Abstract
Arginine adenosine-5′-diphosphoribosylation (ADP-ribosylation) is an enzyme-catalyzed, potentially reversible posttranslational modification, in which the ADP-ribose moiety is transferred from NAD+ to the guanidino moiety of arginine. At 540 Da, ADP-ribose has the size of approximately five amino acid residues. In contrast to arginine, which, at neutral pH, is positively charged, ADP-ribose carries two negatively charged phosphate moieties. Arginine ADP-ribosylation, thus, causes a notable change in size and chemical property at the ADP-ribosylation site of the target protein. Often, this causes steric interference of the interaction of the target protein with binding partners, e.g. toxin-catalyzed ADP-ribosylation of actin at R177 sterically blocks actin polymerization. In case of the nucleotide-gated P2X7 ion channel, ADP-ribosylation at R125 in the vicinity of the ligand-binding site causes channel gating. Arginine-specific ADP-ribosyltransferases (ARTs) carry a characteristic R-S-EXE motif that distinguishes these enzymes from structurally related enzymes which catalyze ADP-ribosylation of other amino acid side chains, DNA, or small molecules. Arginine-specific ADP-ribosylation can be inhibited by small molecule arginine analogues such as agmatine or meta-iodobenzylguanidine (MIBG), which themselves can serve as targets for arginine-specific ARTs. ADP-ribosylarginine specific hydrolases (ARHs) can restore target protein function by hydrolytic removal of the entire ADP-ribose moiety. In some cases, ADP-ribosylarginine is processed into secondary posttranslational modifications, e.g. phosphoribosylarginine or ornithine. This review summarizes current knowledge on arginine-specific ADP-ribosylation, focussing on the methods available for its detection, its biological consequences, and the enzymes responsible for this modification and its reversal, and discusses future perspectives for research in this field.
Keywords: ADP-ribosylation, Arginine, ADP-ribosyltransferase, ADP-ribosylarginine hydrolase, Posttranslational modification
Introduction
ADP-ribosylation of arginine is a reversible posttranslational modification (PTM) of proteins in which the ADP-ribose moiety is transferred from NAD+ to the guanidino group of arginine under release of nicotinamide (Fig. 1). This reaction is catalyzed by a subfamily of ADP-ribosyltransferases (ARTs) that bind NAD+ in an extended conformation, enabling the nucleophilic attack of one of the two terminal nitrogen atoms of the guanidino group of arginine on the β-N-glycosidic bond between nicotinamide and the C1′-atom of the ribose-group (Haag and Koch-Nolte 1997; Jacobson and Jacobson 1989; Margarit et al. 2006; Moss and Vaughan 1990; Tsuge et al. 2008). Nicotinamide is released and a new N-glycosidic bond between arginine and ADP-ribose is generated with an inversion of the conformation at the C1′ atom of ADP-ribose from beta to alpha (Fig. 1). Arginine ADP-ribosylation can be fully reversed by specific enzymes (ADP-ribosylhydrolases). Other acceptor amino acids, such as diphthamide (a modified histidine), cysteine or asparagine, are targeted by other sub-families of ADP-ribosyltransferases via a similar reaction mechanism (Berti et al. 1997; Hottiger et al. 2010; Koch-Nolte et al. 2008; Lang et al. 2010; Locht and Antoine 1995).
Fig. 1.
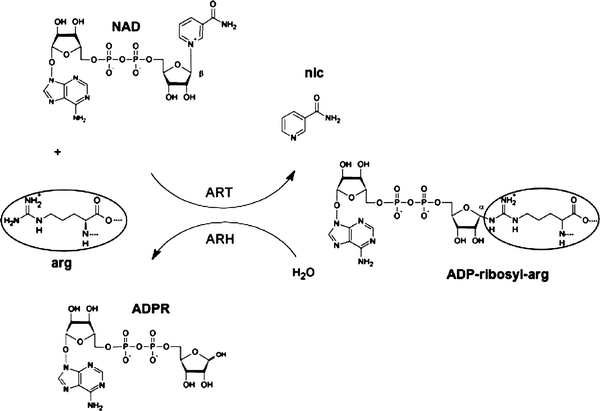
Schematic diagram of the enzyme catalyzed, reversible posttranslational modification of arginine by ADP-ribose. In the active centre of an ADP-ribosyltransferase (ART), NAD+ is brought into an extended conformation that permits the attack of the target arginine on the β-N-glycosidic bond between nicotinamide and the C1′-atom of the ribose group. This leads to the formation of ADP-ribosylarginine with C1′ in α-conformation, while nicotinamide is released. The native arginine can be recovered by the reverse reaction, catalyzed by an ADP-ribosylarginine hydrolase (ARH). This enzyme hydrolyses the α-glycosidic bond, releasing ADP-ribose
Methods for detecting arginine ADP-ribosylation
Several NAD+ analogues have been employed to visualize arginine ADP-ribosylation. Using radioactively labelled [32P]-NAD+ as a substrate, radiolabelled target proteins can be detected by SDS-PAGE autoradiography (Fig. 2) (Koch-Nolte et al. 1996; Vandekerckhove et al. 1987; Zolkiewska and Moss 1993). In a similar way, the use of biotinylated NAD+ or etheno-NAD+ (an NAD+ analogue with an additional penta-ring, i.e. etheno-group, on the adenosine moiety) allows detection of the target protein by western blot analyses using streptavidin or the etheno-adenosine-specific monoclonal antibody 1G4, respectively (Krebs et al. 2003). These NAD-analogues can be used also to detect ADP-ribosylated proteins on the surface of intact cells by flow cytometry. However, they do not allow the detection of endogenously ADP-ribosylated proteins and do not provide information on the identity of the acceptor amino acid.
Fig. 2.
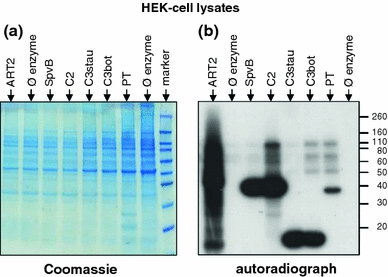
Monitoring ADP-ribosylation by specific and promiscuous ARTs by SDS-PAGE autoradiography. HEK-cell lysates were incubated for 10 min at 37°C with arginine-specific ARTs (ART2.2, SpvB, C2) or non-arginine-specific ARTs (C3stau2, C3bot, PT) in the presence of [32P]-NAD+. Proteins were size-fractionated by SDS-PAGE and visualized by Coomassie staining (a). SDS-resistant, i.e. covalently incorporated, radioactivity was detected by autoradiography (b). While most bacterial ARTs modify a single prominent band, mouse ART2.2 (lane 1) modifies many target proteins. Enzymes used for ADP-ribosylation: Mouse ART2.2 (ART2); Salmonella enterica SpvB toxin (SpvB); Clostridium botulinum C2 toxin (C2); Staphylococcus aureus exoenzyme C3stau2 (C3stau); Clostridium botulinum C3 toxin (C3bot); Pertussis toxin (PT)
Several attempts have been made to generate antibodies that recognize ADP-ribosylated arginine residues, analogous to those that have been used for detecting phospho-tyrosine or phospho-serine residues (Meyer and Hilz 1986; Osago et al. 2008; Schwab et al. 2000). Most of the antibodies described so far, however, lack specificity for arginine ADP-ribose, i.e. show cross reactivity with other mono- or poly-ADP-ribosylated amino acid residues. Similarly, a naturally occurring ADP-ribose-binding domain has been used to detect and purify ADP-ribosylated proteins. This domain was found to bind to proteins ADP-ribosylated on arginine and other amino acid residues (Dani et al. 2009).
ADP-riboslyation of arginine can be distinguished from that at other amino acid residues by its higher sensitivity to hydrolysis by hydroxylamine (Cervantes-Laurean et al. 1993), or by arginine-specific ADP-ribosylhydrolase ARH1 (see below). Moreover, ADP-ribosylation of arginine can be blocked by arginine analogues such as agmatine or meta-iodobenzylguanidine (MIBG) that function as competitive inhibitors (Fig. 3). Both, agmatine and MIBG, can themselves be ADP-ribosylated by arginine-specific ARTs, a reaction that can be monitored by thin layer chromatography and autoradiography (Koch-Nolte et al. 1996; Loesberg et al. 1990).
Fig. 3.
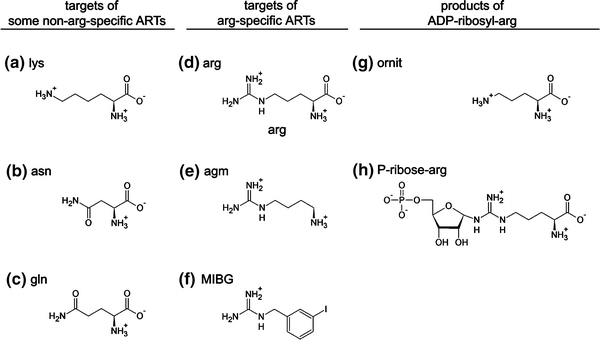
Schematic diagrams of molecules relevant to ADP-ribosylation. a–c Target amino acids with a terminal nitrogen group that can be modified by non-arginine specific ARTs; d–f guanidino group-containing targets of arginine-specific ARTs; g, h products of enzymatic and non-enzymatic hydrolysis of ADP-ribosylated-arginine. a lysine; b asparagine; c glutamine; d arginine; e agmatine; f MIBG (meta-iodobenzylguanidine); g ornithine; h phospho-ribosylarginine
Specific ADP-ribosylated arginine residues can be identified by mass spectrometry (MS) and/or sequencing. To this end, the purified ADP-ribosylated protein is subjected to protease digestion. The resulting peptides are fractionated by high-performance liquid chromatography (HPLC) and subsequently identified by Edman sequencing or MS (Margarit et al. 2006; Paone et al. 2002; Pope et al. 1985; Terashima et al. 1992; Zhou et al. 1996). Recently, two new techniques have been developed to facilitate the identification of target residues by MS. In one approach, the fragmentation of ADP-ribose-containing tryptic peptides generates characteristic breakdown products of ADP-ribose or ADP-ribosylarginine that can be used to trace the ADP-ribosylated peptides (Hengel et al. 2009; Osago et al. 2009). In a second approach, tryptic fragments containing ADP-ribose moieties were specifically enriched using a titan oxide affinity matrix before MS analysis (Lang et al. 2010).
Specific ADP-ribosylated arginine residues have also been identified successfully by site-directed mutagenesis followed by an ADP-ribosylation assay, e.g. in transfected cells or with purified proteins (Adriouch et al. 2008; Ganesan et al. 1998; Ganesan et al. 1999a; Hochmann et al. 2006; Lupi et al. 2000).
Structural and functional consequences of arginine ADP-ribosylation
Arginine ADP-ribosylation causes a notable change in size and chemical property at the ADP-ribosylation site of the target protein. While the average size of one amino acid is approximately 110 Da, the attached ADP-ribose group is a large bulky group that is nearly five times as big (approx. 540 Da). In contrast to arginine, which, at neutral pH, is positively charged, ADP-ribose carries two negatively charged phosphate moieties. Thus, the attachment of ADP-ribose results in a relatively large increase in molecular weight and in the introduction of two new negative charges to the acceptor molecule. By comparison, phosphorylation leads to an increase of approximately 80 Da and to the introduction of a single additional negative charge.
ADP-ribosylation at arginine residues can affect the function of target proteins by different mechanisms and the resulting effect on protein function can be activating or inactivating. In principle, the introduction of the bulky ADP-ribose moiety could sterically block the interaction with a binding partner, create a new docking site for ADP-ribose binding domains in other proteins, or induce a conformational change. Steric blockade is expected to be inactivating, whereas the creation of interaction sites with molecular partners or the induction of a conformational change will often be activating, as in case of phosphorylation.
Steric hindrance has been shown by X-ray crystallography to be the mechanism by which the ADP-ribosylation of G-actin at Arg177 inhibits actin polymerisation (Margarit et al. 2006). Steric hindrance has also been implicated in the reduced binding of the small G protein ras to its guanine nucleotide exchange factor Cdc25 upon ADP-ribosylation of ras at Arg41 (Ganesan et al. 1999b), and in altered DNA recognition by E. coli RNA polymerase upon ADP-ribosylation at Arg265 (Depping et al. 2005; Westblade et al. 2008).
ADP-ribosylation can also result in the creation of molecular interaction sites. The macro module found in the histone macroH2A1.1 and several other proteins is an ADP-ribose binding domain (Till and Ladurner 2009). An archaebacterial macro domain has been used successfully in pulldown experiments to purify arginine ADP-ribosylated proteins (Dani et al. 2009). While histone macroH2A1.1 has recently been shown to bind to poly-ADP-ribosylated nuclear proteins in live cells (Timinszky et al. 2009), it remains to be determined whether arginine-ADP-ribosylated proteins similarly interact with macro domains or other protein modules in vivo.
The most thoroughly studied example for the activation of a target protein by the induction of a conformational change is the gating of the P2X7 ion channel by ADP-ribosylation (Seman et al. 2003). P2X7 is an adenosine-5′-triphosphate (ATP) gated cation channel expressed on haematopoietic cells that mediates a multitude of pro-inflammatory effects. Triggering of P2X7 by its soluble ligand, ATP, causes an influx of calcium and an efflux of potassium ions, resulting in membrane changes (externalisation of phosphatidylserine, blebbing) and, ultimately, in cell death by apoptosis. Activation of P2X7 generally requires high (upper micromolar range) concentrations of extracellular ATP. However, on naive murine T lymphocytes, which carry ART2.2, P2X7 is activated by low micromolar concentrations of extracellular NAD+ via ADP-ribosylation on Arg 125 (Adriouch et al. 2008; Schwarz et al. 2009). This leads to rapid apoptotic death of naïve T lymphocytes carrying ART2.2, a phenomenon termed NAD-induced cell death (NICD) (Seman et al. 2003). It has been proposed that this provides space for the expansion of activated effector T cells, which are rendered resistant to NICD upon shedding of ART2.2 (Koch-Nolte et al. 2006).
ART2-deficient mice (Ohlrogge et al. 2002) are viable and fertile, but are resistant to NICD (Adriouch et al. 2008; Seman et al. 2003). Depending on the expression levels of ART2.2 and on other factors, different T cell subpopulations show varying degrees of sensitivity to NICD. Thus, in a mouse model for insulin-dependent type 1 diabetes (T1D), the absence of ART2 leads to the enhanced survival of a population of invariant Natural Killer-T (iNKT) cells that exert a negative regulatory influence on disease progression. Here, genetic deletion or pharmacological blockade of ART2 are associated with a decreased incidence of the disease (Scheuplein et al. 2010).
Scope of arginine ADP-ribosylation
How many different target proteins can be modified by ADP-ribosylation? This question is difficult to answer, but several lines of evidence indicate that the target proteins identified to date represent only the tip of an iceberg. In principle, the scope of arginine ADP-ribosylation is determined by the relative location of ARTs and their target proteins, and by the local availability of the substrate NAD+. The concentration of NAD+ is two to three orders of magnitude higher in the intracellular than in the extracellular compartment. All vertebrate ARTs identified to date are membrane-bound or secreted ectoenzymes with the capacity to modify several different proteins (promiscuous), while most bacterial toxin ARTs affecting vertebrate organisms act inside the cell and modify only a single target (monospecific) (Koch-Nolte et al. 2008). The incubation of intact mouse T lymphocytes (that carry cell-surface ART2) with radioactive NAD results in the labelling of numerous bands, of which only a minority has been identified. Although no intracellular arginine-specific ARTs have been characterized to date, arginine ADP-ribosylation of intracellular proteins including Gβ, actin, and histones has been detected by biochemical means (Corda and Di Girolamo 2003). Little is known about the availability of NAD+ in different subcellular compartments, but an elegant recent study using poly ADP-ribose polymers generated by poly-ADP-ribosylpolymerase-1 (PARP-1) targeted to different cellular compartments provided evidence for distinct NAD+ pools in mitochondria, peroxisomes, and in the ER and Golgi (Dolle et al. 2010). In addition to cell-surface and intracellular substrates, it is at present impossible to estimate the scope of secreted soluble proteins such as cytokines (Saxty et al. 2001) or defensins (Paone et al. 2002) that may serve as targets for ADP-ribosylation.
A better assessment of the scope of endogenously ADP-ribosylated target proteins should become possible with improvements of affinity isolation techniques for purifying arginine-ADP-ribosylated proteins before analysis by mass spectrometry. The finding that arginine-specific ADP-ribosyltransferases can employ etheno-NAD or biotin-NAD as substrates opens the possibilities of using immobilized monoclonal antibody 1G4 directed against etheno-adenosine or immobilized avidin to affinity purify etheno-ADP-ribosylated or biotin-ADP-ribosylated target proteins, respectively (Krebs et al. 2003). Similarly, immobilized macro-domains may be suited to purify the proteome of ADP-ribosylated target proteins (Dani et al. 2009). Neither of these procedures, however, is specific for arginine-ADP-ribosylation. This bottleneck might be surmounted only with novel affinity purification tools based on ADP-ribosyl-arginine specific antibodies.
Arginine-specific ADP-ribosyltransferases
Arginine residues are ADP-ribosylated by arginine-specific ADP-ribosyltransferases (ARTs). These enzymes represent a well-characterized subfamily of the larger family of structurally related proteins that includes ARTs which ADP-ribosylate other amino acid residues, DNA, tRNA and antibiotics (Koch-Nolte et al. 2008). On the basis of the constellation of amino acid residues at three positions essential for NAD-binding, ARTs can be divided into two major subclasses, ARTC (Cholera toxin-like) carrying the R-S-E triad motif and ARTD (Diphtheria toxin-like) carrying the H-Y-E triad motif (Hottiger et al. 2010). Arginine-specific ARTs belong to the ARTC subclass and can be distinguished from other members of this subclass on the basis of a characteristic variant R-S-EXE triad motif (Fig. 4). Arginine-specific ARTs have been isolated and molecularly characterized from phages, bacteria and vertebrates (Table 1). Target arginines have been identified in numerous target proteins. In many but not all cases, the responsible ARTs have been identified by molecular cloning.
Fig. 4.
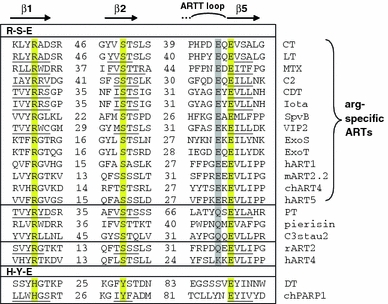
Sequence alignment of arginine- and non-arginine-specific ARTs. The regions surrounding the three catalytically important residues on the β1, β2, and β5 strands are aligned (Hottiger et al. 2010; Koch-Nolte et al. 2001; Otto et al. 2005). The R-S-E and H-Y-E motifs characterizing the major ART families are highlighted in yellow. For the R-S-E ARTs, the residue in position −2 relative to the catalytic glutamic acid in β5 is highlighted in grey. Specificity for arginine is determined by the presence of glutamic acid at this position (R-S-EXE motif). Enzymes shown in the alignment: cholera toxin (CT); Escherichia coli heat labile enterotoxin (LT); Bacillus sphaericus mosquitocidal toxin (MTX); Clostridium botulinum C2 toxin (C2); Clostridium difcile ADP-ribosylating toxin (CDT); Clostridium perfringens iota toxin (Iota); Salmonella enterica SpvB toxin (SpvB); Bacillus cereus VIP2 toxin (VIP2); Pseudomonas aeruginosa exoenzyme S (ExoS); Pseudomonas aeruginosa exoenzyme T (ExoT); human ART1 (hART1); mouse ART2.2 (mART2.2); chicken ART4 (chART4); human ART5 (hART5); pertussis toxin (PT); Pieris rapae pierisin-1 (pierisin); Staphylococcus aureus exoenzyme C3stau2 (C3stau2); rat ART2 (rART2); human ART4 (hART4); diphtheria toxin (DT); chicken poly(ADP-ribose)transferase-1 (chPARP-1). Underlined sequences indicate residues confirmed to be in β-strands in the respective crystal-structures. The ARTT (ADP-ribosyltransferase turn-turn) loop is involved in target protein recognition and contributes to catalysis (Koch-Nolte et al. 2001; Menetrey et al. 2008)
Table 1.
Arginine-specific ARTs and their target specificities
| Enzyme | Specificitya | Target(s) | Residue(s)b | Reference(s) |
|---|---|---|---|---|
| Microbial | ||||
| E.coli T-phages toxins (ALT/MOD) | p | RNA-polymerase and others | R265 | (Goff 1974; Depping et al. 2005) |
| Rhodospirillum rubrum DRAT | s | Dinitrogenase reductase | R101 | (Pope et al. 1985; Ludden 1994) |
| Vibrio Cholerae toxin (CT) | s | Gαs, Gαt | R187 | (Moss et al. 1977; Spangler 1992) |
| Escherichia coli toxin (LT1/LT2) | s | Gαs, Gαt | R187 | (Moss et al. 1979; Spangler 1992) |
| Bacillus sphaericus toxin (MTX) | p | EF-Tu and others | Unknown | (Schirmer et al. 2002) |
| Clostridium botulinum toxin (C2) | s | Actin | R177 | (Aktories et al. 1986; Vandekerckhove et al. 1988) |
| Clostridium difficile toxin (CDT) | s | Actin | R177 | (Stubbs et al. 2000) |
| Clostridium perfringens iota toxin | s | Actin | R177 | (Vandekerckhove et al. 1987; Boquet et al. 1998) |
| Salmonella enterica toxin (SpVB) | s | Actin | R177 | (Tezcan-Merdol et al. 2001; Margarit et al. 2006) |
| Bacillus cereus toxin (VIP2) | s | Actin | R177 | (Han et al. 1999) |
| Pseudomonas exotoxin S (ExoS) | p | ras and others | R41/R128 | (Ganesan et al. 1998) |
| Pseudomonas exotoxin T (ExoT) | p | Crk and others | Unknown | (Sun and Barbieri 2003) |
| Vertebrate | ||||
| ART1 | p |
α7 integrin, HNP-1 and others |
Unknown R14/R24 |
(Zolkiewska and Moss 1993; Paone et al. 2002; Paone et al. 2006) |
| ART2.1/ART2.2 | p | P2X7 and others | R125/R133 | (Seman et al. 2003; Adriouch et al. 2008) |
| Turkey erythrocyte ART | p | Actin | R95, R372 | (Just et al. 1995) |
| Chicken heterophil ART | p | Tuftsin, actin and others |
R4 R28, R206 |
(Terashima et al. 1995; Terashima et al. 1997) |
| Rabbit muscle cell ART | p | Desmin | R48, R62 | (Huang et al. 1993; Zhou et al. 1996) |
| Chinese hamster ART | p | Gβ | R129 | (Lupi et al. 2000) |
| Hen liver nuclear ART | p | Kemptide | R2, R3 | (Matsuura et al. 1988) |
aSpecific (s) or promiscuous (p)
bPreferred sites are in bold
Factors determining the specificity of certain ARTs for arginine
The factors determining the specificity of arginine-specific ARTs are still poorly understood. In addition to their target proteins, many arginine-specific ARTs can also ADP-ribosylate poly-l-arginine and small arginine-analogues that contain a guanidino group such as agmatine or MIBG (Fig. 3). However, they do not ADP-ribosylate other terminal nitrogen-containing amino acid side chains such as asn, gln or lys, which do serve as targets for other ARTs (e.g. C. botulinum C3, P. luminescence TccC5, and human PARP-1, respectively) (Altmeyer et al. 2009; Lang et al. 2010; Sekine et al. 1989).
Several lines of evidence indicate that arginine specificity is governed by a glutamic acid residue at the edge of the catalytic cleft. This acidic amino acid residue is positioned two amino acids upstream of the catalytic glutamic acid and defines the variant R-S-EXE triad motif (Koch-Nolte et al. 2008). Figure 4 shows five examples, where pairs of closely related ARTs differ at this position. In each case only the EXE containing ART modifies arginine. Cholera toxin (EXE) ADP-ribosylates Gαs at Arg187, whereas the related pertussis toxin (QXE) ADP-ribsylates Gαi at cysteine (Corda and Di Girolamo 2003). MTX (EXE) ADP-ribosylates elongation factor Tu and other proteins on arginine, the related pierisin (QXE) ADP-ribosylates guanosine in DNA (Carpusca et al. 2006). C2 and SpvB (EXE) ADP-ribosylate actin at Arg177 (Aktories et al. 1986; Margarit et al. 2006; Vandekerckhove et al. 1988), the related C3 enzymes (QXE) ADP-ribosylate rho at asparagine 41 (Aktories and Barth 2004; Sekine et al. 1989). Mouse ART2 (EXE) ADP-ribosylates P2X7 at Arg125 and Arg133 and other proteins at arginine residues (Adriouch et al. 2008; Koch-Nolte et al. 1996), rat ART2 (QXE) does not modify target proteins but hydrolyzes NAD+ to ADP-ribose and nicotinamide (Haag et al. 1995). Chicken ART4 (EXE) modifies several proteins at arginine residues; human ART4 (KXE) does not display any detectable ART activity (Glowacki et al. 2002; Grahnert et al. 2008). Direct experimental evidence for the importance of the EXE-motif was provided by site directed mutagenesis of ART2 and C3, in which creating or disrupting this motif conferred or destroyed the ability to ADP-ribosylate arginine residues (Hara et al. 1996; Maehama et al. 1996; Vogelsgesang and Aktories 2006).
Factors determining the preference of promiscuous ARTs for certain arginines
It is a striking observation that some arginine-specific ARTs show a high degree of specificity for a certain target protein, while others are capable of modifying many different targets. ARTs such as SpvB and C2 ADP-ribosylate only a specific target protein at a specific arginine residue, i.e. actin at Arg177 (Fig. 2, lanes 3 and 4). Other ARTs appear more promiscuous, e.g. mouse ART2, which ADP-ribosylates many proteins, albeit only at arginine residues (Fig. 2, lane 1). However, even in the case of this “promiscuous” ART, the identification of two specific arginine residues in the target protein P2X7 shows that only certain arginine residues are chosen for modification, i.e. Arg125 and Arg133, but not 16 other arginine residues exposed on the surface of the protein (Fig. 5). This raises the question of what governs the selection of target proteins, and within them, of target residues? Target specificity is presumably regulated on several levels.
Fig. 5.
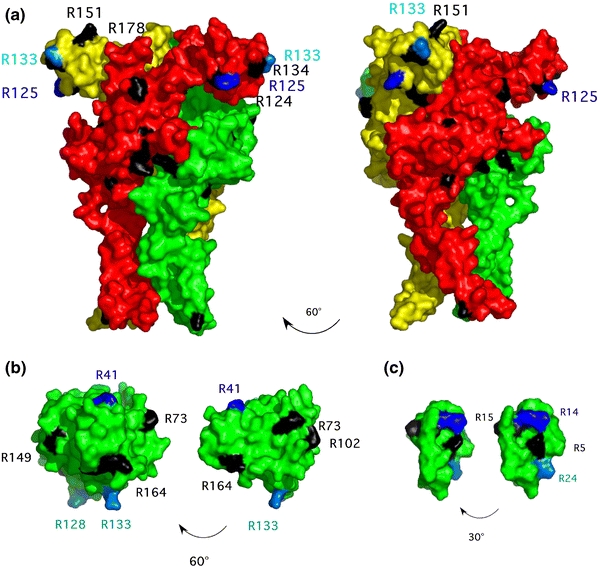
Three-dimensional models of P2X7 (a), ras (b), and HNP-1 (c), highlighting arginine residues that are targets for ADP-ribosylation. Preferred (primary) ADP-ribosylation sites are highlighted in blue, secondary target residues in cyan. Other arginine residues that are not targets for ADP-ribosylation are shown in black. Two views of each molecule, rotated as indicated by the arrow, are shown. The representations were made using the PyMol Molecular Viewer software and are based on the following structures in the PDB database: 2KHT (HNP-1) and 121P (ras). For P2X7, arginine residues were projected onto the recently solved structure of zebrafish P2X4 (PDB: 3I5D)
In the case of the specific ARTs, target specificity is likely determined by structural complementarity between the enzyme and its target. In the recently elucidated 3D structure of co-crystallized Clostridium perfringens iota toxin and actin, five loops in the toxin flanking the NAD-binding crevice interact substantially with actin (Tsuge et al. 2008). The target arginine 177 of actin thereby is positioned close to the first glutamate residue of the EXE motif. It is reasonable to assume that other specific ARTs such as DRAT, T4 and CT undergo similar extensive interactions with their target proteins. The EXE motif is contained within a prominent loop, designated ADP-ribosyltransferase turn turn or ARTT loop, preceding and including the catalytic glutamic acid residue (Fig. 4) (Han and Tainer 2002; Koch-Nolte et al. 2001). This loop has been shown to participate in target recognition also in the 3D structures of other distantly related ARTs with their targets, including PARP-1 and ADP-ribose (Ruf et al. 1998), rifampin ADP-ribosyltransferase and rifampin (Baysarowich et al. 2008), and Pseudomonas aeruginosa exotoxin A (ETA) and elongation factor 2 (Jorgensen et al. 2008).
In the case of the promiscuous ARTs, binding of target proteins evidently is much less stringently controlled by surface complementarity. Nevertheless, the ARTT loop seems to play a certain role also in target selection by the more promiscuous ARTs. The ART domains of exotoxins S and T of P. aeruginosa are closely related, yet differ in specificity for target molecules. Barbieri and coworkers elegantly demonstrated that specificity for target proteins could be transferred by grafting defined loops from one enzyme onto the other (Sun et al. 2004). In the case of these promiscuous ARTs and the closely related vertebrate ARTs, other factors such as cellular compartmentalization and topological constraints presumably regulate accessibility of arginine residues in target proteins. In this respect, promiscuous ARTs resemble certain protein kinases which are promiscuous toward a wide range of artificial substrates, but nevertheless exhibit a certain degree of target specificity in vivo (Woodgett et al. 1986).
A first level of specificity is achieved by regulation of the access of the enzyme to the target molecule. The plasma membrane and intracellular membranes present natural barriers that could restrict the access of ARTs to their targets. Exoenzymes S and T (ExoS and exoT) of Pseudomonas aeruginosa are injected into the cytosol of eukaryotic cells by the type III secretion apparatus of P. aeruginosa. These enzymes thus may have access mainly to cytosolic proteins (Barbieri and Sun 2004). Conversely, most of the known vertebrate arginine-specific ARTs are anchored in the outer leaflet of the plasma membrane by covalent linkage to glysosylphosphatidylinositol (GPI) (Glowacki et al. 2001). These ecto-enzymes thus may have access only to the ecto-domains of other membrane-associated or secreted proteins. Moreover, the GPI anchor governs association of ARTs with specialized membrane microdomains, designated lipid rafts, which may further restrict the access of these enzymes to other raft-associated proteins (Bannas et al. 2005). However, it is conceivable that ecto-ARTs may translocate across the membrane and thereby gain access to cytosolic targets such as Gβ, which is ADP-ribosylated at Arg129 (Lupi et al. 2000). An additional level of specificity may be provided by the topological constraint of the membrane anchor, which may allow access of the ART only to those arginine residues of its target that are at the same distance from the membrane as the catalytic site. This has been hypothesized to determine the choice of target residues modified by ART1 in integrin α7 (Zolkiewska 2005).
It is conceivable that the accessibility of arginine residues for ADP-ribosylation is influenced also by the conformation of the target protein and/or its interaction with other proteins. Integrins and other identified targets of membrane anchored ARTs, for example, are known to undergo large conformational alterations, e.g. after binding to ligands on other cells. Such conformational rearrangements conceivably could uncover or hide particular arginine ADP-ribosylation sites (Zolkiewska 2005). Moreover, binding of the target protein to other interaction partners may sterically interfere with access of ARTs to potential target arginines. This has been demonstrated, for example, in the case of antibodies that sterically interfere with ADP-ribosylation of LFA-1 (Nemoto et al. 1996) and P2X7 (our own observations).
Promiscuous ARTs often modify their target molecules at more than one site, but seem to show preference for certain sites (Table 1). These sites often are modified preferentially or exclusively at low concentrations of the substrate NAD+. Elevation of the substrate concentration may lead to the modification at additional sites, sometimes changing the biological consequences of ADP-ribosylation. An example is the modification of integrin α7 by ART1. At low concentrations of NAD+, integrin α7 is modified exclusively within the stalk region of the molecule, leading to an increased binding to its substrate laminin. At higher concentrations of NAD+, integrin α7 is additionally modified within the ligand-binding region, which inhibits binding to laminin (Zhao et al. 2005; Zolkiewska 2005). Moreover, ADP-ribosylation at one site may influence the accessibility of other arginines in the same protein for ADP-ribosylation. For example, ras is ADP-ribosylated by exoS preferentially at R41 but also at R128. When both sites are mutated to lysine, a third site R135 is ADP-ribosylated (Ganesan et al. 1998; Ganesan et al. 1999a).
Crystal structures of ART target proteins allow a comparison of the location and structural contexts of arginines that serve as targets versus those that do not. Figure 5 illustrates the location of the preferred and secondary ADP-ribosylation sites on P2X7, ras, and human neutrophil peptide-1 (HNP-1). HNP-1, a small secretory protein of 30 amino acids, is preferentially modified by human ART1 on R14 with an additional site at R24 (Paone et al. 2006; Paone et al. 2002; Stevens et al. 2009). Mouse P2X7 is modified by ART2.2 at R125 and at R133 (Adriouch et al. 2008) (Fig. 5). Of note, in both of these cases as well as in the case of the preferred site in HNP-1, the modified arginine is part of a di-arginine (RR) motif. In two of these sites (HNP-1 R14 and P2X7 R133) the first arginine within the motif is the acceptor, whereas in P2X7 R125 the second arginine is modified. The artificial substrate kemptide, a synthetic heptapeptide (L-R-R-A-S-L-G) also contains two consecutive arginines that are the target sites for ADP-ribosylation by both, bacterial and eukaryotic ARTs (Kharadia and Graves 1987; Matsuura et al. 1988). Interestingly, when kemptide is modified by cholera toxin, the first arginine is the preferred modification site (Kharadia and Graves 1987), whereas when kemptide is modified by hen liver ART, the second arginine within the motif is the preferred modification site (Matsuura et al. 1988). However, ART1 and ART2 can also modify single arginines, i.e. that are not part of a di-arginine motif (Paone et al. 2006) (and our own unpublished observations). Thus, for promiscuous arginine-specific ARTs, the target arginines identified so far are not located within any evident common sequence or structural motif.
Reversion of arginine ADP-ribosylation
Like many posttranslational modifications with regulatory functions, mono-ADP-ribosylation of arginine is a potentially reversible process. Examples have been reported for both, complete removal of ADP-ribose and restoration of protein function, as well as for the processing of ADP-ribosylarginine to other PTMs (Fig. 6).
Fig. 6.
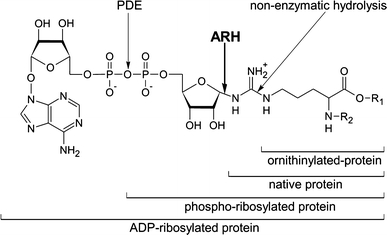
Enzymatic and non-enzymatic processing of ADP-ribosylarginine. See text for details. ARH ADP-ribosylhydrolase, PDE phosphodiesterase
Complete removal of ADP-ribose
Full restoration of protein function by complete removal of the ADP-ribose moiety is illustrated by the ADP-ribosylation cycle described in the bacterium Rhodospirillum rubrum. These photosynthetic bacteria use reversible ADP-ribosylation to regulate nitrogen metabolism (Ludden 1994). In conditions of darkness or ammonium saturation, an ADP-ribosyltransferase (dinitrogenase reductase ADP-ribosyltransferase, DRAT) is activated to ADP-ribosylate dinitrogenase reductase at Arg101, leading to the inactivation of this key enzyme of nitrogen fixation. Under conditions of light, an ADP-ribosyl-arginine hydrolase (dinitrogenase reductase ADP-ribosylhydrolase, DRAG) is activated that removes the regulatory ADPR moiety, regenerating and reactivating the unmodified enzyme (Fig. 1). A homologue of DRAG, ADP-ribosylhydrolase-1 (ARH1; gene name ADPRH) was identified in mammals (Moss et al. 1988). It is expressed in the cytoplasm and has the same enzymatic activity as DRAG, i.e. it removes ADP-ribose from modified arginine residues. As ARH1 is localized in the intracellular compartment and the known arginine-specific ARTs in mammals modify extracellular proteins, ecto-ARTs and ARH1 could function as partners in ADP-ribosylation cycles only if at least one of these proteins could translocate across the membrane. Of note, the β-subunit of heterotrimeric G proteins (G) is ADP-ribosylated on Arg129 by an as yet uncharacterized ART, and is de-ADP-ribosylated by a cytosolic hydrolase, providing an example for an ADP-ribosylation cycle in mammalian cells (Lupi et al. 2000). It has been suggested that ARH1 may also reverse ADP-ribosylation of proteins targeted by arginine-specific bacterial ARTs. Consistent with this hypothesis, cells from ARH1-deficient mice were found to be more susceptible to the cytotoxic effects of cholera toxin (Kato et al. 2007).
Processing of ADP-ribosyl-arginine
Partial reversal of extracellular arginine ADP-ribosylation reactions may result from the action of extracellular phosphodiesterases that cleave adenosine-5′-monophosphate (AMP) from ADP-ribose, leaving phosphoribose attached to the target arginine. This has been described for the α7 integrin on skeletal muscle cells (Zolkiewska and Moss 1995). This modification removes the bulk of the ADP-ribose moiety, possibly relieving steric hindrance. However, the phosphoribose moiety still contains a negative charge and precludes re-ADP-ribosylation of the target protein at that residue.
A second possibility for reverting arginine ADP-ribosylation has recently been described for HNP-1, i.e. the non-enzymatic hydrolysis at the guanidino carbon, resulting in replacement of ADP-ribosylarginine by ornithine (Stevens et al. 2009). This modification also precludes re-ADP-ribosylation at this site.
Interestingly, these “secondary” modifications of ADP-ribosylated arginine residues create new posttranslational modifications in their own right. However, it remains to be determined what effects, if any, the decoration of a protein with a phosphoribose moiety or the replacement of arginine with ornithine has on the function of the protein.
Interaction of arginine-ADP-ribosylation with other PTMs
Many proteins carry multiple posttranslational modifications. This is exemplified by P2X7 and HNP-1, the targets of ADP-ribosylation discussed above. Posttranslational modifications described in the literature for the two proteins are summarized in Table 2. In addition to ADP-ribosylation at Arg125 and Arg133, P2X7 is glycosylated at various asparagine residues in the extracellular domain, and may be subject to intracellular tyrosine phosphorylation (Kim et al. 2001) and/or palmitoylation at different residues within the C-terminal cytoplasmic tail (Gonnord et al. 2009). HNP-1 is synthesized as a prepropeptide of 94 amino acids, which is processed via several proteolytical cleavage steps to the functional final peptide, which contains the 30 C-terminal amino acids. The peptide is also a substrate for phosphorylation on tyrosine 21 of the mature peptide (Rikova et al. 2007). In addition, ADP-ribosylated arginine at position 14 may be replaced by ornithine (Stevens et al. 2009).
Table 2.
Posttranslational modifications of P2X7 and HNP-1
| mP2X7 | hHNP-1 |
|---|---|
| Posttranslational modification (P_UniMod accession number) | |
| ADP-ribosylation (P_213) | ADP-ribosylation (P_213) |
| R125, R133 | R14, R24 |
| N-linked glycosylation | “Ornithinylation” (P_372) |
| N74, N187, N202, N213, N241 | R14 |
| Phosphorylation (P_21) | Phosphorylation (P_21) |
| Y343 | Y21 |
| Palmitoylation (P_47) | |
| C unknown | |
| Proteolysis (Uniprot) | |
| AA 1-595 | AA 1-19 signaling peptide |
| AA 20-38 propeptide | |
| AA 39-94 prepeptide | |
| AA 65-94 peptide | |
Individual PTMs may exert effects on protein function on their own, but they can also interact to modulate protein function. An example is given by P2X7. The localization of this receptor to lipid rafts is controlled by palmitoylation within the C-terminal cytoplasmic tail of the receptor (Gonnord et al. 2009). The recruitment of the receptor to lipid rafts, in turn, determines its accessibility to ADP-ribosylation by ART2, and thus its activation via extracellular NAD+. ADP-ribosylation of arginines may influence other posttranslational modifications of target proteins. Thus, proteolytic cleavage by trypsin is blocked at arginine residues modified by ADP-ribosylation (Paone et al. 2002; Westblade et al. 2008).
Perspectives
The molecular characterization of enzymes catalyzing arginine ADP-ribosylation and the identification of target proteins constitute important advances in the understanding of this posttranslational modification. Delineating the scope of this modification and its functional consequences in vivo remain major challenges. Several recent technological advances should facilitate this objective in the coming years. Improved tools for the detection of ADP-ribosylated proteins in complex biological mixtures, such as body fluids or cell lysates, include recombinant macro-domains (Dani et al. 2009; Till and Ladurner 2009) and antibodies specific for ADP-ribosyl arginine (Meyer and Hilz 1986; Osago et al. 2008; Schwab et al. 2000). Improved techniques for enriching ADP-ribosylated peptides and for detecting ADP-ribosylated residues will facilitate identification of novel target proteins by mass spectrometry (Hengel et al. 2009; Lang et al. 2010; Osago et al. 2009). Furthermore, improved structure–function prediction algorithms may allow the identification of new ARTs, including novel pathogen-associated toxins (Fieldhouse and Merrill 2008). Finally, the development of specific inhibitors of arginine ADP-ribosylation, such as the blocking single-domain antibodies (nanobodies) against ART2.2 and the Salmonella virulence protein SpvB (Koch-Nolte et al. 2007; Wesolowski et al. 2009) open new avenues to probe the functional role of this modification in vivo, and may open new therapeutic options for the treatment of inflammatory and toxin-mediated diseases.
Conclusions
Arginine ADP-ribosylation is a posttranslational modification that introduces a large, bulky group carrying two negative charges onto the target protein. This modification can affect protein functions by sterically blocking interactions with partner molecules, by inducing conformational changes, or by creating docking sites for new interaction partners. Enzymes catalyzing this reaction have been described in phages, bacteria, and vertebrates, but seem to be lacking in plants, insects, worms, yeast and other unicellular eukaryotes. Arginine-specific ARTs are characterized by the R-S-EXE signature motif of catalytically important amino acid residues. Like other PTMs affecting protein function, arginine-specific ADP-ribosylation is potentially reversible. ADP-ribosylarginine specific hydrolases (ARHs) can restore target protein function by hydrolytic removal of the entire ADP-ribose moiety. In some cases, ADP-ribosylarginine can be processed into secondary PTMs, e.g. phosphoribosylarginine or ornithine, whose functional significance has yet to be determined.
ADP-ribosylation is but one example of how posttranslational modifications can profoundly affect the function of proteins. In the era of microarray technology, it is increasingly important to recognize that knowledge of gene expression levels alone is insufficient to characterize the functional roles of proteins within a given biological context, e.g. a disease process (Jungblut et al. 2008; Schluter et al. 2009). Beyond the description of any individual modification, however, the interplay between different posttranslational modifications also should be taken into account.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grant No310/6 to FH and FKN and by stipends from the Studienstiftung des Deutschen Volkes to SL and MU.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Adriouch S, Bannas P, Schwarz N, Fliegert R, Guse AH, Seman M, Haag F, Koch-Nolte F. ADP-ribosylation at R125 gates the P2X7 ion channel by presenting a covalent ligand to its nucleotide binding site. FASEB J. 2008;22:861–869. doi: 10.1096/fj.07-9294com. [DOI] [PubMed] [Google Scholar]
- Aktories K, Barth H. Clostridium botulinum C2 toxin—new insights into the cellular up-take of the actin-ADP-ribosylating toxin. Int J Med Microbiol. 2004;293:557–564. doi: 10.1078/1438-4221-00305. [DOI] [PubMed] [Google Scholar]
- Aktories K, Barmann M, Ohishi I, Tsuyama S, Jakobs KH, Habermann E. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannas P, Adriouch S, Kahl S, Braasch F, Haag F, Koch-Nolte F. Activity and specificity of toxin-related mouse T cell ecto-ADP-ribosyltransferase ART2.2 depends on its association with lipid rafts. Blood. 2005;105:3663–3670. doi: 10.1182/blood-2004-08-3325. [DOI] [PubMed] [Google Scholar]
- Barbieri JT, Sun J. Pseudomonas aeruginosa ExoS and ExoT. Rev Physiol Biochem Pharmacol. 2004;152:79–92. doi: 10.1007/s10254-004-0031-7. [DOI] [PubMed] [Google Scholar]
- Baysarowich J, Koteva K, Hughes DW, Ejim L, Griffiths E, Zhang K, Junop M, Wright GD. Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc Natl Acad Sci USA. 2008;105:4886–4891. doi: 10.1073/pnas.0711939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti PJ, Blanke SR, Schramm VL. Transition state structure for the hydrolysis of NAD catalyzed by diphtheria toxin. J Am Chem Soc. 1997;119:12079–12088. doi: 10.1021/ja971317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpusca I, Jank T, Aktories K. Bacillus sphaericus mosquitocidal toxin (MTX) and pierisin: the enigmatic offspring from the family of ADP-ribosyltransferases. Mol Microbiol. 2006;62:621–630. doi: 10.1111/j.1365-2958.2006.05401.x. [DOI] [PubMed] [Google Scholar]
- Cervantes-Laurean D, Minter DE, Jacobson EL, Jacobson MK. Protein glycation by ADP-ribose: studies of model conjugates. Biochemistry. 1993;32:1528–1534. doi: 10.1021/bi00057a017. [DOI] [PubMed] [Google Scholar]
- Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, Corda D, Di Girolamo M. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc Natl Acad Sci USA. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depping R, Lohaus C, Meyer HE, Ruger W. The mono-ADP-ribosyltransferases Alt and ModB of bacteriophage T4: target proteins identified. Biochem Biophys Res Commun. 2005;335:1217–1223. doi: 10.1016/j.bbrc.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Dolle C, Niere M, Lohndal E, Ziegler M. Visualization of subcellular NAD pools and intra-organellar protein localization by poly-ADP-ribose formation. Cell Mol Life Sci. 2010;67:433–443. doi: 10.1007/s00018-009-0190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieldhouse RJ, Merrill AR. Needle in the haystack: structure-based toxin discovery. Trends Biochem Sci. 2008;33:546–556. doi: 10.1016/j.tibs.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Ganesan AK, Frank DW, Misra RP, Schmidt G, Barbieri JT. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- Ganesan AK, Mende-Mueller L, Selzer J, Barbieri JT. Pseudomonas aeruginosa exoenzyme S, a double ADP-ribosyltransferase, resembles vertebrate mono-ADP-ribosyltransferases. J Biol Chem. 1999;274:9503–9508. doi: 10.1074/jbc.274.14.9503. [DOI] [PubMed] [Google Scholar]
- Ganesan AK, Vincent TS, Olson JC, Barbieri JT. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J Biol Chem. 1999;274:21823–21829. doi: 10.1074/jbc.274.31.21823. [DOI] [PubMed] [Google Scholar]
- Glowacki G, Braren R, Cetkovic-Cvrlje M, Leiter EH, Haag F, Koch-Nolte F. Structure, chromosomal localization, and expression of the gene for mouse ecto-mono(ADP-ribosyl)transferase ART5. Gene. 2001;275:267–277. doi: 10.1016/S0378-1119(01)00608-4. [DOI] [PubMed] [Google Scholar]
- Glowacki G, Braren R, Firner K, Nissen M, Kuhl M, Reche P, Bazan F, Cetkovic-Cvrlje M, Leiter E, Haag F, Koch-Nolte F. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002;11:1657–1670. doi: 10.1110/ps.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff CG. Chemical structure of a modification of the Escherichia coli ribonucleic acid polymerase alpha polypeptides induced by bacteriophage T4 infection. J Biol Chem . 1974;249:6181–6190. [PubMed] [Google Scholar]
- Gonnord P, Delarasse C, Auger R, Benihoud K, Prigent M, Cuif MH, Lamaze C, Kanellopoulos JM. Palmitoylation of the P2X7 receptor, an ATP-gated channel, controls its expression and association with lipid rafts. FASEB J. 2009;23:795–805. doi: 10.1096/fj.08-114637. [DOI] [PubMed] [Google Scholar]
- Grahnert A, Richter S, Siegert F, Berndt A, Hauschildt S. The orthologue of the “acatalytic” mammalian ART4 in chicken is an arginine-specific mono-ADP-ribosyltransferase. BMC Mol Biol. 2008;9:86. doi: 10.1186/1471-2199-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag F, Koch-Nolte F. ADP-Ribosylation in animal tissues: structure, function and biology of mono(ADP-Ribosyl)transferases and related enzymes, vol 419. New York: Plenum Press; 1997. [DOI] [PubMed] [Google Scholar]
- Haag F, Andresen V, Karsten S, Koch-Nolte F, Thiele H. Both allelic forms of the rat T cell differentiation marker RT6 display nicotinamide adenine dinucleotide (NAD)-glycohydrolase activity, yet only RT6.2 is capable of automodification upon incubation with NAD. Eur J Immunol. 1995;25:2355–2361. doi: 10.1002/eji.1830250835. [DOI] [PubMed] [Google Scholar]
- Han S, Tainer JA. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int J Med Microbiol. 2002;291:523–529. doi: 10.1078/1438-4221-00162. [DOI] [PubMed] [Google Scholar]
- Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. Evolution and mechanism from structures of an ADP‐ribosylating toxin and NAD complex. Nat Struct Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- Hara N, Tsuchiya M, Shimoyama M. Glutamic acid 207 in rodent T-cell RT6 antigens is essential for arginine-specific ADP-ribosylation. J Biol Chem. 1996;271:29552–29555. doi: 10.1074/jbc.271.47.29552. [DOI] [PubMed] [Google Scholar]
- Hengel SM, Shaffer SA, Nunn BL, Goodlett DR. Tandem mass spectrometry investigation of ADP-ribosylated kemptide. J Am Soc Mass Spectrom. 2009;20:477–483. doi: 10.1016/j.jasms.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochmann H, Pust S, von Figura G, Aktories K, Barth H. Salmonella enterica SpvB ADP-ribosylates actin at position arginine-177-characterization of the catalytic domain within the SpvB protein and a comparison to binary clostridial actin-ADP-ribosylating toxins. Biochemistry. 2006;45:1271–1277. doi: 10.1021/bi051810w. [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35(4):208–219 [DOI] [PubMed]
- Huang HY, Graves DJ, Robson RM, Huiatt TW. ADP‐ribosylation of the intermediate filament protein desmin and inhibition of desmin assembly in vitro by muscle ADP‐ribosyltransferase. Biochem Biophys Res Commun. 1993;197:570–577. doi: 10.1006/bbrc.1993.2517. [DOI] [PubMed] [Google Scholar]
- Jacobson MK, Jacobson EL. ADP-ribose transfer reactions: mechanisms and biological significance. New York: Springer; 1989. [Google Scholar]
- Jorgensen R, Wang Y, Visschedyk D, Merrill AR. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 2008;9:802–809. doi: 10.1038/embor.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungblut PR, Holzhutter HG, Apweiler R, Schluter H. The speciation of the proteome. Chem Cent J. 2008;2:16. doi: 10.1186/1752-153X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just I, Sehr P, Jung M, van Damme J, Puype M, Vandekerckhove J, Moss J, Aktories K. ADP‐ribosyltransferase type A from turkey erythrocytes modifies actin at Arg‐95 and Arg‐372. Biochemistry. 1995;34:326–333. doi: 10.1021/bi00001a040. [DOI] [PubMed] [Google Scholar]
- Kato J, Zhu J, Liu C, Moss J. Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice. Mol Cell Biol. 2007;27:5534–5543. doi: 10.1128/MCB.00302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharadia SV, Graves DJ. Relationship of phosphorylation and ADP-ribosylation using a synthetic peptide as a model substrate. J Biol Chem. 1987;262:17379–17383. [PubMed] [Google Scholar]
- Kim M, Jiang LH, Wilson HL, North RA, Surprenant A. Proteomic and functional evidence for a P2X7 receptor signalling complex. EMBO J. 2001;20:6347–6358. doi: 10.1093/emboj/20.22.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch-Nolte F, Petersen D, Balasubramanian S, Haag F, Kahlke D, Willer T, Kastelein R, Bazan F, Thiele HG. Mouse T cell membrane proteins Rt6-1 and Rt6-2 are arginine/protein mono(ADPribosyl)transferases and share secondary structure motifs with ADP-ribosylating bacterial toxins. J Biol Chem. 1996;271:7686–7693. doi: 10.1074/jbc.271.13.7686. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Reche P, Haag F, Bazan F. ADP-ribosyltransferases: plastic tools for inactivating protein and small molecular weight targets. J Biotechnol. 2001;92:81–87. doi: 10.1016/S0168-1656(01)00356-X. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Adriouch S, Bannas P, Krebs C, Scheuplein F, Seman M, Haag F. ADP-ribosylation of membrane proteins: unveiling the secrets of a crucial regulatory mechanism in mammalian cells. Ann Med. 2006;38:188–199. doi: 10.1080/07853890600655499. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Reyelt J, Schossow B, Schwarz N, Scheuplein F, Rothenburg S, Haag F, Alzogaray V, Cauerhff A, Goldbaum FA. Single domain antibodies from llama effectively and specifically block T cell ecto-ADP-ribosyltransferase ART2.2 in vivo. FASEB J. 2007;21:3490–3498. doi: 10.1096/fj.07-8661com. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Kernstock S, Mueller-Dieckmann C, Weiss MS, Haag F. Mammalian ADP-ribosyltransferases and ADP-ribosylhydrolases. Front Biosci. 2008;13:6716–6729. doi: 10.2741/3184. [DOI] [PubMed] [Google Scholar]
- Krebs C, Koestner W, Nissen M, Welge V, Parusel I, Malavasi F, Leiter EH, Santella RM, Haag F, Koch-Nolte F. Flow cytometric and immunoblot assays for cell surface ADP-ribosylation using a monoclonal antibody specific for ethenoadenosine. Anal Biochem. 2003;314:108–115. doi: 10.1016/S0003-2697(02)00640-1. [DOI] [PubMed] [Google Scholar]
- Lang AE, Schmidt G, Schlosser A, Hey TD, Larrinua IM, Sheets JJ, Mannherz HG, Aktories K. Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science. 2010;327:1139–1142. doi: 10.1126/science.1184557. [DOI] [PubMed] [Google Scholar]
- Locht C, Antoine R. A proposed mechanism of ADP-ribosylation catalyzed by the pertussis toxin S1 subunit. Biochimie. 1995;77:333–340. doi: 10.1016/0300-9084(96)88143-0. [DOI] [PubMed] [Google Scholar]
- Loesberg C, van Rooij H, Smets LA. Meta-iodobenzylguanidine (MIBG), a novel high-affinity substrate for cholera toxin that interferes with cellular mono(ADP-ribosylation) Biochim Biophys Acta. 1990;1037:92–99. doi: 10.1016/0167-4838(90)90106-P. [DOI] [PubMed] [Google Scholar]
- Ludden PW. Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes. Mol Cell Biochem. 1994;138:123–129. doi: 10.1007/BF00928453. [DOI] [PubMed] [Google Scholar]
- Lupi R, Corda D, Di Girolamo M. Endogenous ADP-ribosylation of the G protein beta subunit prevents the inhibition of type 1 adenylyl cyclase. J Biol Chem. 2000;275:9418–9424. doi: 10.1074/jbc.275.13.9418. [DOI] [PubMed] [Google Scholar]
- Maehama T, Hoshino S, Katada T. Increase in ADP-ribosyltransferase activity of rat T lymphocyte alloantigen RT6.1 by a single amino acid mutation. FEBS Lett. 1996;388:189–191. doi: 10.1016/0014-5793(96)00568-6. [DOI] [PubMed] [Google Scholar]
- Margarit SM, Davidson W, Frego L, Stebbins CE. A steric antagonism of actin polymerization by a salmonella virulence protein. Structure. 2006;14:1219–1229. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Matsuura R, Tanigawa Y, Tsuchiya M, Mishima K, Yoshimura Y, Shimoyama M. Preferential ADP-ribosylation of arginine-3 in synthetic heptapeptide Leu-Arg-Arg-Ala-Ser-Leu-Gly. Biochem J. 1988;253:923–926. doi: 10.1042/bj2530923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetrey J, Flatau G, Boquet P, Menez A, Stura EA. Structural basis for the NAD-hydrolysis mechanism and the ARTT-loop plasticity of C3 exoenzymes. Protein Sci. 2008;17:878–886. doi: 10.1110/ps.073398508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Hilz H. Production of anti-(ADP-ribose) antibodies with the aid of a dinucleotide-pyrophosphatase-resistant hapten and their application for the detection of mono(ADP-ribosyl)ated polypeptides. Eur J Biochem. 1986;155:157–165. doi: 10.1111/j.1432-1033.1986.tb09471.x. [DOI] [PubMed] [Google Scholar]
- Moss J, Vaughan M. ADP-ribosylating toxins and G proteins: insights into signal transduction. Washington, DC: American Society for Microbiology; 1990. [DOI] [PubMed] [Google Scholar]
- Moss J, Garrison S, Oppenheimer NJ, Richardson SH. NAD‐dependent ADP-ribosylation of arginine and proteins by Escherichia coli heat‐labile enterotoxin. J Biol Chem. 1979;254:6270–6272. [PubMed] [Google Scholar]
- Moss J, Tsai SC, Adamik R, Chen HC, Stanley SJ. Purification and characterization of ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1988;27:5819–5823. doi: 10.1021/bi00415a063. [DOI] [PubMed] [Google Scholar]
- Nemoto E, Yu Y, Dennert G. Cell surface ADP-ribosyltransferase regulates lymphocyte function-associated molecule-1 (LFA-1) function in T cells. J Immunol. 1996;157:3341–3349. [PubMed] [Google Scholar]
- Ohlrogge W, Haag F, Lohler J, Seman M, Littman DR, Killeen N, Koch-Nolte F. Generation and characterization of ecto-ADP-ribosyltransferase ART2.1/ART2.2-deficient mice. Mol Cell Biol. 2002;22:7535–7542. doi: 10.1128/MCB.22.21.7535-7542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osago H, Terashima M, Hara N, Yamada K, Tsuchiya M. A new detection method for arginine-specific ADP-ribosylation of protein—a combinational use of anti-ADP-ribosylarginine antibody and ADP-ribosylarginine hydrolase. J Biochem Biophys Methods. 2008;70:1014–1019. doi: 10.1016/j.jprot.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Osago H, Yamada K, Shibata T, Yoshino K, Hara N, Tsuchiya M. Precursor ion scanning and sequencing of arginine-ADP-ribosylated peptide by mass spectrometry. Anal Biochem. 2009;393:248–254. doi: 10.1016/j.ab.2009.06.028. [DOI] [PubMed] [Google Scholar]
- Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, Moss J. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci USA. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paone G, Stevens LA, Levine RL, Bourgeois C, Steagall WK, Gochuico BR, Moss J. ADP-ribosyltransferase-specific modification of human neutrophil peptide-1. J Biol Chem. 2006;281:17054–17060. doi: 10.1074/jbc.M603042200. [DOI] [PubMed] [Google Scholar]
- Pope MR, Murrell SA, Ludden PW. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenosine diphosphoribosylation of a specific arginine residue. Proc Natl Acad Sci USA. 1985;82:3173–3177. doi: 10.1073/pnas.82.10.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Ruf A, Rolli V, de Murcia G, Schulz GE. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J Mol Biol. 1998;278:57–65. doi: 10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
- Saxty BA, Yadollahi-Farsani M, Upton PD, Johnstone SR, MacDermot J. Inactivation of platelet-derived growth factor-BB following modification by ADP-ribosyltransferase. Br J Pharmacol. 2001;133:1219–1226. doi: 10.1038/sj.bjp.0704187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuplein F, Rissiek B, Driver JP, Chen YG, Koch-Nolte F, Serreze DV. A recombinant heavy chain antibody approach blocks ART2 mediated deletion of an iNKT cell population that upon activation inhibits autoimmune diabetes. J Autoimmun. 2010;34:145–154. doi: 10.1016/j.jaut.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer J, Wieden HJ, Rodnina MV, Aktories K. Inactivation of the elongation factor Tu by mosquitocidal toxin‐catalyzed mono‐ADP‐ribosylation. Appl Environ Microbiol. 2002;68:4894–4899. doi: 10.1128/AEM.68.10.4894-4899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter H, Apweiler R, Holzhutter HG, Jungblut PR. Finding one’s way in proteomics: a protein species nomenclature. Chem Cent J. 2009;3:11. doi: 10.1186/1752-153X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab CJ, Colville MJ, Fullerton AT, McMahon KK. Evidence of endogenous mono-ADP-ribosylation of cardiac proteins via anti-ADP-ribosylarginine immunoreactivity. Proc Soc Exp Biol Med. 2000;223:389–396. doi: 10.1046/j.1525-1373.2000.22355.x. [DOI] [PubMed] [Google Scholar]
- Schwarz N, Fliegert R, Adriouch S, Seman M, Guse AH, Haag F, Koch-Nolte F. Activation of the P2X7 ion channel by soluble and covalently bound ligands. Purinergic Signal. 2009;5:139–149. doi: 10.1007/s11302-009-9135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine A, Fujiwara M, Narumiya S. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J Biol Chem. 1989;264:8602–8605. [PubMed] [Google Scholar]
- Seman M, Adriouch S, Scheuplein F, Krebs C, Freese D, Glowacki G, Deterre P, Haag F, Koch-Nolte F. NAD-induced T cell death: ADP-ribosylation of cell surface proteins by ART2 activates the cytolytic P2X7 purinoceptor. Immunity. 2003;19:571–582. doi: 10.1016/S1074-7613(03)00266-8. [DOI] [PubMed] [Google Scholar]
- Spangler BD. Structure and function of cholera toxin and the related Escherichia coli heat‐labile enterotoxin. Microbiol Rev. 1992;56:622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LA, Levine RL, Gochuico BR, Moss J. ADP-ribosylation of human defensin HNP-1 results in the replacement of the modified arginine with the noncoded amino acid ornithine. Proc Natl Acad Sci USA. 2009;106:19796–19800. doi: 10.1073/pnas.0908381106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. Production of actin‐specific ADP‐ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186:307–312. doi: 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- Sun J, Barbieri JT. Pseudomonas aeruginosa ExoT ADP‐ribosylates CT10 regulator of kinase (Crk) proteins. J Biol Chem. 2003;278:32794–32800. doi: 10.1074/jbc.M304290200. [DOI] [PubMed] [Google Scholar]
- Sun J, Maresso AW, Kim JJ, Barbieri JT. How bacterial ADP-ribosylating toxins recognize substrates. Nat Struct Mol Biol. 2004;11:868–876. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- Terashima M, Mishima K, Yamada K, Tsuchiya M, Wakutani T, Shimoyama M. ADP-ribosylation of actins by arginine-specific ADP-ribosyltransferase purified from chicken heterophils. Eur J Biochem. 1992;204:305–311. doi: 10.1111/j.1432-1033.1992.tb16638.x. [DOI] [PubMed] [Google Scholar]
- Terashima M, Yamamori C, Shimoyama M. ADP‐ribosylation of Arg28 and Arg206 on the actin molecule by chicken arginine‐specific ADP‐ribosyltransferase. Eur J Biochem. 1995;231:242–249. doi: 10.1111/j.1432-1033.1995.tb20693.x. [DOI] [PubMed] [Google Scholar]
- Terashima M, Hara N, Badruzzaman M, Shimoyama M, Tsuchiya M. ADP-ribosylation of tuftsin suppresses its receptor‐binding capacity and phagocytosis-stimulating activity to murine peritoneal macrophages. FEBS Lett. 1997;412:227–232. doi: 10.1016/S0014-5793(97)00784-9. [DOI] [PubMed] [Google Scholar]
- Tezcan‐Merdol D, Nyman T, Lindberg U, Haag F, Koch‐Nolte F, Rhen M. Actin is ADP‐ribosylated by the Salmonella enterica virulence‐associated protein SpvB. Mol Microbiol. 2001;39:606–619. doi: 10.1046/j.1365-2958.2001.02258.x. [DOI] [PubMed] [Google Scholar]
- Till S, Ladurner AG. Sensing NAD metabolites through macro domains. Front Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Tsuge H, Nagahama M, Oda M, Iwamoto S, Utsunomiya H, Marquez VE, Katunuma N, Nishizawa M, Sakurai J. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin. Proc Natl Acad Sci USA. 2008;105:7399–7404. doi: 10.1073/pnas.0801215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J, Schering B, Barmann M, Aktories K. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 1987;225:48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J, Schering B, Barmann M, Aktories K. Botulinum C2 toxin ADP-ribosylates cytoplasmic beta/gamma-actin in arginine 177. J Biol Chem. 1988;263:696–700. [PubMed] [Google Scholar]
- Vogelsgesang M, Aktories K. Exchange of glutamine-217 to glutamate of Clostridium limosum exoenzyme C3 turns the asparagine-specific ADP-ribosyltransferase into an arginine-modifying enzyme. Biochemistry. 2006;45:1017–1025. doi: 10.1021/bi052253g. [DOI] [PubMed] [Google Scholar]
- Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol. 2009;198:157–174. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westblade LF, Minakhin L, Kuznedelov K, Tackett AJ, Chang EJ, Mooney RA, Vvedenskaya I, Wang QJ, Fenyo D, Rout MP, et al. Rapid isolation and identification of bacteriophage T4-encoded modifications of Escherichia coli RNA polymerase: a generic method to study bacteriophage/host interactions. J Proteome Res. 2008;7:1244–1250. doi: 10.1021/pr070451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett JR, Gould KL, Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986;161:177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Gruszczynska-Biegala J, Zolkiewska A. ADP-ribosylation of integrin alpha7 modulates the binding of integrin alpha7beta1 to laminin. Biochem J. 2005;385:309–317. doi: 10.1042/BJ20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Huiatt TW, Robson RM, Sernett SW, Graves DJ. Characterization of ADP-ribosylation sites on desmin and restoration of desmin intermediate filament assembly by de-ADP-ribosylation. Arch Biochem Biophys. 1996;334:214–222. doi: 10.1006/abbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- Zolkiewska A. Ecto-ADP-ribose transferases: cell-surface response to local tissue injury. Physiology (Bethesda) 2005;20:374–381. doi: 10.1152/physiol.00028.2005. [DOI] [PubMed] [Google Scholar]
- Zolkiewska A, Moss J. Integrin alpha 7 as substrate for a glycosylphosphatidylinositol-anchored ADP-ribosyltransferase on the surface of skeletal muscle cells. J Biol Chem. 1993;268:25273–25276. [PubMed] [Google Scholar]
- Zolkiewska A, Moss J. Processing of ADP-ribosylated integrin alpha 7 in skeletal muscle myotubes. J Biol Chem. 1995;270:9227–9233. doi: 10.1074/jbc.270.16.9227. [DOI] [PubMed] [Google Scholar]


