Abstract
We have determined the structure of the β–carbonic anhydrase from the dicotyledonous plant Pisum sativum at 1.93 Å resolution, using a combination of multiple anomalous scattering off the active site zinc ion and non-crystallographic symmetry averaging. The mol– ecule assembles as an octamer with a novel dimer of dimers of dimers arrangement. Two distinct patterns of conservation of active site residues are observed, implying two potentially mechanistically distinct classes of β–carbonic anhydrases. The active site is located at the interface between two monomers, with Cys160, His220 and Cys223 binding the catalytic zinc ion and residues Asp162 (oriented by Arg164), Gly224, Gln151, Val184, Phe179 and Tyr205 interacting with the substrate analogue, acetic acid. The substrate binding groups have a one to one correspondence with the functional groups in the α–carbonic anhydrase active site, with the corresponding residues being closely superimposable by a mirror plane. Therefore, despite differing folds, α- and β–carbonic anhydrase have converged upon a very similar active site design and are likely to share a common mechanism.
Keywords: β-carbonic anhydrase/convergent evolution/X-ray crystallography/zinc enzyme
Introduction
β–carbonic anhydrase (β–CA; EC 4.2.1.1) is an enzyme that catalyses the reversible hydration of carbon dioxide. This enzyme has been found in species from all three domains of life, with representatives in several eubacterial species including Escherichia coli (Guilloton et al., 1992), in the thermophilic archaeote Methanobacterium thermo– autotrophicum (Smith and Ferry, 1999) and in a variety of higher plants and algae. Homologous sequences of uncertain function are found in the fungus Saccharomyces cerevisiae and in the nematode Caenorhabditis elegans. In addition to β–CA, there exist two other enzymes that share the same catalytic function. γ–carbonic anhydrase (γ–CA) was first identified in 1994 in the archaeote Methanosarcina thermophila, and homologous sequences have also been found in plants and eubacteria (Alber and Ferry, 1994). Thus far, little is known about the details of γ–CA biochemistry although the enzyme's structure has been solved, revealing a highly unusual fold, a trimer of left-handed β–helices (Kisker et al., 1996). α–carbonic anhydrase (α–CA) was the first CA discovered, and is often simply referred to as the CA. It is found across all domains of life, but is best studied in animals where it occurs in a number of isozymic forms (numbered I–VII) differing widely in their kinetic properties and tissue distribution. Among the best characterized isozymes is α–CAII, which is found in many cell types, including erythrocytes, where it facilitates rapid exchange of CO2 in the respiratory cycle. α–CAs also play a role in other physiological processes such as tissue mineralization and intra-ocular pressure regulation. Much of the clinical interest in this enzyme stems from the latter role where it is the target of the sulfonamidyl glaucoma drugs (Liljas et al., 1994; Lindskog, 1997).
The reaction catalysed by α–CA proceeds via a two-stage ping pong mechanism:
EZn2+OH– + CO2 + H2O ⇌ EZn2+OH2 + HCO3– (1)
EZn2+H2O + B ⇌ EZn2+OH– + BH+ (2)
where E is the enzyme and B is a buffer molecule. The value of kcat/Km may be obtained from equilibrium isotope exchange kinetics, and reaches a value of 1.8 × 109 M–1s–1 for the α–CAII reaction between CO2 and hydroxide, essentially at the limit of diffusion control. The rate of the overall reaction may be measured by monitoring the rate of evolution of protons in a saturated solution of CO2. This parameter reflects the rate limiting proton transfer step, as shown by a strong hydrogen isotope effect on kcat and the dependence of the reaction rate on the amount and nature of the buffer present (Ren and Lindskog, 1992). kcat for the fast α–CAII isozyme is 1 × 106 s–1. The affinity of the enzyme for the substrate CO2 is very low, with spectroscopic studies placing Km at ∼10 mM. Crystallographic studies of α–CA reveal a 30 kDa monomeric protein folded in an extended, predominantly antiparallel β–sheet that wraps around to form a 15 Å deep conical depression housing the active site (Liljas et al., 1972).
β–CA has been most intensively studied in higher plants because of the critical supporting role it plays in the physiology of photosynthesis. The primary role of this enzyme is to minimize resistance to the diffusion of CO2 from the stomatal air spaces, where CO2 is initially absorbed, to the chloroplast stroma where carbon is fixed by the enzyme ribulose bisphosphate carboxylase/oxygenase (RuBisCO). In C4 plants the protein is expressed predominantly in the cytoplasm of mesophyll cells, where by converting CO2 to HCO3– it provides substrate for phosphoenolpyruvate carboxylase and is thus an integral component of the CO2 concentrating mechanism. In the more common C3 plants, β–CA is a major component of leaf protein (0.5–2% of the total) and is localized primarily in the stroma of the chloroplast, although significant activity is also found in the cytoplasm of photosynthetic cells (Badger and Price, 1994). In the stroma, where the alkaline pH stabilizes bicarbonate relative to CO2, the presence of β–CA in association with the Calvin cycle enzyme complex, which includes RuBisCO, promotes efficient photosynthesis by providing a CO2 source in close proximity to the CO2 sink (Jebanathirajah and Coleman, 1998). Inhibition of β–CA production by antisense expression can to some degree be compensated for by the plant by increasing stomatal conductance, at the cost, however, of excessive water loss (Majeau et al., 1994).
Historically, the study of the biochemistry of β–CA has developed as a sideline to studies of the α–CA enzymes, and indeed, it was not until the first plant sequences were determined that scientists realized that the two enzymes were non-homologous (Fawcett et al., 1990). Consequently, understanding of the β–CA enzymes has advanced quickly by drawing on the experimental techniques and functional models developed in the study of α–CA, with experimental results generally assessed against the α–CA benchmarks. Kinetic characterization of the β–CA enzyme has shown that the reaction has much in common with that of α–CA. kcat/Km for the pea enzyme is 1.8 × 108 M–1s–1, while kcat is 4 × 105 s–1 (Johansson and Forsman, 1993); therefore, this enzyme is almost as fast and as efficient as α–CAII, the fastest of the α–CA isozymes. The reaction is activated by increasing pH, but the behaviour is more complicated than in α–CA, preventing the assignment of a single pKa value (Johansson and Forsman, 1993). The observed strong hydrogen isotope effect again indicates the presence of a rate limiting proton transfer step, presumably corresponding to step 2 of the reaction outlined above for α–CA (Johansson and Forsman, 1994; Rowlett et al., 1994). The mature β–CA monomer has a mol. wt of ∼25 kDa (chloroplastic β–CAs are generally nuclear encoded and so are initially expressed with a two-component signal presequence of ∼100 amino acids to direct them to the chloroplast's stroma; Johansson and Forsman, 1992). The reported oligomeric nature of the enzyme encompasses a large variety of states; published estimates of molecular weights indicate octamers (Kandel et al., 1978; Rumeau et al., 1996), hexamers, tetramers (Hiltonen et al., 1998; Smith and Ferry, 1999) and dimers. The molecule has been shown to bind one zinc molecule per subunit, and a combination of extended X-ray absorption fine structure (EXAFS) and mutagenesis studies in the spinach enzyme and mutagenesis studies in the pea enzyme have led to the proposal that the metal ligands are Cys160, His220 and Cys223; the same studies also concluded that zinc appears essential for catalysis (Provart et al., 1993; Bracey et al., 1994; Rowlett et al., 1994).
In summary, biochemical characterization of the β–CA enzyme has revealed a number of close parallels with α–CA, but understanding of the details of the mechanism has been hampered by the lack of structural context in which to interpret these results. Here, we report the structure of the β–CA from the common pea, Pisum sativum, at 1.93 Å in complex with the inhibitor acetic acid.
Results and discussion
The structure was determined by multiple anomalous dispersion (MAD) off the active site zinc ions combined with 8–fold non-crystallographic symmetry (NCS) averaging and phase extension. The asymmetric unit contains two hemi-octamers (the first containing molecules A–D, the second molecules E–H), with each hemi-octamer being one half of an octamer located on a crystallographic 2–fold axis. Residues N–terminal to amino acid 120, although partially stabilized for some monomers by crystal contacts, do not appear integral to the enzyme's structure, instead forming a flexible linker between the molecule and the site of post-stromal-import proteolytic processing.
One peculiarity of these crystals is that layers of molecules with high mean temperature (B)–factors (58.7, 65.6, 52.7 and 55.9 for an average of 58 Å2) alternate with layers of molecules with low mean B-factors (39.5, 30.6, 35.6 and 31.7 for an average of 34 Å2) in the lattice c direction. Not all of the independent molecules in the asymmetric unit are therefore equally well defined, with the four molecules with low B–factors showing the expected level of detail, and the other four having weaker density and resembling molecules solved at lower resolution.
Structure of β–CA
Comparison of the β–CA structure with the database of existing structures using the program DALI revealed that while there are elements of the fold that resemble previously known structures, the overall fold is novel. As was anticipated from the absence of sequence conservation, the β–CA fold resembles neither that of α–CA nor that of γ–CA. It therefore confirms that there are indeed (at least) three protein folds that have independently evolved the CA function.
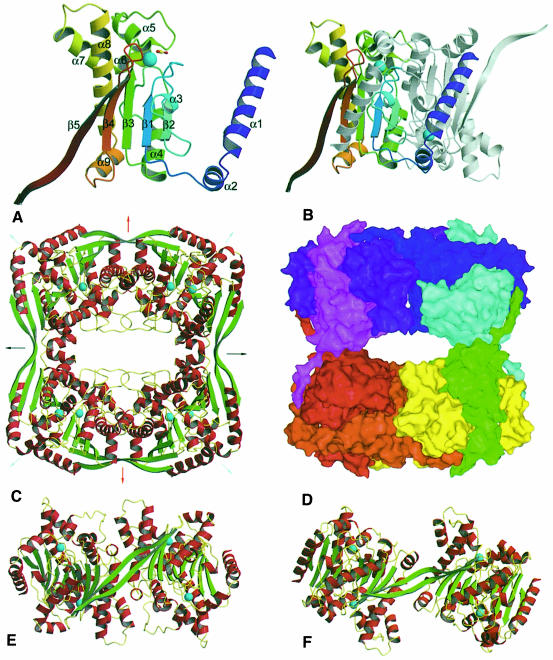
The β–CA monomer has an unusually non-compact appearance, with a wedge-shaped core flanked by two protruding motifs that mediate interactions involved in oligomerization (Figure 1A). As predicted by circular dichroism (Johansson and Forsman, 1993), the structure is predominantly α–helical, and is built around the core of a dinucleotide-binding fold motif with a four-stranded parallel β–sheet ordered 2–1–3–4 with α–helices forming right-handed crossover connections. Found in the loop between β3 and β4 are five helices (α5–α9), which form a compact subdomain. At the end of strand β4 the chain doubles back into a fifth C-terminal antiparallel β–strand. Pairs of monomers are joined up to form dimers through an extensive interface that buries 27% (3255 of 12 119 Å2) of the monomer's surface area (Figure 1B), a substantial portion of which is mediated by helices α1 and α2, which form a finger that wraps most of the way around the second molecule. Strands β2 from each of the two monomers cross at a 60° angle in an antiparallel fashion forming a pair of hydrogen bonds between residues 179 and 179′ (numbering as in Provart et al., 1993).
Fig. 1. Fold and oligomeric organization of β–CA. (A) Ribbon of the β–CA monomer, with secondary structure nomenclature as indicated. The colour is graded from blue to red, N-terminus to C-terminus. (B) Ribbon diagram of the β–CA dimer. One monomer is coloured as in (A), the other is in white. Extensive contacts are mediated by helices α1 and α2 as they wrap around the secondary monomer. (C) Ribbon diagram of the β–CA octamer. The octamer is assembled as a dimer of dimers of dimers. Arrows indicate the approximate position of the symmetry elements: red arrows, crystallographic 2–fold symmetry elements; black arrows, NCS elements that apply to the whole octamer; and cyan arrows, local non-crystallographic 2–fold axes that apply only to one dimer. Interactions between dimers are primarily mediated through strand β5. (D) Molecular surface of the β–CA octamer. Each monomer is coloured differently to highlight the complex interweaving of molecules that occurs in octamer assembly, with each monomer contacting five other monomers. (E) View of half an octamer (the other half omitted for clarity) down the crystallographic 2–fold axis. At the interface formed here, the dimer–dimer interaction buries a substantial portion of the molecular surface, although most of the interactions are mediated by water molecules. (F) View of half an octamer as seen down a non-crystallographic 2–fold axis. Here, far less surface area is buried and almost all of the interactions are mediated by strand β5. The configuration of the monomers as seen here may be mapped onto that in Figure 1E by a 54° twist around a vector approximately colinear with the β5 strand.
This dimer is then the basic building block stitched together by interactions mediated by strand β5 into a loosely packed octamer with a diamond-shaped channel of ∼35 × 15 Å running through the centre (Figure 1C and D). The octamer does not possess the usual 422 (dimer of tetramers) point group symmetry previously inferred from electron micrographs of the chickpea enzyme (Aliev et al., 1986), but rather has 222 (dimer of dimers) symmetry with the above described dimer forming the repeating unit. This oligomerization arrangement is novel, with no precedent in the Research Collaboratory for Structural Bioinformatics (RBSC) protein structure database. The assembly of a complex with 222 point group symmetry from an object that possesses intrinsic 2–fold symmetry necessitates that one molecular surface mediates two distinct types of interactions, much as is observed in the assembly of hexameric and pentameric units in a viral capsid from a single structural protein. This phenomenon is seen here, where different interactions are observed at the interfaces mediated by the crystallographic and the non-crystallographic 2–fold axes (Figure 1C–F). Almost all protein–protein interactions responsible for forming the octamer from dimers are mediated through the interaction of strand β5, which pairs up in an antiparallel fashion with its equivalent in the second dimer in an interaction strongly reminiscent of a strand exchange event. There is, however, no evidence that compact versions of the enzyme, where strand β5 folds back to form a sixth strand, exist in any natural β–CA. The dimer–dimer packing along the NCS axis is mediated almost entirely by strand β5, with only 342 of the total 2504 Å2 buried at this interface (total surface area for a pair of dimers is 33 263 Å2) being mediated by other interactions. Although the interface generated by the crystallographic 2–fold axis buries a substantial proportion of the monomer's surface (compare Figure 1E and F), only 1472 Å2 is rendered solvent inaccessible (this is beyond the β5–β5′ interaction, which buries a further 2011 Å2), as almost all interactions between the molecules are mediated through ordered water molecules. One type of interface can be generated from the other by fixing one dimer and rotating the second 54° around an axis roughly colinear with the β–strands. This movement is accommodated by a slight twisting of the β5–β5′ strand at residues Leu319 and Leu323, where the β5–β5′ pair separates from β4.
The second half of β5, the motif that mediates most of the oligomerization interactions, appears unique to dicotyledonous plants (Figure 2). Given that the active site utilizes elements from two monomers (see below), functional monomeric β–CAs seem unlikely, but it is plausible that the other reported oligomeric forms, including tetramers and hexamers, could be assembled using some other structural motif to mediate oligomerization. With regard to possible hexameric enzymes, it is interesting to note that the β–CA from Zea mays is constrained to be a 3 × N multimer as it exists as a head-to-tail fusion of three β–CA copies. As in the pea enzyme structure, the C-terminus of β5 would contact the N-terminus of a second molecule as it wraps around a third molecule that is sandwiched between them. A complex may be assembled by interdigitating two fusion molecules, yielding a dimer containing six β–CA subunits.
Fig. 2. Multiple sequence alignment of a subset of putative β–CAs. Numbering is according to the Pisum sativum species, with secondary structural elements indicated underneath. Amino acids in the active site are boxed. Sequences shown include the dicotyledonous angiosperms Pisum sativum (PISAT), Spinachia olearchia (SPOLE) and Flaveria bidentis (FLBID), the monocotyledonous angiosperms Oriza sativa (ORSAT), Hordeum vulgare (HOVUL) and Zea mays (ZEMAY) (second of three head-to-tail fusion repeats), the eubacteria Escherichia coli (ESCOL), Bacillus subtilis (BASUB) and Mycobacterium tuberculosis (MYTUB), the yeast Saccharomyces cerevisiae (SACER), the red alga Porphyridium purpureum (POPUR) (N–terminal repeat), the cyanobacterium Synechococcus PCC7942 (SYNSP) and the archaeote Methanobacterium thermoautotrophicum (METHE). Sequences were aligned using ClustalX (Thompson et al., 1994) and displayed with ALSCRIPT (Barton, 1993).
The active site
The active site cleft is located at the interface of two monomers and consists of a cavity largely sequestered from solution. As predicted from sequence conservation, EXAFS and mutagenesis studies, the zinc ligands are Cys160, His220 and Cys223 (Provart et al., 1993; Bracey et al., 1994; Rowlett et al., 1994). It has been shown in α–CA that the fine tuning of metal ion energetics depends in part upon the chemical nature of the groups interacting with the metal ion ligands, which for β–CA include no sidechain atoms: His220 hydrogen bonds with the carbonyl oxygen of residue 221; Cys223 Sγ hydrogen bonds with the amide nitrogen of residue 225 and a water molecule; and Cys160 Sγ hydrogen bonds with the amide nitrogens of residues 162 and 185. Asp162 presents its Oδ1 to the active site cavity and is locked into place by forming two hydrogen bonds with the guanidinium group of Arg164. The sidechain of Gln151′ also lines the active site pocket, with hydrogen bonds from the sidechain oxygen and backbone amide nitrogen of the residue Ser163 binding the Gln151′ Oɛ1, leaving the amide nitrogen free for interacting with bound ligands. Val184, Tyr205′ and Phe179′ together present a continuous hydrophobic surface in the binding pocket.
Because of the two cysteine ligands of the catalytic zinc ion, the β–CA active site is somewhat more negatively charged than that of α–CA. These extra full charges would seem to be at least partially compensated for by dipole effects: not only is the active site located at the N-terminus of a helix, but also almost every other backbone amide in the region of the active site is oriented with the nitrogen pointing into the active site cavity.
A narrow, hydrophilic channel that passes a bottleneck between Tyr205′, Gln151′, Gly224 and Asp162 is the only access to the active site of β–CA from bulk solvent. It is too narrow to allow the passage of anything larger than a water molecule, indicating the need for some sort of rearrangement in the course of the enzyme's catalytic cycle. Highly suggestive in this regard is the deep finger-like hydrophobic pocket lining the far edge of Tyr205′. This empty space would allow the widening of the solvent access channel by rotating the Tyr205′ sidechain 30° around the Cα–Cβ bond towards Ile243 without causing any steric clashes. Such a rearrangement would also be consistent with the binding of the larger known inhibitors such as sulfonamides, which would otherwise be too bulky to be accommodated in β–CA's active site.
β–CAs occur in two basic classes
Of the residues in the active site, only the three zinc ligands and the Asp/Arg pair are conserved in every β–CA sequence known (Figure 2). The pattern of conservation of the other key active site residues (Gln151, Ser161, Ser163, Phe179, Val184 and Tyr205) is interesting because in a subset of sequences, including all known sequences of higher plants, all these residues are conserved, while in other organisms, each of these residues displays a set of parallel substitutions. Thus, in M.thermoautotrophicum, Mycobacterium tuberculosis and Bacillus subtilis (among others; see Smith and Ferry, 1999 for further examples), Gln151 is substituted with Pro, Ser161 with Met, Ser163 with Thr/Ala, Phe179 with Lys, Val184 with Ala and Tyr205 with Val. This implies a substantial redesign of the active site with the interactions with the substrate at atoms other than the zinc-bound oxygen mediated in a radically different manner. We therefore conclude that there are two distinct groups of β–CA, differing in their pattern of sequence conservation, active site design and possibly also in their mechanism. For convenience, we designate the group that includes the plant sequences as the ‘plant’ type and those with the altered active site as the ‘Cab’ type after the M.thermoautotrophicum enzyme.
Ligand binding
β–CA was crystallized in the presence of 400 mM acetate, a weak inhibitor (maize β–CA displays 15% of maximal activity in 100 mM sodium acetate; Hatch, 1991). The presence of a flat, trilobed piece of density closely associated with the catalytic zinc ion in each active site is consistent with a bound acetate molecule. The shape and position of this density, however, vary significantly from active site to active site, forcing a case by case interpretation.
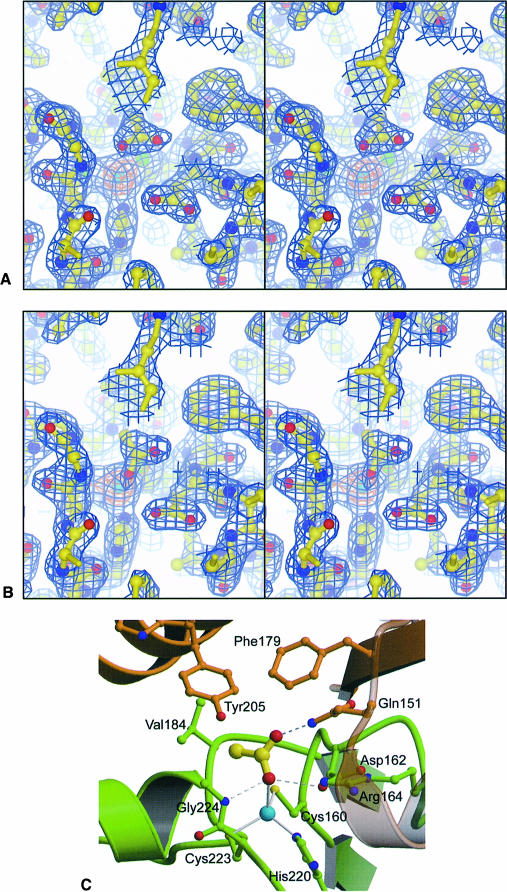
In the active site of molecule D (Figure 3A), two of the three acetate atoms are well positioned to form hydrogen bonds, and are therefore interpreted as being the oxygen atoms (at 1.93 Å resolution methyl groups are indistinguishable from oxygen atoms; consequently, interpretation of the symmetric density in terms of particular atoms is based upon the chemical environment occupied by that atom). The first of these oxygen atoms, designated O1, forms a hydrogen bond with Gly224 N (2.9 Å); the second, designated O2 forms a hydrogen bond with Gln151′ Nɛ2 (3.1 Å). The third ‘atom’ makes contacts only with hydrophobic residues, and is assumed to be a methyl group. O1 is the acetate atom closest to the zinc, and refines to a zinc–oxygen distance of 3.6 Å with the plane of the acetate molecule perpendicular to the oxygen–zinc bond. O1 is placed 3.8 Å from Asp162 Oδ1, indicating that no hydrogen bond exists with this group. There is also no density present that would indicate a zinc-bound water molecule in the active site. The pH at which this experiment was performed is sufficiently close to the pKa of acetate that although the anionic form will dominate, appreciably high concentrations of the neutral, protonated species should also be present. This difference is not resolvable by direct observation at this resolution, as one cannot distinguish charged from uncharged states or the presence from the absence of protons. Because O1 is significantly displaced away from the hydrogen bond acceptor Asp162, and no compensating interaction is formed, we interpret the ligand in this active site to be the unprotonated acetate ion.
Fig. 3. Ligand binding in various of the active sites of β–CA. (A) Stereo picture of the active site of molecule D showing σA-weighted electron density for the acetate ion. The map is contoured at 1.5σ (blue) and 8σ (orange). (B) Stereo picture of the active site of molecule G showing σA-weighted electron density for the acetic acid molecule. The map is contoured at 1.5σ (blue) and 8σ (orange). (C) Binding mode of acetic acid in the β–CA active site, with zinc–ligand interactions shown as solid lines and hydrogen bonds as dashed lines. The binding site is composed of two molecules, with one molecule shown in green and the other one in brown.
Similar density is observed in the active site of molecule C, where this density coexists with a strong (2.0σ) density peak closely associated with the zinc ion. The acetate density lends itself to the same interpretation as above, with essentially the same binding geometry, while the extra peak, occupying the fourth tetrahedral zinc position and well resolved from the acetate density, is interpreted as a water molecule/hydroxide ion. Refined as a water molecule, it occupies a position where it can form hydrogen bonds with both Gly224 N (3.3 Å) and Asp162 Oδ1 (2.9 Å).
In the active sites of molecules G (Figure 3B) and H, the inhibitor's O1 is 2.5 Å from the zinc ion, completing the canonical tetrahedral geometry with respect to the other zinc ligands. This oxygen atom forms hydrogen bonds with both Asp162 Oδ1 (3.0 Å) and Gly119 N (3.1 Å) with good tetrahedral geometry. O2 hydrogen bonds with Gln151′ Nɛ2 (2.9 Å) and its second free electron pair points at the edge of the ring of Phe179′ (3.4 Å). The methyl group makes van der Waals interactions with the sidechains of Phe179′, Val184 and Tyr205′. Since in order to form the hydrogen bond with both Asp162 Oδ1 and Gly119 N, O1 would need to be protonated and sp3 hybridized, we infer that this molecule is acetic acid.
It should be noted that the zinc–oxygen distance observed in molecules G and H is significantly longer than that generally observed for zinc–oxygen bonds (1.95–2.1 Å). Although this may be real, perhaps a consequence of the neutral nature of the ligating species, it is more likely that the density in these active sites is a superposition of two different conformations, a major one with acetic acid bound closely to the zinc, and a minor one with acetate bound, similar to molecule D. The inhibitor would then refine to a position that is the weighted average of these two, resulting in the zinc–O1 distance being overestimated. In general, the ligand position may be biased away from the zinc, and distances observed for these two active sites should be interpreted with caution.
The higher temperature factors associated with the other four molecules in the asymmetric unit cause the density to be less defined. Generally, the inhibitor molecules in these active sites refine to positions equivalent to those in molecules G and H.
The observation of distinct binding modes in different active sites in the asymmetric unit implies the existence of some mechanism by which residues in a given active site can sense their crystallographic environment (as otherwise there would be no correlation between succes– sive unit cells and one would simply observe the average density). Superposition of all four well defined binding sites, however, reveals no significant differences consistently associated with a particular binding mode. Having a zinc ion with only three strong ligands, as is observed here in monomer D, would be anticipated to be relatively unstable and has never been reported for any of the α–CA or γ–CA complex structures. Possibly the ionic acetate is able to compete with acetic acid for binding in part due to stabilizing electrostatic interactions with the zinc ion, but it seems likely that it is only the high concentration of the acetate ion at this pH that allows this binding mode to be observed.
Proposed model for β–CA catalysis
Before discussing potential mechanisms for β–CA, it is useful to review what several decades of detailed studies have taught us about the same reaction as it occurs in the analogous enzyme α–CA. Here, the most important functional group is the zinc ion, which is coordinated by three histidyl ligands with the fourth position available for the binding of water. The zinc-bound water also hydrogen bonds with a second water molecule and Thr199 Oγ1. The latter group acts as a hydrogen bond donor with respect to Glu106, and therefore allows only atoms capable of acting as hydrogen bond donors to bind to the zinc ion with tetrahedral geometry. Thr199 acts as a filter and is often called the ‘gatekeeper’ residue. Together with Glu106, it is absolutely conserved and enzymes mutated in these positions show 100–fold reduced activity. The zinc ion acts as a Lewis acid, which, with some help from the hydrogen bond provided by Thr199, lowers the pKa of water from 15.8 to ∼7, allowing the generation of a stable OH– ion at physiological pH. The water molecule, which forms the third ligand for the zinc-bound water, is the first member of a proton wire conducting a proton to His64, which in turn acts as a temporary way station for the proton prior to its transfer to a buffer molecule. This residue adopts alternative conformations with the imidazole group close to the zinc for accepting the proton and then more solvent exposed to allow transfer to a buffer species in bulk solution; for this reason, it is known as the proton shuttle. His64 is found only in a subset of α–CAs, including α–CAII, and is critical for fast turnover. The OH– ion is the nucleophile that attacks the electrophilic CO2 carbon. CO2 binding is very weak and the mode of interaction is not well understood. The extensive hydrophobic patch consisting of Val121, Val143, Leu198 and Trp209 has been proposed to play a role, and the backbone amide nitrogen of Thr199, which forms a hydrogen bond to bicarbonate analogues, is believed to also hydrogen bond with CO2 helping to electrophilically activate the CO2 molecule. The product, bicarbonate, is bound through the same interactions: the zinc-bound oxygen is protonated and hydrogen bonds with Thr199 Oγ1 and a water molecule; the second oxygen hydrogen bonds with Thr199 N; and the third oxygen is buried in the hydrophobic pocket (Liljas et al., 1994; Lindskog, 1997).
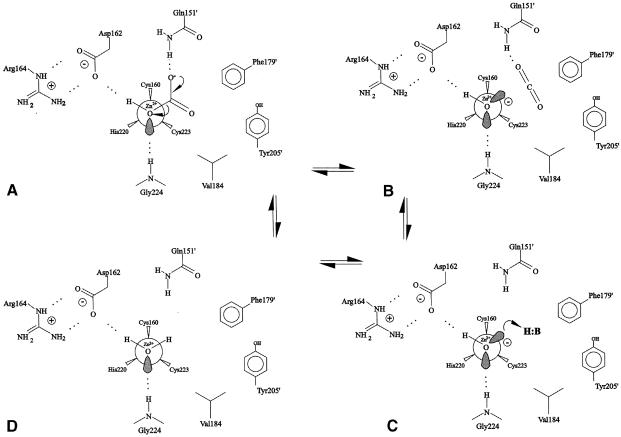
Since acetic acid and bicarbonate are isoelectronic except at the methyl group, which, in any case, is buried in a hydrophobic pocket, the interactions made by the two species in the active site are highly similar and so acetic acid binding (Figure 3C) would seem a reasonable model for bicarbonate binding (although bicarbonate is likely to bind closer to the zinc than the refined position of acetic acid). A bicarbonate anion bound at the tetrahedral zinc position in the same orientation as is adopted by acetic acid in active sites G and H would then, like the acetic acid molecule, have the zinc-ligating oxygen hydrogen bonding with Gly224 N and Asp162 Oδ1, the second oxygen hydrogen bonding Gln151′ and the third in the hydrophobic pocket. These interactions, helped perhaps through an interaction between O3 and the zinc ion, stabilize the molecule as it splits into CO2 and hydroxide as a normal mode vibration of the O–C bond, leaving CO2 free to diffuse out of the active site (Figure 4A). The hydroxide would remain at the tetrahedral zinc position, as seen in active site C, still hydrogen bonding with Asp162 and Gly224 (Figure 4B). Stabilization of the hydroxide ion would be facilitated by the zinc ion and perhaps also by the hydrogen bond from Asp162.
Fig. 4. Proposed mechanism for β–CA. (A) Bicarbonate bound in the active site, in a position similar to that seen for the acetic acid molecule, forming hydrogen bonds with Gly224, Asp162 and Glu151. (B) This molecule decomposes into CO2, which continues to interact with Gln151′ and hydroxide. (C) CO2 diffuses out of the active site leaving a hydroxide ion bound to the zinc. This ion then accepts a proton from a buffer molecule in bulk solvent, leaving water bound at the active site zinc (D).
Clearly, the derivation of this mechanism is influenced by the pre-established mechanism for α–CA, but there is also much independent evidence for it. Both the aspartate residue and the residue locking it into position by a pair of hydrogen bonds, Arg164, are absolutely conserved in all β–CA sequences. Also, the Asp162Asn mutation in the spinach enzyme, constructed to test the possibility that this residue may be a zinc ligand, has enzymatic activity two orders of magnitude lower than the wild-type enzyme, a similar effect to that seen in Thr199 mutants in α–CA. The role of Asp162 in β–CA could be seen as that of a ‘gatekeeper’ residue in analogy with Thr199 in α–CA, preventing non-protonated atoms from binding effectively (as is seen with the binding of the ionized acetate), ensuring that the hydroxide ion is correctly oriented on the zinc for nucleophilic attack and also, perhaps, through the hydrogen bond it provides, helping to lower the hydroxide pKa. β–CA could also be said to possess a second filter, Gly224 N, which as a hydrogen bond donor to the same zinc-bound atom effectively restricts zinc binding to atoms with at least one proton and at least two free electron pairs. Gln151 is also conserved in all plant type β–CA sequences, as is Ser163, which orients it, and the hydrophobic nature of the rest of the active site pocket residues.
The second step of the reaction involves the regeneration of water from the zinc-bound hydroxide ion (Figure 4C and D). In β–CA the dependence of the reaction rate on buffer concentration implies that a buffer molecule serves as the ultimate proton acceptor, as indeed it must, as the reaction could proceed no faster than 103–104 s–1 with water acting in this role (Lindskog, 1997). The route taken by the proton out of the active site, however, is not clear. There are no residues apparent that could act as a proton shuttle in analogy to His64 in α–CAII. His209 was proposed as a candidate for this role (Björkbacka et al., 1999), but the structure shows this residue to be 10 Å from the zinc water position, completely solvent exposed and confined to a single conformation. The need to guide the movement of protons over several angstroms in α–CA seems to arise as a consequence of the active site being located at the bottom of a 15 Å deep funnel; in β–CA, with its active site much closer to the protein surface, efficient proton transfer might not require such a device. Whereas in α–CA the water molecule, which serves as one of the hydroxyl ligands, acts as the first link in the proton wire, in β–CA the analogous position is occupied by Gly224. Therefore, only the electron pair formerly mediating the bond to CO2 is available to accept the proton. This geometry also prevents the two steps of the reaction from occurring in a concerted fashion. The proton-accepting free electron pair points then into the active site cavity and away from the channel, which connects the active site with bulk solvent. Since work is required to order each water molecule in a proton conducting wire, long paths for proton transfer are incompatible with reactions at the near diffusion limited rates seen in CAs (Silverman, 1995). This implies that the most plausible route for proton passage is one that leads fairly directly to bulk solvent, i.e. in the direction of Tyr205′ OH. This path would be consistent with the observation that mutations in His209 result in a much lower effective Kbuffer (Björkbacka et al., 1999), as this residue is located near Tyr205 and may form part of a non-specific buffer binding site.
Structural convergence and non-convergence among CAs
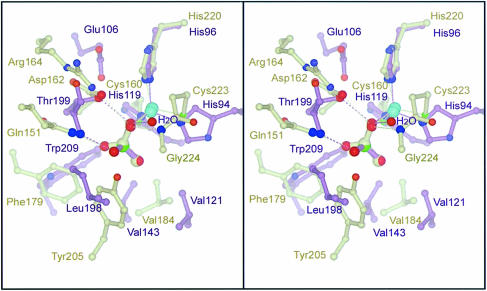
The binding of the acetic acid molecule in the active site of β–CA, when compared with the binding of bicarbonate-mimicking inhibitors such as bisulfite to α–CA, reveals many strong parallels. The zinc ligand inhibitor oxygen atom also forms hydrogen bonds with both a proton donor and an obligate proton acceptor, the second inhibitor oxygen acts as a proton acceptor, while the third atom is in a hydrophobic pocket. The zinc-bound oxygen atom is in a chiral environment defined by the remainder of the inhibitor molecule, the zinc ion and the hydrogen bond donating and accepting groups. A simple rotational superposition of the active sites of α–CA and β–CA produces a very poor overlay. Creating the mirror image of one molecule and then superimposing the two active sites reveals that functionally equivalent groups in the two active sites possess a very similar geometric arrangement, as shown by the overlay in Figure 5. Superimposed atoms (using β–CA molecules G and H) include the respective zinc ions, the respective protein atoms ligating zinc, atoms Thr199 Oγ1 (α–CA) and Asp162 Oδ1 (β–CA), Thr199 N and Gln147′ Nɛ2, the zinc hydroxide binding water mol– ecule and Gly224, and atoms O1 and O1, O3 and CH3, S and C of a bisulfite ion bound to α–CA and acetic acid bound to β–CA, respectively. The root-mean-square (r.m.s.) deviation for these 11 atom pairs is <0.4 Å. Furthermore, the residues forming the hydrophobic patch for the two enzymes also overlay quite closely, although without a one-to-one correlation of atoms.
Fig. 5. Active site overlay of α–CA and β–CA. Superimposition of the mirror image of the active site of α–CA with bisulfite bound (RCSB id code 5cac) (Håkansson et al., 1992) in plum onto the active site of β–CA with acetic acid bound in beige. The atoms used in the overlay include the respective zinc divalent cations; the zinc ligands His96 Nɛ2, His94 Nɛ2 and His119 Nδ2 (α–CA) versus His120 Nɛ2, Cys223 Sγ and Cys160 Sγ (β–CA); bisulfite atoms S, O2 and O3 and the acetic acid atoms C, O1 and CH3; among inhibitor/substrate interacting moieties, Thr199 Oγ1 on Asp162 Oδ2 and Thr199 N on Gln147′ Nɛ2, water on Gly224 N. This superposition also maps the residues of the respective hydrophobic patches, Phe179′, Val184 and Tyr205′ versus Val121, Val143, Leu198 and Trp209, approximately onto one another, although for these atoms there are not the same clear one-to-one-correspondences.
The third fold family of CAs, γ–CA, does not appear to use an obligate hydrogen bond accepting group as a ‘gatekeeper’ for the zinc-ligating atom, and in general, we anticipate that this enzyme employs a somewhat different molecular mechanism in catalysis. Although the zinc-binding ligands and bound substrate/inhibitor are somewhat similar in their spatial arrangement, the surrounding residues appear quite dissimilar in their functionality to those in the active sites of α- and β–CA, and no meaningful superimposition onto β–CA appears possible in either hand.
The close structural similarities in the active sites of α- and β–CAs lead us to conclude that these two enzymes have convergently evolved to a remarkable degree. Coupled with the close matches in the biochemical data available for the two enzymes, we further conclude that the enzymes are also likely to utilize a very similar mechanism. They should then serve as excellent models for one another and detailed comparisons of the results of experiments performed in parallel on the two should help advance understanding of both. Having such a pair of ‘twins’ is extremely useful, as experiments difficult to carry out in one system may prove more tractable in the other. For example, it has been suggested that a hydrogen bond between CO2 and Thr199 N in α–CA is needed for electrophilic activation of CO2 (Håkansson et al., 1992), but as the interaction is mediated by a mainchain atom not only are sequence conservation arguments inapplicable, but also the issue is unresolvable by mutagenesis studies. Not only does the finding of a similar motif in the (conserved) Gln151 sidechain amide of β–CA help lend credence to the idea that such a group has some functional role, but also the fact that the analogous substrate–enzyme interaction is mediated by a sidechain means that the exact functional role should now be amenable to experimental inquiry.
Similar motifs found in a pair of convergently evolved enzymes highlight them as being of potential functional importance in much the same way as sequence conservation within an enzyme family does. Here, one feature both conserved in and converged upon by α–CA and plant type β–CA (and also to a lesser degree in γ–CA) is the extensive active site hydrophobic patch. In α–CA, these residues have been proposed to perform many functions, including forming a CO2 binding site or in some way helping turnover (by excluding solvent, stabilizing the transition state, etc.). However, given that α–CA binds CO2 with 103- to 104–fold lower affinity than RuBisCO, although the hydrophobic residues may bind CO2 they are not likely to be optimized for this function. While it is true that mutagenesis of these amino acids to charged or larger amino acids is very disruptive to α–CAII turnover, some mutations to hydrophilic residues of similar volume are only moderately disruptive. For example, the Val121Ser, Val143Asn and Leu198Ser mutants show approximately 3-, 3- and 5–fold reduction in CO2 hydration activity, respectively (Fierke et al., 1991; Nair et al., 1991; Krebs et al., 1993). Since some α–CA isozymes are 104–fold less active than α–CAII, disruptions of this magnitude should be accommodated in the α–CA active site, yet even in the least active isozymes the hydrophobic nature of these residues is absolutely conserved. This implies a very strong selective pressure against hydrophilic residues in these positions, with selective pressure not being primarily for maximal turnover. We propose that substrate discrimination is what is exerting this pressure.
Most enzymes act on substrates with a large number of atoms, most of which are not affected during the catalytic cycle and so afford multiple potential strong and unaltering interactions, which allows for clear energetic discrimin– ation between substrates and non-substrates. Not only is CA's substrate small, but also the charge and hydrogen bonding behaviour of almost every atom changes during the catalytic cycle. This means that there are very few functionalities, and even fewer consistent ones, with which the enzyme can make the strong interactions that would allow energetic discrimination between the substrate mol– ecules and other, potentially inhibitory molecules. The fact that α–CA and β–CA (and to a lesser degree γ–CA) display appreciable inhibition constants for almost any small anion, including many present at appreciable levels in normal cellular environments (Johansson and Forsman, 1993; Alber and Ferry, 1994; Liljas et al., 1994), attests to the fact that CAs have not fully solved this problem, and as such the overall productivity of the enzyme in vivo is likely to be less constrained by the enzyme's maximal potential turnover (as measured in in vitro assays) than by competitive inhibition by the anions present in the cellular environment. By minimizing the number of potential hydrogen bond acceptors and donors around the zinc ion, the presence of the hydrophobic residues helps ensure that the binding energy of such molecules is as unfavourable as possible. Weak substrate binding is one necessary consequence of this, but since OH– is intrinsically highly nucleophilic and CO2 is intrinsically highly electrophilic, the bicarbonate–CO2 equilibration step remains faster than proton transfer. Other structural features shared by CAs would also seem to play a role in maximizing discrimin– ation. The gatekeeper residue, Thr199 in α–CA and Asp162 in β–CA, is set up as an obligatory hydrogen bond acceptor near the zinc ion, forcing unprotonated inhibitors to bind to the zinc ion with suboptimal geometry and therefore lower energy. Also, in all three CAs the active site is confined and sequestered from bulk solvent. Since maximal turnover is limited by how fast protons can be passed to buffer molecules in bulk solvent, this might seem counterproductive, but forcing molecules to approach the active site through a restricted diameter passage could help, by steric means, to filter out larger molecular anions that might otherwise also be inhibitory.
Materials and methods
Crystallization, data collection and processing
Protein was purified and crystals grown as previously described (M.S.Kimber, J.R.Coleman and E.F.Pai, submitted). Briefly, the protein was crystallized by vapour diffusion against 16% polyethylene glycol (PEG) 4000 MW (Fluka), 400 mM ammonium acetate, 50 mM dithiothreitol and 100 mM sodium citrate pH 5.0 at 4°C. All data were collected with crystals flash-frozen at 100 K in artificial mother liquor with 25% PEG 4000 and 30% ethylene glycol as cryoprotectant. Crystals were of the orthorhombic space group C222 with cell parameters a = 136.9, b = 143.3, c =202.1 Å and α = β = γ = 90°. The crystals were generally highly anisotropic in their scattering. A three wavelength MAD data set was collected at the National Synchrotron Light Source, beamline X8–C, using an MAR345 area detector. The 1.93 Å high resolution data set was collected at the Advanced Photon Source beamline 14C (BioCARS) with a Quantum Q4 CCD detector. All data were reduced using DENZO and scaled using SCALEPACK (Otwinowski and Minor, 1997) (Table I).
Table I. Data collection statistics.
| λ1 (peak) | λ2 (edge) | λ3 (remote) | High resolution | |
|---|---|---|---|---|
| Wavelength (Å) | 1.281323 | 1.282716 | 1.00000 | 1.0000 |
| Resolution (Å) | 30–2.7 | 30–2.8 | 30–2.7 | 40–1.93 |
| No. of observations | 253 785 | 144 276 | 166 306 | 779 081 |
| No. unique | 49 623 | 43 192 | 48 177 | 138 484 |
| Rsyma | 0.066 | 0.048 | 0.053 | 0.067 |
| (last shell) | 0.309 | 0.179 | 0.212 | 0.277 |
| I/σ | 12.2 | 15.6 | 13.8 | 13.0 |
| (last shell) | 4.4 | 5.1 | 3.8 | 3.7 |
| Completeness | 0.908 | 0.876 | 0.877 | 0.932 |
| (last shell) | 0.839 | 0.722 | 0.679 | 0.785 |
Rsym = ΣhklΣi∣Ii – <I>∣/ΣhklΣi<I> where Ii is the ith measurement of the reflection intensity I and <I> is the weighted mean of all measurements of I.
Phase determination
The structure was determined by MAD using the signal from the intrinsic zinc atoms only. The program SOLVE (Terwilliger and Berendzen, 1999) was used to find a solution to the heavy atom substructure using the three wavelength MAD dataset at 3.8 Å. Eight zinc ions were found with high occupancies (0.85–1.05), yielding a figure of merit (FOM) of 0.58. While elements of secondary structure were visible in the MAD map, connectivity was relatively poor and the map was unsuitable for chain tracing. The disposition of the zinc sites allowed initial determin– ation of the location and orientation of two of the NCS 2–fold axes. Phase improvement and extension to 2.7 Å was then performed in the CCP4 program DM (Cowtan and Main, 1996) using techniques including solvent flattening and 3–fold NCS averaging with a sphere being used as the NCS mask. The improvement in the map was sufficient to allow much of the secondary structure to be recognized in a skeletonization and the accurate determination of the remaining NCS elements together with the definition of a more accurate mask. Eight–fold symmetry averaging, coupled with solvent flattening and histogram matching in DM produced a high quality map with an overall FOM of 0.78. The protein was then fully traced into this map using the molecular graphics program O (Jones et al., 1991).
Refinement
Simulated annealing torsion angle refinement against amplitude maximum likelihood targets was performed in CNS version 0.5 (Brünger et al., 1998). Individual B–factor refinement was also used in the same program. Rounds of refinement were interspersed with rounds of rebuilding in O. The model was initially refined using strict NCS with the dimer as asymmetric unit, which was later released to eight molecules with strong harmonic restraints. No NCS restraints were used in the final rounds of refinement. The current model contains residues 120–329, 117–329, 116–329, 120–329, 118–329, 116–329, 110–329 and 119–329 for molecules A–H respectively, with no chain breaks. The asymmetric unit also contains eight Zn2+ ions, eight Cl– ions, four Cu2+ ions (at half occupancy, a tentative interpretation of a strong density peak between Cys166 and Cys166′ at the monomer–monomer interface), eight acetate molecules and 894 water molecules (Table II). Figures were prepared using SPOCK and MOLSCRIPT (Kraulis, 1991) and rendered using Raster3D (Merritt and Murphy, 1994).
Table II. Refinement statistics.
| Resolution range (Å) | 40–1.93 |
| Rcryst (%)a | 22.7 |
| Rfree (%)b | 25.2 |
| R.m.s. deviations | |
| bond lengths (Å) | 0.012 |
| bond angles (°) | 1.5 |
| torsion angles (°) | 22 |
| improper torsion angles (°) | 1.0 |
aRcryst = Σhkl∣Fobs – Fcalc∣/Fobs where Fobs and Fcalc are the observed and the calculated structure factors, respectively, and the summation is over the 99% of reflections used for model refinement.
bRfree as for Rcryst except summed only over the 1% of reflections not used for model refinement.
Acknowledgments
Acknowledgements
The authors wish to thank Dr John R.Coleman for bringing this problem to our attention, for his generous gift of the pCA plasmid-containing cells and for helpful discussions. We are grateful to the staff at BNL X8–C and APS BioCARS for assistance with data collection. This research was supported by the National Science and Engineering Council of Canada through an Industrial Research Chair to E.F.P. M.S.K. is the recipient of a Medical Research Council of Canada post-graduate student scholarship and a University of Toronto Open Scholarship. A joint grant from the Medical Research and National Sciences and Engineering Councils of Canada enabled use of beamline X8-C at the National Synchrotron Light Source. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-Eng-38. Atomic coordinates have been deposited at the RCSB structure database, id 1EKJ.
References
- Alber B.E. and Ferry, J.G. (1994) A carbonic anhydrase from the archaeon Methanosarcina thermophila.Proc. Natl Acad. Sci. USA, 91, 6909–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliev D.A., Guliev, N.M., Mamedov, A.M. and Tsuprun, V.L. (1986) Physico-chemical properties and quaternary structure of carbonic anhydrase from Cicer arietinum leaves. Biokhimiya, 51, 1785–1794. [Google Scholar]
- Badger M.R. and Price, G.D. (1994) The role of carbonic anhydrase in photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol., 45, 369–392. [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Björkbacka H., Johansson, I.M. and Forsman, C. (1999) Possible roles for His 208 in the active-site region of chloroplast carbonic anhydrase from Pisum sativum.Arch. Biochem. Biophys., 361, 17–24. [DOI] [PubMed] [Google Scholar]
- Bracey M.H., Christiansen, J., Tovar, P., Cramer, S.P. and Bartlett, S.G. (1994) Spinach carbonic anhydrase: investigation of the zinc-binding ligands by site-directed mutagenesis, elemental analysis and EXAFS. Biochemistry, 33, 13126–13131. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998)Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cowtan K.D. and Main, P. (1996) Phase combination and cross-validation in iterated density modification calculations. Acta Crystallogr. D, 52, 43–48. [DOI] [PubMed] [Google Scholar]
- Fawcett T.W., Browse, J.A., Volokita, M. and Bartlett, S.G. (1990) Spinach carbonic anhydrase primary structure deduced from the sequence of a cDNA clone. J. Biol. Chem., 265, 5414–5417. [PubMed] [Google Scholar]
- Fierke C.A., Calderone, T.L. and Krebs, J.F. (1991) Functional consequences of engineering the hydrophobic pocket of carbonic anhydrase II. Biochemistry, 30, 11054–11063. [DOI] [PubMed] [Google Scholar]
- Guilloton M.B., Korte, J.J., Lamblin, A.F., Fuchs, J.A. and Anderson, P.M. (1992) Carbonic anhydrase in Escherichia coli. A product of the cyn operon. J. Biol. Chem., 267, 3731–3734. [PubMed] [Google Scholar]
- Håkansson K., Carlsson, M., Svensson, L.A. and Liljas, A. (1992) Structure of native and apo carbonic anhydrase II and structure of some of its anion–ligand complexes. J. Mol. Biol., 227, 1192–1204. [DOI] [PubMed] [Google Scholar]
- Hatch M.D. (1991) Carbonic anhydrase assay: strong inhibition of the leaf enzyme by CO2 in certain buffers. Anal. Biochem., 192, 85–89. [DOI] [PubMed] [Google Scholar]
- Hiltonen T., Bjorkbacka, H., Forsman, C., Clarke, A.K. and Samuelsson, G. (1998) Intracellular β–carbonic anhydrase of the unicellular green alga Coccomyxa. Cloning of the cDNA and characterization of the functional enzyme overexpressed in Escherichia coli.Plant Physiol., 117, 1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebanathirajah J.A. and Coleman, J.R. (1998) Association of carbonic anhydrase with a Calvin cycle enzyme complex in Nicotiana tabacum.Planta, 204, 177–182. [DOI] [PubMed] [Google Scholar]
- Johansson I.M. and Forsman, C. (1992) Processing of the chloroplast transit peptide of pea carbonic anhydrase in chloroplasts and in Escherichia coli. Identification of two cleavage sites. FEBS Lett., 314, 232–236. [DOI] [PubMed] [Google Scholar]
- Johansson I.M. and Forsman, C. (1993) Kinetic studies of pea carbonic anhydrase. Eur. J. Biochem., 218, 439–446. [DOI] [PubMed] [Google Scholar]
- Johansson I.M. and Forsman, C. (1994) Solvent hydrogen isotope effects and anion inhibition of CO2 hydration catalysed by carbonic anhydrase from Pisum sativum.Eur. J. Biochem., 224, 901–907. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W. and Kjeldgaard, M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kandel M., Gornall, A.G., Cybulsky, D.L. and Kandel, S.I. (1978) Carbonic anhydrase from spinach leaves. Isolation and some chemical properties. J. Biol. Chem., 253, 679–685. [PubMed] [Google Scholar]
- Kisker C., Schindelin, H., Alber, B.E., Ferry, J.G. and Rees, D.C. (1996) A left-hand β-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila.EMBO J., 15, 2323–2330. [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Krebs J.F., Rana, F., Dluhy, R.A. and Fierke, C.A. (1993) Kinetic and spectroscopic studies of hydrophilic amino acid substitutions in the hydrophobic pocket of human carbonic anhydrase II. Biochemistry, 32, 4496–4505. [DOI] [PubMed] [Google Scholar]
- Liljas A. et al. (1972)Crystal structure of human carbonic anhydrase C. Nature New Biol., 235, 131–137. [DOI] [PubMed] [Google Scholar]
- Liljas A., Håkansson, K., Jonsson, B.H. and Xue, Y. (1994) Inhibition and catalysis of carbonic anhydrase. Recent crystallographic analyses. Eur. J. Biochem., 219, 1–10. [DOI] [PubMed] [Google Scholar]
- Lindskog S. (1997) Structure and mechanism of carbonic anhydrase. Pharmacol. Ther., 74, 1–20. [DOI] [PubMed] [Google Scholar]
- Majeau N., Arnoldo, M.A. and Coleman, J.R. (1994) Modification of carbonic anhydrase activity by antisense and over-expression constructs in transgenic tobacco. Plant Mol. Biol., 25, 377–385. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy, M.E.P. (1994) Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Nair S.K., Calderone, T.L., Christianson, D.W. and Fierke, C.A. (1991) Altering the mouth of a hydrophobic pocket. Structure and kinetics of human carbonic anhydrase II mutants at residue Val-121. J. Biol. Chem., 266, 17320–17325. [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Provart N.J., Majeau, N. and Coleman, J.R. (1993) Characterization of pea chloroplastic carbonic anhydrase. Expression in Escherichia coli and site-directed mutagenesis. Plant Mol. Biol., 22, 937–943. [DOI] [PubMed] [Google Scholar]
- Ren X. and Lindskog, S. (1992) Buffer dependence of CO2 hydration catalyzed by human carbonic anhydrase I. Biochim. Biophys. Acta, 1120, 81–86. [DOI] [PubMed] [Google Scholar]
- Rowlett R.S., Chance, M.R., Wirt, M.D., Sidelinger, D.E., Royal, J.R., Woodroffe, M., Wang, Y.F., Saha, R.P. and Lam, M.G. (1994) Kinetic and structural characterization of spinach carbonic anhydrase. Biochemistry, 33, 13967–13976. [DOI] [PubMed] [Google Scholar]
- Rumeau D., Cuine, S., Fina, L., Gault, N., Nicole, M. and Peltier, G. (1996) Subcellular distribution of carbonic anhydrase in Solanum tuberosum L. leaves: characterization of two compartment-specific isoforms. Planta, 199, 79–88. [DOI] [PubMed] [Google Scholar]
- Silverman D.N. (1995) Proton transfer in carbonic anhydrase measured by equilibrium isotope exchange. Methods Enzymol., 249, 479–503. [DOI] [PubMed] [Google Scholar]
- Smith K.S. and Ferry, J.G. (1999) A plant-type (β-class) carbonic anhydrase in the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum.J. Bacteriol., 181, 6247–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen, J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]