Abstract
The laminins are large heterotrimeric glycoproteins with fundamental roles in basement membrane architecture and function. The C–terminus of the laminin α chain contains a tandem of five laminin G–like (LG) domains. We report the 2.0 Å crystal structure of the laminin α2 LG4–LG5 domain pair, which harbours binding sites for heparin and the cell surface receptor α–dystroglycan, and is 41% identical to the laminin α1 E3 fragment. LG4 and LG5 are arranged in a V–shaped fashion related by a 110° rotation about an axis passing near the domain termini. An extended N–terminal segment is disulfide bonded to LG5 and stabilizes the domain pair. Two calcium ions, one each in LG4 and LG5, are located 65 Å apart at the tips of the domains opposite the polypeptide termini. An extensive basic surface region between the calcium sites is proposed to bind α–dystroglycan and heparin. The LG4–LG5 structure was used to construct a model of the laminin LG1–LG5 tandem and interpret missense mutations underlying protein S deficiency.
Keywords: dystroglycan/extracellular matrix/laminin E3 fragment/X-ray crystallography
Introduction
Basement membranes are specialized sheet-like extracellular matrices that underlie epithelial and endothelial cell sheets and surround muscle cells, fat cells and peripheral nerve axons. The supramolecular structure of basement membranes derives largely from extensive protein networks formed by collagen IV and laminin (Timpl and Brown, 1996). The laminins constitute a family of heterotrimeric (αβγ) glycoproteins with diverse and crucial functions in basement membrane assembly and function (Aumailley and Smyth, 1998). Five α, three β and three γ chains have been identified to date, which associate to form at least 12 laminin isoforms (Aumailley and Smyth, 1998; Iivanainen et al., 1999; Koch et al., 1999). Laminin 1 (α1β1γ1) and the closely related laminin 2 (α2β1γ1) are cross-shaped molecules (Beck et al., 1990). All three chains contribute to the α–helical coiled-coil that forms the long arm of the cross, whereas the short arms are composed of one chain each. The N–terminal globular domains at the tips of the short arms mediate the calcium-dependent polymerization of laminins 1 and 2 into a polygonal network (Yurchenco and Cheng, 1993). The α chains are unique in that they contain at their C–termini a tandem of five laminin G–like (LG) domains, LG1–LG5. The C–terminal LG4–LG5 pair, which in the case of the α1 chain can be released by limited proteolysis (E3 fragment), harbours binding sites for heparin, sulfatides and the cell surface receptor dystroglycan (Ott et al., 1982; Ervasti and Campbell, 1993; Gee et al., 1993; Smalheiser, 1993; Sung et al., 1993; Talts et al., 1999).
α–dystroglycan (α–DG), the extracellular portion of the dystroglycan transmembrane complex (Henry and Campbell, 1999), plays an important role in basement membrane assembly by binding and organizing soluble laminin at the cell surface, thereby facilitating the polymerization process (Henry and Campbell, 1998; Colognato et al., 1999; Montanaro et al., 1999). Laminin 1 is present characteristically in epithelial tissues (Aumailley and Smyth, 1998), where α–DG additionally may bind to the LG domains of the major basement membrane proteoglycan perlecan (Talts et al., 1999). In muscle, the α–DG–laminin 2 complex provides a crucial linkage of the cytoskeleton to the extracellular matrix, and mutations in the laminin α2 chain result in muscular dystrophy (Henry and Campbell, 1999). In nerve cells, α–DG supports Schwann cell adhesion to laminins 1 and 2 (Yamada et al., 1994, 1996). The α–DG–laminin 2 complex also serves as an attachment site for Mycobacterium leprae during Schwann cell penetration (Rambukkana et al., 1998).
Structure–function studies of laminins have long been hampered by the difficulty of obtaining recombinant fragments of the LG1–LG5 tandem. We have recently been able to obtain individual LG domains, as well as LG tandems, in recombinant form by expression in mammalian cells (Talts et al., 1998; Andac et al., 1999). This has allowed a precise mapping of binding sites on the LG4–LG5 tandem. In the laminin α1 chain, the binding sites for heparin, sulfatides and α–DG are contained within LG4, and site-directed mutagenesis of basic residues has identified several regions as important for ligand binding (Andac et al., 1999). In contrast, in the related α2 chain, only the LG4–LG5 pair, but not the individual LG domains, supports high-affinity α–DG binding. Heparin binds to LG5 alone, albeit less strongly than to the LG4–LG5 pair (Talts et al., 1999).
We have recently solved the crystal structure of the laminin α2 chain LG5 domain (α2LG5), which revealed a 14–stranded β–sandwich with similarity to the pentraxins (Hohenester et al., 1999). The calcium dependence of α–DG binding could be explained by the presence of a conserved calcium site on α2LG5, which was suggested to bind directly to the anionic sugar moieties of α–DG. In addition, several lysine residues in LG5 were found to be important for α–DG and heparin binding to the laminin α2 chain (Hohenester et al., 1999). The structure of a single LG domain from the neuronal cell recognition molecule neurexin Iβ was also reported recently (Rudenko et al., 1999). This LG domain is similar to α2LG5, but lacks the calcium-binding site conserved in laminins, agrin and perlecan (Hohenester et al., 1999).
To understand how the LG4 and LG5 domains in laminin 2 cooperate in α–DG binding, we have now determined the structure of the laminin α2 chain LG4–LG5 pair (α2LG4–5). The apparently rigid α2LG4–5 tandem is seen to use an extensive basic surface region bounded by the conserved calcium-binding sites to bind α–DG. Our structure also provides the foundation to understand LG tandems in other proteins.
Results and discussion
Structure determination
The recombinant α2LG4–5 fragment used in this study spans residues 2729–3118 of the mature murine laminin α2 chain and additionally contains a vector-derived APLA sequence at the N–terminus (Talts et al., 1998). The boundaries of the construct were chosen to match the well characterized proteolytic E3 fragment of the homologous laminin α1 chain (Ott et al., 1982; Deutzmann et al., 1988). Thus, the LG4 module in α2LG4–5 is preceded by an N–terminal segment of ∼30 residues containing a single cysteine, Cys2747. The α2LG4–5 crystal structure was solved by a combination of molecular replacement (MR) and single isomorphous replacement (SIR) and refined to 2.0 Å resolution (Tables I and II). The first and last residue defined by the electron density are His2744 and Thr3117, respectively. The first 19 residues including the APLA sequence are disordered in the crystal. The α2LG4–5 tandem bears an N-linked glycan attached to Asn2889 in LG4 (Talts et al., 1998). This residue is located in a mobile loop segment and, as only weak density is observed for the sugar moieties, no attempts were made to include the glycan in the model.
Table I. Data collection and phasing statisticsa.
| Native 1 | Native 2 | Sm(NO3)3 | |
|---|---|---|---|
| Resolution range (Å) | 20–2.6 | 20–2.0 | 20–2.8 |
| Beamline | DESY X31 | SRS 9.6 | DESY X31 |
| Unique reflections | 15 308 | 31 704 | 13 474 |
| Multiplicity | 3.3 | 3.4 | 2.0 |
| Rmergeb | 0.060 (0.179) | 0.056 (0.135) | 0.065 (0.322) |
| Completeness (%) | 99.6 (99.8) | 94.8 (93.6) | 98.8 (98.0) |
| Rderivc | 0.238 | ||
| RCullisd (centric/acentric) | 0.77/0.85 | ||
| Phasing powere (centric/ acentric) | 0.84/1.04 |
aValues in parentheses are for reflections in the highest resolution bin.
bRmerge = ΣhΣi∣Ii(h) – <I(h)>∣/ΣhΣi Ii(h), where Ii(h) is the i-th measurement of reflection h and <I(h)> is the weighted mean of all measurements of h.
cRderiv = Σh∣∣FPH∣ – ∣FP∣∣/Σh∣FP∣, where FP and FPH are the native and derivative structure factors, respectively. Only data to 3.0 Å resolution were used in phasing.
dRCullis = Σh∣∣∣FPH∣ – ∣FP∣∣ – ∣FH∣∣/Σh∣∣FPH∣ – ∣FP∣∣, where FH is the calculated heavy atom structure factor.
eThe phasing power is defined as (r.m.s. FH/r.m.s. lack of closure) summed over all reflections used in the heavy atom refinement.
Table II. Refinement statistics.
| Resolution range (Å) | 20–2.0 |
| Reflections (working set) | 29 432 |
| Reflections (test set) | 2244 |
| Protein atoms | 2863 |
| Metal ions | 2 Ca2+ |
| Solvent sites | 252 |
| Rcrysta | 0.233 |
| Rfreea | 0.261 |
| R.m.s.d. bond lengths (Å) | 0.007 |
| R.m.s.d. bond angles (°) | 1.5 |
| R.m.s.d. B-factors (Å2) | 2.2 |
aR = Σh∣Fobs – Fcalc∣/Σh Fobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rcryst and Rfree were calculated using the working and test set, respectively.
The structure of α2LG4–5
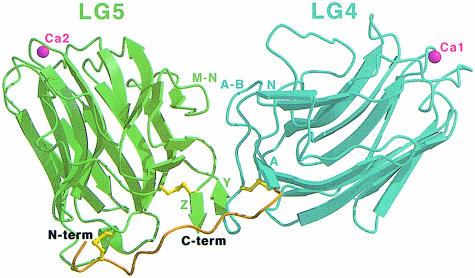
The α2LG4–5 structure has approximate overall dimensions of 35 × 40 × 80 Å. Both LG domains, LG4 and LG5, are built up from the typical 14–stranded antiparallel β–sandwich with a disulfide bridge near the domain termini and a calcium-binding site at the opposite edge of the sandwich (Hohenester et al., 1999; Rudenko et al., 1999). In the α2LG4–5 tandem, the two domains are arranged in a V–shaped fashion such that their termini are located close to the domain interface at the bottom of the V, while the calcium-binding sites are situated at the tips of the domains at 65 Å from one another (Figure 1). LG4 and LG5 are related by an almost pure rotation of 110° about an axis passing near the domain termini. This arrangement is forced by the close proximity of the domain termini and is unusual for tandemly repeated domains, which commonly have their termini at opposite ends of the domain (Bork et al., 1996). The portion of the N–terminal segment that is defined by the electron density extends ∼30 Å away from LG4, running alongside LG5 and forming a disulfide bridge to the α–helical turn in LG5 situated opposite the calcium site (Cys2747–Cys3017). This unexpected structural feature results in the pairing of all cysteines in α2LG4–5. The N–terminal segment also greatly strengthens the association between LG4 and LG5, making the α2LG4–5 tandem a rigid structure with limited potential for domain flexing. The disordered portion of the N–terminus, which is unusual in that it contains many prolines and hydrophobic residues, is likely to interact with some part of LG1–LG3 in the complete laminin LG1–LG5 tandem.
Fig. 1. Cartoon drawing of the α2LG4–5 structure. The N–terminal segment (residues 2744–2758) is in brown, LG4 (2759–2933) in light blue and LG5 (2934–3117) in green. Disulfide bridges are in yellow and calcium ions are shown as pink spheres. The N- and C–termini are labelled. Both LG domains are 14–stranded antiparallel β–sandwiches. β–strands and loop regions involved in the inter-domain interface are labelled. The β–strands Y and Z in LG5 are not formed in the structure of the isolated LG5 domain (Hohenester et al., 1999).
Our previous α2LG5 structure (Hohenester et al., 1999) allowed the intra-domain disulfide bridges in the laminin LG1–LG5 tandem to be assigned with confidence (two in LG1 and one each in the other LG domains). The present α2LG4–5 structure has revealed an unusual inter-domain bridge between the LG3–LG4 linker and LG5. Thus, we can account for all 14 cysteines present in the laminin α1 and α2 chain LG1–LG5 tandems, finally clarifying a confusing issue.
The interface between LG4 and LG5
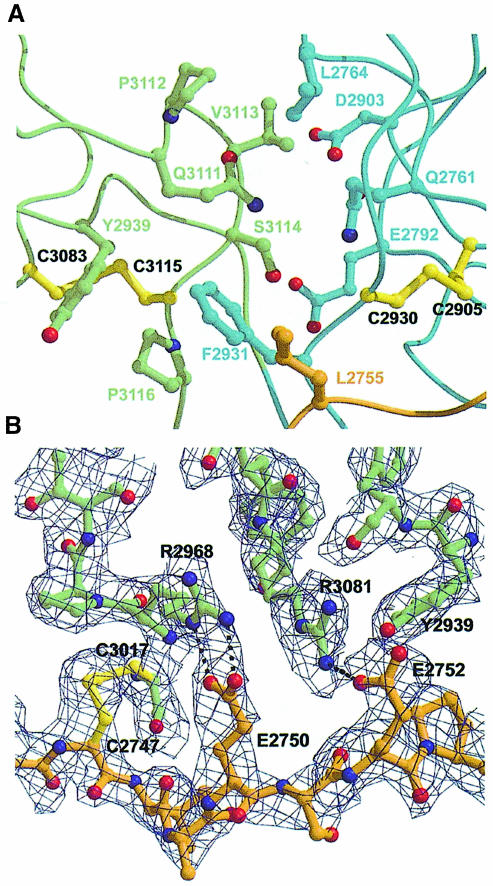
Neglecting the N–terminal segment (Ser2729–Gly2758), 430 Å2 of accessible surface area are buried in the LG4–LG5 interface (Figure 2A). The interface is constructed entirely from the N- and C–terminal portions of both LG domains. LG4 contributes strand A, the A–B turn and the loop following strand N, whereas LG5 contributes the M–N turn, strands Y and Z and their associated turns. The interface is formed from both hydrophobic and polar residues, but there is only one direct hydrogen bond between LG4 and LG5, involving Glu2792 and the amide nitrogen atom of Ser2936. The most prominent hydrophobic contact involves Phe2931, which packs against Gln3111, Pro3116 and Tyr2939 of LG5. In addition, Leu2764 in the LG4 A–B turn contacts Pro3112 and Val3113 of LG5. It is noteworthy that almost all of the key interface residues cluster near the conserved intra-domain disulfide bridges of both LG4 and LG5. At the back of the interface (in the view of Figure 2A), there is a shallow cavity lined by polar residues, among them Gln2761, Glu2792, Asp2903 and Ser3114, which contains several ordered water molecules mediating indirect contacts between LG4 and LG5. In the α2LG4–5 tandem, the first and last residues of LG5 form a small antiparallel β–sheet. The two strands, termed Y and Z, were not observed in the structure of the isolated LG5 domain (Hohenester et al., 1999), even though the corresponding residues (2933–2935 and 3115–3117, respectively) were present in the construct. Evidently, their conformation is dependent on the stabilizing influence of the adjacent LG4 domain, and the small Y–Z β–sheet should thus be regarded as a feature of the LG4–LG5 domain interface.
Fig. 2. ( A) The LG4–LG5 interface. The colour scheme is the same as in Figure 1. Residues involved in inter-domain contacts are shown as ball-and-stick models and are labelled. The most prominent contact is centred on Phe2931 (see the text). ( B) Interactions between the N–terminal segment (in brown) and LG5 (in green). Dashed lines indicate hydrogen bonds. The electron density shown is a simulated annealing 2Fobs – Fcalc omit map at 2.0 Å resolution (1.5σ contouring), in which the N–terminal segment and Cys3017 in LG5 were excluded from the phasing model.
The interactions between the N–terminal segment and LG5 bury a further 380 Å2 of accessible surface area (Figure 2B). Two hydrophobic residues preceding strand A in LG4, Pro2753 and Leu2755, contribute to the major hydrophobic region centred on Phe2931. Additionally, the residues flanking Leu2755 and Phe2931 form two main chain hydrogen bonds in an antiparallel β–sheet-like fashion. Further away from the LG4–LG5 interface and directly adjacent to the Cys2747–Cys3017 disulfide bridge, two hydrogen-bonded ion pairs (Glu2750–Arg2968 and Glu2752–Arg3081) are formed between the N–terminal segment and the LG5 domain. The N–terminal segment thus appears to be well integrated into the α2LG4–5 structure, consistent with earlier results obtained by limited proteolysis of the laminin α1 chain (Ott et al., 1982; Deutzmann et al., 1988).
Comparison of LG4 and LG5
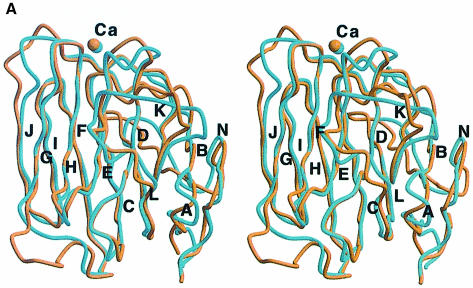
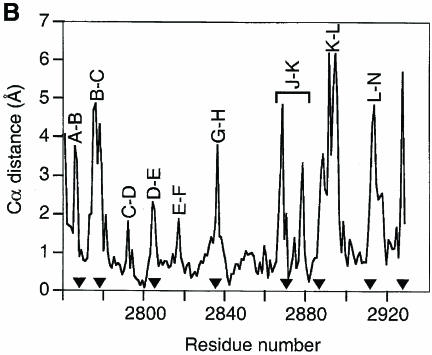
Despite a sequence identity of just under 20%, LG4 and LG5 are remarkably similar, and 133 Cα atoms (∼75% of the domains) can be superimposed with a root-mean-square deviation (r.m.s.d.) of 1.1 Å (Figure 3). With one notable exception, β–strands are conserved throughout and the structural differences between LG4 and LG5 are confined to loop regions. Significant differences are seen in the conformations of the B–C and L–M loops, which interact with one another. The LG4 B–C loop contains an imperfect α–helical turn, whereas the corresponding stretch in LG5 is in a fully extended conformation. The L–M loop is shorter in LG4 and, despite maintaining an extended conformation, the LG4 segment corresponding to strand M in LG5 is not involved in direct hydrogen bonding to strand L, but interacts with it via two water molecules. This deviation from conventional antiparallel hydrogen bonding is due to the presence of a proline, Pro2915, in the LG4 segment corresponding to strand M. The long K–L loop that folds over the concave face of the β–sandwich assumes different conformations in LG4 and LG5 and contains the glycosylation site at Asn2889 in LG4. Furthermore, the short α–helix in LG5, which is disulfide bonded to the N–terminal segment, is absent in LG4. Finally, some differences can be seen in the A–B, D–E and J–K loops. Overall, however, the high degree of structural conservation is most striking and reinforces our confidence in being able to make useful extrapolations from our structure to other members of the LG domain superfamily.
Fig. 3. ( A) Stereoview of a superposition of LG4 (light blue) and LG5 (brown). Common β–strands are labelled A–N. A total of 133 Cα atoms was used in the superposition (r.m.s.d. 1.1 Å). ( B) Cα distance plot corresponding to the superposition of LG4 and LG5 shown in (A). The sequence numbering corresponds to LG4, which was used as the reference structure. Filled triangles indicate the positions of insertions or deletions. Selected loop regions are labelled.
Calcium-binding sites in α2LG4–5
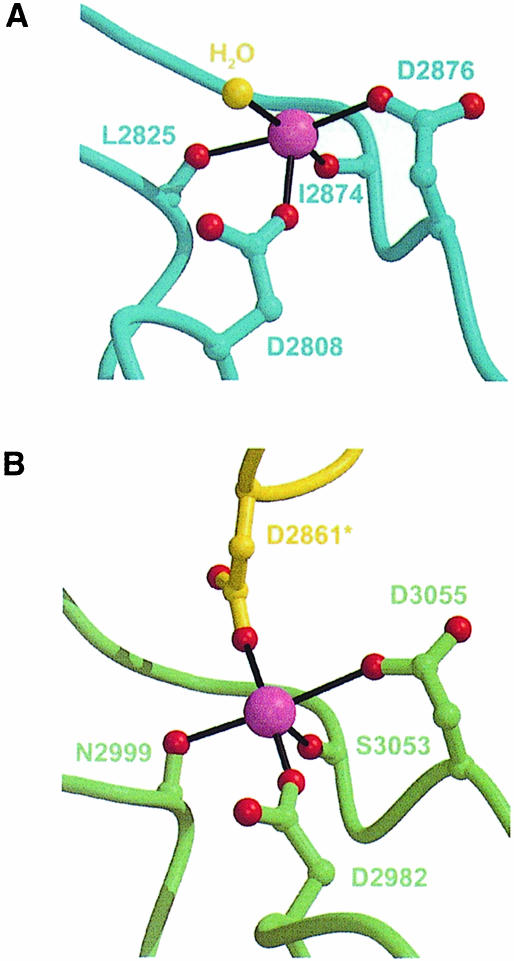
The α2LG4–5 structure contains two calcium ions, calcium 1 and 2, both apparently bound with full occupancy. The calcium-binding site in LG5 was known to exist and that in LG4 was predicted from sequence comparison (Hohenester et al., 1999). LG4 binds calcium 1 with two aspartic acid side chains, Asp2808 and Asp2876, and two main chain carbonyl oxygen atoms (Figure 4A). A water molecule occupies the fifth coordination site, leaving one apex of the coordination octahedron empty. Weak, roughly spherical electron density indicates the presence of a sixth calcium 1 ligand, which we have refined as water molecule 4003, although the large distance from the metal ion (3.8 Å) suggests that the ligand may be a HEPES buffer molecule. Excluding the unidentified ligand, the mean calcium–ligand distance in LG4 is 2.4 Å. LG5 binds calcium in a manner equivalent to LG4, the calcium 2 ligands being Asp2982, Asp3055 and two carbonyl oxygen atoms (Figure 4B). The calcium 2 coordination is augmented by the side chain of Asp2861 originating from the LG4 I–J turn of another α2LG4–5 molecule in the crystal, resulting in a mean calcium–ligand distance of 2.3 Å. In the previously reported crystal structure of α2LG5, a buffer-derived sulfate ion was bound to calcium 2 (Hohenester et al., 1999). The α2LG4–5 crystals were grown in the absence of sulfate ions, but an acidic residue from a neighbouring protein molecule is found in place of the calcium-bound sulfate ion. The strong preference of calcium 2 to complete its coordination sphere by recruiting additional anionic ligands implies a high electrophilicity of the calcium site in LG5. This adds further support to our previous proposal that acidic sugar moieties on α–DG may bind directly to the calcium ions present in LG domains (Hohenester et al., 1999). It remains to be shown whether α–DG binding to α2LG4–5 requires calcium 1, calcium 2 or both ions. Since the calcium sites are situated on the same face of the α2LG4–5 domain pair, all three alternatives are structurally feasible.
Fig. 4. Calcium-binding sites in α2LG4–5. ( A) Calcium 1 in LG4. The calcium ligands are shown as ball-and-stick models and are labelled. Calcium–ligand bonds are shown as black sticks. A metal-bound water molecule is shown in yellow. ( B) Calcium 2 in LG5. The calcium coordination is similar to calcium 1, but Asp2861 from a packing-related molecule in the crystal (in yellow) occupies the fifth coordination site (see the text).
Ligand-binding sites in α2LG4–5
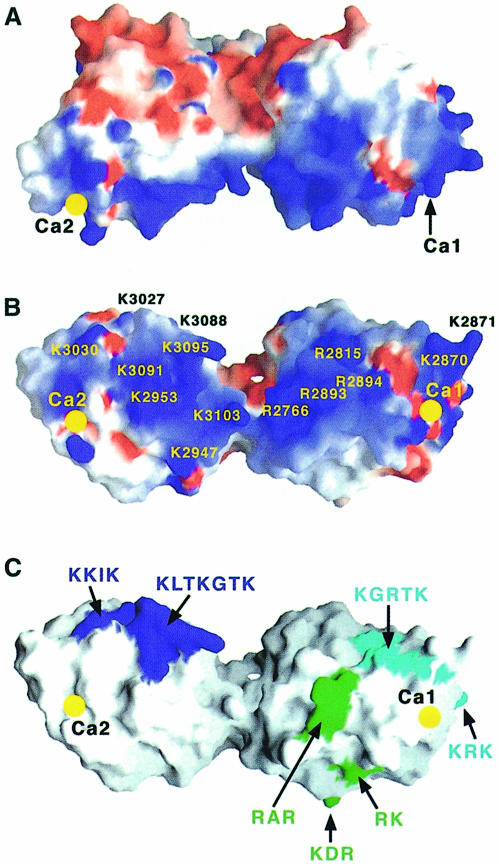
The α2LG4–5 fragment contains major binding sites for α–DG, heparin and sulfatides (Talts et al., 1999). Binding to α–DG, but not to the other ligands, is strictly calcium dependent (Ervasti and Campbell, 1993; Gee et al., 1993; Smalheiser, 1993; Talts et al., 1999). High-affinity binding appears to require LG4 and LG5 to be present in tandem arrangement, although mutations of key residues in LG5 alone have been shown to reduce dramatically the affinity of α2LG4–5 for α–DG and heparin (Hohenester et al., 1999). An electrostatic surface representation of the α2LG4–5 structure shows a striking segregation of positive and negative potential (Figure 5). The wide cleft between the LG4 and LG5 domains contains numerous basic residues, which, together with the calcium sites at the extremities of the α2LG4–5 tandem, form an extensive region of positive potential, well suited for binding the large acidic ligands α–DG and heparin. As discussed before, at least one of the calcium sites must be directly involved in α–DG binding to account for the metal dependence of the interaction. Calcium 2 in LG5 would appear to be the most likely candidate because of its electrophilic nature. Previous site-directed mutagenesis has shown four residues in LG5 (lysines 3027, 3030, 3088 and 3091) to be involved in the binding of α–DG and heparin (Hohenester et al., 1999). The regions containing these residues make up only a fraction of the total basic surface area (KKIK and KLTKGTK in Figure 5C), however, suggesting that additional basic residues may be involved in ligand binding. An extended α–DG-binding site in α2LG4–5 might also include lysines 2947, 2953 and 3103 on LG5, as well as a number of arginines on LG4 (Figure 5B).
Fig. 5. ( A and B) Electrostatic surface representations of α2LG4–5. The two views are related by a rotation of 70° about the horizontal axis. LG5 is on the left, LG4 on the right. Positive and negative potential is indicated by blue and red colouring, respectively, at the 5 kT level. The positions of the calcium sites are indicated by yellow filled circles. Also indicated are the approximate positions of basic residues suggested to form the extensive α–DG- and heparin-binding sites of α2LG4–5 (see the text). ( C) Mapping of regions implicated in ligand binding onto a solvent-accessible surface of α2LG4–5. The view direction is the same as in (B). The dark blue sequence regions in LG5 are involved in α–DG and heparin binding to the laminin α2 chain (Hohenester et al., 1999; Talts et al., 1999). The light blue regions in LG4 are involved in both α–DG and heparin binding to the laminin α1 chain, whereas the green regions contribute only to α–DG binding (Andac et al., 1999).
In the LG4–LG5 tandem of the laminin α1 chain, which corresponds to the proteolytic E3 fragment (Ott et al., 1982), the LG4 domain alone is sufficient to support high-affinity α–DG and heparin binding (Andac et al., 1999). Thus, although laminin 1 and 2 bind α–DG and heparin through a structurally highly conserved LG4–LG5 domain pair, the relative contributions of the individual LG domains have diverged during evolution, with LG4 apparently predominant in the α1 chain and LG5 in the α2 chain. Basic regions implicated in ligand binding to α1LG4 (Andac et al., 1999) map to the surroundings of the calcium 1 site in LG4 (KGRTK, KRK, RAR, KDR and RK in Figure 5C). To gain insight into the different binding properties of LG tandems in the laminin α1 and α2 chains, we constructed a homology model of α1LG4–5 based on the α2LG4–5 structure (41% sequence identity; Figure 6). The electrostatic surface properties of this model are remarkably similar to those of α2LG4–5, with an extended basic region encompassing both LG4 and LG5 (not shown). This was unexpected, as the residues imparting the basic character are generally not conserved between the two isoforms. It also poses a problem for understanding the specific binding to the large anionic α–DG glycoprotein. Since both laminin isoforms present similar basic surfaces covering the entire LG4–LG5 tandem and the calcium sites are conserved between α1LG4–5 and α2LG4–5, additional specific interactions must account for the observed isoform differences in α–DG binding. It is not obvious from our structure what these interactions might be, but we are now in a position to address this question experimentally by site-directed mutagenesis.
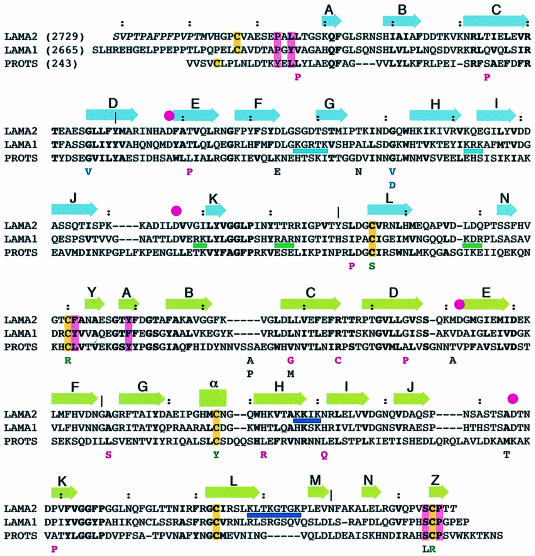
Fig. 6. Structure-based sequence alignment: LAMA2, mouse laminin α2; LAMA1, mouse laminin α1; PROTS, human protein S. The sequence numbers correspond to the mature polypeptide chains. Ser2665 in LAMA1 corresponds to the N–terminus of the proteolytic E3 fragment (Ott et al., 1982; Deutzmann et al., 1988). In LAMA2, every tenth residue is marked by a colon and every one hundredth residue by a vertical bar. Secondary structure elements of the α2LG4–5 structure are indicated above the alignment, in the colour scheme introduced in Figure 1. Residues in italics are disordered in the α2LG4–5 structure. Pink filled circles indicate calcium-binding residues. Cysteines are shaded yellow and residues contributing to the LG4–LG5 domain interface (see the text) are shaded pink. Sequence regions involved in α–DG and heparin binding are underlined in the colours used in Figure 5C. Missense mutations resulting in protein S deficiency (Villoutreix et al., 1997, and references therein) are indicated below the protein S sequence, as are their likely effects on the structure: red, introduction of proline into regions with main chain hydrogen bonding; green, mutation of cysteines in disulfide bridges; pink, perturbation of hydrophobic side chain packing; light blue, replacement of structurally important glycines.
Implications for other LG tandems
We have determined the first structure of an LG domain tandem, and it is interesting to consider whether the relative domain orientation observed in α2LG4–5 is maintained in other, more distantly related tandems. Uninterrupted tandems of more than two LG domains are only found in the laminin α chains, whereas pairs of domains are more widespread. The anticoagulant protein S (PS) (Lundvall et al., 1986) and the tyrosine receptor kinase ligand Gas6 (Manfioletti et al., 1993; Stitt et al., 1995; Varnum et al., 1995) contain an LG pair, as does sex hormone-binding globulin (SHBG) (Joseph and Baker, 1992). Neurexins, which form a large family of neuron-specific cell recognition molecules, contain both single and paired LG domains, separated from one another by EGF-like (EG) domains (Missler et al., 1998). The LG domains in the basement membrane proteoglycans perlecan and agrin are all separated by one or two EG domains (Timpl and Brown, 1996).
We predict uninterrupted LG pairs to have a similar relative domain orientation to that of α2LG4–5, i.e. a pure rotation of ∼120° between successive domains. Such an arrangement would appear to be dictated by the close proximity of the domain termini and a relative constant linker length of 5–10 residues; it probably does not depend greatly on the detailed nature of the interface. The V–shaped arrangement of paired LG domains may be disadvantageous in certain settings, which could explain the frequent occurrence of EG domains as spacer units. The stabilization of an LG pair by an N–terminal segment participating in an inter-domain disulfide bridge is unique to the laminin LG4–LG5 pair and its close relatives, PS, Gas6 and SHBG. These domain pairs are likely to be rigid and function as a single unit, whereas other LG pairs lacking this stabilizing feature may be more flexible.
In laminin α chains, the LG1–LG5 tandem is interrupted between LG3 and LG4 by a long linker (∼50 residues in laminin α2), a significant part of which is now seen to contribute to the stabilization of the LG4–LG5 pair. Because of the unexpected path of the linker, LG5 is likely to be closer in space to the LG1–LG3 portion than LG4. There are only 10 and six residues, respectively, connecting LG1 to LG2 and LG2 to LG3 (counting from the C–terminal cysteine to the first residue of strand A of the following LG domain). These short linkers must place severe constraints on the domain arrangement. We have, therefore, constructed a rough model of the LG1–LG3 tandem in the following way. We assumed that the α2LG4–5 structure with the N–terminal segment removed is a reasonable approximation to the LG1–LG2 tandem. By applying the 110° rotation that maps LG4 onto LG5 a second time, we then obtained an orientation for the third LG module in LG1–LG3. The resulting model structure of LG1–LG3, which has the shape of a cloverleaf with a diameter of ∼90 Å, has several striking features. First, LG3 does not clash with either LG1 or LG2, yet the domains are sufficiently close to one another to form sizable interfaces. Secondly, the C–terminus of LG2 and the N–terminus of LG3 are close and the gap can be spanned readily by a short linker, as required. Thirdly, the circumference of the cloverleaf is formed by the edges of the LG β–sandwiches that are known to harbour the polysaccharide and protein interaction sites of other LG domains (Hohenester et al., 1999; Rudenko et al., 1999). Both the N- and C–termini of LG1–LG3 emerge near the centre of the cloverleaf. The connection to LG4–LG5 is afforded by a long linker, while the connection to the α–helical coiled-coil, which forms the rod of the long arm of laminin, is much shorter. Assuming that the first cysteine following the α chain heptad repeats actually belongs to LG1 and takes part in a second intra-domain disulfide bond (Hohenester et al., 1999), the rod–LG1 linker is predicted to consist of only a few residues. Hence, there may be substantial interactions between the rod and LG1–LG3, perhaps involving the disulfide-bonded C–termini of the β and γ chains. This could explain why certain activities associated with the LG1–LG3 tandem, such as cell adhesion and spreading, as well as neurite outgrowth-promoting properties, require the presence of an intact rod in addition to the LG domains (Deutzmann et al., 1990; Sung et al., 1993; Lallier et al., 1994). In summary, our model of the laminin LG tandem, which was derived largely from the α2LG4–5 structure, is consistent with a substantial body of previous experimental data and, furthermore, agrees well with low-resolution images of laminin obtained by electron microscopy (Beck et al., 1990).
PS is a vitamin K-dependent plasma protein that functions as a cofactor to activated protein C in blood coagulation. PS deficiency has been established as a risk factor for thrombosis (Dahlbäck, 1991). PS is composed of a Gla module, a thrombin-sensitive region, four EG domains and a C–terminal pair of LG domains with ∼20% sequence identity to laminin α2LG4–5. The LG pair mediates the binding of PS to C4b-binding protein (C4BP), which abolishes the anticoagulant activity (He et al., 1997; Evenas et al., 1999). A sequence alignment of the LG pair of PS and the laminin α1 and α2 chains is shown in Figure 6. Evidently, the C–terminal portion of PS adopts the same structure as α2LG4–5. Not only are the individual LG domains conserved, but we find that the similarities extend to key interface residues and to the N–terminal segment that is disulfide bonded to the second LG domain in the pair. The affinity of PS for C4BP is enhanced 65-fold by calcium ions, which have been suggested to bind to the LG domains of PS (He et al., 1997). Our analysis rules out calcium-binding sites in PS corresponding to those in α2LG4–5, but other sites may exist of course. Villoutreix et al. (1997) have compiled 27 missense mutations in humans that result in PS deficiency and have attempted to rationalize their effects, but the analysis was severely limited by the lack of three-dimensional information on the LG structure. While it is not our intention to repeat in detail their analysis in the light of our structure, several general points can be made (Figure 6). We find that mutations are detrimental because they introduce prolines into regions of extended conformation with β–sheet hydrogen bonding (six cases), affect cysteines involved in disulfide bridges (four cases), introduce charges or holes into the hydrophobic core (five cases) or affect structurally important glycines (two cases). One PS mutation (Ser624Leu) is interesting as it affects a conserved residue in the LG domain interface (Ser3114 in α2LG4–5). The remaining nine mutations are difficult to classify and may affect PS glycosylation or binding to C4BP.
No reliable structure–function data exist on PS binding to C4BP (Villoutreix et al., 1997). Similarly, little detailed information is available regarding the binding of Gas6 to tyrosine kinase receptors of the Tyro 3/Axl family (Stitt et al., 1995; Varnum et al., 1995) and of SHBG to steroid hormones (Joseph and Baker, 1992). It will be interesting to see whether these binding sites are located in the same region as the established α–DG- and heparin-binding sites on the laminin LG4–LG5 tandem.
Materials and methods
Crystallization and data collection
The recombinant fragment α2LG4–5 was expressed in human embryonic kidney cells (293-EBNA) and purified as described (Talts et al., 1998). Crystals were obtained by the hanging drop vapour diffusion method at room temperature. Equal volumes (1.5 μl each) of protein solution (8 mg/ml in 10 mM Tris–HCl pH 8.0, 100 mM NaCl) and mother liquor [100 mM Na–HEPES pH 7.5, 2–8% (v/v) 2–propanol, 19–23% (w/v) PEG 4000, 10 mM CaCl2] were mixed and disposed over a reservoir of 1 ml of mother liquor. Crystals grew within 4–6 days and belong to space group C2221 with unit cell dimensions a = 70.6, b = 111.5 and c = 124.8 Å. There is one copy of α2LG4–5 in the asymmetric unit, resulting in a solvent content of ∼60%.
For data collection, crystals were soaked for ∼1 min in artificial mother liquor supplemented with 15% glycerol, suspended in nylon loops and flash-frozen in the cold nitrogen stream of an Oxford cryostream cooling device. To obtain a heavy atom derivative, crystals were soaked in mother liquor supplemented with 10 mM Sm(NO3)3 for 2 days. Native 1 and derivative data were collected at 100 K to resolutions of 2.6 and 2.8 Å, respectively, on station X31 at DESY, Hamburg (λ = 0.91 Å) using a MARRESEARCH image plate detector. A further native data set to 2.0 Å resolution (native 2) was collected at 100 K on station 9.6 at the SRS, Daresbury (λ = 0.87 Å) using an ADSC Quantum–4 CCD detector. In an attempt to reduce the high degree of mosaicity of α2LG4–5 crystals resulting from conventional flash-freezing, we used the crystal annealing procedure described by Harp et al. (1998). A crystal was removed from the cold nitrogen stream and re-soaked for a further 2 min at room temperature in cryoprotectant solution before being flash-frozen a second time. This procedure resulted in a significant improvement of the crystalline order. All diffraction data were processed with MOSFLM (Leslie, 1994) and reduced using programs of the CCP4 suite (Collaborative Computing Project No. 4, 1994). Data collection statistics are summarized in Table I.
Structure determination and refinement
The structure of α2LG4–5 was solved by a combination of MR and SIR. Using the entire structure of the isolated α2LG5 domain (Hohenester et al., 1999) as a search model in the CCP4 program AMoRe, a single outstanding solution corresponding to LG5 in α2LG4–5 was obtained. LG4 was then located by a partial translation function, in which LG5 was held fixed and the search model truncated to regions conserved between LG4 and LG5 (Hohenester et al., 1999). Following rigid-body refinement of an α2LG4–5 model consisting of a partial LG4 domain and the complete LG5 domain, a correlation coefficient of 0.46 and an R–factor of 0.46 were obtained (20–3.0 Å). Additional phase information was obtained from a samarium derivative. Heavy atom sites were deduced from difference Patterson maps and validated by difference Fourier maps calculated with phases from the MR model. Two major and one minor sites were identified and refined in the CCP4 program MLPHARE (Table I). The MR and SIR phases to 3.0 Å resolution were combined with the CCP4 program SIGMAA, and the resulting electron density map was subjected to density modification with the CCP4 program DM. The modified electron density map enabled the core residues of LG4 to be built, but the chain termini of α2LG4–5 and several loop regions remained ambiguous. Successive cycles of rebuilding with O (Jones et al., 1991) and combination of model and SIR phases allowed the model to be completed.
The α2LG4–5 structure was first refined with X-PLOR (Brünger, 1992) at 2.6 Å resolution using the native 1 data. When the high-resolution native 2 data became available, a round of simulated annealing from 3000 K was performed to remove model bias, and the structure was refined further at 2.0 Å resolution. A bulk solvent correction was applied. The final refined model has an R–factor of 0.233 (Rfree = 0.261). The geometry of the refined structure was analysed with PROCHECK (Laskowski et al., 1993). This identified 88.0% of the amino acid residues in the most favoured regions of the Ramachandran plot, 9.8% in allowed regions, 0.6% in generously allowed regions and 1.6% in disallowed regions. All seven residues with unfavourable main chain conformations are well defined by the electron density and are either involved in calcium binding or form part of the heparin-binding site of α2LG4–5. Refinement statistics are summarized in Table II. The α2LG4–5 coordinates have been deposited with the Protein Data Bank (entry 1dyk) and will be held for 1 year.
Preparation of figures
Figures 1–4 were made with BOBSCRIPT (Kraulis, 1991; Esnouf, 1997) and Raster3D (Merritt and Murphy, 1994). Figure 5 was made with GRASP (Nicholls et al., 1991).
Acknowledgments
Acknowledgements
We thank the staff at DESY beamline X31 and SRS beamline 9.6 for help with data collection, and Peter Brick for helpful criticism on the manuscript. This work was supported by a Wellcome Trust Senior Research Fellowship to E.H. and by the Deutsche Forschungsgemein– schaft (R.T.). J.F.T. acknowledges support from the Kungl. Fysiografiska sällskapet i Lund.
References
- Andac Z., Sasaki, T., Mann, K., Brancaccio, A., Deutzmann, R. and Timpl, R. (1999) Analysis of heparin, α–dystroglycan and sulfatide binding to the G domain of laminin α1 chain by site-directed mutagenesis. J. Mol. Biol., 287, 253–264. [DOI] [PubMed] [Google Scholar]
- Aumailley M. and Smyth, N. (1998) The role of laminins in basement membrane function. J. Anat., 193, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K., Hunter, I. and Engel, J. (1990) Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J., 4, 148–160. [DOI] [PubMed] [Google Scholar]
- Bork P., Downing, A.K., Kieffer, B. and Campbell, I.D. (1996) Structure and distribution of modules in extracellular proteins. Q. Rev. Biophys., 29, 119–167. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR Version 3.1: A System for Crystallography and NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Collaborative Computing Project No. 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Colognato H., Winkelmann, D.A. and Yurchenco, P.D. (1999) Laminin polymerization induces a receptor–cytoskeleton network. J. Cell Biol., 145, 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlbäck B. (1991) Protein S and C4b-binding protein: components involved in the regulation of the protein C anticoagulant system. Thromb. Haemost., 66, 49–61. [PubMed] [Google Scholar]
- Deutzmann R., Huber, J., Schmetz, K.A., Oberbäumer, I. and Hartl, L. (1988) Structural study of long arm fragments of laminin. Eur. J. Biochem., 177, 35–45. [DOI] [PubMed] [Google Scholar]
- Deutzmann R., Aumailley, M., Wiedemann, H., Psyny, W., Timpl, R. and Edgar, D. (1990) Cell adhesion, spreading and neurite stimulation by laminin fragment E8 depends on maintenance of secondary and tertiary structure in its rod and globular domain. Eur. J. Biochem., 191, 513–522. [DOI] [PubMed] [Google Scholar]
- Ervasti J.M. and Campbell, K.P. (1993) A role for the dystrophin–glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol., 122, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MOLSCRIPT which includes greatly enhanced colouring facilities. J. Mol. Graph., 15, 132–134. [DOI] [PubMed] [Google Scholar]
- Evenas P., Garcia de Frutos, P., Linse, S. and Dahlbäck, B. (1999) Both G-type domains of protein S are required for the high-affinity interaction with C4b-binding protein. Eur. J. Biochem., 266, 935–942. [DOI] [PubMed] [Google Scholar]
- Gee S.H., Blacher, R.W., Douville, P.J., Provost, P.R., Yurchenco, P.D. and Carbonetto, S. (1993) Laminin-binding protein 120 from brain is closely related to the dystrophin-associated glycoprotein, dystroglycan and binds with high affinity to the major heparin binding domain of laminin. J. Biol. Chem., 268, 14972–14980. [PubMed] [Google Scholar]
- Harp J.M., Timm, D.E. and Bunick, G.J. (1998) Macromolecular crystal annealing: overcoming increased mosaicity associated with cryocrystallography. Acta Crystallogr. D, 54, 622–628. [DOI] [PubMed] [Google Scholar]
- He X., Shen, L., Malmborg, A.-C., Smith, K.J., Dahlbäck, B. and Linse, S. (1997) Binding site for C4b-binding protein in vitamin K-dependent protein S is fully contained in carboxy–terminal laminin-G-type repeats: a study using recombinant factor IX–protein S chimeras and surface plasmon resonance. Biochemistry, 36, 3745–3754. [DOI] [PubMed] [Google Scholar]
- Henry M.D. and Campbell, K.P. (1998) A role for dystroglycan in basement membrane assembly. Cell, 95, 859–870. [DOI] [PubMed] [Google Scholar]
- Henry M.D. and Campbell, K.P. (1999) Dystroglycan inside and out. Curr. Opin. Cell Biol., 11, 602–607. [DOI] [PubMed] [Google Scholar]
- Hohenester E., Tisi, D., Talts, J.F. and Timpl, R. (1999) The crystal structure of a laminin G–like module reveals the molecular basis of α–dystroglycan binding to laminins, perlecan and agrin. Mol. Cell, 4, 783–792. [DOI] [PubMed] [Google Scholar]
- Iivanainen A., Morita, T. and Tryggvason, K. (1999) Molecular cloning and tissue-specific expression of a novel murine laminin γ3 chain. J. Biol. Chem., 274, 14107–14111. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.-Y., Cowan, S.W. and Kjeldgaard, M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Joseph D.R. and Baker, M.E. (1992) Sex hormone-binding globulin, androgen-binding protein and vitamin K-dependent protein S are homologous to laminin A, merosin and Drosophila crumbs protein. FASEB J., 6, 2477–2481. [DOI] [PubMed] [Google Scholar]
- Koch M., Olson, P.F., Albus, A., Jin, W., Hunter, D.D., Brunken, W.J., Burgeson, M.F. and Champliaud, M.F. (1999) Characterization and expression of the laminin γ3 chain: a novel, non-basement membrane-associated, laminin chain. J. Cell Biol., 145, 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lallier T., Deutzmann, R., Perris, R. and Bronner-Fraser, M. (1994) Neural crest cell interactions with laminin: structural requirements and localization of the binding site for α1β1 integrin. Dev. Biol., 162, 451–464. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1994) MOSFLM Users Guide. MRC-LMB Cambridge, UK. [Google Scholar]
- Lundvall Å., Dackowski, W., Cohen, E., Shaffer, M., Dahlbäck, B., Stenflo, J. and Wydro, R. (1986) Isolation and sequence of the cDNA for human protein S, a regulator of blood coagulation. Proc. Natl Acad. Sci. USA, 83, 6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfioletti G., Brancolini, C., Avanzi, G. and Schneider, C. (1993) The protein encoded by a growth arrest-specific gene (gas6) is a new member of the vitamin K-dependent proteins related to protein S, a negative coregulator in the blood coagulation cascade. Mol. Cell. Biol., 13, 4976–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy, M.E.P. (1994) Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Missler M., Fernandez-Chacon, R. and Südhof, T.C. (1998) The making of neurexins. J. Neurochem., 71, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Montanaro F., Lindenbaum, M. and Carbonetto, S. (1999) α–dystroglycan is a laminin receptor involved in extracellular matrix assembly on myotubes and muscle cell viability. J. Cell Biol., 145, 1325–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Ott U., Odermatt, E., Engel, J., Furthmayr, H. and Timpl, R. (1982) Protease resistance and conformation of laminin. Eur. J. Biochem., 123, 63–72. [DOI] [PubMed] [Google Scholar]
- Rambukkana A., Yamada, H., Zanazzi, G., Mathus, T., Salzer, J.L., Yurchenco, P.D., Campbell, K.P. and Fischetti, V.A. (1998) Role of α–dystroglycan as a Schwann cell receptor for Mycobacterium leprae. Science, 282, 2076–2079. [DOI] [PubMed] [Google Scholar]
- Rudenko G., Nguyen, T., Chelliah, Y., Südhof, T.C. and Deisenhofer, J. (1999) The structure of the ligand-binding domain of neurexin Iβ: regulation of LNS domain function by alternative splicing. Cell, 99, 93–101. [DOI] [PubMed] [Google Scholar]
- Smalheiser N.R. (1993) Cranin interacts specifically with the sulfatide-binding domain of laminin. J. Neurosci. Res., 36, 528–538. [DOI] [PubMed] [Google Scholar]
- Stitt T.N. et al. (1995)The anticoagulant factor protein S and its relative, GAS6, are ligands for the Tyro 3/Axl family receptor tyrosine kinases. Cell, 80, 661–670. [DOI] [PubMed] [Google Scholar]
- Sung U., O'Rear, J.J. and Yurchenco, P.D. (1993) Cell and heparin binding in the distal long arm of laminin: identification of active and cryptic sites with recombinant and hybrid glycoprotein. J. Cell Biol., 123, 1255–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talts J.F., Mann, K., Yamada, Y. and Timpl, R. (1998) Structural analysis and proteolytic processing of recombinant G domain of mouse laminin α2 chain. FEBS Lett., 426, 71–76. [DOI] [PubMed] [Google Scholar]
- Talts J.F., Andac, Z., Göhring, W., Brancaccio, A. and Timpl, R. (1999) Binding of G domains of laminin α1 and α2 chains and perlecan to heparin, sulfatides, α–dystroglycan and several extracellular matrix proteins. EMBO J., 18, 863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl R. and Brown, J.C. (1996) Supramolecular assembly of basement membranes. BioEssays, 18, 123–132. [DOI] [PubMed] [Google Scholar]
- Varnum B.C. et al. (1995)Axl receptor tyrosine kinase stimulated by the vitamin K-dependent protein encoded by growth arrest-specific gene 6. Nature, 373, 623–626. [DOI] [PubMed] [Google Scholar]
- Villoutreix B.O., Garcia de Frutos, P., Lövenklev, M., Linse, S., Fernlund, P. and Dahlbäck, B. (1997) SHBG region of the anticoagulant cofactor protein S: secondary structure prediction, circular dichroism spectroscopy and analysis of naturally occurring mutations. Proteins, 29, 478–491. [PubMed] [Google Scholar]
- Yamada H., Shimizu, T., Tanaka, T., Campbell, K.P. and Matsumura, K. (1994) Dystroglycan is a binding protein of laminin and merosin in peripheral nerve. FEBS Lett., 352, 49–53. [DOI] [PubMed] [Google Scholar]
- Yamada H. et al. (1996)Dystroglycan is a dual receptor for agrin and laminin-2 in Schwann cell membrane. J. Biol. Chem., 271, 23418–23423. [DOI] [PubMed] [Google Scholar]
- Yurchenco P.D. and Cheng, Y.–S. (1993) Self-assembly and calcium-binding sites in laminin. J. Biol. Chem., 268, 17286–17299. [PubMed] [Google Scholar]