Abstract
Pathogenic Escherichia coli often carry determinants for several different adhesins. We show a direct communication between two adhesin gene clusters in uropathogenic E.coli: type 1 fimbriae (fim) and pyelonephritis-associated pili (pap). A regulator of pap, PapB, is a key factor in this cross-talk. FimB recombinase turns on type 1 fimbrial expression, and PapB inhibited phase transition by FimB in both off-to-on and on-to-off directions. On-to-off switching requiring FimE was increased by PapB. By analysis of FimB– and FimE–LacZ translational fusions it was concluded that the increase in on-to-off transition rates was via an increase in FimE expression. Inhibition of FimB-promoted switching was via a different mechanism: PapB inhibited FimB-promoted in vitro recombination, indicating that FimB activity was blocked at the fim switch. In vitro analyses showed that PapB bound to several DNA regions of the type 1 fimbrial operon, including the fim switch region. These data show that Pap expression turns off type 1 fimbriae expression in the same cell. Such cross-talk between adhesin gene clusters may bring about appropriate expression at the single cell level.
Keywords: adhesion/cross-talk/fimbriae/phase switch/regulation
Introduction
Fimbriae-mediated adherence is important for the virulence of Escherichia coli in the urinary tract (Svanborg et al., 1994). Uropathogenic E.coli often express several different adhesins that mediate specific adherence. Expression of the individual adhesin determinants is regulated in response to growth conditions, and most are subject to phase variation. Over 80% of uropathogenic E.coli express type 1 fimbriae, and E.coli isolates associated with acute pyelonephritis almost always express P pili. P fimbriae enhance the virulence of uropathogenic strains through specific adherence and increased induction of mucosal inflammation (Svanborg et al., 1994). Mutations in the pap gene cluster encoding P fimbriae reduce bacterial persistence in the mouse urinary tract (Hagberg et al., 1983). Mutational inactivation of the papG adhesin in a urinary tract pathogen dramatically decreases colonization and inflammation in the kidneys of monkeys (Roberts et al., 1994). Type 1 fimbriae may be important first in the establishment and then in the persistence of such infections (Schaeffer et al., 1987; Connell et al., 1996; Donnenberg, 1996; Mulvey et al., 1998). A recent study using a mouse model demonstrated that antibodies to the adhesion moiety of type 1 fimbriae (FimH) can successfully block colonization of the bladder by uropathogenic E.coli (Langermann et al., 1997). It was previously reported that the virulence of a P-fimbriated uropathogenic E.coli strain can be reduced by inactivation of a second fimbrial type (e.g. type 1 fimbriae) (Connell et al., 1996). However, how the expression of different types of fimbriae is co-ordinated remains largely unknown. Earlier studies have shown that E.coli strain KS71, carrying genes for either P, type 1C, or type 1 fimbriae, mainly express a single type of fimbriae on individual cells (Nowicki et al., 1984). Only 9% of the cells carry more than one fimbrial type and there is a rapid phase variation in fimbrial synthesis (Nowicki et al., 1984).
The expression of type 1 fimbriae is phase variable and depends on the orientation of an invertible 314 bp DNA switch (Abraham et al., 1985). This switch contains a promoter (Olsen and Klemm, 1994) for fimA, which encodes the main fimbrial subunit that is expressed only when the switch is in the ‘on’ but not the ‘off’ orientation (Abraham et al., 1985). The inversion process is carried out by the recombinases FimB (on-to-off and off-to-on) and FimE (on-to-off only) (Klemm, 1986; McClain et al., 1991, 1993; Gally et al., 1996). In addition to the fim recombinases, normal inversion of the fim switch in vivo requires the presence of integration host factor (IHF) (Dorman and Higgins, 1987; Eisenstein et al., 1987; Blomfield et al., 1997) and leucine-responsive regulatory protein (Lrp) (Blomfield et al., 1993; Gally et al., 1994). The histone-like protein H-NS is also required for normal switching rates and is considered to exert an indirect effect on switching by repressing the transcription of fimB and fimE (Kawula and Orndorff, 1991; Olsen and Klemm, 1994).
PapB is a transcriptional regulator of the pap operon, and stimulates pap expression at a low level but represses expression at high levels (Forsman et al., 1989). The PapB protein can recognize a DNA structure including a 9 bp repeat sequence containing T/A triplets at a conserved position and binds DNA in an oligomeric fashion perhaps through minor groove contact (Xia et al., 1998). This novel DNA binding mode could be important for PapB homologous proteins to function as transcriptional regulators in different fimbrial adhesin systems and for the potential cross-talk between them. Here we report our studies on the cross-regulation between pap and type 1 (fim) fimbriae operons. PapB can act to increase the on-to-off phase transition frequency of type 1 fimbriae by increasing FimE expression. Moreover, PapB blocks the activity of FimB at the fim switch and thus prevents the switching on of type 1 fimbrial expression. This cross-talk would effectively make expression of the two adhesin clusters mutually exclusive, except during transition from one type to the other.
Results
Effects of pap genes on type 1 fimbriae expression
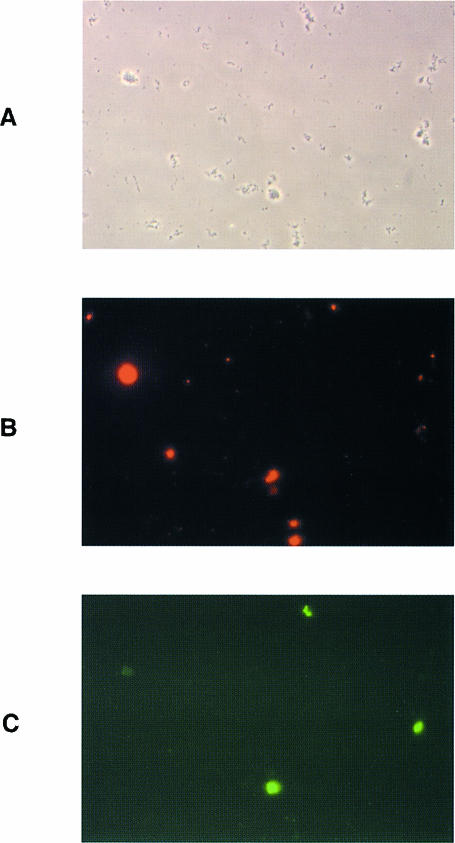
Our preliminary studies indicated that expression of the cloned pap genes in E.coli K12 affected type 1 fimbriae expression (K.Forsman, B.E.Uhlin, J.B.Leathart and D.Gally, unpublished data). The immunofluorescence microscopy test of the uropathogenic E.coli isolate J96 showed that type 1 and Pap fimbriae were not present on the same single cell (Figure 1). On the basis of such observations we therefore decided to test if any of the regulatory genes in the pap determinant (i.e. papB or papI) was responsible. The plasmid pSH2 (Orndorff and Falkow, 1984), which contains the whole type 1 fimbriae operon, was co-transformed into strain HB101 together with plasmids producing PapB (pHMG80) or PapI (pHMG95), or together with the vector control (pBR322). Using electron microscopy, we found that the percentage of piliated bacteria was reduced to only 2% in the presence of overproduced PapB, while ∼80% of the bacteria expressed type 1 fimbriae in the case of the vector control or the PapI-overproducing plasmid. The results of mannose-specific agglutination tests were consistent and confirmed that overproduced PapB essentially abolished the expression of type 1 fimbriae (Table I). To analyse this regulatory cross-talk, the plasmids producing PapB (pHMG88), PapI (pHMG98), the plasmid containing the pap operon (pHMG845), and the vector control (pACYC184) were introduced into two fimA–lacZYA transcriptional fusion strains, AAEC198A (fimB+E+) and AAEC374A (fimB–E–). AAEC374A has stop codons engineered into both fimB and fimE so that no functional recombinases are produced. In addition, the switch is locked in the phase-on orientation in this strain so that any changes in expression from the fimA–lacZYA reporter should reflect only transcription initiation levels at the fimA promoter (Gally et al., 1993). No changes were detected (Table II), indicating that PapB does not have a direct effect on RNA polymerase activity at the fimA promoter. In contrast, β–galactosidase levels in AAEC198A reflect both fimA promoter activity and the phase state of the population. Under many in vitro conditions this means only 1% of the bacteria are phase-on (Gally et al., 1993) due to the overriding on-to-off switching activity of FimE. However, even under these conditions the level of β–galactosidase activity was reduced to 40% in the presence of expressed papB. This result indicated that papB altered fim phase-switching frequencies.
Fig. 1. Immunofluorescence staining of E.coli J96 cells. Bacteria grown in LB at 37°C were prepared for microscopic analysis as described in Materials and methods. (A) Phase-contrast micrograph of bacterial cells. (B) Anti-Pap–lissamine–rhodamine staining. (C) Anti-type-1–fluorescein isothiocyanate staining.
Table I. Inhibition of type 1 fimbriae production by PapB.
| Strain | Plasmid | HA phenotypea |
HA titreb | EMc % piliated bacteria | |
|---|---|---|---|---|---|
| –mannose | +mannose | ||||
| HB101 | – | – | nd | 0 | |
| HB101 | pSH2 | +++ | – | 1/128 | 77 |
| HB101 | pSH2 + pBR322 | ++ | – | 1/128 | 78 |
| HB101 | pSH2 + pHMG80 | – | – | 1/16 | 2 |
| HB101 | pSH2 + pHMG95 | ++ | – | 1/128 | 82 |
aThe haemagglutination (HA) test was performed with human erythrocytes as described in Materials and methods.
bThe HA titre was determined as the highest dilution of the bacterial culture with which the haemagglutination with guinea pig erythrocytes could still be clearly observed (nd, not determined).
cThe EM test was performed as described in Materials and methods.
Table II. Effects of papB on fimA, fimB and fimE expression.
| Strains and fim–lac fusions | Relative β-galactosidase activity (units)a |
|||
|---|---|---|---|---|
| pACYC184 (vector) | pHMG98 (papI+) | pHMG88 (papB+) | pRHU845 (papA+-K+) | |
| AAEC198A (wt) (fimA–lacZ) | 1.0 (132.8) | 1.01 (134.1) | 0.40 (52.6) | 0.69 (91.8) |
| AAEC374A ‘switch locked on’ (fimA–lacZ) | 1.0 (1887.1) | 1.09 (2049.3) | 1.09 (2054.5) | 1.09 (2050.0) |
| BGEC056 (FimB–LacZ) | 1.0 (64.1) | 1.15 (73.9) | 0.71 (45.6) | 0.74 (47.7) |
| BGEC088 (FimE–LacZ) | 1.0 (38.0) | 1.21 (46.0) | 2.03 (77.0) | 1.59 (59.9) |
aThe β-galactosidase activity was measured as described by Miller (1972). All data represent the average values obtained from at least three separate experiments. The experiments were performed in the presence of IPTG (final concentration 1 mM) and the relative β-gal activity level obtained with co-resident vector control pACYC184 was set to 1.0 for each strain.
Effects of papB on FimB- and FimE-promoted phase switching
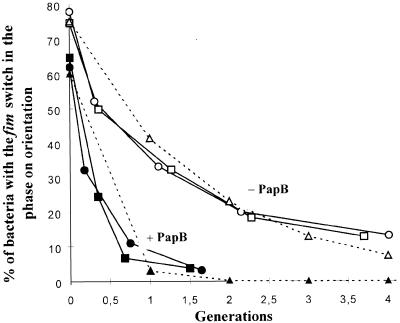
To determine if papB could change FimB- or FimE-promoted phase-switching frequencies, the plasmid producing PapB (pHMG88) and a vector control (pACYC184) were introduced into strains AAEC370A (fimB+fimE–) or AAEC198A (fimB+fimE+). The FimB- and FimE-promoted switching frequencies were measured and calculated as described in Materials and methods. For FimE switching, the frequency was increased >2-fold in the presence of papB, from 0.45 per cell per generation to >0.9 (Table III), giving rise to a much lower percentage of ‘switched on’ bacteria (Figure 2). On the other hand, the FimB-promoted switching (both on-to-off and off-to-on), when papB was induced, was locked. The figures in Table III are listed as <8 × 10–5, although the values are likely to be even lower as not a single phase transition was detected among thousands of colonies analysed by the method used to measure FimB switching frequencies (Materials and methods). These switching data show that the induction of PapB essentially inhibited FimB switching (>50–fold reduction). It prevented any phase switching to the on orientation and increased the phase switching to the off orientation mediated by FimE.
Table III. Effects of papB on the inversion of the fim switcha.
| Type of inversion and genotype | Frequency of inversionb in strain with: |
||
|---|---|---|---|
| No plasmid | pACYC184 (vector) | pHMG88 (papB+) | |
| Off-to-on, fimB+E– (AAEC370A) | 1.5 × 10–3 (± 0.3) | 3.3 × 10–3 (± 0.4) | <3.3 × 10–5 (± 1.6) |
| On-to-off, fimB+E– (AAEC370A) | 1.8 × 10–3 (± 0.8) | 2.1 × 10–3 (± 0.7) | <8 × 10–5 |
| On-to-off, fimB+E+ (AAEC198A) | 0.40c,d | 0.45c | >0.9c |
aStrains were grown in defined rich medium and switching frequencies (standard deviation within parentheses) were determined as described in Materials and methods.
bInversion frequency per cell per generation.
cDetermined by best fit to model curves (Gally et al., 1993).
dData referenced from Leathart and Gally (1998).
Fig. 2. Effect of papB on the on-to-off switching of strain AAEC198A (fimB+fimE+fimA–lacZYA) in defined rich medium at 37°C. The data from two separate experiments with the strain carrying the PapB-producing plasmid pHMG88 (closed symbols) or the vector plasmid pACYC184 (open symbols) are shown. Model rates of 0.9 (▴) and 0.45 (▵) per cell generation are shown by the dotted lines. The frequencies determined from these data are shown in Table III.
Effects of papB on fimB and fimE expression
To test if this effect on FimB- and FimE-promoted switching frequencies was due to changes in fimB and fimE expression levels, plasmids expressing PapB or PapI were introduced into strains AAEC200, AAEC261A, BGEC056 and BGEC088. The β–galactosidase activity from the chromosomal FimB–LacZ translational fusion was slightly reduced (to ∼70%) due to papB, while the β–gal activity from the FimE–LacZ translational fusion was increased almost 2–fold in the presence of papB as compared with the vector control (Table II). There was no effect seen in the case of papI. The results with transcriptional fusions were similar, but the effect of papB was weaker (data not shown). According to previous studies, FimB is the only recombinase that can switch from off-to-on and FimE functions dominantly in the on-to-off direction (Klemm, 1986; McClain et al., 1991, 1993; Gally et al., 1996). The increased level of fimE expression correlated well with the 2–fold increase in FimE-promoted switching activity (Figure 2). However, while the slight decrease in fimB expression may contribute to the silenced FimB activity, it is unlikely to be the primary mechanism.
In vitro recombination at fim
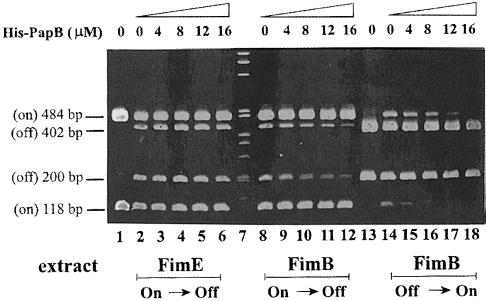
To determine if FimB-promoted recombination was being blocked by PapB directly at the level of the fim switch, in vitro switching experiments were carried out in the absence and presence of purified His-PapB protein (see Materials and methods). As shown in Figure 3, the FimB-promoted switching (both on-to-off and off-to-on) was clearly inhibited by His-PapB. However, addition of His-PapB did not alter FimE-promoted recombination at the fim switch. The lack of any effect on FimE switching acted as a good control for this experiment, which clearly showed that PapB could directly inhibit FimB- but not FimE-promoted phase switching. This result using the in vitro switching assay demonstrated that PapB inhibition of FimB activity was by inhibition of the recombination process at the fim switch and not primarily a result of the slight reduction in FimB expression. The converse was true for the PapB effect on FimE switching.
Fig. 3. Effect of purified His-PapB on the in vitro recombination in bacterial extracts containing either FimB or FimE. Recombination assays were set up as described in Materials and methods. Lane 1: control extract (pET11 in NEC26) plus pMM36 (switch starts in the on orientation). Lane 2: FimE extract (pIB382 in NEC26) plus pMM36. Lane 3: FimE extract with 4 μM His-PapB plus pMM36. Lane 4: FimE extract with 8 μM His-PapB plus pMM36. Lane 5: FimE extract with 12 μM His-PapB plus pMM36. Lane 6: FimE extract with 16 μM His-PapB plus pMM36. Lane 7: 1 kb marker. Lane 8: FimB extract (pIB378 in NEC26) plus pMM36. Lane 9: FimB extract with 4 μM His-PapB plus pMM36. Lane 10: FimB extract with 8 μM His-PapB plus pMM36. Lane 11: FimB extract with 12 μM His-PapB plus pMM36. Lane 12: FimB extract with 16 μM His-PapB plus pMM36. Lane 13: control extract (pET11 in NEC26) plus pJL2 (switch in the off orientation). Lane 14: FimB extract plus pJL2. Lane 15: FimB extract with 4 μM His-PapB plus pJL2. Lane 16: FimB extract with 8 μM His-PapB plus pJL2. Lane 17: FimB extract with 12 μM His-PapB plus pJL2. Lane 18: FimB extract with 16 μM His-PapB plus pJL2.
PapB binding to the fim switch DNA and the promoter regions of fimB and fimE
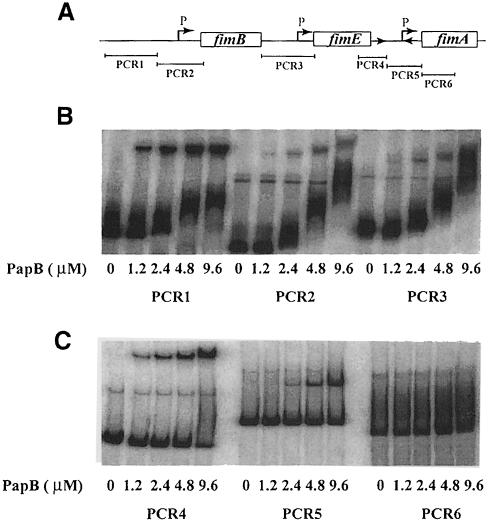
Our findings suggested that PapB could inhibit FimB activity at the fim switch and alter FimB and FimE expression by binding within the fim regulatory region. To test if PapB can bind to fim operon regions, radiolabelled DNA fragments that include sequences adjacent to and within the fim switch, and regions upstream of the fimB and fimE genes, were obtained by PCR amplification (Figure 4A; DNA fragments denoted PCR1–6). The gel retardation assays showed that the fragments PCR1 (upstream of fimB promoter region), PCR4 (upstream of the invertible element) and PCR5 (the invertible region) were shifted such that distinct bands representing more slowly migrating species appeared in the presence of increasing amounts of PapB (Figure 4B and C). For PCR2 (fimB promoter region) and PCR3 (fimE promoter region), the ladder pattern of DNA shift suggested that the binding affinity of PapB for these fragments was not as strong and specific as for the above mentioned fragments. There were gradual increases in shifted bands throughout the range of protein concentrations tested (1.2–9.6 μM). For comparison it could be mentioned that in such analyses with pap DNA target fragments, ∼4–6 μM PapB protein could form a distinct and rather defined complex (Xia and Uhlin, 1999). PCR6, encompassing the segment downstream of the invertible element and ∼360 bp into the fimA gene, was not retarded by PapB under those conditions (Figure 4B and C). Inspection of the nucleotide sequences of these regions revealed that there are potential PapB binding sites (T/A triplets repeated with 9 bp periodicity) located in the type 1 fimbriae operon, corresponding to fragments PCR1–5 in particular. These results suggest that binding of PapB in the promoter regions of fimB and fimE could produce the slight changes in expression measured (Table II) and that the inhibition of FimB but not FimE recombination at the fim switch may also be through binding of PapB in this region. The actual mechanism of FimB inhibition by PapB remains to be determined.
Fig. 4. PapB binding to the fim regulatory DNA region. (A) Outline of the fimB, fimE, fimA regions and positions of the promoters (P). The relative positions and lengths of different PCR fragments are also indicated. (B and C) Gel mobility shift assays of His-PapB binding to DNA fragments (PCR1–6). The assays were carried out as described in Materials and methods.
Discussion
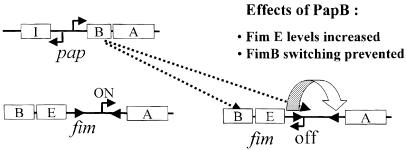
The present study provides the first evidence for cross-talk between two of the most ubiquitous and well characterized adhesin gene clusters in E.coli, type 1 fimbriae and P-specific pili. Our studies demonstrated that the regulatory protein PapB could inhibit type 1 fimbriae expression from the fim operon by affecting the fim phase-switching system as illustrated schematically in Figure 5. Our results are consistent with the previous observations that uropathogenic E.coli cells rarely carry more than one fimbrial type despite the fact that they contain multiple fimbrial gene clusters (Nowicki et al., 1984). The cross-talk between different fimbrial-adhesin gene systems is presumably important for pathogens to survive under changing environmental conditions. The bacteria can save energy by only expressing the fimbriae required at the time to interact with host cells. The potential complications with chaperones and ushers can be prevented and the immune recognition can be reduced. The single cell monofimbriate expression is also advantageous in case one fimbrial type would prevent proper functioning of the other. While we present a model for cross-talk from the pap to the fim gene cluster, we have no evidence for communication in the reverse direction.
Fig. 5. Schematic illustration of the cross-talk mediated by PapB between the pap and fim gene clusters.
We tested if PapB could influence the fim switch by determining FimB-promoted switching in fimB+fimEfimA–lacZYA strains. In the presence of papB, both the FimB-promoted on-to-off and off-to-on switches were almost completely blocked (Table III). However, the FimEpromoted on-to-off switching frequency was increased ∼2–fold by papB (Table III; Figure 2). One potential criticism of the findings is that papB was studied in multicopy affecting fim in single copy at its wild-type position in the chromosome. However, our preliminary data indicated that the levels of PapB produced by the plasmid constructs used were within 2-fold of those produced from single-copy wild-type cells, i.e. the uropathogenic E.coli isolate J96 (Y.Xia, unpublished data). Therefore, we consider that the wild-type situation was mimicked appropriately during the in vivo switching experiments. Results from the in vitro recombination assay gave further support for the role of PapB in the FimB-promoted switching: both on-to-off and off-to-on switching were clearly inhibited by the addition of purified His-PapB protein (Figure 3). The inhibition of FimB activity at the fim switch in the presence of PapB could follow three routes. First, PapB may bind to the switch in such a way as to inhibit FimB access but not FimE; there are slight differences in DNA occupancy by the two recombinases (Gally et al., 1996). Alternatively, PapB binding to the switch may interfere with an as yet unidentified cofactor or complex formation specific for FimB recombination. Thirdly, PapB may directly interact with FimB via protein–protein interaction to prevent FimB binding and/or activity.
Several potential PapB binding sites are found in the type 1 fimbriae operon, ranging from upstream of the promoter regions of the fimB and fimE genes to the invertible element. The gel mobility shift assays showed that PapB protein could bind to all these regions in vitro (Figure 4), but the binding was not strong enough to reveal any clearly protected region as identified by DNase I footprinting analyses (data not shown). There was a slight effect by papB on the expression of both fimB (down to ∼70%) and fimE (up 2–fold) (Table II). The effects could be due to the weak binding of PapB upstream of fimB and fimE, or indirectly via another regulator of their expression such as H-NS. The E.coli H-NS protein has been shown to bind to the promoter regions of fimB and fimE and downregulate the expression of these two genes in a temperature-dependent manner (Olsen and Klemm, 1994; Donato et al., 1997; Olsen et al., 1998).
In addition to the fim recombinases, normal inversion of the fim switch in vivo requires the presence of Lrp. Lrp binds with high affinity to sites 1 and 2 to increase both FimB and FimE recombination in vivo (Gally et al., 1994). Recently, it was reported that Lrp binding to site 3 inhibits recombination (Roesch and Blomfield, 1998). IHF is another protein required for the fim switch (Dorman and Higgins, 1987; Eisenstein et al., 1987). IHF binds both adjacent to (site 1) and within (site 2) the fim switch to stimulate FimB- and FimE-promoted switching (Blomfield et al., 1997). Since PapB can bind to the upstream and invertible element, it may compete with IHF and/or Lrp for DNA binding and disrupt the structure required for recombination. However, the mechanism is unclear. To clarify the mechanism, the variants in PapB homologues and how they interact with each other and systems such as type 1 fimbriae are currently being investigated. This regulation makes sense in the context of pathogenicity islands and gene transfer, as operon-specific regulators can immediately assert control over other housekeeping and virulence-associated genes without recourse to modification of the activity of global regulators. Potentially, this kind of regulator could be an important precedent for other clusters for which ‘specific’ regulators may have other functions at other sites.
Materials and methods
Plasmids, bacterial strains, media and growth conditions
Plasmids and bacterial strains are listed in Table IV. Media included L broth (LB), and L agar (LA), which consisted of L broth containing 1.5% agar. MOPS [3-(N–morpholino) propanesulfonic acid] minimal medium was used for liquid growth experiments in which switching frequencies were determined and for expression of FimB and FimE. MOPS media were supplemented with 10 μM thiamine and 0.4% glucose. For growth on agar media, 1.5% agar was added to MOPS media. Indicator media were minimal MOPS glucose plates supplemented with X-gal (40 μg/ml) and isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM). Liquid cultures were grown at the temperature indicated. Antibiotics used were carbenicillin (Cb; 100 μg/ml), chloramphenicol (Cm; 10 μg/ml) and tetracycline (Tc; 10 μg/ml).
Table IV. Strains and plasmids used in this study.
| Strain/plasmid | Genotype/phenotype | Reference/source |
|---|---|---|
| E.coli strain | ||
| HB101 | F– lacY1 recA13 | Boyer and Roulland-Dussoix (1969) |
| JM109 | F′ lacIq recA1 | Yanisch-Perron et al. (1985) |
| AAEC198A | MG1655 ΔlacZYA fimA–lacZYA (switch locked on) | Blomfield et al. (1991, 1993) |
| AAEC370A | MG1655 ΔlacZYA fimA–lacZYA fimE-am18 (switch starts on) | Blomfield et al. (1993) |
| AAEC374A | MG1655 ΔlacZYA fimA–lacZYA fimB-am6 fimE-am18 | Blomfield et al. (1993) |
| BGEC056 | MG1655 fimB–lacZYA FimB–LacZ | I.C.Blomfield (unpublished) |
| BGEC088 | MG1655 Δlac fimE–lacYA FimE–LacZ | I.C.Blomfield (unpublished) |
| NEC026 | BL21 (DE3) Δfim | Gally et al. (1996) |
| Plasmid | ||
| pQE30 | expression vector, Cbr | Qiagen manufacturer's instruction |
| pYN1 | wt papB gene cloned into pQE30, Cbr | Xia et al. (1998) |
| pSH2 | pACYC184 J96 fim B-H, Cmr | Orndorff and Falkow (1984) |
| pBR322 | cloning vector, Cbr Tcr | Bolivar et al. (1978) |
| pHMG80 | lacP(UV5) papB+/pBR322 | this laboratory (unpublished) |
| pHMG95 | lacP(UV5) papI+/pBR322 | this laboratory (unpublished) |
| pACYC184 | low-copy-number cloning vector, Tcr Cmr | Chang and Cohen (1978) |
| pHMG98 | lacIQ lacP(UV5) papI+ /pACYC184 | Forsman et al. (1989) |
| pHMG88 | lacIQ lacP(UV5) papB+ /pACYC184 | Forsman et al. (1989) |
| pRHU845 | pap gene cluster cloned in pACYC184 | Norgren et al. (1984) |
| pET11 | T7 promoter expression vector, Cbr | Linn and St. Pierre (1990) |
| pIB378 | pET11, fimB | Gally et al. (1996) |
| pIB382 | pET11, fimE | Gally et al. (1996) |
| pMM36 | fim cloned between transcriptional terminator rrnBT1T2 and T4 gene 32 terminator, Cmr (switch on) | McClain et al. (1991) |
| pJL2 | fim cloned between transcriptional terminator rrnBT1T2 and T4 gene 32 terminator, Cmr (switch off) | McClain et al. (1991) |
Agglutination tests
The adhesion phenotype of bacteria grown on LA plates at 37°C was tested by haemagglutination assays, in the presence or absence of mannose, using human erythrocytes (Hull et al., 1981). The haemagglutination titre with guinea pig erythrocytes was determined by dilution of bacterial cultures grown statically in LB at 37°C.
Electron microscopy
Bacterial suspensions in buffer (10 mM Tris–HCl pH 7.5, 10 mM magnesium chloride) were allowed to sediment on copper grids coated with thin films of 2% Formvar. After negative staining with 1% sodium silicotungstate pH 6.0, the grids were examined in a JEOL 1003 microscope.
Immunofluorescence microscopy
A bacterial overnight culture grown in LB at 37°C (static growth) was washed and diluted in phosphate-buffered saline (PBS). Bacterial suspension (100 μl; 105–106 cells/ml) was incubated with an equal volume of anti-Pap and anti-type-1 antisera at 37°C for 1 h. The reaction mixture was then washed three times with PBS containing 10% glycerol, and incubated in the same buffer with fluorescence-conjugated second antibody at 37°C for 1 h. The reaction mixture was then washed again three times with PBS containing 10% glycerol. Bacterial suspension (10 μl) was loaded on a glass slide and observed with an immunofluorescence microscope.
β-galactosidase assay
The specific activity of β-galactosidase was assayed as described (Miller, 1972). Bacteria harbouring the corresponding plasmids were grown in LB at 37°C; 1 mM IPTG was added at mid-logarithmic phase to induce the expression of the proteins. All data represent the mean values obtained from at least three separate experiments.
DNA techniques
Plasmid isolation, gel electrophoresis, transformation, amplification of DNA by PCR, and DNA labelling were performed by standard procedures (Maniatis et al., 1982). Restriction endonuclease digestions and DNA ligation reactions were performed under conditions recommended by the manufacturers (Boehringer Mannheim, New England Biolabs Inc.).
Expression and purification of His-PapB
The plasmid containing wt-papB gene (based on pQE30) was introduced into JM109, and protein expression was induced by addition of IPTG (final concentration 1 mM) during the logarithmic phase. The His-tagged PapB protein was purified as previously described (Xia et al., 1998).
Analysis of protein–DNA interactions
Gel mobility shift assays to detect protein–DNA interaction were performed as previously described (Forsman et al., 1989; Göransson et al., 1989). DNA fragments containing the different regions of the type 1 fimbriae operon were obtained by PCR amplification. Plasmid pSH2 (Orndorff and Falkow, 1984) was used as the template, while primers fim7 (5′–AACAAAACCAGATTTGCAAT–3′) and fim9 (5′–TAGTGGCTATTATCATGCTA–3′) were used to obtain the fragment containing the upstream region of the fimB gene (PCR1); primers fim8 (5′–TAGCATGATAATAGCCACTA–3′) and fim10 (5′–TTCTTCATC– GTTTTTCCCTT–3′) were used to obtain the fragment containing the promoter region of the fimB gene (PCR2); primers fim11 (5′–GAG– ATACCAGGGATGGTGTT–3′) and fim12 (5′–CTGCATCATGGC– CTGAACTT–3′) were used to obtain the fragment containing the promoter region of the fimE gene (PCR3); primers fim1 (5′–CTC– GGGCATCGAAATATTCG–3′) and fim4 (5′–GATGCTTTTTGTCGT– TTTTTAATATTTTTATGCTTGAGAAA–3′) were used to obtain the fragment containing the upstream region of the invertible element (PCR4); primers fim3 (5′–TTTCTCAAGCATAAAAATATTAAAAAA– CGACAAAAAGCATC–3′) and fim6 (5′–AATTTTCATGCTGCTTTC– CTTTCAAAAAACTATTTCTAAAT–3′) were used to obtain the fragment containing the invertible element (PCR5); and primers fim5 (5′–ATTTAGAAATAGTTTTTTGAAAGGAAAGCAGCATGAAAATT–3′) and fim2 (5′–CTGCAGAGCCAGAACGTTGG–3′) were used to obtain the fragment containing the downstream region of the invertible element which was used as a negative control (PCR6). The PCR products were end-labelled with [γ–32P]ATP and T4 polynucleotide kinase. The purified His-PapB protein was mixed with labelled DNA fragments (5000–10 000 c.p.m.) in the presence of 0.5 μg of poly(dI–dC) and 50 mM KCl in buffer B [25 mM HEPES pH 7.5, 0.1 mM EDTA, 5 mM dithiothreitol (DTT), 10% glycerol] in a final volume of 10 μl. The reaction mixtures were incubated at 25°C for 15 min and then immediately loaded onto 8% polyacrylamide-bis (37.5:1) for electrophoresis.
FimB and FimE switching frequencies in vivo
FimB- and FimE-promoted switching frequencies were calculated using fimA–lacZYA strains cultured on MOPS minimal medium agar plates as described previously (Gally et al., 1993). Tetracycline (10 μg/ml), X-gal (40 μg/ml) and IPTG (0.5 mM) were added as necessary. FimB switching was calculated by determining the proportion of bacteria (AAEC370A, fimB+fimE–fimA–lacZYA) that switched from one phase to the other during a measured number of generations. FimB rates of transition are usually low (1 × 10–3 per cell per generation), i.e. one cell per thousand switching per generation. As a consequence, individual bacteria produce colonies that reflect the phase state of the initial bacterium on media containing the indicator X-gal. Blue colonies reflect an initially phase-on bacterium and white colonies an initially phase-off bacterium. For example, to measure FimB off-to-on switching in the presence of PapB, AAEC370A containing either pHMG88 (PapB overproducing) or pACYC184 (vector control) were grown up on minimal MOPS Tet X-gal IPTG plates at 37°C. At least seven phase-off (white colonies) were selected, suspended and diluted in MOPS buffer to give 1000–2000 bacteria/ml and spread (100 μl) onto MOPS minimal Tet X-gal agar plates at 37°C. After incubation for 24 h the proportion of blue (phase-on) and white (phase-off) colonies was determined. This reflects the proportion of phase-on and -off bacteria in the original colonies. In addition, the number of generations through which the original colonies had been cultured was calculated from the total number of bacteria. As each colony analysed originated from a switch-off bacterium, the proportion of bacteria with the switch in the on orientation in the resulting colonies (after 20–23 generations) is a reflection of the switching rate. This frequency is calculated using probability theory and presented as per cell per generation (Gally et al., 1993). The mean switching frequencies were calculated from the values obtained from at least seven colonies for both the control and papB vectors in AAEC370A. This method was repeated for switching in the on-to-off direction, this time starting with blue (phase-on) colonies. The method is accurate for low switching frequencies (<1 × 10–2).
In order to measure the higher FimE-switching frequencies, a variation of this method was used. Previous work had demonstrated that growth of AAEC198A, fimB+fimE+fimA–lacZYA at a higher temperature (41°C) and in minimal medium reduces FimE switching frequencies so that the phase status of the colony (blue or white) again reflects the phase orientation of the initial organism (Gally et al., 1993). These defined growth conditions are also used to produce a culture with a high proportion of phase-on bacteria, which can then be transferred to appropriate media (Rich MOPS + IPTG and X-gal) at 37°C to measure the rate of on-to-off transition. The proportion of on or off bacteria present in the population at any time is determined by plating back onto MOPS minimal indicator medium at the higher temperature (41°C). That this proportion is an accurate reflection of the switch status in the population has been confirmed by PCR and restriction digestion of the fim switch. The actual rates are then estimated by best fit to model curves based on FimE switching probabilities and back-switching effects introduced by a low rate of FimB switching (Gally et al., 1993). The FimE-switching rates were determined as above in the presence and absence of induced papB.
In vitro recombination assay
The FimB and FimE proteins were expressed as described (Gally et al., 1996). Maximum solubilization of FimB was achieved at 28°C with a short (2 h) induction time, using 0.4 mM IPTG in minimal MOPS medium. Production of soluble FimE was increased by carrying out induction in minimal MOPS medium at lower temperatures (23–28°C) overnight, also using 0.4 mM IPTG. Extracts were prepared by sonication, the induced culture was centrifuged and washed with 100 mM NaCl, 10 mM Tris–HCl pH 7.6. The culture was resuspended in 1/40th volume of sonication buffer, which contained 10 mM NaCl, 50 mM Tris–HCl pH 7.6, 1 mM EDTA, 0.1 mM DTT. The bacteria were then lysed by sonication. The sonicate was centrifuged to remove insoluble cell debris, and supernatant fractions were stored at –20°C with 50% glycerol.
The in vitro recombination assay was carried out as described (Gally et al., 1996). The basic assay system (10 μl) was composed of 2 μl of 5× binding buffer (100 mM Tris–glycine pH 9.4, 500 mM NaCl, 5 mM EDTA, 20 mM spermidine and 0.005% sodium azide), 1 μl of supercoiled pMM36 or pJL2 (50 ng), 5–6 μl of 125 mM leucine and 1–2 μl of bacterial extract (FimB, FimE or control). Purified His-PapB (2–8 μl; 1000–4000 ng) was added in the reaction system to test the influence of PapB on FimB- and FimE-controlled phase switch. The supercoiled plasmid substrates were prepared using Qiagen midiprep columns. Reactions were incubated at 37°C for 1–4 h. After appropriate incubation periods, the reactions were stopped by heating the samples to 70°C for 10 min, and then placing them on ice. Samples (1 μl) were amplified by PCR using two primers situated on either side of the switch, resulting in a fragment of 602 bp, which was then cleaved with HinfI, a restriction enzyme that cuts within the switch. The switch in the on-orientation produces two products of 484 and 118 bp, while the off-orientation products are 200 and 402 bp. The digests were separated on a 6% acrylamide gel, stained with ethidium bromide.
Acknowledgments
Acknowledgements
This work was supported by grants from the Swedish Natural Science Research Council, the Swedish Medical Research Council, the Göran Gustafsson Foundation for Research in Natural Science and Medicine, the British Medical Research Council, and the British Biotechnology and Biological Sciences Research Council.
References
- Abraham J.M., Freitag, C.S., Clements, J.R. and Eisenstein, B.I. (1985) An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl Acad. Sci. USA, 82, 5724–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield I.C., McClain, M.S., Princ, J.A., Calie, P.J. and Eisenstein, B.I. (1991) Type 1 fimbriation and fimE mutants of Escherichia coli K–12. J. Bacteriol., 173, 5298–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield I.C., Calie, P.J., Eberhardt, K.J., McClain, M.S. and Eisenstein, B.I. (1993) Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol., 175, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield I.C., Kulasekara, D.H. and Eisenstein, B.I. (1997) Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol. Microbiol., 23, 705–717. [DOI] [PubMed] [Google Scholar]
- Bolivar F. (1978) Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene, 4, 121–136. [DOI] [PubMed] [Google Scholar]
- Boyer H.W. and Roulland-Dussoix, D. (1969) A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol., 41, 459–472. [DOI] [PubMed] [Google Scholar]
- Chang A.C. and Cohen, S.N. (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol., 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell I., Agace, W., Klemm, P., Schembri, M., Marild, S. and Svanborg, C. (1996) Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl Acad. Sci. USA, 93, 9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato G.M., Lelivelt, M.J. and Kawula, T.H. (1997) Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J. Bacteriol., 179, 6618–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg M.S. and Welch,R.A. (1996) Virulence determinants of uropathogenic Escherichia coli. In Mobley,L.T. and Warren,J.W. (eds), Urinary Tract Infections. American Society for Microbiology Press, Washington, DC, pp. 135–174. [Google Scholar]
- Dorman C.J. and Higgins, C.F. (1987) Fimbrial phase variation in Escherichia coli: dependence on integration host factor and homologies with other site-specific recombinases. J. Bacteriol., 169, 3840–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein B.I., Sweet, D.S., Vaughn, V. and Friedman, D.I. (1987) Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc. Natl Acad. Sci. USA, 84, 6506–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman K., Göransson, M. and Uhlin, B.E. (1989) Autoregulation and multiple DNA interactions by a transcriptional regulatory protein in E.coli pili biogenesis. EMBO J., 8, 1271–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally D.L., Bogan, J.A., Eisenstein, B.I. and Blomfield, I.C. (1993) Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol., 175, 6186–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally D.L., Rucker, T.J. and Blomfield, I.C. (1994) The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J. Bacteriol., 176, 5665–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally D.L., Leathart, J. and Blomfield, I.C. (1996) Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol. Microbiol., 21, 725–738. [DOI] [PubMed] [Google Scholar]
- Göransson M., Forsman, K., Nilsson, P. and Uhlin, B.E. (1989) Upstream activating sequences that are shared by two divergently transcribed operons mediate cAMP-CRP regulation of pilus-adhesin in Escherichia coli. Mol. Microbiol., 3, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hagberg L., Hull, R., Hull, S., Falkow, S., Freter, R. and Svanborg Eden, C. (1983) Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect. Immun., 40, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R.A., Gill, R.E., Hsu, P., Minshew, B.H. and Falkow, S. (1981) Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun., 33, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawula T.H. and Orndorff, P.E. (1991) Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histonelike protein H-NS. J. Bacteriol., 173, 4116–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. (1986) Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J., 5, 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langermann S. et al. (1997)Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science, 276, 607–611. [DOI] [PubMed] [Google Scholar]
- Leathart J.B. and Gally, D.L. (1998) Regulation of type 1 fimbrial expression in uropathogenic Escherichia coli: heterogeneity of expression through sequence changes in the fim switch region. Mol. Microbiol., 28, 371–381. [DOI] [PubMed] [Google Scholar]
- Linn T. and St. Pierre, R. (1990) Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol., 172, 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- McClain M.S., Blomfield, I.C. and Eisenstein, B.I. (1991) Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol., 173, 5308–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain M.S., Blomfield, I.C., Eberhardt, K.J. and Eisenstein, B.I. (1993) Inversion-independent phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol., 175, 4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Mulvey M.A., Lopez-Boado, Y.S., Wilson, C.L., Roth, R., Parks, W.C., Heuser, J. and Hultgren, S.J. (1998) Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science, 282, 1494–1497. [DOI] [PubMed] [Google Scholar]
- Norgren M., Normark, S., Lark, D., O'Hanley, P., Schoolnik, G., Falkow, S., Svanborg-Eden, C., Båga, M. and Uhlin, B.E. (1984) Mutations in E.coli cistrons affecting adhesion to human cells do not abolish Pap pili fiber formation. EMBO J., 3, 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki B., Rhen, M., Vaisanen-Rhen, V., Pere, A. and Korhonen, T.K. (1984) Immunofluorescence study of fimbrial phase variation in Escherichia coli KS71. J. Bacteriol., 160, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P.B. and Klemm, P. (1994) Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett., 116, 95–100. [DOI] [PubMed] [Google Scholar]
- Olsen P.B., Schembri, M.A., Gally, D.L. and Klemm, P. (1998) Differential temperature modulation by H-NS of the fimB and fimE recombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol. Lett., 162, 17–23. [DOI] [PubMed] [Google Scholar]
- Orndorff P.E. and Falkow, S. (1984) Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J. Bacteriol., 159, 736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.A. et al. (1994)The Gal(α1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl Acad. Sci. USA, 91, 11889–11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch P.L. and Blomfield, I.C. (1998) Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol. Microbiol., 27, 751–761. [DOI] [PubMed] [Google Scholar]
- Schaeffer A.J., Schwan, W.R., Hultgren, S.J. and Duncan, J.L. (1987) Relationship of type 1 pilus expression in Escherichia coli to ascending urinary tract infections in mice. Infect. Immun., 55, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svanborg C., Orskov,F. and Orskov,I. (1994) Fimbriae and disease. In Klemm,P. (ed.), Bacterial Fimbriae. CRC Press, Inc., Boca Raton, FL, pp. 253–266. [Google Scholar]
- Xia Y. and Uhlin, B.E. (1999) Mutational analysis of the PapB transcriptional regulator in Escherichia coli: Regions important for DNA binding and oligomerization. J. Biol. Chem., 274, 19723–19730. [DOI] [PubMed] [Google Scholar]
- Xia Y., Forsman, K., Jass, J. and Uhlin, B.E. (1998) Oligomeric interaction of the PapB transcriptional regulator with the upstream activating region of pili adhesin gene promoters in Escherichia coli. Mol. Microbiol., 30, 513–523. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira, J. and Messing, J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33, 103–119. [DOI] [PubMed] [Google Scholar]