Abstract
Plasmepsin I (PMI) is one of the four vacuolar pepsin-like proteases responsible for hemoglobin degradation by the malarial parasite Plasmodium falciparum, and the only one with no crystal structure reported to date. Due to substantial functional redundancy of these enzymes, lack of inhibition of even a single plasmepsin can defeat efforts in creating effective antiparasitic agents. We have now solved crystal structures of the recombinant PMI as apoenzyme and in complex with the potent peptidic inhibitor, KNI-10006, at the resolution of 2.4 and 3.1 Å, respectively. The apoenzyme crystallized in the orthorhombic space group P212121 with two molecules in the asymmetric unit and the structure has been refined to the final R-factor of 20.7%. The KNI-10006 bound enzyme crystallized in the tetragonal space group P43 with four molecules in the asymmetric unit and the structure has been refined to the final R-factor of 21.1%. In the PMI-KNI-10006 complex, the inhibitors were bound identically to all four enzyme molecules, with the opposite directionality of the main chain of KNI-10006 relative to the direction of the enzyme substrates. Such a mode of binding of inhibitors containing an allophenylnorstatine-dimethylthioproline insert in the P1-P1’ positions, previously reported in a complex with PMIV, demonstrates the importance of satisfying the requirements for the proper positioning of the functional groups in the mechanism-based inhibitors towards the catalytic machinery of aspartic proteases, as opposed to binding driven solely by the specificity of the individual enzymes. A comparison of the structure of the PMI-KNI-10006 complex with the structures of other vacuolar plasmepsins identified the important differences between them and may help in the design of specific inhibitors targeting the individual enzymes.
Keywords: Malaria, Plasmepsin, Aspartic Protease, Crystal structure, Enzyme inhibition
1. Introduction
Malaria, the most prevalent human disease of parasitic origin, is responsible for the death every year of nearly two million people, most of them children (Banerjee et al., 2002; Greenwood et al., 2005). The deadliest form of malaria is caused by the parasite Plasmodium falciparum, which invades erythrocytes and degrades a significant fraction of host cell hemoglobin (Kolakovich et al., 1997; Xiao et al., 2007). This process takes place in the food vacuole of the parasite (Francis et al., 1997). Four plasmepsins (PM) present in the food vacuole of P. falciparum, PMI, PMII, histo-aspartic protease (HAP), and PMIV, have been shown to be directly involved in the process of hemoglobin degradation (Banerjee et al., 2002; Coombs et al., 2001). These enzymes belong to the pepsin-like family of aspartic proteases and their sequences are highly homologous. The sequence identity between PMI and PMII, PMIV, and HAP is 73%, 68%, and 63%, respectively. However, sequence identity between PMI and cathepsin D, a more distantly related human aspartic protease, is only 35% (Francis et al., 1994). Similarly to other pepsin-like aspartic proteases, the active sites of PMI, PMII, and PMIV include two crucial aspartic acid residues, Asp32 and Asp215 (residue numbers used throughout this paper correspond to the numbers in the catalytically active form of mammalian pepsin), whereas Asp32 is replaced by histidine in HAP (Berry et al., 1999).
It has been postulated that the process of hemoglobin degradation is initiated by PMI cleaving the Phe33-Leu34 bond in the α-globin chain of native hemoglobin (Moon et al., 1997). It has been shown that pepstatin A, a general tight-binding aspartic protease inhibitor, as well as SC-50083, a specific inhibitor of PMI, kill cultured P. falciparum parasites, most probably by blocking hemoglobin degradation (Bailly et al., 1992; Francis et al., 1994; Liu et al., 2009). Several other selective inhibitors of PMI, such as Ro40-4388 and Ro40-5576, have also been shown to possess similar antiparasitic activity (Moon et al., 1997), although it cannot be excluded that their activity was due to interactions with targets other than vacuolar plasmepsins (Moura et al., 2009). Nevertheless, these results suggest that inhibition of PMI, as well as other plasmepsins, might provide an avenue for the development of novel drugs against malaria.
Recombinant expression of active plasmepsins has been generally challenging, as exemplified by many failures to purify enzymatically active HAP (Xiao et al., 2006). Similarly to HAP, expression of active recombinant PMI has also presented major difficulties (Coombs et al., 2001; Ersmark et al., 2006; Luker et al., 1996; Moon et al., 1997) that were overcome only recently through reengineering of the expression construct (Fig. 1) (Xiao et al., 2007). Although recombinant PMI was expressed in the past, attempts to crystallize it were unsuccessful (Moon et al., 1997). However, the enzyme has been extensively studied using biochemical techniques (Liu et al., 2009).
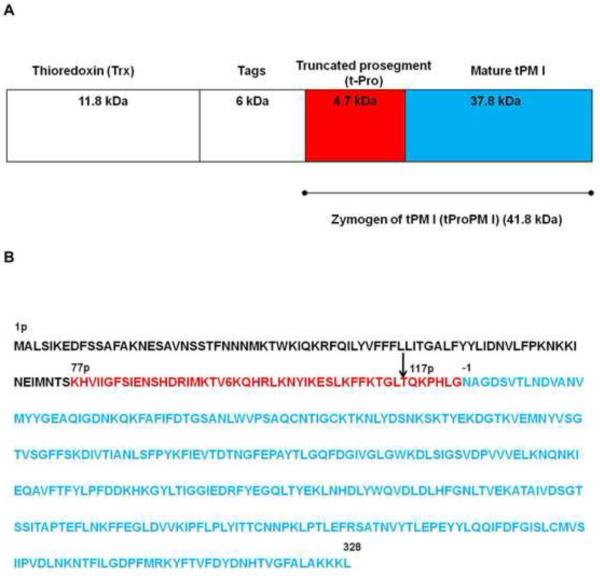
Figure 1.
Schematic representation of the construct of the recombinant Trx-tPMI fusion protein (A) and the amino acid sequence of PMI (B). (A) Diagram showing the expression construct of the recombinant truncated PMI thioredoxin fusion protein (Trx-tPMI). Zymogen of tPMI (tProPM I) was generated by removing the 76 N-terminal amino acids from the prosegment region which was linked to a highly soluble thioredoxin and tags. The tag region contains His-Tag, S-Tag, thrombin cleavage site, and enterokinase cleavage site. The zymogen of truncated PMI (tProPMI) was incorporated after the enterokinase cleavage site and the first 7 residues (AMAISDP) of tProPMI were derived from the plasmid, pET32b (+) (Xiao et al., 2007). (B) Amino acid sequence of PMI. The truncated prosegment (K77p-G123p, where p denotes the prosegment) is shown in red and mature PMI shown in blue (N1-L329). The soluble fusion protein was capable of autocatalytic activation at pH 4.2, which occurred between L116p-T117p (indicated by an arrow), seven residues upstream of the native cleavage site at G123p-N1 (Gluzman et al., 1994).
The inhibitor of PMI utilized in this study was KNI-10006, previously shown to be a potent inhibitor of this enzyme (Nezami et al., 2003). The KNI series of inhibitors was originally designed and developed primarily for the purpose of inhibiting HIV-1 protease (HIV-1 PR) (Kiriyama et al., 1993; Kiso, 1993; Kiso et al., 1999; Mimoto et al., 1991). These inhibitors have been designed based on the concept of “substrate transition-state mimicry” (Kiso, 1996), utilizing the central core made of an α-hydroxy-β-amino acid derivative, allophenylnorstatine (Apns), which contains a hydroxymethylcarbonyl isostere, followed by dimethylthioproline (Dmt) (Bhaumik et al., 2009; Nguyen et al., 2008a). This particular core was designed to mimic the unique Phe-Pro cleavage site found in GagPol polyprotein of HIV-1. A number of inhibitors containing such a core were synthesized and they exhibited high potency for inhibition of HIV-1 PR (Kiso, 1995), with considerable selectivity over human aspartic proteases (Clemente et al., 2006). It has also been shown that inhibitors belonging to this series exhibit good bioavailability and low toxicity (Kiriyama et al., 1996). Further optimization of the KNI inhibitors, supported by extensive structural and biochemical studies, resulted in the synthesis of many new compounds shown to be potent inhibitors of retroviral enzymes, such as HIV-1 and HTLV-1 PRs (Abdel-Rahman et al., 2004; Kimura et al., 2007; Maegawa et al., 2004; Nguyen et al., 2008b; Zhang et al., 2008a; Zhang et al., 2008b). Subsequent experiments have shown that selected KNI inhibitors are also effective against proteases expressed in Plasmodium parasites (Nezami et al., 2003). KNI-10006 was shown to be relatively potent inhibitor of PMI, PMII, PMIV, and HAP, with IC50 values of 0.28, 0.039, 0.69 and 0.015 μM, respectively (Bhaumik et al., 2009; Nezami et al., 2003). Since the outer parts of the molecule are joined by rotatable bonds to its core, they can easily adapt to the varied binding sites present in plasmepsins (Nezami et al., 2003), thus explaining their potency against different members of the family. Modeling studies have been previously undertaken in order to explain the mode of binding of KNI-10006 to PMII (Kiso et al., 2004; Nezami et al., 2003). The mode of binding of this inhibitor to HAP was investigated in a previous crystallographic study which has shown that it was bound in the active site of this enzyme with the central hydroxyl pointing away from the catalytic aspartates (Bhaumik et al., 2009), making it even more interesting to study its effects on other plasmepsins.
In this study, we describe crystallization of PMI and report crystal structures of its apo form and of a complex with KNI-10006. The two structures of PMI are compared with each other and with those of other plasmepsins and their inhibitor complexes.
2. Materials and methods
2.1. Synthesis of KNI-10006 and KNI-10772
The inhibitor KNI-10006 was synthesized as described previously (Kiso et al., 2004). KNI-10772 was synthesized in a similar manner, however allophenylnorstatine was replaced by phenylnorstatine [(2R,3S)-3-amino-2-hydroxy-4-phenylbutyric acid]. Briefly, (1S,2R)-1-amino-2-indanol was coupled with N-Boc-protected (R)-5,5-dimethyl-1,3-thiazolidinecarboxylic acid using benzotriazol-1-yloxy-tris(dimethylamino)phosphonium hexafluorophosphate (BOP) and triethylamine (Et3N) in dimethylformamide (DMF), followed by Boc-deprotection with 4 N HCl/dioxane and anisole to give an intermediate. The intermediate was coupled with N–Boc-protected phenylnorstatine using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC·HCl), 1-hydroxybenzotriazole (HOBt) and Et3N in DMF, followed by subsequent Boc-deprotection with HCl/dioxane, and EDC-HOBt coupling with 2,6-dimethylphenoxyacetic acid to give the crude product. Purification by preparative HPLC yielded KNI-10772 with >98% purity; 1H NMR (DMSO-d6) δ (ppm): 7.96 (d, J = 8.7 Hz, 1H), 7.82 (d, J = 9.3 Hz, 1H), 7.32–7.13 (m, 9H), 7.03–6.89 (m, 3H), 5.21 (dd, J = 8.4 Hz, 4.8 Hz, 1H), 4.81 (s, 2H), 4.59–4.49 (m, 2H), 4.46–4.42 (m, 1H), 4.40–4.34 (m, 1H), 4.13 (d, J = 14.1 Hz, 1H), 3.98 (d, J = 14.4 Hz, 1H), 3.08–2.87 (m, 3H), 2.84–2.75 (m, 1H), 2.16 (s, 6H), 1.50 (s, 3H), 1.43 (s, 3H); MS (ESI) m/z: 632 [M + H]+.
2.2. Protein expression and purification
Expression and purification of the recombinant PMI was performed with a modification of the previously published method (Xiao et al., 2007). In order to express soluble and properly folded enzyme, a unique construct of PMI was generated using a strategy similar to the one previously employed for PMII (Hill et al., 1994) and HAP (Xiao et al., 2006) (Fig. 1). The details of cloning of PMI and construction of the expression plasmid were described previously (Xiao et al., 2007). Since the expression of active recombinant PMI has been challenging, we elected a strategy involving expression of a construct containing a truncated zymogen coupled with a fusion protein to improve the solubility. Unlike prosegments of most aspartic proteases which are ~50 amino acid residues long (Koelsch et al., 1994), prosegments of PMs have long N-terminal prosegment (PS) (>120 residues). Since zymogen forms of other aspartic proteases have been successfully expressed in the past (Tanaka and Yada, 1996), we prepared constructs of a comparable form of PMI (tPMI) (Lys77p-Leu329, p denotes residues of the PS and the numbering is relative to the amino terminus) by removing 76 N-terminal amino acid residues, thus excluding the trans-membrane region and reducing the length of PS to 46 amino acid residues.
Since thioredoxin (Trx) fusion-based expression is a useful means to produce correctly folded, soluble, heterologous proteins in E. coli (LaVallie and McCoy, 1995), tPMI was fused with the highly soluble thioredoxin in the pET32b(+) vector (Novagen, Mississauga, ON, Canada). The fusion partner was followed by a thrombin cleavage site, (His6) tag, S-Tag, and an enterokinase cleavage site (total of ~6 kDa), added in order to increase the efficiency of protein purification.
In the present study we further optimized the previously described expression and purification steps (Xiao et al., 2007) to produce sufficient amount of recombinant protein for structural studies. Overnight culture of E. coli Rosetta-gami B (DE3) pLysS cells (Novagen, Mississauga, ON, Canada) transformed with the recombinant plasmid was used to inoculate 1 l of LB medium supplemented with 50 μg ml−1 ampicillin, 12.5 μg ml−1 tetracycline, 12.5 μg ml−1 kanamycin, and 34 μg ml−1 chloramphenicol. Cells were grown to an OD600 of 1.0 at 37 °C, 250 rpm and were induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After 12 h of incubation at 30 °C, cells were harvested by centrifugation at 4,500g for 10 min at 4 °C and then frozen at −20 °C.
Frozen cells were thawed at 4 °C overnight and suspended in 25 mL of 50 mM Tris-HCl buffer (pH 7.4), containing Benzonase (Novagen, Mississauga, ON, Canada) and chicken lysozyme (Sigma Aldrich Co., St. Louis, MO, USA). Suspensions were incubated at room temperature for 4 h with gentle shaking. Lysed cells were centrifuged at 20,000 g for 40 min at 4 °C.
Protein purification was performed using an AKTA™ FPLC system (GE Healthcare, Piscataway, NJ, USA). The supernatant was applied onto a Co-MAC cartridge (Novagen, Mississauga, ON, Canada) equilibrated with 0.3 M NaCl in 50 mM Tris-HCl (pH 7.4). The column was then washed with a loading buffer and the recombinant protein sample was eluted with 0.3 M NaCl, 0.25 M imidazole in 50 mM Tris-HCl (pH 7.4). Fractions containing truncated plasmepsin I fusion protein (Trx-tPMI) were dialyzed in 20 mM Tris-HCl (pH 7.4) and concentrated using an Amicon® Ultra-15 30 kDa NMWL centrifugal filter device (Millipore Corp., Bedford, MA, USA).
Mature tPMI was obtained by activating Trx-tPMI in 0.1 M sodium acetate (pH 4.2) at 37 °C for 2 h. The sample was subsequently washed and concentrated in 20 mM Tris-HCl (pH 7.4) using an Amicon® Ultra-15 10 kDa NMWL centrifugal filter device, applied onto a HR 5/5 mono Q anion exchange column and then eluted with a linear gradient of 1 M NaCl in 20 mM Tris-HCl (pH 7.4). The sample was further purified by gel filtration with a Superose™ 12 10/300 GL column (GE Healthcare, Piscataway, NJ, USA). The elution peak of the gel filtration chromatograph corresponds to the monomeric form of the mature tPMI, with molecular weight of 37.8 kDa. The fraction of pure tPMI was concentrated in 20 mM Tris-HCl buffer (pH 7.4) using an Amicon® Ultra-15 10 kDa NMWL centrifugal filter device.
The N-terminal amino acid sequences were determined by the Edman degradation method at the Hospital for Sick Children (Toronto, ON, Canada). Autocatalytic cleavage occurred between Leu116p-Thr117p, seven residues upstream of the native cleavage site at Gly123p-Asn1 (Gluzman et al., 1994). This cleavage site is the same as reported in previous studies (Moon et al., 1997; Xiao et al., 2007). Purity of the protein sample was monitored by 12% or 15% gel SDS-PAGE according to the method of Laemmli (Laemmli, 1970) in a Mini-Protean II electrophoresis cell (Bio-Rad, Hercules, CA, USA). The separated protein was stained with Gel-Code Blue Stain reagent (Canada Thermo Scientific, ON, Canada). Protein concentration was determined using UV absorbance at 280 nm and the Bio-Rad Dc protein assay (Bio-Rad Laboratories, Hercules, CA, USA), for which standard curves were generated using bovine serum albumin.
2.3. Crystallization
For crystallization of the PMI-KNI-10006 complex, a sample of purified PMI was transferred to 20 mM Tris buffer pH 8.0 and concentrated to 8 mg ml−1. Concentrated DMSO solution of KNI-10006 (Ki 11.0 nM) was mixed with the protein sample to yield the final inhibitor concentration of 0.3 mM in the mixture. The sample was subsequently incubated for four hours and centrifuged. Since relatively small amount of purified PMI protein was available for crystallization, only a single crystallization screen was set up using sitting-drop vapor-diffusion method at 293 K, using Qiagen PEG Suite screen solutions, with a few conditions producing small crystals. The best crystals appeared in the drop containing 0.3 μl protein solution and 0.3 μl reservoir solution, equilibrated against 75 μl reservoir solution. The reservoir solution contained 20% PEG 2000 MME and 0.1 M MES buffer at pH 6.5.
The crystal of apo-PMI used in this study was grown using the mixture of purified PMI (7.0 mg ml−1) in 20 mM Tris buffer, pH 7.4, and 1.1 mM KNI-10772 (Ki 46.0 nM). The crystallization screen was set up using sitting-drop vapor-diffusion method at 293 K, using Qiagen PEG Suite screen solutions. Only one crystal appeared in the drop containing 0.3 μl protein solution and 0.3 μl reservoir solution, equilibrated against 75 μl reservoir containing 25% PEG 6000 and 0.1 M sodium acetate buffer at pH 4.6. It was subsequently determined that this crystal did not contain bound inhibitor.
2.4. Data collection
X-ray diffraction data for the PMI-KNI-10006 complex crystal were collected to 3.1 Å resolution using a MAR300CCD detector at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL. The data set was collected at 100 K, using 20% (v/v) glycerol added to reservoir solution as cryoprotectant. The crystals are tetragonal in space group P43 with unit cell parameters of a=b=93.7, c=160.1 Å. Data for the apo-PMI crystal were collected using Cu Kα radiation generated by a Rigaku MicroMax 007HF X-ray source operated at 40 kV and 30 mA, equipped with a MAR345dtb detector. A data set was collected at 100 K, using 30% (v/v) ethylene glycol added to the reservoir solution as cryoprotectant. The crystal belongs to the orthorhombic space group P212121 with unit cell parameters of a=72.3, b=93.4, c=108.9 Å. All data sets were indexed and integrated using the program XDS (Kabsch, 1993). Integrated intensities were converted to structure factors with modules F2MTZ and CAD of CCP4 (CCP4, 1994). Randomly selected 5.0% of the observed unique reflections were reserved for calculation of Rfree. The statistics of data collection are presented in Table 1.
Table 1.
Data collection and refinement statistics
| A. Data collection statistics a | PMI–KNI-10006 | Apo-PMI |
| Space group | P43 | P212121 |
| Unit cell parameters a, b, c (Å) | 93.7, 93.7, 160.1 | 72.3, 93.4, 108.9 |
| Temperature (K) | 100 | 100 |
| Wavelength (Å) | 1.0000 | 1.5418 |
| Resolution (Å) | 40.0-3.10 (3.20-3.10) | 40.0-2.40 (2.50-2.40) |
| Rmerge (%)b | 14.4 (163.6) | 20.5 (131.3) |
| Completeness (%) | 99.0 (98.5) | 99.5 (98.3) |
| I/σ(I) | 11.8 (1.2) | 9.1 (1.6) |
| Unique reflections | 24 816 (2227) | 29306 (3268) |
| Redundancy | 5.8 (5.8) | 7.1 (7.2) |
| No. of molecules/asymmetric unit | 4 | 2 |
| B. Refinement statistics | ||
| Resolution (Å) | 30.0-3.10 | 30.0-2.40 |
| Working set: number of reflections | 23562 | 27830 |
| Rfactor (%) | 21.1 | 20.7 |
| Test set: number of reflections | 1240 | 1465 |
| Rfree (%) | 29.9 | 28.3 |
| Protein atoms | 10 410 | 5191 |
| No. of inhibitor molecules | 4 | - |
| No. of glycerol molecules | 7 | - |
| No. of water molecules | - | 186 |
| C. Geometry statistics | ||
| rmsd (bond distance) (Å) | 0.020 | 0.020 |
| rmsd (bond angle) (deg.) | 2.16 | 2.19 |
| Ramachandran plotc | ||
| Most favored region (%) | 80.8 | 86.0 |
| Additionally allowed regions (%) | 17.2 | 12.9 |
| Generously allowed regions (%) | 1.3 | 0.9 |
| Disallowed regions (%) | 0.7 | 0.2 |
| D. PDB code | 3QS1 | 3QRV |
The values in parentheses are for the highest resolution shell
Rmerge = Σh Σi |<I>h – I h,i| / ΣhΣiIh,i.
As defined by PROCHECK.
2.5. Structure solution and refinement
PMI exhibits high level of sequence identity with plasmepsins II (73.0%) and IV (68.0%). Several crystal structures of both plasmepsins are available in the PDB, providing a wide choice of starting models for molecular replacement. We first solved the structure of the PMI–KNI-10006 complex using the coordinates of PMII (PDB code 2BJU) as the search model.
The Matthews coefficient (Matthews, 1968) for these crystals is 2.37 Å3 Da−1, assuming the presence of four molecules in the asymmetric unit. Automated search by PHASER (Read, 2001) revealed the correct placement of the first two molecules in the asymmetric unit and unambiguously fixed the space group as P43. These two molecules were subsequently used as a search model allowing us to find the correct orientation of the two remaining molecules in the asymmetric unit. Initially, a few cycles of refinement using REFMAC5 (Murshudov et al., 1997) and rebuilding using COOT (Emsley and Cowtan, 2004) were performed for the protein only. Subsequent analysis of the Fo-Fc electron density map indicated that each active site of the four monomers was occupied by an inhibitor molecule. After inclusion of inhibitor molecules, iterative cycles of refinement in REFMAC5 and model building in the electron density maps using COOT were carried out. Very tight NCS restraints were used during all refinement cycles. The overall anisotropy was modeled with TLS parameters by dividing each molecule into five TLS groups, comprising residues 0-194, 195-201, 203-229, 230-234, and 235-328. Although the mature PMI polypeptide chain contains seven extra amino acid residues (Fig. 1) that belong to the propeptide, those residues are not visible in the final electron density map. The final model also lacks residues −1 and 329 of molecules A and B, and residues −1, 0 and 329 from molecules C and D, which could not be built because of the lack of electron density.
The structure of apo-PMI was solved by molecular replacement with PHASER using molecule A of the PMI-KNI-10006 complex as a search model. Two independent molecules were found in the asymmetric unit. The initial model was refined for several cycles using REFMAC5. Although PMI was crystallized in the presence of high concentration of KNI-10772, analysis of the Fo-Fc electron density map in the active sites did not show any interpretable density for the inhibitor. Iterative cycles of refinement in REFMAC5 and model building in the electron density maps using COOT were carried out. Very tight NCS restraints were used in the initial stages of refinement, but slowly the restraints were released as the model was becoming more complete; medium NCS restraints were applied for the final refinement cycles. Water molecules were progressively introduced at peaks of electron density higher than 3σ in Fo-Fc maps while monitoring the decrease of Rfree. Proper hydrogen bonding was required for placement of all solvent molecules. The overall anisotropy was modeled with TLS parameters by dividing each molecule into two TLS groups, comprising residues 0–190 and 191–328. The final apo-PMI model lacks residues −1 and 73-77 of molecule A that could not be built because of the lack of electron density.
The refinement statistics for the refined structures are presented in Table 1. The structures were analyzed using programs PROCHECK (Laskowski et al., 1993) and COOT (Emsley and Cowtan, 2004). Structural superpositions were performed with programs SSM (Krissinel and Henrick, 2004) and ALIGN (Cohen, 1997) and the figures were generated with the program PYMOL (DeLano, 2002).
3. Results
3.1. Solution and quality of the refined structures
The crystal structures of the apo form of the recombinant PMI (Fig. 2A), as well as of its complex with KNI-10006 (Fig. 2B), have been solved by molecular replacement and refined at the resolution of 2.4 and 3.1 Å, respectively (Table 1). The structure of the complex was solved first, using the coordinates of PMII to initiate molecular replacement. The presence of four crystallographically independent molecules (A-D) in the asymmetric unit of the PMI-KNI-10006 complex, although initially complicating the process of structure solution, provided verification of the overall correctness of the final structure despite its relatively low resolution, due to the similarity of the molecules and of the mode of binding of the inhibitor. Apo-PMI structure was solved using molecule A of its inhibitor complex as a search model. Although several loops in the dimeric structure of apo-PMI were different than their counterparts in the inhibited enzyme, a complete model could be built and refined with good statistics. The stereochemical parameters of the final refined models are summarized in Table 1. The overall quality of the models, as assessed by PROCHECK (Laskowski et al., 1993) and by other parameters such as R-factors, meets the acceptable criteria for structures solved with comparably limited resolution of the diffraction data (Jaskolski et al., 2007; Wlodawer et al., 2008). All main chain atoms and most of the side chains fit well into the 2Fo-Fc electron density map. The quality of the final Fo-Fc electron density map in the active sites of the complex with KNI-10006 (Fig. 3A), calculated based on phases that did not include the contribution from the inhibitor molecules (Fig. 3B) is also quite good, defining their conformation in an unambiguous manner.
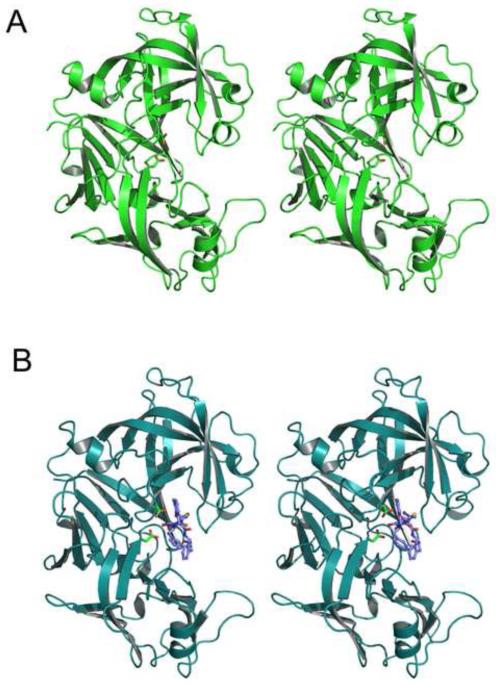
Figure 2.
Cartoons showing the overall fold of PMI. (A) A molecule of apo-PMI. (B) A molecule of PMI complexed with KNI-10006. The two catalytic aspartic acid residues (Asp32 and Asp215) are shown as sticks and the inhibitor (in panel B only) is shown in ball-and-stick representation.
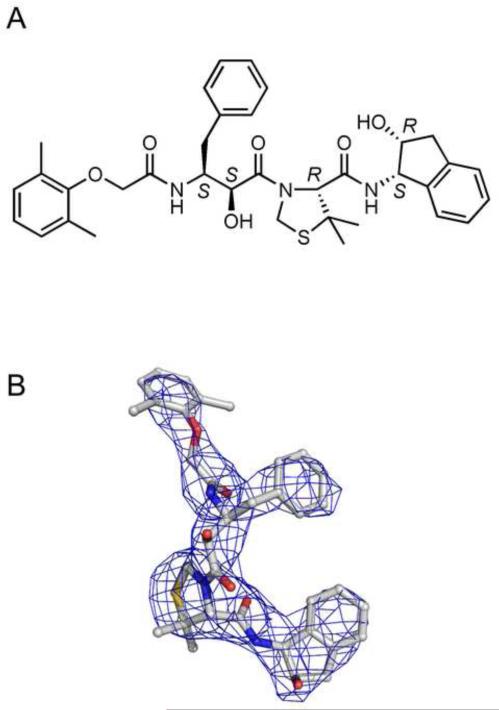
Figure 3.
KNI-10006 inhibitor. (A) A diagram showing the chemical structure of KNI-10006. (B) An initial Fo-Fc omit electron density map of KNI-10006 bound to molecule A of PMI contoured at 2.0σ level, with the final model superimposed.
3.2. Overall description of the structure
The overall fold of PMI (Fig. 2A) is typical for pepsin-like enzymes, as was expected due to the high sequence homology of PMI and other plasmepsins with other members of the aspartic protease family. Structures of mammalian enzymes, such as chymosin, renin, and cathepsin D, or the fungal enzymes rhizopuspepsin, endothiapepsin, and penicillopepsin (reviewed in Ref. (Dunn, 2002)), are available. Most importantly, the structures of three other plasmepsins, P. falciparum PMII (Asojo et al., 2002; Asojo et al., 2003; Prade et al., 2005; Robbins et al., 2009; Silva et al., 1996) and HAP (Bhaumik et al., 2009), as well as P. falciparum, P. malariae, and P. vivax PMIV (Bernstein et al., 1999; Bernstein et al., 2003; Clemente et al., 2006), have also been reported. The mature molecule of PMI (329 residues) is folded into two topologically similar N- and C-terminal domains. A large substrate-binding cleft is located between them, delineating an active site with two catalytic aspartates, Asp32 and Asp215 (pepsin numbering) and nucleophilic water molecule bound between them. The N-terminal domain contains the “flap”, another important structural element of the catalytic machinery. The flap is a flexible β-loop that carries residues important for catalysis and which undergoes conformational changes upon ligand binding. The amino and carboxyl ends of the two domains of PMI are assembled into a characteristic six-stranded interdomain β-sheet which serves to link the two domains together and is conserved in the aspartic protease family.
Two molecules are present in the asymmetric unit of the apo-PMI crystals, forming a non-crystallographic dimer. The surface area buried upon formation of the dimer is 1210 Å2 . Both molecules are quite similar, with the exception of two regions that include residues 106-114 and 127-133, respectively, which are significantly different. The r.m.s. deviation between 319 Cα atoms of molecule A and the corresponding atoms in molecules B is 0.73 Å. The flap is fully ordered in molecule B and assumes a closed conformation, although the density at its tip is rather weak. On the other hand, five residues from the flap in molecule A (73-77) are not visible in the electron density map due to apparent disorder, and the remaining part of the flap indicates that in this molecule the flap assumes slightly more open conformation than in molecule B. Both flaps are not making contacts with symmetry-related molecules and the difference in their order may reflect small differences in the local interactions.
Four molecules of PMI are present in the asymmetric unit of the tetragonal crystals of the PMI-KNI-10006 complex, forming a unique non-crystallographic tetramer (Fig. 4). The most extensive intermolecular interactions within the tetramer involve the loops comprising residues 278a-282 in each monomer. The surface area buried upon formation of the tetramer is 947 Å2. One loop packs against its counterparts from other monomers forming an assembly resembling a barrel and assumes different conformation in the four monomers. Altogether, these loops form the central part of the tetramer. The r.m.s. deviation between 326 Cα atoms of molecule A and the corresponding atoms in molecules B, C, and D is 0.32, 0.54, and 0.56 Å, respectively. The tetramer present in the asymmetric unit of the PMI crystals can be described as a dimer of dimers A-B and C-D. The relationship between the molecules within each of these dimers resembles the structural arrangement of two PMII molecules seen in a crystallographic dimer of PMII complexed with the inhibitor RS370 (Protein Data Bank (PDB) ID 1LF2) (Asojo et al., 2002), although intermolecular interactions are not as tight. The loop region comprised of residues 239-244 in PMI in one monomer is found in close proximity to the inhibitor molecule occupying the active site of another monomer, bringing Phe242 from the former molecule close to 2-aminoindanol group of the inhibitor (Fig. 5). Similar hydrophobic interactions have also been reported for PMII complexed with inhibitor RS370 (Asojo et al., 2002). As seen in most structures of plasmepsins complexed with inhibitors, the flap in the inhibited form of PMI assumes a closed conformation (open conformations of the flaps have only been reported in HAP complexed with KNI-10006 (Bhaumik et al., 2009) and in PMII complexed with an achiral inhibitor (Prade et al., 2005)).
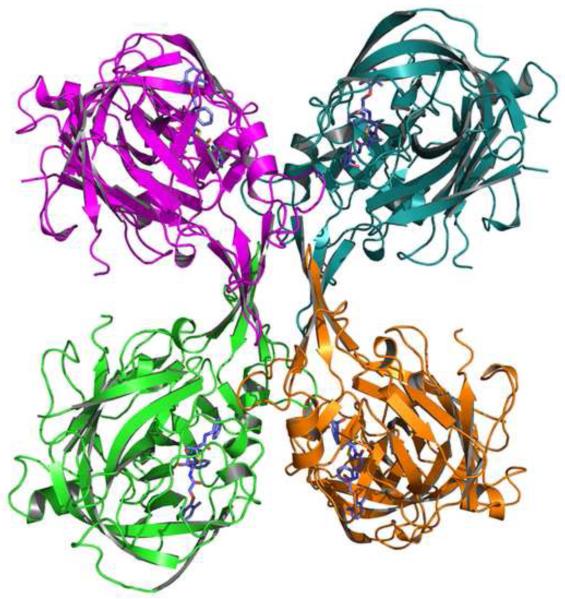
Figure 4.
Structure of the tetramer seen in the crystals of the PMI–KNI-10006 complex. The main chains of the protein are shown as cartoons and the inhibitor molecules are shown in stick representation.
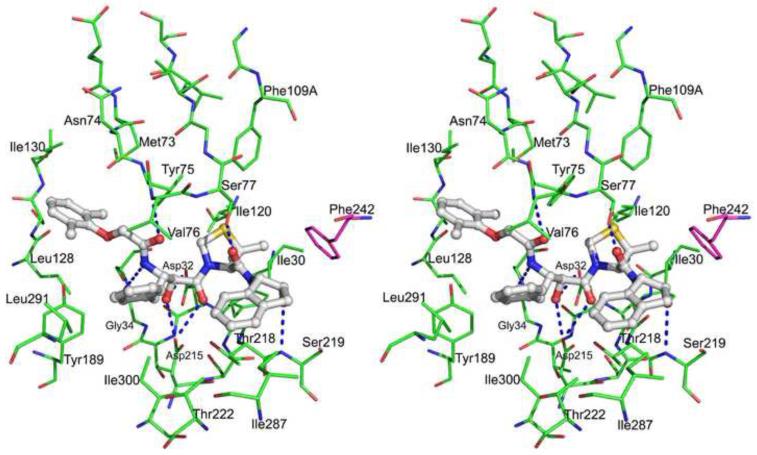
Figure 5.
A stereoview showing the active site of the PMI-KNI-10006 complex. The inhbitor is shown in ball-and-stick model (light gray). The amino acids are shown as lines, molecule A (green) and molecule B (magenta). Hydrogen bonds between the inhibitor and the protein are shown as dotted lines.
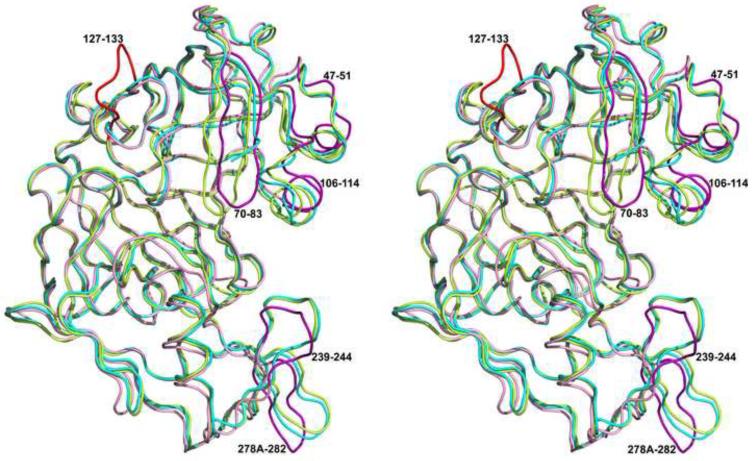
A comparison of the A and B subunits of the apo-PMI with the four subunits of the PMI-KNI-10006 complex shows a few structural differences between the inhibited and the ligand-free forms of PMI. Superposition of 318 Cα atoms the A monomer of apo-PMI structure with the A, B, C and D subunits of PMI-KNI-10006 complex resulted in r.m.s. deviation of 0.97, 0.97, 1.01 and 1.02 Å, respectively, with most of the differences limited to the loops 47-51, 71-83, 106-115, 239-244, and 278a-282. The r.m.s. deviation between 322 Cα atoms of the B monomers of the apo-PMI structure with the corresponding atoms in the A, B, C and D subunits of PMI-KNI-10006 complex structure is 1.25, 1.25, 1.24 and 1.29 Å, respectively. The higher r.m.s. deviation in the latter superposition is predominantly due to the significantly different conformation of the loop containing residues 127-133 in the B subunit of apo-PMI compared to the A subunit. The differences in the conformations of the loops 47-51, 106-114, 239-244, and 278a-282 in both subunits of apo-PMI compared to the PMI-KNI-10006 complex are comparable (Fig. 6).
Figure 6.
A superposition of the Cα traces of the A (cyan) and B (green) molecules of the apo-PMI onto the C molecule of the PMI-KNI-10006 complex (pink). The loop containing residues 127-133 in the B subunit of the apo-PMI is red. Five loop regions (47-51, 70-83, 106-114, 239-244, and 278a-282) which differ between the apoenzyme and the inhibitor complex are highlighted in magenta.
3.3. Comparisons with other eukaryotic aspartic proteases
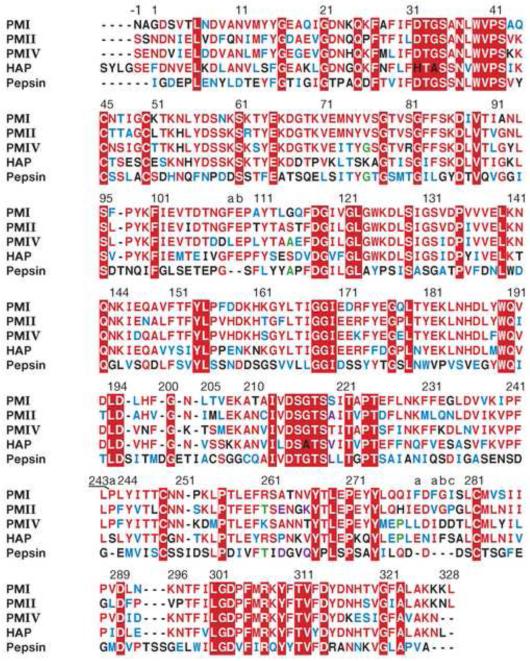
The apo-PMI structure was compared with the structures of unliganded pepsin (PDB ID 4PEP) and PMII (PDB ID 1LF4), resulting in the r.m.s. deviations of 1.4 (307 Cα pairs) and 1.5 Å (316 Cα pairs), respectively. Structures of inhibitor complexes of pepsin, PMII, PMIV, and HAP were compared with the PMI-KNI-10006 complex, yielding r.m.s.d. of 1.4 Å for the pepsin-pepstatin A complex, 1.01 Å for PMIV-KNI-764 complex (318 Cα pairs), 1.42 Å for HAP-KNI-10006 complex (315 Cα pairs) and 0.85 Å, 1.04 Å and 1.15 Å for pepstatin A complexes of PMII (321 Cα pairs), PMIV (315 Cα pairs) and HAP (315 Cα pairs), respectively. Comparison of the structures of inhibitor-bound PMII and PMIV with the PMI-KNI-10006 complex shows that the largest deviations are observed for the loops containing residues 239-244 and 278a-282. Superpositions of the PMI-KNI-10006 complex onto the complexes of PMII, HAP, PMIV, and pepsin with pepstatin A have been used to generate the structurally based sequence alignment shown in Fig. 7.
Figure 7.
Structure-based sequence alignment of PMI, PMII, PMIV, HAP, and pepsin. Strictly conserved residues are shown in white on red background, limited identities are red and green, similarities are blue, and differences are black. Inserted residues are labeled with a preceding number followed by letters a, b, etc.
3.4. Active site of the apoenzyme
The overall architecture of the active site of apo-PMI is similar to that observed in other pepsin-like aspartic proteases (Dunn, 2002). The active site which includes two catalytically active aspartate residues is located in the cleft formed in between the N- and C-terminal domains. In the active site of molecule A, the nucleophilic water molecule (Wat103) is present and located between Asp32 and Asp215, together with another water molecule, Wat179, found close to Asp32. The latter water molecule makes a hydrogen bond with the carbonyl oxygen of Gly34. A very similar arrangement of two water molecules that superimpose well on Wat103 and Wat179 has also been observed in the apo-PMII active site (PDB-ID 1LF4) (Asojo et al., 2003). The flap covering the active site which carries the important residue Tyr75 is partially disordered in molecule A, with residues 73-77 missing in the final model. By contrast, the flap is well ordered in molecule B and is in a closed conformation, positioning the side chain of Tyr75 close to the active site. The conformations of Trp39 also differ correspondingly between two molecules. In molecule B, Trp39 is found in the conformation that it usually adopts in the structures of aspartic proteases with closed flap conformation, similar to what has been reported for the majority of liganded enzymes, as well as for apo-PMII (Asojo et al., 2003) and for the apo form of pepsin (4PEP (Sielecki et al., 1990)). In this conformation, Trp39 forms a hydrogen bond with the flap residue Tyr75, maintaining the conserved hydrogen bonded network that connects the flap to the catalytic Asp32. However, in molecule A Trp39 adopts a second conformation, similar to what was found in the apoenzyme of HAP. In that conformation Trp39 is also involved in a hydrogen bonded network with Asp32, but the interaction is via Ser35. Thr218 occupies a similar position in the active sites of both molecules and forms hydrogen bonds with Asp215. Structure-based superposition of the two molecules also shows a different conformation of the loop containing residues 127-133. This loop comes close to the flap in molecule A, resulting in its significant shift compared to the position in molecule B. It is also important to note that the conformations of the loop containing residues 106-114 are different in the two molecules. This loop brings Phe109A close to the active site of molecule A, leaving no space for the Trp39 to flip, whereas Phe109A occupies a different position in molecule B, allowing Trp39 to rotate away from the active site.
3.5. Inhibition of PMI by KNI-10006
KNI-10006 is a comparatively small inhibitor of aspartic proteases which incorporates 2,6-dimethylphenyloxyacetyl at P2, allophenylnorstatine (Apns) at P1, dimethylthioproline (Dmt) at P1’, and 2-aminoindanol at P2’ (Fig. 3A). All four molecules of PMI that are present in the asymmetric unit of the crystal contain electron density in their active sites that can be interpreted as corresponding to KNI-10006. The conformation of the inhibitor molecules and their environment are similar in all four active sites. Fig. 3B shows an omitmap Fo-Fc electron density map of the inhibitor bound to molecule A of PMI. In the PMI-KNI-10006 complex, the hydroxyl group of the Apns moiety of the inhibitor is placed in between the two catalytic aspartates (Fig. 5). This central hydroxyl group of the inhibitor forms hydrogen bonds with the Oδ2 atoms Asp32 and Asp215.
The directionality of the main chain of KNI-10006 bound in the active site of PMI is opposite to that of other peptidomimetic inhibitors bound to pepsin-like aspartic proteases, as well as to the putative direction of a polypeptide substrate. As a result of such reverse binding, the P1 Apns group of the inhibitor is bound in the S1’ pocket of the enzyme, and, conversely, the P1’ Dmt group is bound in the S1 pocket (the assignment of the binding pockets in the active site of PM1 follows the accepted nomenclature (Davies, 1990)). The only inhibitor that has been reported to bind in such orientation is KNI-764 complexed to PMIV (Clemente et al., 2006). Whereas the reverse mode of binding was correlated with the protonation states of Asp32 and Asp215 through a computational analysis (Gutierrez-de-Teran et al., 2006), the reasons for this phenomenon were not elucidated.
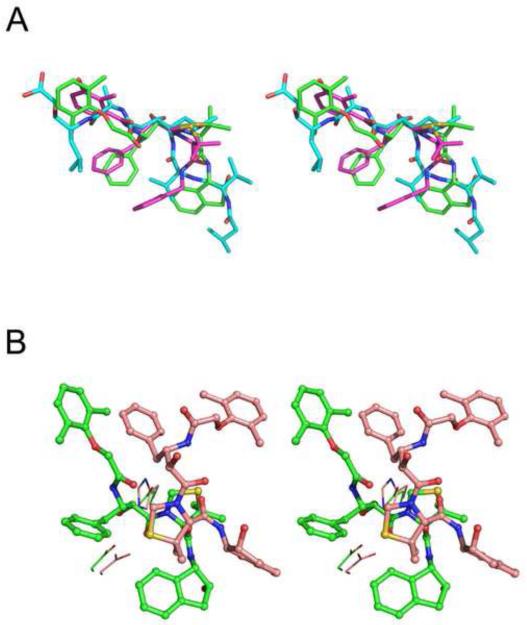
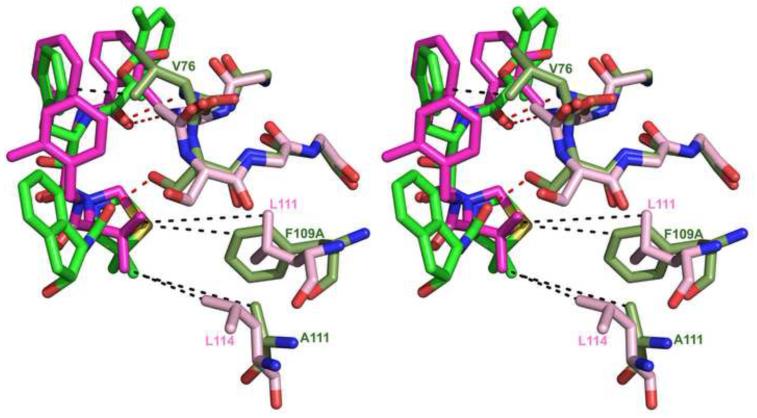
Structural superposition of the Cα atoms of PMI-KNI-10006 complex with those of the PMIV-KNI-764 and PMII-pepstatin A complexes gave r.m.s.d. of 1.04 Å and 0.85 Å, respectively. KNI-10006 is bound to the PMI active site in an extended conformation, similar to that of pepstatin A and KNI-764 in PMIV (Fig. 8A), but different than in HAP (Fig. 8B – also see below). Although the overall binding mode of the two KNI inhibitors is the same, there are some differences in the interactions between KNI-764 and KNI-10006 and the active site residues of PMIV and PMI, respectively. These differences may be due to the different nature of some key residues in the vicinity of the active sites of these two enzymes. In particular, the 2,6-dimethylphenyloxyacetyl moiety of the KNI-10006 is pushed closer to the flap compared to the 3-hydroxy-2-methyl benzyl group of KNI-764 due to the presence of bulkier Tyr189 in PMI at the position of the corresponding Phe192 in PMIV. These two terminal moieties of the inhibitors are in contact with similar hydrophobic residues, with the exception of Met73 in PMI, substituted by isoleucine in PMIV. The phenyl group of the Apns moiety of KNI-10006 makes additional hydrophobic interactions with Val76 which is present at the tip of the flap in PMI, as opposed to Gly78 that is found in the corresponding location in PMII (Fig. 9). The puckering of the ring of the Dmt moiety of KNI-10006 is different than in KNI-764. Differences in the conformations of these rings can be attributed in part to the two substitutions in the structurally equivalent residues that interact with them in two plasmepsins. Whereas the larger side chain of Phe109A in PMI is substituted by the smaller Leu111 in PMIV, the smaller Ala111 in PMI is substituted by the larger side chain of Leu114 in PMIV (Fig. 9). Another apparent reason for the differences in the conformation of these rings is a substitution of the 2-methylbenzylamine moiety of KNI-764 by the 2-aminoindanol group of KNI-10006. However, molecular dynamics simulation of KNI-764 complexed to PM-IV (Gutierrez-de-Teran et al., 2006) also showed that the 2-methylbenzylamine moiety of KNI-764 should adapt the same conformation as the 2-aminoindanol group of KNI-10006 in the PMI complex.
Figure 8.
Conformation of KNI-10006 and other plasmepsin inhibitors. (A) A comparison of the conformation of KNI-10006 (green) bound to PMI, pepstatin A (cyan) bound to PMII, and KNI-764 (magenta) bound to PMIV. (B) A comparison of the conformation of KNI-10006 bound to PMI (green) and to HAP (light pink). The inhibitor is shown in ball and stick representation and the active site residues are shown as lines.
Figure 9.
Variations in the sequences of PMI (moss) and PMIV (pink) affect the binding mode of KNI inhibitors. KNI-10006 is shown in green and KNI-764 in magenta. Dashed lines indicate hydrophobic contacts (black) and hydrogen bonds (red).
Docking studies of KNI-10006 into the active site of PMII were previously conducted in order to predict the mode of binding of this compound in the active site of plasmepsins (Kiso et al., 2004; Nezami et al., 2003)). It was proposed that 2,6-dimethylphenyloxyacetyl moiety at P2 faces the tip of the flap and the conserved region containing residues 216-218 (PMIV numbering) (Nezami et al., 2003). The model postulated that the 2-aminoindanol moiety of the inhibitor should be bound in the hydrophobic pocket, forming a hydrogen bond with Tyr192. Since this model was created before any experimental structures of the KNI inhibitors with plasmepsins were available, it did not anticipate the reverse direction of binding that positions the 2,6-dimethylphenyloxyacetyl moiety in the hydrophobic pocket and causing the other end of the inhibitor, containing 2-aminoindanol moiety, to interact with the conserved region comprised of residues 217-219.
In its complex with PMI, KNI-10006 makes multiple contacts of a different nature with the active site residues. The phenyl group of Apns makes hydrophobic contacts with the side chains of Leu291 and Ile300, as well as with Val76 from the flap. The 2,6-dimethylphenyloxyacetyl group of the inhibitor is placed in a hydrophobic pocket of the active site, making apolar contacts with Met73, Tyr75, Leu128, Ile130, and Tyr189. The oxygen atom (OAE) of the carbonyl group of 2,6-dimethylphenyloxyacetyl is an acceptor of a hydrogen bond with the main chain amide of Val76. The carbonyl oxygen atom of Gly34 is hydrogen bonded to the NH (NAZ) group of Apns. The carbonyl oxygen atom (OAG) of Apns also forms a hydrogen bond with the Oδ2 atom of Asp215. The Dmt moiety of the inhibitor is docked in a hydrophobic pocket formed by Ile30, Tyr75, Phe109A, and Ile120. The side chain of Ser77, which is present at the tip of the flap, forms a hydrogen bond with the carbonyl group located between the Dmt and 2-aminoindanol moieties. The orientation of 2-aminoindanol moiety is supported by the formation of a hydrogen bond between its hydroxyl group and the main chain NH group of Ser219. It is important to note that the 2-aminoindanol group is also involved in hydrophobic interactions with the side chain of Phe242 from another molecule. Similar hydrophobic interactions were reported in the PMII-EH58 complex (Asojo et al., 2003).
3.6. Comparison of the binding modes of KNI-10006 to PMI and HAP
An unusual mode of binding of KNI-10006 was previously observed in its complex with HAP, another member of the plasmepsin family (Bhaumik et al., 2009). While the overall superposition of 315 main chain Cα atoms of PMI-KNI-10006 complex (molecule A) with the HAP-KNI-10006 complex (molecule A) resulted in r.m.s.d. 1.42 Å, indicating a similar fold of these enzymes, the binding mode of KNI-10006 inhibitors in the active sites of two enzymes was found to be completely different (Fig. 8B). A sequence comparison highlights a number of different residues in the active sites of HAP and PMI (Fig. 7); the most crucial change is the replacement of the catalytic Asp32 by histidine. This substitution (His in HAP vs Asp in all other plasmepsins) is the most likely cause for the hydroxyl in the central core of KNI-10006 to point away from the catalytic residues. In the PMI-KNI-10006 complex, however, the central hydroxyl group of KNI-10006 is bound between Asp32 and Asp215. The 2,6-dimethylphenyloxyacetyl moiety of the KNI inhibitor is bound in a hydrophobic pocket which is located close to the surface of the PMI molecule, whereas the same moiety of the inhibitor is bound in a different area, the so called “flap pocket” of HAP (Bhaumik et al., 2009), which is located deep inside the protein molecule. The flap is found in the open conformation in the HAP-KNI-10006 complex. The flap is in the closed conformation in the PMI-KNI-10006 structure, with Trp39 and Phe109A moving towards the flap pocket, leading to its closure. Closure of the flap in the PMI-KNI-10006 complex blocks the gate to the flap pocket due to the movement of Met73 and Tyr75 closer to the active site. The corresponding residues in HAP, i.e. leucine and serine, are less bulky. The structures of these two complexes clearly show that KNI-10006 is a very good inhibitor of plasmepsins, since it can adapt upon its binding different conformations depending on the architecture of the active site of an individual enzyme.
4. Discussion
PMI is the first of the four vacuolar plasmepsins in P. falciparum that initiates hydrolysis of hemoglobin, and it is also the last of the four for which structural data were not available prior this study. As expected, the enzyme exhibits an overall fold of pepsin-like aspartic proteases and contains a flexible flap covering the active site, which changes its conformation in both the apo- and inhibited enzyme molecules.
As is often seen when the structures of the free and inhibited forms of enzymes are compared, binding of an inhibitor to PMI also induces conformational changes in the enzyme molecule. A structural comparison of the apo-PMI with its complex with KNI-10006 indicates that the conformation of the loops containing residues 239-244 and 278a-282 differ between these structures. In the PMI-KNI-10006 complex, the loop containing residues 278a-282 packs against its counterpart and forms the central part of the tetramer. Two small helical fragments in the N-terminal domain of PMI, comprising residues 47-51 and 106-114, also move upon the inhibitor binding in a manner similar to what was observed in the structure of HAP (Bhaumik et al., 2009) and other aspartic proteases. Very large differences in the conformation of the loop 127-133 in the two molecules of the apoenzyme molecules could be attributed to the differences in the crystal contacts.
A comparison of the binding sites in PMI with other plasmepsins from food vacuole (Table 2) revealed their remarkable similarity due to the high level of sequence identity. PMI and PMII are the closest among the four compared plasmepsins. Only the S1’ pocket of PMII has two residues larger than their counterparts in PMI; the residues in the other binding sites of PMII are identical to those that interact with KNI-10006 in PMI. In general, the nature of the binding sites in PMI is predominantly of hydrophobic character with the exception of subsites S2 and S2’. Our structural data are in a very good agreement with the findings of the subsite preferences for the PMI (Phe/Leu for P1/P1’ and Ser/Gln for P2/P2’) (Liu et al., 2009). The binding sites in PMIV, and particularly in HAP, contain more variations compared to the sites in PMI, therefore possible variations in the specificity requirements are expected. The availability of structural data for all four plasmepsins is crucial for the design of specific inhibitors of these redundant enzymes in order to prevent hemoglobin digestion and incapacitate the parasite.
Table 2.
Residues forming the subsites of the substrate binding pockets in plasmepsins. Residues identical in all P. falciparum vacuolar plasmepsins are white on red background, and those identical in some, but not all, are red. Residues that differ between the plasmepsins are black.
| Binding |
PMI | PMII | PMIV | HAP |
|---|---|---|---|---|
| S1 | Ile30 | Ile30 | Ile30 | Leu30 |
| Tyr75 | Tyr75 | Tyr75 | Ser75 | |
| Ser77 | Ser77 | Ser77 | Ala77 | |
| Phe109A | Phe109A | Leu109A | Phe109A | |
| Ile120 | Ile120 | Ile120 | Val120 | |
| Gly217 | Gly217 | Gly217 | Ala217 | |
| S2 | Thr218 | Thr218 | Thr218 | Thr218 |
| Thr222 | Thr222 | Thr222 | Thr222 | |
| Ile300 | Ile300 | Ile300 | Val300 | |
| Ser219 | Ser219 | Ser219 | Ser219 | |
| Ile287 | Ile287 | Leu287 | Val287 | |
| S1’ | Val76 | Val76 | Gly76 | Lys76 |
| Tyr189 | Tyr189 | Phe189 | Met189 | |
| Leu291 | Phe291 | Ile291 | Leu291 | |
| Ile300 | Ile300 | Ile300 | Val300 | |
| Val289 | Leu289 | Val289 | Ile289 | |
| S2’ | Ser35 | Ser35 | Ser35 | Ser35 |
| Tyr75 | Tyr75 | Tyr75 | Ser75 | |
| Leu128 | Leu128 | Leu128 | Leu128 | |
| Tyr189 | Tyr189 | Phe189 | Met189 | |
| S3’ | No interactions |
|||
| S4’ | Val76 | Val76 | Gly76 | Lys76 |
| Ile130 | Ile130 | Ile130 | Ile130 |
The structure of the complexes of PMI with KNI-10006 was the second one that reported an unorthodox binding mode of KNI inhibitors to plasmepsins. The KNI compounds are mechanism-based inhibitors of aspartic proteases with a non-hydrolysable transition state isostere in place of the normal hydrolysable P1-P1’ amide bond. Similarly to pepstatin, a general inhibitor of aspartic proteases (Davies, 1990; Umezawa et al., 1970), these inhibitors contain in their central core a tetrahedral carbon atom with a single hydroxyl that occupies the position of nucleophilic water molecule in the apoenzyme. Although the configuration of this hydroxyl is S in both the KNI and statine-based inhibitors, the directionality of the latter in all the structures of various aspartic proteases known to date was consistently opposite to that of the KNI inhibitors in PMI and PMIV. Both KNI-764 and KNI-10006 are identical in their P1-P1’ region, thus it is very likely that this reverse mode of binding must be related to the properties of inhibitor core. These features can be narrowed down by comparing the structures described above with the structures of PMII complexed to the inhibitors RS367 and RS370 (PDB codes 1LEE and 1LF2, respectively). The N-terminal halves of the latter two inhibitors are identical with their KNI counterparts with the exception of a single group in the central core: CH2 in the former is substituted by C=O in the latter. As a result of such a substitution, the orientation of the hydroxyl, while preserving the nominal S configuration, has changed in KNI-10006. On the other hand, the orientation of hydroxyl and the binding mode of RS367 and RS370 are identical to pepstatin and other peptidomimetic inhibitors. KNI-10772, which differs from KNI-10006 only by the R chirality of the core, is a weaker inhibitor and we did not manage to obtain its cocrystals.
It is thus likely that proper alignment of the hydroxyl groups is more important to the mode of binding than the exact fit of the side chains of the inhibitors in the side-chain binding pockets of the enzyme. Incidentally, this phenomenon could not be noticed for the fully symmetric retroviral enzymes since, in that case, the definition of the primed and unprimed pockets simply followed the nominal definition of the respective groups of the inhibitors. However, the reverse binding of the KNI inhibitors to plasmepsins may provide the basis for the unambiguous assignment of the unique direction of attack by the nucleophilic water on the peptide bond of the substrate in pepsin-like aspartic proteases. A good analogy is the attack on either the re or si faces of the scissile bond by serine and cysteine proteases, dictated by the strictly conserved differences in the orientations of the reactive groups in the catalytic residues (serine and cysteine) in the active sites of the corresponding enzymes (Bode et al., 1987; Luo et al., 2001).
Acknowledgements
We would like to thank Drs. Sergei Pletnev and Zbyszek Dauter for their help in data collection. Diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID, located at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This project was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Financial support from the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs Program is also gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PDB accession numbers
Atomic coordinates and structure factors for the apo and inhibited form of PMI have been deposited in the PDB with the codes 3QRV and 3QS1, respectively.
References
- Abdel-Rahman HM, Kimura T, Hidaka K, Kiso A, Nezami A, et al. Design of inhibitors against HIV, HTLV-I, and Plasmodium falciparum aspartic proteases. Biol. Chem. 2004;385:1035–1039. doi: 10.1515/BC.2004.134. [DOI] [PubMed] [Google Scholar]
- Asojo OA, Afonina E, Gulnik SV, Yu B, Erickson JW, et al. Structures of Ser205 mutant plasmepsin II from Plasmodium falciparum at 1.8 Å in complex with the inhibitors rs367 and rs370. Acta Crystallogr. 2002;D58:2001–2008. doi: 10.1107/s0907444902014695. [DOI] [PubMed] [Google Scholar]
- Asojo OA, Gulnik SV, Afonina E, Yu B, Ellman JA, Haque TS, Silva AM. Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparum. J. Mol. Biol. 2003;327:173–181. doi: 10.1016/s0022-2836(03)00036-6. [DOI] [PubMed] [Google Scholar]
- Bailly E, Jambou R, Savel J, Jaureguiberry G. Plasmodium falciparum: differential sensitivity in vitro to E-64 (cysteine protease inhibitor) and Pepstatin A (aspartyl protease inhibitor) J Protozool. 1992;39:593–599. doi: 10.1111/j.1550-7408.1992.tb04856.x. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Liu J, Beatty W, Pelosof L, Klemba M, Goldberg DE. Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc. Natl. Acad. Sci. USA. 2002;99:990–995. doi: 10.1073/pnas.022630099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein NK, Cherney MM, Loetscher H, Ridley RG, James MN. Crystal structure of the novel aspartic proteinase zymogen proplasmepsin II from Plasmodium falciparum. Nat. Struct. Biol. 1999;6:32–37. doi: 10.1038/4905. [DOI] [PubMed] [Google Scholar]
- Bernstein NK, Cherney MM, Yowell CA, Dame JB, James MN. Structural insights into the activation of P. vivax plasmepsin. J Mol. Biol. 2003;329:505–524. doi: 10.1016/s0022-2836(03)00444-3. [DOI] [PubMed] [Google Scholar]
- Berry C, Humphreys MJ, Matharu P, Granger R, Horrocks P, et al. A distinct member of the aspartic proteinase gene family from the human malaria parasite Plasmodium falciparum. FEBS Lett. 1999;447:149–154. doi: 10.1016/s0014-5793(99)00276-8. [DOI] [PubMed] [Google Scholar]
- Bhaumik P, Xiao H, Parr CL, Kiso Y, Gustchina A, Yada RY, Wlodawer A. Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum. J. Mol. Biol. 2009;388:520–540. doi: 10.1016/j.jmb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W, Papamokos E, Musil D. The high-resolution X-ray crystal structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis. Structural analysis, subtilisin structure and interface geometry. Eur. J. Biochem. 1987;166:673–692. doi: 10.1111/j.1432-1033.1987.tb13566.x. [DOI] [PubMed] [Google Scholar]
- CCP4 The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. Collaborative Computational Project, Number 4, 1994. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Govindasamy L, Madabushi A, Fisher SZ, Moose RE, et al. Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor. Acta Crystallogr. 2006;D62:246–252. doi: 10.1107/S0907444905041260. [DOI] [PubMed] [Google Scholar]
- Cohen GE. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Crystallogr. 1997;30:1160–1161. [Google Scholar]
- Coombs GH, Goldberg DE, Klemba M, Berry C, Kay J, Mottram JC. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 2001;17:532–537. doi: 10.1016/s1471-4922(01)02037-2. [DOI] [PubMed] [Google Scholar]
- Davies DR. The structure and function of the aspartic proteinases. Annu. Rev. Biophys. Biophys. Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Dunn BM. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ersmark K, Samuelsson B, Hallberg A. Plasmepsins as potential targets for new antimalarial therapy. Med. Res. Rev. 2006;26:626–666. doi: 10.1002/med.20082. [DOI] [PubMed] [Google Scholar]
- Francis SE, Banerjee R, Goldberg DE. Biosynthesis and maturation of the malaria aspartic hemoglobinases plasmepsins I and II. J Biol. Chem. 1997;272:14961–14968. doi: 10.1074/jbc.272.23.14961. [DOI] [PubMed] [Google Scholar]
- Francis SE, Gluzman IY, Oksman A, Knickerbocker A, Mueller R, et al. Molecular characterization and inhibition of a Plasmodium falciparum aspartic hemoglobinase. EMBO J. 1994;13:306–317. doi: 10.1002/j.1460-2075.1994.tb06263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman IY, Francis SE, Oksman A, Smith CE, Duffin KL, Goldberg DE. Order and specificity of the Plasmodium falciparum hemoglobin degradation pathway. J Clin. Invest. 1994;93:1602–1608. doi: 10.1172/JCI117140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood BM, Bojang K, Whitty CJ, Targett GA. Malaria. Lancet. 2005;365:1487–1498. doi: 10.1016/S0140-6736(05)66420-3. [DOI] [PubMed] [Google Scholar]
- Gutierrez-de-Teran H, Nervall M, Dunn BM, Clemente JC, Aqvist J. Computational analysis of plasmepsin IV bound to an allophenylnorstatine inhibitor. FEBS Lett. 2006;580:5910–5916. doi: 10.1016/j.febslet.2006.09.057. [DOI] [PubMed] [Google Scholar]
- Hill J, Tyas L, Phylip LH, Kay J, Dunn BM, Berry C. High level expression and characterisation of Plasmepsin II, an aspartic proteinase from Plasmodium falciparum. FEBS Lett. 1994;352:155–158. doi: 10.1016/0014-5793(94)00940-6. [DOI] [PubMed] [Google Scholar]
- Jaskolski M, Gilski M, Dauter Z, Wlodawer A. Stereochemical restraints revisited: how accurate are refinement targets and how much should protein structures be allowed to deviate from them? Acta Crystallogr. 2007;D63:611–620. doi: 10.1107/S090744490700978X. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- Kimura T, Nguyen JT, Maegawa H, Nishiyama K, Arii Y, et al. Chipping at large, potent human T-cell leukemia virus type 1 protease inhibitors to uncover smaller, equipotent inhibitors. Bioorg. Med. Chem. Lett. 2007;17:3276–3280. doi: 10.1016/j.bmcl.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Kiriyama A, Mimoto T, Kiso Y, Takada K. Pharmacokinetic study of a tripeptide HIV-1 protease inhibitor, KNI-174, in rats after intravenous and intraduodenal administrations. Biopharm. Drug Dispos. 1993;14:199–207. doi: 10.1002/bdd.2510140303. [DOI] [PubMed] [Google Scholar]
- Kiriyama A, Sugahara M, Yoshikawa Y, Kiso Y, Takada K. The bioavailability of oral dosage forms of a new HIV-1 protease inhibitor, KNI-272, in beagle dogs. Biopharm. Drug Dispos. 1996;17:125–134. doi: 10.1002/(SICI)1099-081X(199603)17:2<125::AID-BDD940>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kiso A, Hidaka K, Kimura T, Hayashi Y, Nezami A, Freire E, Kiso Y. Search for substrate-based inhibitors fitting the S2′ space of malarial aspartic protease plasmepsin II. J. Pept. Sci. 2004;10:641–647. doi: 10.1002/psc.609. [DOI] [PubMed] [Google Scholar]
- Kiso Y. Kynostatin (KNI)-272--a rationally designed tripeptide inhibitor of HIV protease. Nippon. Rinsho. 1993;51(Suppl):139–145. [PubMed] [Google Scholar]
- Kiso Y. Design and synthesis of HIV protease inhibitors containing allophenylnorstatine as a transition-state mimic. Adv. Exp. Med. Biol. 1995;362:413–23. doi: 10.1007/978-1-4615-1871-6_54. 413-423. [DOI] [PubMed] [Google Scholar]
- Kiso Y. Design and synthesis of substrate-based peptidomimetic human immunodeficiency virus protease inhibitors containing the hydroxymethylcarbonyl isostere. Biopolymers. 1996;40:235–244. doi: 10.1002/(sici)1097-0282(1996)40:2<235::aid-bip3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Kiso Y, Matsumoto H, Mizumoto S, Kimura T, Fujiwara Y, Akaji K. Small dipeptide-based HIV protease inhibitors containing the hydroxymethylcarbonyl isostere as an ideal transition-state mimic. Biopolymers. 1999;51:59–68. doi: 10.1002/(SICI)1097-0282(1999)51:1<59::AID-BIP7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Koelsch G, Mares M, Metcalf P, Fusek M. Multiple functions of pro-parts of aspartic proteinase zymogens. FEBS Lett. 1994;343:6–10. doi: 10.1016/0014-5793(94)80596-2. [DOI] [PubMed] [Google Scholar]
- Kolakovich KA, Gluzman IY, Duffin KL, Goldberg DE. Generation of hemoglobin peptides in the acidic digestive vacuole of Plasmodium falciparum implicates peptide transport in amino acid production. Mol. Biochem. Parasitol. 1997;87:123–135. doi: 10.1016/s0166-6851(97)00062-5. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- LaVallie ER, McCoy JM. Gene fusion expression systems in Escherichia coli. Curr. Opin. Biotechnol. 1995;6:501–506. doi: 10.1016/0958-1669(95)80083-2. [DOI] [PubMed] [Google Scholar]
- Liu P, Marzahn MR, Robbins AH, Gutierrez-de-Teran H, Rodriguez D, et al. Recombinant plasmepsin 1 from the human malaria parasite Plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis. Biochemistry. 2009;48:4086–4099. doi: 10.1021/bi802059r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Francis SE, Gluzman IY, Goldberg DE. Kinetic analysis of plasmepsins I and II aspartic proteases of the Plasmodium falciparum digestive vacuole. Mol. Biochem. Parasitol. 1996;79:71–78. doi: 10.1016/0166-6851(96)02651-5. [DOI] [PubMed] [Google Scholar]
- Luo Y, Pfuetzner RA, Mosimann S, Paetzel M, Frey EA, et al. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/s0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- Maegawa H, Kimura T, Arii Y, Matsui Y, Kasai S, Hayashi Y, Kiso Y. Identification of peptidomimetic HTLV-I protease inhibitors containing hydroxymethylcarbonyl (HMC) isostere as the transition-state mimic. Bioorg. Med. Chem. Lett. 2004;14:5925–5929. doi: 10.1016/j.bmcl.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Mimoto T, Imai J, Tanaka S, Hattori N, Kisanuki S, Akaji K, Kiso Y. KNI-102, a novel tripeptide HIV protease inhibitor containing allophenylnorstatine as a transition-state mimic. Chem. Pharm. Bull. (Tokyo) 1991;39:3088–3090. doi: 10.1248/cpb.39.3088. [DOI] [PubMed] [Google Scholar]
- Moon RP, Tyas L, Certa U, Rupp K, Bur D, et al. Expression and characterisation of plasmepsin I from Plasmodium falciparum. Eur. J Biochem. 1997;244:552–560. doi: 10.1111/j.1432-1033.1997.00552.x. [DOI] [PubMed] [Google Scholar]
- Moura PA, Dame JB, Fidock DA. Role of Plasmodium falciparum digestive vacuole plasmepsins in the specificity and antimalarial mode of action of cysteine and aspartic protease inhibitors. Antimicrob. Agents Chemother. 2009;53:4968–4978. doi: 10.1128/AAC.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nezami A, Kimura T, Hidaka K, Kiso A, Liu J, et al. High-affinity inhibition of a family of Plasmodium falciparum proteases by a designed adaptive inhibitor. Biochemistry. 2003;42:8459–8464. doi: 10.1021/bi034131z. [DOI] [PubMed] [Google Scholar]
- Nguyen JT, Hamada Y, Kimura T, Kiso Y. Design of potent aspartic protease inhibitors to treat various diseases. Arch. Pharm. (Weinheim) 2008a;341:523–535. doi: 10.1002/ardp.200700267. [DOI] [PubMed] [Google Scholar]
- Nguyen JT, Zhang M, Kumada HO, Itami A, Nishiyama K, et al. Truncation and non-natural amino acid substitution studies on HTLV-I protease hexapeptidic inhibitors. Bioorg. Med. Chem. Lett. 2008b;18:366–370. doi: 10.1016/j.bmcl.2007.10.066. [DOI] [PubMed] [Google Scholar]
- Prade L, Jones AF, Boss C, Richard-Bildstein S, Meyer S, Binkert C, Bur D. X-ray structure of plasmepsin II complexed with a potent achiral inhibitor. J. Biol. Chem. 2005;280:23837–23843. doi: 10.1074/jbc.M501519200. [DOI] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. 2001;D57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Robbins AH, Dunn BM, Agbandje-McKenna M, McKenna R. Crystallographic evidence for noncoplanar catalytic aspartic acids in plasmepsin II resides in the Protein Data Bank. Acta Crystallogr. 2009;D65:294–296. doi: 10.1107/S0907444908041632. [DOI] [PubMed] [Google Scholar]
- Sielecki AR, Fedorov AA, Boodhoo A, Andreeva NS, James MN. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 A resolution. J. Mol. Biol. 1990;214:143–170. doi: 10.1016/0022-2836(90)90153-D. [DOI] [PubMed] [Google Scholar]
- Silva AM, Lee AY, Gulnik SV, Maier P, Collins J, et al. Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 1996;93:10034–10039. doi: 10.1073/pnas.93.19.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yada RY. Expression of soluble cloned porcine pepsinogen A in Escherichia coli. Biochem. J. 1996;315(Pt 2):443–446. doi: 10.1042/bj3150443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa H, Aoyagi T, Morishima H, Matsuzaki M, Hamada M. Pepstatin, a new pepsin inhibitor produced by Actinomycetes. J. Antibiot. (Tokyo) 1970;23:259–262. doi: 10.7164/antibiotics.23.259. [DOI] [PubMed] [Google Scholar]
- Wlodawer A, Minor W, Dauter Z, Jaskolski M. Protein crystallography for non-crystallographers or how to get the best (but not more) from the published macromolecular structures. FEBS J. 2008;275:1–21. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Sinkovits AF, Bryksa BC, Ogawa M, Yada RY. Recombinant expression and partial characterization of an active soluble histo-aspartic protease from Plasmodium falciparum. Protein Expr. Purif. 2006;49:88–94. doi: 10.1016/j.pep.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Xiao H, Tanaka T, Ogawa M, Yada RY. Expression and enzymatic characterization of the soluble recombinant plasmepsin I from Plasmodium falciparum. Protein Eng Des Sel. 2007;20:625–633. doi: 10.1093/protein/gzm066. [DOI] [PubMed] [Google Scholar]
- Zhang M, Nguyen JT, Kumada HO, Kimura T, Cheng M, Hayashi Y, Kiso Y. Locking the two ends of tetrapeptidic HTLV-I protease inhibitors inside the enzyme. Bioorg. Med. Chem. 2008a;16:6880–6890. doi: 10.1016/j.bmc.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Zhang M, Nguyen JT, Kumada HO, Kimura T, Cheng M, Hayashi Y, Kiso Y. Synthesis and activity of tetrapeptidic HTLV-I protease inhibitors possessing different P3-cap moieties. Bioorg. Med. Chem. 2008b;16:5795–5802. doi: 10.1016/j.bmc.2008.03.055. [DOI] [PubMed] [Google Scholar]