Abstract
The Atp12p protein of Saccharomyces cerevisiae is required for the assembly of the F1 component of the mitochondrial F1F0 ATP synthase. In this report, we show that the F1 α–subunit co-precipitates and co-purifies with a tagged form of Atp12p adsorbed to affinity resins. Moreover, sedimentation analysis indicates that in the presence of the F1 α–subunit, Atp12p behaves as a particle of higher mass than is observed in the absence of the α–subunit. Yeast two-hybrid screens confirm the direct association of Atp12p with the α–subunit and indicate that the binding site for the assembly factor lies in the nucleotide-binding domain of the α–subunit, between Asp133 and Leu322. These studies provide the basis for a model of F1 assembly in which Atp12p is released from the α–subunit in exchange for a β–subunit to form the interface that contains the non-catalytic adenine nucleotide-binding site.
Keywords: Atp12p/F1-ATPase/mitochondria/protein assembly/Saccharomyces cerevisiae
Introduction
ATP synthesis during respiration is catalyzed in eukaryotic cells by the mitochondrial ATP synthase (F1F0 complex) (Penefsky and Cross, 1991; Boyer, 1993). The catalytic unit of the enzyme, F1, is composed of five different types of subunits in the stoichiometric ratio α3β3γδɛ (Penefsky and Cross, 1991; Boyer, 1993). The three-dimensional structures of F1 from bovine heart (Abrahams et al., 1994), rat liver (Bianchet et al., 1998) and yeast mitochondria (Stock et al., 1999) show that the α- and β–subunits are arranged, in alternating fashion, in a hexamer that surrounds the N– and C–termini of the γ–subunit. The interfaces between α- and β–subunits mark the locations of three catalytic and three non-catalytic sites in the enzyme (Abrahams et al., 1994; Bianchet et al., 1998; Stock et al., 1999).
In Saccharomyces cerevisiae, the F1 subunits are encoded by nuclear genes (Takeda et al., 1985, 1986; Guelin et al., 1993; Giraud and Velours, 1994; Paul et al., 1994) and, therefore, are synthesized in the cytoplasm and then imported into mitochondria (Tokatlidis and Schatz, 1999). Like other mitochondrial proteins, the F1 subunits are imported to the matrix compartment as unfolded polypeptide chains, and their folding is facilitated by the Hsp60 and Hsp10 proteins (Hendrick and Hartl, 1993). The final steps in the formation of functional F1 require two proteins called Atp11p and Atp12p (Ackerman and Tzagoloff, 1990; Bowman et al., 1991; Ackerman et al., 1992). Yeast mutants that are deficient for either Atp11p or Atp12p accumulate both the F1 α- and β–subunits in large protein aggregates (Ackerman and Tzagoloff, 1990). Likewise, in yeast deficient for the α–subunit, the β–subunit aggregates, and the α–subunit aggregates in a β–subunit null strain (Ackerman and Tzagoloff, 1990). In contrast, the α- and β–subunits remain soluble in mitochondria of γ (Paul et al., 1994), δ (Giraud and Velours, 1994) or ɛ (Guelin et al., 1993) null mutants, despite the fact that the absence of any of these subunits blocks assembly of functional F1. Interestingly, in the absence of the γ–subunit, there is evidence to suggest that αβ oligomers are formed (Paul et al., 1994). Thus, it appears that aggregation of the F1 α- and β–subunits occurs under conditions in which formation of αβ heterodimers is not possible either because the α- or the β–subunit is missing, or because a protein (i.e. Atp11p or Atp12p) that specifically protects the α- and β–subunits from non-productive interactions is missing.
Atp11p recently has been shown to bind to the β–subunit of F1 (Wang and Ackerman, 2000). Previous work with Atp12p indicated that the functional domain of this protein is located in the C–terminal half (Wang and Ackerman, 1998), but did not provide clues about its mechanism. Here we show that Atp12p interacts specifically with a region of 190 amino acids in the nucleotide-binding domain of the F1 α–subunit, and propose a comprehensive model for the action of both Atp11p and Atp12p in F1 assembly.
Results
Affinity adsorption of biotinylated, His-tagged Atp12p (BtHis-Atp12p)
Yeast strains carrying the plasmid pG57/BTHIS produce a form of Atp12p in which a His6 tag, preceded by a sequence for in vivo biotinylation, is present at the N–terminus of the mature protein. This protein is synthesized in the cytosol as a chimera that includes the mitochondrial targeting sequence of Atp11p at the N–terminus, just proximal to the tag sequences. The migration of BtHis-Atp12p in acrylamide gels indicates that cleavage of the Atp11p leader peptide does not occur (see below). However, even in this precursor form, BtHis-Atp12p provides full respiratory competence to the ATP12 deletion strain aW303ΔATP12.
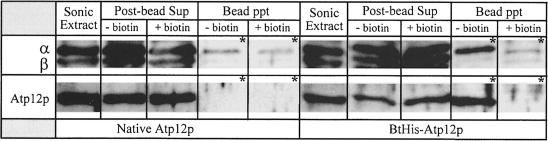
Affinity precipitation of BtHis-Atp12p from mitochondrial extracts of aW303ΔATP12/pG57/BTHIS transformants reveals that the F1 α–subunit is co-precipitated selectively (Figure 1). Neither BtHis-Atp12p nor the F1 α–subunit are precipitated under conditions in which excess biotin is included in the incubation medium.
Fig. 1. Western blots of mitochondrial extracts following affinity precipitation with avidin–Sepharose beads. Protein samples from yeast that produce native Atp12p (control) are shown on the left; protein samples from yeast that produce BtHis-Atp12p are shown on the right. Aliquots of the proteins released following sonic irradiation of mitochondria (Sonic Extract), of the initial supernatants recovered following precipitation of sonic extracts with avidin–Sepharose beads (Post-bead Sup) and of the samples precipitated with avidin beads (Bead Ppt) were loaded on a 12% SDS–polyacrylamide gel, transferred to nitrocellulose and probed either with a mixture of antibodies against the α- and β–subunits or with Atp12p antiserum. To detect even small quantities of F1 α- and β–subunits in the bead precipitates (see *), there was eight times more protein loaded on the gel from these fractions versus the amount loaded to visualize the same proteins in the sonic extract and supernatant fractions. Plus or minus biotin indicates whether or not free biotin was included in the incubation buffer during affinity precipitation.
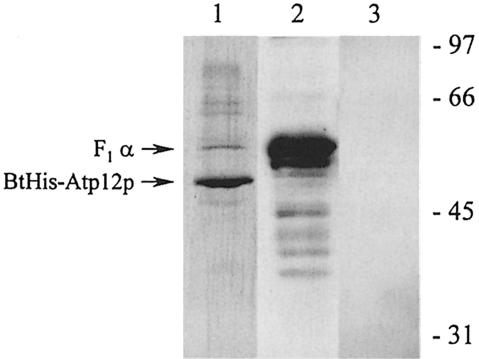
In other work, BtHis-Atp12p was purified from yeast mitochondria by means of two sequential affinity chromatographic steps with Ni-NTA and streptavidin, respectively. The recovered protein was ∼95% pure as judged by SDS–PAGE and silver staining (Figure 2, lane 1). As previously mentioned, BtHis-Atp12p shows an apparent mol. wt of 47 kDa versus the value of 43 kDa expected for the mature form, which suggests that the protein retains its mitochondrial import leader sequence. Western analyses with polyclonal antibody against Atp12p or with an avidin conjugate confirmed the identity of the main component in the preparation as BtHis-Atp12p (data not shown). Western blots probed with antibody against the F1 α–subunit showed that this protein represents the main contaminant in the BtHis-Atp12p preparation (Figure 2, lane 2). Instead, the F1 β,subunit could not be detected using a specific antibody against this protein (Figure 2, lane 3).
Fig. 2. SDS–polyacrylamide gel analysis of BtHis-Atp12p purified from mitochondria. In the final step of purification from mitochondria, BtAtp12p was eluted from streptavidin resin with hot SDS as described in Materials and methods. Aliquots of this fraction (15 μl) were applied to 12% SDS–polyacrylamide gels, after which the gel was either stained with silver (lane 1) or transferred to nitrocellulose in preparation for Western blotting with anti-F1 α–subunit antibody (lane 2) or anti-F1 β–subunit antibody (lane 3). The positions of BtHis-Atp12p and the F1 α–subunit are marked with arrows. The migration of molecular weight standards is shown on the right.
Identification of the α–subunit amino acid sequence that binds Atp12p
A yeast two-hybrid assay was used to screen for binding interactions between Atp12p and the F1 α- and β–subunits. The plasmids employed in this work (Materials and methods, Table II) encode only the mature sequences (i.e. without the mitochondrial leader peptides) of the Atp12p and F1 proteins fused to either the DNA-binding domain or the transcriptional activation domain of Gal4p. Combinations of these plasmids were used to transform a yeast host (Y190) that carries a lacZ reporter gene in the chromosome under transcriptional control of Gal4p. The presence of β–galactosidase activity was used as a reporter of binding between the two Gal4p fusion proteins.
Table II. Recombinant plasmids and encoded proteins.
| Plasmid | Encoded product | Source |
|---|---|---|
| pG57/ST4 | Atp12p, including leader peptide | Bowman et al. (1991) |
| pG57/BTHIS | mature BtHis-Atp12p (G31–Q325), with Atp11p leader peptide | this study |
| pG57/ST22 | mature His-Atp12p (G31–Q325)a | Wang and Ackerman (1998) |
| pAS2-1/ATP12(E289K) | mature His-Atp12p (G31–Q325), with Glu289→Lys289 substitutiona | this study |
| pACT2/ATP2(36–511) | mature F1 β subunit (A36–N511)b | Wang et al. (1999) |
| pACT2/ATP1(35–545) | mature F1 α subunit (L35–F545)b | Wang and Ackerman (2000) |
| pACT2/ATP1(191–545) | F1 α–subunit fragment (D191–F545)b | this study |
| pACT2/ATP1(35–201) | F1 α–subunit fragment (L35–R201)b | this study |
| pACT2/ATP1(35–213) | F1 α–subunit fragment (L35–T213)b | this study |
| pACT2/ATP1(35–322) | F1 α–subunit fragment (L35–L322)b | this study |
| pACT2/ATP1(35–371) | F1 α–subunit fragment (L35–V371)b | this study |
| pACT2/ATP1(133–545) | F1 α–subunit fragment (D133–F545)b | this study |
| pACT2/ATP1(159–545) | F1 α–subunit fragment (D159–F545)b | this study |
| pACT2/ATP1(179–545) | F1 α–subunit fragment (V179–F545)b | this study |
Fusion protein with the DNA-binding domain of Gal4p.
bFusion protein with the activation domain of Gal4p.
The combination of plasmids producing Atp12p and the full-length, mature F1 α–subunit (L35–F545) gave a positive signal in the two-hybrid assay (Figure 3), while there was no β–galactosidase activity in yeast harboring the Atp12p or F1 α–subunit plasmid alone, or in combination with the appropriate partner vector lacking any insert. No evidence of binding between Atp12p and the F1 β–subunit was afforded with this assay (data not shown).
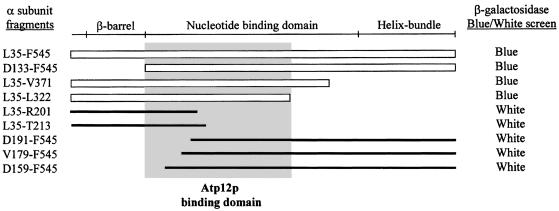
Fig. 3. Yeast two-hybrid screen analysis of the binding interactions between Atp12p and the F1 α–subunit. A protein map of the mature yeast F1 α–subunit (Ala36–Phe545) that shows the positions of three primary domains is given in the upper part of the figure. The designation of the yeast β–barrel domain (56–132), the nucleotide-binding domain (133–416) and the C–terminal helix bundle domain (417–545) is based on information from the homologous α–subunit of beef heart mitochondria (Abrahams et al., 1994). The results from the blue/white screen for β–galactosidase activity are shown on the right. The position and length of the α–subunit fragments that scored positively for binding Atp12p in this assay are shown with open rectangles; negatively scoring fragments are indicated with thick lines. The gray box highlights the span of the α–subunit sequence that contains binding determinants for Atp12p.
Subsequent work employed the two-hybrid system to locate the binding site for Atp12p on the α–subunit. The mature α–subunit has three primary domains: a β–barrel at the N–terminus, a central nucleotide-binding domain and a helix bundle at the C–terminal end of the protein (Abrahams et al., 1994; Bianchet et al., 1998; Stock et al., 1999) (see Figure 3). The fragment D133–F545, which is missing the entire β–barrel, and the fragment L35–V371, which lacks both the helix bundle and a portion of the nucleotide-binding domain, both scored positively for binding Atp12p. The smaller fragment L35–L322 also tested positive, while fragments L35–R201, L35–T213, D191–F545, V179–F545 and D159–F545 were negative. Cumulatively, these studies suggest that the binding site for Atp12p is located in the 190 amino acid region of the α–subunit between Asp133 and Leu322 (gray box, Figure 3). The binding determinants for Atp12p appear to be distributed evenly throughout this region, since fragments harboring only the proximal (L35–R201 and L35–T213) or the distal portion (D191–F545, V179–F545 and D159–F545) do not interact with the assembly factor. Western analysis confirmed that negative scoring fusion proteins were produced in the yeast host (data not shown). The validity of the conclusions derived from these experiments is supported by the observation that the two-hybrid screen provides no evidence of binding between the full-length mature F1 α–subunit and the E289K mutant form of Atp12p. Previous work had shown that this mutant Atp12p is not competent for F1 assembly (Wang and Ackerman, 1998).
The presence of the F1 α–subunit modifies the sedimentation profile of Atp12p
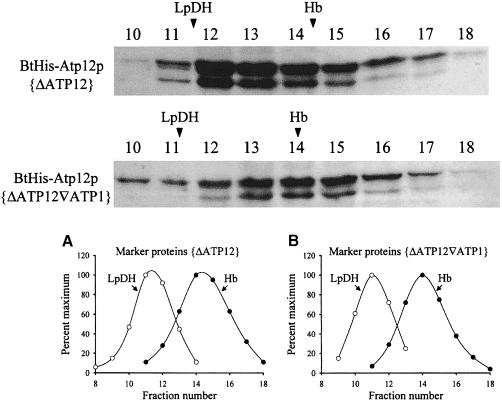
Atp12p from mitochondrial extracts sediments through linear sucrose gradients as a particle with a mass of ∼70–80 kDa (Bowman et al., 1991). Although this size is roughly twice that of the 33 kDa Atp12p monomer, there is strong evidence that Atp12p is not a homodimer (Wang and Ackerman, 1998). To determine whether the sedimentation behavior of Atp12p is due to its interaction with the α–subunit of F1, samples of mitochondrial extracts that contained Atp12p with or without the α–subunit of F1 were analyzed. Plasmid-borne BtHis-Atp12p was used in these experiments to permit visualization of the protein in Western blots with an avidin conjugate. Soluble mitochondrial extracts were prepared from the ΔATP12 (F1 α+) and ΔATP12▿ATP1 (F1 α–) strains harboring the plasmid pG57/BTHIS, and centrifuged through linear sucrose gradients in the presence of molecular weight markers. The sedimentation profile of BtHis-Atp12p in the two samples was probed by Western blot (Figure 4). The sedimentation of the marker proteins in each gradient is indicated above the blots. While in the absence of α–subunit (strain ΔATP12▿ATP1), the BtHis-Atp12p and hemoglobin peaks are almost coincident, in its presence (strain ΔATP12) the BtHis-Atp12p peak is shifted from hemoglobin toward higher sucrose density by ∼1.5 gradient fractions. The difference in the BtHis-Atp12p sedimentation properties in the two samples corresponds to a mass gain of ∼25 kDa and is not large enough to suggest that this protein forms a stable 1:1 complex with the α–subunit (55 kDa), but it may indicate a transient association between these two proteins.
Fig. 4. Sedimentation analysis of Atp12p in the presence/absence of the F1 α–subunit. Soluble mitochondrial extracts were prepared from aW303ΔATP12 and aW303ΔATP12▿ATP1 transformants that produce BtHis-Atp12p and centrifuged through 7–20% linear sucrose gradients in the presence of molecular weight standards (as described in Bowman et al., 1991). Western blots probed with avidin–horseradish peroxidase to visualize BtHis-Atp12p are shown in the upper part of the figure. The arrowheads above the blots show the peak positions of the molecular weight standards, hemoglobin (Hb; Mr = 64 500) and lipoamide dehydrogenase (LpDH; Mr = 100 000) included in the gradients. The plots in the lower part of the figure show the sedimentation profiles for Hb (○) and LpDH (•) in the aW303ΔATP12 (A) and aW303ΔATP12▿ATP1 (B) samples.
Discussion
We have presented several lines of evidence suggesting that Atp12p binds directly to the F1 α–subunit. First, the α–subunit and BtHis-Atp12p can be co-precipitated and co-purified specifically. Failure to detect the F1 β–subunit in these experiments suggests that BtHis-Atp12p either does not interact with the β–subunit or forms only a very weak complex. Secondly, the binding site for Atp12p has been mapped with the yeast two-hybrid screen to a region of 190 amino acids (D133–L322) within the nucleotide-binding domain of the α–subunit (Figure 3). Deletion of residues from either end of this sequence produces fragments that do not show interaction with Atp12p. This lack of binding cannot be attributed to inherent instability of the fragments since Western analysis showed high levels in the cell of the relevant Gal4p fusion proteins. Instead, the requirement for the complete 190 amino acid sequence suggests that other factors, such as correct folding, are required for recognition and binding by Atp12p; the view that Atp12p binds the folded form of the α–subunit is supported by the fact that atp12 mutants accumulate only the mature α–subunit in mitochondria, while hsp60(mif4) mutants, which are deficient in protein folding, accumulate both the precursor and mature forms of mitochondrial matrix proteins (Cheng et al., 1989). Thirdly, sedimentation analyses showed that the presence of the F1 α–subunit influences the sedimentation properties of Atp12p in linear sucrose gradients.
An attractive hypothesis for the function of Atp12p is that this protein shields sequence elements on the F1 α–subunit that would cause abnormal interactions leading to aggregation of the free protein. Prime candidates for such elements are residues that make contact with adjacent β–subunits in the assembled F1, since these amino acids are never intended to be exposed to solvent. The high-resolution X–ray structure of the homologous bovine F1 (Abrahams et al., 1994) is a convenient model to identify the amino acid residues of the yeast α–subunit that are most likely to be protected by Atp12p. The yeast α–subunit sequence D133–L322, which we propose binds Atp12p, is homologous to residues D96–L285 of bovine α (Abrahams et al., 1994). This region extends from β–strand 1 through the end of helix E in the nucleotide-binding domain of the α–subunit, and includes regions of the protein that make contact with the two adjacent β–subunits. The corresponding region of the β–subunit (β–strand 1 through the beginning of helix F) forms the binding site for the Atp11p assembly factor (Wang and Ackerman, 2000), and includes regions of the protein that make contact with the two adjacent α–subunits. Hence, it is conceivable that the αβ interfaces are established through exchange reactions in which a β–subunit replaces α-bound Atp12p and an α–subunit replaces β-bound Atp11p. However, since each α–subunit contacts two different β-subunits and each β–subunit contacts two different α–subunits, it is of mechanistic relevance to determine which β–subunit replaces Atp12p and which α–subunit replaces Atp11p.
Useful information with regard to this point can be derived by analyzing the contributions of the Atp12p-binding domain of the α–subunit and of the Atp11p-binding domain of the β–subunit to each of the two possible αβ interfaces. For this purpose, we have used the coordinates of bovine F1 to calculate the interaction energies between all possible pairs of α- and β–subunits (Table I, columns 1–3), between the Atp12p-binding domain of the α–subunit and the two neighboring β–subunits (columns 4–6, upper half of the table), and between the Atp11p-binding domain of the β–subunit and the two neighboring α–subunits (columns 4–6, lower half of the table); averaged values for each group of αβ interactions are shown in bold face. The fractional contributions of the assembly factor-binding domains (AFBDs) to the overall interaction energy between subunits (the ratios, column by column, between the averaged values in columns 4–6 and 1–3) are reported in columns 7–9. For ease of analysis, the partial energies originating from van der Waals (VDW) interactions (columns 2, 5 and 8) and from electrostatic (ELEC) interactions (columns 3, 6 and 9) are reported to the right of the overall values of the interaction energy (columns 1, 4 and 7). Following the nomenclature of Abrahams et al. (1994), individual rows refer to αβ contacts that form a non-catalytic site (AD, CF and BE subunit pairs) or a catalytic site (AE, BF and CD subunit pairs) (see Figure 5A). Each of the six interfaces of the enzyme is characterized by a different value of interaction energy; the observed variations in the energy terms reflect the fact that the F1 structure employed in the calculations is not symmetric, as one catalytic site is occupied by a triphospho-nucleotide, one is occupied by a diphospho-nucleotide and one is completely empty. Since columns 1–3 represent the interaction energy between entire α- and β–subunits, the same values are obtained whether we consider the binding of α- to β–subunits (e.g. AD row in the upper half of the table) or of β- to α–subunits (e.g. DA row in the lower half of the table) for a given pair. Instead, in columns 4–6, the AD row represents the interaction energy between the Atp12p-binding domain of the αA subunit and the entire βD subunit, while the DA row represents the interaction energy between the Atp11p-binding domain of the βD subunit and the entire αA subunit.
Table I. Interaction energiesa between α- and β–subunits in mitochondrial bovine F1.
| Interaction | Interface | αβ pair | Entire subunit |
Atp12p-binding domain |
AFBDb/entire subunit |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 Total | 2 VDWc | 3 ELECd | 4 Total | 5 VDW | 6 ELEC | 7 Total | 8 VDW | 9 ELEC | |||
| α→β | NCSe | AD | 726.7 | 157.9 | 568.8 | 368.9 | 77.6 | 291.3 | |||
| CF | 720.2 | 150.4 | 569.8 | 383.8 | 75.8 | 308.0 | |||||
| BE | 600.6 | 104.4 | 496.3 | 306.4 | 65.6 | 240.8 | |||||
| 682.5 | 137.6 | 545.0 | 353.0 | 73.0 | 280.0 | 0.52 | 0.53 | 0.51 | |||
| CSf | AE | 368.7 | 119.6 | 249.1 | 103.3 | 22.5 | 80.8 | ||||
| BF | 675.6 | 156.2 | 519.4 | 197.2 | 28.5 | 168.7 | |||||
| CD | 760.0 | 201.4 | 558.5 | 160.7 | 29.0 | 131.6 | |||||
| 601.4 | 159.1 | 442.3 | 153.7 | 26.7 | 127.0 | 0.26 | 0.17 | 0.29 | |||
| Atp11p-binding domain | |||||||||||
| β→α | CS | EA | 368.7 | 119.6 | 249.1 | 130.6 | 77.8 | 52.8 | |||
| DC | 760.0 | 201.4 | 558.5 | 284.8 | 91.8 | 192.9 | |||||
| FB | 675.6 | 156.2 | 519.4 | 296.1 | 97.0 | 199.1 | |||||
| 601.4 | 159.1 | 442.3 | 237.2 | 88.9 | 148.3 | 0.39 | 0.56 | 0.34 | |||
| NCS | EB | 600.6 | 104.4 | 496.3 | 296.2 | 46.0 | 250.2 | ||||
| FC | 720.2 | 150.4 | 569.8 | 269.0 | 55.4 | 213.7 | |||||
| DA | 726.7 | 157.9 | 568.8 | 237.3 | 54.3 | 182.9 | |||||
| 682.5 | 137.6 | 545.0 | 267.5 | 51.9 | 215.6 | 0.39 | 0.38 | 0.40 | |||
aAll interaction energies are expressed in kcal/mol and have a negative sign (attractive interaction).
bAssembly factor-binding domain.
cvan der Waals component of the interaction energy.
dElectrostatic component of the interaction energy.
eNon-catalytic site.
fCatalytic site.
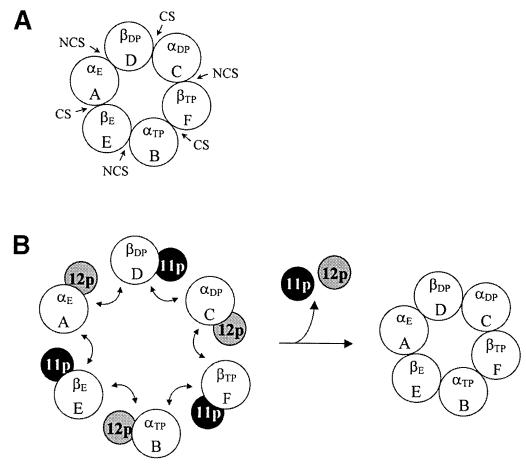
Fig. 5. Model for Atp12p and Atp11p action in mitochondrial F1 assembly. (A) The αβ hexamer of mitochondrial bovine F1. CS, catalytic site; NCS, non-catalytic site. The subscripts TP and DP indicate that the catalytic site is occupied by triphosphate and diphosphate nucleotides, respectively; E indicates an empty catalytic site. (B) Scheme showing the exchange of bound Atp12p for the β–subunit that completes the non-catalytic site, and the exchange of bound Atp11p for the α–subunit that completes the catalytic site.
This analysis shows that the AFBDs do not contribute equally to the interactions of a given α- or β–subunit with each of its neighbors. For example, the Atp12p-binding domain contributes more to the overall interaction between αA and βD than to the interaction between αA and βE (Table I, upper half of columns 7–9). Although the Atp11p-binding domain of the β–subunit appears to provide an overall similar contribution to the interactions with each of the two neighboring α–subunits (Table I, lower half of column 7), there is a clear contribution difference in the hydrophobic component of the interaction (VDW, lower half of column 8), which is stronger for the surfaces involved in the formation of the catalytic sites. These observations suggest that the AFBDs are polarized in such a way that one of the two surfaces available for interaction with the neighboring α- and β–subunit is significantly more hydrophobic than the other. Furthermore, the direction of this polarization is the same in both the α- and the β–subunit: the Atp12p-binding domain contributes primarily to the hydrophobic interaction of the α–subunit with the β–subunit with which it forms a non-catalytic site, and the Atp11p-binding domain contributes primarily to the hydrophobic interaction of the β–subunit with the α–subunit with which it forms a catalytic site. Since exposure of the more hydrophobic parts of the AFBDs may lead to aggregation of free α- and β–subunits, we propose that these are the regions to which Atp12p and Atp11p bind. Thus, assembly of the α3β3 hexamer of F1 could follow a pathway in which β–bound Atp11p is released in exchange for the α–subunit that completes the catalytic site, and α–bound Atp12p is released in exchange for the β–subunit that completes the non-catalytic site (Figure 5B). This model provides a mechanistic frame for the formation of the αβ interfaces, but does not rule out the possibility that other proteins may also be involved in the process of forming a complete hexamer. In particular, the presence of the γ–subunit appears to be necessary to promote the assembly of smaller αβ oligomers into the final complex (Paul et al., 1994). However, no information is available at this moment on the precise step at which the γ–subunit enters the assembly pathway.
Materials and methods
Strains and growth media
All of the biochemical studies described utilized yeast strain aW303ΔATP12 (MATa ade2-1 his3-1,15 leu2-3,112 ura3-1 trp1-1 atp12::LEU2 (Bowman et al., 1991). Sedimentation analyses further employed the strain W303ΔATP12▿ATP1 (MATa ade2-1 his3-1,15 leu2-3,112 ura3-1 trp1-1 atp12::LEU2 atp1::HIS3), which was obtained by selecting for respiratory-deficient, ρ+, Leu+, His+ progeny from a cross between aW303ΔATP12 and W303▿ATP1 (MATα ade2-1 his3-1,15 leu2-3,112 ura3-1 trp1-1 atp1::HIS3); W303▿ATP1 was a generous gift from Dr Alexander Tzagoloff, Columbia University, NY. The presence of two disrupted alleles (ATP1::HIS3 and ATP12::LEU2) in W303ΔATP12▿ATP1 was verified by the fact that the respiratory-deficient phenotype of this strain is not complemented in crosses to yeast harboring a point mutation in either ATP12 or ATP1. Yeast two-hybrid experiments utilized strain Y190 [MAT a ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4Δ gal80Δ cyhr2 LYS2::GAL1UAS-HIS3TATA-HIS3 URA3::GAL1UAS-GAL1TATA-lacZ (Clontech)]. Escherichia coli TB1 {F– araΔ (lac-proAB) rpsL (Strr) [Φ80dlacΔ (lacZ)M15] thi hsdR (rK–mK+)} was the host bacterial strain for the recombinant plasmid constructions. Yeast was grown in the following media: YPD (2% glucose, 2% peptone, 1% yeast extract), YPGal (2% galactose, 2% peptone, 1% yeast extract), EG (2% ethanol, 2% glycerol, 2% peptone, 1% yeast extract), WO [2% glucose, 0.67% yeast nitrogen base without amino acids (Difco)] and SD/–trp,leu,his [2% glucose, 0.67% yeast nitrogen base without amino acids (Difco)], supplemented with all essential amino acids and nucleotides except tryptophan, leucine and histidine). Amino acids and other growth requirements were added at a final concentration of 20–150 μg/ml. The solid media contained 2% agar in addition to the components described above.
Plasmid constructions
The plasmids used in this study are described in Table II. Atp12p is numbered from 1 to 325 [Saccharomyces Genome Database (SGD) locus code: YJL180C], the F1 α–subunit is numbered from 1 to 545 (SGD locus code: YBL099W) and the F1 β–subunit is numbered from 1 to 511 (SGC locus code: YJR121W); in all cases, residue 1 is the initiator methionine in the primary translation products. The plasmids carrying partial segments of a gene, and the encoded products are named according to the codons/amino acids that are retained in the constructs. Plasmid pG57/BTHIS encodes a 47 kDa chimeric protein that is comprised of the 39 amino acid mitochondrial targeting sequence of Atp11p followed by, respectively, a 74 amino acid biotinylation sequence, a 24 amino acid His6 sequence and the 295 amino acids of Atp12p (G31–Q325) that encompass the mature form of the protein. The construction of this plasmid involved the insertion of a 243 bp fragment, coding for a biotinylation sequence, in the SmaI site at the junction between the sequences for the Atp11p leader peptide and His6-Atp12p in the yeast plasmid pG57/ST22 (Wang and Ackerman, 1998). The DNA for the biotinylation sequence from Propionobacterium shermanii was obtained with PCR using the primers Bio7-f and Bio7-r (Table III), and a plasmid template (YEp352-Bio7) that was a generous gift from Dr Alexander Tzagoloff, Columbia University, NY. Plasmid pAS2-1/ATP12(E289K) was constructed by ligating, in concert, a 519 bp NcoI–FspI fragment from pG57/ST23 (Wang and Ackerman, 1998) and a 470 bp FspI–EcoRI fragment of the mutant atp12 gene cloned from yeast strain E822 (Wang and Ackerman, 1998) with the NcoI and EcoRI sites of pAS2-1 (Clontech). Plasmid pACT2/ATP1(191–545) carries a 1.1 kb HincII–BamHI fragment in the NcoI(blunted)–BamHI sites of pACT2; the ATP1 DNA was prepared from an intermediate plasmid that carries a 1.6 kb EcoRI(blunted)–BamHI fragment in pTRC99a (Pharmacia). The ATP1 inserts in the remaining plasmids were synthesized by PCR using plasmid pACT2/ATP1(35–545) as the template with the primers listed in Table III. The forward and reverse primer pairs used to construct each plasmid are as follows: pACT2/ATP1(35–201), α35–201-f and α35–201-r; pACT2/ATP1(35–213), α35–201-f and α35–213-r; pACT2/ATP1(35–322), α35–201-f and α35–322-r; pACT2/ATP1(35–371), α35–201-f and α35–371-r; pACT2/ATP1(133–545), α133–545-f and α133–545-r; pACT2/ATP1(159–545), α159–545-f and α133–545-r; and pACT2/ATP1(179–545), α179–545-f and α133–545-r. All resultant PCR products were flanked with restriction sites appropriate for subcloning in pACT2 (see Table III).
Table III. Oligonucleotide primers for PCR.
| Primer | Primer sequence (5′→3′)a | ATP1 gene location | Restriction sites |
|---|---|---|---|
| Bio7-f | AAGCTTGCACCCGGGCAGGTCGA | –b | SmaI |
| Bio7-r | GTGCGTCACCCGGGCTTGATGA | –c | SmaI |
| α35-201-f | CCACCAAACCCAAAAAAAG | –d | EcoRI |
| α35-201-r | CGAGCTCGAGCTATCTTTGACCTCTACCGATAG | complementary to +584 to +603 | XhoI |
| α35-213-r | CGAGCTCGAGCTAAGTCTTACCTGTTTGACGAT | complementary to +620 to +639 | XhoI |
| α35-322-r | CGAGCTCGAGCTACAACAACAAAGATAATTGACGGT | complementary to +944 to +966 | XhoI |
| α35-371-r | CGAGCTCGAGTCTAGACATCACCACCTTGGGTT | complementary to +1095 to +1114 | XhoI |
| α133–545-f | CATATGGCCATGGATGTCCCAGTCGGTCC | +397 to +413 | NcoI |
| α133–545-r | AAATTGAGATGGTGCACGATGCA | –e | XhoI |
| α159–545-f | CATATGGCCATGGACGCTGCCGGTCGTTC | +475 to +491 | NcoI |
| α179–545-f | CATATGGCCATGGTCCATGAACCAGTTCAAAC | +535 to +554 | NcoI |
aATP1 nucleotides are shown in bold.
bNucleotides +1 to +23 in YEp352-bio7.
cComplementary to nucleotides +245 to +266 in YEp352-bio7.
dNucleotides +5101 to +5083 in pACT2.
eComplementary to nucleotides +4912 to +4934 in pACT2.
Co-precipitation experiments
Mitochondria were prepared as described (Ackerman et al., 1992) from yeast strains aW303ΔATP12/pG57/ST4 and aW303ΔATP12/pG57/BTHIS, which produce native Atp12p and biotinylated, His-tagged Atp12p (BtHis-Atp12p), respectively, from the 2μ vector YEp352 (Hill et al., 1986). The preparation of soluble mitochondrial extracts and affinity precipitation of these fractions with avidin–Sepharose beads, in the absence or presence of free biotin in the buffer, were performed as described (Wang and Ackerman, 2000).
Purification of BtHis-Atp12p from mitochondria
Mitochondria were prepared from aW303ΔATP12/pG57/BTHIS grown in 18 l of liquid YEPG, suspended to 10 mg/ml in 30 ml of 50 mM Tris–HCl pH 8.0, and sonically irradiated in an ice bath for 1 min at 50% pulse frequency using a power setting of 10 (Branson sonifer, Model 450). Following centrifugation of the disrupted mitochondria at 50 000 r.p.m. in a Beckman 50.1Ti rotor for 30 min at 4°C, 30 ml of the clarified supernatant was applied to a DEAE–Sepharose fast flow column (1 × 10 cm, 4°C). The column was washed with 120 ml of 50 mM Tris–HCl pH 8.0, and proteins were eluted with 30 ml of 50 mM Tris–HCl, 1.2 M NaCl buffer. The eluate from the DEAE column was loaded on an Ni-NTA Superflow (Qiagen) column (1 × 3 cm, 4°C). This column was washed with 10 ml of 50 mM Tris–HCl pH 8.0, followed by 7.5 ml of 50 mM Tris–HCl pH 8.0, 40 mM imidazole, and finally eluted with 10 ml of 50 mM Tris–HCl pH 8.0, 100 mM imidazole. Proteins eluted from the nickel column were applied to a streptavidin (Prozyme) column (0.2 ml, Bio-Rad Poly-Prep column) and the column was washed with 5 ml of 50 mM Tris–HCl pH 8.0, 1.2 M NaCl buffer and then with 2 ml of 20 mM Tris–HCl pH 6.8 buffer containing 2 mM biotin. Owing to the high affinity of streptavidin for biotin (KD = 10–15 M), proteins adsorbed to the resin were eluted by transferring the column material to an Eppendorf tube to which 100 μl of 3× Laemmli-SDS loading buffer was added, and the tube was boiled at 90°C for 10 min. In preparation for gel electrophoresis, the tube was centrifuged to pellet the beads and protein samples were removed from the supernatant.
Calculation of interaction energies
As the structure of yeast F1 has been determined only at low resolution (3.9 Å; Stock et al., 1999), the coordinates of the bovine enzyme (available at the resolution of 2.8 Å, Abrahams et al., 1994) were chosen to study the energetics of the interactions between α- and β–subunits. All calculations were carried out with CNS version 0.5 (Brünger et al., 1998) using the force field of Engh and Huber (1991). In this force field, the hydrogen bond energy term is taken into account implicitly by an appropriate modification of the partial charges at individual atoms and of the van der Waals parameters. Prior to energy calculations, all polar hydrogens were added explicitly and the structure was relaxed by means of 200 cycles of conjugate gradient minimization (Powell, 1977). The minimization step was deemed necessary in order to avoid the potential problem of a few poor contacts dominating the overall value of the interaction energy. The root mean square deviation of the minimized coordinates with respect to the starting coordinates was 0.38 Å. Interaction energies between any two sets of atoms were calculated using a constant dielectric model and including both van der Waals and electrostatic contributions. The following parameterization was adopted for the computation of non-bonded interactions: dielectric constant = 1.0; cut-off for the generation of the list of non-bonded interactions = 7.5 Å; distance at which a shifting function becomes effective = 6.0 Å; distance at which the shifting function forces the non-bonded energy to zero = 6.5 Å.
Miscellaneous procedures
Sedimentation analysis of Atp12p in the presence of hemoglobin (Mr = 64 500) and lipoamide dehydrogenase (Mr = 100 000) molecular weight standards followed the method of Bowman et al. (1991). The yeast two-hybrid screen (Fields and Song, 1989), which included detection of β–galactosidase activity with 5-bromo-4-chloro-3-indoyl β-d-galactopyranoside (X-gal), was performed as described (Wang and Ackerman, 2000). Standard techniques were used for restriction endonuclease analysis of DNA, purification and ligation of DNA fragments, and transformations of and recovery of plasmid DNA from Escherichia coli (Sambrook et al., 1982). Yeast transformations employed the LiAc procedure (Schiestl and Gietz, 1989). The method of Laemmli (1970) was used for SDS–PAGE. Western blotting followed the procedure of Schmidt et al. (1984). Antibodies against Atp12p (Bowman et al., 1991) and the F1 α- and β–subunits (Ackerman and Tzagoloff, 1990) were used at 1:1000, 1:2000 and 1:3000 dilutions, respectively. Monoclonal antibodies against the transcription activation domain and the DNA-binding domain of Gal4p were purchased from Clontech and used at 0.4 and 0.5 μg/ml, respectively. Avidin–horseradish peroxidase conjugate (Bio-Rad) was used at a dilution of 1:3000. Visualization of the protein bands in X–ray film was by chemiluminescence using the ECL system from Amersham. Protein concentrations were estimated by the method of Lowry et al. (1951).
Acknowledgments
Acknowledgements
This work was supported by NIH grant GM48157 to S.H.A.
References
- Abrahams J.P., Leslie, A.G.W., Lutter, R. and Walker, J.E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature, 370, 621–628. [DOI] [PubMed] [Google Scholar]
- Ackerman S.H. and Tzagoloff, A. (1990) Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc. Natl Acad. Sci. USA, 87, 4986–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman S.H., Martin, J. and Tzagoloff, A. (1992) Characterization of ATP11 and detection of the encoded protein in mitochondria of Saccharomyces cerevisiae.J. Biol. Chem., 267, 7386–7394. [PubMed] [Google Scholar]
- Bianchet M.A., Hulihen, J., Pedersen, P.L. and Amzel, L.M. (1998) The 2.8-Å structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis. Proc. Natl Acad. Sci. USA, 95, 11065–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S., Ackerman, S.H. and Tzagoloff, A. (1991) Characterization of ATP12, a yeast nuclear gene required for the assembly of the mitochondrial F1-ATPase. J. Biol. Chem., 266, 7517–7523. [PubMed] [Google Scholar]
- Boyer P.D. (1993) The ATP synthase—a splendid molecular machine. Biochim. Biophys. Acta, 1140, 215–250. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998)Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cheng M.Y., Hartl, F.-U., Martin, J., Pollock, R.A., Kalousek, F., Neupert, W., Hallberg, R.L. and Horwich, A.L. (1989) Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature, 337, 620–625. [DOI] [PubMed] [Google Scholar]
- Engh R.A. and Huber, R. (1991) Accurate bond and angle parameters for X-ray structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Fields S., Song, O.-K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Giraud M.-F. and Velours, J. (1994) ATP synthase of yeast mitochondria. Isolation of the F1 δ subunit, sequence and disruption of the structural gene. Eur. J. Biochem., 222, 851–859. [DOI] [PubMed] [Google Scholar]
- Guelin E., Chevallier, J., Rigoulet, M., Guerin, B. and Velours, J. (1993) ATP synthase of yeast mitochondria. Isolation and disruption of the ATPɛ gene. J. Biol. Chem., 268, 161–167. [PubMed] [Google Scholar]
- Hendrick J.P., Hartl, F.-U. (1993) Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem., 62, 349–384. [DOI] [PubMed] [Google Scholar]
- Hill J.E., Myers, A.M., Koerner, T.J. and Tzagoloff, A. (1986) Yeast/E.coli shuttle vectors with multiple unique restriction sites. Yeast, 2, 163–167. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough, N.J., Farr, A.L. and Randall, R.J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem., 193, 265–275. [PubMed] [Google Scholar]
- Paul M.-F., Ackerman, S., Yue, J., Arselin, G., Velours, J. and Tzagoloff, A. (1994) Cloning of the yeast ATP3 gene coding for the γ–subunit of F1 and characterization of atp3 mutants. J. Biol. Chem., 269, 26158–26164. [PubMed] [Google Scholar]
- Penefsky H.S. and Cross, R.L. (1991) Structure and mechanism of F0F1-type ATP synthases and ATPases. Adv. Enzymol., 64, 173–214. [DOI] [PubMed] [Google Scholar]
- Powell M.J.D. (1977) Restart procedures for the conjugate gradient method. Math. Programming, 12, 241–254. [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2nd edn. [Google Scholar]
- Schiestl R.H. and Gietz, R.D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- Schmidt R.J., Myers, A.M., Gillham, N.W. and Boynton, F.E. (1984) Immunological similarities between specific chloroplast ribosomal proteins from Chlamydomonas reinhardtii and ribosomal proteins from Escherichia coli.Mol. Biol. Evol., 1, 317–334. [DOI] [PubMed] [Google Scholar]
- Stock D., Leslie, A.G.W. and Walker, J.E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science, 286, 1700–1705. [DOI] [PubMed] [Google Scholar]
- Takeda M., Vassarotti, A. and Douglas, M.G. (1985) Nuclear genes coding the yeast mitochondrial adenosine triphosphatase complex. Primary sequence analysis of ATP2 encoding the F1-ATPase β–subunit precursor. J. Biol. Chem., 260, 15458–15465. [PubMed] [Google Scholar]
- Takeda M., Chen, W.-J., Salzgaber, J. and Douglas, M.G. (1986) Nuclear genes encoding the yeast mitochondrial ATPase complex. Analysis of ATP1 coding the F1-ATPase α–subunit and its assembly. J. Biol. Chem., 261, 15126–15133. [PubMed] [Google Scholar]
- Tokatlidis K. and Schatz, G. (1999) Biogenesis of mitochondrial inner membrane proteins. J. Biol. Chem., 274, 35285–35288. [DOI] [PubMed] [Google Scholar]
- Wang Z.-G. and Ackerman, S.H. (1998) Mutational studies with Atp12p, a protein required for assembly of the mitochondrial F1-ATPase in yeast. Identification of domains important for Atp12p function and oligomerization. J. Biol. Chem., 273, 2993–3002. [DOI] [PubMed] [Google Scholar]
- Wang Z.-G. and Ackerman, S.H. (2000) The assembly factor Atp11p binds to the β subunit of the mitochondrial F1 ATPase. J. Biol. Chem., 275, 5767–5772. [DOI] [PubMed] [Google Scholar]
- Wang Z.-G., Schmid, K.J. and Ackerman, S.H. (1999) The Drosophila gene 2A5 complements the defect in mitochondrial F1-ATPase assembly in yeast lacking the molecular chaperone Atp11p. FEBS Lett., 452, 305–308. [DOI] [PubMed] [Google Scholar]