Abstract
Genome-wide association studies have recently identified many new loci associated with human complex diseases. These newly discovered variants typically have weak effects requiring studies with large numbers of individuals to achieve the statistical power necessary to identify them. Likely, there exist even more associated variants, which remain to be found if even larger association studies can be assembled. Meta-analysis provides a straightforward means of increasing study sample sizes without collecting new samples by combining existing data sets. One obstacle to combining studies is that they are often performed on platforms with different marker sets. Current studies overcome this issue by imputing genotypes missing from each of the studies and then performing standard meta-analysis techniques. We show that this approach may result in a loss of power since errors in imputation are not accounted for. We present a new method for performing meta-analysis over imputed single nucleotide polymorphisms, show that it is optimal with respect to power, and discuss practical implementation issues. Through simulation experiments, we show that our imputation aware meta-analysis approach outperforms or matches standard meta-analysis approaches.
Keywords: imputation, meta-analysis, association studies
INTRODUCTION
The genome-wide association study (GWAS) has proven to be a successful method for identifying loci contributing to the genetic basis of complex human diseases. While the list of single nucleotide polymorphisms (SNPs) and genes correlated with phenotypes continues to grow, many of the discovered variants exhibit only a weak-to-moderate effect and account for just a small fraction of the total phenotypic variance. Over 75% of the associations identified by case-control GWAS had reported odds ratios (OR) of less than 1.4 with 39% having less than 1.2. In order to achieve 90% power to capture a SNP with an OR = 1.2, minor allele frequency (MAF) of 0.2, and genome-wide cutoff of 10−6 under a multiplicative model, 15,248 individuals must be collected in a balanced study. Over 82% of discovered loci from completed case-control GWAS are from studies with significantly fewer individuals and are therefore underpowered to reliably discover these associations [Hindorff et al., 2009].
Given this observation, GWAS must be designed with larger numbers of individuals to have sufficient power to identify weaker variants. This requires a large-scale effort to collect potentially tens of thousands of individuals, who are then genotyped at hundreds of thousands of SNPs. Although the cost of genotyping is dropping, it remains difficult to find, screen, and approve individuals suited for a study. For many diseases, especially those with significant impact on global health, multiple groups are performing association studies, each collecting their own case and control cohorts. A natural approach to address the lack of power of each of the individual studies is to combine the cohorts using meta-analysis.
Meta-analysis is a well-studied problem and is currently widely used in the genetics community in the planning and analysis of GWAS. For a review of meta-analysis techniques and pitfalls, see Kavvoura and Ioannidis [2008]. Traditional approaches to meta-analysis combine the statistics at each marker from both studies. This approach requires individuals to be genotyped on the same set of SNPs. Since studies often employ different genotyping platforms and different SNPs pass quality control filters in each study, many markers are not shared between studies and cannot be combined using traditional meta-analysis methods.
Recently, several “imputation” methods have been proposed which use a reference set such as the HapMap [International-HapMap-Consortium, 2005] to estimate the frequency of ungenotyped SNPs in a study [Guan and Stephens, 2008; Li and Abecasis, 2006; Marchini et al., 2007]. Provided that the study population is similar to one of the HapMap populations, these imputation methods are highly accurate for many of the HapMap SNPs. A straightforward approach to combining studies with different marker sets is to impute the ungenotyped SNPs in each study so that all HapMap SNPs are either genotyped or imputed in both studies. A standard meta-analysis method may then be applied to the genotyped and imputed SNPs. Indeed, several recent meta-analyses have adopted this approach [Soranzo et al., 2009; Willer et al., 2009; Zeggini et al., 2008] Unfortunately, not all SNPs are imputed with perfect accuracy. In fact, this accuracy may vary greatly from SNP to SNP. Most meta-analyses do not take this into account and this uncertainty leads to a loss of power.
Recently, de Bakker et al. [2008] have analyzed issues relating to conducting meta-analysis in the context of GWAS. In particular, they suggested incorporating estimates of imputation accuracy into the meta-analysis statistic by using an imputed SNP information measure. While this heuristic is intuitive, the exact statistic that maximizes meta-analysis study power remains unknown. In this work, we develop a new statistic, which takes this approach, correcting for potential inaccuracies of imputation by weighting results from each association study based on the accuracy of the imputation at each marker. In brief, results with large studies and accurate imputation are given more weight than smaller studies with inaccurate imputation. Furthermore, we analytically derive an optimal set of weights for combining results from each study in order to maximize power. We show that it can result in a significant increase in power compared to the standard weighted sum of Z-scores (WSoZ) approach used, for example, in three recent meta-analyses [Soranzo et al., 2009; Willer et al., 2009; Zeggini et al., 2008]. Unfortunately, the optimal weights cannot be computed directly from the data since they require knowledge about the true accuracy of the imputation. There are several methods for estimating the accuracy and we examine the application of one developed by Li and Abecasis [2006] in the context of our imputation aware meta-analysis statistic. We conduct several experiments showing that our new method for handling imputed genotypes from distinct SNP sets improves the power of meta-analysis.
METHODS
CASE-CONTROL STUDIES
In this work, we consider meta-analyses performed over several case-control studies, although our method can be adapted to handle continuous phenotypes. We begin with a description of a case-control study in order to introduce some notation. In a case-control study, individuals are collected from two groups, the cases and the controls. The individuals in each group differ along a phenotype of interest, such as disease state, but are otherwise members of the same population. The individuals are genotyped on a set of SNPs, and the allele frequency of each SNP si is measured in the cases and in the controls . Assuming a study with N/2 cases and N/2 controls where the true SNP frequencies in the population, cases, and controls are pi, , and , respectively, the Z-score statistic Zi in Equation (1) is computed for each SNP. It is normally distributed with mean equal to the non-centrality parameter (NCP) and variance 1. Those SNPs with statistic Zi>ϕ−1(1α/2), where ϕ−1(x) is the quantile function of the standard normal distribution and α is the significance threshold, are considered significant and maybe linked to a causal variant for the phenotype.
| (1) |
TRADITIONAL META-ANALYSIS
In order to combine data from several case-control studies, one of many standard meta-analysis approaches maybe employed. One common approach, taken by a growing number of GWAS meta-analyses is to take a WSoZ from each of the independent studies [Soranzo et al., 2009; Willer et al., 2009; Zeggini et al., 2008]. The data required from each study are the statistics for each SNP i in each study j, and the number of individuals Nj in each study j. We assume an equal number of cases and controls, although our methods can easily be adapted to unbalanced association studies.
For each SNP si in the studies, a meta-analysis statistic Mi, which is a WSoZ defined in Equation (2), is computed.
| (2) |
Mi is defined for any weights which are positive and with at least one greater than zero. The statistical power of using Mi to detect associations depends on the weights and is maximized when the weights . Intuitively, larger weights are assigned to studies with more individuals, and therefore with more power to detect an association. The optimality of these weights is shown with a direct application of the Cauchy Schwartz inequality . Under the fixed effects assumption of the WSoZ approach, for all j and there is equality when .
IMPUTATION
Unfortunately, the set of SNPs genotyped in a GWAS, or “tag” SNPs, are not identical between studies, so the required for meta-analysis are not immediately available. Furthermore, the set of tag SNPs is much smaller than the total number of SNPs in the population and it is likely that the causal variants are not contained in the tag SNP set. Recently, several methods have been developed to leverage existing data sets with millions of genotyped SNPs, such as the HapMap, to improve the power of association studies. If the study population is closely matched to a HapMap population, then it is possible to measure statistics over SNPs not included in the set of tag SNPs. In addition to improving the power of association studies, imputation methods can be used to aid meta-analysis of association studies that used different sets of tag SNPs by computing statistics at SNPs missing from either study but contained in the HapMap. Meta-analysis is performed by imputing the missing SNPs in each study and computing a statistic for each SNP i in the HapMap and each study j. This procedure will provide the required statistics to perform meta-analysis at all SNPs in both studies as well as all HapMap SNPs not contained in either study.
While imputation methods are accurate for a large number of SNPs, they are by no means perfect, and so statistics computed over imputed SNPs are not identical to those computed over the genotyped tag SNPs. The NCP at a tag SNP is a function of its relative risk, disease model, MAF, study size, and correlation coefficient to the causal variant. Let be the NCP of tag SNP si in a case-control study. Imputing si instead of genotyping it directly will alter the NCP of the resulting statistic. We define ri,j as the correlation coefficient between the imputed genotypes and the true genotypes of SNP si in study j. Intuitively, if ri,j is close to 1 then SNP is imputed well and the NCP will be to , and if ri,j is close to 0 then little information is known about the true genotypes of si and the NCP will be close to 0. The NCP of an imputed SNP is equal to , a function of the NCP of the SNP it is imputing as well as the correlation coefficient between the imputed and true genotypes. Current methods ignore this difference between imputed and genotyped SNPs; below, we show that this can lead to a reduction in power, and we present a new method to address this issue.
IMPUTATION AWARE META-ANALYSIS
The statistic computed for an imputed SNP does not necessarily share NCP across studies. The assumption that from the simple meta-analysis described above is still valid. However, the correlation between the imputed and true genotypes may vary from study to study affecting the NCP. Consider the situation in which two different studies with different tag sets impute a HapMap SNP sH. The linkage patterns between sH and the two different tag sets may give, for example, a correlation coefficient rH,1 = 0:7 for the first study and rH,2 = 0:95 for the second study. If both studies have N individuals, then the NCPs will be in the first study and in the second study. Given this result, the derivation for Mi in the simple case above no longer holds. Treating the statistics as the equivalent of directly genotyped SNPs may weaken the meta-analysis power. Our objective is to develop a new meta-analysis statistic, which accounts for the imputation error.
Adopting the same framework as the WSoZ method we wish to find a set of weights such that a weighted combination of the from each study will maximize Mi. The we propose is . Since , this is equivalent to . In this case, we consider not only study size but also the quality of the imputed genotypes. Provided that the imputed genotypes are accurate estimates of the probability of the true genotype given the observed tag SNP genotypes, poorly imputed SNPs will have low NCPs because their ri,j will be close to zero. A large study with poorly imputed genotypes for a SNP will not alter the meta-analysis statistic significantly if there exists a smaller study that genotypes the SNP directly. The proof of optimality once again follows from a direct application of the Cauchy Schwartz inequality.
To understand the effect of this new statistic consider a SNP si in a two study meta-analysis where each study has N/2 cases and N/2 controls. Suppose study 1 genotypes the SNP directly and that in study 2 the SNP is imputed, that is, ri,1 = 1 and ri,2 = r. Then in order to maximize power, we must maximize the NPC of the meta-analysis statistic Mi. We set and and get NCP of . If instead we choose to follow the standard WSoZ method for meta-analysis and set for all j, then we would get NCP of . In this case, if then the meta-analysis will have even less power than either study alone. If both studies impute the SNP then the potential for loss of power compared to our method is even greater.
ESTIMATING IMPUTATION CORRELATION
We showed that the correlation between the true and imputed genotypes ri,j are the weights which maximize the power of the meta-analysis. Unfortunately, these weights cannot be computed directly since the true genotypes of the imputed SNPs are unknown.
Several estimates of imputation quality relying solely on the imputed genotypes have been proposed. One such estimate of ri,j proposed by Li and Abecasis [2006] is called r2. It is the ratio of the empirical variance of the imputed genotypes to the expected variance given the imputation estimate of the MAF p̂.
| (3) |
Provided that the imputed genotypes are the expected dosages given the observed genotypes, then this will be the expected correlation coefficient.
Differences between the study population and the HapMap, the genotyping density and the finite size of the HapMap can effect this estimate of correlation [Zaitlen et al., 2009]. We examine the relation between the true ri,j and this estimate of imputation quality over several data sets. We show that the correlation is estimated closely enough to warrant the use of our new meta-analysis statistic over the WSoZ method when combining imputed genotypes.
RESULTS
POWER SIMULATIONS
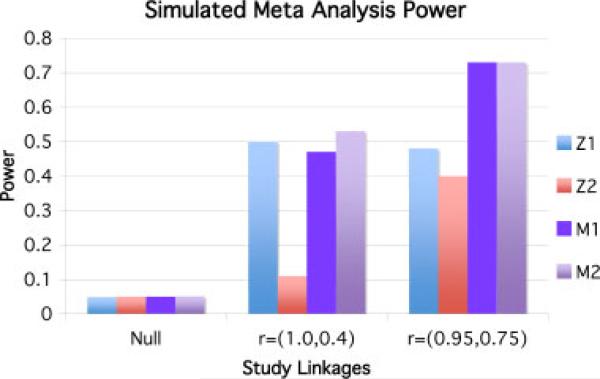
The difference in power between using a standard WSoZ and our imputation aware meta-analysis method is explored by simulating pairs of case-control studies. For every pair, we record the power of each study as well as the power of each type of meta-analysis. Figure 1 shows the results of three such simulations. In each of these simulations, both studies contain 2,000 individuals with equal numbers of cases and controls. The disease model is multiplicative with an OR of 1.203 and a causal SNP MAF of 0.05, giving an expected power of 50%. The genotypes in each study are generated as conditional binomial random variables with some correlation coefficient r to the causal variant. An r of 1 means that the causal variant and the generated genotypes are identical. For each study, we compute the Z-score and if the corresponding P-value is less than 0.05 we consider it successful. We also compute the weighted combination of the Z-scores from both studies according to the traditional method and our imputation aware method. This process is repeated 1,000 times and the power of the four methods is computed as the fraction of times a successful test occurred with an α = 0:05. In each simulation, our imputation aware meta-analysis statistic matched or beat the power of the traditional method. The difference between the methods is especially large when the quality of imputation is poor. In some circumstances, traditional meta-analysis power can be even lower than the power of an individual study, but this is never the case for the imputation aware statistic. Filtering poorly imputed SNPs has been suggested as means for addressing this issue [Zeggini et al., 2008]. This may prevent power loss beyond each of the individual studies if the threshold is high enough, but it will not prevent a power loss compared to the imputation aware statistic.
Fig. 1.
Power of simulated studies. Z1 is the power of study 1, Z2 is the power of study 2, M1 is the power of the WSoZ method, and M2 is the power of the imputation aware meta-analysis method. In the Null example, the genotypes are completely unlinked to the causal variants in both studies 1 and 2. In the second example, study 1 genotypes the causal variant directly and study 2 imputes it with r = 0.4. In the third example, study 1 and study two both impute the SNP with r = 0.95 and r = 0.75, respectively. Notice that the imputation aware meta-analysis method matches or beats the power of the traditional method in each case, and that in the second example the power actually drops in the traditional method due to poor imputation quality that is not accounted for in the second study. SNP, single nucleotide polymorphism; WSoZ, weighted sum of z-scores.
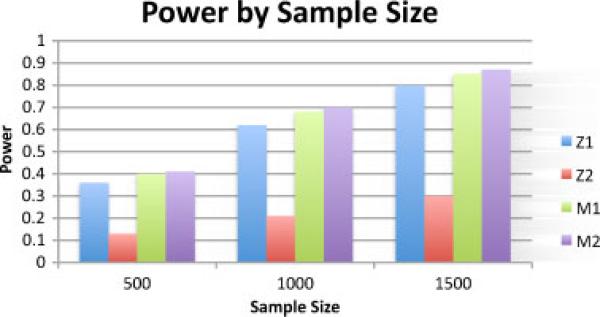
To further explore the difference between the WSoZ approach, we repeated the above experiments varying sample size instead of correlation coefficient. The correlation between the genotypes and the causal variant was fixed at 0.8 and 0.4 for the first and second study, respectively. We simulated balanced studies with 500, 1,000, and 1,500 cases. The results are presented in Figure 2. Again our imputation meta-analysis statistic outperformed the WSoZ approach.
Fig. 2.
Power of simulated meta-analysis studies under various sample sizes. Z1 is the power of study 1, Z2 is the power of study 2, M1 is the power of the WSoZ method, and M2 is the power of the imputation aware meta-analysis method. The genotypes are linked to the causal variants with r = 0.4 in study 1 and r = 0.8 in study 2. The sample size is the number of cases in balanced case-control study. For the entire range, the imputation aware meta-analysis beats the power of the WSoZ method showing the result is robust across variations in sample size. SNP, single nucleotide polymorphism; WSoZ, weighted sum of z-scores.
CORRELATION COEFFICIENT ESTIMATES
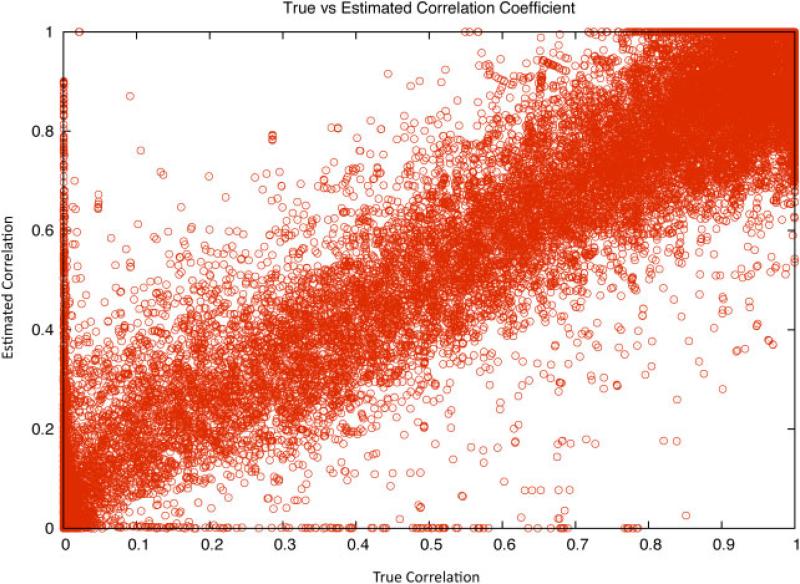
The optimal weighting of the Z-scores from individual studies cannot be computed from the data since the true genotypes of the imputed SNPs are unknown. Instead, the correlation between the true and imputed genotypes must be estimated. We examine the estimate r2 defined by Li and Abecasis [2006] over real genotype data in order to asses the feasibility of using our imputation aware meta-analysis method without access to the true value of . Using the controls from the Wellcome Trust Case-Control Consortium (WTCCC), we randomly removed one quarter of the genotyped SNPs producing new data sets for chromosomes 1, 2, and 22. For each data set, we imputed the removed SNPs with EMINIM [Kang et al., 2010] and computed the true value of for each SNP. We then estimated this correlation coefficient using r2. The results are shown in Figure 3. For all but the SNPs with low MAF, the value of r2 very closely approximates the true . In this data, which is still less dense than commercially available genotyping chips, the correlation exceeded 0.95.
Fig. 3.
Plot of the true correlation coefficient r2 versus the estimated correlation coefficient r2 of imputed SNPs in the WTCCC controls. The estimated and true correlation coefficients are highly correlated with r = 0.95 showing that the estimate is accurate. For SNPs with low minor allele frequency, the estimates of r2 are not reliable. SNP, single nucleotide polymorphism; MAF, minor allele frequency.
We repeated the experiments shown in Figures 1 and 2 with values of r sampled from the error observed in Figure 3. Since the estimates of r2 are tightly correlated with the true r2, there was no noticeable difference in the performance of our imputation aware meta-analysis. Thus, even without access to the optimal weights our method is still more powerful than traditional meta-analysis.
DISCUSSION
Currently, meta-analysis of genome-wide association studies is commonly performed using a WSoZ approach. This well-established method linearly combines the results of each study weighting them by their size. In this way, larger studies are up-weighted relative to smaller ones and their results have greater influence in the final meta-analysis statistic. GWAS do not necessarily contain the same set of genotyped SNPs and so additional work must be done before meta-analysis can be conducted. Specifically, an imputation method is used to estimate the genotypes of SNPs absent from either study. Typically, Z-scores over these imputed SNPs are then combined between studies using the traditional method.
Although the traditional method is optimal under certain reasonable assumptions, it does not take into account errors from imputation of genotypes. Thus, a large study that poorly imputes a genotype will be given more weight than a smaller study that imputes it perfectly. In this work, we introduce a novel meta-analysis statistic to deal with this issue of imputed genotypes in meta-analysis. Specifically, we adjust the weighting scheme of the traditional method to take into account the accuracy of the imputed genotypes. The new weights are function of both sample size and the correlation coefficient between the imputed and true genotypes. We show that our method is optimal under the same set of assumptions as the traditional approach. In addition, we show that for many cases our new statistic not only improves the meta-analysis power but also prevents a loss in power compared to each individual study that can occur when SNPs are poorly imputed.
Unfortunately, the optimal weights in our statistic are not computable from the results of GWAS and imputation. However, there exist several techniques for estimating them either directly from the imputed data or with a secondary data set such as the HapMap. We performed several experiments to examine the accuracy of one approach and found that although there are slight differences in accuracy depending on MAF and tag set density, for most current studies, the approach is accurate enough to estimate the weights effectively. That is, the power of the meta-analysis will still be improved using our new method with estimated correlation coefficients compared to using the previous method, which ignores imputation issues altogether.
ACKNOWLEDGMENTS
N.Z. and E.E. are supported by the National Science Foundation Grants No. 0513612, No. 0731455 and No. 0729049, and National Institutes of Health Grant No. 1K25HL080079. Part of this investigation was supported using the computing facility made possible by the Research Facilities Improvement Program Grant Number C06 RR017588 awarded to the Whitaker Biomedical Engineering Institute, and the Biomedical Technology Resource Centers Program Grant Number P41 RR08605 awarded to the National Biomedical Computation Resource, UCSD, from the National Center for Research Resources, National Institutes of Health. Additional computational resources were provided by the California Institute of Telecommunications and Information Technology (Calit2), and by the UCSD FWGrid Project, NSF Research Infrastructure Grant Number EIA-0303622. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- de Bakker PIW, Ferreira MAR, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. DOI: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Stephens M. Practical issues in imputation-based association mapping. PLoS Genet. 2008;4:e1000279. doi: 10.1371/journal.pgen.1000279. DOI: 10.1371/journal.pgen.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L, Junkins H, Mehta J, Manolio T. A catalog of published genome-wide association studies. 2009 Available from: www.genome.gov/26525384.
- International-HapMap-Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. DOI: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen N, Eskin E. EMINIM: an adaptive and memory efficient algorithm for genotype imputation. J Comput Biol. 2010;17:547–560. doi: 10.1089/cmb.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavvoura FK, Ioannidis JPA. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. DOI: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- Li Y, Abecasis G. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. DOI: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, Hammond N, Stolk L, Nica A, Inouye M, Hofman A, Stephens J, Wheeler E, Arp P, Gwilliam R, Jhamai PM, Potter S, Chaney A, Ghori MJR, Ravindrarajah R, Ermakov S, Estrada K, Pols HAP, Williams FM, McArdle WL, van Meurs JB, Loos RJF, Dermitzakis ET, Ahmadi KR, Hart DJ, Ouwehand WH, Wareham NJ, Barroso I, Sandhu MS, Strachan DP, Livshits G, Spector TD, Uitterlinden AG, Deloukas P. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. DOI: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJF, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KTT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O'Rahilly S, Purmann C, Rees MG, Ridderstrle M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YCL, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Consortium WTCC, Witteman JCM, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MRR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HEE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN, ANthropometric Traits Consortium GI Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. DOI: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Min KH, Eskin E. Linkage effects and analysis of finite sample errors in the hapmap. Hum Hered. 2009;68:73–86. doi: 10.1159/000212500. DOI: 10.1159/000212500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PIW, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrm KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJJ, Doney ASF, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jrgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CNA, Payne F, Perry JRB, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjgren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Consortium WTCC, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. DOI: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]