Abstract
Understanding the brain requires a broad range of approaches and methods from the domains of biology, psychology, chemistry, physics, and mathematics. The fundamental challenge is to decipher the “neural choreography” associated with complex behaviors and functions, including thoughts, memories, actions, and emotions. This demands the acquisition and integration of vast amounts of data of many types, at multiple scales in time and in space. Here, we discuss the need for neuroinformatics approaches to accelerate progress, using several illustrative examples. The nascent field of ‘connectomics’ aims to comprehensively describe neuronal connectivity at either a macroscopic level (long-distance pathways for the entire brain) or a microscopic level (axons, dendrites, synapses in a small brain region). The Neuroscience Information Framework encompasses all of neuroscience and facilitates integration of existing knowledge and databases of many types. These examples illustrate the opportunities and challenges of data mining across multiple tiers of neuroscience information and underscore the need for cultural and infrastructure changes if neuroinformatics is to fulfill its potential to advance our understanding of the brain.
Deciphering the workings of the brain is the domain of neuroscience, one of the most dynamic fields of modern biology. Over the last few decades, our knowledge about the nervous system has advanced at a remarkable pace. These advances are critical for understanding the mechanisms underlying the broad range of brain functions, from controlling breathing to forming complex thoughts. They are also essential for uncovering the causes of the vast array of brain disorders whose impact on humanity is staggering (1). To accelerate progress, it is vital to develop more powerful methods for capitalizing on the amount and diversity of experimental data generated in association with these discoveries.
The human brain contains ~80 billion neurons that communicate with each other via specialized connections or synapses (2). A typical adult brain has ~150 trillion synapses (3). The point of all this communication is to orchestrate brain activity. Each neuron is a piece of cellular machinery that relies on neurochemical and electrophysiological mechanisms to integrate complicated inputs and communicate information to other neurons. But no matter how accomplished, a single neuron can never perceive beauty, feel sadness or solve a mathematical problem. These capabilities emerge only when networks of neurons work together. Ensembles of brain cells, often quite far flung, form integrated neural circuits, and the activity of the network as a whole supports specific brain functions such as perception, cognition or emotions. Moreover, these circuits are not static. Environmental events trigger molecular mechanisms of “neuroplasticity” that alter the morphology and connectivity of brain cells. The strengths and pattern of synaptic connectivity encode the “software” of brain function. Experience, by inducing changes in that connectivity, can significantly alter the function of specific circuits during development and throughout the lifespan.
A grand challenge in neuroscience is to elucidate brain function in relation to its multiple layers of organization that operate at different spatial and temporal scales. Central to this effort is tackling “neural choreography” -- the integrated functioning of neurons into brain circuits--their spatial organization, local and long-distance connections, their temporal orchestration, and their dynamic features, including interactions with their glial cell partners. Neural choreography cannot be understood via a purely reductionist approach. Rather, it entails the convergent use of analytical and synthetic tools to gather, analyze and mine information from each level of analysis, and capture the emergence of new layers of function (or dysfunction) as we move from studying genes and proteins, to cells, circuits, thought, and behavior.
The Need for Neuroinformatics
The profoundly complex nature of the brain requires that neuroscientists use the full spectrum of tools available in modern biology - genetic, cellular, anatomical, electrophysiological, behavioral, evolutionary and computational. The experimental methods involve many spatial scales, from electron microscopy to whole brain human neuroimaging, and time scales ranging from microseconds for ion channel gating to years for longitudinal studies of human development and aging. An increasing number of insights emerge from integration and synthesisacross these spatial and temporal domains. However, such efforts face impediments related to the diversity of scientific subcultures and differing approaches to data acquisition, storage, description, and analysis and, even the language in which they are described. It is often unclear how best to integrate the linear information of genetic sequences, the highly visual data of neuroanatomy, the time-dependent data of electrophysiology, and the more global level of analyzing behavior and clinical syndromes.
The great majority of neuroscientists carry out highly focused, hypothesis-driven research that can be powerfully framed in the context of known circuits and functions. Such efforts are complemented by a growing number of projects that provide large datasets aimed not at testing a specific hypothesis but instead enabling data-intensive discovery approaches by the community at large. Notable successes include gene expression atlases from the Allen Institute for Brain Sciences (4) and the GENSAT project (5), and disease-specific human neuroimaging repositories (6). However, the neuroscience community is not yet fully engaged in exploiting the rich array of data currently available, nor is it adequately poised to capitalize on the forthcoming data explosion.
Below we highlight several major endeavors that provide complementary perspectives on the challenges and opportunities in neuroscience data mining. One is a set of “connectome” projects that aim to comprehensively describe neural circuits at either the macroscopic or the microscopic level. Another, the Neuroscience Information Framework (NIF), encompasses all of neuroscience and provides access to existing knowledge and databases of many types. These and other efforts provide fresh approaches to the challenge of elucidating neural choreography.
Connectomes – Macroscopic and Microscopic
Brain anatomy provides a fundamental three-dimensional framework around which many types of neuroscience data can be organized and mined. Decades of effort have revealed immense amounts of information about local and long-distance connections in animal brains. A wide range of tools (e.g. immunohistochemistry, in situ hybridization) have characterized the biochemical nature of these circuits that are studied electrophysiologically, pharmacologically and behaviorally (7). Several ongoing efforts aim to integrate anatomical information into searchable resources that provide a backbone for understanding circuit biology and function (8, 9, 10). The challenge of integrating such data will dramatically increase with the advent of high throughput anatomical methods, including those emerging from the nascent field of connectomics.
A ‘connectome’ is a comprehensive description of neural connectivity for a specified brain region at a specified spatial scale (11,12). Connectomics currently includes distinct subdomains for studying the macro-connectome (long-distance pathways linking patches of gray matter) and the micro-connectome (complete connectivity within a single gray matter patch).
The Human Connectome Project
Until recently, methods for charting neural circuits in the human brain were sorely lacking (13). This situation has changed dramatically with the advent of noninvasive neuroimaging methods. Two complementary modalities of MRI (magnetic resonance imaging) provide the most useful information about long-distance connections. One modality uses diffusion imaging to determine the orientation of axonal fiber bundles in white matter, based on preferential diffusion of water molecules parallel to these fiber bundles. Tractography is an analysis strategy that uses this information to estimate long-distance pathways linking different gray-matter regions (14, 15). A second modality, resting-state functional MRI (R-fMRI), is based on slow fluctuations in the standard fMRI ‘BOLD’ signal that occur even when subjects are at rest. The time courses of these fluctuations are correlated across gray-matter locations, and the spatial pattern of the resultant ‘functional connectivity’ correlation maps are closely related but not identical to the known pattern of direct anatomical connectivity (16, 17). Diffusion imaging and R-fMRI each have important limitations, but together they offer powerful and complementary windows on human brain connectivity.
To address these opportunities, NIH recently launched the Human Connectome Project (HCP) and awarded grants to two consortia (18). The consortium led by Washington University in St. Louis and the University of Minnesota (19) aims to characterize whole-brain circuitry and its variability across individuals in 1,200 healthy adults (300 twin pairs and their non-twin siblings). Besides diffusion imaging and R-fMRI, task-based fMRI data will be acquired in all subjects, along with extensive behavioral testing; 100 subjects will also be studied using magnetoencephalography (MEG) and electroencephalography (EEG). Acquired blood samples will enable genotyping or full-genome sequencing of all subjects near the end of the 5-year project. Currently, data acquisition and analysis methods are being extensively refined using pilot datasets. Data acquisition from the main cohort will commence in mid-2012.
Neuroimaging and behavioral data from the HCP will be made freely available to the neuroscience community via a database (20) and a platform for visualization and user-friendly data mining. This informatics effort involves major challenges owing to the large amounts of data (expected to be ~1 petabyte), the diversity of data types, and the many possible types of data mining. Some investigators will drill deeply by analyzing a high-resolution connectivity maps between all gray matter locations. Others will explore a more compact ‘parcellated connectome’ among all identified cortical and subcortical parcels. Data mining options will reveal connectivity differences between sub-populations that are selected by behavioral phenotype (e.g., high vs. low IQ) and various other characteristics (Fig. 1). The utility of HCP-generated data will be enhanced by close links to other resources containing complementary types of spatially organized data, such as the Allen Human Brain Atlas (21), which contains neural gene expression maps.
Fig. 1.
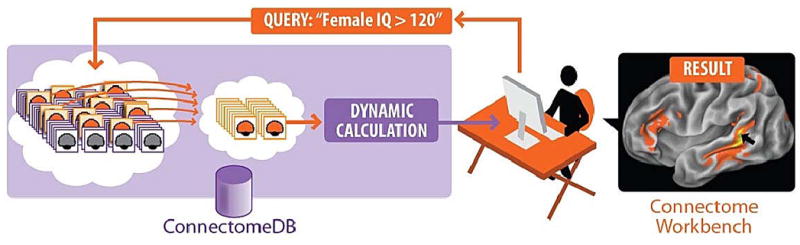
Schematic illustration of online data mining capabilities envisioned for the Human Connectome Project. Investigators will be able to pose a wide range of queries (e.g., connectivity patterns of a particular brain region of interest averaged across a group of individuals based on behavioral criteria) and view the search results interactively on 3-D brain models. Datasets of interest will be freely available for downloading and additional offline analysis.
Micro-connectomes
Recent advances in serial section electron microscopy, high-resolution optical imaging methods, and sophisticated image segmentation methods enable detailed reconstructions of the microscopic connectome at the level of individual synapses, axons, dendrites, and glial proceses (22-24). Current efforts focus on reconstruction of local circuits, such as small patches of cerebral cortex or retina, in laboratory animals. As such datasets begin to emerge, a fresh set of informatics challenges will arise in handling petabyte amounts of primary and analyzed data, and in providing data mining platforms that enable neuroscientists to navigate complex local circuits and examine interesting statistical characteristics.
Micro- and macro-connectomes exemplify distinct data types within particular tiers of analysis that will eventually need to be linked. Effective interpretation of both macro- and micro-connectomic approaches will require novel informatics and computational approaches that enable these two types of data to be analyzed in a common framework and infrastructure. Efforts such as the Blue Brain Project (25) represent an important initial thrust in this direction, but the endeavor will entail decades of effort and innovation.
Powerful and complementary approaches such as ‘optogenetics’ operate at an intermediate (‘meso-connectome’) spatial scale by directly perturbing neural circuits in vivo or in vitro with light-activated ion channels inserted into selected neuronal types (26) . Other optical methods, such as calcium imaging with two-photon laser microscopy, enable analysis of the dynamics of ensembles of neurons in microcircuits (27, 28), and can lead to new conceptualizations of brain function (29). Such approaches provide an especially attractive window on neural choreography as they assess or perturb the temporal patterns of macro-or micro -circuit activity.
The Neuroscience Information Framework
Connectome-related projects illustrate ways in which neuroscience as a field is evolving at the level of neural circuitry. Other discovery efforts include genome-wide gene expression profiling (e.g. (30)) or epigenetic analyses across multiple brain regions in normal and diseased brains. This wide range of efforts results in a sharp increase in the amount and diversity of data being generated, making it unlikely that neuroscience will be adequately served by only a handful of centralized databases, as is largely the case for the genomics and proteomics community (31). How, then, can we access and explore these resources more effectively to support the “data intensive discovery” envisioned in the Fourth Paradigm (32)?
Tackling this question was a prime motivation behind the Neuroscience Information Framework (33). The NIF was launched in 2005 to survey the current ecosystem of neuroscience resources (databases, tools, materials) and to establish a resource description framework and search strategy for locating, accessing and utilizing digital neuroscience-related resources (34).
The NIF catalog, a human curated registry of known resources, currently includes more than 3500 such resources, and new ones are added daily. Over 2,000 of these resources are databases that range in size from hundreds to millions of records. Many were created at considerable effort and expense, yet most of them remain underutilized by the research community.
Clearly, it is inefficient for individual researchers to sequentially visit and explore thousands of databases, and conventional online search engines are inadequate, insofar as they do not effectively index or search database content. To promote discovery and use of on-line databases, the NIF created a portal through which users can search not only the NIF registry, but the content of multiple databases simultaneously. The current NIF federation includes more than 65 databases accessing ~30 million records (35) in major domains of relevance to neuroscience (Fig. 2). Besides very large genomic collections, there are nearly 1 million antibody records; 23,000 brain connectivity records; and >50,000 brain activation coordinates. Many of these areas are covered by multiple databases, which NIF knits together into a coherent view. While impressive, this represents only the tip of the iceberg. Most individual databases are underpopulated because of insufficient community contributions. Entire domains of neuroscience (e.g. electrophysiology, behavior) are underrepresented compared to genomics and neuroanatomy.
Fig 2.
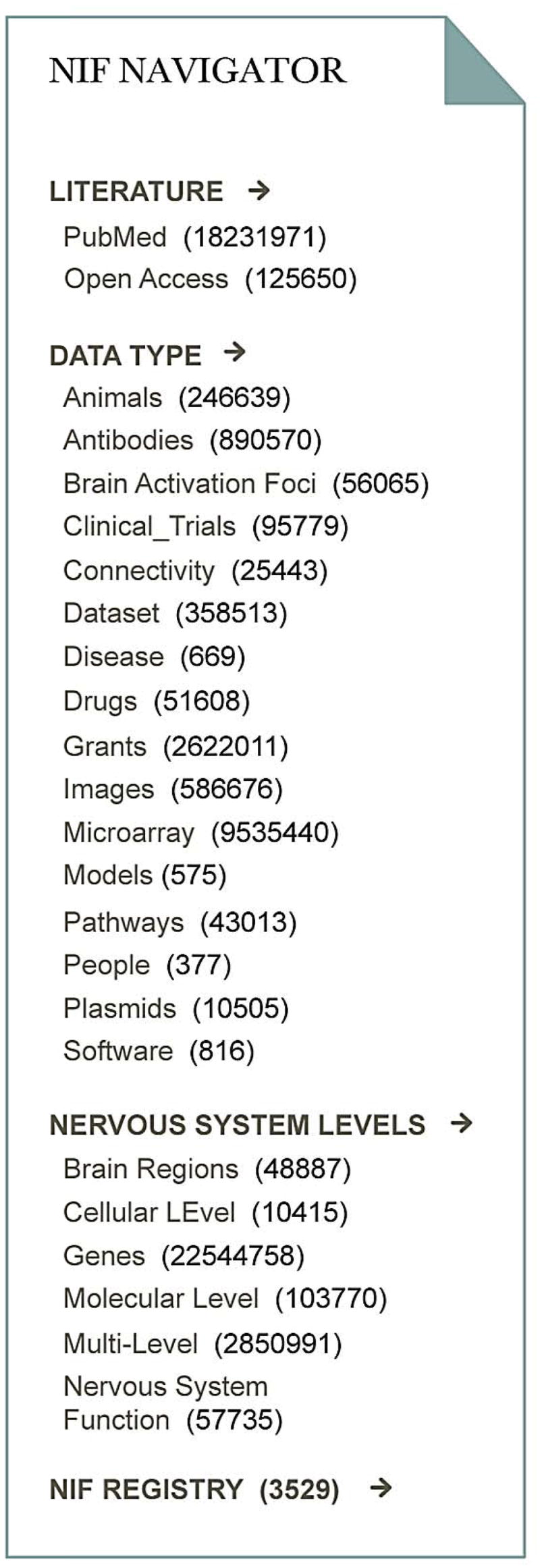
Current contents of the NIF. The NIF navigation bar displays the current contents of the NIF data federation organized by data type and level of the nervous system. The number of records in each category is displayed in parentheses.
Ideally, NIF users should be able not only to locate answers that are known, but to mine available data in ways that spur new hypotheses regarding what is not known. Perhaps the single biggest roadblock to this higher order data mining is the lack of standardized frameworks for organizing neuroscience data. Individual investigators often use terminology or spatial coordinate systems customized for their own particular analysis approaches. This customization is a significant barrier to data integration, requiring considerable human effort to access each resource, understand the context and content of the data, and determine the conditions under which they can be compared to other datasets of interest.
To address the terminology problem, NIF has assembled an expansive lexicon and ontology covering the broad domains of neuroscience by synthesizing open access community ontologies (36). The Neurolex and accompanying NIFSTD ontologies provide definitions of over 50,000 concepts using formal languages to represent brain regions, cells, subcellular structures, molecules, diseases and functions, and the relations among them. When users search for a concept through NIF, it automatically expands the query to include all synonymous or closely related terms. For example, a query for “striatum” will include “neostriatum, dorsal striatum, caudoputamen, caudate putamen” and other variants.
Neurolex terms are accessible through a wiki (37) that allows users to view, augment and modify these concepts. The goal is to provide clear definitions of each concept that can be utilized not only by humans but by automated agents, such as NIF, to navigate the complexities of human neuroscience knowledge. A key feature is the assignment of a unique resource identifier to make it easier for search algorithms to distinguish among concepts that share the same label. For example, nucleus (part of cell) and nucleus (part of brain) are distinguished by unique ID’s. Using these identifiers in addition to natural language to reference concepts in databases and publications, while conceptually simple, is an especially powerful means for making data maximally discoverable and useful.
These efforts to develop and deploy a semantic framework for neuroscience, spearheaded by NIF and by the International Neuroinformatics Coordinating Facility (38) are complemented by projects related to brain atlases and spatial frameworks (39–41) providing tools for referencing data to a standard coordinate system based on brain anatomy of a given organism.
Neuroinformatics as a Prelude to New Discoveries
How might improved access to multiple tiers of neurobiological data help us understand the brain? Imagine that we are investigating the neurobiology of bipolar disorder, an illness in which moods are normal for long periods of time, yet are labile and sometimes switch to mania or depression without an obvious external trigger. While highly heritable, this disease appears to be genetically very complex and possibly quite heterogeneous (42). We may discover numerous genes that impart vulnerability to the illness. Some may be ion channels, others synaptic proteins, or transcription factors. How will we uncover how disparate genetic causes lead to a similar clinical phenotype? Are they all affecting the morphology of certain cells, the dynamics of specific microcircuits, for example within the amygdala, the orchestration of information across regions, for example between the amygdala and the prefrontal cortex? Can we create genetic mouse models of the various mutated genes and show a convergence at any of these levels? Can we capture the critical changes in neuronal and/or glial function (at any of the levels) and find ways to prevent the illness? Discovering the common thread for such a disease will surely benefit from tools that facilitate navigation across the multiple tiers of data—genetics, gene expression/ epigenetics, changes in neuronal activity and differences in dynamics at the micro and macro levels depending on the mood state. No single focused level of analysis will suffice to achieve a satisfactory understanding of the disease. In neural choreography terms, we need to identify the dancers, define the nature of the dance and uncover how the disease disrupts it.
Recommendations
Need for a cultural shift
To meet the grand challenge of elucidating neural choreography, we need increasingly powerful scientific tools to study brain activity in space and in time, to extract the key features associated with particular events, and to do so on a scale that reveals commonalities and differences between individual brains. This requires an informatics infrastructure that has built-in flexibility to incorporate new types of data and navigate across tiers and domains of knowledge.
The NIF currently provides a platform for integrating and systematizing existing neuroscience knowledge and has been working to define best practices for those producing new neuroscience data. Good planning and future investment is needed to broaden and harden the overall framework for housing, analyzing and integrating future neuroscience knowledge. The International Neuroinformatics Coordinating Facility (INCF) plays an important role in coordinating and promoting this framework at a global level.
But can neuroscience evolve so that neuroinformatics becomes integral to how we study the brain? This would entail a cultural shift in the field regarding the importance of data sharing and mining. It would also require recognition that neuroscientists produce data not just for consumption by readers of the conventional literature, but for automated agents that can find, relate, and begin to interpret data from databases as well as the literature. Search technologies are advancing rapidly, but the complexity of scientific data continues to challenge. To make neuroscience data maximally interoperable within a global neuroscience information framework, we encourage the neuroscience community and the associated funding agencies to consider the following set of general and specific suggestions:
Neuroscientists should, as much as is feasible, share their data in a form that is machine accessible, i.e., through a web-based database or some other structured form that benefits from increasingly powerful search tools.
Databases spanning a growing portion of the neuroscience realm need to be created, populated, and sustained. This effort needs adequate support from federal and other funding mechanisms.
Because databases become more useful as they are more densely populated (43), adding to existing databases may be preferable to creating customized new ones. NIF, INCF and other resources provide valuable tools for finding existing databases.
Data consumption will increasingly involve machines first and humans second. Whether creating database content or publishing journal articles, neuroscientists should annotate content using community ontologies and identifiers. Coordinates, atlas, and registration method should be specified when referencing spatial locations.
Some types of published data (e.g., brain coordinates in neuroimaging studies) should be reported in standardized table formats that facilitate data mining.
Investment needs to occur in interdisciplinary research to develop computational, machine learning, and visualization methods for synthesizing across spatial and temporal information tiers.
Educational strategies from undergraduate through postdoctoral levels are needed to ensure that neuroscientists of the next generation are facile with data mining and data sharing tools of the future.
Cultural changes are needed to promote widespread participation in this endeavor. These ideas are not just a way to be responsible and collaborative; they may serve a vital role in attaining a deeper understanding brain function and dysfunction.
With such efforts, and some luck, the machinery that we have created, including powerful computers and associated tools, may provide us with the means to comprehend this “most unaccountable of machinery” (44), our own brain.
Acknowledgments
Funded in part by NIH 5P01-DA021633-02, Office of Naval Research (N00014-02-1-0879) and the Pritzker Neuropsychiatric Research Consortium (to H.A); the Human Connectome Project (1U54MH091657-01)from the 16 NIH Institutes and Centers that Support the NIH Blueprint for Neuroscience Research (to DVE); the Neuroscience Information Framework is supported by NIH Neuroscience Blueprint Contract HHSN271200800035C via NIDA to MEM. We thank D. Marcus, R. Poldrack, S. Curtiss and S.J. Watson for discussions and comments on the manuscript.
References and Notes
- 1.W. H. O. Report. Mental health and development: targeting people with mental health conditions as a vulnerable group (released September 2010) [Google Scholar]
- 2.Azevedo FA, et al. J Comp Neurol. 2009 Apr 10;513:532. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 3.Pakkenberg B, et al. Exp Gerontol. 2003 Jan–Feb;38:95. doi: 10.1016/s0531-5565(02)00151-1. [DOI] [PubMed] [Google Scholar]
- 4.http://www.alleninstitute.org/.
- 5.http://www.gensat.org/.
- 6.http://adni.loni.ucla.edu.
- 7.Bjorklund A, Hokfelt T. Handbook of Chemical Neuroanatomy Book Series-Elsevier. 1–21. pp. 1983–2005. [Google Scholar]
- 8.http://cocomac.org/home.asp
- 9.http://brancusi.usc.edu/bkms/
- 10.http://brainmaps.org/
- 11.http://en.wikipedia.org/wiki/Connectome.
- 12.Sporns O, Tononi G, Kotter R. PLoS Compu Biol. 2005;1:e2. doi: 10.1371/journalpcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crick F, Jones E. Nature. 1993 Jan 14;361:109. doi: 10.1038/361109a0. [DOI] [PubMed] [Google Scholar]
- 14.Johansen-Berg H, Behrens TEJ. Diffusion MRI: From quantitative measurement to in-vivo neuroanatomy. 1. Academic Press; 2009. p. 490. [Google Scholar]
- 15.Johansen-Berg H, Rushworth MF. Annu Rev Neurosci. 2009;32:75. doi: 10.1146/annurev.neuro.051508.135735. [DOI] [PubMed] [Google Scholar]
- 16.Vincent JL, et al. Nature. 2007 May 3;447:83. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D, Snyder AZ, Shimony JS, Fox MD, Raichle ME. Cereb Cortex. 2010 May;20:1187. doi: 10.1093/cercor/bhp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.http://humanconnectome.org/consortia.
- 19.http://humanconnectome.org.
- 20.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. Neuroinformatics. 2007 Spring;5:11. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 21.http://human.brain-map.org/.
- 22.Briggman KL, Denk W. Curr Opin Neurobiol. 2006 Oct;16:562. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Lichtman JW, Livet J, Sanes JR. Nature Reviews Neuroscience. 2008;9:417. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SJ. Curr Opin Neurobiol. 2007;17:601. doi: 10.1016/j.conb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markram H. Nat Rev Neurosci B. 2006;153:153. doi: 10.1038/nrn1848. [DOI] [PubMed] [Google Scholar]
- 26.Deisseroth K. Nat Methods. 2011;8:26. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grewe BF, Helmchen F. Curr Opin Neurobiol. 2009;19:520–529. doi: 10.1016/j.conb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Watson BO, et al. Front Neurosci. 2010;15 doi: 10.3389/fnins.2010.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buzsaki G. 2010;68:362. doi: 10.1016/j.neuron.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernard R, et al. Mol Psychiatry. 2010 Apr 13; epub ahead of print. [Google Scholar]
- 31.Martone ME, et al. Nature Neurosci. 2004;7:467. doi: 10.1038/nn1229. [DOI] [PubMed] [Google Scholar]
- 32.Hey T, Tansler S, Tolle K, editors. The Fourth Paradigm: Data Intensive Scientific Discovery. Microsoft Research Publishing; 2009. [Google Scholar]
- 33.http://neuinfo.org.
- 34.Gardner D, et al. Neuroinformatics. 2008;6:149. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta A, et al. Neuroinformatics. 2008 Sep;6:205. doi: 10.1007/s12021-008-9033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bug WJ, et al. Neuroinformatics. 2008 Sep;6:175. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.http://neurolex.org.
- 38.http://incf.org.
- 39.http://www.brain-map.org.
- 40.http://wholebraincatalog.org.
- 41.http://incf.org/core/programs/atlasing.
- 42.Akil H, et al. Science. 2010;327:1580. doi: 10.1126/science.1188654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ascoli GA. Nature Rev Neurosci. 2006;7:318. doi: 10.1038/nrn1885. [DOI] [PubMed] [Google Scholar]
- 44.Nicolson N, Trautmann J, editors. The Letters of Virginia Woolf. V. Harcourt, Brace & Co; 1982. pp. 1932–1935. [Google Scholar]


