Abstract
Tourette disorder (TD) is a childhood onset neuropsychiatric syndrome defined by persistent motor and vocal tics. Despite a long-standing consensus for a strong genetic contribution, the pace of discovery compared to other disorders of similar prevalence has been slow, due in part to a paucity of studies and both clinical heterogeneity and a complex genetic architecture. However, the potential for rapid progress is high. Recent rare variant findings have pointed to the importance of copy number variation, the overlap of risks among distinct diagnostic entities, the contribution of novel molecular mechanisms, and the value of family based studies. Finally, analysis of a cohort of sufficient size to identify common polymorphisms of plausible effect is underway, promising key information regarding the contribution of common alleles to TD.
Introduction
Tourette disorder is defined by the combination of persistent motor and phonic tics. These are unwanted, rapid, repetitive and stereotyped movements or vocalizations. The natural history of the disorder includes onset in childhood, a waxing and waning course, and, for many individuals, symptom reduction in adulthood. Current diagnostic approaches dictate that only individuals with the combination of unwanted movements and vocalizations meet criteria for TD. However tics in only one of these domains often occur and, if persistent, are categorized as either chronic motor tics (CMT) or chronic vocal tics (CVT). These are thought to represent a TD spectrum of disorders that also includes the co-occurrence of tics ands obsessive-compulsive disorder (OCD).
TD was once thought to be rare; but estimates now converge on a world -wide prevalence of 0.3–1%, though study samples have tended to be small and many investigations have not met the highest standards for contemporary large scale epidemiological studies[1–3]. In addition, despite the observation that as many as 1 percent of the population meets diagnostic criteria for TD, a minority of affected individuals present to clinic with tics as a primary complaint. Moreover, it is typically the coincidence of chronic tics with other psychiatric syndromes, including OCD, depression, and attention deficit hyperactivity disorder that leads individuals and families to seek medical attention. Estimates of comorbidity among TD and these disorders are, accordingly, quite high.
The molecular, cellular and anatomical bases of tics and TD remain in question. However there has been a long-standing consensus regarding the contribution of genetic factors [4,5]. From the earliest descriptions of the syndrome, a high degree of heritability has been noted. Indeed, TD was initially thought to represent a single gene, Mendelian disorder [6]. Presently there is a consensus that overall, TD has a far more complex allelic architecture, one that appears to be similar to other common neuropsychiatric syndromes, involving both a high degree of locus and allelic heterogeneity and polygenic inheritance.
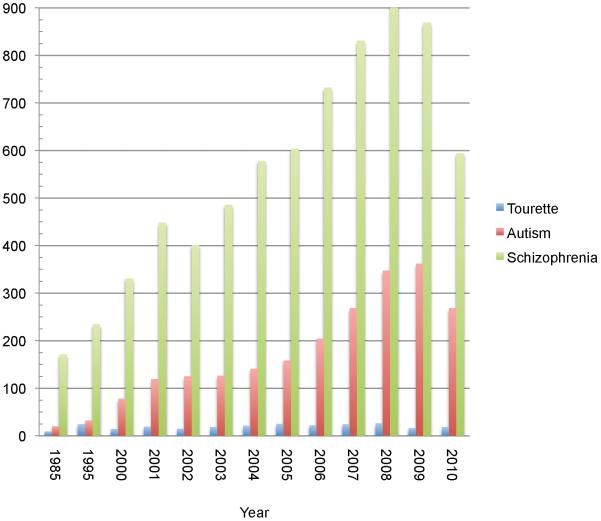
The tentative quality of this description is a consequence of the fact that after a highly productive early era characterizing the heritability and familiality of TD, progress in genetics and genomics over the past decade has been halting. The field is just now beginning to analyze data from what would be considered sufficiently large samples to power reliable studies of common variation and there is, to date, only a single published report of genome wide detection of rare copy number variation (CNV), conducted in a sample of 111 probands and 73 controls [7]. Indeed when one queries Pubmed for Tourette genetics, fewer than 25 primary research papers are found annually for the last decade. When compared to other complex multi-genic neuropsychiatric syndromes such as autism or schizophrenia, both the differences in the volume of data currently available as well as the rate of growth of the field, based on this crude metric, is striking (Figure 1). Not surprisingly then, answers to key questions that have begun to be addressed in other areas of psychiatric genetics, regarding the overall contribution of common versus rare variation; the importance of de novo versus transmitted alleles, and the identity of definitive risk genes are all on the horizon.
Figure 1. Annual publication number in the genetics of Tourette syndrome, autism and schizophrenia.
A Pubmed search was conducted using the terms “Tourette genetics”; “autism genetics” or “schizophrenia genetics.” The searches were performed for the dates January 1-Dec 30th of the year noted in the graph. Articles annotated as reviews in pubmed were subtracted before tallying the total number of publications per year.
Studies to date have comprehensively and rigorously explored and rejected the hypothesis of single gene inheritance and cumulatively point to limits on the effect sizes of contributing common alleles. Moreover over the last several years rare variant finding have pointed to novel hypotheses regarding pathogenic mechanisms and possibly new avenues for treatment, and recent data suggests the TD may follow a pattern emerging in the study of other neuropsychiatric disorders in which specific sequence or structural variations increase risk for range of outcomes that previously would have been considered distinct.
A brief history of TD genetics
Based on the largest twin study reported to date (N= 43 twin pairs) concordance rates are 50%–77% for monozygotic twins compared to 10%–23% for dizygotic twins [8], with the range dependent on whether TD alone or spectrum conditions, such as CMT or CVT are considered. Similar to other developmental neuropsychiatric syndromes, there is a strong male predominance (approximately 4:1) [2,9] However, family studies demonstrate that within TD pedigrees, if one includes OCD as affected status, the risk to male and female relatives of a TD proband approaches 1:1, with females relatives more likely to show obsessions and compulsions [10].
Early gene discovery efforts focused on large multigenerational pedigrees that were hypothesized to reflect single gene autosomal dominant inheritance [6,11–13]. However, over time, as the techniques for mapping Mendelian disorders reached maturity and no TD locus was identified, this hypothesis was abandoned. Subsequent segregation analyses [14–16] led to the conceptualization of TD as a genetically complex multigenic disorder.
Importantly, it has become clear that TD pedigrees often demonstrate bilineal inheritance [15,17,18]. Depending on ascertainment approaches, between 25–40 percent of probands have a history of either TD or OCD on both the maternal and paternal lines. This finding provides some explanation for the thwarted early efforts at parametric linkage in what appeared to be highly promising dominant pedigrees and point to the continued importance of comprehensive phenotypic assessment in pedigrees showing putative Mendelian inheritance.
By the late 1990s, the lack of results from parametric linkage analyses resulted in a shift toward nonparametric approaches[19,20]. While these efforts are theoretically capable of identifying the contribution of either common or rare variation, they are not robust to the combination of small sample size and marked locus heterogeneity. As a limited number of TD sibling pairs have so far been studied, in practice these analyses have investigated the contribution of common variation of moderate to large effect. Based on cumulative recent experience in other complex disorders, it is not surprising that such loci have proven difficult to characterize, and, in general, these alleles would seem increasingly unlikely to constitute a significant proportion of the spectrum of variation contributing to TD.
Similar to other psychiatric syndromes, the lions' share of studies in TD genetics has focused on the contribution of common variants. Since 2000, 37 such studies are referenced in Pubmed, compared to 11 cytogenetic mapping and 10 parametric linkage papers. The majority has assessed association of one or a small number of candidate gene single nucleotide polymorphisms (SNPs) using either a case control or transmission disequilibrium test. Based on the results of genome wide association studies for a wide range of common, complex disorders, even the most recent TD studies have reported on sample sizes that would be considered unlikely to support the detection of common variant risks of plausible magnitude[21–27].
Rare variants and TD
While the search for common alleles has predominated, there has nonetheless been a steady parallel effort to evaluate the contribution of rare variants These have included cytogenetics, parametric linkage in individual pedigrees or isolated populations, targeted sequencing and analysis of copy number variation. As noted, the earliest of these studies were conducted in the context of a widely held belief that TD was a Mendelian disorder. However, since the late 1990s, these have largely shifted to an “outlier approach” to gene discovery, based on the notion that while mutations of large effect may represent only a fraction of the allelic spectrum underlying TD, they may nonetheless provide a valuable point of traction with regard to the molecular pathophysiology of the disorder (for review see [28])
Cytogenetics
Mapping of approximately a dozen TD probands or families with chromosomal abnormalities have been reported over the past decade. These include several cases of TD coincident with known genetic disorders including Smith Magenis [29] and 22q11 deletion syndrome [30]. One individual with 22q11 duplication was also ecently described [31], though the question remains whether this CNV carries any risk for neurodevelopmental phenotypes. Several mapping studies have pointed to novel candidate genes or regions[32–36]. As a follow-up to these findings, mutation screening or sequencing studies have been reported for IMMP2L [37] and several transcripts within a candidate region of chromosome 18q[34]. However, no pathogenic mutations have been identified among the small number of patients that have been screened.
To date, there has been only a single instance in which cytogenetic mapping has led to the identification of deleterious rare mutations in a nearby gene. In 2005, our laboratory [38] reported a de novo chromosome 13 inversion in a sporadic TD pedigree. Sequencing of SLIT and TRK like family member 1 (SLITRK1), the gene mapping nearest to one of two breakpoints, revealed a single base frameshift deletion as well as two independent occurrences of a very rare mutation (var321) in a highly conserved base within the SLITRK1 3’UTR, corresponding to the binding site for the microRNA hsa-miR-189. This variant was found to be associated with TD based on a comparison with 4296 control chromosomes.
The biology of SLITRK1 has subsequently been pursued on several fronts. Following previous evidence that the protein is involved in the regulation of neurite outgrowth [39] our initial study demonstrated that over-expression of SLITRK1 in cortical neurons promotes dendritic growth, while the deletion/frameshift mutation does not. SLITRK1 expression was confirmed in cortical striatal circuits, regions long implicated in TD pathology [40]. The regulation of neurite outgrowth was later determined to be mediated by binding to 14-3-3 molecules [41]; and the mouse knockout was found to have an anxiety phenotype and evidence for increased noradrenergic neurotransmission[42], recapitulating results from early human CSF studies in TD [43]. A particularly intriguing result has been the very recent finding of a striking obsessive-compulsive phenotype and impaired striatal dendritic morphology resulting from the mouse knockout of the closely related molecule, SLITRK5 [44].
However, on balance, the contribution of SLITRK1 to TD remains in question. Resequencing efforts, though of modest scale, have not revealed additional pathogenic coding mutations [45–47] and subsequent association studies have been inconclusive. Two publications specifically evaluated var321 [48,49] using tests of transmission within families. Given the low allele frequency (<.001 in the Caucasian population) neither cohort approached the sample size necessary to conduct a meaningful statistical analysis. However both suggested that stratification, particularly among the Ashkenazi population, may have accounted for the initial finding.
Our laboratory recently tested this hypothesis [50] using a combination of genome-wide genotyping, a multi-dimensional scaling analysis and dense haplotype mapping and found no evidence to support this contention. Instead the data further confirmed the initial observation that the initial var321 mutations were not present on a common haplotype, indicating they were either independent events or reflected an ancient allele shared by unrelated affected individuals, with either alternative providing support for the association with TD.
Finally, two cytogenetic findings have been notable for their overlap with the other psychiatric and developmental syndromes: An insertion of chromosome 2p21-p23 at 7q35-q36 was found to disrupt the gene coding for Contactin Associated Protein 2 (CNTNAP2) in three affected individuals from a single family [51] and a transmitted deletion in the gene Neuroligin 4X [52] was found in a proband with autism and motor tics; both his sibling with TD and ADHD and his mother, suffering from a learning disorder, anxiety, and depression, also carried the deletion. These two genes have been strongly implicated in intellectual disability and autism spectrum disorders [53–60] and emerging data points to a possible association with schizophrenia[61–63] (Table 1). Large-scale sequencing of TD probands has not yet been reported for either transcript.
Table 1.
Overlap of variants/genes identified in Tourette syndrome, autism, schizophrenia and attention deficit hyperactivity disorder (ADHD)
| Tourette syndrome | Autism* | Schizophrenia* | ADHD | |
|---|---|---|---|---|
| Neuroligin 4 | Exonic deletion in a single affected family [52] | Molecular Cytogenetics; sequencing, parametric linkage [53–54] | ||
| Contactin Associated Protein 2 | Complex chromosomal rearrangement in a single pedigree [51] | Molecular cytogenetics (de novo inversion), sequencing, homozygosity mapping, common variant association [55–59] | CNV, common variant association [60–63] | Copy number variation (one occurrence, intronic) [86] |
| Neurexin 1 | CNV (2 occurrences) [7] | CNV study, molecular cytogenetics, homozygosity mapping [79–83] | CNV, sequencing [75–78] | |
| 1q21 deletion | CNV (1 occurrence) [7] | CNV [87] | CNV[88–91] | |
| IMMPL2 | Molecular Cytogenetic mapping of a de novo duplication [32] | Common variant analysis [84] and CNV [85] | CNV (1 occurrence) [86] |
references with regard to autism and schizophrenia provide illustrative data and are not intended to be comprehensive; Not all observation reported in the various disorders reflect confirmed associations
Copy Number Variation
The theme of overlapping risks among diagnostically distinct syndromes has been further supported by a recent genome wide CNV study [7]. The authors addressed potential confounds that have often been overlooked in studies of structural variation, including population stratification and batch effects. While they did not find an overall increase in any category of CNV among cases versus controls, they did find an interesting overlap with CNVs previously implicated in autism spectrum disorders and schizophrenia, including involving NRNX1 and 1q21 (Table 1). As the authors note, these findings are preliminary and, given the small sample size (111 cases and 73 controls), they were not able to firmly establish the overall contribution of rare structural variation, determine the role of de novo CNVs, or demonstrate a clear association of particular variants with TD.
Parametric linkage
After the initial failure to map a single gene mutation in very large pedigrees, parametric linkage efforts turned to studying smaller families based on the notion that the identification of any gene carrying a variation of major effect might help illuminate the biology of TD. Several of these studies have reached or approached genome wide significance [64–69]. To date they have not led to the identification of a likely deleterious sequence variation within the linkage interval(s), with one exception: In this case, parametric linkage of a family consisting of a father and 8 offspring and no evidence of bilineal inheritance was reported by our group. Traditional mapping efforts revealed a single region of the genome reaching the maximum theoretical LOD score (Lod =2.1). Sequencing of all known genes in the interval led to the finding of one nonsense mutation, in the gene L-histidine Decarboxylase (HDC), the rate-limiting enzyme in histamine biosynthesis [70].
The result points to an interesting mechanistic link to prior hypotheses regarding the involvement of dopaminergic pathways in TD: histaminergic (HA) neurotransmission is mediated by three of four known G-protein coupled histamine receptors (H1-H4). Both histamine 2 (H2R) and histamine 3 (H3R) receptors are significantly enriched in the human and rodent striatum [71]. H3R is of particular interest as it acts: 1) as a presynaptic auto-receptor on HA projection neurons; 2) as a pre-synaptic receptor on non-HA containing neurons regulating a variety of neurotransmitters, including dopamine and serotonin; and 3) as a post-synaptic receptor that co-localizes with and modulates dopamine signaling through both D1 and D2 receptors in the striatum. Finally, HDC null mice show decreased brain HA and increased sensitivity to stereotypic behaviors upon administration of DA agonists [72], These repetitive behaviors have previously been proposed as a model of human tics [73].
The convergence of the human genetic and model systems data and the potential availability of clinically-useful H3R compounds being studied in related psychiatric conditions [74] suggests several promising avenues to further evaluate the generalizability of the biology implicated by gene discovery in this single outlier pedigree.
Conclusions
Tourette disorder is a tremendously interesting and surprisingly common syndrome for which there is long standing evidence of a genetic contribution. The limited number of published reports and, in retrospect, the small scale of study cohorts likely accounts for the relatively slow rate of progress compared to other neuropsychiatric disorders. However, despite these obstacles, recent investigations have pointed in promising and unexpected directions, including suggesting the contribution of impaired dendritic growth in the striatum, an overlap of genetic risks with a range of other developmental disorders, and a possible role for histamine in the genesis, exacerbation and, potentially, treatment of some cases of TD.
In fact, TD may be particularly amenable to a broad range of gene discovery efforts. The developmental time course, which typically improves by early adulthood, would at least suggest that selection against TD alleles might be moderated and, consequently, that common alleles of modest effect may account for a proportion of genetic risk. Given the maturity of genome wide common variant methods, this question will soon be answered by the ongoing a large cohort GWAS currently underway by the Tourette Syndrome International Consortium on Genetics. Alternatively, despite the general skepticism regarding parametric linkage among psychiatric geneticists through much of the late 90s and early 2000s, it is striking that multiple promising multigenerational pedigrees have been identified with TD and mapped during this time. The ability to identify multiplex families and the increasing availability of next generation sequencing technologies promises to further invigorate these family based gene discovery efforts. Finally, the results from copy number variation studies in other areas of medicine and the earliest data from the TD field suggest that this is likely to be productive avenue of inquiry and may reveal surprising biological links between TD and what are now conceptualized as distinct neuropsychiatric conditions.
Acknowledgements
Dr. State is supported in part by a grant from the National Institutes of Neurological Disorder and Stroke, NS056276
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robertson MM, Eapen V, Cavanna AE. The international prevalence, epidemiology, and clinical phenomenology of Tourette syndrome: a cross-cultural perspective. J Psychosom Res. 2009;67:475–483. doi: 10.1016/j.jpsychores.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 2*.Robertson MM. The prevalence and epidemiology of Gilles de la Tourette syndrome. Part 1: the epidemiological and prevalence studies. J Psychosom Res. 2008;65:461–472. doi: 10.1016/j.jpsychores.2008.03.006. [DOI] [PubMed] [Google Scholar]; This is a very useful summary of the extant epidemiological data on Tourette from around the world.
- 3.Robertson MM. The prevalence and epidemiology of Gilles de la Tourette syndrome. Part 2: tentative explanations for differing prevalence figures in GTD, including the possible effects of psychopathology, aetiology, cultural differences, and differing phenotypes. J Psychosom Res. 2008;65:473–486. doi: 10.1016/j.jpsychores.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Pauls DL, Cohen DJ, Heimbuch R, Detlor J, Kidd KK. Familial pattern and transmission of Gilles de la Tourette syndrome and multiple tics. Arch Gen Psychiatry. 1981;38:1091–1093. doi: 10.1001/archpsyc.1981.01780350025002. [DOI] [PubMed] [Google Scholar]
- 5.Kidd KK, Prusoff BA, Cohen DJ. Familial pattern of Gilles de la Tourette syndrome. Arch Gen Psychiatry. 1980;37:1336–1339. doi: 10.1001/archpsyc.1980.01780250022001. [DOI] [PubMed] [Google Scholar]
- 6.Pauls DL, Leckman JF. The inheritance of Gilles de la Tourette's syndrome and associated behaviors. Evidence for autosomal dominant transmission. N Engl J Med. 1986;315:993–997. doi: 10.1056/NEJM198610163151604. [DOI] [PubMed] [Google Scholar]
- 7**.Sundaram SK, Huq AM, Wilson BJ, Chugani HT. Tourette syndrome is associated with recurrent exonic copy number variants. Neurology. 2010;74:1583–1590. doi: 10.1212/WNL.0b013e3181e0f147. [DOI] [PMC free article] [PubMed] [Google Scholar]; This carefully controlled study was the first to report on the contribution of copy number variation in Tourette syndrome and suggested an overlap of risks with other developmental neuropsychiatric disorders.
- 8.Price RA, Kidd KK, Cohen DJ, Pauls DL, Leckman JF. A twin study of Tourette syndrome. Arch Gen Psychiatry. 1985;42:815–820. doi: 10.1001/archpsyc.1985.01790310077011. [DOI] [PubMed] [Google Scholar]
- 9.Freeman RD, Fast DK, Burd L, Kerbeshian J, Robertson MM, Sandor P. An international perspective on Tourette syndrome: selected findings from 3,500 individuals in 22 countries. Dev Med Child Neurol. 2000;42:436–447. doi: 10.1017/s0012162200000839. [DOI] [PubMed] [Google Scholar]
- 10.Pauls DL, Towbin KE, Leckman JF, Zahner GE, Cohen DJ. Gilles de la Tourette's syndrome and obsessive-compulsive disorder. Evidence supporting a genetic relationship. Arch Gen Psychiatry. 1986;43:1180–1182. doi: 10.1001/archpsyc.1986.01800120066013. [DOI] [PubMed] [Google Scholar]
- 11.Baron M, Shapiro E, Shapiro A, Rainer JD. Genetic analysis of Tourette syndrome suggesting major gene effect. Am J Hum Genet. 1981;33:767–775. [PMC free article] [PubMed] [Google Scholar]
- 12.Jagger J, Prusoff BA, Cohen DJ, Kidd KK, Carbonari CM, John K. The epidemiology of Tourette's syndrome: a pilot study. Schizophr Bull. 1982;8:267–278. doi: 10.1093/schbul/8.2.267. [DOI] [PubMed] [Google Scholar]
- 13.Curtis D, Robertson MM, Gurling HM. Autosomal dominant gene transmission in a large kindred with Gilles de la Tourette syndrome. Br J Psychiatry. 1992;160:845–849. doi: 10.1192/bjp.160.6.845. [DOI] [PubMed] [Google Scholar]
- 14.Walkup JT, LaBuda MC, Singer HS, Brown J, Riddle MA, Hurko O. Family study and segregation analysis of Tourette syndrome: evidence for a mixed model of inheritance. Am J Hum Genet. 1996;59:684–693. [PMC free article] [PubMed] [Google Scholar]
- 15.Kurlan R, Eapen V, Stern J, McDermott MP, Robertson MM. Bilineal transmission in Tourette's syndrome families. Neurology. 1994;44:2336–2342. doi: 10.1212/wnl.44.12.2336. [DOI] [PubMed] [Google Scholar]
- 16.Hasstedt SJ, Leppert M, Filloux F, van de Wetering BJ, McMahon WM. Intermediate inheritance of Tourette syndrome, assuming assortative mating. Am J Hum Genet. 1995;57:682–689. [PMC free article] [PubMed] [Google Scholar]
- 17.Hanna PA, Janjua FN, Contant CF, Jankovic J. Bilineal transmission in Tourette syndrome. Neurology. 1999;53:813–818. doi: 10.1212/wnl.53.4.813. [DOI] [PubMed] [Google Scholar]
- 18.McMahon WM, van de Wetering BJ, Filloux F, Betit K, Coon H, Leppert M. Bilineal transmission and phenotypic variation of Tourette's disorder in a large pedigree. J Am Acad Child Adolesc Psychiatry. 1996;35:672–680. [PubMed] [Google Scholar]
- 19.Genome scan for Tourette disorder in affected-sibling-pair and multigenerational families. Am J Hum Genet. 2007;80:265–272. doi: 10.1086/511052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A complete genome screen in sib pairs affected by Gilles de la Tourette syndrome. The Tourette Syndrome Association International Consortium for Genetics. Am J Hum Genet. 1999;65:1428–1436. doi: 10.1086/302613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Crane J, Fagerness J, Osiecki L, Gunnell B, Stewart SE, Pauls DL, Scharf JM. Family-based genetic association study of DLGAP3 in Tourette Syndrome. Am J Med Genet B Neuropsychiatr Genet. 2010 doi: 10.1002/ajmg.b.31134. [DOI] [PMC free article] [PubMed] [Google Scholar]; The gene under investigation, DLGAP3 (aka SAPA3) It is one of only a handful for which the mouse knockout demonstrates an excessive grooming phenotype; one that is responsive to treatment with a selective serotonin reuptake inhibitor. The other two, HoxB8 and SLITRK5, are equally intriguing.
- 22.Herzberg I, Valencia-Duarte AV, Kay VA, White DJ, Muller H, Rivas IC, Mesa SC, Cuartas M, Garcia J, Bedoya G, et al. Association of DRD2 variants and Gilles de la Tourette syndrome in a family-based sample from a South American population isolate. Psychiatr Genet. 2010;20:179–183. doi: 10.1097/YPG.0b013e32833a215a. [DOI] [PubMed] [Google Scholar]
- 23.Chou IC, Lin HC, Wang CH, Lin WD, Lee CC, Tsai CH, Tsai FJ. Polymorphisms of interleukin 1 gene IL1RN are associated with Tourette syndrome. Pediatr Neurol. 2010;42:320–324. doi: 10.1016/j.pediatrneurol.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Miranda DM, Wigg K, Kabia EM, Feng Y, Sandor P, Barr CL. Association of SLITRK1 to Gilles de la Tourette Syndrome. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:483–486. doi: 10.1002/ajmg.b.30840. [DOI] [PubMed] [Google Scholar]
- 25.Kindler J, Schosser A, Stamenkovic M, Schloegelhofer M, Leisch F, Hornik K, Aschauer H, Gasche C. Tourette's syndrome is not associated with interleukin-10 receptor 1 variants on chromosome 11q23.3. Psychiatry Res. 2008;157:235–239. doi: 10.1016/j.psychres.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 26.Miranda DM, Wigg K, Feng Y, Sandor P, Barr CL. Association study between Gilles de la Tourette Syndrome and two genes in the Robo-Slit pathway located in the chromosome 11q24 linked/associated region. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:68–72. doi: 10.1002/ajmg.b.30580. [DOI] [PubMed] [Google Scholar]
- 27.Riviere JB, St-Onge J, Gaspar C, Diab S, Dion Y, Lesperance P, Tellier G, Richer F, Chouinard S, Dube MP, et al. Genome-wide TDT analysis in French-Canadian families with Tourette syndrome. Can J Neurol Sci. 2010;37:110–112. doi: 10.1017/s0317167100009744. [DOI] [PubMed] [Google Scholar]
- 28.State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shelley BP, Robertson MM, Turk J. An individual with Gilles de la Tourette syndrome and Smith-Magenis microdeletion syndrome: is chromosome 17p11.2 a candidate region for Tourette syndrome putative susceptibility genes? J Intellect Disabil Res. 2007;51:620–624. doi: 10.1111/j.1365-2788.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- 30*.Robertson MM, Shelley BP, Dalwai S, Brewer C, Critchley HD. A patient with both Gilles de la Tourette's syndrome and chromosome 22q11 deletion syndrome: clue to the genetics of Gilles de la Tourette's syndrome? J Psychosom Res. 2006;61:365–368. doi: 10.1016/j.jpsychores.2006.06.011. [DOI] [PubMed] [Google Scholar]; This report presages more recent results suggesting that identical structural variants can lead to a wide range of diagnoses formerly considered entirely distinct.
- 31.Clarke RA, Fang ZM, Diwan AD, Gilbert DL. Tourette syndrome and klippel-feil anomaly in a child with chromosome 22q11 duplication. Case Report Med. 2009;2009:361518. doi: 10.1155/2009/361518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petek E, Windpassinger C, Vincent JB, Cheung J, Boright AP, Scherer SW, Kroisel PM, Wagner K. Disruption of a novel gene (IMMP2L) by a breakpoint in 7q31 associated with Tourette syndrome. Am J Hum Genet. 2001;68:848–858. doi: 10.1086/319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroisel PM, Petek E, Emberger W, Windpassinger C, Wladika W, Wagner K. Candidate region for Gilles de la Tourette syndrome at 7q31. Am J Med Genet. 2001;101:259–261. doi: 10.1002/1096-8628(20010701)101:3<259::aid-ajmg1374>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 34.State MW, Greally JM, Cuker A, Bowers PN, Henegariu O, Morgan TM, Gunel M, DiLuna M, King RA, Nelson C, et al. Epigenetic abnormalities associated with a chromosome 18(q21-q22) inversion and a Gilles de la Tourette syndrome phenotype. Proc Natl Acad Sci U S A. 2003;100:4684–4689. doi: 10.1073/pnas.0730775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford FC, Ait-Ghezala G, Morris M, Sutcliffe MJ, Hauser RA, Silver AA, Mullan MJ. Translocation breakpoint in two unrelated Tourette syndrome cases, within a region previously linked to the disorder. Hum Genet. 2003;113:154–161. doi: 10.1007/s00439-003-0942-4. [DOI] [PubMed] [Google Scholar]
- 36.Cuker A, State MW, King RA, Davis N, Ward DC. Candidate locus for Gilles de la Tourette syndrome/obsessive compulsive disorder/chronic tic disorder at 18q22. Am J Med Genet A. 2004;130A:37–39. doi: 10.1002/ajmg.a.30066. [DOI] [PubMed] [Google Scholar]
- 37.Petek E, Schwarzbraun T, Noor A, Patel M, Nakabayashi K, Choufani S, Windpassinger C, Stamenkovic M, Robertson MM, Aschauer HN, et al. Molecular and genomic studies of IMMP2L and mutation screening in autism and Tourette syndrome. Mol Genet Genomics. 2007;277:71–81. doi: 10.1007/s00438-006-0173-1. [DOI] [PubMed] [Google Scholar]
- 38**.Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]; The paper was the first to identify rare functional sequence mutations in individuals with TD, pointed for the first time to the SLITRK family of molecules in human developmental psychopathology, and provided evidence for the relevence of microRNA mediated regulation of gene expression in this process.
- 39.Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci. 2003;24:117–129. doi: 10.1016/s1044-7431(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 40.Stillman AA, Krsnik Z, Sun J, Rasin MR, State MW, Sestan N, Louvi A. Developmentally regulated and evolutionarily conserved expression of SLITRK1 in brain circuits implicated in Tourette syndrome. J Comp Neurol. 2009;513:21–37. doi: 10.1002/cne.21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Kajiwara Y, Buxbaum JD, Grice DE. SLITRK1 binds 14-3-3 and regulates neurite outgrowth in a phosphorylation-dependent manner. Biol Psychiatry. 2009;66:918–925. doi: 10.1016/j.biopsych.2009.05.033. [DOI] [PubMed] [Google Scholar]; The study used biochemical methods to study the function of SLITRK1 in the central nervous system. The authors found evidence that the molecule is cleaved and secreted and that phosphorylation of the intracellular domain regulates the demonstated effect on neurite outgrowh
- 42*.Katayama K, Yamada K, Ornthanalai VG, Inoue T, Ota M, Murphy NP, Aruga J. Slitrk1-deficient mice display elevated anxiety-like behavior and noradrenergic abnormalities. Mol Psychiatry. 2010;15:177–184. doi: 10.1038/mp.2008.97. [DOI] [PubMed] [Google Scholar]; An animal model of SLITRK1 deficiency shows altered noradrenergic function and an anxiety phenotype that is resued by alpha-agonists, which are used in the treatment of Tourette syndrome
- 43.Leckman JF, Goodman WK, Anderson GM, Riddle MA, Chappell PB, McSwiggan-Hardin MT, McDougle CJ, Scahill LD, Ort SI, Pauls DL, et al. Cerebrospinal fluid biogenic amines in obsessive compulsive disorder, Tourette'ssyndrome, and healthy controls. Neuropsychopharmacology. 1995;12:73–86. doi: 10.1038/sj.npp.1380241. [DOI] [PubMed] [Google Scholar]
- 44**.Shmelkov SV, Hormigo A, Jing D, Proenca CC, Bath KG, Milde T, Shmelkov E, Kushner JS, Baljevic M, Dincheva I, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. doi: 10.1038/nm.2125. 591p following 602. [DOI] [PMC free article] [PubMed] [Google Scholar]; An elegant study of the SLITRK5 knock-out mouse. The behavioral phenotype of excessive grooming has only been reported with regard to two other knockouts (HoxB8 and SAPAP3). The link between the observed phenotype and obsessive compulsive disorder is further supported by the response of the knockout to serotonin reuptake inhibitors. The authors go on to show abnormal dendritic development in the striatum and altered glutamateric activity in cortical striatal circuits.
- 45.Zimprich A, Hatala K, Riederer F, Stogmann E, Aschauer HN, Stamenkovic M. Sequence analysis of the complete SLITRK1 gene in Austrian patients with Tourette's disorder. Psychiatr Genet. 2008;18:308–309. doi: 10.1097/YPG.0b013e3283060f6f. [DOI] [PubMed] [Google Scholar]
- 46.Chou IC, Wan L, Liu SC, Tsai CH, Tsai FJ. Association of the Slit and Trk-like 1 gene in Taiwanese patients with Tourette syndrome. Pediatr Neurol. 2007;37:404–406. doi: 10.1016/j.pediatrneurol.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 47.Deng H, Le WD, Xie WJ, Jankovic J. Examination of the SLITRK1 gene in Caucasian patients with Tourette syndrome. Acta Neurol Scand. 2006;114:400–402. doi: 10.1111/j.1600-0404.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- 48.Scharf JM, Moorjani P, Fagerness J, Platko JV, Illmann C, Galloway B, Jenike E, Stewart SE, Pauls DL. Lack of association between SLITRK1var321 and Tourette syndrome in a large family-based sample. Neurology. 2008;70:1495–1496. doi: 10.1212/01.wnl.0000296833.25484.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Keen-Kim D, Mathews CA, Reus VI, Lowe TL, Herrera LD, Budman CL, Gross-Tsur V, Pulver AE, Bruun RD, Erenberg G, et al. Overrepresentation of rare variants in a specific ethnic group may confuse interpretation of association analyses. Hum Mol Genet. 2006;15:3324–3328. doi: 10.1093/hmg/ddl408. [DOI] [PubMed] [Google Scholar]; The report raised the important issue of the potential confound of population stratification in studies of rare variants. The authors hypothesize that the reported association of SLITRK1 is a result of occult overrepresentation of Ashkenazi cases versus controls in the original report from your group.
- 50•.O'Roak BJ, Morgan TM, Fishman DO, Saus E, Alonso P, Gratacos M, Estivill X, Teltsh O, Kohn Y, Kidd KK, et al. Additional support for the association of SLITRK1 var321 and Tourette syndrome. Mol Psychiatry. 2010;15:447–450. doi: 10.1038/mp.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper rebuts the concerns raised in Keen Kim et al and Scharf et al regarding the contribution of population stratification to the finding of association of the gene SLITRK1 with Tourette disorder.
- 51**.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]; This case report was the first to identify Contactin Associated Protein 2 in human CNS pathology and subsequently provided some of the earliest evidence for an overlap in genetic risks between TD and other developmental neuropsychiatric disorders
- 52*.Lawson-Yuen A, Saldivar JS, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 2008;16:614–618. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]; This case report was one of the first to suggest an overlap between genes implicated in autism spectrum disorders and those contributing to Tourette syndrome.
- 53.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 56.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, Mumford JA, Poldrack RA, Dapretto M, Geschwind DH, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2:56–80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, Bijlsma EK, Oortveld MA, Ekici AB, Reis A, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124:192–199. doi: 10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Blake DJ, Forrest M, Chapman RM, Tinsley CL, O'Donovan MC, Owen MJ. TCF4, schizophrenia, and Pitt-Hopkins Syndrome. Schizophr Bull. 2010;36:443–447. doi: 10.1093/schbul/sbq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 64*.Breedveld GJ, Fabbrini G, Oostra BA, Berardelli A, Bonifati V. Tourette disorder spectrum maps to chromosome 14q31.1 in an Italian kindred. Neurogenetics. 2010;11:417–423. doi: 10.1007/s10048-010-0244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This and the following five reports highlight the potential for mapping of Mendelian forms of Tourette syndrome. This is particularly promising given the rapid adoption of whole-exome and whole genome sequencing approaches.
- 65*.Merette C, Brassard A, Potvin A, Bouvier H, Rousseau F, Emond C, Bissonnette L, Roy MA, Maziade M, Ott J, et al. Significant linkage for Tourette syndrome in a large French Canadian family. Am J Hum Genet. 2000;67:1008–1013. doi: 10.1086/303093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Laurin N, Wigg KG, Feng Y, Sandor P, Barr CL. Chromosome 5 and Gilles de la Tourette syndrome: Linkage in a large pedigree and association study of six candidates in the region. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:95–103. doi: 10.1002/ajmg.b.30779. [DOI] [PubMed] [Google Scholar]
- 67*.Knight S, Coon H, Johnson M, Leppert MF, Camp NJ, McMahon WM. Linkage analysis of Tourette syndrome in a large utah pedigree. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.31035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Verkerk AJ, Cath DC, van der Linde HC, Both J, Heutink P, Breedveld G, Aulchenko YS, Oostra BA. Genetic and clinical analysis of a large Dutch Gilles de la Tourette family. Mol Psychiatry. 2006;11:954–964. doi: 10.1038/sj.mp.4001877. [DOI] [PubMed] [Google Scholar]
- 69*.Curtis D, Brett P, Dearlove AM, McQuillin A, Kalsi G, Robertson MM, Gurling HM. Genome scan of Tourette syndrome in a single large pedigree shows some support for linkage to regions of chromosomes 5, 10 and 13. Psychiatr Genet. 2004;14:83–87. doi: 10.1097/01.ypg.0000107927.32051.f5. [DOI] [PubMed] [Google Scholar]
- 70**.Ercan-Sencicek AG, Stillman AA, Ghosh AK, Bilguvar K, O'Roak BJ, Mason CE, Abbott T, Gupta A, King RA, Pauls DL, et al. L-histidine decarboxylase and Tourette's syndrome. N Engl J Med. 2010;362:1901–1908. doi: 10.1056/NEJMoa0907006. [DOI] [PMC free article] [PubMed] [Google Scholar]; The paper reports on a rare nonsense mutation that provides evidence for a role for histaminergic neurotransmission in the etiology of modulation of Tourette syndrome and related disorders and suggests potential avenues for treatment based on this molecular mechanism.
- 71.Haas HL, Sergeeva OA, Selbach O. Histamine in the nervous system. Physiol Rev. 2008;88:1183–1241. doi: 10.1152/physrev.00043.2007. [DOI] [PubMed] [Google Scholar]
- 72.Kubota Y, Ito C, Sakurai E, Watanabe T, Ohtsu H. Increased methamphetamine-induced locomotor activity and behavioral sensitization in histamine-deficient mice. J Neurochem. 2002;83:837–845. doi: 10.1046/j.1471-4159.2002.01189.x. [DOI] [PubMed] [Google Scholar]
- 73.Saka E, Graybiel AM. Pathophysiology of Tourette's syndrome: striatal pathways revisited. Brain Dev. 2003;25(Suppl 1):S15–19. doi: 10.1016/s0387-7604(03)90002-7. [DOI] [PubMed] [Google Scholar]
Additional references for table 1
- 74.Esbenshade TA, Fox GB, Cowart MD. Histamine H3 receptor antagonists: preclinical promise for treating obesity and cognitive disorders. Mol Interv. 2006;6:77–88. 59. doi: 10.1124/mi.6.2.5. [DOI] [PubMed] [Google Scholar]
- 75.Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, Mukaddes NM, Yoo SY, Hanson E, Hundley R, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kirov G, Rujescu D, Ingason A, Collier DA, O'Donovan MC, Owen MJ. Neurexin 1 (NRXN1) deletions in schizophrenia. Schizophr Bull. 2009;35:851–854. doi: 10.1093/schbul/sbp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shah AK, Tioleco NM, Nolan K, Locker J, Groh K, Villa C, Stopkova P, Pedrosa E, Lachman HM. Rare NRXN1 promoter variants in patients with schizophrenia. Neurosci Lett. 2010;475:80–84. doi: 10.1016/j.neulet.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan J, Noltner K, Feng J, Li W, Schroer R, Skinner C, Zeng W, Schwartz CE, Sommer SS. Neurexin 1alpha structural variants associated with autism. Neurosci Lett. 2008;438:368–370. doi: 10.1016/j.neulet.2008.04.074. [DOI] [PubMed] [Google Scholar]
- 81.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maestrini E, Pagnamenta AT, Lamb JA, Bacchelli E, Sykes NH, Sousa I, Toma C, Barnby G, Butler H, Winchester L, et al. High-density SNP association study and copy number variation analysis of the AUTD1 and AUTD5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry. 2010;15:954–968. doi: 10.1038/mp.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pagnamenta AT, Bacchelli E, de Jonge MV, Mirza G, Scerri TD, Minopoli F, Chiocchetti A, Ludwig KU, Hoffmann P, Paracchini S, et al. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol Psychiatry. 2010;68:320–328. doi: 10.1016/j.biopsych.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D'Arcy M, deBerardinis R, Frackelton E, Kim C, et al. Rare structural variants found in attentiondeficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, Franke A, Malafosse A, Genton P, Thomas P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.keda M, Aleksic B, Kirov G, Kinoshita Y, Yamanouchi Y, Kitajima T, Kawashima K, Okochi T, Kishi T, Zaharieva I, et al. Copy number variation in schizophrenia in the Japanese population. Biol Psychiatry. 2010;67:283–286. doi: 10.1016/j.biopsych.2009.08.034. [DOI] [PubMed] [Google Scholar]
- 89.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]