Abstract
Gonadotrophin-releasing hormone (GnRH) neurones control the onset and maintenance of fertility. Aberrant development of the GnRH system underlies infertility in Kallmann syndrome [KS; idiopathic hypogonadotropic hypogonadism (IHH) and anosmia]. Some KS patients harbour mutations in the fibroblast growth factor receptor 1 (Fgfr1) and Fgf8 genes. The biological significance of these two genes in GnRH neuronal development was corroborated by the observation that GnRH neurones were severely reduced in newborn transgenic mice deficient in either gene. In the present study, we hypothesised that the compound deficiency of Fgf8 and its cognate receptors, Fgfr1 and Fgfr3, may lead to more deleterious effects on the GnRH system, thereby resulting in a more severe reproductive phenotype in patients harbouring these mutations. This hypothesis was tested by counting the number of GnRH neurones in adult transgenic mice with digenic heterozygous mutations in Fgfr1/Fgf8, Fgfr3/Fgf8 or Fgfr1/Fgfr3. Monogenic heterozygous mutations in Fgfr1, Fgf8 or Fgfr3 caused a 30–50% decrease in the total number of GnRH neurones. Interestingly, mice with digenic mutations in Fgfr1/Fgf8 showed a greater decrease in GnRH neurones compared to mice with a heterozygous defect in the Fgfr1 or Fgf8 alone. This compounding effect was not detected in mice with digenic heterozygous mutations in Fgfr3/Fgf8 or Fgfr1/Fgfr3. These results support the hypothesis that IHH/KS patients with digenic mutations in Fgfr1/Fgf8 may have a further reduction in the GnRH neuronal population compared to patients harbouring monogenic haploid mutations in Fgfr1 or Fgf8. Because only Fgfr1/Fgf8 compound deficiency leads to greater GnRH system defect, this also suggests that these fibroblast growth factor signalling components interact in a highly specific fashion to support GnRH neuronal development.
Keywords: fibroblast growth factor receptor, fibroblast growth factor 8, gonadotrophin-releasing hormone neurones, hypogonadotropic hypogonadism, Kallmann syndrome
Centrally, reproductive fitness is determined by gonadotrophin-releasing hormone (GnRH) neurones, which are localised in the preoptic area and hypothalamus (1–3). Disruption of GnRH neuronal development results in severe clinical consequences that are primarily associated with reproductive failure. In humans, GnRH system deficiency is a hallmark of Kallmann syndrome (KS), which is a form of idiopathic hypogonadotropic hypogonadism (IHH) associated with anosmia. As expected, KS/IHH patients have nondetectable to low gonadotrophin and gonadal steroid hormone levels, ultimately resulting in the absence of pubertal onset and reproductive failure.
To date, KS has been causally linked to loss-of-function mutations in six genes: anosmin-1, fibroblast growth factor receptor 1 (Fgfr1), fibroblast growth factor 8 (Fgf8), prokineticin receptor-2, prokineticin-2 or Chd-7 (4–10). Of these gene mutations, the loss-of-function mutations in Fgf genes document the deleterious impact of fibroblast growth factor (FGF) signalling deficiency on the GnRH system and reproductive performance (11–13). Recently, we demonstrated that fibroblast growth factor receptor (FGFR) 1 is the FGF receptor especially critical for mouse GnRH neuronal development (11). Fgfr1 hypomorphic mice, which express 30–60% lower levels of functional Fgfr1 mRNA (14), showed approximately 88% reduction in the total number of GnRH neurones compared to wild-type littermates (11). By contrast, the GnRH neuronal system was unaffected in newborn Fgfr3 knockout mice (11). Furthermore, studies showed that Fgf8 hypomorphic mice, which exhibit a 65% reduction in Fgf8 mRNA levels, suffered a loss in GnRH neurones similar to Fgfr1 hypomorphs (11). The convergence of phenotype between Fgf8 and Fgfr1 hypomorphs suggests that FGFR1, not FGFR3, is the cognate receptor mediating the actions of FGF8 in the genesis of GnRH neurones (11).
A fraction of KS/IHH patients was shown to harbour heterozygous loss-of-function mutations in multiple genes (9, 15). For example, a proband who harboured digenic heterozygous mutations in Fgfr1 and Fgf8 was diagnosed with absent puberty (IHH). Of interest, the proband's father, who carried a monogenic heterozygous Fgfr1 mutation, exhibited only delayed puberty (9). The most likely explanation for this observation is that the digenic presence of heterozygous mutations in Fgfr1 and Fgf8 are additive during development, thereby impacting the GnRH neuronal system more deleteriously. To test this, we examined the GnRH system in transgenic mice harbouring several Fgf deficiencies. First, we examined whether the GnRH neuronal system is compromised in transgenic mice with monogenic heterozygous mutations in Fgfr1, Fgfr3 or Fgf8. Second, we examined whether mice harbouring digenic heterozygous mutations in Fgfr1/Fgf8, Fgfr3/Fgf8 or Fgfr1/Fgfr3 exhibited compound deleterious effects on the GnRH neuronal system. The results obtained in the present study should allow us to more clearly define individual and additive roles of each of these three FGF signalling components in the development and maintenance of the GnRH system.
Materials and methods
Transgenic animals and crossbreeding
Fgfr1 hypomorphs (129sv/CD-1; obtained from Canadian Mutant Mouse Repository, Toronto, Ontario, Canada), Fgfr3 knockout mice (Swiss-Webster/C57Bl/6; obtained from Jackson Laboratories, Bar Harbor, ME, USA) and Fgf8 hypomorphs (129P2/OlaHsd*CD-1; obtained from Mouse Regional Resource Centers, Davis, CA, USA) (14, 16, 17) were housed in our animal facility under a 12 : 12 h light/dark cycle and fed ad libitum. Initially, transgenic mice were bred through heterozygote : heterozygote mating to obtain: wild-type and heterozygous (+/−) Fgfr1 hypomorphic mice, Fgfr3 knockout mice or Fgf8 hypomorphic mice. Subsequently, Fgfr1 +/− were crossbred with Fgf8 +/− mice to generate F1 mice offspring harbouring digenic heterozygous mutations in Fgfr1 and Fgf8 (Fgfr1 +/− / Fgf8 +/−). Similarly, we crossbred Fgfr3 +/− mice with Fgf8 +/− mice, and Fgfr1 +/− mice with Fgfr3 +/− mice, to yield F1 mice with digenic heterozygous mutations in Fgfr3 and Fgf8 (Fgfr3 +/− / Fgf8 +/−)or Fgfr1 and Fgfr3 (Fgfr1 +/− / Fgfr3 +/−). Within each crossbreeding, wild-type, Fgfr1 +/−, Fgfr3 +/− or Fgf8 +/− mice were also generated. All comparisons were made among siblings (wild-type, monogenic, digenic) with the same hybrid genetic background to minimise the effect on the GnRH system as a result of strain differences. A polymerase chain reaction was performed on genomic DNA isolated from tail biopsies to identify genotype. All animal procedures complied with protocols as approved by the Institutional Animal Care and Use Committee at the University of Colorado.
GnRH immunocytochemistry
Adult mouse brains (2 months of age) were immersion-fixed in 4% paraformaldehyde/0.1 m phosphate buffer for 24 h at 4 °C. GnRH neurones were detected using the rabbit anti-GnRH polyclonal antibody LR5 (generously provide by Dr R. Benoit, Montreal General Hospital, Quebec, Canada) as previously described (11). Briefly, serial coronal sections of adult brains (40 μm) were incubated with LR-5 (1 : 10 000) diluted in 0.1 m phosphate buffered-saline/0.4% triton-X containing 4% normal donkey serum and 10% normal horse serum for 4 days at 4 °C. Subsequently, sections were incubated with a biotinylated-donkey anti-rabbit antibody (dilution 1 : 500; Jackson Laboratories, West Grove, PA, USA), avidin and biotin-coupled horseradish peroxidase (ABC kit; Vector Laboratories, Burlingame, CA, USA), and reacted with 0.05% 3,3′-diaminobenzidine/0.01% H2O2. Sections were mounted on gelatin-coated glass slides, dehydrated in increasing concentrations of ethanol, delipified with Histoclear (Fisher Scientific, Fair Lawn, NJ, USA), and covers-lipped with Permount (Fisher Scientific).
GnRH neuronal counts and statistical analysis
The number of GnRH-immunoreactive (IR) neurones was specifically quantified in serial coronal 40-μm sections from the anterior preoptic region to the posterior hypothalamus. Although this analysis did not include the rostral brain regions that are in the direct vicinity of the olfactory bulbs, the population of GnRH neurones residing within the regions quantified encompassed the vast majority of the mouse GnRH neuronal system. All slides were coded and scored by an individual who was blind to their identities. A GnRH-IR neurone was scored positive only if it also showed a clear cell nucleus. GnRH neuronal counts from both males and females were pooled in our analysis because the number of GnRH-IR neurones did not differ between the sexes in our hands. To analyse the distribution of GnRH neurones, the beginning of the organum vasculosum lamina terminalis (OVLT) was designated as section 0. Subsequently, 20 sections prior to and after section 0 were included in the analysis. One-way anova followed by the Student–Newman–Keuls test or Student's t-test was used to test for significant differences. P < 0.05 was considered statistically significant.
Results
GnRH system deficit in Fgfr1 +/−, Fgfr3 +/− and Fgf8 +/− mice
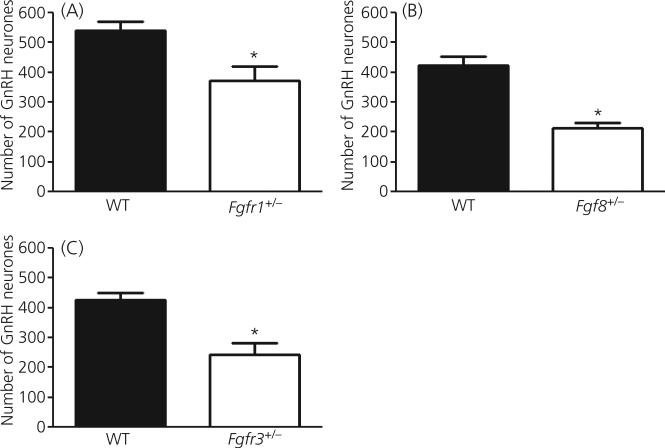
Student t-tests indicated that adult mice harbouring monogenic heterozygous mutations in Fgfr1, Fgfr3 or Fgf8 had significantly reduced GnRH-IR neurones in the preoptic/hypothalamus region compared to wild-type. The total number GnRH-IR neurones in Fgfr1 +/− mice (372 ± 48.4; n = 7) mice was significantly lower than in wild-type Fgfr1 mice (539 ± 30.1; n = 8; P = 0.046; Fig. 1). Similarly, Fgf8 +/− mice (214 ± 18.3; n = 4) have significantly less GnRH-IR neurones than wild-type Fgf8 mice (424 ± 29.1; n = 4; P < 0.01; Fig. 1b), which replicated our earlier findings (11). The number of GnRH-IR neurones was also significantly lower in Fgfr3 +/− mice (245 ± 36.8; n = 5) than in wild-type Fgfr3 mice (426 ± 24.9; n = 6; P = 0.01; Fig. 1c). No gross morphological aberrations in the brain of these transgenic animals were detected (data not shown). Moreover, no marked reduction in fertility was detected in adult Fgfr1 +/−, Fgfr3 +/− or Fgf8 +/− mice at 2 months of age (data not shown).
Fig. 1.
The number of gonadotrophin-releasing hormone (GnRH) neurones in the preoptic/hypothalamus region of mice with monogenic heterozygous defects in fibroblast growth factor signalling components. (a) Fgfr1 +/− mice (n = 7) have significantly fewer GnRH neurones than wild-type (WT) mice (n = 8; P = 0.046). (B) Fgf8 +/− mice (n = 4) have significantly fewer GnRH neurones than wild-type mice (n = 4; P < 0.02). (c) Fgfr3 +/− mice (n = 5) have significantly fewer GnRH neurones than wild-type mice (n = 6; P = 0.01). Asterisks represent significant differences. Each bar represents the mean ± SEM.
Exacerbation of GnRH system deficit in Fgfr1 +/− / Fgf8 +/− mice
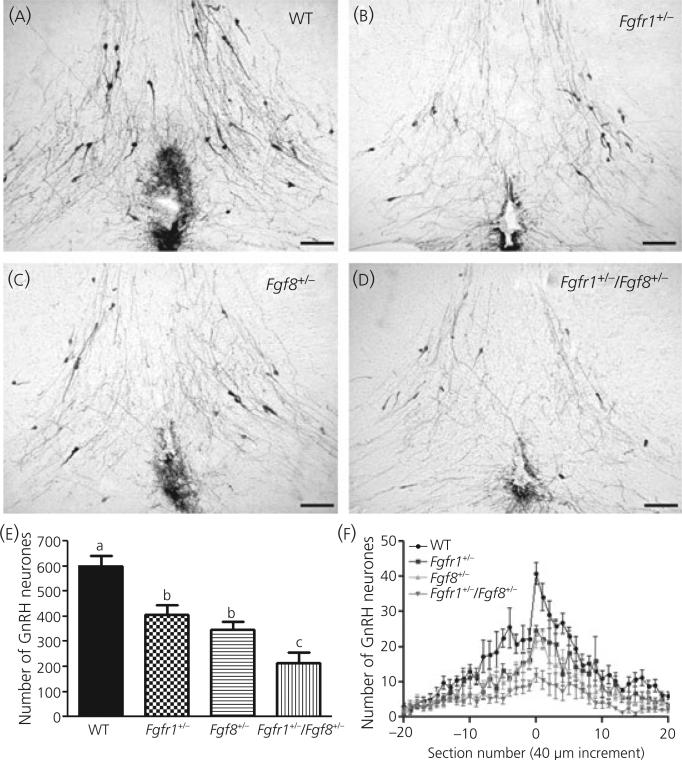
One-way anova showed that the total number of GnRH-IR neurones in the preoptic/hypothalamic region of mice with monogenic or digenic heterozygous mutations in Fgfr1 and/or Fgf8 was significantly different (i.e. higher or lower) among genotypes (i.e. wild-type, Fgfr1 +/−, Fgf8 +/− and Fgfr1 +/− / Fgf8 +/−; P < 0.0001; Fig. 2a–e). Post-hoc analysis showed that the number of GnRH-IR neurones was significantly lower in Fgfr1 +/− (407 ± 40.0; n = 5), Fgf8 +/− (347 ± 32.5; n = 7) or Fgfr1 +/− / Fgf8 +/− (215 ± 39.0; n = 5) mice compared to wild-type (601 ± 41.5; n = 7; Fig. 2e). Fgfr1+/− / Fgf8 +/− mice had more GnRH neuronal loss compared to Fgfr1 +/− or Fgf8 +/− mice (Fig. 2e). There was no significant difference between Fgfr1 +/− and Fgf8 +/− mice. No gross morphological aberrations in the brain of these transgenic animals were detected (data not shown). The distribution of GnRH neurones in the fore-brain in mice of all genotypes retained the basic pattern of having more GnRH neurones near the OVLT (Fig. 2f).
Fig. 2.
Gonadotrophin-releasing hormone (GnRH) system in Fgfr1 +/−, Fgf8 +/− or Fgfr1 +/− / Fgf8 +/− mice. Representative photomicrographs showing GnRH-immunoreactive (IR) neurones at the level of the organum vasculosum lamina terminalis (OVLT) of (a) wild-type (WT) (n = 7), (b) Fgfr1 +/− (n = 5), (c) Fgf8 +/− (n = 7) and (d) Fgfr1 +/− / Fgf8 +/− (n = 5) mice. Note that the number of GnRH-IR neurones is reduced in Fgfr1 +/−, Fgf8 +/− and Fgfr1 +/− / Fgf8 +/− mice compared to wild-type mice. Moreover, the reduction in the number of GnRH-IR neurones in Fgfr1 +/− / Fgf8 +/ mice is visually more prominent compared to Fgfr1 +/− or Fgf8 +/− mice. Scale bar = 100 μm. (e, f) Total number and distribution of GnRH neurones in mice deficient in Fgfr1 and/or Fgf8.(e) anova followed by post-hoc test showed the total number of GnRH-IR neurones in the preoptic/hypothalamus was significantly lower in Fgfr1 +/− (n = 5), Fgf8 +/− (n = 7) or Fgfr1 +/− / Fgf8 +/− (n = 5) mice than in wild-type (n = 7) mice (P < 0.0001 by anova). Moreover, the loss of GnRH neurones was greater in Fgfr1 +/− / Fgf8 +/− mice than in Fgfr1 +/− and Fgf8 +/− mice. (f) The rostral–caudal distribution of GnRH neurones in wild-type, Fgfr1 +/−, Fgf8 +/− and Fgfr1 +/− / Fgf8 +/− mice showed that mice of all genotypes retained the basic pattern of having more GnRH neurones near the OVLT. Each data point represents the mean ± SEM.
GnRH system deficit was not exacerbated in Fgfr3+/− / Fgf8+/− mice
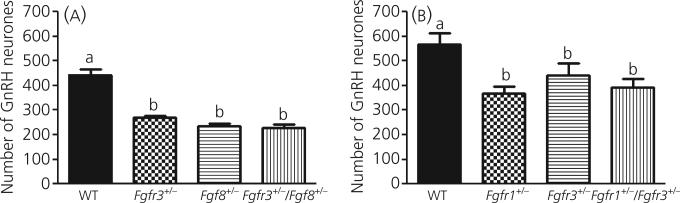
One-way anova showed that the total number of GnRH-IR neurones in the preoptic/hypothalamic region of mice with monogenic or digenic heterozygous mutations in Fgfr3 and/or Fgf8 was different among genotypes (P < 0.0001; Fig. 3a). Post-hoc analysis showed that the total number of GnRH-IR neurones in the preoptic/hypothalamic region was significantly lower in Fgfr3 +/− (270 ± 8.1; n = 6), Fgf8 +/− (234 ± 9.4; n = 5) and Fgfr3 +/− / Fgf8 +/− (227 16.3; n = 7) mice compared to wild-type (443 ± 21.9; n = 7; Fig. 3a). There were no differences among Fgfr3 +/−, Fgf8+/− and Fgfr3 +/− / Fgf8 +/− mice. No gross morphological aberrations in the brain of these transgenic animals were detected (data not shown).
Fig. 3.
The number of gonadotrophin-releasing hormone (GnRH) neurones in the preoptic/hypothalamus region of mice with digenic or monogenic heterozygous deficiencies in various fibroblast growth factor signalling components. (a) anova followed by post-hoc test showed GnRH neuronal number was significantly lower in Fgfr3 +/− (n = 6), Fgf8 +/− (n = 5) or Fgfr3 +/− / Fgf8 +/− (n = 7) mice compared to wild-type mice (WT) (n = 7; P < 0.0001 by anova). (b) GnRH neuronal number was significantly lower in Fgfr1 +/− (n = 6), Fgfr3 +/− (n = 5) or Fgfr1 +/− / Fgfr3 +/− (n = 5) mice compared to wild-type mice (n = 5; P < 0.0001 by anova). Different letters represent significant differences. Each bar represents the mean ± SEM.
GnRH system deficit was not exacerbated in Fgfr1 +/− / Fgfr3 +/− mice
One-way anova showed that the total number of GnRH-IR neurones in the preoptic/hypothalamic regions of mice with monogenic or digenic heterozygous mutations in Fgfr1 and/or Fgfr3 was different among genotypes (P < 0.05; Fig. 3b). Post-hoc analysis showed that the total number of GnRH-IR neurones in the preoptic/hypothalamic region was significantly lower in Fgfr1 +/− (367 ± 27.7; n = 6), Fgfr3 +/− (442 ± 47.9; n = 5) and Fgfr1 +/− / Fgfr3 +/− (394 ± 33.5; n = 5) mice compared to wild-type (566 ± 47.5; n = 5; Fig. 3b). There were no differences among Fgfr1 +/−, Fgfr3 +/− and Fgfr1 +/− / Fgfr3 +/− mice. No gross morphological aberrations in the brain of these transgenic animals were detected (data not shown).
Discussion
Two novel findings emerged from the present study. First, monogenic Fgfr3 haploinsufficiency reduced approximately one-half of the GnRH neuronal population in adult mice. Second, contrary to what one might be predicted, not all compound deficiencies in FGF signalling lead to more deleterious effects on the GnRH system than monogenic deficiencies alone. A greater GnRH system defect was seen only when Fgfr1 and Fgf8 deficiencies were combined. The latter finding supports the hypothesis that the presence of digenic mutation in Fgfr1 and Fgf8 may actually result in a further loss of GnRH neurones, ultimately contributing to the reported increase in the severity of reproductive symptoms (9, 15).
In the present study, we found that the number of GnRH neurones was higher in wild-type mice when they contained the 129sv/CD-1 background (Fgfr1 hypomorphic mice). This discrepancy is likely a result of the inherently higher number of GnRH neurones in this hybrid background. To minimise the effect on the GnRH system as a result of strain differences, we limited our comparisons between siblings (wild-type, monogenic, digenic) who shared similar hybrid genetic background. We acknowledge that even littermates may have variable genetic background in this hybrid breeding scheme but, given that the phenotypes associated with FGF signalling deficiency are highly penetrant (14, 16, 17), and that our data are consistent and statistically significant, it is unlikely that reduced GnRH neurones in FGF signalling-deficient mice resulted by chance simply from differences in their genetic background.
We previously showed that the GnRH system was unaffected in newborn homozygous Fgfr3 knockout mice (Fgfr3−/−) (11); thus, the current observation that adult Fgfr3 +/− mice have a significant reduction in the number of GnRH-IR neurones is unexpected. At present, complete data on the GnRH system of adult Fgfr3−/− and newborn Fgfr3 +/− mice are unavailable; thus, it is difficult to fully explain the role of Fgfr3. However, two possibilities exist. First, there may be an age-dependent decline of GnRH neurones in mice harbouring Fgfr3 deficiency. This decline is only manifested shortly after birth, and thus the diminished GnRH neuronal population was only seen in adults (i.e. present study) but not in newborn mice (9) deficient in Fgfr3 signalling. In this regard, Fgfr3 could be more involved in the postnatal survival and/or differentiation of GnRH neurones than their development. Supporting this hypothesis, transgenic mice with reduced FGF responsiveness in GnRH neurones (13) have been shown to undergo a postnatal age-dependent decline in the number of GnRH neurones (18). Second, when Fgfr3 is genetically ablated, as in the case of Fgfr3−/− mice, the expression of other Fgfrs such as Fgfr1 may developmentally up-regulate to compensate. This compensation could lead to an absence of GnRH system defect in Fgfr3−/− mice as observed previously (11). Regardless, the data obtained in the present study are the first to show that Fgfr3 haploinsufficiency significantly compromises the adult GnRH system. Fgfr3 thus becomes the third FGF signalling gene that, when mutant, leads to an aberrant GnRH system in the mouse (11, 13). These data provide a strong impetus for exploring the role of Fgfr3 as a candidate gene for IHH/KS.
A study in one X-linked KS foetus showed that GnRH neurones do not reach the preoptic/hypothalamus regions as a result of migratory failure (19). However, previous data indicated migratory failure is unlikely the cause for GnRH system defects in mice with Fgf8 and Fgfr1 deficiencies. Indeed, extensive analysis of the newborn nasal and brain region of Fgfr1−/− and Fgf8−/− hypomorphs did not detect aberrantly localised GnRH neurones (11). Furthermore, transgenic mice with reduced FGF responsiveness in GnRH neurones exhibited normal GnRH neuronal migration into the fore-brain (13). Instead, we showed that GnRH neurones simply did not emerge from the olfactory placode of Fgf8−/− hypomorphic embryos, suggesting Fgf8-dependent activation of Fgfr1 was required for the genesis of GnRH neurones (11). Together, these results strongly suggest that the reduced GnRH-IR neurones in Fgfr1 +/−, Fgf8 +/− or Fgfr1 +/− / Fgf8 +/− mice result from the developmental elimination of GnRH neurones rather than migratory failure.
In the present study, we found that approximately 60–70% of GnRH neurones are missing in the preoptic area/hypothalamus of Fgfr1- and/or Fgf8-deficient mice. It is unknown how this GnRH neuronal loss impacts the function of the postnatal mouse GnRH system, and whether GnRH neuronal loss in specific brain regions differentially affects reproductive function. Previous studies in rodents have shown that GnRH neurones at the level of the OVLT are especially important for the preovulatory luteinising hormone/follicle-stimualting hormone (LH/FSH) surge (20–22). Indeed, destruction of the OVLT, which results in a striking reduction in GnRH content in the OVLT and median eminence, also causes a dramatic attenuation of the preovulatory LH/FSH surge (23). In our laboratory, adult Fgfr1 +/− or Fgf8 +/− mice do not exhibit a marked reduction in fertility, although this does not necessarily indicate the preovulatory LH/FSH surge is not compromised. Currently, we are investigating whether the preovulatory LH/FSH surge in Fgfr1 +/−, Fgf8 +/−, and Fgfr1 +/− / Fgf8 +/− female mice exhibit marked deficits.
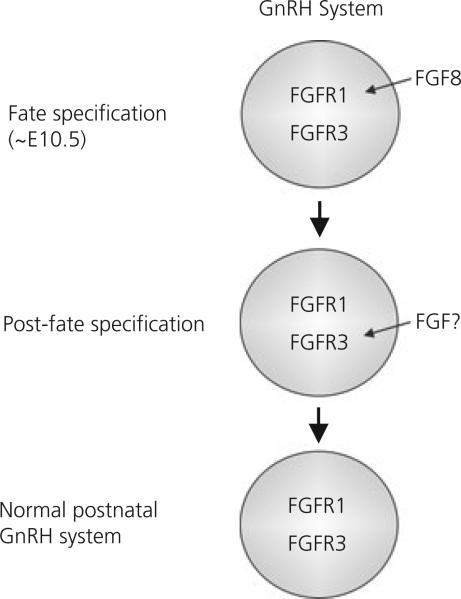
On the basis of our data, we have constructed a working model that may explain why only the Fgfr1/Fgf8 combination was effective in exacerbating GnRH system defects (Fig. 4). First, we hypothesise that the simultaneous presence of heterozygous deficiencies in Fgfr1 and Fgf8 impact the same GnRH precursor population at the same time to enhance the severity of GnRH neuronal fate specification defects, leading to additive effects when both are mutant. This hypothesis is well supported by our previous findings that Fgf8 acts through Fgfr1 to stimulate the genesis of GnRH neurones (11). Second, we hypothesise that Fgf8 and Fgfr3 deficiencies are not additive because they affect the same GnRH neuronal population at different times. In other words, Fgf8 deficiency obliterates a population of GnRH precursor cells that subsequently becomes Fgfr3-dependent after fate specification. Because this population is already lost when Fgf8 is deficient, an additional reduction in Fgfr3 signalling has no further effect. The second hypothesis is based strictly on our present observations and requires future validation. However, the second hypothesis is consistent with our present data suggesting a later involvement of Fgfr3 in the postnatal maintenance of the GnRH system. Similarly, Fgfr1 and Fgfr3 deficiencies are not additive because they may impact the same GnRH neuronal population at different times, with the former affecting GnRH precursors before fate specification and the latter after fate specification. This assumption again requires further validation. Overall, our model proposes that different combinations of Fgfr/Fgf exert different effects on the GnRH neuronal system in a sequential and temporally specific fashion. This specific nature of FGF action was supported by the findings that FGF ligands and receptors are sequentially expressed in the olfactory region during a developmental time frame consistent with the emergence and migration of GnRH neurones (24, 25).
Fig. 4.
Hypothetical model illustrating the temporal sequence in which fibroblast growth factor (FGF) signalling components may affect gonadotrophin-releasing hormone (GnRH) neuronal development. In this model, we hypothesise that Fgf8-dependent activation of fibroblast growth factor receptor (FGFR) 1 is required for GnRH neuronal fate specification [i.e. approximately embryonic day (E) 10.5]. After fate specification, the same population of GnRH neurones becomes dependent on Fgfr3 for maintenance (11). Thus, deficiencies in Fgfr1, Fgf8 or Fgfr3 alone could compromise the GnRH system by affecting genesis (Fgfr1 and Fgf8) or maintenance (Fgfr3). Fgfr3 mutation in addition to Fgfr1 or Fgf8 did not further worsen the GnRH system defect because the target population was already absent as a result of earlier fate specification failure. Conversely, because Fgfr1 and Fgf8 both act on the fate specification stage, their effects can be additive. At present, it is unclear which FGF ligand(s) is required for the activation of Fgfr3 at the post-fate specification stage. Cumulatively, the data obtained in the present study suggest that the proper development of the mouse GnRH system requires the intricate orchestration of multiple FGF/FGFR signals in a time-specific manner.
The olfactory placode, from where mouse GnRH neurones are derived, expresses Fgfr1, FGFR2 and Fgfr3 proteins (26). Nonetheless, previous studies have shown that fate specification of GnRH precursor cells requires Fgf8 signalling that is mediated by Fgfr1 instead of Fgfr3 (11). Fgf8-dependent Fgfr1 activation may enhance the survival rate of GnRH precursor cells. Indeed, previous studies have shown that FGFRs induce a myriad of intracellular signal transduction pathways including the PI3K/AKT pathway, which protects against apoptotic cell death (27, 28). Therefore, the loss of GnRH neurones in mice harbouring monogenic heterozygous Fgfr1/Fgf8 deficiencies may be the result of a decrease in cell survival, and this defect may be further enhanced in mice with digenic heterozygous Fgfr1/Fgf8 deficiencies. A similar mechanism may also cause the loss of GnRH neurones found in Fgfr3 deficient mice, albeit after GnRH neurones have become fate specified.
In conclusion, the present study has produced two major findings. First, Fgfr3 is important for the normal development and/or maintenance of the mouse GnRH neuronal population, possibly by exerting its effects after GnRH neuronal fate specification. Consequently, Fgfr3 is a candidate gene that may be involved in the pathogenesis of IHH/KS. Second, not all compound deficiencies in FGF signalling lead to the exacerbation of the deleterious effects on the GnRH system compared to monogenic deficiencies alone. Only the combination of Fgfr1 and Fgf8 deficiencies caused greater deleterious effects on the GnRH neuronal population. The latter finding illustrates the highly complex and specific nature in which FGF signalling components interact to promote the emergence and maintenance of the GnRH system.
Acknowledgements
This study was supported by NIH RO1 HD042634, the Endocrine Society Bridge Award to P.S.T and NIH K99HD058044 to W.C.J.C.
Footnotes
The authors declare that there are no conflicts of interest.
References
- 1.Livne I, Gibson MJ, Silverman AJ. Biochemical differentiation and intercellular interactions of migratory gonadotropin-releasing hormone (GnRH) cells in the mouse. Dev Biol. 1993;159:643–656. doi: 10.1006/dbio.1993.1271. [DOI] [PubMed] [Google Scholar]
- 2.Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 3.Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, Brown CJ, Willard HF, Lawrence C, Persico MG, Camerino G, Ballabio A. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 5.Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardelin JP, Julliard AK, Moniot B, Soussi-Yanicostas N, Verney C, Schwanzel-Fukuda M, Ayer-Le Lievre C, Petit C. Anosmin-1 is a regionally restricted component of basement membranes and interstitial matrices during organogenesis: implications for the developmental anomalies of X chromosome-linked Kallmann syndrome. Dev Dyn. 1999;215:26–44. doi: 10.1002/(SICI)1097-0177(199905)215:1<26::AID-DVDY4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 7.Dode C, Levilliers J, Dupont JM, De Paepe A, Le Du N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pecheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 8.Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falardeau J, Chung WCJ, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, Quinton R, Na S, Hall JE, Huot C, Alois N, Pearce SH, Cole LW, Hughes V, Mohammadi M, Tsai P, Pitteloud N. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 11.Chung WCJ, Moyle SS, Tsai PS. Fibroblast growth factor 8 signaling through fibroblast growth factor receptor 1 is required for the emergence of gonadotropin-releasing hormone neurons. Endocrinology. 2008;149:4997–5003. doi: 10.1210/en.2007-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill JC, Tsai PS. Expression of a dominant negative FGF receptor in developing GNRH1 neurons disrupts axon outgrowth and targeting to the median eminence. Biol Reprod. 2006;74:463–472. doi: 10.1095/biolreprod.105.046904. [DOI] [PubMed] [Google Scholar]
- 13.Tsai PS, Moenter SM, Postigo HR, El Majdoubi M, Pak TR, Gill JC, Paruthiyil S, Werner S, Weiner RI. Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol Endocrinol. 2005;19:225–236. doi: 10.1210/me.2004-0330. [DOI] [PubMed] [Google Scholar]
- 14.Partanen J, Schwartz L, Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev. 1998;12:2332–2344. doi: 10.1101/gad.12.15.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 17.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 18.Rochester JR, Chung WCJ, Tsai P-S. Restoration of the GnRH System in Aging Male Transgenic Animals Paired with Females. Society for Neuroscience; Chicago, IL: 2009. [Google Scholar]
- 19.Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Mol Brain Res. 1989;6:311–326. doi: 10.1016/0169-328x(89)90076-4. [DOI] [PubMed] [Google Scholar]
- 20.Zoeller RT, Young WS., III Changes in cellular levels of messenger ribonucleic acid encoding gonadotropin-releasing hormone in the anterior hypothalamus of female rats during the estrous cycle. Endocrinology. 1988;123:1688–1689. doi: 10.1210/endo-123-3-1688. [DOI] [PubMed] [Google Scholar]
- 21.Park OK, Gugneja S, Mayo KE. Gonadotropin-releasing hormone gene expression during the rat estrous cycle: effects of pentobarbital and ovarian steroids. Endocrinology. 1990;127:365–372. doi: 10.1210/endo-127-1-365. [DOI] [PubMed] [Google Scholar]
- 22.Porkka-Heiskanen T, Urban JH, Turek FW, Levine JE. Gene expression in a subpopulation of luteinizing hormone-releasing hormone (LHRH) neurons prior to the preovulatory gonadotropin surge. J Neurosci. 1994;14:5548–5558. doi: 10.1523/JNEUROSCI.14-09-05548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samson WK, McCann SM. Effects of lesions in the organum vasculosum lamina terminalis on the hypothalamic distribution of luteinizing hormone-releasing hormone and gonadotropin secretion in the ovariectomized rat. Endocrinology. 1979;105:939–946. doi: 10.1210/endo-105-4-939. [DOI] [PubMed] [Google Scholar]
- 24.Bachler M, Neubuser A. Expression of members of the Fgf family and their receptors during midfacial development. Mech Dev. 2001;100:313–316. doi: 10.1016/s0925-4773(00)00518-9. [DOI] [PubMed] [Google Scholar]
- 25.Ford-Perriss M, Abud H, Murphy M. Fibroblast growth factors in the developing central nervous system. Clin Exp Pharmacol Physiol. 2001;28:493–503. doi: 10.1046/j.1440-1681.2001.03477.x. [DOI] [PubMed] [Google Scholar]
- 26.Gill JC, Moenter SM, Tsai PS. Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology. 2004;145:3830–3839. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- 27.Boilly B, Vercoutter-Edouart AS, Hondermarck H, Nurcombe V, Le Bourhis X. FGF signals for cell proliferation and migration through different pathways. Cytokine Growth Factor Rev. 2000;11:295–302. doi: 10.1016/s1359-6101(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 28.Volosin M, Song W, Almeida RD, Kaplan DR, Hempstead BL, Friedman WJ. Interaction of survival and death signaling in basal forebrain neurons: roles of neurotrophins and proneurotrophins. J Neurosci. 2006;26:7756–7766. doi: 10.1523/JNEUROSCI.1560-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]