Abstract
Background
The aim of this study was to create polymeric materials with known properties in order to study the preconditions for complement activation.
Methods
Initially, 22 polymers were screened for complement activating capacity. Based on these results six polymers (P1–P6) were characterized regarding physico-chemical parameters e.g.: composition; surface area; pore size; and protein adsorption from human EDTA-plasma.
Results
P2, P4 and reference particles of polystyrene and polyvinyl chloride, were hydrophobic, bound low levels of protein and were poor complement activators. Their accessible surface was limited to protein adsorption in that they had pore diameters smaller than most plasma proteins. P1 and P3 were negatively charged and adsorbed IgG and C1q. A ten-fold difference in complement activation was attributed to the fact that P3 but not P1 bound high amounts of C1-inhibitor. The hydrophobic P5 and P6 were low complement activators. They selectively bound apolipoproteins AI and AIV (and vitronectin), which probably limited the binding of complement activators to the surface.
Conclusion
We demonstrate the usefulness of the modus operandi to employ a high-throughput procedure to synthesize a great number of novel substances, assay their physico-chemical properties with the aim to study the relationship between the initial protein coat on a surface and subsequent biological events. Data obtained from the six polymers characterized here, suggest that a complement-resistant surface should be hydrophobic, uncharged, and have a small available surface, accomplished by nanostructured topography. Additional attenuation of complement can be achieved by selective enrichment of inert proteins and inhibitors.
Keywords: Polymers, biomaterials, complement, plasma protein adsorption hemocompatibility
INTRODUCTION
Modern medicine uses a range of biomaterials e.g. in implants, biosensors, and scaffolds, with or without drug release etc, and there is consequently an increasing need for new, biocompatible materials. Medical science has reached the stage when most materials can be used clinically, albeit in combination with systemic anticoagulants (heparin, warfarin, platelet receptor inhibitors etc). Thus, primary activation of platelets and coagulation is not the main problem. By contrast, inflammation, e.g., during hemodialysis where complement is alimental in initiating adverse reactions remains one of the major problems. For instance, inflammation is likely a major cause of arthrosclerosis development frequently seen in this treatment. Generation of C5a has been demonstrated to contribute to hemodialysis-associated thrombosis by inducing tissue factor (a key initiator of coagualtion in vivo) and cytokines in peripheral blood neutrophils 1. Using clinically relevant models for hemodialysis it was demonstrated that inhibition of complement at the level of C3 efficiently reduced the production of tissue factor and cytokines by neutrophils 1. This is but one of many recently identified mechanisms for crosstalk between the different cascade systems and cellular elements of the blood 2.
Most biomaterials come into contact with blood, either transiently during implantation or permanently. When an artificial surface is exposed to blood, plasma proteins adsorb and form a monolayer on the surface of the material within seconds. The composition of the adsorbed protein layer, as well as the conformation of the proteins, are unique for each biomaterial and is determined by the chemical properties of the surface of that material 3, 4. Proteins which are known to undergo a surface-induced change in conformation are complement component C3 4 and IgG 5, both of which can induce an activation of the complement system on the biomaterial surface and on top of the adsorbed protein layer, ultimately leading to recruitment, activation and binding of leukocytes to the surface 6–8. Other examples are adsorbed Factor XII 9 and fibrinogen 10, which have been shown to cause contact activation of the coagulation system and activation of platelets, respectively. Reactions at all these stages contribute to the compatibility of a material 6, 11.
In several reports the composition of the protein layer, either on modified model surfaces 12, 13) or authentical biomaterials, e.g., from membranes after hemodialysis or ex vivo perfusion 14, 15, have been investigated but the study protocols and results have differed considerably, which makes it difficult to correlate the structure of a given material with a biological outcome. Historically, such studies have preferentially employed techniques, which reflect the topography of the protein layer, which includes studies using antibodies detected by immunochemical or optical techniques 5, 12, 13. In more recent studies, the total amount of bound proteins have been desorbed and analyzed by SDS-PAGE followed by identification by Western blotting and/or mass spectrometry 15–17. The reason for variations in the results may be due to that the topography of the protein layer differs from that of the total layer (desorption) and perhaps even more importantly the source of plasma proteins.
Plasma proteins have been provided either in purified form or as serum or plasma (diluted or undiluted), or as whole blood anticoagulated with heparin, citrate, hirudin or EDTA, or without any added anticoagulantia (4, 5, 7, 9, 10, 12–17). This makes the results widely dispersed due to the fact that the cascade systems are active or inhibited dependent on the additive or the source of plasma protein. In the present paper we wanted to focus on the initial protein adsorption, which determines the outcome of the material-blood interaction. For that reason we have selected to use EDTA in the protein adsorptions studies, since it totally inhibits both complement and coagulation activation and leaves most plasma proteins in a state similar to when they initially contacted the material surface.
The objective of this study was to synthesize polymers with different physico-chemical properties in order to study the impact on protein adsorption, to be used as a model system with the primary aim to understand which proteins that are adsorbed on a hemocompatible surface, and secondly to implement this knowledge to construct tomorrows biomaterials with improved hemocompatibility compared to those used in the clinic today.
The aim is that the novel polymers described here ultimately should interact with biological tissue, including blood, e.g., in the form of nanoparticles or immune columns with clearance functions, as drug release modalities, or as injectable polymers 18–20. Therefore, we only used monomers and crosslinkers, commonly used in polymer synthesis, that were soluble in water or ethanol in order to minimize future toxic effects and to allow better polymerization in contact with tissues. We synthesized an array of 22 polymers by copolymerization of monomers and crosslinkers (selected to provide a range of functionalities: acidic, basic, neutral, hydrophobic) since this approach enables synthesis of a large number of polymers with great variability regarding physico-chemical properties in a comparatively easy and rapid way.
Complement activation is a clinically relevant biological property and one of the parameters included in the criteria for testing of the hemocompatibility of biomaterials defined in ISO 10993-4 21. The polymers were therefore screened for activation of the complement system using plasma, with the specific thrombin inhibitor hirudin, which leaves complement function intact 22, 23, and six polymers with heterogenous composition and complement activation properties were chosen for further characterization.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Diacryloylpiperazine (DAP, >99%) and 2-hydroxyethyl methacrylate (HEMA, >99%) were obtained from Fluka. Divinylbenzene (DVB, 80%), ethylene glycol dimethacrylate (EGDMA, 98%), N-isopropyl acrylamide (IPAAm, 97%), methacrylic acid (MAA, 99%), styrene (99%), ammonium persulfate (APS, 98%) and N,N,N,′N′-tetramethylene diamine (TEMED, 99%) were obtained from Sigma-Aldrich (St Louis, MO, USA). Soda glass was purchased from Kebo, polystyrene flakes (PS) were obtained from Alfa Aesar, and polyvinyl chloride (PVC ≤ 75 μm) from Polysciences. Ethanol (99.7%) was obtained from Solveco. 2,2′-azobisisobutyronitrile (AIBN, 98%) was obtained from Janssen Chimica and was recrystallized from methanol before use. EGDMA and DVB were purified by extraction with 0.1 M NaOH, dried over MgSO4, filtered, and passed through activated Al2O3 prior to use. MAA was purified by vacuum distillation, and styrene was passed through activated Al2O3 prior to use.
Polyclonal antibodies recognizing the following human proteins were obtained from Dako (Glostrup, Denmark): α-2-macroglobulin, C1q, fibrinogen, C3 (C3c), C4 (C4c), C5, IgG, haptoglobulin, transferrin, human serum albumin (HSA), antithrombin, hemopexin, and apolipoprotein AI (ApoAI). Polyclonal antibodies recognizing human C1-inhibitor (C1-INH), factor H, factor I, vitronectin, C4b-binding protein (C4BP), and factor XII were purchased from The Binding Site (Birmingham, UK), and anti-human apolipoprotein AIV (ApoAIV) was obtained from Abcam (Cambridge, UK).
Synthesis of polymer particles
In a typical polymer synthesis, functional monomer and crosslinker were dissolved in the porogen by sonication. Solutions were then purged with N2 for 2 min before the addition of initiator. Polymerization was carried out in a 55°C water bath for up to 24 h to achieve complete polymerization. The obtained polymer monoliths were ground and sieved through a 63-μm-mesh sieve, followed by sedimentation from acetone (75 mL, 3 × 20 min) to remove any fine particles. The resulting polymers were sieved a second time (25 μm), and the fraction containing the 25- to 63-μm particles was collected, air-dried overnight and stored at room temperature until use.
Endotoxin determination
The polymer particles were tested for the presence of endotoxins using an Enodafe PTS Portable Test System with Limulus Amebocyte Lysate (LAL) Test Cartridge (Charles River Laboratories, Charleston, SC, USA).
Blood
Pools of human serum, hirudin-plasma and EDTA-plasma were prepared from blood drawn from six healthy donors into Vacutainer™ tubes (Becton, Dickinson and Co., Plymouth, UK). To prepare the serum pool, the blood was allowed to clot at room temperature for 45 min in Vacutainer™ glass tubes without additives. The hirudin-plasma pool was prepared by manual addition of the recombinant form of the thrombin inhibitor hirudin, lepirudin (Refludan™, Aventis Pharma, final concentration 50 μg/mL), via a fine syringe into a Vacutainer™ glass tube without other additives; pre-filled EDTA plastic tubes were used to collect the EDTA-plasma pool. The supernatants were collected and pooled after centrifugation at 3450 × g for 25 min at 20°C. The pools were stored at −80°C prior to use. This study was performed with the consent of the Ethical Committee of the University Hospital of Linköping, Sweden.
Complement-activating ability of the polymers
In order to decide on polymers to study in greater detail, the complement-activating properties of the synthesized materials were analyzed and used as a selection tool (as a supplement to the composition data). A 5-mg sample of each polymer was incubated with hirudin-plasma (which contains an active complement system, and has been shown to be suitable for in vitro complement activation experiments 22, 23) in heparinized 2-mL Eppendorf tubes at 37°C for 30 min with rotation. After incubation, the samples were centrifuged in a tabletop centrifuge, the plasma was collected, and EDTA was added (final concentration 10 mM, to avoid any further activation of the complement system). The remaining polymer particles were treated with 2% SDS (in PBS) for 20 min at 37°C in order to elute any adsorbed C3a molecules. The particles were then centrifuged, and the eluates were collected. The samples were stored at −80°C prior to C3a analysis. As a control, an aliquot of plasma was incubated in the same manner as described above, but without the addition of polymer.
C3a ELISA
Generation of the anaphylatoxin C3a was used as an indicator of complement activation caused by the polymers. The levels of C3a were measured in both the plasma samples and the eluates by sandwich ELISA, using mAb 4SD17.3, which is specific for a neo-epitope which is expressed in C3a and hydrolyzed C3 (C3H2O) but not in native, unactivated C3, as the capture antibody and biotinylated anti-human C3a, together with HRP-conjugated streptavidin (GE Healthcare, Uppsala, Sweden), for detection. Staining was performed with o-phenylendiamine dihydrochloride (OPD, 0.25 mg/mL, Sigma) dissolved in citrate–phosphate buffer (0.1 M, pH 5) containing H2O2 (0.5 μL/mL buffer). The absorbance was measured at 490 nm. Zymosan-activated serum calibrated against a solution of purified C3a was used as a standard; values are given in ng/mL, and pooled zymosan-activated serum from blood donors was diluted 1:500 and used as a control 24.
Selection of polymers
Six polymers were selected for further detailed examination. These six materials varied in terms of their composition, complement-activating ability, and C3a adsorption capacity. The selected polymers, all of which had a crosslinker-to-monomer ratio of 80:20, were designated P1 (MAA/DAP), P2 (MAA/DVB), P3 (MAA/EGDMA), P4 (IPAAm/EGDMA), P5 (Styrene/EGDMA), and P6 (HEMA/EGDMA). These six polymers were also synthesized at crosslinker:monomer ratios of 60:40 and 90:10 to investigate whether the crosslinking ratio affected the complement activation induced by the polymer. Reference materials of the same size range were prepared as described above from commercially obtained PS, PVC, and glass beads.
Physical characterization of the polymer particles
Scanning electron microscope (SEM) micrographs of the polymers (P1–P6, PS, PVC and glass) were prepared using a high vacuum SEM (JSM 840, JEOL) at 10 kV. BET surface area analysis was performed by N2 adsorption on a Micromeritics ASAP 2400 instrument. Elemental analyses (C, H) were performed by Mikrokemi AB (Uppsala, Sweden). FT-IR spectra were recorded using polymer samples dispersed in KBr on a Nicolet Avatar 320 FT-IR instrument by diffuse reflectance IR spectroscopy.
Incubation of polymer particles prior to protein analyses
Polymer particles were incubated in plasma (4 mg/mL) containing 10 mM EDTA (which inhibits both the complement and the coagulation system, and is therefore preferable for protein adsorption studies) for 30 min at 37°C with continuous rotation. After the incubation, the samples were centrifuged in a tabletop centrifuge, and the plasma was discarded. The particles were carefully washed three times with PBS-Tween (PBS containing 0.05% Tween 20) prior to the determination of the total amount protein, gel electrophoresis, COPAS, and the particle ELISA experiments.
Quantification of the total protein adsorbed to the synthesized polymer particles
After incubation of the particles in EDTA-plasma, as described above, the adsorbed proteins were eluted from the particles by the addition of 2% SDS (in PBS), followed by incubation for 20 min at 37°C. The samples were centrifuged, and the eluates were collected. Protein levels were measured both in the eluate fractions, as well as on the remaining particles in order to detect any protein that might have been left behind as a result of incomplete elution. Measurements were achieved with BCA Assay Kit according to the manufacturer’s recommendations (detergent-compatible, Pierce, Rockford, IL, USA), with bovine serum albumin as a standard. The absorbance at 560 nm was determined for the eluate fractions as well as the supernatants from the assayed particle samples.
Visualization of adsorbed protein pattern by SDS-PAGE
Two milligrams of each particle were incubated in EDTA-plasma, hirudin-plasma, or serum and washed with PBS-Tween as described above. 5X SDS-electrophoresis buffer (12 mM Tris, 0.4% SDS, pH 6.8) was added to the particles, and the samples were heated to 99 °C for 5 min. The particles were then loaded onto 10% SDS-PAGE gels, and after separation, the proteins were stained with Coomassie brilliant blue (BDH Biochemical, England).
Identification of enriched proteins by western blot analysis
To identify proteins that showed clear enrichment on the SDS-PAGE gel (compared to the levels in plasma) the adsorbed proteins from 0.5 mg particles were separated on SDS-PAGE gels as described above. After separation the proteins were transferred to PVDF Immun-Blot membranes (BioRad) and the membranes were blocked with 1% bovine serum albumin in PBS-Tween, and developed using antibodies against a broad selection of plasma proteins for identification. The selection of antibodies was primarily based on results from previously performed experiments, where plasma proteins adsorbed to a PS surface were eluted, separated by SDS-PAGE, and identified by peptide mass fingerprinting with the MALDI-TOF technique. These identified proteins included HSA, C3, C4, α2-macroglobulin, IgG, Apolipoproteins (Apo) ApoAI and ApoAIV, transferrin, fibrinogen, and haptoglobulin. In addition, we included antibodies against vitronectin and C1q in attempts to identify proteins with molecular weights ≈75 kDa and ≈410 kDa, respectively. All antibodies were biotinylated using biotin-amidocaproate N-hydroxysuccimide ester, and HRP-conjugated streptavidin was used for antibody detection, followed by staining with 3,3′-diaminobenzidine tetrahydrochloride (Sigma, Steinheim, Germany) dissolved in PBS (1 mg/mL with 0.5 μL H2O2/mL).
COPAS analysis
Complex Object Parametric Analyzer and Sorter (COPAS) is a commercial available flow cytometer-like instrument enabling fluorescence measurement of large particles and objects (20–1,500 μm, Union Biometrica, Somerville, MA, USA). Prior to the flow cytometer analysis, antibodies were FITC-labeled using a standard protocol. The polymer particles were incubated in EDTA-plasma and washed with PBS-Tween as described above, then labeled with one of the following FITC-labeled antibodies: anti-C3c, anti-C1q, anti-ApoAI, or anti-vitronectin. As a negative control, a FITC-labeled rabbit IgG fraction from non-immunized animals was used. Unbound antibodies were removed by washing the particles with PBS-Tween, and the samples were kept in the dark, on ice until COPAS-analysis.
A minimum of 1,500 particles were collected in each COPAS measurement. The flow cytometer was equipped with a 488/514 multiline laser, and the acquired data were processed with FCS Express Software (De Novo Software, CA, US) yielding histograms and mean fluorescence intensity (MFI) values. Each experiment was performed four times, and the negative control was subtracted from each value.
Quantification of individual proteins
To quantify the levels of specific plasma proteins that were adsorbed to the surface of each polymer, an ELISA-like method was developed. After incubation with EDTA-plasma and washing of the particles as described above, antibodies were added to the polymers, and the samples were incubated in 2-mL Eppendorf® polypropylene tubes (Sarstedt, Nümbrecht, Germany). The antibodies (based on the MALDI and western blot experiments described above and supplemented with some selected abundant plasma proteins) were recognizing the complement components C3, C4, C5, C1q, including the regulators of complement activations (RCAs) C1-inhibitor (C1-INH), factor H, factor I, vitronectin, and C4-binding protein (C4BP); the coagulation proteins factor XII, antithrombin, and fibrinogen; the lipoproteins apolipoprotein AI and AIV; the transport proteins HSA, haptoglobulin, hemopexin, and transferrin; and IgG and alpha-2-macroglobulin, respectively. As a negative control, a purified IgG fraction from a non-immunized rabbit was used. All antibodies were biotinylated, and HRP-conjugated streptavidin was used for detection. Staining was performed with OPD, the samples were centrifuged and the absorbance of the supernatants was measured at 490 nm. Each experiment was performed in triplicate, and the negative control was subtracted from each value.
Statistical analysis
All experiments were performed three to six separate times (as stated in the figure legends), and the data are presented as mean values + SEM. Statistical significance was calculated using Kruskal-Wallis test with Dunn’s post-hoc test. Significance was tested using a Pearson correlation test.
RESULTS
Selection of polymers
The polymeric materials (n=22) were first synthesized with crosslinker:monomer ratios of 80:20, then ground and sieved to yield polymer particles in the size range of 25–63 μm. When the complement-activating ability (indicated by the generation of C3a) of the materials was investigated in hirudin-plasma, the results varied greatly, with values ranging from close to background levels (≈0 ng/mL) to dramatically high levels of C3a (>12,000 ng/mL) after subtraction of the control value. The polymers also exhibited variable adsorption capacities for the generated C3a, ranging from 15 to 95% of the total C3a measured in each sample (see Table 1). This complement-activation data, together with the composition of the material, was used to select six polymers to be characterized in detail; P1 (MAA/DAP), P2 (MAA/DVB), P3 (MAA/EGDMA), P4 (IPAAm/EGDMA), P5 (Styrene/EGDMA), and P6 (HEMA/EGDMA). (Table 2, Figure 1). The chosen polymers were heterogeneous, being variable in terms of composition, complement-activating ability, and C3a adsorption capacity. The polymer particles were characterized with regard to their physical properties, e.g. pore size and surface area and the micro-topography of each polymer surface was visualized by scanning electron microscopy (SEM). In addition, we determined the total protein adsorption ability, and performed detailed mapping of the adsorbed protein patterns on the polymers using a complex objects parametric sorter and analyzer (COPAS), followed by a particle-ELISA. A flow chart describing the experimental setup is shown in Figure 2.
Table 1.
Generation of the complement activation fragment C3a, after incubation of the polymer particles in hirudin-plasma. The results are shown as mean values (n=4) ± SEM, and the amount of C3a adsorbed to each polymer is shown as a percentage of the total C3a. This analysis was used as a selection tool in order to choose polymers with heterogeneous properties (i.e., complement-activating capacity and adsorption ability) for further detailed studies.
| Polymer composition | C3a total ng/mL (% adsorbed) | Polymer composition | C3a total ng/mL (% adsorbed) | Polymer composition | C3a total ng/mL (% adsorbed) |
|---|---|---|---|---|---|
| DAP + water: | DVB + EtOH: | EGDMA + EtOH: | |||
| MAA (* P1) | 12,600±590 (45) | MAA (* P2) | ≈ 0 ±88 (48) | MAA (* P3) | 1,100 ±170 (95) |
| MA | 12,500±380 (20) | MA | 1,400 ±250 (20) | MA | 900 ±320 (30) |
| AAm | 12,700±590 (20) | AAm | 2,900 ±490 (15) | AAm | 2,000 ±270 (25) |
| IPAAm | 7,400 ±580 (20) | IPAAm | 1,600 ±760 (20) | IPAAm (* P4) | 2,000 ±200 (15) |
| HEMA | 13,700±700 (20) | Styrene | 740 ±250 (20) | Styrene (* P5) | 1,000 ±60 (25) |
| AMPSA | 5,000 ±220 (85) | HEMA | ≈ 0 ±360 (20) | HEMA (* P6) | 2,100 ±200 (40) |
| 4-VP | ≈ 0 ±160 (15) | 4-VP | 4,300 ±440 (85) | ||
| AMPSA | ** | AMPSA | ** | ||
| Polystyrene (PS) | 850 ±260 (30) | Polyvinyl chloride (PVC) | 3,000 ±200 (15) | Glass | 950 ±160 (30) |
N,N-diacryloylpiperazine (DAP); divinylbenzene (DVB); ethylene glycol dimethacrylate (EGDMA); methacrylic acid (MAA), N-isopropyl acrylamide (IPAAm), 2-hydroxyethyl methacrylate (HEMA), styrene, methallyl alcohol (MA), acrylamide (AAm), 4-vinylpyridine (4-VP) or 2-acrylamido-2-methylpropane sulfonic acid (AMPSA).
Polymers selected for further surface studies.
Not fully polymerized (excluded)
Table 2.
Composition and predicted chemical properties at physiological pH of the polymers selected for further study. The properties of the polymers were predicted based on the chemical characteristics of each component presented in Figure 1.
| Polymer | Monomer (20%) | Crosslinker (80%) | Predicted properties of the polymer at physiological pH | Functional group |
|---|---|---|---|---|
| P1 | MAA | DAP | Hydrophilic Negatively charged | −COO− |
| P2 | MAA | DVB | Hydrophobic Negatively charged | −COO− |
| P3 | MAA | EGDMA | Hydrophobic Negatively charged | −COO− |
| P4 | IPAAm | EGDMA | Hydrophobic | −CONH2CH(CH3)2 |
| P5 | Styrene | EGDMA | Hydrophobic | −C6H5 |
| P6 | HEMA | EGDMA | Hydrophobic | −OH |
| PS | Styrene | __ | Hydrophobic | − C6H5 |
| PVC | Vinyl chloride | __ | Slightly hydrophobic | −Cl |
| Glass | __ | __ | Hydrophilic Negatively charged | −SiOH/SiO(−) |
Figure 1.
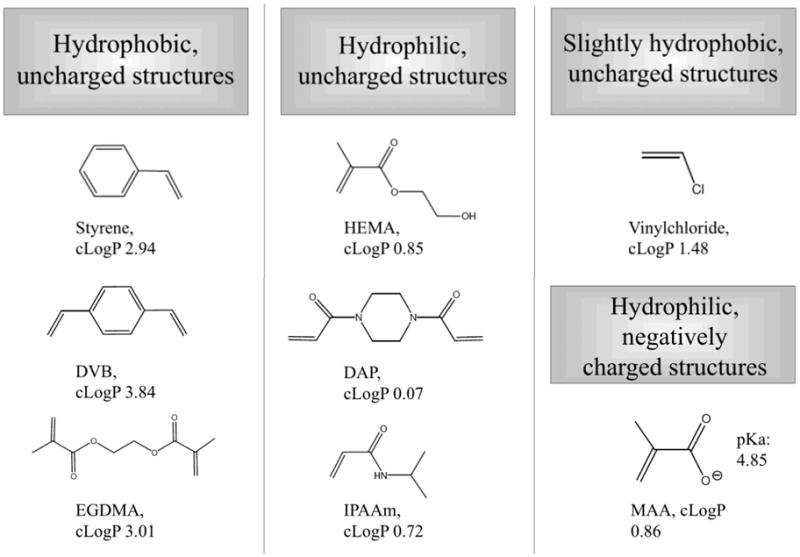
Molecular structures and properties of the monomers and crosslinkers used to synthesize the six selected polymers. The calculated octanol/water partition coefficients (cLogP values) were obtained with MarvinSketch 4.1.5, Copyright © 1998–2007 ChemAxon Ltd.
Figure 2.
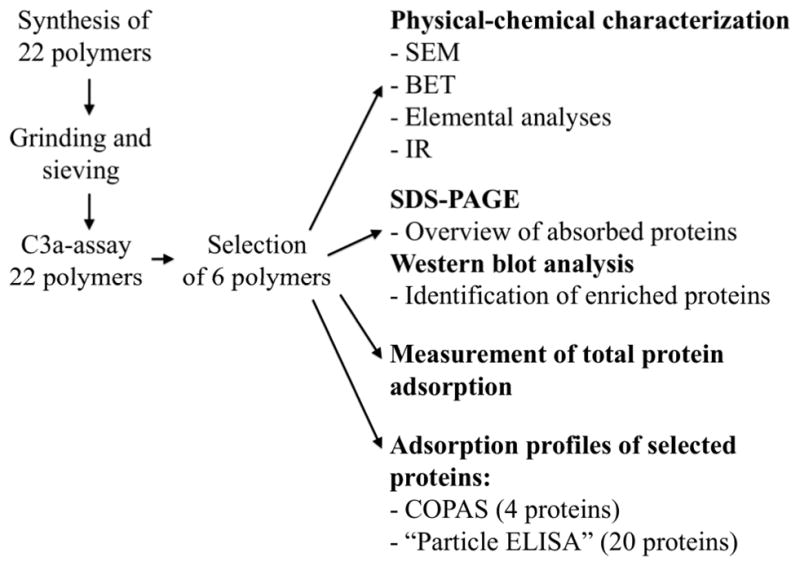
Flow-chart of the experimental set-up in this project.
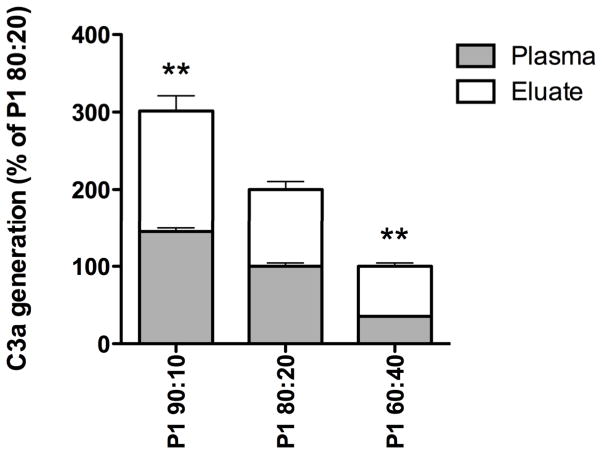
The six polymers (P1–P6) were also synthesized with crosslinker:monomer ratios of 60:40 and 90:10 (in addition to 80:20, described above), and the levels of generated C3a were analyzed after incubation in hirudin-plasma. No significant relationship between the C3a levels and the crosslinker:monomer ratio was seen for polymers P2–P6 (data not shown). However, polymer P1 (DAP/MAA) displayed a dose-dependent correlation between the increasing generation of C3a and increasing amounts of the crosslinker DAP (Figure 3, p<0.01). Further studies were performed with a crosslinker:monomer ratio of 80:20 for all the polymers.
Figure 3.
Generation of the complement activation fragment C3a after incubation of the polymer P1 (DAP/MAA, at three different crosslinker:monomer ratios) in hirudin-plasma. Adsorbed C3a molecules were eluted from the polymers with 2% SDS. Decreasing amounts of the crosslinker DAP resulted in less activation of the complement system. The results are presented as % C3a generation compared to the crosslinker:monomer ratio 80:20, which here corresponds to 100% for plasma and eluate, respectively. The results are shown as mean values +SEM (n=4); the results for the ratios 90:10 and 60:40 were significantly higher and lower, respectively, than the value for the 80:20 ratio (p<0.01).
Endotoxin determination
The endotoxin content of all tested polymer particles were found to be below the detection limit of the assay i.e., 0.01 EU/mL or 1 pg/mL. The value for the negative control was also <0.01 EU/mL and the value for the positive control included in the kit was 0.12 EU/mL.
Physical and chemical properties of polymer particles
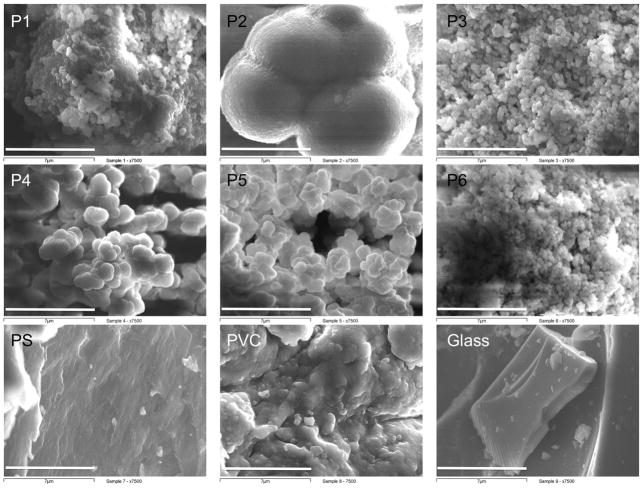
The SEM micrographs (magnification x7500) show large differences of the polymer morphologies, as observed in the μm-range. P1, P3 and P6 were found to have rough surfaces, while the surfaces of P2, PS and glass were shown to be much smoother. P4 and P5 exhibit an intermediate roughness (Figure 4). BET analysis of the polymers revealed large differences in their gas-accessible surface areas, with values ranging from 6.1 m2/g for glass to 188 m2/g for P1. The average pore diameter also varied greatly, from 28 Å for P2 to 134 Å for P1 (and >500 Å for glass; Table 3). FT-IR analyses showed peaks (O-H, C-H, C=O, C-O, C-N) characteristic of each polymer type (Table S1). Elemental analysis of the synthesized polymers (C, H) corresponded well with the theoretical values of polymer composition (Table S2).
Figure 4.
Scanning electron micrographs of the synthesized polymers P1-P6 and the reference polymers PS, PVC and soda glass (bar = 7 μm, magnification = x7500).
Table 3.
Surface area and the average pore size of the polymer particles, as determined by BET studies (N2 adsorption).
| MAA/DAP (P1) | MAA/DVB (P2) | MAA/EGDMA (P3) | IPAAm/EGDMA (P4) | Styrene/EGDMA (P5) | HEMA/EGDMA (P6) | PS | PVC | Glass | |
|---|---|---|---|---|---|---|---|---|---|
| Surface area (m2/g) | 188 | 161 | 203 | 130 | 25 | 105 | 11 | 17 | 6.1 |
| Average pore diameter (Å) | 133.5 | 27.6 | 84.4 | 32.1 | 101.6 | 116.6 | 45.4 | 40.9 | 512 |
Quantification of total protein adsorbed to the synthesized polymer particles
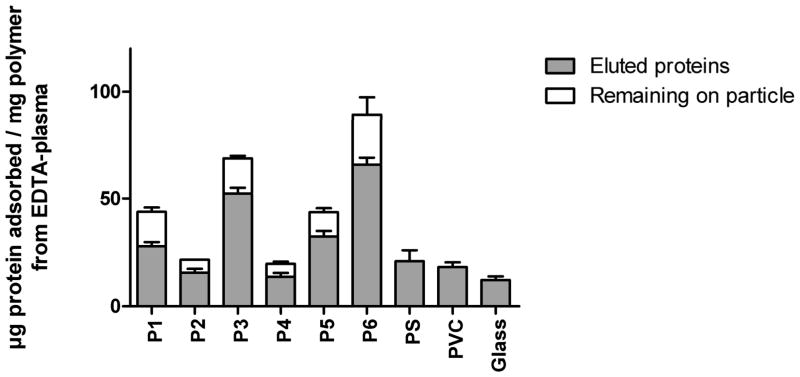
The total amount of protein adsorbed to the polymers was determined after incubation in EDTA-plasma and elution with 2% SDS. Protein levels were measured in the eluted fraction as well as on the polymers after the elution to allow us to detect any potential remaining proteins that had not been completely eluted. The amount of protein bound per mg of polymer was variable, from ~20 μg/mg polymer for P2, P4, and the control particles to values as much as four times higher for P6; P6 bound the highest level of proteins and was closely followed by P3. The effectiveness of the elution step was found to be polymer-dependent, with incomplete elution occurring from the polymers that displayed the highest total binding of protein. No detectable residual protein was seen in the case of the polymers with a lower total protein binding (Figure 5). There was a strong correlation (p<0.02) between the pore size and the level of protein binding of the polymers. In contrast, there was no co-variation between the surface area and the pore size of the polymers under used conditions.
Figure 5.
Protein adsorbed to 1-mg polymer particles after a 30-min incubation in EDTA-plasma. After incubation, the particles were carefully washed, and the proteins were eluted with 2% SDS. The amount of protein in the eluate as well as the protein remaining on the particles was measured to account for any incomplete elution. The results shown are means + SEM (n=6).
SDS-PAGE and identification of enriched proteins by Western blot analysis
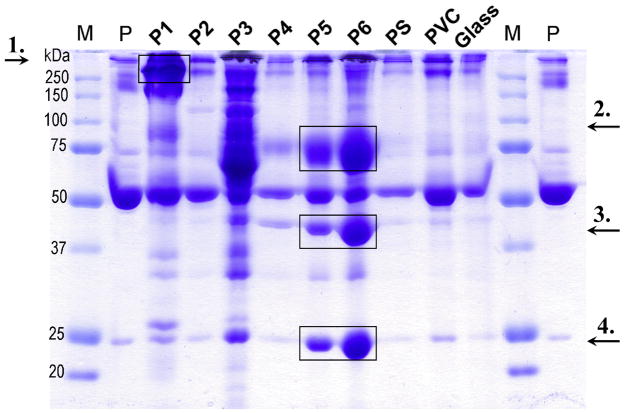
To visualize the plasma proteins adsorbed to 2-mg polymer particle samples, non-reducing SDS-PAGE was stained with Coomassie brilliant blue (see Figure 6). The gel revealed great differences in the properties of the polymers, both in terms of the total amount of protein adsorbed as well as the composition of the adsorbed protein pattern. P1, P3, and P6 showed a high adsorption capacity (consistent with the results above), with a pattern very different from the plasma reference (hirudin plasma diluted 1/50). P1 had a high affinity for a high-molecular weight protein, which was identified as the complement recognition molecule C1q (≈410 kDa) by Western blot analysis. P5 and P6 displayed a similar binding pattern, with a considerable enrichment of three proteins that were just faintly visible in plasma reference lane. These proteins were identified as vitronectin (≈75 kDa), apolipoprotein AIV (≈45 kDa), and apolipoprotein AI (≈25 kDa). In contrast, HSA (migrating at ≈55–60 kDa) adsorbed in similar amounts to all surfaces. C3, C4, α2-macroglobulin, IgG, transferrin, fibrinogen, and haptoglobulin were detected on the particles and all proteins were further quantified as described below. Similar macroscopic results were obtained after incubation in EDTA-plasma as well as serum (data not shown).
Figure 6.
Patterns of plasma proteins adsorbed onto 2 mg of polymer particles after a 30-min incubation in hirudin-plasma. The gel shows, from the far left: molecular marker (M), hirudin plasma (P) diluted 1/50; P1 through P6 are the synthesized polymers, followed by the reference polymers PS, PVC, and glass. To identify the proteins that were enriched on polymers P1, P5, and P6, Western blot analysis was used, and the results showed that P1 had a high affinity for C1q (≈410 kDa, arrow 1), whereas P5 and P6 were enriched in vitronectin (≈75 kDa, arrow 2), apolipoprotein AIV (≈45 kDa, arrow 3), and apolipoprotein AI (≈25 kDa, arrow 4) on their surfaces. In contrast, HSA (migrating at ≈55–60 kDa) was seen in similar amounts to all surfaces. Comparable results were obtained when the particles were incubated in EDTA-plasma or serum (data not shown).
COPAS analysis
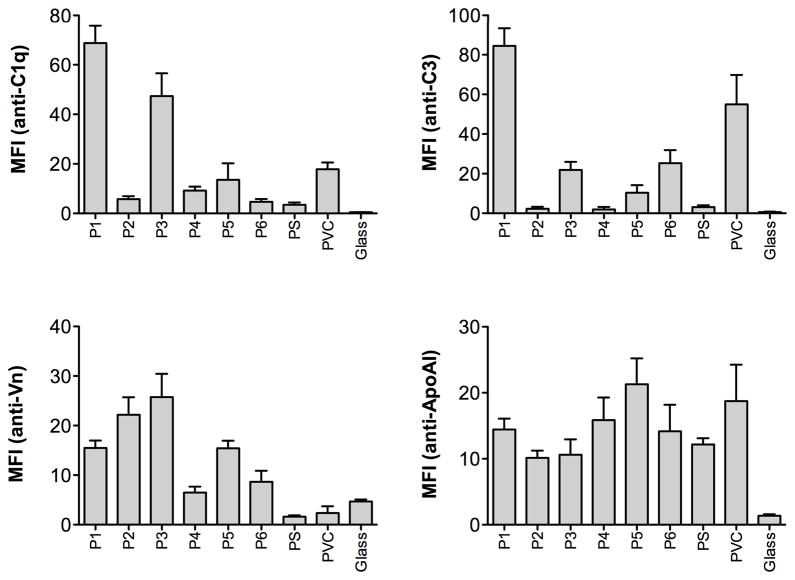
COPAS analysis was used to determine the degree of adsorption of four selected proteins (C1q, C3, vitronectin, and ApoAI). The results showed extensive differences in the amount of surface-exposed protein on the various materials. C1q was confirmed to bind preferentially to P1 and also P3. High levels of vitronectin were found on P1, P2, and P3, intermediate levels on P5, while lower levels of surface exposed vitronectin were detected on P4 and P6 together with the reference materials. Polymer P5 bound the highest levels of ApoAI, while P1, P3, and P6 adsorbed the highest levels of C3 (See Figure 7, and compare to Table 4).
Figure 7.
Adsorption profiles for C1q, C3, vitronectin (Vn) and apolipoprotein AI (ApoAI) on the synthesized polymers and the reference materials, measured with COPAS flow cytometry, after incubation in EDTA-plasma. The results are shown as mean values of mean fluorescence intensity (MFI) + SEM (n=4). In each experiment a minimum of 1,500 particles were collected and analyzed.
Table 4.
The levels of adsorbed plasma proteins on the polymer surfaces were measured after incubation with human EDTA-plasma for 30 min. The surface-bound proteins were identified with an ELISA-like method in which the adsorbed proteins were measured directly on the particles, as described in Experimental Procedures. The results are shown as mean OD-values on the supernatants. Each experiment was performed in triplicate, and the negative control was subtracted from each value.
| Identified protein | MAA/DAP (P1) | MAA/DVB (P2) | MAA/EGDMA (P3) | IPAAm/EGDMA (P4) | Styrene/EGDMA (P5) | HEMA/EGDMA (P6) | PS | PVC | Glass |
|---|---|---|---|---|---|---|---|---|---|
| α-2-macroglobulin | 0.38 | 0.06 | 0.34 | 0.11 | 0.08 | 0.21 | 0.04 | 0.05 | 0.06 |
| C4BP | 1.3 | 0.12 | 0.65 | 0.08 | 0.29 | 0.30 | 0.05 | 0.09 | 0.07 |
| C1q | 1.5 | 0.11 | 1.6 | 0.17 | 0.26 | 0.63 | 0.09 | 0.06 | 0.07 |
| Fibrinogen | 0.36 | 0.40 | 0.62 | 0.48 | 0.57 | 0.45 | 0.24 | 0.12 | 0.09 |
| C4 | 0.99 | 0.16 | 0.95 | 0.25 | 0.60 | 1.4 | 0.11 | 0.10 | 0.05 |
| C3 | 1.4 | 0.20 | 0.75 | 0.12 | 0.25 | 1.1 | 0.08 | 0.09 | 0.05 |
| C5 | 0.74 | 0.07 | 0.35 | 0.08 | 0.10 | 0.37 | 0.06 | 0.05 | 0.04 |
| Factor H | 1.4 | 0.34 | 1.5 | 0.16 | 0.31 | 0.52 | 0.09 | 0.08 | 0.06 |
| IgG | 0.67 | 0.08 | 0.54 | 0.07 | 0.1 | 0.15 | 0.05 | 0.08 | 0.05 |
| Factor XII | 0.72 | 0.11 | 1.3 | 0.14 | 0.19 | 0.54 | 0.08 | 0.05 | 0.16 |
| Haptoglobulin | 1.4 | 0.11 | 0.69 | 0.20 | 0.15 | 0.39 | 0.06 | 0.08 | 0.05 |
| C1-inhibitor | 0.58 | 0.13 | 1.6 | 0.58 | 0.58 | 1.3 | 0.11 | 0.08 | 0.09 |
| Factor I | 0.40 | 0.06 | 0.22 | 0.07 | 0.11 | 0.21 | 0.04 | 0.06 | 0.05 |
| Vitronectin | 0.57 | 0.94 | 1.5 | 0.32 | 0.28 | 0.60 | 0.14 | 0.08 | 0.16 |
| Transferrin | 0.54 | 0.09 | 0.40 | 0.06 | 0.06 | 0.14 | 0.06 | 0.07 | 0.05 |
| Albumin | 0.41 | 0.44 | 0.81 | 0.10 | 0.30 | 0.35 | 0.17 | 0.09 | 0.06 |
| Antithrombin | 0.55 | 0.25 | 1.2 | 0.12 | 0.16 | 0.59 | 0.07 | 0.16 | 0.07 |
| Hemopexin | 0.61 | 0.16 | 0.88 | 0.10 | 0.14 | 0.26 | 0.05 | 0.06 | 0.05 |
| ApoAIV | 0.07 | 0.06 | 0.23 | 0.06 | 0.07 | 0.18 | 0.05 | 0.06 | 0.13 |
| ApoAI | 0.10 | 0.27 | 0.25 | 0.07 | 0.17 | 0.78 | 0.08 | 0.06 | 0.05 |
Quantification of individual proteins
An ELISA-like method was developed in order to detect a large number of plasma proteins adsorbed to the polymers. Mapping of the individual plasma proteins on the surface of 1-mg samples of each polymer was performed after incubation in EDTA-plasma. Antibodies recognizing complement- and coagulation-associated proteins, including several RCAs, as well as other abundant plasma proteins, were used to identify the adsorbed proteins. This technique was less time-consuming than the COPAS analysis. The results were well correlated (p<0.0001), despite the differences in quantifying the particles (by weight in the particle ELISA and by counting events in the COPAS).
The results revealed widely heterogeneous binding of proteins to the various polymers. All of the analyzed proteins were positively indentified on each of the polymers as well as on the control particles. Both P1 and P3 were found to adsorb high levels of the complement initiating molecules IgG and C1q. Both polymers bound high and similar amounts of the RCA factor H, while P1 bound much less of the two inhibitors C1INH and vitronectin compared to P3.
The results also showed an enrichment of the two apolipoproteins (AI and AIV) on P6. Polymer P3 bound the majority of the examined proteins to a high degree, while P2 and P4 showed a generally low level of protein binding. The results are presented as mean OD-values/mg of polymer after subtraction of values for the negative control for each polymer (see Table 4, compare to Figure 7).
DISCUSSION
In the present work we have synthesized an array (n=22) of polymers using combinations of three cross-linkers and eight different monomers (selected to provide a range of functionalities; acidic, basic, neutral, hydrophobic) in order to study how the initial adsorption of plasma proteins influences the biological response. In this study we chose to work with polymer particles rather than planar polymers surfaces because this approach: 1) provides a higher surface/plasma ratio which enables identification of proteins binding in low amounts; 2) enables the use of the flow cytometry based COPAS technique to visualize protein binding on the polymer surfaces; 3) was the basis to develop the particle ELISA (using Tween to minimize the nonspecific binding of proteins, including detecting antibodies) which provides the basis for a higher throughput of analysis of the generated polymers compared to what would be possible using planar surfaces. We found that there was an apparent association between the physicochemical properties of the novel polymers, the amount of adsorbed proteins and the composition of the initially formed protein monolayer and how the polymers elicited complement activation.
The complement activation induced by the novel polymers in hirudin-anticoagulated plasma, was shown to vary widely. In general, all polymers containing the hydrophilic crosslinker DAP were found to activate the complement system to a high or very high degree, while the polymers that contained either one of the hydrophobic crosslinkers DVB or EGDMA only induced a low or moderate complement activation (in hirudin plasma where the complement system is fully active). The complement activating data was used to select six polymers for further detailed studies with the goal to select as heterogeneous materials as possible, both regarding material composition and complement activating ability. The monomer MAA is commonly used in polymer syntheses and was therefore selected with all three crosslinkers resulting in the negatively charged polymers P1–P3. P3–P6 consisted of the crosslinker EGDMA, together with four monomers, giving rise to hydrophobic polymers that were uncharged, with the exception of P3, which was hydrophobic and negatively charged.
The six polymers were further characterized with regard to deposition of the initial monolayer of protein on the surfaces. These studies were performed without interference of the cascade systems in EDTA-plasma, where both the complement and the coagulation system were totally inactive. The unique protein adsorption fingerprints of each polymer that were obtained after contact with plasma or serum were visualized with SDS gel electrophoresis. These results are consistent with the results obtained from the quantification of total adsorbed protein, with higher levels of proteins adsorbed onto P1, P3, P5 and P6 and significantly lower levels onto P2 and P4, which resembled the reference polymers PS, PVC, and glass.
The low complement activating properties of P2 and P4 and the control polymers is most likely explained by the low protein adsorption. This is caused by the fact that the pore sizes of P2 and P4 as well as PS and PVC were three times smaller than those of the other polymers (30–40Å vs. >100Å), based on the BET analysis data. Smaller pores limit the total accessible surface area for the proteins by sterically hindering the binding of many proteins of medium molecular size (50–100 kDa, such as HSA (≈69 kDa), with dimensions of 80×80×30Å 25 or high molecular size (>100 kDa, such as IgG (≈150 kDa), with dimensions of 100×140×45Å 26. This notion was supported by the absence of correlation between the surface area accessible to N2 and the total amount of proteins adsorbed to the P1–P6. Others have previously identified pore size to be of great importance for the adsorption of proteins to a biomaterial 27, as well as for the biological response elicited by the material 28, 29. The pore size in the glass particles with an estimated diameter of >500Å is by far larger than the diameter of most plasma proteins and can be regarded as a change in topography and is not to be expected to interfere with protein binding. The three reference particles, which had the smallest surface area, all showed low protein binding.
It was found that polymer P1, which was the most potent complement activator, bound high levels of the complement recognition protein C1q. C1q is a key protein in the activation of the classical pathway of the complement cascade, recognizing and binding structures such as antigen-bound IgG and IgM, as well as some viruses and gram-negative bacteria. The plasma concentration of this large protein (≈410 kDa) is 100–180 mg/L. C1q is one of the most highly positive charged (cationic; pI ~ 9.3) proteins in human plasma 30 and it provides multivalent attachment to the surface via its collagen-like arms. The enrichment of C1q on P1 probably reflects a combination of the chemical nature of the polymer, its large pore size, and the high level of IgG bound to the particles. The high binding of C1q and the gammaglobulin IgG might be explained by electrostatic interactions between the charged proteins and the high density of electrons contributed by the amide functionality of the crosslinker. In addition, P3, which has similar pore size, showed similar high binding of C1q as P1 but the chemical basis for this is not clear. Our results indicate that the nitrogen-containing crosslinker DAP probably also played an important role, since all the DAP-crosslinked polymers tested (six polymers in total) proved to be specifically enriched in C1q to a high degree (data not shown). This explanation is also consistent with the dose-dependent increase in generated C3a that was observed in the initial screenings, when the ratio of crosslinker (DAP) to monomer was increased for polymer P1. Although both P1 and P3 bound significant amounts of IgG and C1q, there was a more than 10-fold difference in complement activation between the polymers (12,600 ng/mL of C3a for P1 vs. 1,100 ng/mL for P3). A plausible explanation for this difference may be found in their binding of RCAs: the levels of factor H were slightly lower for P1, but the most pronounced discrepancy was seen regarding C1INH where P1 bound approximately 1/3 of the amount compared to P3 (Table 4). It is therefore likely that the amount of C1INH and factor H associated with P3 is sufficient to attenuate complement activation induced by this polymer while there are too low amounts of RCAs on P1, resulting in massive complement activation.
P5 and P6, which were poor activators of complement, had similar and distinctive adsorption patterns with a significant enrichment of vitronectin (75 kDa), ApoAIV (45 kDa), and ApoAI (25 kDa) when analysed by SDS-PAGE. The binding of these proteins detected by COPAS and particle ELISA was lower, most likely due to the fact that these techniques are limited to measuring molecules exposed on the surface of the polymer, whereas the electrophoresis-based data includes all proteins adsorbed onto and inside the porous particles. Vitronectin is a multifunctional protein that has, among other roles, been shown to act as a complement regulator that decreases cytolytic activity by interfering with the membrane attack complex. Furthermore, the protein is known to bind to biomaterial surfaces and to decrease the complement activation on polystyrene and on bacterial surfaces 31–33. Apolipoprotein AI is the major component in high-density lipoproteins (HDL) and can be found both incorporated into HDL and in free form in plasma. Apolipoprotein AIV is an acidic plasma glycoprotein which has low affinity for lipoproteins and is the most hydrophilic of all the human apolipoproteins 34. Several groups have reported that apolipoproteins are adsorbed to biomaterial surfaces in significant amounts, but the biological importance of this binding is still unclear (17, 35). P2, which induced negligible complement activation in the initial screening, bound a higher amount of factor H and vitronectin compared to P4 which induced moderate activation (2000 ng C3a/ml), suggesting that the difference could be due to a more active down regulation on P2. Alternatively, the lack of activation might be related to binding of albumin and fibrinogen. Of all the tested particles, P2 bound the highest proportion of albumin. In addition, the protein bound to both P2 and P4 contained a high proportion of fibrinogen. When adsorbed to surfaces, both albumin and fibrinogen can attenuate complement activation by failing to provide binding sites for generated C3b. Consequently; lower amounts of alternative pathway complexes are formed, attenuating the amplification of complement activation by the alternative pathway 7, at least at the initial stage of blood exposure.
In conclusion we have by using a model system consisting of newly synthesized polymers in which we can change the charge, the wetability and the functionally available surface etc., defined some of the preconditions for complement activation on material surfaces in contact with blood. 1) The material should not be negatively charged since this tends to lead to deposition of IgG and C1q and thus induce complement activation (P1 & P3). 2) It should preferentially be hydrophobic with the capacity to bind apolipoproteins, possibly in the form of HDL (P5 & P6). 3) It should present a low available surface area, which is made possible by the introduction of a nanostructure of pores that are small enough to exclude most plasma proteins, (P2, P4, PS and PVC).
These results in part confirm earlier conclusions obtained using less defined systems (8 and references therein). Future studies of these polymers in the form of planar surfaces will possibly require additional surface analyses, to confirm that the surface properties of the polymers does not significantly differ from the bulk compositions. The result demonstrates that this polymer model is a powerful and comprehensive system to define the requirements for the activation of innate immune and inflammatory systems in blood in vitro and in the future, in vivo. The overall aim for these approaches is to construct novel potential biomaterials with hemocompatibility superior to those used in the clinic today.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Research Council (VR) 2009-4675, 2009-4462, 2006-6041, Carl Trygger’s Foundation, and grants # EB003968 and AI068730 from the National Institutes of Health (USA) and the Linnæus University. We thank Dr. Deborah McClellan for excellent editorial assistance.
References
- 1.Kourtzelis I, Markiewski MM, Doumas M, Rafail S, Kambas K, Mitroulis I, et al. Complement anaphylatoxin C5a contributes to hemodialysis-associated thrombosis. Blood. 2010;116(4):631–9. doi: 10.1182/blood-2010-01-264051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markiewski MM, Nilsson B, Ekdahl KN, Mollnes TE, Lambris JD. Complement and coagulation: strangers or partners in crime? Trends Immunol. 2007;28(4):184–92. doi: 10.1016/j.it.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Horbett T. The role of adsorbed proteins in tissue response to biomaterials. In: Ratner BDHA, Schoen FJ, Lemons JE, editors. Biomaterials science, an introduction to materials in medicine. 2. London: Elsevier Academic Press; 2004. pp. 237–46. [Google Scholar]
- 4.Andersson J, Ekdahl KN, Larsson R, Nilsson UR, Nilsson B. C3 adsorbed to a polymer surface can form an initiating alternative pathway convertase. J Immunol. 2002;168(11):5786–91. doi: 10.4049/jimmunol.168.11.5786. [DOI] [PubMed] [Google Scholar]
- 5.Wettero J, Bengtsson T, Tengvall P. C1q-independent activation of neutrophils by immunoglobulin M-coated surfaces. J Biomed Mater Res. 2001;57(4):550–8. doi: 10.1002/1097-4636(20011215)57:4<550::aid-jbm1201>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 6.Gorbet MB, Sefton MV. Biomaterial-associated thrombosis: roles of coagulation factors, complement, platelets and leukocytes. Biomaterials. 2004;25(26):5681–703. doi: 10.1016/j.biomaterials.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 7.Andersson J, Ekdahl KN, Lambris JD, Nilsson B. Binding of C3 fragments on top of adsorbed plasma proteins during complement activation on a model biomaterial surface. Biomaterials. 2005;26(13):1477–85. doi: 10.1016/j.biomaterials.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson B, Ekdahl KN, Mollnes TE, Lambris JD. The role of complement in biomaterial-induced inflammation. Mol Immunol. 2007;44(1–3):82–94. doi: 10.1016/j.molimm.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Bäck J, Huber-Lang M, Elgue G, Kalbitz M, Sanchez J, Ekdahl KN, et al. Distinctive regulation of contact activation by antithrombin and C1-inhibitor on activated platelets and material surfaces. Biomaterials. 2009;30(34):6573–80. doi: 10.1016/j.biomaterials.2009.07.052. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Simonovsky FI, Ratner BD, Horbett TA. The role of adsorbed fibrinogen in platelet adhesion to polyurethane surfaces: a comparison of surface hydrophobicity, protein adsorption, monoclonal antibody binding, and platelet adhesion. J Biomed Mater Res A. 2005;74(4):722–38. doi: 10.1002/jbm.a.30381. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson B, Korsgren O, Lambris JD, Ekdahl KN. Can cells and biomaterials in therapeutic medicine be shielded from innate immune recognition? Trends Immunol. 2009;31(1):32–8. doi: 10.1016/j.it.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekdahl KN, Nilsson B, Gölander CG, Elwing H, Lassen B, Nilsson UR. Complement activation on radio frequency plasma modified polystyrene surfaces. J Coll Interface Sci. 1993;158:121–8. [Google Scholar]
- 13.Tengvall P, Askendal A, Lundstrom I, Elwing H. Studies of surface activated coagulation: antisera binding onto methyl gradients on silicon incubated in human plasma in vitro. Biomaterials. 1992;13(6):367–74. doi: 10.1016/0142-9612(92)90042-m. [DOI] [PubMed] [Google Scholar]
- 14.Francoise Gachon AM, Mallet J, Tridon A, Deteix P. Analysis of proteins eluted from hemodialysis membranes. J Biomater Sci Polym Ed. 1991;1(4):263–76. doi: 10.1163/156856291x00160. [DOI] [PubMed] [Google Scholar]
- 15.Bonomini M, Pavone B, Sirolli V, Del Buono F, Di Cesare M, Del Boccio P, et al. Proteomics characterization of protein adsorption onto hemodialysis membranes. J Proteome Res. 2006;5(10):2666–74. doi: 10.1021/pr060150u. [DOI] [PubMed] [Google Scholar]
- 16.Rosengren A, Pavlovic E, Oscarsson S, Krajewski A, Ravaglioli A, Piancastelli A. Plasma protein adsorption pattern on characterized ceramic biomaterials. Biomaterials. 2002;23(4):1237–47. doi: 10.1016/s0142-9612(01)00244-7. [DOI] [PubMed] [Google Scholar]
- 17.Magnani A, Barbucci R, Lamponi S, Chiumento A, Paffetti A, Trabalzini L, et al. Two-step elution of human serum proteins from different glass-modified bioactive surfaces: a comparative proteomic analysis of adsorption patterns. Electrophoresis. 2004;25(14):2113–424. doi: 10.1002/elps.200305826. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino Y, Koide H, Urakami T, Kanazawa H, Kodama T, Oku N, et al. Recognition, neutralizaton, and clearance of target peptides in the bloodstream of living mice by molecularly imprinted polymer nanoparticles: A plastic antibody. Journal of American chemcial society. 2010;132:6644–5. doi: 10.1021/ja102148f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moghimi SM, Andersen AJ, Hashemi SH, Lettiero B, Ahmadvand D, Hunter AC, et al. Complement activation cascade triggered by PEG-PL engineered nanomedicines and carbon nanotubes: the challenges ahead. J Control Release. 2010;146(2):175–81. doi: 10.1016/j.jconrel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Lopez A, Persson C, Hilborn J, Engqvist H. Synthesis and characterization of injectable composites of poly[D, L-lactide-co-(epsilon-caprolactone)] reinforced with beta-TCP and CaCO3 for intervertebral disk augmentation. J Biomed Mater Res B Appl Biomater. 2010;95(1):75–83. doi: 10.1002/jbm.b.31685. [DOI] [PubMed] [Google Scholar]
- 21.Seyfert UT, Biehl V, Schenk J. In vitro hemmocompatibility testing of biomaterials according to the ISO 10993-4. Biomolecular Engineering. 2002;19:91–6. doi: 10.1016/s1389-0344(02)00015-1. [DOI] [PubMed] [Google Scholar]
- 22.Mollnes TT, Brekke OL, Fung M, Fure H, Christiansen D, Bergseth G, et al. Essential role of the C5a receptor in E coli-induced oxidative burst and phagocytosis revealed by a novel lepirudin-based human whole blood model of inflammation. Blood. 2002;100(5):1869–77. [PubMed] [Google Scholar]
- 23.Bexborn F, Engberg AE, Sandholm K, Mollnes TE, Hong J, Nilsson Ekdahl K. Hirudin versus heparin for use in whole blood in vitro biocompatibility models. J Biomed Mater Res A. 2009;89(4):951–9. doi: 10.1002/jbm.a.32034. [DOI] [PubMed] [Google Scholar]
- 24.Ekdahl KN, Nilsson B, Pekna M, Nilsson UR. Generation of iC3 at the interface between blood and gas. Scand J Immunol. 1992;35(1):85–91. doi: 10.1111/j.1365-3083.1992.tb02837.x. [DOI] [PubMed] [Google Scholar]
- 25.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999;12(6):439–46. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 26.You HX, Lowe CR. AFM studies of protein adsorption .2. Characterization of immunoglobulin G adsorption by detergent washing. Journal of Colloid and Interface Science. 1996;182(2):586–601. [Google Scholar]
- 27.Williams DF. On the mechanisms of biocompatibility. Biomaterials. 2008;29(20):2941–53. doi: 10.1016/j.biomaterials.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Ferraz N, Carlsson J, Hong J, Ott MK. Influence of nanoporesize on platelet adhesion and activation. J Mater Sci Mater Med. 2008 Sep;19(9):3115–21. doi: 10.1007/s10856-008-3449-7. [DOI] [PubMed] [Google Scholar]
- 29.Ferraz N, Nilsson B, Hong J, Karlsson Ott M. Nanoporesize affects complement activation. J Biomed Mater Res A. 2008;87(3):575–81. doi: 10.1002/jbm.a.31818. [DOI] [PubMed] [Google Scholar]
- 30.Loos M, Trinder P, Kaul M. Components and Reactivity. In: Rother K, Till GO, Hänsch GM, editors. The Complement System. Berlin Heidelberg: Springer-Verlag; 1998. [Google Scholar]
- 31.Hayman EG, Pierschbacher MD, Suzuki S, Ruoslahti E. Vitronectin - a major cell attachment-promoting protein in fetal bovine serum. Exp Cell Res. 1985;160(2):245–8. doi: 10.1016/0014-4827(85)90173-9. [DOI] [PubMed] [Google Scholar]
- 32.Lundberg F, Lea T, Ljungh A. Vitronectin-binding staphylococci enhance surface-associated complement activation. Infect Immun. 1997;65(3):897–902. doi: 10.1128/iai.65.3.897-902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallstrom T, Blom AM, Zipfel PF, Riesbeck K. Nontypeable Haemophilus influenzae protein E binds vitronectin and is important for serum resistance. J Immunol. 2009;183(4):2593–601. doi: 10.4049/jimmunol.0803226. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg RB. Differences in the hydrophobic properties of discrete alpha-helical domains of rat and human apolipoprotein A-IV. Biochim Biophys Acta. 1987;918(3):299–303. doi: 10.1016/0005-2760(87)90234-7. [DOI] [PubMed] [Google Scholar]
- 35.Cornelius RM, Archambault J, Brash JL. Identification of apolipoprotein A-I as a major adsorbate on biomaterial surfaces after blood or plasma contact. Biomaterials. 2002;23(17):3583–7. doi: 10.1016/s0142-9612(02)00083-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.