 2005). Genomic regions enriched in ISWI were identified using the peak score function that takes into account the length and intensity of the normalized raw log2(ChIPISWI/input) signals (see Supplementary data). ISWI ChIP-on-chip data derive from three biological replicates.
Motif discovery analysis
We have studied the correlation existing between the number of occurrences of a motif M in a data set D of sequences obtained from the ISWI ChIP data, and the peak score corresponding to the same sequences. In particular, after we identified the most significant motifs with MDscan (Liu et al, 2002) or MEME (Bailey et al, 2006), we correlated the number of occurrences of the motifs within the sequence with the peak score of the sequence to which the motif belongs to (see Supplementary data). Putative ISWI motif binding factors were identified querying the TRANSFAC and JASPAR databases to look for D. melanogaster homologues.
Nucleosome positioning array hybridization and data analysis
Salivary gland mononucleosomal DNA was purified and hybridized competitively against total larval GenDNA on a custom-tiled array (see Supplementary data).
Labelled genomic and mononucleosomal DNA were hybridized over a custom-tiled arrays containing 7.7 Mbp of the Drosophila genome, in 385 000 spots made of 50mer overlapped by 30 bp. Microarray hybridizations were performed according to NimbleGen protocols. Microarray scanning and data extraction were conducted using NimbleScan software (Kim et al, 2005). Nucleosome position data were derived from data coming from three biological replicates each for wild-type and for ISWI mutants. All raw data files relative to the nucleosome-tiled array experiments are available at http://math.unipa.it/pinello/iswi/nucleosome_raw.zip.
Identification of nucleosome position differences between wild-type and ISWI mutants
In order to find regions containing changes in nucleosome positions between wild-type and ISWI mutant chromatin, we conducted an analysis using both the raw signal as well as the nucleosome classification of the MLM (see Supplementary data). Differences in the raw signal were identified calculating the Z-score of the ratio between the wild-type and the ISWI mutant data tracks (Supplementary Figure S13). Differences encompassing at least three probes with a Z-score value ⩾2 were extracted. These parameters are set to identify differences equal to a shift of half a nucleosome.
Analysis of expected nucleosome occupancy near the TSS and at the 3′-end
To characterize the average nucleosome occupancy level at the TSS and 3′-end of the genes present in the nucleosome-tiled array, four lists of genes have been considered: all genes covered by the microarray, all genes repressed or overexpressed in ISWI mutant, all genes nearby the binding sites of ISWI. Fragments of length of 1000 bp centred at the TSS or at the 3′-end of each genes have been extracted from each one of the four lists. The average of nucleosomes occupancy of such extracted fragments was calculated and represented as a raw signal or MLM output.
Supplementary Material
Acknowledgments
We thank members of the laboratory, and Peter Becker for his precious feedbacks and comments on the manuscript. We also thank S Rosalia for her inspiring vision of our work. AS was supported by a FIRC Fellowship. MT and AMRI were supported by a Telethon Fellowship. This work was supported by grants from Fondazione Telethon, Giovanni Armenise Harvard Foundation, MIUR-FIRB, HFSP, AIRC and EMBO YIP to DFVC.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arancio W, Onorati MC, Burgio G, Collesano M, Ingrassia AM, Genovese SI, Fanto M, Corona DF (2010) The nucleosome remodeling factor ISWI functionally interacts with an evolutionarily conserved network of cellular factors. Genetics 185: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgio G, La Rocca G, Sala A, Arancio W, Di Gesu D, Collesano M, Sperling AS, Armstrong JA, van Heeringen SJ, Logie C, Tamkun JW, Corona DF (2008) Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI. PLoS Genet 4: e1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Armstrong JA, Tamkun JW (2004) Genetic and cytological analysis of Drosophila chromatin-remodeling factors. Methods Enzymol 377: 70–85 [DOI] [PubMed] [Google Scholar]
- Corona DF, Clapier CR, Becker PB, Tamkun JW (2002) Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Langst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell 3: 239–245 [DOI] [PubMed] [Google Scholar]
- Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW (2007) ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol 5: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW (2004) Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta 1677: 113–119 [DOI] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, Tsukiyama T, Wu C, Pimpinelli S, Tamkun JW (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5: 355–365 [DOI] [PubMed] [Google Scholar]
- Di Gesu V, Lo Bosco G, Pinello L, Yuan GC, Corona DF (2009) A multi-layer method to study genome-scale positions of nucleosomes. Genomics 93: 140–145 [DOI] [PubMed] [Google Scholar]
- Dirscherl SS, Krebs JE (2004) Functional diversity of ISWI complexes. Biochem Cell Biol 82: 482–489 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB (2004) ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci 117: 3707–3711 [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T (2005) Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev 19: 942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof A (2006) Epigenetic regulators and histone modification. Brief Funct Genomic Proteomic 5: 222–227 [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J, Jin Y, Walker DL, Wang D, Wang Y (1999) Chromatin structure and nuclear remodeling. Crit Rev Eukaryot Gene Expr 9: 267–277 [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Woo CJ, Tolstorukov MY, Kingston RE, Park PJ (2008) Nucleosome positioning in human HOX gene clusters. Genome Res 18: 1554–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI (2007) MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28: 121–133 [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS (2008) NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS (2002) An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20: 835–839 [DOI] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT (2005) Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166 [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2007) Mechanisms of epigenetic inheritance. Curr Opin Cell Biol 19: 266–272 [DOI] [PubMed] [Google Scholar]
- Park Y, Kuroda MI (2001) Epigenetic aspects of X-chromosome dosage compensation. Science 293: 1083–1085 [DOI] [PubMed] [Google Scholar]
- Quivy JP, Becker PB (1996) The architecture of the heat-inducible Drosophila hsp27 promoter in nuclei. J Mol Biol 256: 249–263 [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2006) Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Siriaco G, Deuring R, Chioda M, Becker PB, Tamkun JW (2009) Drosophila ISWI regulates the association of histone H1 with interphase chromosomes in vivo. Genetics 182: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Allis CD, Lucchesi JC (2001) Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem 276: 31483–31486 [DOI] [PubMed] [Google Scholar]
- Stephens GE, Craig CA, Li Y, Wallrath LL, Elgin SC (2004) Immunofluorescent staining of polytene chromosomes: exploiting genetic tools. Methods Enzymol 376: 372–393 [DOI] [PubMed] [Google Scholar]
- Stopka T, Skoultchi AI (2003) The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci USA 100: 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GH, Elgin SC (1988) Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J 7: 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C (1995) Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T (2007) Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ (2005) Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.