SUMMARY
Membrane budding is a key step in vesicular transport, multivesicular body and exosome biogenesis, and enveloped virus release. Coated vesicle formation, which is usually involved in budding towards cytosol, represents a protein-driven pathway of membrane budding suited to its function in intracellular protein sorting. Certain instances of cell entry by viruses and toxins, and microdomain-dependent multivesicular body biogenesis in animal cells, are examples of a mainly lipid-driven paradigm. Caveolae biogenesis, HIV-1 budding, and perhaps ESCRT-catalyzed multivesicular body biogenesis involve aspects of both the protein scaffold and membrane microdomain paradigms. Some of these latter events involve budding away from cytosol, and this unusual topology involves novel mechanisms. Progress in the structural and energetic bases of these different paradigms will be discussed.
Eukaryotic cells are defined by their compartmentalization into membrane-delimited structures. The protein and lipid content of these membranes is maintained and regulated by a constant flux of vesicular trafficking. Each vesicular trafficking event involves the budding of a membrane vesicle from the donor membrane, typically followed by its regulated transport, docking to, and fusion with, an acceptor membrane. Many viruses are membrane enveloped and escape from host cells by membrane budding events.
Our laboratory has been characterizing the unusual membrane budding reaction promoted by the ESCRTs, which has led us to take a fresh look at how membrane lipid properties might make protein-dependent, energetically expensive, reactions, easier. Several excellent reviews have covered the way proteins induce curvature in biological membranes (Farsad and De Camilli, 2003; McMahon and Gallop, 2005; Voeltz and Prinz, 2007), and the physical principles of membrane curvature (Zimmerberg and Kozlov, 2006). This review will take a different viewpoint, and consider the comparative roles of proteins and lipids in select examples of vesicular budding events (Fig. 1); similarities and differences in budding events in synthetic vs. cellular contexts; the potential roles of proteins in orchestrating lipid phase changes, and the roles of lipids in recruiting and regulating proteins; and the implications of the above for cell physiology. This article is not intended to be a comprehensive review of all cellular budding events. Rather, we consider how emerging mechanistic thinking in multivesicular body formation, virus budding, and lipid phase separation-induced budding puts more classical coated vesicle budding mechanisms into perspective, and vice versa.
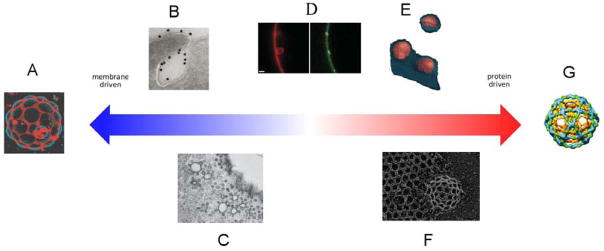
Figure 1. Proteins and lipid microdomains in membrane budding.
From left to right, A. Budding of phase separated lipid microdomains from GUVs composed of synthetic lipids. Reproduced by permission from (Baumgart et al., 2003), an example of membrane budding in the absence of any proteins. B. Shiga toxin (gold label) induces membrane buds acting from outside the plasma membrane. Reproduced by permission from (Romer et al., 2007) and example of a protein triggering a primarily microdomain-driven budding event. C. Caveolae, a hybrid between a membrane microdomain and a protein coat. Reproduced by permission from (Parton and Simons, 2007). D. ESCRT-I and – II induced buds in synthetic GUVs. Reproduced by permission from (Wollert and Hurley, 2010). Proteins organize these structures, but do not form a coat, suggesting a possible role for microdomains. E. HIV-1 buds visualized by EM tomography (Carlson et al., 2010). The bud is organized by the HIV-1 capsid protein, heavily enriched in raft lipids, and cleaved by ESCRT proteins. F. Deep etch visualization of clathrin coated pits. Reproduced by permission from (Heuser et al., 1987). Clathrin assembles into baskets in the absence of membranes, but is too flexible to deform membranes on its own. For this, clathrin needs help from other membrane-deforming proteins and possibly from lipids. G. Molecular model of the COP II cage. Reproduced by permission from (Russell and Stagg, 2010), an example of a protein structure that can form in the absence of lipids and can impose its shape on any simple bilayer-forming lipid mixture.
ENERGETICS OF VESICLE BUDDING
The formation of spherical vesicles from a flat membrane of typical biological composition and no intrinsic propensity to curve entails a membrane bending free energy (Helfrich, 1973) of ΔG = 8πκ ~ 250–600 kBT, given κ ~ 10–25 kBT, where kBT is thermal energy (Bloom et al., 1991). This is important for biology, because events that require a hundred-fold or more times thermal energy do not occur spontaneously. Biophysical studies of membrane budding, which offer the promise of accounting for energetics, are typically carried out in vesicles that are much larger than the biological size scale. Fortunately, the energetic cost of bud formation is, to a first approximation, independent of the size of the bud. In pure lipid mixtures used in biophysical studies, phase separation over size scales of microns occurs readily and spreads the energetic cost over on the order of many x 106 lipid molecules. In cells, however, membrane buds are on the size scale of 20–100 nm, thus involving as few as ~ 103 – 104 lipid molecules. The molecular cell biology of membranes poses the question how a modest number of protein-lipid interactions can sum to the needed free energy, or alternatively, how lipids themselves can contribute to lowering the energy barrier.
COATED VESICLE BUDDING
Clathrin
Coated vesicle formation is the dominant mechanism of membrane budding into the cytosol and the paradigm for protein-directed budding (Fig. 1F–G, 2). Clathrin is the archetypal vesicular coat. Clathrin coated vesicles (CCVs) are typically 60 – 100 nm in diameter (Bonifacino and Lippincott-Schwartz, 2003; Brodsky et al., 2001). Clathrin can form baskets in vitro in the absence of membranes that resemble the coats clathrin coated vesicles (CCVs), and the basket structure has been characterized in molecular detail (Fotin et al., 2004). Clathrin itself binds neither membranes nor cargo, but relies on adaptors for this function. The AP-2 complex is the best studied of these adaptors (Robinson and Bonifacino, 2001), and functions in clathrin mediated endocytosis (CME) at the plasma membrane. The AP-2 adaptor complex opens up in the presence of cargo and the lipid phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2 ) to form a flat platform capable of binding multiple PI(4,5)P2 and cargo molecules (Jackson et al., 2010). The established role for PI(4,5)P2 in this pathway is to recruit AP-2 and other proteins to the site of budding. A role for PI(4,5)P2 clustering into microdomains has been suggested on theoretical grounds (Liu et al., 2006), but has yet to be directly visualized.
Figure 2. Coated vesicle budding.
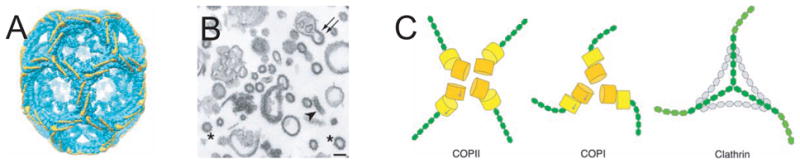
A. EM structure of clathrin basket reproduced by permission form (Fotin et al., 2004) B. COP II vesicles produced from purified components, reproduced by permission from (Lee et al., 2005). C. Structural parallels between clathrin, COP I, and COP II. Reproduced by permission from (Lee and Goldberg, 2010).
Clathrin is absolutely required for the budding of AP-2- and cargo -rich plasma membrane domains, which remain flat in its absence (Hinrichsen et al., 2006). However, clathrin monomers are flexible, which gives it the ability to form different types of lattices and to adapt to various cargoes (Ehrlich et al., 2004). Given the flexibility of clathrin monomers, the energy of clathrin polymerization has been proposed on theoretical grounds to be insufficient on its own to bend the membrane into a bud (Nossal, 2001). However, this concept has yet to be confirmed experimentally and is not universally accepted. Cholesterol is important for CME by many (though not all) accounts (Rodal et al., 1999; Subtil et al., 1999), although CME is less sensitive to cholesterol depletion than most coat-independent budding pathways (Sandvig et al., 2008). Clathrin, cargo adaptors, and PI(4,5)P2 are necessary but not sufficient on their own to induce membrane curvature. The essential early endocytic factor epsin wedges its amphipathic helix α0 into the membrane upon PI(4,5)P2 binding, promoting positive curvature (Ford et al., 2002). The muniscin proteins FCHo1/2 (Syp1 in yeast) are BAR domain containing proteins that bind cargo and promote positive curvature very early in endocytosis (Henne et al., 2010; Reider et al., 2009; Stimpson et al., 2009; Traub and Wendland, 2010). In principle, the reagents and concepts would appear to be in place to reconstitute clathrin-dependent membrane budding. Reconstitution of CME using synthetic lipids and purified proteins would be an important step in determining whether clathin, AP-2, one or more amphipathic helix and/or BAR domain proteins, and PI(4,5)P2 really constitute the minimum requirements for membrane bud formation in this pathway.
The scission of the clathrin-coated bud to form a detached vesicle is a complex process in its own right, and the reader is referred to recent reviews (Pucadyil and Schmid, 2009). Finally, following scission, the clathrin coat is removed by the ATP-dependent action of Hsc70 and its cofactor auxillin (Eisenberg and Greene, 2007). It is only following nucleotide hydrolysis that the energetic cost of clathrin-induced membrane deformation ΔG = 8πκ is finally paid, making the full reaction cycle- from flat membrane to uncoated vesicle- thermodynamically irreversible.
COP I and COP II
Vesicles carrying cargo from the ER to the Golgi are coated by the COP II complex, which, like clathin, can form membrane-free baskets in vitro with vesicle-like dimensions (Stagg et al., 2006). COP II vesicles have a preferred size, but as with clathrin, the flexibility of the COP II subunits allows formation of expanded lattices that can accommodate large cargoes such as procollagen and chylomicrons (Stagg et al., 2008). COP II vesicle budding has been reconstituted in vitro from purified proteins and synthetic lipids (Lee et al., 2005; Matsuoka et al., 1998). COP II consists of the Sec23/24 subcomplex, which binds lipids and cargo via a gently curved face (Bi et al., 2002); the Sec13/31 subcomplex, which forms an outer cage around the vesicle, and the membrane bending GTPase Sar1. The Sec23/24 and Sec13/31 subcomplexes in combination are sufficient to form buds, with Sar1 strictly required only for the scission of the buds. GTP hydrolysis by Sar1 provides energy input into the system, making the overall process culminating in uncoated cargo-loaded vesicles thermodynamically irreversible. A membrane consisting only of synthetic unsaturated phospholipids was capable of supporting budding (Matsuoka et al., 1998). COP I-coated vesicles are responsible for retrograde traffic from the Golgi to the ER, and this reaction has also been reconstituted from purified proteins and synthetic lipids. The budding reaction requires the coatomer complex, Arf1-GTP, and protein cargo tails tethered to the membrane, but has no special lipid requirements (Bremser et al., 1999). Budding occurs even from vesicles composed of the pure synthetic phospholipid DOPC doped with small amounts of a lipopeptide model cargo. Very recently, a composite crystallographic structure of αβ’ε cage-forming component of coatomer was determined, and shown to resemble the clathrin triskelion (Lee and Goldberg, 2010). COP I and COP II provide some of the purest examples of protein-directed membrane budding, in which the protein coat imposes its shape upon the membrane with minimal dependence on its lipid composition.
MEMBRANE MICRODOMAINS AND BUDDING
Lipid phase separation as a budding mechanism
In contrast to the protein-dominated paradigm of coated vesicle budding, phase separation in simple lipid mixtures can drive budding on a micron scale in synthetic model membranes, in the absence of proteins (Baumgart et al., 2003) (Fig. 1A, 3). Membrane bilayers can adopt either a solid or liquid phase, with the translational and conformational order of the lipid chains depending on their composition and the temperature. The liquid phase is the more relevant to biology, and can be subdivided into liquid disordered (Ld) and liquid ordered (Lo) phases. Lipids in the Ld phase have higher conformational freedom and diffusion coefficients than in the Lo phase. At biological temperatures, the Ld and Lo phases can coexist in membranes of mixed composition (Elson et al., 2010; Garcia-Saez and Schwille, 2010). In general, phospholipids with unsaturated chains prefer the Ld phase, while cholesterol, sphingolipids and phospholipids with saturated chains prefer the Lo phase (Lingwood and Simons, 2010). Typically, the energetic cost for contact between dissimilar lipids is small, ~ 0.5 kBT (Garcia-Saez and Schwille, 2010), but becomes significant when summed over many lipids. The higher acyl chain order in the Lo phase results in their elongation to their maximum extent, hence Lo membrane domains are thicker than Ld domains. The height mismatch at the phase boundary is energetically unfavorable, because it forces the polar headgroup region of the Ld domain into contact with the hydrophobic portion of the Lo domain. The free energy cost per unit length is known as the line tension, and has units of force. In order to minimize the free energy associated with line tension, membrane domains will coalesce with one another into circular zones. When circular domains reach a critical size at which the line tension energy term exceeds the Helfrich energy of membrane deformation, the membrane will deform out of plane in order to minimize the zone of contact (Lipowsky, 1992). If the line tension is high enough, the neck connecting the membrane bud can be severed, leading to the formation of detached vesicles. In addition to line tension effects, membrane microdomain formation can bend membranes by concentrating lipids with distinct intrinsic curvatures, and the contents of such microdomains can not only drive budding but dictate its direction (Bacia et al., 2005).
Figure 3. Membrane microdomains and budding.
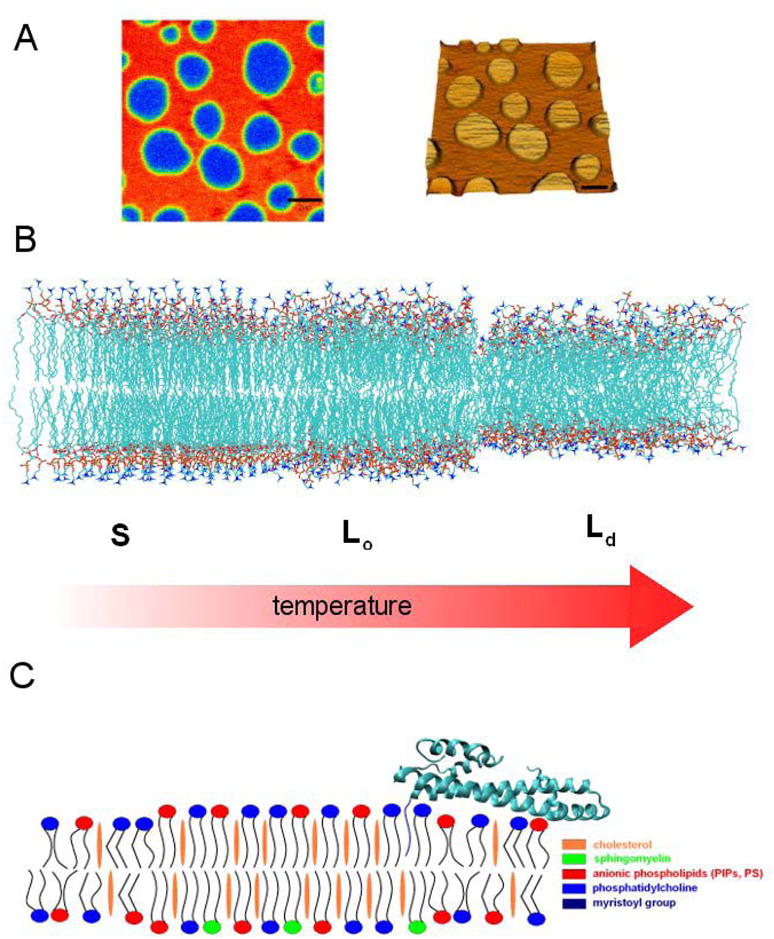
A. Coexistence of phases in model membranes visualized by AFM in a supported bilayer. Reproduced by permission from (Chiantia et al., 2006). B. Phase transitions in a single-lipid membrane analyzed by molecular dynamics simulations. Reproduced by permission from (Heller et al., 1993). C. Schematic model of a raft-type membrane microdomain, including a model for a myristoylated ESCRT-III subunit Vps20 as an example of protein anchoring to rafts.
The complex lipid mixture of the plasma membrane supports phase separation in micron sized domains when reconstituted in giant unilamellar vesicles (Baumgart et al., 2007). However, in living cells, membrane microdomains are heterogeneous, highly dynamic, nanoscale structures (Hancock, 2006; Lingwood and Simons, 2010; Pike, 2006). In the most up-to-date biophysical view, these nanoscale structure likely correspond to critical fluctuations (Veatch et al., 2007). While the concepts of the Lo and Ld phases are oversimplifications of the variety of dynamic membrane substructures that exist in cells (Lingwood and Simons, 2010), they will be used in this review because they are useful intuitive handles, deeply ingrained in the literature, and helpful in relating model membrane studies to biology. Most, but not all, of the membrane microdomains implicated in cellular budding are the sterol- and sphingolipid-rich domains known as “rafts”. Why don’t rafts and other microdomains coalesce on the micron scale in living cells, as they do in model membranes? The answer is not known, but the action of the cytoskeleton and membrane traffic, and the large fraction of protein in cellular membranes, is usually invoked. Indeed, it is to be expected that cells would have mechanisms to block the unchecked growth of microdomains, as the ensuing spontaneous vesiculation of cell membranes would be disastrous.
Soluble and lumenally-anchored cargoes, viruses, and toxins are selectively transported in vesicular carriers even though they have no direct communication with the cytosol to signal their packaging and sorting. In some cases, transmembrane sorting receptors serve as adaptors to link cargo to conventional cytosolic coat complexes. In other cases, membrane rafts make the link. Simian virus 40 (SV40) and cholera toxin enter cells by binding to multiple molecules of the ganglioside GM1 (Damm et al., 2005; Kirkham et al., 2005), a raft-favoring lipid. The cholera toxin B subunit (Merritt et al., 1994) and the SV40 VP1 protein (Neu et al., 2008) both bind to GM1 as pentamers. Cholera toxin pentamer binding clusters GM1 (Fig. 4) and so induces formation of an Lo microdomain in model membranes (Hammond et al., 2005) and, in turn, budding (Bacia et al., 2005; Ewers et al.). Shiga toxin B subunit binds the glycolipid Gb3, and appears to operate by a similar paradigm. In this case tubular vesicles are formed, and lipid compression favoring negative curvature is thought to be the driving force (Romer et al., 2007). In each of these examples, it is clear that clustering of lipids leads to important changes in membrane structure, and so to budding. The proposed physical mechanisms remain speculative, however. These mechanisms remain a profound challenge to experimentalists, and this is an area where the growing ability of computer simulations to tackle membrane dynamics on a realistic time scale should be very helpful.
Figure 4. Protein structures that nucleate microdomains by clustering raft lipids.
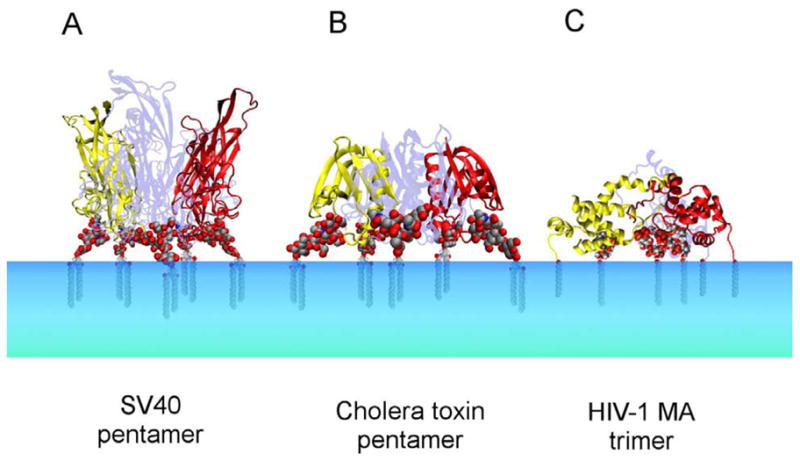
A. SV 40 VP1 pentamer bound to GM1 headgroup (Neu et al., 2008). B. Cholera toxin B subunit pentamer bound to GM1 (Merritt et al., 1994). C. Composite model of the myristoylated HIV-1 MA trimer bound to PI(4,5)P2 (Hill et al., 1996; Saad et al., 2008; Saad et al., 2006). Lipid tails were modeled in each case. Figures were generated with VMD 1.8.6.
Caveolae
Caveolae (“little caves”) are flask-shaped 60–80 nm invaginations of the plasma membrane that consist of raft lipids, caveolins 1–3, and the caveolin-associated cavins 1–4 (Hansen and Nichols, 2010). Caveolins are multiply palmitoylated, after which they are constitutively associated with cholesterol and other raft lipids. Caveolins are pentahelical proteins. Two of the caveolin helices insert deeply into the membrane, almost but not completely spanning the bilayer, while the other three helices are amphipathic and are thought to wedge themselves into the interfacial region of the membrane (Parton et al., 2006). Caveolin structure has analogies to the reticulons and DP1/Yop1 proteins that keep ER membrane tubules curved (Hu et al., 2008; Shibata et al., 2009) and to another plasma membrane raft protein, flotillin (Bauer and Pelkmans, 2006). Caveolins undergo phosphoregulation by multiple protein kinases (Pelkmans and Zerial, 2005). Curvature induction by caveolin-1 is switched off when it is phosphorylated at Ser80, which adjoins one of the predicted interfacial α-helices. The energetic book-keeping of caveolar curvature has not been worked out, but clearly must differ from the picture for conventional externally coated vesicles. Insertion of caveolin into the membrane presumably shifts the intrinsic curvature of the membrane such that the positively curved bud is the low energy state and the flat caveolin microdomain is the high energy state. Thus, once the caveolin microdomain is formed, energy input is probably needed to flatten the membrane rather than to curve it. ATP hydrolysis by protein kinases that phosphorylate caveolin might provide the thermodynamic driving force for membrane flattening. Dephosphorylation by protein phosphatases would, in this speculative scheme, allow the membrane to spring back to its low energy state.
Caveolae contain a quantized number of caveolin molecules, ~144, consistent with the formation of a highly organized coat (Pelkmans and Zerial, 2005). Cavins are soluble proteins rich in predicted coiled-coil structure and basic residues, but otherwise structurally uncharacterized. They seem to be important for caveolar structure, but the precise role of these recently discovered factors in structuring the caveolar coat is not clear. From the perspective of their quantized caveolin protein content, caveolae could be viewed as highly organized assemblies whose specialized structure and distinct curvature is caveolin-driven but lipid-stabilized. From the alternative perspective of their lipid content, caveolae could be viewed instead as a specialized, morphologically distinct membrane microdomain, whose formation is driven by lipids but stabilized by caveolin (Parton and Simons, 2007). The hybrid nature of caveolae, seemingly at once both coated vesicle and membrane microdomain, makes them a particularly fascinating example of the interplay between proteins and lipids in membrane budding.
Tetraspanin enriched microdomains
Tetraspanin-enriched microdomains (TEMs) have been implicated as another potential example of a membrane microdomain involved in budding, based mainly on their enrichment in exosomes and in the intralumenal vesicles (ILVs) of immune cell multivesicular bodies (MVBs) (Pols and Klumperman, 2009). Tetraspanins are a family of at least 32 proteins in mammals, and are defined by the presence of four transmembrane-spanning α-helices (Hemler, 2005). Tetraspanins have two extracellular domains; the second such domain, EC2, is the larger of the two. The structure of the EC2 region of CD81 has been determined, revealing an extensive dimerization interface (Kitadokoro et al., 2001). The minimal functional tetraspanin oligomer is probably a homodimer. These proteins are multiply palmitoylated on their short intracellular loop and N- and C-terminal extensions, and these palmitoylations are central to their ability to form (TEMs). Tetraspanins bind to a wide range of potential cargo proteins (Hemler, 2005), potentially coupling them to TEMs and thereby to microdomain-mediated budding. More extensive mechanistic analysis of the budding mechanism responsible for TEM traffic will be eagerly awaited.
MULTIVESICULAR BODIES
The sorting of unneeded, damaged, or dangerous plasma membrane proteins to the lysosome for degradation is carried out by endosomes (Sorkin and von Zastrow, 2009). This pathway also is central to the biogenesis of the lysosome (or yeast vacuole), as it carries newly synthesized lysosomal enzymes from the trans-Golgi to their destination. In the metazoa, the endosomal pathways have many additional roles, with the most pertinent to this review being the biogenesis of lysosome related organelles (LROs) (Raposo and Marks, 2007) and exosomes. MVBs (also known as multivesicular endosomes; Fig. 5) are key intermediates in endolysosomal transport (Gruenberg and Stenmark, 2004; Piper and Katzmann, 2007). MVBs are formed by the invagination and scission of buds from the limiting membrane of the endosome into the lumen. The mechanism of MVB biogenesis adds a new perspective to cellular membrane budding, because it is the main physiological example of membrane budding away from the cytosol.
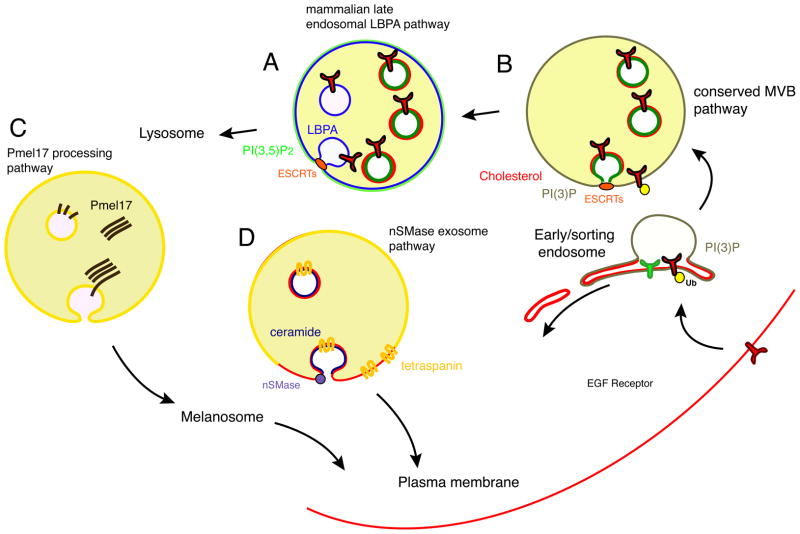
Figure 5. Different types of MVBs in animal cells make use of different paradigms for coatless budding away from cytosol.
A. ESCRT- and LBPA-dependent MVBs form from late endosomes in animal cells. LBPA is shown in blue. B. The conserved ESCRT-dependent MVB biogenesis pathway from early endosomes in yeast and animal cells. PI(3)P (green) has been directly visualized in these MVBs. Cholesterol (red) has been visualized in animal cell MVBs but it has not been directly confirmed whether these are ESCRT-dependent or not. C. Specialized formation of MVBs containing polymerized Pmel17. D. Ceramide dependent MVBs bud from raft-like and tetraspanin-enriched microdomains in animal cells. Ceramide is shown in purple.
ESCRTs and multivesicular bodies
Yeast (S. cerevisiae) has a single MVB pathway that drives the internalization of ubiquitinated transmembrane proteins into early endosomes (Piper and Katzmann, 2007). The pathway is initiated by the presence of the lipid phosphatidylinositol 3-phosphate (PI(3)P) and membrane-tethered ubiquitin moieties on the endosome surface. PI(3)P is synthesized by the class III PI 3-kinase Vps34, an enzyme essential for the progression of the endolysosomal pathway. PI(3)P is the defining marker of early endosomes, autophagosomes, and in mammalian cells, phagosomes. PI(3)P signals are recognized by FYVE and PX domain containing proteins (Misra et al., 2001). In the MVB pathway, the key FYVE domain protein is a subunit of the ESCRT-0 complex. ESCRT-0 contains five ubiquitin binding domains (UBDs) (Ren and Hurley, 2010) and clusters ubiquitinated cargo in vitro (Wollert and Hurley, 2010). Recruitment of ESCRT-0 to the early endosomal membrane initiates the recruitment of the ESCRT-I, -II, and – III complexes (Saksena et al., 2007; Williams and Urbe, 2007). Based on in vitro reconstitution, ESCRT-I and – II drive membrane budding, while ESCRT-III cleaves the bud necks to form intralumenal vesicles (ILV) (Hurley and Hanson, 2010; Wollert and Hurley, 2010; Wollert et al., 2009). The in vitro ESCRT budding reactions have been carried out with a mixture of saturated and unsaturated phospholipids and cholesterol (Wollert and Hurley, 2010), but the precise lipid requirements for the reaction have yet to be analyzed in detail.
Strikingly, ESCRT-I, -II, and III all localize to the bud neck (Wollert and Hurley, 2010). ESCRT-III subunits assemble into tubular structures in vitro and when overexpressed (Bajorek et al., 2009; Hanson et al., 2008; Lata et al., 2008). The ESCRT-III proteins coat the interior of lipid tubes created in vitro (Lata et al., 2008) and have diameters of 40–50 nm for lipid-free tubes, and ~100 nm for lipid-coated tubes. These tubes exceed the narrowest dimensions of bud necks in cells, based on just a few observations that suggest a size closer to ~20 nm (Murk et al., 2003). However, the tubes taper to a dome at their end (Fabrikant et al., 2009), and the narrowing dome at the end of the tube may represent its most important functional feature. Lipid tube extrusion by ESCRT-III seems to have no special lipid requirements, as it can be supported in vitro by a simple mixture of the unsaturated phospholipids SOPC and DOPS (Lata et al., 2008). Indeed, while most ESCRTs are unique to the eukarya, ESCRT-III is conserved in a subset of Archaea, where its functions in the membrane abscission step of cell division (Lindas et al., 2008; Samson et al., 2008). Thus the Archaeal ESCRT-III orthologs can presumably function in membrane scission with Archaeal lipids, which are radically different from eukaryotic lipids, and rich in rigid, bilayer-spanning tetraether linkages (Koga and Morii, 2005). It is thought on theoretical grounds that membrane tubes are induced by the binding of the curved ESCRT-III polymer to the membrane (Lenz et al., 2009).
ESCRT-III polymerization governs the late stage of neck development leading to scission, but it is not likely to be the main factor in the initial budding event. The initial formation of the bud is driven by the assembly of ESCRT-I and – II with one another and with the endosome membrane (Wollert and Hurley, 2010). The structure of this assembly is unknown, and the nature of the assembly is a pressing question in the field. Composite structures of the ESCRT-I and – II complexes have been developed on the basis of crystal structures of the separate components together with hydrodynamic information of the complete complexes in solution (Im and Hurley, 2008; Kostelansky et al., 2007). These structures show that multiple membrane and ESCRT-III attachment sites are separated by rigid spacers of up to 18 nm across, suggesting a mechanism to induce or at least stabilize formation of a membrane neck of roughly those dimensions. Subsequent recruitment and polymerization of ESCRT-III into spiral domes (Fabrikant et al., 2009) would then narrow and sever the neck in the current model (Hurley and Hanson, 2010). The observation that the ESCRT complexes localize to the bud neck explains how they bud membranes away from the cytosol without themselves being consumed in the bud. This mechanism stands in sharp contrast to the familiar budding of coated vesicles towards cytosol, described above. The thermodynamic driving force for the pathway is the coupling of ESCRT-III solubilization and recycling to ATP hydrolysis by the dodecameric AAA ATPase Vps4 (Babst et al., 1998; Wollert et al., 2009). While the overall thermodynamic driving force is clear, the energetic trajectory of neck-directed bud formation is currently unknown. Theoretical analysis of the membrane mechanics of this process is urgently needed, as is a better understanding of the roles of lipids.
All four ESCRT complexes are conserved between yeast and metazoa. In its broad outlines, the ESCRT dependent conversion of early endosomes into MVBs is the same in yeast and metazoa (Raiborg and Stenmark, 2009). ILVs in mammalian cells are highly enriched in cholesterol and tetraspanins (Mobius et al., 2003; van der Goot and Gruenberg, 2006). At least some of the cholesterol and tetraspanin rich ILVs in mammalian cells belong to a separate pathway from the ESCRTs, however (Simons and Raposo, 2009). Raft markers such as long-chain sphingomyelins transit through MVBs (Koivusalo et al., 2007). Consistent with a possible ESCRT-sterol connection, defects in ESCRT function block endosomal cholesterol transport in mammalian cells (Bishop and Woodman, 2000; Peck et al., 2004). In yeast, ergosterol and, more speculatively, Sna3 (Piper and Katzmann, 2007), might replace the roles of cholesterol and tetraspanins in microdomain formation. Given that ESCRTs bud membrane without a coat, and that most other coatless budding mechanisms rely on membrane microdomains of some sort, it is tempting to speculate that ESCRT-mediated budding could involve tetraspanin and cholesterol-rich domains. Very little is known about how ESCRTs might couple to such microdomains. Ubiquitination of tetraspanins (Lineberry et al., 2008) and, in yeast, Sna3 (Stawiecka-Mirota et al., 2007) has been reported. The ESCRT-III subunit Vps20 must be myristoylated for full function (Babst et al., 2002; Yorikawa et al., 2005). Yet even unmyristoylated Vps20 has a tens of nM affinity for membranes, dropping to low nM when bound to ESCRT-II (Im et al., 2009), suggesting that myristoylation is required for another reason than membrane targeting alone. The myristoyl moiety is saturated and favors Lo phase microdomain association, but single myristoylation is by itself a weak membrane anchor (Resh, 2006), and so well-adapted when reversible association is important. Another important question surrounds the nature of the PI(3)P lipid that binds to ESCRT complexes through its headgroup. Substantial levels of PI(3)P are found in the MVB lumen (Gillooly et al., 2000). A critical gap in understanding ILV formation is the lack of data on the tail compositions of the total endosomal and ILV pools of PI(3)P. The concept of an ESCRT-microdomain link is speculative. In the absence of other explanations for the unusual coatless budding by the ESCRTs, these issues call for further investigation.
Animal cells have more than one kind of MVB
Animal cells have additional pathways of MVB formation not found in yeast. The mammalian late endosomal and lysosomal lipidome contains up to 20 % of the unusual lipid lysobisphosphatidic acid (LBPA), which is not found in other organelles or in yeast. Mammalian cells have a late endosomal MVB pathway that seems to depend on LBPA microdomains that are probably induced on the lumenal leaflet by acidic pH (Matsuo et al., 2004). The ultimate thermodynamic basis for membrane curvature in the LBPA pathway would presumably be in the energy expended to pump protons into the lumen of the endosome. This late endosomal pathway also involves ESCRT proteins (Falguieres et al., 2008). The late endosomal MVB pathway should not, however, be confused with the canonical early endosomal ESCRT pathway described above, which does not involve LBPA. MVB formation is involved in the biogenesis of LROs, of which melanosomes are the most intensively studied (Raposo and Marks, 2007). In melanosome biogenesis, Pmel17 is sorted into ILVs in an ESCRT-independent reaction (Theos et al., 2006). Pmel17 is a special cargo in that its lumenal domain forms fibers, and may be an example of the lumenal assembly of a cargo helping to drive its own inward budding into the endosome.
Exosomes are 50–100 nm vesicles released from cells by the fusion of MVBs with the plasma membrane (Simons and Raposo, 2009). At least one population of exosomes is produced by an ESCRT-independent pathway in which neutral sphingomyelinase, acting from the cytosolic face of the membrane, hydrolyzes sphingomyelin to ceramide (Trajkovic et al., 2008). The formation of ILVs by sphingomyelinase has been reconstituted in vitro using GUVs with pre-existing phase separation (Trajkovic et al., 2008). Sphingomyelinase cleavage of the phosphodiester bond between ceramide and the SM headgroup provides a potential mechanism to put energy into this budding pathway and make it thermodynamically irreversible. Ceramide-induced ILVs bud exclusively from the Lo phase (Trajkovic et al., 2008). Ceramide has several special properties, including a small headgroup that would favor its presence in the inner leaflet of the ILV, and an ability to self-associate through headgroup hydrogen bonding. It is not clear which properties of ceramide are most important for ILV formation. Exosomes produced by the sphingomyelinase pathway are highly enriched in the tetraspanin CD63, suggestive of a coupling between TEMs and ceramide domains. Of the three pathways described above, the latter two are, based on current knowledge, ESCRT-independent. It will be interesting to see if there are ever circumstances under which the ESCRTs cooperate with the melanosome or ceramide pathways.
VIRAL BUDDING
Enveloped virus budding: with ESCRTs and without
Membrane budding is an essential part of the life cycle of enveloped viruses. Most, but not all, enveloped viruses bud from cells by co-opting the host ESCRT machinery (Bieniasz, 2006; Morita and Sundquist, 2004; Welsch et al., 2007), whose role in budding of vesicles in MVBs was described above (Fig. 6). Virus budding, like MVB formation, involves budding away from cytosol. In the well-studied example of HIV-1, formation of the initial plasma-membrane attached bud is driven by the energetically favorable self-assembly of the CA domain of Gag into hexamers (Briggs et al., 2009; Wright et al., 2007). CA does not bind directly to membranes, so the energy of CA self-assembly is transduced to the membrane through the membrane-binding MA domain, part of the same polypeptide chain at this stage in HIV-1 assembly (Hill et al., 1996). Recombinant HIV-1 Gag constructs lacking a part of the membrane-binding MA domain and all of the ESCRT-binding p6 domain are able to assemble with RNA to form spherical shells in vitro, in the absence of membranes (Campbell et al., 2001). These lipid-free shells are slightly smaller than authentic immature HIV-1 virions, with the differences accounted for by the absence of membrane and the MA domain (Briggs et al., 2009). The shells assemble via CA domain hexamers, which cannot pack into a sphere, and therefore a few gaps remain in an otherwise almost complete lattice (Briggs et al., 2009). However, in authentic released HIV-1 particles, the Gag shells are only 60 % complete on average (Carlson et al., 2008). Could such an incomplete shell scaffold bud formation? Below, the role of membrane microdomains as another key contributor to HIV-1 bud formation will be described.
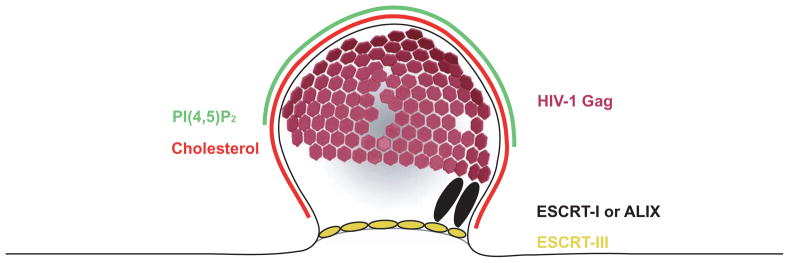
Figure 6. Lipids and ESCRTs in HIV-1 Assembly.
Apart from viral proteins, the release of HIV-1 requires both specific cellular lipids and proteins, which are recruited to the budding site by the viral Gag protein. Gag assembles to an imperfect hexagonal lattice (purple) on the plasma mebrane. It binds the plasma membrane marker PI(4,5)P2 through a specific binding site in its N-terminus. PI(4,5)P2 (green), cholesterol (red) and certain other raft lipids are enriched in the viral membrane compared to the plasma membrane. Through its C-terminus, Gag recruits the ESCRT proteins to the budding site. Gag can bind both ESCRT-I and ALIX (black), which will both recruit ESCRT-III (yellow) to the budding site.
In contrast to the self-encoded ability of HIV-1 to form attached buds, the release of these buds from the host cell requires the cooptation of the host cell ESCRT machinery. ESCRT-recruiting late domain motifs have been identified in most genera of enveloped viruses (Bieniasz, 2006; Chen and Lamb, 2008; Freed, 2002; Morita and Sundquist, 2004). HIV-1 is the prototypical example of an ESCRT-dependent virus. HIV-1 engages the ESCRT-I complex though a PTAP motif in the p6 region of its Gag protein (Huang et al., 1995), and interference with this interaction dramatically reduces HIV-1 release (Demirov et al., 2002a; Demirov et al., 2002b; Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001). To make matters more complicated, efficient release can be rescued by overexpressing the ESCRT-associated protein ALIX, which binds to another motif in Gag p6, YPXnL (Fisher et al., 2007; Usami et al., 2007). Defects in both of these interactions can be rescued by overexpression of HECT domain ubiquitin ligases (Chung et al., 2008; Jadwin et al.; Usami et al., 2008). All of these interaction serve the same ultimate purpose of recruiting ESCRT-III to the nascent viral bud for scission, which is thought to be carried out by the same process as for cleavage of ILVs in the lumen of MVBs (Hurley and Hanson, 2010).
If HIV-1 is the archetype of a virus dependent on host cell membrane scission machinery, other viruses seem to carry out both budding and scission entirely with virally encoded proteins. The membrane-associated matrix protein of Newcastle disease virus (NDV, a paramyxovirus) induces both bud formation and scission when assembled on model membranes (Shnyrova et al., 2007). The release of virus-like particles is stimulated by negatively charged lipids and cholesterol. NDV contains a late domain motif identical to that of the closely related ESCRT-dependent paramyxovirus SV5 (Schmitt et al., 2005). The function, if any, of ESCRTs in NDV release might be to accelerate a vesiculation which the virus already is capable of performing. The matrix protein of vesicular stomatitis virus (VSV) is capable of inducing membrane buds in vitro (Solon et al., 2005). In vitro VSV budding occurs in a simple mixture of acidic phospholipids, and appears to be driven by self-assembly of the matrix protein. The in vitro buds are not cleaved by the matrix protein, indicating the requirement for additional scission factors. Indeed, VSV budding from cells requires an ESCRT-I-binding late domain (Irie et al., 2004). Why does the matrix protein of one putatively ESCRT-dependent virus, NDV, support both budding and scission on its own, while another, that of VSV, supports only formation of attached buds? It is too soon to say if these are intrinsic differences between these viruses, or relate merely to experimental differences.
Even for the archetypal ESCRT-dependent virus HIV-1, there seem to be circumstances in which ESCRT-dependence can be circumvented. The effect of mutating its two ESCRT-interacting late domains depends on the cell type, with primary monocyte-derived macrophages and the Jurkat T-cell line retaining >20% particle release even when both domains were inactivated (Fujii et al., 2009). Further, replacing the C-terminal part of Gag, including the RNA-binding nucleocapsid (NC) domain and the late domain-containing p6 domain, with a leucine zipper motif preserves efficient particle release despite absence of ESCRT-interacting motifs (Zhang et al., 1998). Deleting part of NC and the flanking p1 sequence has the same effect of making HIV-1 release independent of a functional ESCRT machinery (Popova et al., 2010). All in all, these findings show that a baseline of ESCRT-independent HIV-1 release exists, and that this level can be raised by subtle alterations in the Gag protein. The studies mentioned above quantified the amount of virus released on a timescale of 16–72 h, and it is still possible that the microscopic kinetics of the budding process, which takes place on the timescale of 5–25 min (Ivanchenko et al., 2009; Jouvenet et al., 2008), could have been more severely compromised. The ESCRT-independent scission observed for NDV in vitro and for certain HIV-1 variants in vivo suggests that in some cases the role of ESCRTs is merely to speed up the final stage of release. In other cases, such as wild-type HIV-1, the ESCRTs seem to have a deeper role in viral morphogenesis(Carlson et al., 2008).
Membrane microdomains and influenza budding
The influenza virus is the best-characterized case of enveloped virus budding without an ESCRT. Influenza neither has a typical late domain sequence, nor is it inhibited by overexpressing a dominant negative Vps4 (Bruce et al., 2009; Chen et al., 2007). Influenza virus is one of the most studied examples of a lipid raft-associated virus. Its hemagglutinin (HA) and neuraminidase (NA) proteins are associated with lipid rafts through their transmembrane domains (Barman et al., 2004), and the membrane of released influenza virions has a pronounced raft character with higher order than that of non-raft associated enveloped virus (Polozov et al., 2008; Scheiffele et al., 1999). This raft association serves to cluster HA on the plasma membrane, thus increasing its concentration on the released particles (Barman et al., 2004; Takeda et al., 2003), and it is further involved in the sorting of HA and NA to the apical face of polarized cells (Barman et al., 2004). A budding mechanism for influenza which reconciles its ESCRT-independence and raft-association was recently assigned to the viral ion channel M2 (Rossman, 2010). This protein has a conserved amphipathic helix which is sufficient for vesicle scission in a minimal in vitro system, where is predominantly acts at the border between Ld and cholesterol-enriched Lo domains. M2 was further localized to the neck of budding influenza particles by immuno-EM, and mutations disrupting its amphipathic helix appeared to increase the number of virus buds still being associated with the cell. This is the first detailed description of an ESCRT-independent viral budding mechanism, and it will be interesting to see if it is parallelled in other systems.
How HIV-1 uses raft and non-raft lipids to bud from cells
The HIV-1 membrane is highly ordered (Aloia et al., 1993; Lorizate et al., 2009), with elevated levels of cholesterol and certain other raft lipids (GM3 and ceramide) compared to the plasma membrane from which they bud (Brugger et al., 2006; Chan et al., 2008). Cholesterol depletion blocks HIV-1 particle release by inhibiting membrane binding and multimerization of Gag (Ono et al., 2007). Thus, the lipid segregation at HIV-1 budding sites clearly has a functional role in the formation and release of HIV-1 particles. What, precisely, is this role? It is tempting to speculate that microdomain formation not only contributes to the normal HIV-1 budding pathway, but facilitates the ESCRT-independent budding noted above for unusual HIV-1 Gag constructs. However, note that cholesterol depletion actually promotes HIV-1 budding in the case of the PTAP-defective virus that buds independent of the ESCRTs (Ono and Freed, 2001). This suggests that as with the ESCRT-independent budding of influenza, cholesterol has multiple roles.
HIV-1 and other retroviruses use protein-lipid interactions to target their assembly to the plasma membrane. The N-terminal matrix domain (MA) of HIV-1 Gag has a basic surface (Hill et al., 1996) and a covalently bound myristyl fatty acid chain which is necessary for virus release (Ono and Freed, 1999). The “myristyl switch” model describes how this myristyl moiety is in a buried conformation in the monomeric cytosolic protein, and becomes exposed upon Gag oligomerization (Saad et al., 2008; Saad et al., 2006; Tang et al., 2004). Thus, the membrane binding of the Gag protein is linked to its multimerization and assembly into a lattice. The weak membrane affinity of the MA myristate and non-specific interactions between the basic face of MA with bulk acidic phospholipids are not sufficient for efficient HIV-1 particle release. For release to occur, the particle assembly must be targeted either to the plasma membrane, or to membranous compartments which can fuse with the plasma membrane, leading to virion release. PI(4,5)P2, described above as a key factor in clathrin coated vesicle formation, is the defining lipid marker of the plasma membrane (McLaughlin et al., 2002). The matrix domain of HIV-1 Gag targets specifically to the plasma membrane by binding tightly to the phosphoinositide PI(4,5)P2, and this interaction is required for Gag assembly and HIV-1 budding (Ono et al., 2004).
How can the raft dependence of HIV-1 Gag assembly be reconciled with its dependence on PI(4,5)P2? PI(4,5)P2 is generally considered a non-raft lipid, although the microscopic analysis of the tail composition of different pools of PI(4,5)P2 is not elaborated to the point where this can be said with certainty for all PI(4,5)P2. The apparent answer to this question highlights the frightening ingenuity of HIV-1 in co-opting cellular systems. The binding of PI(4,5)P2 to Gag triggers the myristyl switch, leading to exposure of the buried myristoyl group (Saad et al., 2006). In the solution structure of the myristoylated MA complex bound to a short-chain PI(4,5)P2, the myristoyl and the 1′ fatty acid tail of PI(4,5)P2 extend into the lipid bilayer, whereas the 2′ fatty acid tail of PI(4,5)P2 becomes buried in a pocket in MA vacated by ejection of the myristate (Saad et al., 2006). In the current view of this mechanism, the 1′ tail is preferentially saturated and the 2′ preferentially unsaturated. Thus the MA-PI(4,5)P2 complex would in this scheme expose two saturated chains, transforming it into a raftophile. It will be interesting to see if any cellular budding proteins-perhaps including the myristoylated ESCRT-III protein Vps20- use similar mechanisms to bridge raft and non-raft lipids. HIV-1 release, with its exploitation of so many of the physiological budding paradigms described in a single event, is one of the most remarkable illustrations of how the dance between proteins and lipids leads to membrane buds.
CONCLUDING REMARKS
We hope to have provided a few examples of how the geometry, topology, and energetics of some selected membrane budding events in cells are adapted to their biological functions. Transport through cytosolic vesicular carriers of membrane proteins that have cytosolic tails is carried out most often through vesicles coated by the clathrin, COP I, and COP II complexes, which we now know to have structural similarities to one another (Lee and Goldberg, 2010). The cytosolic tails provide the signal for assembly, coat proteins scaffold the membrane, amphipathic helix and BAR domain factors help bend the membrane, and uncoating-coupled hydrolysis of ATP or GTP provides the thermodynamic driving force. In the evolution of coats, the benefits of flexibility have been traded off against scaffolding power, with clathrin apparently optimized for flexibility, while COP II is optimized as a more potent membrane-curving scaffold.
Viruses and toxins often enter cells by engaging with host transmembrane proteins and co-opting coat-dependent budding mechanisms, but the defensive evolution of host organisms combats this. Lipid-based entry through the induction of membrane microdomains, as exemplified by SV40, Shiga toxin, and cholera toxin illustrate one way pathogens avoid having to rely on mutable surface proteins of the host. The physical basis of this entry mechanism uses completely different principles to the same functional end. Caveolae present a fascinating hybrid of a protein scaffolding and membrane microdomain mechanisms. The real cellular function(s) of caveolae are enigmatic, leaving us for now in the dark as to the evolutionary drive for such unusual structures.
The ESCRT system, the main interest of our laboratory, is adapted for budding away from the cytosol in the opposite topology of conventional coated vesicles. The ESCRT system evolved to avoid the use of a protein coat because of this unusual topology. The unique mechanism by which ESCRTs stabilize and sever membrane buds has become much clearer over the past year. However, the pathway of early bud formation, before the bud neck has contracted enough for the ESCRT proteins to bridge across it, is still obscure. This led us to ask if membrane microdomains might have a role in ESCRT-mediated bud formation. If this were the case, membrane microdomains might serve as a unifying principle connecting the diverse types of ESCRT-dependent and microdomain-dependent MVBs in animal cells. The various ESCRT- and microdomain-dependent flavors of enveloped virus budding mirror the distinct varieties of animal cell MVBs. This is not surprising given that these two processes share the same unusual property of budding away from cytosol. Microdomains and ESCRTs have the same advantage for budding away from cytosol, in that cytosolic coat proteins need not be irreversibly consumed in the process.
Membrane budding and the related topic of membrane tubulation have become exceptionally vibrant fields, driven by advances in technology. Computational resources now allow sophisticated simulations of budding (Reynwar et al., 2007). Reconstitution of budding events from completely defined systems (Bremser et al., 1999; Matsuoka et al., 1998; Romer et al., 2007; Wollert and Hurley, 2010) has established molecular mechanisms in several cases, and opened the door to more sophisticated biophysical analysis (Bassereau, 2010). Electron microscopy has been the foundation of our understanding of membrane budding in cells since the beginning. Looking forward, advanced electron tomography will undoubtedly shape our future views of how membranes bud, as classical electron microscopy has in the past and present. As in other areas of cell biology, rapid advances in live cell imaging are making powerful and ever-increasing contributions. Membrane budding is a required part of the life cycle of two of the most dangerous human pathogens, HIV and influenza, and insights into the fundamental nature of these budding events is perhaps the most urgently needed of all.
Acknowledgments
We thank E. Freed, G. Raposo, W. Prinz, M. Marks, J. Gruenberg, and J. Bonifacino for comments on drafts of the manuscript, and many colleagues for stimulating discussions. Research in the Hurley laboratory is supported the Intramural program of the National Institutes of Health, NIDDK and IATAP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aloia RC, Tian HR, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci U S A. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacia K, Schwille P, Kurzchalia T. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc Natl Acad Sci U S A. 2005;102:3272–3277. doi: 10.1073/pnas.0408215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WMB, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–U795. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S, Adhikary L, Chakrabarti AK, Bernas C, Kawaoka Y, Nayak DP. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza A virus neuraminidase in raft association and virus budding. J Virol. 2004;78:5258–5269. doi: 10.1128/JVI.78.10.5258-5269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassereau P. Division of labour in ESCRT complexes. Nat Cell Biol. 2010;12:422–423. doi: 10.1038/ncb0510-422. [DOI] [PubMed] [Google Scholar]
- Bauer M, Pelkmans L. A new paradigm for membrane-organizing and -shaping scaffolds. FEBS Lett. 2006;580:5559–5564. doi: 10.1016/j.febslet.2006.08.077. [DOI] [PubMed] [Google Scholar]
- Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci U S A. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom M, Evands E, Mouritsen OG. Physical properties of the fluid lipid-bilayer component of cell membranes: a perspective. Q Rev Biophys. 1991;24:293–397. doi: 10.1017/s0033583500003735. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Lippincott-Schwartz J. Coat proteins: shaping membrane transport. Nat Rev Mol Cell Biol. 2003;4:409–414. doi: 10.1038/nrm1099. [DOI] [PubMed] [Google Scholar]
- Bremser M, Nickel W, Schweikert M, Ravazzola M, Amherdt M, Hughes CA, Sollner TH, Rothman JE, Wieland FT. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Briggs JAG, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: Formation and function of clathrin- coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Bruce EA, Medcalf L, Crump CM, Noton SL, Stuart AD, Wise HM, Elton D, Bowers K, Digard P. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krasslich HG. The HIV lipidome: A raft with an unusual composition. Proc Natl Acad Sci U S A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci U S A. 2001;98:10875–10879. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Brigg JAG, Glass B, Riches JD, Simon MN, Johnson MC, Muller B, Grunwald K, Krausslich HG. Three-Dimensional Analysis of Budding Sites and Released Virus Suggests a Revised Model for HIV-1 Morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, De Marco A, Oberwinkler H, Habermann A, Briggs JAG, Krausslich HG, Grunewald K. Cryo Electron Tomography of Native HIV-1 Buding Sites. PLoS Path. 2010 doi: 10.1371/journal.ppat.1001173. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R, Uchil PD, Jin J, Shui GH, Ott DE, Mothes W, Wenk MR. Retroviruses Human Immunodeficiency Virus and Murine Leukemia Virus Are Enriched in Phosphoinositides. J Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiantia S, Kahya N, Ries J, Schwille P. Effects of ceramide on liquid-ordered domains investigated by simultaneous AFM and FCS. Biophys J. 2006;90:4500–4508. doi: 10.1529/biophysj.106.081026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm EM, Pelkmans L, Kartenbeck J, Mezzacasa A, Kurzckalia T, Helenius A. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J Cell Biol. 2005;168:477–488. doi: 10.1083/jcb.200407113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002a;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002b;76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Boll W, van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Greene LE. Multiple roles of auxilin and Hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- Elson EL, Fried E, Dolbow JE, Genin GM. Phase Separation in Biological Membranes: Integration of Theory and Experiment. In Annual Review of Biophysics. 2010;39:207–226. doi: 10.1146/annurev.biophys.093008.131238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H, Romer W, Smith AE, Bacia K, Dmitrieff S, Chai WG, Mancini R, Kartenbeck J, Chambon V, Berland L, et al. GM1 structure determines SV40-induced membrane invagination and infection. Nat Cell Biol. 2010;12:11–U36. doi: 10.1038/ncb1999. [DOI] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JAG, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comp Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falguieres T, Luyet PP, Bissig C, Scott CC, Velluz MC, Gruenberg J. In Vitro Budding of Intralumenal Vesicles into Late Endosomes Is Regulated by Alix and Tsg101. Mol Biol Cell. 2008;19:4942–4955. doi: 10.1091/mbc.E08-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsad K, De Camilli P. Mechanisms of membrane deformation. Curr Opin Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai QT, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng YF, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Freed EO. Viral late domains. J Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Saez AJ, Schwille P. Stability of lipid domains. FEBS Lett. 2010;584:1653–1658. doi: 10.1016/j.febslet.2009.12.036. [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nature Reviews Molecular Cell Biology. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Hammond AT, Heberle FA, Baumgart T, Holowka D, Baird B, Feigenson GW. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc Natl Acad Sci U S A. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nature Reviews Molecular Cell Biology. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Nichols BJ. Exploring the caves: cavins, caveolins and caveolae. Trends Cell Biol. 2010;20:177–186. doi: 10.1016/j.tcb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- Heller H, Schaefer M, Schulten K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal phases. J Phys Chem. 1993;97:8343–8360. [Google Scholar]
- Hemler ME. Tetraspanin functions and associated microdomains. Nature Reviews Molecular Cell Biology. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo Proteins Are Nucleators of Clathrin-Mediated Endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Keen JH, Amende LM, Lippoldt RE, Prasad K. Deep etch visualization of 27S clathrin- A tetrahedral tetramer. J Cell Biol. 1987;105:1999–2009. doi: 10.1083/jcb.105.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc Natl Acad Sci U S A. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen L, Meyerhoiz A, Groos S, Ungewickell EJ. Bending a membrane: How clathrin affects budding. Proc Natl Acad Sci U S A. 2006;103:8715–8720. doi: 10.1073/pnas.0600312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JJ, Shibata Y, Voss C, Shemesh T, Li ZL, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- Huang MJ, Orenstein JM, Martin MA, Freed EO. P6(Gag) Is Required for Particle-Production from Full-Length Human-Immunodeficiency-Virus Type-1 Molecular Clones Expressing Protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT complexes: It’s all in the neck. Nature Reviews Molecular and Cell Biology. 2010 doi: 10.1038/nrm2937. advance online publication, June 30, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14:902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Wollert T, Boura E, Hurley JH. Structure and Function of the ESCRT-II-III Interface in Multivesicular Body Biogenesis. Dev Cell. 2009;17:234–243. doi: 10.1016/j.devcel.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Licata JM, Jayakar HR, Whitt MA, Bell P, Harty RN. Functional analysis of late-budding domain activity associated with the PSAP motif within the vesicular stomatitis virus M protein. J Virol. 2004;78:7823–7827. doi: 10.1128/JVI.78.14.7823-7827.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko S, Godinez WJ, Lampe M, Kräusslich HG, Eils R, Rohr K, Bräuchle C, Müller B, Lamb DC. Dynamics of HIV-1 assembly and release. PloS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Honing S, Evans PR, Owen DJ. A Large-Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the AP2 Clathrin Adaptor Complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadwin JA, Rudd V, Sette P, Challa S, Bouamr F. Late Domain-Independent Rescue of a Release-Deficient Moloney Murine Leukemia Virus by the Ubiquitin Ligase Itch. J Virol. 2010;84:704–715. doi: 10.1128/JVI.01319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M, Fujita A, Chadda R, Nixon SJ, Kurzchalia TV, Sharma DK, Pagano RE, Hancock JF, Mayor S, Parton RG. Ultrastructural identification of uncoated caveolin-independent early endocytic vehicles. J Cell Biol. 2005;168:465–476. doi: 10.1083/jcb.200407078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y, Morii H. Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci, Biotechnol, Biochem. 2005;69:2019–2034. doi: 10.1271/bbb.69.2019. [DOI] [PubMed] [Google Scholar]
- Koivusalo M, Jansen M, Somerharju P, Ikonen E. Endocytic trafficking of sphingomyelin depends on its acyl chain length. Mol Biol Cell. 2007;18:5113–5123. doi: 10.1091/mbc.E07-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky MS, Schluter C, Tam YYC, Lee S, Ghirlando R, Beach B, Conibear E, Hurley JH. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger H, Weissenhorn W. Helical Structures of ESCRT-III are Disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Goldberg J. Structure of Coatomer Cage Proteins and the Relationship among COPI, COPII, and Clathrin Vesicle Coats. Cell. 2010;143:123–132. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MCS, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Lenz M, Crow DJG, Joanny JF. Membrane Buckling Induced by Curved Filaments. Phys Rev Lett. 2009;103 doi: 10.1103/PhysRevLett.103.038101. [DOI] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proceedings of the National Academy of Sciences USA. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineberry N, Su L, Soares L, Fathman CG. The single subunit transmembrane E3 ligase gene related to anergy in lymphocytes (GRAIL) captures and then ubiquitinates transmembrane proteins across the cell membrane. J Biol Chem. 2008;283:28497–28505. doi: 10.1074/jbc.M805092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid Rafts As a Membrane-Organizing Principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Lipowsky R. Budding of membranes induced by intramembrane domains. Journal of Phys II France. 1992;2:1825–1840. [Google Scholar]
- Liu J, Kaksonen M, Drubin DG, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc Natl Acad Sci U S A. 2006;103:10277–10282. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorizate M, Brugger B, Akiyama H, Glass B, Muller B, Anderluh G, Wieland FT, Krausslich HG. Probing HIV-1 Membrane Liquid Order by Laurdan Staining Reveals Producer Cell-dependent Differences. J Biol Chem. 2009;284:22238–22247. doi: 10.1074/jbc.M109.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul M, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Wang JY, Gambhir A, Murray D. PIP2 AND proteins: Interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Sarfaty S, Vandenakker F, Lhoir C, Martial JA, Hol WGJ. Crystal structure of cholera toxin B-pentamer bound to receptor GM1 pentasaccharide. Protein Sci. 1994;3:166–175. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Miller GJ, Hurley JH. Recognizing phosphatidylinositol 3-phosphate. Cell. 2001;107:559–562. doi: 10.1016/s0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- Mobius W, van Donselaar E, Ohno-Iwashita Y, Shimada Y, Heijnen HFG, Slot JW, Geuze HJ. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic. 2003;4:222–231. doi: 10.1034/j.1600-0854.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Murk JLAN, Humbel BM, Ziese U, Griffith JM, Posthuma G, Slot JW, Koster AJ, Verkleij AJ, Geuze HJ, Kleijmeer MM. Endosomal compartmentalization in three dimensions: Implications for membrane fusion. Proceedings of the National Academy of Sciences of the USA. 2003;100:13332–13337. doi: 10.1073/pnas.2232379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu U, Woellner K, Gauglitz G, Stehle T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc Natl Acad Sci U S A. 2008;105:5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossal R. Energetics of clathrin basket assembly. Traffic. 2001;2:138–147. doi: 10.1034/j.1600-0854.2001.020208.x. [DOI] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Binding of human immunodeficiency virus type 1 Gag to membrane: Role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Waheed AA, Freed EO. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology. 2007;360:27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Hanzal-Bayer M, Hancock JF. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J Cell Sci. 2006;119:787–796. doi: 10.1242/jcs.02853. [DOI] [PubMed] [Google Scholar]
- Parton RG, Simons K. The multiple faces of caveolae. Nature Reviews Molecular Cell Biology. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- Peck JW, Bowden ET, Burbelo PD. Structure and function of human Vps20 and Snf7 proteins. Biochem J. 2004;377:693–700. doi: 10.1042/BJ20031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Zerial M. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature. 2005;436:128–133. doi: 10.1038/nature03866. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polozov IV, Bezrukov L, Gawrisch K, Zimmerberg J. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat Chem Biol. 2008;4:248–255. doi: 10.1038/nchembio.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MS, Klumperman J. Trafficking and function of the tetrspanin CD63. Exp Cell Res. 2009;315:1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Popova E, Popov S, Gottlinger HG. Human Immunodeficiency Virus Type 1 Nucleocapsid p1 Confers ESCRT Pathway Dependence. J Virol. 2010;84:6590–6597. doi: 10.1128/JVI.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL. Conserved Functions of Membrane Active GTPases in Coated Vesicle Formation. Science. 2009;325:1217–1220. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS. Melanosomes - dark organelles enlighten endosomal membrane transport. Nature Reviews Molecular Cell Biology. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XF, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29:1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- Reynwar BJ, Illya G, Harmandaris VA, Muller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Rodal SK, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, Aly MRE, Fraisier V, Florent JC, Perrais D, et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–U673. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010 doi: 10.1016/j.cell.2010.08.029. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C, Stagg SM. New Insights into the Structural Mechanisms of the COPII Coat. Traffic. 2010;11:303–310. doi: 10.1111/j.1600-0854.2009.01026.x. [DOI] [PubMed] [Google Scholar]
- Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, Soheilian F, Freed EO, Summers MF. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, Torgersen ML, Raa HA, Van Deurs B. Clathrin-independent endocytosis: from nonexisting to an extreme degree of complexity. Histochemistry and Cell Biology. 2008;129:267–276. doi: 10.1007/s00418-007-0376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Hu JJ, Kozlov MM, Rapoport TA. Mechanisms Shaping the Membranes of Cellular Organelles. Annu Rev Cell Dev Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- Shnyrova AV, Ayllon J, Mikhalyov II, Villar E, Zimmerberg J, Frolov VA. Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J Cell Biol. 2007;179:627–633. doi: 10.1083/jcb.200705062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Raposo G. Exosomes - vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Solon J, Gareil O, Bassereau P, Gaudin Y. Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system. J Gen Virol. 2005;86:3357–3363. doi: 10.1099/vir.0.81129-0. [DOI] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nature Reviews Molecular Cell Biology. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- Stagg SM, LaPointe P, Razvi A, Gurkan C, Potter CS, Carragher B, Balch WE. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, Urban-Grimal D, Morsomme P. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic. 2007;8:1280–1296. doi: 10.1111/j.1600-0854.2007.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson HEM, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-Arriving Syp1p and Ede1p Function in Endocytic Site Placement and Formation in Budding Yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil A, Gaidarov I, Kobylarz K, Lampson MA, Keen JH, McGraw TE. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci U S A. 1999;96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci U S A. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci U S A. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos AC, Truschel ST, Tenza D, Hurbain I, Harper DC, Berson JF, Thomas PC, Raposo G, Marks MS. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev Cell. 2006;10:343–354. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular Endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Traub LM, Wendland B. Cell Biology: How to don a coat. Nature. 2010;465:556–557. doi: 10.1038/465556a. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot FG, Gruenberg J. Intra-endosomal membrane traffic. Trends Cell Biol. 2006;16:514–521. doi: 10.1016/j.tcb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Veatch SL, Soubias O, Keller SL, Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc Natl Acad Sci U S A. 2007;104:17650–17655. doi: 10.1073/pnas.0703513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA. Sheets, ribbons and tubules - how organelles get their shape. Nature Reviews Molecular Cell Biology. 2007;8:258–264. doi: 10.1038/nrm2119. [DOI] [PubMed] [Google Scholar]
- Welsch S, Muller B, Krausslich HG. More than one door - Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nature Reviews Molecular Cell Biology. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–873. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Qian HY, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nature Reviews Molecular Cell Biology. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]