Abstract
We have created a novel enzyme reactor using electric field-mediated orientation and immobilization of proteolytic enzymes (trypsin/chymotrypsin) on biocompatible PVDF membranes in a continuous flow-through chamber. Using less than 5 minutes, this reactor in various enzyme combinations can produce enhanced rapid digestion for standardized prototypic proteins, hydrophilic proteins and hydrophobic transmembrane proteins when compared to in-solution techniques. With improved digestive efficiency, our reactor improved the overall functional analysis of lipid raft proteomes by identifying more closely functionally linked proteins and elucidated a richer set of biological processes and pathways linked to the proteins than traditional in-solution methods.
Keywords: Enzyme reactor, Rapid digestion, Membrane proteins, Lipid rafts, Neurotransmission, Bioinformatics data extraction
With advances in data confidence using the statistical analysis of increased sample replicates, high-throughput mass spectrometry (MS)-based shotgun proteomics has the potential to transit from a discovery technology to a declarative scoring process [1–4]. Such advances would greatly facilitate the creation of global and dynamic views of simultaneous functional protein interactions in biological settings. One of the most challenging steps in high-throughput format experiments is the rapid and efficient generation of peptides for such MS-based proteomics analysis. The challenges are particularly stringent for membrane proteins regulating central nervous system transmission as they demonstrate poor solubilization, low abundance and relative paucities of tryptic cleavage sites, e.g. ligand-gated ion channels or G protein-coupled receptors (GPCRs), which are usually under-represented in the MS discovery process [5].
Less specific proteases such as proteinase K [6], elastase[7] and pepsin[8] have been used for the proteomic analysis of such under-represented proteins. Non-specific proteases can produce large numbers of peptides but have demonstrated a low degree of specificity for local sites in certain structural motifs [9]. Specific protease digestion in a high-throughput format for under-represented membrane proteins is highly desirable for rapid sample processing, targeted cleavage, and fast database interrogation. To fulfill such requirements, immobilized enzyme reactors (IMERs) with specific proteases have the potential to provide rapid and enhanced digestion, which is also essential to the automation, reproducibility, efficiency of protein identification and improved proteomic coverage. IMERs rely upon efficient immobilization of functional enzymes to a variety of solid supports: resin beads, membranes, the inner surface of fused silica capillaries and monolithic materials. IMERs can be generated using several mechanisms including sol-gel entrapment [10], layer-by-layer assembly [11], covalent binding or physical adsorption [12]. IMERs have demonstrated excellent processing properties with high enzymatic activity and are highly synergistic with high-throughput MS analysis.[13] Traditional strategies for IMER preparations however are extremely time-consuming and involve many steps that may degrade enzyme bioactivity. Moreover, extraction of biologically-relevant MS data for under-represented transmembrane (TM) proteins is often difficult compared to extraction of soluble protein data using current technology.
To address these issues, we describe a rapid, efficient and facile strategy to achieve functional enzyme immobilization at the surface of a biocompatible PVDF membrane using electric-field-orientation (EFO: the details of the setup are in Supplementary data under “Materials and Methods”, Figure S1 A–D and Figure S2) [14–16]. Using a controllable electric field, enzyme dipoles can be fixed at the surface of the biocompatible membrane in a continuous flow-through chamber within only 15 minutes. Due to our rapid reactor generation, we can employ proteolytic enzymes in their most active formats. When dissolving trypsin in ABC (ammonium bicarbonate) as compared to acetic acid for immobilization, we observed substantial increases in BSA sequence coverage from 26% (acetic acid) to 94% (ABC) (data not shown). This elevated coverage is suggestive that the resultant conformation of an immobilized enzyme is crucial for its function. Dissolution and immobilization of an enzyme at its pH optimum may help to sustain suitable conformation and charge properties for active protein digestion. Although the use of proteolytic enzymes in their inactive forms may decrease enzyme autolysis, the rapid preparation process of EFO-IMER and increased stability from immobilized enzymes may attenuate the possibility of enzyme autolysis. Another advantage of the EFO-IMER process is the ability, through its simple and rapid physical construction, to accommodate multiple endoproteases in one reactor. This aspect can then increase digestion efficiency in a rapid and reliable manner using multiple ambient temperatures.
To demonstrate the experimental usefulness of our EFO-IMER, we first employed the prototypic protein BSA (bovine serum albumin). BSA (1 mg/mL) was used to test the efficiency of EFO-IMER-immobilized trypsin at multiple infusion rates. We performed all MS analyses for resultant digests on multiple LXQ or LTQ mass spectrometers (ThermoFinnigan), essentially as described previously[17] (supplementary Methods). As judged by SDS-PAGE and LC-MS/MS analysis of BSA peptides, we determined that the optimized flow rate for chamber digestion was 2 μL/min (Figure S1E). The reproducibility for the immobilization of the target protease(s) could be precisely controlled by input enzymes concentration, electric field strength and application time (n=5, CV=2.1%). Our results demonstrated that the enzyme was very stable. Using BSA, we found that the CV of BSA coverage was <5% within one day (n=6), and we achieved a CV <22% within 10 days (n=10). No significant cross-contamination was found after thorough washing with ABC. Since non-specific cleavages and missed cleavages are very important for successful protein ID generation, we compared trypsin efficiency on these two aspects between in solution and after immobilization on PVDF membranes, aided by EFO. The presence of peptides with non-specific cleavages was 16% in the EFO-IMER based dataset and 30% in the in-solution dataset, suggesting that rapid digestion facilitated specific digestion. These two methods yielded similar percentages of missed cleavage sites in the total set of identified peptides (Figure S3). It is interesting to note that a much better coverage as well as more peptides could be identified using EFO-IMER. To test whether the EFO-IMER was compatible with chaotropic agents, we used 8M urea to reconstitute BSA. The coverage in this condition was 86% using the EFO-IMER, compared to 67% using in-solution digestion, suggesting that the EFO-IMER can also work with chaotropic agents of high concentration.
Indicating the flexibility of our EFO-IMER system, i.e. it easily accommodated multiple endoproteases in a single reactor. We next assessed its performance, side-by-side, with in-solution protocols using trypsin, chymotrypsin and a mixture of trypsin and chymotrypsin respectively (Figure S2A). We used chymotrypsin as a model protease because it can produce more proteolytic peptides from membrane proteins than other individual proteases due to its hydrophobic cleavage sites [18, 19]. To compare and illustrate resultant peptide information, we designed a 6-set Venn diagram software program (VENNTURE) and a sequence alignment program (using C++), to visualize the unique and overlapping peptides generated from our multiple applied digestive methods (Figure S2B, S2C). We tested multiple digestive paradigms comparing standard digestive techniques to our EFO-IMER, using both trypsin and chymotrypsin. The EFO-IMER reactor configuration employing trypsin and chymotrypsin produced the highest number of detected peptides (155) and the best sequence coverage (94%) of BSA, compared to the other 5 paradigms tested (Figure S2A, S2C) (n=3, CV<5.6%). Using an additional prototypic protein, α-casein, we also observed a similar superiority of the EFO-IMER process compared to standard digestive techniques (Figure S3A–C). EFO-IMER-derived peptide mixtures were also efficiently handled by standard LC MS/MS systems, as well as commonly used database search algorithms. As different digestion methods can create idiosyncratic peptide outputs, the integration of results from multiple preparative processes could lead to greater sequence coverage and eventually a deeper biological appreciation of the input sample (Figures S2C,3C).
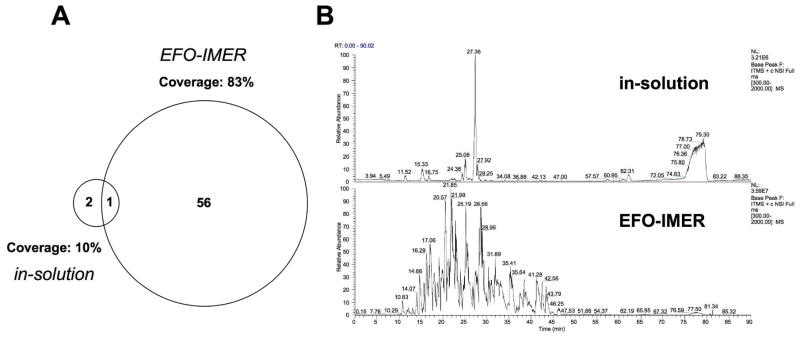
We next employed a protein prototype from a group (transmembrane proteins) commonly under-represented in MS analysis. Bacteriorhodopsin, a heptahelical TM protein, was chosen as an archetypical hydrophobic and poorly digestible protein to further assess EFO-IMER digestive efficiency and its range of applicability. EFO-IMER-mediated digestion of bacteriorhodopsin, compared to classical overnight in-solution digestion, resulted in a greater number of recovered peptides, a higher sequence coverage and a greatly reduced processing time (EFO-IMER, 0.5h; in-solution, 18h: Figure 1). As many important TM proteins usually demonstrate a low abundance, compared to cytosolic proteins, we also investigated the ability of the EFO-IMER to assist analysis of multiple protein levels of bacteriorhodopsin. The EFO-IMER demonstrated an ability to efficiently extract protein information from considerably lower bacteriorhodopsin levels than the in-solution process. EFO-IMER-mediated digestion of a 1 mg/mL concentration of bacteriorhodopsin yielded 57 unique peptides (83% coverage, 80% hydrophobic peptides) compared to only 3 peptides (10% coverage, 0% hydrophobic peptides) recovered using in-solution digestion of 1 mg/mL bacteriorhodopsin. EFO-IMER-mediated digestion of 0.1 or 0.01 mg/mL bacteriorhodopsin yielded 22 peptides (41% coverage, 60% hydrophobic peptides) and 1 peptide (9% coverage, 100% hydrophobic peptides) respectively, while no peptides were recovered from in-solution digestion of these lower bacteriorhodopsin concentrations. We reasoned that hydrophobic peptides generated after proteolysis would not be retained on the PVDF membranes of the chamber and could be efficiently analyzed by LC-MS/MS. One of the possible reasons for this is, that after enzyme immobilization with the aid of EFO, the surfaces of PVDF were saturated by enzymes and were changed from hydrophobic to hydrophilic. Therefore some hydrophilic proteins, instead of hydrophobic proteins, could be lost during the EFO-IMER process.
Figure 1.
A) Proportional Venn diagram illustrating the number of isolated peptides and percentage coverage of bacteriorhodopsin digestion using EFO-IMER or in-solution digestion by LC-MS/MS analysis and MASCOT database searches. B) Representative chromatograms of in-solution-digested bacteriorhodopsin (upper panel) or EFO-IMER-digested bacteriorhodopsin (lower panel) indicating the considerably greater peptide extraction from the hydrophobic protein.
We also compared the EFO-IMER with a proprietary immobilized trypsin (Promega, Cat.# V9012). Sequence coverage of digested bacteriohodopsin was only 12% using immobilized trypsin compared to 82% using EFO-IMER immobilized with trypsin and chymotrypsin. There are several possible reasons for this interesting result: more homogeneous immobilization aided by EFO; more active enzyme due to fresh preparation in the EFO-IMER format; and more efficient cleavage of membrane proteins by the combination of trypsin and chymotrypsin.
While the proteomic-MS analysis of prototypic proteins is aimed at generating maximal accuracy of peptide identification and coverage, the extraction of biologically-relevant functional data is the main priority for MS-based proteomic analysis of complex biological samples [20]. We therefore investigated whether the EFO-IMER could facilitate the enhanced extraction of biologically-relevant data from proteomic analyses of highly complex, hydrophobic protein extracts. We used neuronal lipid rafts as model samples. Lipid rafts are small, high-density lipid areas of the plasma membrane that contain multiple signaling proteins crucial for synaptic neurotransmission [21]. Their high hydrophobicity and relatively low protein content pose significant problems of solubilization, digestion and data extraction, which may hinder proteomic-MS investigation of the important membrane proteins present in lipid rafts [22]. Lipid rafts were extracted from mouse cortex and the presence of the neuronal lipid raft marker, flotillin-1, in fractions was assessed (Figure S4). Flotillin-rich fractions (fractions 2–5) were then pooled for each individual animal (n=3).
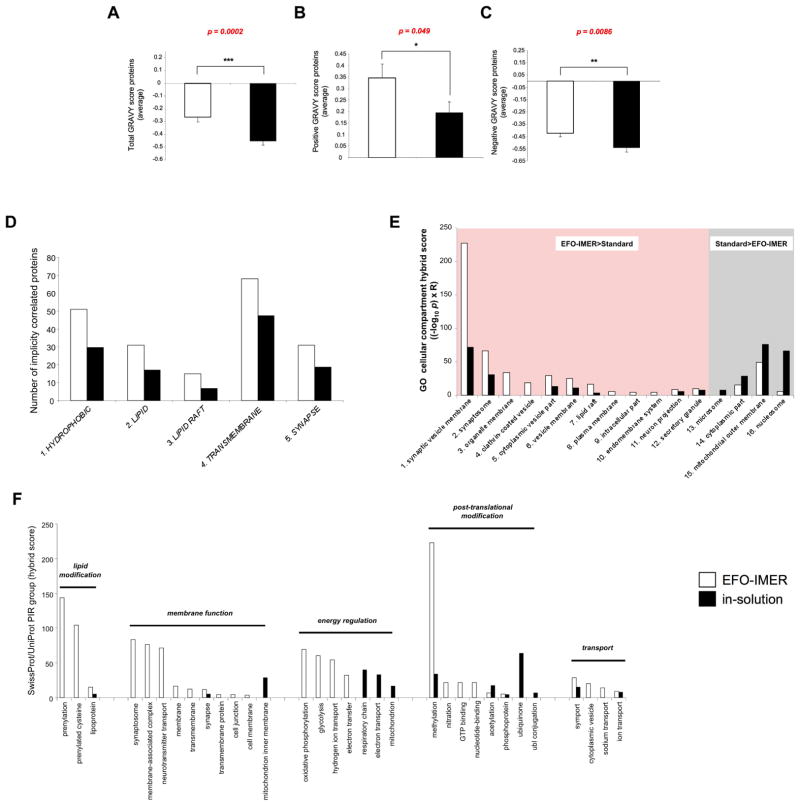
Using EFO-IMER or in-solution digestion protocols, we were able to identify multiple proteins from cortical lipid raft extracts (Table S1, S2 and S3). The functional signaling relevance of the extracted protein data for both scenarios was then directly compared using multiple complementary un-biased bioinformatic techniques (Figure S5). Using ProtParam (http://expasy.org/tools/protparam.html) to calculate the grand average of hydropathy (GRAVY), the EFO-IMER-derived protein set, possessed more proteins with a higher GRAVY score than the in-solution process. Mean scores for the total extracted proteins and the positive or negative GRAVY-scoring proteins demonstrated a significantly greater hydrophobicity of the EFO-IMER-derived protein set compared to the in-solution set (Figure 2A–C). While the EFO-IMER process identified a greater number of proteins with a greater hydrophobic nature than the in-solution process, we did however identify a greater ‘total’ number of proteins with the in-solution process. Therefore, with respect to the nature of the extracted proteins the in-solution process identified proteins with a more hydrophilic nature compared to the EFO-IMER. One possible explanation for this may be the relative depletion of the more common hydrophilic proteins from the EFO-IMER chamber due to their physical interaction with the considerable area of hydrophilic surfaces of the reaction chamber (facilitated by enzyme saturation) compared to that which may occur with in-solution digestion. Future reduction of this ‘hydrophilic’ protein loss, via chamber volume reduction, or different protein enrichment techniques may therefore assist future IMER preparation.
Figure 2.
ProtParam, GeneIndexer, GO-cellular component term protein set analysis and PIR keyword analysis. GRAVY scores were generated for each protein in the EFO-IMER (white bars) or in-solution (black bars) digestion of murine cortical neuron raft extracts. A) Mean (± standard error) GRAVY scores for all raft proteins extracted using both digestion techniques (statistical significance calculated with un-paired two-tailed Student’s t-test: GraphPad Prism v.5). B) Mean (± standard error) GRAVY scores for accumulated proteins from each extraction technique that possessed net positive GRAVY scores. C) Mean (± standard error) GRAVY scores for accumulated proteins from each extraction technique that possessed net negative GRAVY scores. D) GeneIndexer (Computable Genomix) was employed generate interrogation term-specific subsets of proteins from the primary input EFO-IMER or in-solution datasets. Correlation of input proteins with interrogation terms (in capitals, 1–5) required at least two interrogation term cross-correlations with an implicit correlation score of ≥ 0.1. Interrogation term (1–5) correlation for protein sets from EFO-IMER- (white bar) or in-solution- (black bar) derived datasets. For each membrane-related interrogation term (1–5) the subsets of correlated proteins were greater for the EFO-IMER process compared to in-solution. E) Gene Ontology cellular component (GOcc) analysis of EFO-IMER (white bars) or in-solution (black bars) digestion protein datasets. Magnitude of GOcc term population (1–16) was measured by hybrid score ((−log10 enrichment probability (p)) x enrichment ratio). GOcc terms with a higher hybrid score for the EFO-IMER-derived compared to in-solution-derived protein set (1–12) are highlighted in red. GOcc terms that were significantly populated to a greater extent by the in-solution-derived protein set (13–16) are denoted in grey. F) SwissProt/UniProt protein information resource (PIR) group analysis of EFO-IMER- or in-solution digestion-derived protein sets from cortical lipid rafts. A hybrid score ((−log10 enrichment probability (p)) x enrichment ratio) was generated for each specific significantly-populated group. PIR groups were then clustered according to functional relationships indicated by horizontal bars.
Lipid rafts, in-part, act to recruit and stabilize the functional interactions between membrane proteins crucial for synaptic neurotransmission. We next investigated the functional biases of EFO-IMER- or in-solution-derived protein sets using a novel data-mining process, as described previously [23, 24], i.e. latent semantic indexing (LSI: GeneIndexer, Computable Genomix). LSI allows the relative quantitative measurement of the correlation of identity of multiple proteins in a set with a given input text interrogation term. We noted that more proteins from the EFO-IMER versus in-solution datasets were implicitly correlated (correlation score ≥ 0.1 is considered indicative of an implicit correlation) with membrane-focused interrogation terms such as ‘HYDROPHOBIC’, ‘LIPID’, ‘LIPID RAFT’, ‘TRANSMEMBRANE’ and ‘SYNAPSE’ (Figure 2D). Using Gene Ontology-cellular component (GOcc) analysis to identify the subcellular compartments where the constituents of the EFO-IMER or in-solution dataset proteins are most statistically likely (p<0.05) to reside, we noted that considerably more membrane-related GOcc term groups were more strongly populated by the EFO-IMER dataset compared to the in-solution extracted dataset (Figure 2E). We next compared the structural and functional grouping of these different extracted datasets using NIH-DAVID (Database for Annotation, Visualization and Integrated Discovery version 6.7: (http://david.abcc.ncifcrf.gov/home.jsp) to interrogate the SwissProt/Uniprot Protein Information Resource (PIR: http://pir.georgetown.edu/) group keywords databases. Using these analyses of the EFO-IMER or in-solution extracted protein datasets, we found that EFO-IMER-mediated extracts were able to better generate protein datasets indicative of trans-synaptic areas and cellular signaling processes (Figure 2F). Post-synaptic lipid rafts contain many important signaling proteins and a considerable number of these hydrophobic raft proteins are receptors. We therefore assessed whether the EFO-IMER or in-solution extracted raft datasets demonstrated any transmembrane receptor-signaling bias. Using Ingenuity Pathway Analysis to study the enrichment of receptor-signaling pathways generated by either EFO-IMER or in-solution protein sets we noted a strong bias in the receptor-based relationships between the two experimental datasets (Figure S6). The EFO-IMER-extracted dataset possessed a much more profound receptor-signaling footprint (greater number and higher scoring receptor-controlled signaling pathways), compared to a similar analysis of the in-solution protein set (Figure S6). Using these multiple, un-biased, integrative bioinformatic analyses, our data suggests that extraction of protein datasets using the EFO-IMER process yields datasets that are more representative of the input sample and that display a greater biologically-relevant functional output than classical in-solution techniques.
In conclusion, we have employed the EFO-IMER to enhance digestion of hydrophilic (BSA, α-casein) and hydrophobic prototypic proteins (bacteriorhodopsin). In addition we have shown that EFO-IMER-mediated processing can extract more useful biologically-relevant and biologically-representative data from complex hydrophobic protein mixtures (lipid rafts). The streamlined EFO-IMER-based sample preparation workflow is compatible with automation and may assist future high-throughput proteomic analysis of under-represented protein groups. This could benefit biomarker verification and validation using targeted mass spectrometry or a label-free method for biomarker detection.
Supplementary Material
Acknowledgments
This work was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1.Cravatt BF, Simon GM, Yates JR., III The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 2.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–7. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 3.Durr E, Yu J, Krasinska KM, Carver LA, Yates JR, III, Testa JE, et al. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat Biotechnol. 2004;22:985–92. doi: 10.1038/nbt993. [DOI] [PubMed] [Google Scholar]
- 4.Qian WJ, Jacobs JM, Liu T, Camp DG, II, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol Cell Proteomics. 2006;5:1727–44. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu B, McClatchy DB, Jin YK, Yates JR., III Strategies for shotgun identification of integral membrane proteins by tandem mass spectrometry. Proteomics. 2008;8:3947–55. doi: 10.1002/pmic.200800120. [DOI] [PubMed] [Google Scholar]
- 6.Wu CC, MacCoss MJ, Howell KE, Yates JR., III A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–8. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 7.Rietschel B, Arrey TN, Meyer B, Bornemann S, Schuerken M, Karas M, et al. Elastase digests: New ammunition for shotgun membrane proteomics. Mol Cell Proteomics. 2009;8:1029–43. doi: 10.1074/mcp.M800223-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rietschel B, Bornemann S, Arrey TN, Baeumlisberger D, Karas M, Meyer B. Membrane protein analysis using an improved peptic in-solution digestion protocol. Proteomics. 2009;9:5553–7. doi: 10.1002/pmic.200900532. [DOI] [PubMed] [Google Scholar]
- 9.Gilmore JM, Washburn MP. Advances in shotgun proteomics and the analysis of membrane proteomes. Journal of Proteomics. 2010;73:2078–91. doi: 10.1016/j.jprot.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Dulay MT, Baca QJ, Zare RN. Enhanced proteolytic activity of covalently bound enzymes in photopolymerized sol gel. Anal Chem. 2005;77:4604–10. doi: 10.1021/ac0504767. [DOI] [PubMed] [Google Scholar]
- 11.Salditt T, Schubert US. Layer-by-layer self-assembly of supramolecular and biomolecular films. J Biotechnol. 2002;90:50–70. doi: 10.1016/s1389-0352(01)00049-6. [DOI] [PubMed] [Google Scholar]
- 12.Girelli AM, Mattei E. Application of immobilized enzyme reactor in on-line high performance liquid chromatography: A review. J Chromatogr, B: Anal Technol Biomed Life Sci. 2005;819:3–16. doi: 10.1016/j.jchromb.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 13.Krenkova J, Svec F. Less common applications of monoliths: IV. Recent developments in immobilized enzyme reactors for proteomics and biotechnology. J Sep Sci. 2009;32:706–18. doi: 10.1002/jssc.200800641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talasaz AAH, Nemat-Gorgani M, Liu Y, Ståhl P, Dutton RW, Ronaghi M, et al. Prediction of protein orientation upon immobilization on biological and nonbiological surfaces. Proc Natl Acad Sci U S A. 2006;103:14773–8. doi: 10.1073/pnas.0605841103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heuts J, Salber J, Goldyn AM, Janser R, Möller M, Klee D. Bio-functionalized star PEG-coated PVDF surfaces for cytocompatibility- improved implant components. J Biomed Mater Res, Part A. 2010;92:1538–51. doi: 10.1002/jbm.a.32478. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Shen H, Yi T, Wen D, Pang N, Liao J, et al. Synergistic design of electric field and membrane in facilitating continuous adsorption for cleanup and enrichment of proteins in direct ESI-MS analysis. Anal Chem. 2008;80:8920–9. doi: 10.1021/ac800816k. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5:1214–23. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 18.Lu X, Zhu H. Tube-Gel Digestion: A Novel Proteomic Approach for High Throughput Analysis of Membrane Proteins. 2005. pp. 1948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 20.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nature Reviews Genetics. 2009;10:617–27. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 21.Maudsley S, Martin B, Luttrell LM. G protein-coupled receptor signaling complexity in neuronal tissue: Implications for novel therapeutics. Curr Alzheimer Res. 2007;4:3–19. doi: 10.2174/156720507779939850. [DOI] [PubMed] [Google Scholar]
- 22.Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer's mice: Understanding the interface between physiology and disease. PLoS ONE. 2008;3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadwick W, Zhou Y, Park SS, Wang L, Mitchell N, Stone MD, Becker KG, Martin B, Maudsley S. Minimal peroxide exposure of neuronal cells induces multifaceted adaptive responses. PLoS One. 2010;5:e14352. doi: 10.1371/journal.pone.0014352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chadwick W, Brenneman R, Martin B, Maudsley S. Complex and multidimensional lipid raft alterations in a murine model of Alzheimer's disease. Int J Alzheimers Dis. 2010:604792. doi: 10.4061/2010/604792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.