Abstract
Proteins perform their function in cells where macromolecular solutes reach concentrations of >300 g/L and occupy >30% of the volume. The volume excluded by these macromolecules will stabilize globular proteins because the native state occupies less space than the denatured state. Theory predicts that crowding can increase the ratio of folded to unfolded protein by a factor of 100, amounting to 3 kcal/mol of stabilization at room temperature. We tested the idea that volume exclusion dominates the crowding effect in cells with a variant of protein L, a 7-kDa globular protein with seven lysine residues replaced by glutamic acids. Eighty-four percent of the variant molecules populate the denatured state in dilute buffer at room temperature, compared to 0.1% for the wild-type protein. We then used in-cell nuclear magnetic resonance spectroscopy to show that the cytoplasm of Escherichia coli does not overcome even this modest (~1 kcal/mol) free energy deficit. The data are consistent with the idea that non-specific interactions between cytoplasmic components can overcome the excluded volume effect. Evidence for these interactions is provided by the observation that adding simple salts folds the variant in dilute solution, but increasing the salt concentration inside E. coli does not fold the protein. Our data are consistent with other studies of protein stability in cells, and suggest that stabilizing excluded volume effects, which must be present under crowded conditions, can be ameliorated by non-specific interactions between cytoplasmic components.
The effects of high macromolecule concentrations on equilibrium properties arise from two phenomena. The first, excluded volume, is the result of the impenetrable nature of atoms. The volume excluded by the crowding molecules is unavailable to the test protein. The native, folded state of a globular protein takes up less space than the denatured state. Application of Le Chatelier’s principle leads to the conclusion that volume exclusion favors the native state because it occupies less space.1
The other phenomenon involves specific and non-specific intermolecular chemical interactions. If the crowding molecule interacts with only the native state, the effect is stabilizing. If the crowder has an affinity for protein in general, the effect is destabilizing. These opposing effects are reminiscent of ligand binding and urea denaturation. Binding pulls the equilibrium between the native and denatured state toward the native state because the crowder binds this state. Urea pulls the equilibrium toward the denatured state because that state exposes more surface area. Although high urea concentrations also introduce an excluded volume effect,2 this contribution is smaller than that from chemical interactions with the denatured state. Thus, one cannot know, a priori, how crowding will affect globular protein stability.
Until recently, the stabilization afforded by volume exclusion was thought to dominate both in vitro and in cells, although there were hints of compensation.3–5 We have shown that non-specific, non-covalent intermolecular interactions and excluded volume effects compete to affect diffusion.6 Here, we test this idea in terms of protein stability in cells.
We chose as our test protein the Immunoglobulin G binding domain of protein L from the mesophile Streptococcus magnus. For many years,7 ProtL has been used as a model for protein folding.8 This well studied 7-kDa protein has the properties expected for a protein from a mesophilic organism9 and exhibits reversible unfolding at 25 °C in dilute solution via a two-state reaction with a stability of 4.3 kcal/mol.9 Changing seven of its lysines to glutamic acids lowers the stability, such that the majority of the protein molecules are in the denatured state in dilute buffer. The destabilization arises from the variant’s decreased hydrophobic surface area, not the increase in negative charge.9 The variant does fold, however, on adding Na+ salts9 or, as described below, K+ salts. If excluded volume effects dominate crowding, then the fraction of the variant molecules in the native state should increase in cells relative to dilute solution. We tested this idea.
ProtL locations were determined by osmotically shocking E. coli to separate the cytoplasm and periplasm.10 SDS-PAGE results show an increase in ProtL expression over 4 h of induction (Figure S1, in Supporting Information [SI]). The variant is located almost entirely in the cytoplasm through 4 h. The wild-type protein is primarily located in the cytoplasm from 0 to 1 h. Later, it is present in both compartments. At all times, the level of the wild-type protein is greater than that of the variant. These observations are consistent with the idea that increasing expression causes cytoplasmic proteins to migrate to the periplasm.11 For all studies of the wild-type protein, we limited the expression time such that the protein is in the cytoplasm.
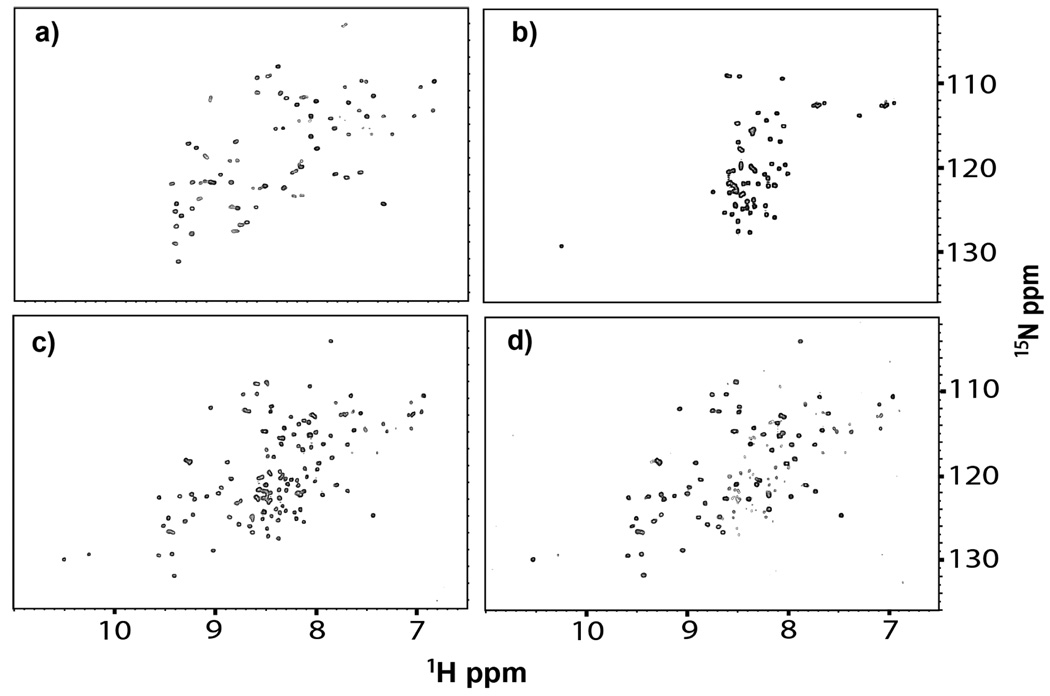
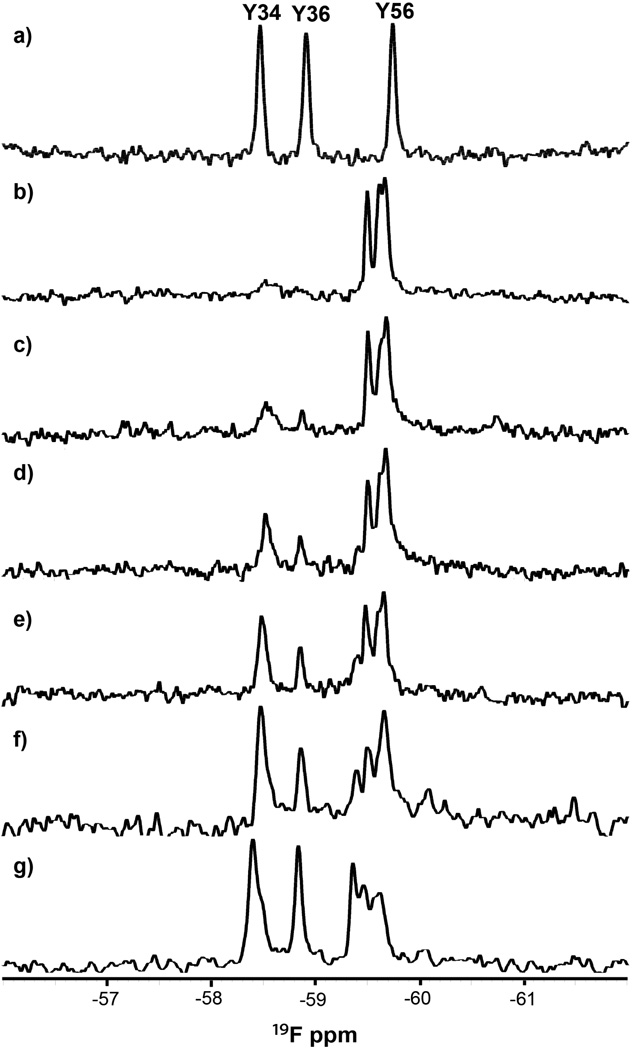
First, we monitored wild-type ProtL by using the 1H-15N heteronuclear single-quantum coherence (HSQC) experiment. The protein is folded (Figure 1a) as indicated by the large dispersion of 1H chemical shifts. The fold was also assessed by using 19F NMR. ProtL contains three tyrosines, so 3-fluorotyrosine (3-FY) substitution12 should yield three resonances. This prediction is borne out (Figure 2a). The resonances are well separated as expected for a folded protein.
Figure 1.
HSQC spectra (20 mM phosphate buffer, pH 6.0, 25 °C) of the 15N-enriched a) wild-type protein and the Kx7E variant in b) 0 M, c) 0.3 M, and d) 0.8 M NaCl.
Figure 2.
19F spectra (20 mM phosphate buffer, pH 6.0, 37 °C) of the 19F-labeled a) wild-type protein and the Kx7E variant in b) 0 M, c) 0.1 M, d) 0.2 M, e) 0.3 M, f) 0.5 M, and g) 1 M NaCl. Assignments were made by using mutagenesis (Figure S2, SI).
The purified, 15N-enriched Kx7E variant was dissolved in 20 mM phosphate buffer solutions, pH 6.0, containing 0 to 1 M NaCl. The limited 1H chemical shift dispersion in the HSQC spectrum (Figure 1b) indicates that the variant is unfolded in buffer alone.9,13–15 Upon increasing the NaCl concentration to 0.3 M, the dispersion increases to that of a folded protein, although crosspeaks from the unfolded form are visible (Figure 1c). In 0.8 M NaCl, almost all the variant is folded (Figure 1d) and the crosspeak pattern is similar to that of wild-type ProtL (Figure 1a).
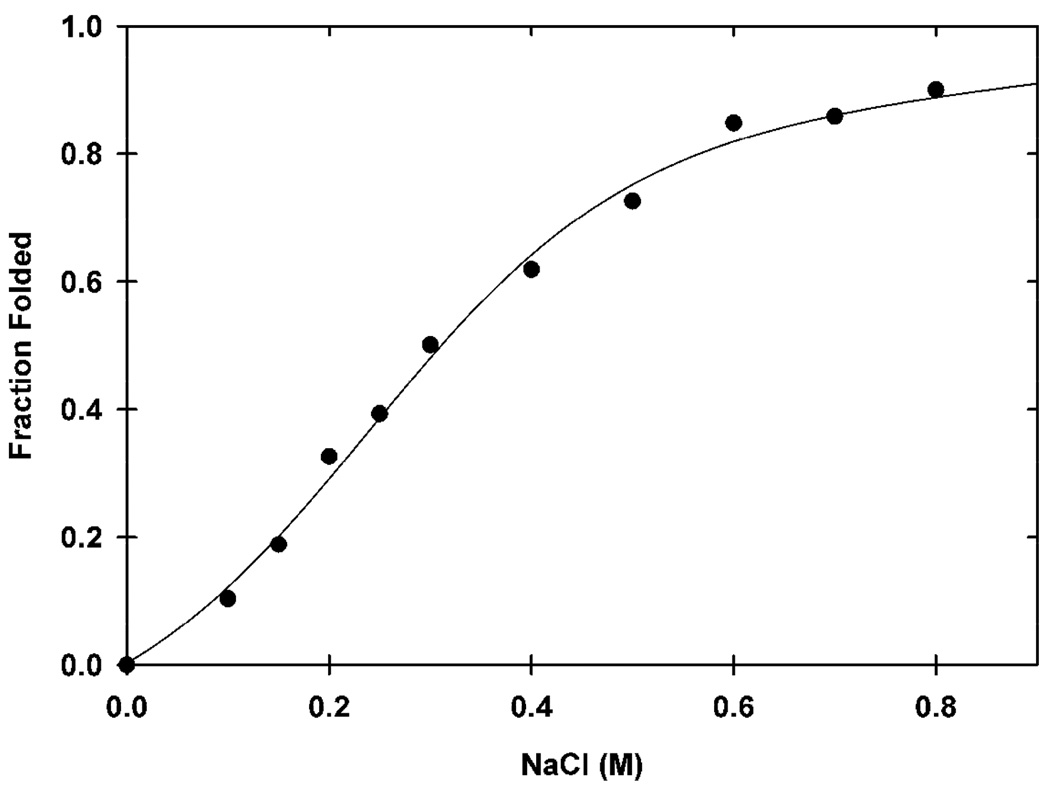
The average intensities of the unfolded and folded Kx7E crosspeaks at each NaCl concentration (Table S1, SI) were used to produce a titration curve (Figure 3). The transition is 50% complete at 0.3 M NaCl and reaches a maximum at 0.8 M NaCl. The data were fit to a two-state model.16 The unfolding free energy at 0 M NaCl is −1.0 kcal/mol (RMSD of 2%) and represents the free energy required to fold half the variant molecules in the absence of NaCl. The salt-induced folding is reversible.9
Figure 3.
Titration curve showing the fraction of folded Kx7E variant (20 mM phosphate buffer, pH 6.0, 25 °C) as a function of NaCl concentration.
We also monitored the fold of the variant by using 19F NMR. The purified 19F-labeled protein was dissolved in 20 mM phosphate buffer solutions, pH 6.0, containing NaCl. The spectrum in the absence of added NaCl (Figure 2b) shows the three expected resonances in a narrow chemical shift range, indicative of an unfolded protein. As the NaCl concentration increases, two resonances move downfield, and the intensities of the resonances attributed to the unfolded protein decrease (Figure 2c–g). At 1 M NaCl, most of the protein is folded, although resonances from the unfolded protein remain at low intensities. The chemical shifts from the variant in 1 M NaCl are consistent with those of the wild-type protein (Figure 2a). The increasing fraction of folded protein with increasing NaCl concentration is consistent with the HSQC data (Figure 1), although not enough data points were acquired for quantification.
We used in-cell NMR to assess wild-type ProtL in E. coli. The HSQC spectrum of a cell slurry containing the 15N-enriched wild-type protein is shown in Figure 4a. The spectrum is the same as that of cells without an expression vector (Figure S3, SI). That is, the spectrum of the protein is unobservable; the crosspeaks are from 15N-enriched metabolites.17 These observations are consistent with results showing that 1H-15N HSQC spectra from many globular proteins are not detectable in E. coli.18–21 After lysing the cells, the wild-type protein spectrum (Figure 1a) is detected (Figure 4b). Furthermore, the spectrum is unchanged in the presence of 1 M NaCl (Figure 4c), as expected for a mesophilic protein. The spectrum of the cell supernatant shows no protein crosspeaks (Figure S4, SI), confirming that the protein is in the cells. To detect the protein in cells we turned to 19F NMR.21
Figure 4.
HSQC spectra (25 °C) of the 15N-enriched wild-type protein and the Kx7E variant in E. coli. a) Cell slurry expressing the wild-type protein. b) Lysate of cells from a). c) Lysate of cells shown in a) upon adding NaCl to a final concentration of 1 M. d) Cell slurry expressing the Kx7E variant. e) Lysate of cells from d). f) Lysate of cells shown in d) upon adding NaCl to a final concentration of 1 M.
The in-cell experiments were repeated with the 19F-labeled wild-type protein. Three broad resonances from the protein were detected in the 19F spectrum of the cell slurry (Figure 5a). The fourth resonance, from unincorporated 3-FY, is sharper and overlaps the most upfield protein resonance. Although broad, the resonances are well separated and similar to those from the in vitro spectrum (Figure 2a). The supernatant spectrum also shows a sharp resonance from unincorporated 3-FY (Figure 5b) and three additional small, sharp resonances. The positions of these resonances match those of wild-type ProtL in dilute solution (Figure 2a), indicating that a small amount of protein (<10%) leaks from the cells. These data confirm that wild-type ProtL is folded in E. coli and show that it can be detected by 19F NMR.
Figure 5.
19F spectra (37 °C) of the 19F-labeled wild-type protein or the Kx7E variant in E. coli. a) Cell slurry expressing wild-type protein. b) Supernatant from the sample used in a). c) Cell slurry expressing the Kx7E variant. d) Supernatant from the sample used in c). e) Cell slurry expressing the Kx7E variant grown and induced in hyperosmotic media. Asterisks denote the resonance from free 3-FY.
Disordered proteins are detectable in cells by 1H-15N HSQC NMR because of their internal motion.18–20 The HSQC spectrum of the cell slurry at 25 °C exhibits a narrow chemical shift dispersion in the 1H dimension (Figure 4d), characteristic of the unfolded protein (Figure 1b). The ability to detect the HSQC spectrum of the variant in cells shows that the protein is soluble in the cytoplasm. The spectrum of the cell supernatant shows no protein crosspeaks (Figure S4, SI), indicating that the spectrum in Figure 4d arises from the variant inside cells. The protein remains unfolded upon cell lysis (Figure 4e) but folds upon increasing the NaCl concentration (Figure 4f) or the KCl concentration (Figure S5, SI). These results demonstrate that, although the variant is foldable, the E. coli cytoplasm is unable to fold Kx7E.
The 15N results were confirmed by using 19F NMR. The 19F spectrum of the Kx7E cell slurry displays an envelope of broad resonances in a narrow chemical shift range and a sharp peak from unincorporated 3-FY (Figure 5c). The position of the envelope is inconsistent with the resonances from the folded variant (Figures 2a, 5a) but consistent with the position of the resonances from the unfolded variant (Figure 2b). The absence of the downfield resonance from the folded variant is especially evident. No resonances from the variant are detected in the spectrum of the supernatant, indicating that the protein does not leak (Figure 5d).
It has been known for over 40 years that the E. coli intracellular K+ concentration can be manipulated by adjusting the osmolality of the medium.22 An attempt was made to fold the variant in E. coli by increasing the intracellular concentration of K+ by growth in hyperosmotic (1.05 Osm) media.3,23–25 The 19F spectrum of the Kx7E cell slurry in hyperosmotic minimal media (Figure 5e), however, shows no resonances from the folded protein. The the upfield shift of the resonance envelope of the unfolded protein is probably caused by the increase in K+ concentration.
Many in vitro studies of crowding focus on volume exclusion. In these experiments, the environment comprises only one or a few different types of crowders. Furthermore, it is usually assumed that the crowding molecules have little interaction with the test protein, unless their van der Waals surfaces attempt to overlap, at which point the repulsive forces increase exponentially. Recent work, however, demonstrates the importance of nonspecific intermolecular interactions in both experiments and simulations.5,6,26,27 Thus, one cannot ignore chemical interactions in studies of macromolecular crowding.
We examined the unstable Kx7E variant of protein L. The variant folds in the presence of high concentrations of salt,9 prompting the question whether the crowded E. coli cytosol would have a similar effect.
We acquired HSQC spectra of the variant in vitro by adding increasing concentrations of NaCl. The spectra allowed construction of a folding titration curve. The unfolding free energy of Kx7E in the absence of NaCl is −1.0 kcal/mol. This NMR-derived value compares favorably to those derived by Tadeo et al. (−0.4 to −0.5 kcal/mol) from guanidinium chloride and urea titration experiments with CD detection.9 Therefore, the E. coli cytosol would have to deliver between 0.4 and 1.0 kcal/mol of stabilization to fold 50% of the variant molecules.
In-cell NMR data (Figures 4, 5) show that wild-type ProtL is folded while the variant remains unfolded. We conclude that the E. coli cytosol does not provide sufficient free energy to fold Kx7E. In other words, the volume exclusion provided by the highly crowded intracellular environment is insufficient to overcome the unfavorable free energy of folding.
We also tested whether increasing the K+ concentration in cells could fold the variant. The cytoplasmic K+ concentration in E. coli is ~0.2 M23,25 for cells grown in minimal media (0.1 Osm). Nearly all of the cytoplasmic K+ in such E. coli, however, is associated with polyanions such as DNA and RNA.24 Thus, not enough salt is available to fold the variant in E. coli.
We increased the cytoplasmic K+ concentration by increasing the osmolality of the medium from 0.16 Osm to 1.05 Osm. Under these conditions the cytoplasmic K+ concentration reaches between 0.5 and 0.9 M.23,25 Importantly, at least half of this upshift is due to an increase in free cytoplasmic K+.23 Therefore, the concentration of free cytoplasmic K+ under hyperosmotic conditions is at least 0.3 M, which leads to easily detectable levels of the folded protein in vitro (Figures 2, 3). Even under these conditions we do not detect the folded form in cells (Figure 5e). Furthermore, it is unlikely that a significant amount of the unfolded form is associated with chaperones because the observation of crosspeaks in Figure 4d is inconsistent with the molecular weight of a Kx7E-chaperone or proteasome complex.28
Our results suggest that any stabilizing excluded volume effect, which must exist because of the high concentration of macromolecules in the cytoplasm, is more than offset by a destabilizing effect. The destabilization probably arises from non-specific protein-protein interactions. Wang et al. show that rotational diffusion of the globular protein chymotrypsin inhibitor 2 is slowed beyond that expected by viscosity alone in crowded protein solutions and E. coli lysates, and that the difference is due to nonspecific intermolecular interactions between the crowders and test protein.6 These non-specific interactions in cells can lead to irreversible denaturation,29 explaining why the Kx7E variant is unfolded in cells. Such non-specific protein-protein interactions may also explain why the variant does not fold completely in vitro even at high salt concentrations (Figures 2g, 3).
Although volume exclusion is expected to stabilize the native state of globular proteins,30 the E. coli cytosol comprises biologically active molecules. These molecules chemically interact with the protein under study, and these interactions can be stabilizing or destabilizing. Therefore, it is not clear, a priori, how macromolecular crowding in cells will affect protein stability. The few available in-cell studies of globular protein stability indicate no change or a slight destabilization in E. coli compared to dilute solution.3,4 Although dilute solution data may be physiologically relevant for stable globular proteins, our data show that nonspecific intermolecular interactions can overcome volume exclusion-induced compaction, causing globular proteins to remain unfolded. The situation is different for intrinsically disordered proteins where the crowded environment in cells can result in compaction.31,32 The dissimilar behavior is probably caused by the different amino acid compositions of these two protein classes.33
Given that KCl folds the variant in vitro, it is striking that increasing the intracellular K+ concentration fails to have the same effect. This discrepancy indicates that non-specific intermolecular interactions may prevent refolding of globular proteins even when solution conditions favor it.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health grant 5DP1OD783 and the National Science Foundation grant MCB 1051819. We thank Marc ter Horst and Gregory Young for spectrometer maintenance, and Elizabeth Pielak for helpful comments.
Footnotes
Supporting Information Available: Material and methods, Table S1, and Figures S1–S5. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cheung MS, Klimov D, Thirumalai D. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4753. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schellman JA. Biophys. J. 2003;85:108. doi: 10.1016/S0006-3495(03)74459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghaemmaghami S, Oas TG. Nat. Struct. Biol. 2001;8:879. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 4.Ignatova Z, Krishnan B, Bombardier JP, Marcelino AM, Hong J, Gierasch LM. Peptide Sci. 2007;88:157. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGuffee SR, Elcock AH. PLoS Comput. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YQ, Li C, Pielak GJ. J. Am. Chem. Soc. 2010;132:9392. doi: 10.1021/ja102296k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wikström M, Drakenberg T, Forsén S, Sjöbring U, Björck L. Biochemistry. 1994;33:14011. doi: 10.1021/bi00251a008. [DOI] [PubMed] [Google Scholar]
- 8.Yi Q, Baker D. Protein Sci. 1996;5:1060. doi: 10.1002/pro.5560050608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tadeo X, Lopez-Mendez B, Trigueros T, Lain A, Castano D, Millet O. PLoS Biol. 2009;7:e1000257. doi: 10.1371/journal.pbio.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shevchik VE, Condemine G, Robert-Baudouy J. EMBO J. 1994;13:2007. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes CO, Pielak GJ. Proteins: Struct., Funct., Bioinf. 2011;79:347. doi: 10.1002/prot.22906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan F, Kuprov I, Craggs TD, Hore PJ, Jackson SE. J. Am. Chem. Soc. 2006;128:10729. doi: 10.1021/ja060618u. [DOI] [PubMed] [Google Scholar]
- 13.Zhang O, Forman-Kay JD. Biochemistry. 1995;34:6784. doi: 10.1021/bi00020a025. [DOI] [PubMed] [Google Scholar]
- 14.Dyson HJ, Wright PE. Annu. Rev. Phys. Chem. 1996;47:369. doi: 10.1146/annurev.physchem.47.1.369. [DOI] [PubMed] [Google Scholar]
- 15.McDonald CC, Phillips WD. J. Am. Chem. Soc. 1969;91:1513. doi: 10.1021/ja01034a039. [DOI] [PubMed] [Google Scholar]
- 16.Santoro MM, Bolen DW. Biochemistry. 1988;27:8063. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 17.Sharaf NG, Barnes CO, Charlton LM, Young GB, Pielak GJ. J. Magn. Reson. 2010;202:140. doi: 10.1016/j.jmr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, Pielak GJ. J. Am. Chem. Soc. 2008;130:6310. doi: 10.1021/ja801020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnes CO, Monteith WB, Pielak GJ. ChemBioChem. 2011;12:390. doi: 10.1002/cbic.201000610. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Wang GF, Wang YQ, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RAS, Mehl RA, Pielak GJ. J. Am. Chem. Soc. 2010;132:321. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pielak GJ, Li C, Miklos AC, Schlesinger AP, Slade KM, Wang GF, Zigoneanu IG. Biochemistry. 2009;48:226. doi: 10.1021/bi8018948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epstein W, Schultz SG. J. Gen. Physiol. 1965;49:221. doi: 10.1085/jgp.49.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richey B, Cayley DS, Mossing MC, Kolka C, Anderson CF, Farrar TC, Record MT., Jr J. Biol. Chem. 1987;262:7157. [PubMed] [Google Scholar]
- 24.Cayley S, Lewis BA, Guttman HJ, Record MT. J. Mol. Biol. 1991;222:281. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- 25.Konopka MC, Weisshaar JC, Record MT., Jr Methods Enzymol. 2007;428:487. doi: 10.1016/S0076-6879(07)28027-9. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Pielak GJ. J. Am. Chem. Soc. 2009;131:1368. doi: 10.1021/ja808428d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miklos AC, Li C, Sharaf NG, Pielak GJ. Biochemistry. 2010;49:6984. doi: 10.1021/bi100727y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. Nature. 2010;467:868. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- 29.Plaza del Pino IM, Ibarra-Molero B, Sanchez-Ruiz JM. Proteins. 2000;40:58. doi: 10.1002/(sici)1097-0134(20000701)40:1<58::aid-prot80>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 30.Zhou HX, Rivas G, Minton AP. Annu. Rev. Biophys. 2008;37:353. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNulty BC, Young GB, Pielak GJ. J. Mol. Biol. 2006;355:893. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 32.Dedmon MM, Patel CN, Young GB, Pielak GJ. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12681. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunker AK, Silman I, Uversky VN, Sussman JL. Curr. Opin. Struct. Biol. 2008;18:756. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.