Abstract
The de novo design of protein-protein interfaces is a stringent test of our understanding of the principles underlying protein-protein interactions and would enable new approaches to biological and medical challenges. Here we describe a novel motif-based method to computationally design protein-protein complexes with native-like interface composition and interaction density. Using this method we designed a pair of proteins, Prb and Pdar, that heterodimerize with a Kd of 130 nM, 1,000-fold tighter than any previously designed de novo protein-protein complex. Directed evolution identified two point mutations that improve affinity to 180 pM. Crystal structures of complexes containing designed and evolved proteins reveal binding is entirely through the designed interface, making use of specific designed interactions. Surprisingly, in the evolved complex one of the partners is rotated 180 degrees relative to the design model. This work demonstrates that current understanding of protein-protein interfaces is sufficient to rationally design interfaces de novo, and underscores remaining challenges.
Keywords: Computational design, directed evolution, protein-protein interface, protein interaction, hotspot, Ankyrin Repeat, protein docking
Introduction
Protein-protein interfaces are at the crossroads of a wide variety of biological processes from cellular adhesion to immune function. A better understanding of the physical basis for protein interactions would not only advance our knowledge of a process fundamental to many aspects of biology, but would also enable the rational design of novel protein interactions for research and therapeutic purposes. A stringent test of current understanding is the rational design of a novel high-affinity protein starting with two non-interacting natural proteins. Efforts to probe the relative importance of energetic features in natural protein interfaces have been convoluted by the evolutionary pressures that may subtly shape or constrain an interface. In these cases, it can be difficult to distinguish how the geometry and composition of a given protein interface has been optimized for binding kinetics, specificity, or overall affinity throughout its evolutionary history. The creation of a novel protein complex could therefore inform our basic understanding of macromolecular interactions. In addition to illuminating basic principles, de novo design could serve as a starting point for building pairs of proteins which interact with one another but are devoid of competing interactions with endogenous cellular proteins; the importance of such “orthogonal protein pairs” as building blocks for creating interaction networks has been recently highlighted (Mandell and Kortemme, 2009b; Reinke et al., 2010).
Directed evolution techniques have been used to successfully create protein interfaces never before seen in nature (Binz et al., 2005); however, these methods currently allow the redesign of only one partner at a time, and are thus unable to generate an interface that is simultaneously co-optimized on both sides. Computational design algorithms are potentially capable of co-optimizing a protein interface, but the direct application of simple computational methods for simultaneous sequence optimization of two proteins arranged next to one another does not lead to interaction pairs that resemble naturally occurring non-obligate dimers.
A popular descriptive model for protein interfaces is that of an “O-ring,” with a tightly packed hydrophobic core surrounded by a ring of polar interactions. The lynchpin of the O-ring is one or more interaction “hotspots,” residues that contribute much more than surrounding amino acids to the overall interaction energy of the complex. By definition, mutation of a hotspot greatly diminishes or abrogates formation of the protein complex. Collectively, this qualitative description captures two key properties that distinguish natural protein interfaces from earlier computational designs resembling protein cores: a high density of favorable interaction in the center of the interface, and a spatial separation of hydrophobic and polar regions of the interface.
We have developed a computational approach to design protein interfaces with these properties, and used it to design both sides of novel interfaces between two proteins with no natural affinity for one another. The resulting complexes are therefore “de novo” designs, never before seen in nature and created entirely using computational techniques. While previous studies have successfully redesigned existing interactions for altered specificities (Kortemme et al., 2004; Shifman and Mayo, 2002) or created low-affinity complexes (Huang et al., 2007; Jha et al., 2010), we now describe a novel protein complex with sub-nanomolar affinity obtained by integrating our new protein interface design approach with directed evolution.
Results
Computational Approach
Previous approaches to de novo protein interface design by us and others (Huang et al., 2007), involved first positioning the two component proteins near one another in space via computational docking. For each resultant orientation the interfacial residues were identified, and the sequence at these positions was optimized using standard computational design methodology. Our initial efforts using this approach did not lead to models containing interaction density comparable to that observed in natural protein interfaces, even after incorporation of methodology that included sampling intramolecular backbone degrees of freedom (Mandell and Kortemme, 2009a). In the context of designing stable monomeric proteins, an explicit representation of backbone flexibility has proven critical in achieving appropriate interaction density through tight packing of the protein core (Kuhlman et al., 2003). For protein-protein interface design, however, we and others learned that the constraint of maintaining the structure of the constituent proteins made it difficult to achieve design models with appropriate interaction density (Jha et al., 2010).
To overcome these obstacles, we developed a “motif-based” design methodology. In order to achieve a high density of interactions in the center of the interface, one or more templated interactions (“motifs”) are required; we define these to be tryptophan or tyrosine sidechains which are simultaneously well-packed and engaged in a hydrogen bond. The remaining interfacial residues are designed subsequently in order to ensure their compatibility with the central motifs. Although the details of this methodology are rationalized on the basis of established properties of naturally occurring protein interfaces, our method does not “graft” residues from existing protein interfaces. Instead, we draw upon the general properties of natural protein complexes to build new interfaces de novo. This strategy is outlined below and details of the computational implementation are available in the Supplemental Materials.
The first step in our protocol for designing new interfaces is to choose a pair of proteins for redesign. Each of these initial natural proteins represents a “scaffold” upon which the designs are built. The ankyrin repeat (AR) protein superfamily is a general protein-binding module which is used by nature in a wide variety of contexts. Distinct ARs associate with an assorted array of protein partners in higher eukaryotes (Letunic et al., 2006) and are present as a diverse family of bacterial effector proteins (Pan et al., 2008), demonstrating the broad potential specificity of this scaffold. Crystal structures of several protein complexes involving ARs reveal diverse binding orientations and surface sequence compositions of their protein partners, further underscoring the ability of the ARs to bind a wide variety of partners (Mosavi et al., 2004). We paired each of several AR proteins with each member of a set of 37 structurally diverse thermostable proteins (see Supplemental Materials) and used a surface feature-matching approach (Schneidman-Duhovny et al., 2003) followed by rigid-body docking to generate a large set of bound orientations that exhibited shape complementarity at the backbone level (Figure 1a).
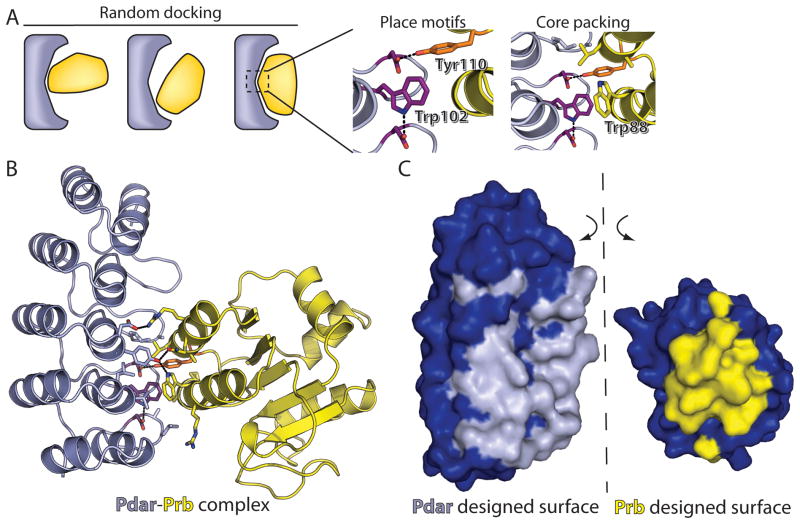
Figure 1. Computational Design Approach.
The Ankyrin Repeat protein (redesigned to become Pdar) is colored gray and its partner protein (redesigned from PH1109 to become Prb) is colored yellow. (A) The surfaces of each protein are first matched by general shape complementarity and local docking. Promising rigid body orientations are used in an attempt to place a central hydrogen-bonding tyrosine or tryptophan motif, followed by local design to enforce hydrophobic packing around the motif. (B) Overview of interfacial amino acids in the designed complex of Prb-Pdar. The central motifs are colored as in A. (C) Open-book surface view of the completely designed Prb-Pdar interface. Non-designed residues are colored blue, and designed interfacial residues are colored either gray (Pdar) or yellow (Prb). All figures containing molecular graphics were generated using PyMOL (DeLano, 2004).
Our strategy for designing interfaces that capture the high interaction density seen in naturally evolved interfaces is to work outward from the center of the interface. First we introduce residues forming a pair of aromatic interaction motifs, then surround these with an inner layer of tightly packed hydrophobic sidechains, and finally protect this core from bulk solvent using an outer layer of polar sidechains (Figure 1a). We anticipate that the central motifs will provide a large contribution to the overall affinity of the complex, meeting the criterion of hotspot residues in natural protein interfaces. We used as our guide the observation that hotspot residues have a distinctive amino acid composition that favors tyrosine and tryptophan (Bogan and Thorn, 1998; Moreira et al., 2007), and further that the energetic contribution of these residues is enhanced when involved in a hydrogen bond (Guharoy, 2005). Building on the observation that pre-ordered sidechain “anchor” residues are often present near hotspots in naturally occurring interfaces (Li et al., 2004; Rajamani et al., 2004), we selected a row of structurally conserved aspartate residues at the back of the AR binding groove to serve as hydrogen bond acceptors for hotspot residues at the center of the designed interface. These aspartates are pre-ordered by hydrogen bonds to the AR backbone, an interaction seen in nearly every AR crystal structure, making them ideal candidates to serve as anchor residues.
We developed a protocol to rapidly screen all tyrosine and tryptophan rotamers at all interfacial backbone positions on both partners for the ability to make a hydrogen bond to one of the (fixed) anchoring aspartate sidechains, and applied this protocol to each of the bound orientations generated earlier. Orientations were discarded from further consideration if two non-overlapping motifs could not be identified; we further required that at least one of these motifs contain an intermolecular hydrogen bond. The search was carried out in a computationally efficient manner by screening for a single intermolecular motif first, then searching for a second motif only if the initial search was successful. For each pair of motifs, the hydrophobic layer surrounding the motifs was completed by applying RosettaDesign to select optimal aliphatic amino acids at the backbone positions in the immediate vicinity of these two hydrogen-bonded aromatic sidechains. Only cases with suitably tight packing in this layer were carried further.
In the final step of our protocol, RosettaDesign was used to optimize residue identities at the interface periphery holding fixed the hydrophobic inner layer at the center of the interface. To produce design models with global electrostatic complementarity, a favorable bias was applied for acidic residues on the AR protein and basic residues on its partner. Since the bonus was applied at all backbone positions without consideration of structure, this aspect of the design protocol is intended to play a role in modulating binding affinity, rather than contribute to specific contacts and modes of interaction.
Finally, a series of sequential filters were applied to the resultant design models, eliminating from consideration models that did not conform to quantitative structural descriptors generated from a survey of the Protein Data Bank. Filters were applied to ensure “native-like” interface size, tight packing (assessing direct interactions as well as searching for void volume), and a lack of steric clashes. Design models which did not satisfy the hydrogen bonding potential of the conserved aspartate residues in the back of the AR binding groove via direct hydrogen bonds or implicit hydrogen bonds to solvent were also removed. To ensure that the overarching design goal of high interaction density was met, we applied an additional filter based on the intermolecular interaction energy divided by the surface area of the complex. This filter was specifically inspired by our observation that native protein-protein interfaces in general had higher interaction density than our early design models, indicating that this filter should be particularly useful in eliminating one of the most common classes of designs that were not sufficiently “native-like”: those with interfaces that achieved high calculated interaction energies through a large number of non-cooperative, broadly interspersed contacts.
Every step of the protocol described above was only designed to produce models with favorable interactions in the context of the complexed structure. Notably, “negative design” was not explicitly used at any stage of the protocol – neither to disfavor binding to alternative binding partners, nor to disfavor binding of the designed proteins in an orientation that differs from that of the designed interface.
Prb-Pdar: A Computationally Designed Complex That Binds with Sub-Micromolar Affinity
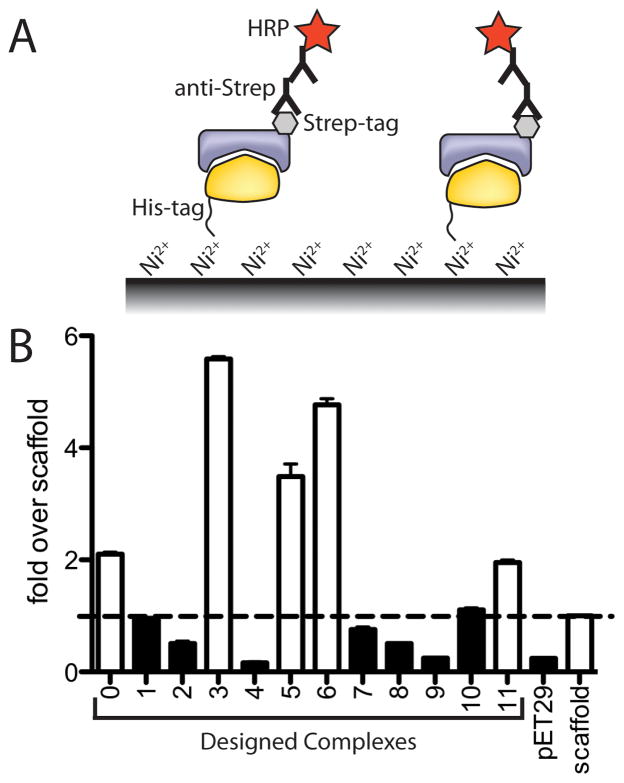
The twelve designed pairs remaining after computational filtering (Supplemental Figure S1) were expressed in E. coli and screened via ELISA for stable interactions (see Methods). To avoid potentially unstable designs, the ELISA screen was designed to only identify proteins that are both solubly expressed and interact with one another. Five of the twelve yielded a signal more than two-fold over background nonspecific binding of a StrepII-tagged noncognate protein (Figure 2).
Figure 2. Experimental screening for interacting computationally designed complexes.
A) An ELISA screen for stably-associating designed protein complexes. Both partners, one His6-tagged and the other Strep-tagged, were co-expressed in E. coli. Complexes were then captured on a Nickel-derivatized surface detected with an anti-Strep antibody. B) Nearly half of the computationally designed protein-protein complexes (open bars) displayed signal more than two-fold over a control that replaces one side of Complex #11 with the wild-type protein used as a scaffold for the design (arbitrary identifiers 0–11 were assigned to the 12 designed pairs remaining after computational filtering).
We have carried out a retrospective quantitative comparison to explain why some complexes showed detectable binding in our initial screen while others did not. We examined descriptors to quantify shape complementarity, packing, electrostatics, solvation, sidechain entropy and a variety of other features, yet each found that the designs were indistinguishable from native protein-protein interfaces. We additionally applied a machine learning-based approach which has proven very successful at distinguishing native complexes from incorrectly predicted structures (London and Schueler-Furman, 2008); this method could not discriminate between design models. Our computational design methodology is therefore capable of creating interfaces indistinguishable from those found in natural protein complexes. However, it is still unclear what metric might discriminate binding designs from those that do not bind, highlighting an unexpected deficiency in currently accepted metrics to discriminate protein interfaces. As these design models represent a stringent discrimination challenge for the protein-protein docking community, we have made them publicly available for download (see Supplemental Materials).
Of the promising protein pairs emerging from our ELISA screen, complex #11 was selected for further in vitro characterization since it had the most favorable interface interaction density (Supplemental Table S1), and was therefore the best exemplar of our design goals. Complex #11 corresponds to a consensus ankyrin repeat (PDB code 1MJ0) paired with PH1109, a Pyrococcus horikoshii coenzyme A (CoA)-binding protein (PDB codes 2D59 and 2D5A). We henceforward refer to the redesigned proteins as Prb (PH1109 protein redesigned for binding) and Pdar (Prb-binding designed ankyrin repeat).
The interface in the model has been completely designed (Figure 1), leaving surfaces that bear little resemblance to the original wild-type proteins. Indeed, Prb harbors 17 mutations relative to PH1109 and Pdar differs from the consensus AR at 23 positions. In the Prb-Pdar design model, the aromatic residues comprising the two Tyr/Trp central motifs (see “Computational Approach”) are Tyr110 of Prb and Trp102 of Pdar; these sidechains are anchored via hydrogen bonds to Asp98 and Asp131 of Pdar. The central motif Trp102 of Pdar is oriented perpendicular to an aromatic group on Prb, Trp88, which initiates a small hydrogen bond network to Tyr77 of Pdar then Gln116 of Prb. These motifs are buttressed by a layer of aliphatic and aromatic sidechains, which are in turn surrounded by a rim of polar residues.
To explore whether our design protocol had recapitulated a backbone orientation already observed in nature, we applied the MM-align and TM-align algorithms (Mukherjee and Zhang, 2009; Zhang and Skolnick, 2005) to search for similar arrangements amongst both multiple chain complexes and monomers in the Protein Data Bank. The highest scoring results for both searches consist of AR proteins bound to structurally unrelated partners, demonstrating the uniqueness of the designed complex relative to all structures of natural interfaces solved to date.
While the overall interface bears little resemblance to any known natural interface, the use of motifs in the design protocol led to specific sets of interactions that are strongly reminiscent of naturally occurring counterparts. In the structure of the human growth hormone bound to its receptor, for example, much of the binding affinity derives from two tryptophan sidechains on the receptor, Trp104 and Trp169 (Clackson and Wells, 1995). Like the motifs in the Prb-Pdar design model, each of these is tightly packed in a complementary hydrophobic pocket on the hormone surface (Clackson et al., 1998). Trp104 of the receptor is additionally engaged in an intermolecular hydrogen bond to the hormone (analogous to Tyr110 of Prb), while Trp169 donates a hydrogen bond to an intramolecular acceptor (analogous to Trp102 of Pdar). Structures of both natural and evolved AR complexes show further examples in which tightly-packed tryptophan and tyrosine sidechains form both intermolecular and intramolecular hydrogen bonds to the aspartate sidechains at the back of the AR binding groove (Batchelor et al., 1998; Kohl et al., 2005).
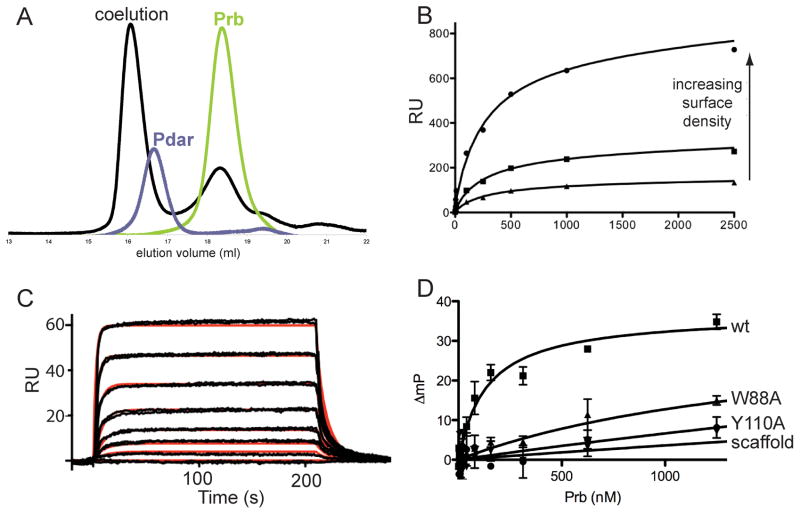
To further characterize the interaction between Prb and Pdar, each protein was separately expressed and purified. Each monomer was separately analyzed by size exclusion chromatography to determine its characteristic elution volume. Notably, a stoichiometric mix of the monomers elutes at an earlier volume than either protein alone, suggesting that Prb and Pdar form a stable complex (Figure 3a). Light scattering analysis indicates that a mixture of the Prb and Pdar proteins is monodisperse and has a radius of gyration consistent with formation of a stable 1:1 complex, with a calculated total molecular weight matching that expected for the heterodimer.
Figure 3. Prb and Pdar form a hotspot-based high affinity protein complex with kinetics similar to those observed in natural protein assemblies.
A) Prb and Pdar stably associate with one another to co-elute from a gel filtration column. B) Equilibrium surface plasmon resonance (SPR) measurements indicate that Prb and Pdar associate with Kd 60–90 nM. C) Kinetic SPR measurements at low surface density indicate that Prb and Pdar associate with kon of 7–9×105 M−1 s−1 and koff of 0.05–0.1 s−1, which is in the range of natural protein-protein complexes and corresponds to Kd approximately 50–150 nM. Duplicate measurements are shown in black, and a 1:1 Langmuir fit is shown in red. D) Fluorescence polarization confirms that Prb and Pdar bind one another with a Kd of 135 nM. Furthermore, Pdar does not associate with wild-type PH1109, the protein scaffold used as a starting point in the Prb design. Mutation of key designed interfacial residues abrogates complex formation.
The co-elution of Prb and Pdar by size exclusion chromatography suggests that they possess a reasonably strong affinity for one another. To further characterize the association of these designed proteins, we utilized surface plasmon resonance (SPR) to estimate the binding affinity and kinetics of the Prb-Pdar complex (see Methods). Fitting a simple model of total binding to the equilibrium phase binding response yielded a dissociation constant (Kd) between 60 and 90 nM across three different Pdar surface densities (Figure 3b). Independently fitting kinetic on- and off-rate parameters to the association and dissociation phases of the SPR data from multiple Pdar surface densities with a 1:1 Langmuir binding model yielded a kon of 7–9×105 M−1s−1 and koff of 0.05–0.1 s−1 (Figure 3c), corresponding to a Kd of 90–120 nM, consistent with the equilibrium phase SPR analysis. The relatively strong binding affinity between Prb and Pdar is consistent with their stable co-elution observed via size exclusion chromatography, and is approximately 1,000-fold tighter than that of any previous de novo computationally designed protein complex (Huang et al., 2007; Jha et al., 2010).
To independently confirm the affinity of the Prb-Pdar interaction, we utilized fluorescence polarization binding measurements (see Methods). Fitting change in polarization to a simple total binding model yields a Kd of 135 ± 38 nM (Figure 3d). The consistency between the dissociation constants measured by two SPR methods as well as fluorescence polarization confirms that Prb and Pdar do indeed stably interact, with a dissociation constant of ~100 nM. Furthermore, Pdar does not bind the wild-type PH1109 protein used as a scaffold for building Prb (Figure 3d), indicating that the association of Pdar with Prb is not due to some inherent affinity of the redesigned ankyrin for PH1109.
The Tyr/Trp central motifs introduced early in the design process form the lynchpin of the Prb-Pdar interface. These residues were introduced with the expectation that their interactions would be critical for the overall affinity of the interface, akin to hotspots in naturally occurring protein complexes (Bogan and Thorn, 1998; Clackson and Wells, 1995). To experimentally determine how these important interactions in the design model contribute to affinity, we made two variants of Prb. The first is a direct hotspot knockout in which we mutated the Prb central motif, Tyr110, to alanine. The other is an indirect hotspot knockout in which we substituted alanine in place of Prb Trp88, which is predicted to play an important role in stabilizing the Pdar central motif (see Figure 1a). We determined the affinities of each of these variants by fluorescence polarization, and found that neither alanine mutant forms a detectable complex with Pdar (Figure 3d). Thus, Prb-Pdar represents the first computationally designed de novo protein complex to capture several key features of naturally occurring protein complexes, including high affinity and incorporation of hotspot residues. Because the design protocol was predicated upon surrounding Tyr/Trp motifs with an inner layer of tightly packed hydrophobic groups and an outer ring of polar sidechains, we postulate that complementary pairs of such protein surfaces are sufficient to encode affinity within an artificial protein complex.
Affinity Maturation of the Prb-Pdar Complex
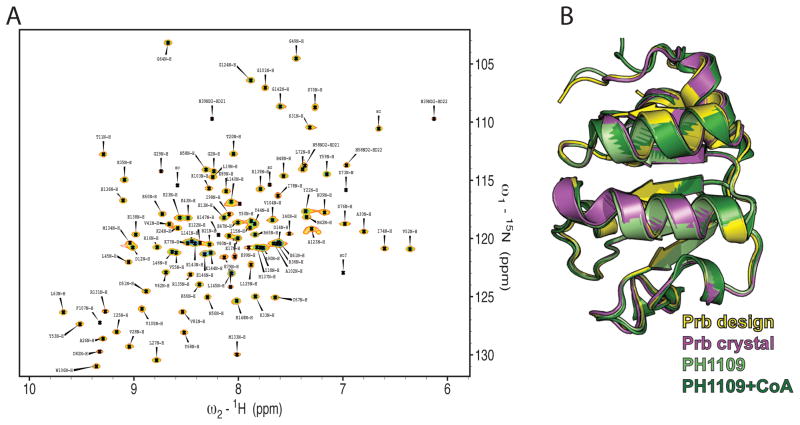
Despite extensive attempts to crystallize the Prb-Pdar complex for structural studies, no high-quality crystals were obtained. While attempting to use NMR to map the Prb-Pdar interface, we discovered that, although all visible peaks in an 1H-15N HSQC NMR spectrum of Prb could be assigned, nearly thirty peaks for residues located at the putative interface were missing entirely from the spectrum (Figure 4a). These absent peaks are not due to general instability of the design, since Prb is well-folded and nearly as thermostable as the wild-type PH1109 protein (Supplemental Figure S2). We solved a 1.9 Å crystal structure of apo Prb, and found it to have an RMSD to the design model of 0.54 Å over 814 backbone and sidechain atoms, confirming that the many mutations distinguishing Prb from the PH1109 scaffold do not constitutively distort the monomer (Figure 4b); thus the designed Prb backbone is present in solution to at least some degree. Based on these crystallographic and NMR data, we postulated that local conformational heterogeneity at the designed Prb interface may have caused difficulties in crystallization of the complex. In an attempt to stabilize a unique conformation within the Prb-Pdar complex, we used a combination of phage and yeast display, two library-based affinity-maturation methods, to evolve Prb for tighter binding to Pdar (see Methods).
Figure 4. Interfacial residues of apo Prb can adopt the designed conformation, but may also be mobile.
A) 2D 1H-15N HSQC NMR spectrum of Prb. Peaks are labeled with their respective sequential resonance assignments using the one-letter code of amino acids and the amino acid sequence number. Approximately thirty residues located at the designed interfacial positions are missing from the spectrum, suggesting that they may be conformationally dynamic. B) A crystal structure of the Prb protein (magenta) agrees well with both the design model (yellow, 0.54Å all-atom rmsd) and structures of the PH1109 scaffold either apo (light green, 0.41Å all-atom rmsd) or bound to CoA (dark green, 0.47Å all-atom rmsd).
Using random mutagenesis with mutagenic dNTP analogs (Zaccolo et al., 1996), we created a phage library of 2.9×108 Prb mutants, which we selected for binding to a fixed concentration of Pdar (500 nM) for three rounds. After the third round of phage selection, the remaining Prb mutants were cloned into a yeast display vector. Four additional rounds of selection were performed on the yeast-displayed Prb library by applying fluorescence-activated cell sorting (FACS) to select for yeast cells that exhibited the highest Pdar-binding signals.
Two of the evolved clones, termed PrbC5 and PrbC10, were chosen for further in vitro characterization based on promising behavior during directed evolution. Both PrbC5 (Asp11Gly, Asp83Asn, Val92Ala, Glu119Gly, and Glu135Lys) and PrbC10 (Ile75Val, Asp83Asn, Leu122Pro, and Glu135Lys) bind Pdar with a Kd of approximately 1–4 nM (Supplemental Table S2), representing a ~30-fold improvement over Prb. Half of the mutations within PrbC10 are second-shell hydrophobic changes abutting the center of the designed interface, suggesting that concerted backbone changes caused by second-shell repacking may account for at least some of the increase in affinity. Supporting this idea, CD-monitored thermal melts reveal that the evolved Prb variants are less stable than either the wild-type starting protein (PH1109) or the computationally designed Prb (Supplemental Figure S2), consistent with other studies indicating a trade-off between monomer stability and function (Foit et al., 2009; Hackel et al., 2008; Xu et al., 2002).
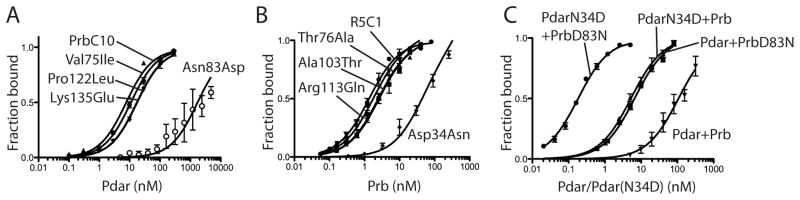
To determine which evolved mutations were most responsible for the increase in affinity, we systematically reverted each mutation to the original designed amino acid and determined the affinity of the resulting protein for Pdar. Though single reversion of each second shell mutation had little effect, reverting the Asp83Asn mutation caused a dramatic reduction in affinity in either the PrbC5 or PrbC10 backgrounds (Figure 5a, Supplemental Table S2). We therefore tested whether the Asp83Asn mutation was sufficient to improve affinity, and found that Prb(Asp83Asn) binds Pdar with a Kd of 5.8 nM (Figure 5c, Supplemental Table S2). This result suggests that the substantial majority of the evolved Prb’s improved affinity for Pdar arises from this single Asp to Asn mutation.
Figure 5. Directed evolution identifies two point mutants that increase the affinity of Prb and Pdar by 720-fold.
A) Reversion of each mutation within the affinity matured PrbC10 clone to its Prb identity indicates that Asn83Asp is responsible for most of PrbC10’s increase in affinity. B) Reversion of each mutation within the affinity matured PdarC1 clone to its Pdar identity indicates that Asn34Asp is responsible for most of PdarC1’s increase in affinity. C) The complex of Prb(Asn83Asp) and Pdar(Asp34Asn) binds with Kd of 180 pM, an increase in affinity of approximately 720-fold through only two mutations.
We next performed directed evolution of the Pdar protein. Given multiple reports of AR proteins selected for binding to a variety of target proteins for which they initially had no natural affinity (Steiner et al., 2008), we wished to exclude the possibility that affinity maturation of Pdar would induce it to bind to a new surface of Prb. We therefore used a negative- and positive-selection strategy to affinity mature Pdar for binding to low concentrations of Prb, while discriminating against adherence to very high concentrations of the PH1109 scaffold protein (see Methods). These selections isolated several clones with core mutations that were predicted by RosettaDesign to destabilize the ankyrin repeat (e.g. Ala75Val, Ala108Val, and Ser141Leu), in addition to peripheral polar mutations. Despite these potentially destabilizing mutations, we observed that the computationally designed and affinity matured proteins are well-folded, and indeed quite thermostable, as measured by circular dichroism.
One of the affinity matured Pdar variants, termed PdarC1 (Asn34Asp, Ala76Thr, Thr103Ala, Gln113Arg), binds Prb with a Kd of 1.3 nM (Figure 5b, Supplemental Table S2). Reversion mutant analysis of PdarC1 suggested that the Asn34Asp mutation was responsible for the majority of the improved affinity for Prb (Figure 5b, Supplemental Table S2). Furthermore, several other clones isolated from the Pdar affinity maturation harbor the Pdar(Asn34Asp) mutation, and introducing this single mutation into Pdar improved affinity approximately twenty-five-fold over the original design (Figure 5c, Supplemental Table S2). Pairing both minimal clones, Prb(Asp83Asn) and Pdar(Asn34Asp), yielded a complex with a dissociation constant of approximately 180 pM (Figure 5c, Supplemental Table S2). Thus, affinity maturation led to a final synthetic complex with binding approximately 1000-fold tighter than the 130 nM dissociation constant of the original Prb and Pdar complex.
Structural Characterization of an Affinity-Matured Prb-Pdar Variant
Neither the Prb(Asp83Asn) nor Pdar(Asn34Asp) mutations make direct interactions within the design model of the Prb-Pdar complex, complicating understanding of their effects. To shed light on the role of these mutations, we attempted to solve crystal structures of several binary complexes between Prb and Pdar variants. Though no crystallization conditions were found for any binary complexes, ternary complexes containing coenzyme A (CoA) were solved for PrbC5-CoA-Pdar and PrbC10(Lys135Glu)-CoA-Pdar to 2.3 Å and 2.0 Å, respectively (Supplemental Table S3; Prb is based on a CoA-binding protein and hence coenzyme A was included in an attempt to increase the stability of Prb in crystallization trials). Notably, these crystals did not form in the absence of CoA, but the addition of CoA has no effect upon the affinity of the Prb-Pdar complex (Supplemental Table S2). The structures of PrbC5-CoA-Pdar and PrbC10-CoA-Pdar are nearly identical apart from sidechains at which the sequences differ (Supplemental Figure S3), and so we will focus discussion on the higher-resolution crystal structure of the PrbC10-CoA-Pdar complex.
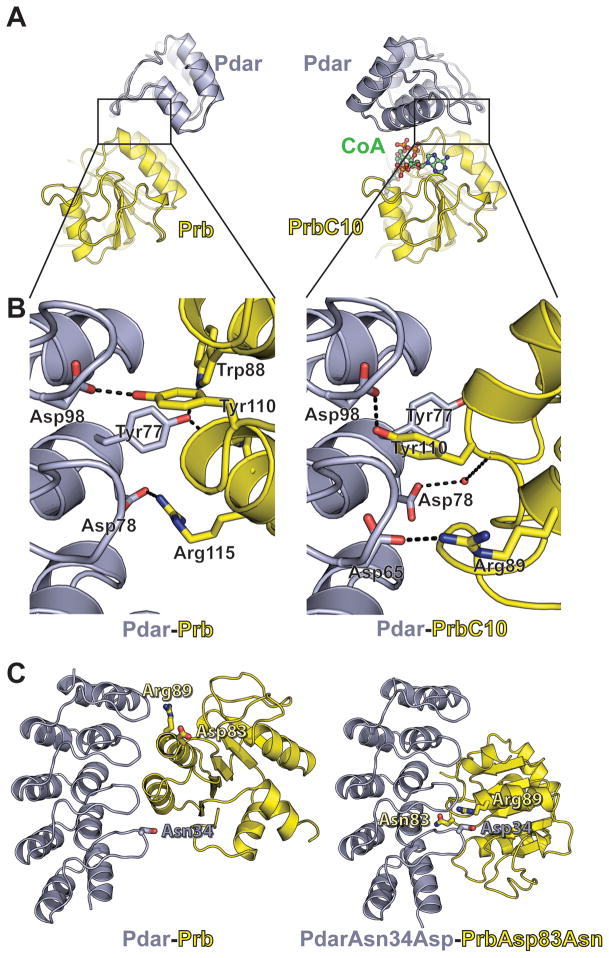
Surprisingly, within the structure of PrbC10-CoA-Pdar, PrbC10 is rotated 180° relative to the designed Prb model, about an axis in line with the Prb(Tyr110) motif that preserves the central designed Prb(Tyr110)-Pdar(Asp98) hydrogen bond (Figure 6a–c). This places Prb(Asp83Asn) within the periphery of the interface, and Pdar(Asn34Asp) within a few angstroms of Prb(Arg89) (Figure 6c). Despite this major difference in orientation, both designed surfaces of PrbC10 and Pdar interact with one another, and the interface is entirely composed of designed residues. Indeed, the designed motifs are central to each interface (Figure 6b), confirming that the computationally encoded sites of high interaction density are central to the complex.
Figure 6. Structural characterization of Prb and a complex evolved from Prb-Pdar.
A) The crystal structure of Prb bound to evolved clone 10 (PrbC10, yellow) bound to coenzyme A (green) and Pdar (gray, right) is rotated 180 degrees relative to the designed Prb (yellow) bound to Pdar (gray, left). B) The evolved interface makes several interactions symmetric to those designed for Prb-Pdar. Pdar is shown in equivalent orientations, highlighting that the complex in the crystal structure is rotated 180 degrees relative to the design model, such that a Prb(Tyr110)/Pdar(Asp98) hydrogen bond is maintained. This generates several similar interactions, such as the Prb(Arg115)/Pdar(Asp78) and PrbC10(Arg89)/Pdar(Asp65) salt bridges. C) The rotated orientation places PrbC10(Arg89) adjacent to Pdar(Asn34), which is mutated to an aspartate in several affinity-matured Pdar variants. Prb(Asp83), which is mutated to asparagine in PrbC10 is brought towards the center of the interface in the rotated orientation. The model of Pdar(Asn34Asp) paired with Prb(Asp83Asn) was generated by computationally mutating relevant amino acids within the Pdar-PrbC10 crystal structure, followed by repacking and energy minimization.
Notwithstanding the unanticipated change in orientation, many of the designed types of interactions are preserved in the rotated crystal structure (Figure 6b). For example, the central PrbC10(Tyr110) motif makes a hydrogen bond to OD2 of Pdar(Asp98), as in the design model, but from the opposite side of the oxygen. The other designed motif residue, Pdar(Trp102), also donates a hydrogen bond to its intended acceptor, Pdar(Asp131), and adopts the intended orientation in order to form extensive intermolecular contacts. As both interfaces are centered on Pdar(Asp98), which extends down from the “fingers” of the AR, both interfaces are capped from the bottom by Pdar(Tyr77). Similarly, Prb(Arg115) forms a salt bridge with Pdar(Asp78) in the design model, while PrbC10(Arg89) forms analogous interactions with Pdar(Asp65) and Pdar(Ser66) in the crystal structure, with Pdar(Asp78) instead forming a water-mediated hydrogen bond to the PrbC10 backbone (Figure 6b).
Because we were unable to obtain a crystal structure of the Prb-Pdar complex prior to affinity maturation, it is unclear whether the original designed complex has the orientation of the design model or of the rotated crystal structure. Since both orientations interact using the same set of residues and with very similar types of interactions, mutants of Prb-Pdar (including those described earlier, Figure 3d) would be expected to affect binding affinity in a similar manner regardless of the orientation.
As noted earlier, in the crystal structure the Prb(Tyr110) motif approaches its Pdar(Asp98) hydrogen bond partner from the opposite side of the oxygen; this is achieved via deeper burial of the Prb(Tyr110) sidechain in the Pdar binding groove (Figure 6b). Despite this difference, the quality of packing around this residue is equally good in both the design model and the crystal structure, whether assessed via interaction energy or other complementary metrics (Sheffler and Baker, 2009). The closer approach of Prb to Pdar leads to an increase in solvent accessible surface area buried by the interface, but the lack of additional strong interactions leads to a decrease in the overall interaction density of the interface in the crystal structure relative to the design model (Supplemental Table S1). The large contribution of charged residue mutations during affinity-maturation (Prb(Asp83Asn) and Pdar(Asn34Asp), (Figure 5c)) and the high salt sensitivity of the binding free energy (results not shown) suggests that electrostatic interactions contribute to the affinity and perhaps the orientational specificity within this synthetic protein-protein interface. Difficulties in accurately modeling backbone movement and modeling electrostatic interactions may have hindered the specification of a unique binding mode.
Discussion
Computational Design and Experimental Optimization of a de novo Protein-Protein Complex
To date, most efforts to study protein-protein interfaces have analyzed natural protein interactions to better understand either a specific pathway or the general properties of protein association. Building on the current knowledge of natural protein complexes, we sought to instead create an artificial protein complex starting with computational design.
Our computational design methodology is akin to in silico co-evolution of a protein complex, in that both sides of the interface are allowed to mutate to promote association. In contrast to standard RosettaDesign, which has proven useful in designing hydrophobic cores for monomeric proteins (Dantas et al., 2007; Dantas et al., 2003; Kuhlman et al., 2003), our method focuses on the unique requirements and characteristics of a dissociable protein-protein interface. From a given orientation of two potential partner proteins, the method begins by placing residues to make ideal interfacial interactions, then building outward to add a hydrophobic layer and finally a periphery. Instead of grafting hot-spot residues from existing natural complexes, we constrain search space by flexibly modeling buried hydrogen bonds, which are a general class of hot-spot interaction. We then rigorously build and filter these hot-spot interactions in a physics-based forcefield. In this way, the computational method recreates the composition of natural protein interfaces within the context of a de novo synthetic interface.
Starting only from the structures of two starting “scaffold” proteins, we computationally generated a de novo complex with a dissociation constant of approximately 100 nM. Each scaffold protein harbors approximately 20 mutations, and we have named the designed proteins Prb and Pdar. The Prb-Pdar complex bears no structural resemblance to any natural interface described to date, and the interacting surface of each partner protein has been completely redesigned. Though computationally designed, the Prb-Pdar complex captures important features of natural protein interfaces including binding kinetics typical of naturally occurring protein complexes (Horn et al., 2009; Schlosshauer and Baker, 2004). This suggests that our computational methodology contains many of the components necessary to encode affinity and can design protein complexes similar to those found in nature. Indeed, computationally-designed complexes are indistinguishable from natural complexes by a wide variety of metrics, including shape complementarity, packing, electrostatics, solvation, and sidechain entropy. The fact that approximately half of our designed complexes did not show detectable binding but were indistinguishable from natural interfaces by commonly-used discriminatory metrics implies unidentified important factors that are critical for affinity. We anticipate that the models of these non-binding designed complexes will serve as a useful benchmark in assessing methods for selecting correct complexes produced by protein docking.
We further explored the interaction of Prb and Pdar by using affinity maturation to isolate mutants of either protein that improve affinity. Directed evolution of either Prb or Pdar separately improved the dissociation constant to approximately 1–4 nM. In both cases, a single mutation is responsible for most of the increase in affinity. Pairing these two point mutants, Prb(Asp83Asn) and Pdar(Asn34Asp), yields a protein-protein complex with a dissociation constant of 180 pM. Compared with any previous computationally-designed de novo protein-protein interaction, the affinities of the initial Prb-Pdar complex and the affinity-matured double mutant are tighter by at least 770- and 560,000-fold, respectively. The synergy between computational design and experimental optimization can therefore be extremely powerful in the de novo creation and optimization of a high-affinity protein interface.
Binding Characteristics of a Synthetic Protein Complex
Prb and Pdar associate and dissociate with one another with rates that lie within the range of those observed for naturally evolved protein complexes. This need not have been the case, since the same dissociation constant could have been obtained with an unnaturally fast on-rate coupled to a fast off-rate, or a slow on-rate coupled to a slow off-rate. Kinetic considerations were not included in the computational design process, which focused exclusively on interface stability. The observation of a designed association rate similar to naturally occurring association rates suggests that in most natural interfaces there has been little evolutionary optimization of association kinetics otherwise natural protein complexes would associate faster than our designed complex. Although some protein-protein complexes do display unusually fast on rates, the creation of a computationally-designed complex with a “normal” on-rate in the absence of explicit kinetic design considerations is consistent with simple diffusional models of protein-protein association (Horn et al., 2009; Schlosshauer and Baker, 2004).
The Orientational Specificity of Protein Complexes
In vitro evolved proteins are frequently found to be promiscuous, exhibiting a wide range of cross-reactivity (Aharoni et al., 2005; Bridgham et al., 2006; Collins et al., 2006; James and Tawfik, 2003; Khersonsky et al., 2006). However, since in vitro evolution methods do not specify the precise arrangement of a synthetic complex, there has been no predictive experiment relating synthetic protein design to orientational plasticity within a single protein-protein interaction pair. We have computationally designed a protein-protein complex that binds with ~130 nM affinity and used directed evolution to identify variants with ~720-fold improved affinity, but discovered that the variants formed a complex in a binding mode that was symmetrically rotated relative to the design model. It is unclear whether this unexpected binding mode emerged as a result of affinity maturation or was indeed the orientation in which the original Prb-Pdar complex interaction was characterized. Intriguingly, the observed rotation occurs about the central, computationally-encoded hot spot motif, and many of the peripheral designed residues make similar types of contacts in the original model and the structure of the affinity matured variant. This may echo naturally evolved protein interfaces, in which central hot spots contribute most of a complex’s binding affinity, while the periphery is mostly energetically neutral.
Collectively, our results indicate that rational design coupled with directed evolution can be highly successful in the de novo introduction of binding affinity, but faces considerable challenges in engineering selectivity. Our computational design protocol is capable of building relatively high affinity de novo protein complexes that exhibit no detectable binding to the scaffold proteins used in the design process. However, we learn that, surprisingly, the challenge of selectivity may extend past that of cross-reactivity between proteins, to alternate orientations within a single complex. This suggests an unexpected generality to lessons learned in the context of designed coiled coils, which can adopt either parallel or antiparallel orientations in a variety of oligomeric states. In the case of coiled-coils, a predictive understanding of the determinants of coiled coil orientation (Apgar et al., 2008) and the factors that lead to cross-reactivity between proteins (Grigoryan and Keating, 2006) together enabled designed interactions that proved selective against a broad range of undesired partners (Grigoryan et al., 2009).
Nature may itself take advantage of ambiguous orientational selectivity in the evolution of natural interfaces. In certain cases, complexes between pairs of related proteins have been found to interact via different sets of contacts (Aloy et al., 2003). Within the CheA-CheY signaling complex, sequence differences between the structurally equivalent E. coli and T. maratima proteins lead to a 90° rotation of the complex around a central aromatic residue reminiscent of Prb(Tyr110) (Park et al., 2004). The Cc-CcP electron transfer complex is so plastic that a single conservative point mutation at the interface can yield extreme differences in orientation (Kang and Crane, 2005). Finally, recent evidence suggests that heterogeneous binding modes of a complex may even exist without mutation, as in the case of Ein-HPr, which samples a 180° flipped orientation that has only a 2 kcal/mol difference to the solved crystal structure (Yu et al., 2009). These examples highlight the fact that even in natural complexes, specificity can be extremely fine-grained, with alternate orientations lying very near canonical crystallographically-observed states in both evolutionary and energetic space.
Conclusions
The computational design of a high affinity, protein-protein complex represents a substantial advance in the rational design of protein interfaces. Our method combining de novo computational design with in vitro affinity maturation is generally applicable to the creation of protein interfaces with arbitrary topologies. The Prb-Pdar complex is, to the best of our knowledge, the first synthetic protein complex that exhibits the high affinity and hotspot-based interactions characteristic of natural protein complexes. In this way, the Prb-Pdar complex highlights the considerations that nature uses in building protein complexes, suggesting that regions of high interaction density are useful in achieving tight binding. However, the observation of a symmetric rotation about the computationally designed hotspot in the affinity matured variant suggests that central interaction density may not be sufficient to enforce a unique orientation. Achieving stringent specificity for a single protein pairing is a significant challenge that may require negative selection, which nature achieves via the evolutionary cost associated with off-target interactions. Since binding orientation is not expected to be a critical functional constraint in many examples of protein binding events, such as carrier-cargo interactions, we surmise that the existence of a single bound arrangement could arise indirectly from optimization to eliminate off-target interactions. Computationally, specificity is best achieved through explicit negative design against a large set of representative off-target proteins, if a suitable set can be identified (Grigoryan et al., 2009). Our results additionally emphasize the importance of including alternative binding arrangements among the set of off-target states. Finally, we note that the successful integration of directed evolution and the computational design process introduced here paves the way for the rational design and evolution of novel high affinity protein-based ligands for natural protein targets, with clear potential for diagnostic and therapeutic applications.
Methods
A complete description of methods are available as supplemental materials.
Supplementary Material
Acknowledgments
We thank Sarel Fleishman for valuable discussions, and Srayanta Mukherjee for assistance using TM-align and MM-align. This work was supported by NIH grant P41 RR011823 (DB), NIH/NIGMS R01 GM065400 (DL), and the Howard Hughes Medical Institute (DB and DL). JK was supported by the Damon Runyon Cancer Research Foundation and the Alfred P. Sloan Fellowship. JEC was supported by the Jane Coffin Childs Memorial Fund. IC was supported by the Ruth L. Kirschstein National Research Service Award. LJ was supported by an NIH Molecular Biophysics Training Grant. NMR studies were carried out as a Community Outreach activity of the NIH Protein Structure Initiative (PSI) Northeast Structural Genomics Consortium (U54-GM094597).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aharoni A, Gaidukov L, Khersonsky O, Mc QGS, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Apgar JR, Gutwin KN, Keating AE. Predicting helix orientation for coiled-coil dimers. Proteins. 2008;72:1048–1065. doi: 10.1002/prot.22118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1681/01.asn.0000926836.46869.e5. [DOI] [PubMed] [Google Scholar]
- Clackson T, Ultsch MH, Wells JA, de Vos AM. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J Mol Biol. 1998;277:1111–1128. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- Clackson T, Wells JA. A Hot-Spot of Binding-Energy in a Hormone-Receptor Interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Collins CH, Leadbetter JR, Arnold FH. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotechnol. 2006;24:708–712. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- Dantas G, Corrent C, Reichow SL, Havranek JJ, Eletr ZM, Isern NG, Kuhlman B, Varani G, Merritt EA, Baker D. High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J Mol Biol. 2007;366:1209–1221. doi: 10.1016/j.jmb.2006.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: Folding and stability of nine completely redesigned globular proteins. J Mol Biol. 2003;332:449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- DeLano WL. PyMol. DeLano Scientific; San Carlos, CA: 2004. [Google Scholar]
- Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JC. Optimizing protein stability in vivo. Mol Cell. 2009;36:861–871. doi: 10.1016/j.molcel.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. J Mol Biol. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guharoy MaPC. Conservation and relative importance of residues across protein-protein interfaces. Proc Natl Acad Sci U S A. 2005;102:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn JR, Sosnick TR, Kossiakoff AA. Principal determinants leading to transition state formation of a protein-protein complex, orientation trumps side-chain interactions. Proc Natl Acad Sci U S A. 2009;106:2559–2564. doi: 10.1073/pnas.0809800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PS, Love JJ, Mayo SL. A de novo designed protein protein interface. Protein Sci. 2007;16:2770–2774. doi: 10.1110/ps.073125207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Tawfik DS. Conformational diversity and protein evolution--a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- Jha RK, Leaver-Fay A, Yin S, Wu Y, Butterfoss GL, Szyperski T, Dokholyan NV, Kuhlman B. Computational Design of a PAK1 Binding Protein. J Mol Biol. 2010;400:257–270. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SA, Crane BR. Effects of interface mutations on association modes and electron-transfer rates between proteins. Proc Natl Acad Sci U S A. 2005;102:15465–15470. doi: 10.1073/pnas.0505176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Kohl A, Amstutz P, Parizek P, Binz HK, Briand C, Capitani G, Forrer P, Pluckthun A, Grutter MG. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure. 2005;13:1131–1141. doi: 10.1016/j.str.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Keskin O, Ma BY, Nussinov R, Liang J. Protein-protein interactions: Hot spots and structurally conserved residues often locate in complemented pockets that pre-organized in the unbound states: Implications for docking. J Mol Biol. 2004;344:781–795. doi: 10.1016/j.jmb.2004.09.051. [DOI] [PubMed] [Google Scholar]
- London N, Schueler-Furman O. Funnel hunting in a rough terrain: Learning and discriminating native energy funnels. Structure. 2008;16:269–279. doi: 10.1016/j.str.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Mandell DJ, Kortemme T. Backbone flexibility in computational protein design. Curr Opin Biotechnol. 2009a;20:420–428. doi: 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nat Chem Biol. 2009b;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- Moreira IS, Fernandes PA, Ramos MJ. Computational alanine scanning mutagenesis--an improved methodological approach. J Comput Chem. 2007;28:644–654. doi: 10.1002/jcc.20566. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Zhang Y. MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming. Nucleic Acids Res. 2009;37:e83. doi: 10.1093/nar/gkp318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Beel BD, Simon MI, Bilwes AM, Crane BR. In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc Natl Acad Sci U S A. 2004;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc Natl Acad Sci USA. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132:6025–6031. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosshauer M, Baker D. Realistic protein-protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 2004;13:1660–1669. doi: 10.1110/ps.03517304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Polak V, Shatsky M, Halperin I, Benyamini H, Barzilai A, Dror O, Haspel N, Nussinov R, et al. Taking geometry to its edge: fast unbound rigid (and hinge-bent) docking. Proteins. 2003;52:107–112. doi: 10.1002/prot.10397. [DOI] [PubMed] [Google Scholar]
- Sheffler W, Baker D. RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18:229–239. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman JM, Mayo SL. Modulating calmodulin binding specificity through computational protein design. J Mol Biol. 2002;323:417–423. doi: 10.1016/s0022-2836(02)00881-1. [DOI] [PubMed] [Google Scholar]
- Steiner D, Forrer P, Pluckthun A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J Mol Biol. 2008;382:1211–1227. doi: 10.1016/j.jmb.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Xu L, Aha P, Gu K, Kuimelis RG, Kurz M, Lam T, Lim AC, Liu H, Lohse PA, Sun L, et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- Yu D, Volkov AN, Tang C. Characterizing dynamic protein-protein interactions using differentially scaled paramagnetic relaxation enhancement. J Am Chem Soc. 2009;131:17291–17297. doi: 10.1021/ja906673c. [DOI] [PubMed] [Google Scholar]
- Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.