Abstract
The phosphotyrosine-binding (PTB) domain of the cell fate determinant Numb is involved in the formation of multiple protein complexes in vivo and can bind a diverse array of peptide sequences in vitro. To investigate the structural basis for the promiscuous nature of this protein module, we have determined its solution structure by NMR in a complex with a peptide containing an NMSF sequence derived from the Numb-associated kinase (Nak). The Nak peptide was found to adopt a significantly different structure from that of a GPpY sequence-containing peptide previously determined. In contrast to the helical turn adopted by the GPpY peptide, the Nak peptide forms a β–turn at the NMSF site followed by another turn near the C–terminus. The Numb PTB domain appears to recognize peptides that differ in both primary and secondary structures by engaging various amounts of the binding surface of the protein. Our results suggest a mechanism through which a single PTB domain might interact with multiple distinct target proteins to control a complex biological process such as asymmetric cell division.
Keywords: asymmetric cell division/nuclear magnetic resonance (NMR) spectroscopy/Numb/PTB domain/structure
Introduction
Protein modules such as the Src homology 2 (SH2) and phosphotyrosine-binding (PTB) domains are found in molecules with diverse biochemical functions, including enzymes, adaptors, transcription factors and cytoskeletal proteins (Pawson, 1995; Pawson and Scott, 1997). Modular domains can fold independently into well defined three-dimensional structures in solution. Isolated domains frequently bind to proteins that contain specific, short sequence motifs with dissociation constants in the nanomolar to micromolar range. These motifs typically consist of a conserved core element required for recognition by a particular class of module and flanking residues that confer specificity for an individual domain. For instance, SH2 domains bind peptides with a core phosphotyrosine (pTyr) residue, while additional specificity is defined by the side chains of residues immediately C–terminal to the pTyr (Songyang et al., 1993). In contrast, PTB domains (Blaikie et al., 1994; Kavanaugh and Williams, 1994; Bork and Margolis, 1995; Gustafson et al., 1995) were described initially as binding core NPX(p)Y motifs, with the affinity of a given PTB domain for a specific peptide being modified further by more N–terminal residues (Trub et al., 1995; van der Geer et al., 1995, 1996; Laminet et al., 1996; Li et al., 1996). For some PTB domains, such as those from Shc and IRS-1, tyrosine phosphorylation is essential for high affinity binding (Trüb et al., 1995; van der Geer et al., 1995). However, other PTB domains, such as those of X11, Fe65 and Disabled, recognize peptide ligands in a fashion that is independent of tyrosine phosphorylation or even the presence of a tyrosine residue (Borg et al., 1996; Zambrano et al., 1997; Zhang et al., 1997; Howell et al., 1999).
Although the significance of modular domains was recognized originally in the context of pTyr signaling, modular protein–protein interactions are also important in many other biological processes (Knoblich et al., 1995; Pawson and Scott, 1997). This is typified by the cell fate determinant Numb, an intrinsic regulator of asymmetric cell division in the nervous system of Drosophila melanogaster (Uemura et al., 1989). Numb is a modular protein capable of specific protein–protein and protein–phospholipid interactions. It contains a membrane-localizing region at its extreme N–terminus followed by a sequence related to the Shc PTB domain and a C–terminal NPF motif that can bind the Eps-homology (EH) domain of Eps15 (Knoblich et al., 1997; Salcini et al., 1997). These elements provide a mechanism through which the Numb protein may interact with other extrinsic or intrinsic determinants of asymmetric cell division. The Numb PTB domain, in particular, appears important both for its asymmetric localization during the division of neural precursors and for inhibition of Notch signaling, and is highly conserved in two mammalian Numb homologs, mNumb and Numb-like (Verdi et al., 1996; Zhong et al., 1996).
Unlike the Shc and IRS-1 PTB domains, which selectively bind to phosphorylated NPXpY motifs (Trüb et al., 1995; van der Geer et al., 1995; Wolf et al., 1995), the Numb PTB domain does not require tyrosine phosphorylation for peptide binding, and appears to have a rather broad binding specificity. In addition to binding to NPXY-containing sequences (Dho et al., 1998), the Numb PTB domain selectively recognizes peptide sequences containing a GPY motif, identified through screening of a tyrosine-oriented synthetic peptide library (Li et al., 1997, 1998). A closely related sequence,YIGQYI, is present in the N–terminal region of the oncoprotein Mdm2, and may mediate binding to the Numb PTB domain in cells (Juven-Gershon et al., 1998).
Additional information concerning the binding properties of the Numb PTB domain has come from the cloning of two Numb-interacting proteins, the Numb-associated kinase (Nak) (Chien et al., 1998) and the partner of Numb (PON) (Lu et al., 1998). Nak is a putative Ser/Thr kinase that interacts with the Numb PTB domain in both in vivo and in vitro binding studies. Genetic experiments have shown that overexpression of Nak in the fly produces phenotypes resembling those obtained from loss-of-function Numb mutations (Chien et al., 1998). Therefore, Nak interacts with Numb not only physically, but also genetically, and in so doing appears to antagonize Numb function during asymmetric cell division in a dose-dependent manner. Interestingly, Nak binds to the Numb PTB domain through a C–terminal site containing an unconventional sequence, GFSNMSFEDFP (Chien et al., 1998). PON co-localizes with Numb and directs Numb asymmetric localization during mitosis of neural and muscle progenitor cells (Lu et al., 1998). PON binds directly to Numb through an interaction involving the Numb PTB domain and an N–terminal region of PON containing several NPF/Y sites (Lu et al., 1998). Provocatively, the Numb PTB domain may interact directly with the cytoplasmic region of Notch, although the precise binding sequence has not been derived (Guo et al., 1996; Zhong et al., 1996). Collectively, these studies show that the Numb PTB domain is a rather promiscuous protein module capable of binding to multiple peptides that share no strong sequence identity.
How does the Numb PTB domain recognize such a diverse array of peptides? Our previous studies demonstrated that the Numb PTB domain possesses a large hydrophobic surface groove that potentially can accommodate peptides longer than the YIGPpYL ligand (Li et al., 1998). Here we present the NMR structure of the Numb PTB domain in complex with a peptide derived from Nak, a physiological ligand of Numb. Comparison of the two Numb PTB domain–peptide complexes demonstrates that the Numb PTB domain recognizes different peptides using a common hydrophobic groove. The versatility of the Numb PTB domain appears to stem from a flexible usage of this groove, which in turn is determined by the characteristics of the peptide ligands. Our results provide a molecular mechanism by which a single PTB module might interact successively with distinct partners, and thereby possess multiple functional roles.
Results
Binding of the Numb PTB domain to Nak peptides
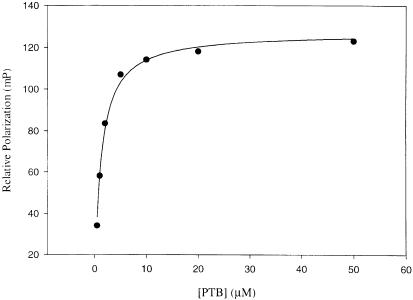
A peptide (GFSNMSFEDFP, designated Nak–c) derived from the Numb PTB domain-binding site in Nak was labeled with fluorescein to examine its affinity for the Numb PTB domain in a fluorescence polarization study. As shown in Figure 1, the Numb PTB domain expressed as a GST fusion bound to fluorescent peptide Nak–c in a concentration-dependent manner, whereas GST alone did not show appreciable binding (data not shown). The measured dissociation constant (Kd) of 1.7 μM for the Numb PTB domain–Nak–c complex is in agreement with that estimated from an in vitro binding study by Chien et al. (1998) employing the full-length Nak protein.
Fig. 1. Binding of the Numb PTB domain to fluorescein-labeled peptide Nak–c as measured by fluorescence polarization. Repetitive measurements typically produced Kd (dissociation constant) values with deviations of <5% from each other.
To explore the relative contributions of individual residues in the Nak–c peptide to PTB domain binding, we performed alanine scanning substitutions on the peptide and analyzed the relative affinities of the resulting peptides for the dNumb PTB domain in a competition assay. As shown in Table I, alanine substitutions had a range of effects on peptide affinity. While certain residues, such as Gly-6, Glu+1 and Phe+3, were indifferent to alanine substitutions, replacement of each of the other residues in Nak–c by alanine either greatly reduced or significantly enhanced binding to the Numb PTB domain (Table I). Notably, peptides N–3A and F0A displayed negligible PTB binding, and the affinities of peptides D+2A and F–5A were substantially decreased compared with peptide Nak–c, suggesting that these residues play important roles in mediating high affinity Numb PTB domain binding. However, substitutions of residues Ser-4, Met-2 or Ser-1 by alanine augmented affinity. This unexpected observation contrasts with similar studies performed on the YIGPpYL peptide, where Ala substitutions invariably led to reduced binding affinities (Li et al., 1997). We also synthesized a peptide bearing double alanine substitutions at residues Ser-1 and Met-2 (Nak-AA, Table I). Interestingly, the relative affinity of this peptide is increased by a factor of 15, compared with increases by factors of 4–8 produced by the singly alanine-substituted peptides. The fact that one can engineer peptide agonists with significantly enhanced binding affinities over the parent peptide by simple amino acid substitutions implies that the native peptide (Nak–c), and hence the naturally occurring protein (Nak), is not optimized for binding to its physiological partner, Numb.
Table I. Relative affinitiesa of Nak peptides for the dNumb PTB domain.
| Peptide | Sequence | Rel. affinity (%) |
|---|---|---|
| –6–4–20+2+4 | ||
| Nak–c | G-F-S-N-M-S-F-E-D-F-P | 100 |
| G-6A | A-F-S-N-M-S-F-E-D-F-P | 135 |
| F-5A | G-A-S-N-M-S-F-E-D-F-P | 66 |
| S-4A | G-F-A-N-M-S-F-E-D-F-P | 250 |
| N–3A | G-F-S-A-M-S-F-E-D-F-P | N/D |
| M-2A | G-F-S-N-A-S-F-E-D-F-P | 400 |
| S-1A | G-F-S-N-M-A-F-E-D-F-P | 851 |
| F0A | G-F-S-N-M-S-A-E-D-F-P | 3.5 |
| E+1A | G-F-S-N-M-S-F-A-D-F-P | 82 |
| D+2A | G-F-S-N-M-S-F-E-A-F-P | 27 |
| F+3A | G-F-S-N-M-S-F-E-D-A-P | 92 |
| Nak-AA | G-F-S-N-A-A-F-E-D-F-P | 1500 |
| NakΔC | G-F-S-N-M-S-F-E | <1 |
| Nak▿N | S-A-K-T-G-F-S-N-M-S-F-E-D-F-P | 95 |
| N–TrkA/ | H-I-I-E-N-M-S-F-E-D-F-P | <1 |
| C–Nak |
aRelative affinities were calculated according to the IC50 (concentration for 50% inhibition of binding) value of an individual peptide when competing against fluorescein-labeled peptide Nak–c for binding to the dNumb PTB domain. Relative affinities were normalized to that of the unlabeled Nak–c peptide, set to 100%. Repetition of the measurements produced values with <5% deviation from those reported.
To examine the collective effect of residues C–terminal to the NMSF motif, we synthesized a peptide lacking the three most C–terminal amino acids. This peptide, NakΔC, completely lost its affinity for the Numb PTB domain, a much more severe effect than produced by single residue substitutions, indicating that these residues together play an important role in Numb PTB binding. Since the Nak–c peptide has a relatively short N–terminus compared with other high affinity PTB ligands, we synthesized two peptides with extended N–termini. The first peptide, Nak▿N contains a four residue N–terminal extension corresponding to the native sequence in Nak. This peptide had a similar affinity for the Numb PTB domain to that of the shorter Nak–c peptide (Table I), indicating that the Nak–c peptide probably fully occupies the ligand-binding site on the Numb PTB domain. The second peptide, N–TrkA/C–Nak, was constructed with the N–terminal residues from a TrkA peptide sequence, a high affinity ligand for the Shc PTB domain, and the C–terminal portion of the Nak–c peptide. However, this peptide was unable to bind to the Numb PTB domain (Table I), suggesting that the specific N–terminal residues of Nak–c are important for Numb PTB binding and that enriching the N–terminus with hydrophobic residues does not promote affinity.
Structure of the Numb PTB domain–Nak peptide complex
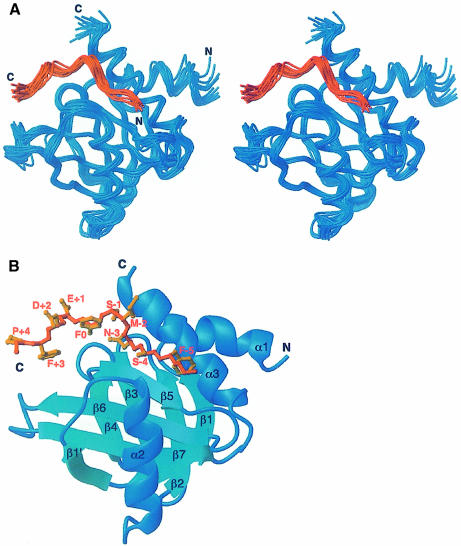
To gain further insight into the molecular basis for the unusual binding properties of the Numb PTB domain, we determined the structure of a 1:1 complex of the PTB domain of Drosophila Numb with the Nak–c peptide by multidimensional NMR spectroscopy. The structure of the complex (Figure 2) was calculated using the ARIA protocol (Nilges et al., 1998) from a total of 2028 NOE distance, 50 hydrogen bond and 94 chemical shift-derived dihedral restraints. The final ensemble of 20 calculated structures is displayed in Figure 2A. Statistics of the ensemble of structures are given in Table II.
Fig. 2. Solution structure of the Numb PTB domain–Nak peptide complex. (A) Stereo representation of a superposition of the 20 final structures. (B) Schematic ribbon diagram of the backbone. The protein is shown in blue and the peptide in orange. The side chains of the Nak peptide are labeled. The figure was generated using MOLMOL (Koradi et al., 1996).
Table II. Structural statistics for the 20 final structures of the Numb PTB domain–Nak peptide complex.
| R.m.s. deviations from distance restraints (Å) | ||
| all (2088) | 0.011 ± 0.002 | |
| unambiguous (1539) | 0.011 ± 0.002 | |
| ambiguous (449) | 0.010 ± 0.002 | |
| hydrogen bonds (50) | 0.011 ± 0.003 | |
| Deviations from idealized geometry | ||
| bonds (Å) | 0.0009 ± 0.00005 | |
| angles (°) | 0.279 ± 0.004 | |
| impropers (°) | 0.32 ± 0.02 | |
| Procheck Ramachandran map analysis | ||
| most favored regions | 62.1% | |
| additional allowed regions | 33.5% | |
| generously allowed regions | 3.8% | |
| disallowed regions | 0.6% | |
| Atomic r.m.s.d. (Å) from mean structure | ||
| Backbone | All heavy atoms | |
| PTB (amino acids 67–200) and Nak | 0.70 ± 0.11 | 1.36 ± 0.14 |
| PTB (amino acids 67–200) | 0.73 ± 0.11 | 1.34 ± 0.14 |
| Nak | 0.62 ± 0.22 | 1.09 ± 0.24 |
As previously shown, the structure of the Numb PTB domain is similar to those of the Shc, IRS–1 and X11 PTB domains, which feature a β-sandwich composed of two antiparallel β-sheets arranged nearly orthogonally to each other and capped by a C–terminal α-helix (α3) at one side (Figure 2B). An N–terminal α-helix (α1) is packed against α3 at an angle of ∼40°. A second helix (α2) runs perpendicular to the second sheet of the β-sandwich. The only significant difference between the current PTB structure and the one determined previously is the packing of α1 against α3, which is altered by ∼10°. The r.m.s.d. values for the backbone and for all heavy atoms between the current complex and the Numb PTB domain–GPpY peptide complex are 1.5 and 2.1 Å, respectively.
The Nak peptide adopts a distinct structure upon binding to the Numb PTB domain. In addition to a β–turn formed by the NMSF sequence, somewhat resembling those in the NPX(p)Y motifs recognized by the Shc, IRS-1 and X-11 PTB domains (Zhou et al., 1995, 1996; Eck et al., 1996; Zhang et al., 1997), the C–terminal residues of the peptide (F0 to F+3) are also in a β–turn conformation. Residue F0, which terminates the first β–turn, appears also to initiate the second β–turn. The N–terminus of the peptide, including residues G-6, F-5 and S-4, is in an extended conformation. The peptide-binding site on the protein, which is similar for both the Numb PTB–Nak and the Numb PTB–GPpY peptide complexes, is characterized by a surface groove formed by residues from the β5 strand and the C–terminal α–helix, α3 (Figure 2B).
Comparison of the two Numb PTB domain–peptide complexes
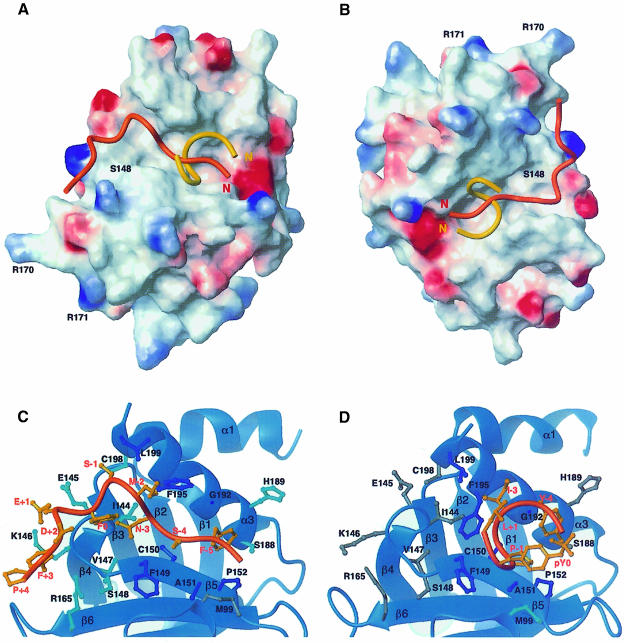
The structure of the Numb PTB domain itself is essentially identical in the two complexes. Significant differences, however, were observed in the structures of the bound peptides. The Nak peptide adopts a structure characterized by two β–turns, whereas the GPpY peptide forms a single helical turn (Figure 3A and B). In addition, the base of the NMSF β–turn in the Nak peptide interacts with helix α3 of the protein, whereas that of the GPpY helical turn is in contact with the β5 strand (Figure 3C and D). The Numb PTB domain can therefore recognize two different peptides that adopt quite distinct conformations.
Fig. 3. (A and B) Surface representations (blue, positive potential; red, negative potential) of the Numb PTB domain structure determined in the current study with both peptides Nak (orange) and GPpY (yellow) shown in Cα traces. The orientation of (A) is the same as for Figure 2 and for (C), while that shown in (B) is from a different angle to emphasize the minimal interactions of the C–terminal residues of the peptide with the binding groove of the PTB domain. (C and D) Interfaces between the PTB domain and the Nak (C) and GPpY (D) peptides. Residues that show contacts with both peptides are colored dark blue; residues that have selective contacts with the displayed peptide are colored cyan, while those that do not are colored gray.
Our previous study of the Numb PTB domain–GPpY peptide complex suggested that the large peptide-binding groove on the surface of the Numb PTB domain potentially could accommodate peptides longer than the GPpY peptide (Li et al., 1998). We show in the current study that this is indeed the case. The Nak peptide is several residues longer than the GPpY peptide, and occupies the same binding groove on the protein (Figure 3A and B). The peptide-binding groove of the Numb PTB domain is characterized by a neutral electrostatic potential and is enriched in hydrophobic residues (Figure 3A). The characteristics of the binding groove dictate that peptide recognition by the Numb PTB domain is unlikely to involve significant charge–charge interactions. In addition to their different conformations, the two peptides bind different amounts of the PTB domain surface. The GPpY peptide engages only a portion of the binding surface available (∼900 Å2 buried surface area), whereas the Nak peptide occupies essentially the entire surface binding groove on the Numb PTB domain (∼1200 Å2 surface area buried) (Figure 3A and B). It is also interesting to note that the GPpY peptide, which is smaller in size and hence more compact, interacts primarily with a patch of hydrophobic residues on the peptide-binding groove of the PTB domain, whereas the Nak peptide interacts with these residues via its N–terminus, but has additional contacts with the rest of the binding groove via residues of its first β–turn (Figure 3C and D). It is therefore striking that the two peptides have comparable binding affinities for the Numb PTB domain and cross-compete effectively against each other in in vitro binding studies despite these differences in modes of binding (Li et al., 1998).
The hydrophobic patch that engages the GPpY peptide is composed of residues from the β5 strand and the α3 helix, including Phe149, Cys150, Ala151, Pro152, Gly192, Phe195 and Leu199. These residues make extensive contacts with residues of the GPpY peptide (Figure 3D) through hydrophobic interactions. Similar hydrophobic interactions are also observed for the Nak peptide (Figure 3C). However, interactions between the Nak peptide and the Numb PTB domain extend beyond this hydrophobic patch. The C–terminal residues of the Nak peptide (Phe0 to Phe+3) make extensive contacts with residues of the β4–β5 loop of the protein. Interestingly, these residues are too far away from the peptide in the Numb PTB–GPpY complex to be involved in the interaction.
The N–terminal residues of the Nak peptide, including residues Gly-6, Phe-5 and Ser-4, are in an extended conformation in the complex. These residues interact primarily with residues Cys150, Ala151, Pro152, Ser188, His189 and Gly192 of the protein (Figure 3C). Asn-3 of the Nak peptide contacts residues Ile144, Cys150 and Phe195 of the PTB domain. It appears to play an important structural role in stabilizing the β–turn formed by the NMSF sequence, as suggested by an observable NOE between its β–proton and ring protons of Phe0. The backbone of residue Met-2 of the Nak peptide is within interaction distance of residues Phe195 and Leu199 of the protein. However, the side chain of Met-2 is largely solvent exposed, which explains why its replacement by an alanine residue, which should decrease the solvent-exposed hydrophobic surface, results in an increase in binding affinity. Also, the substitution should also increase the β-turn-forming potential of the resulting peptide. The side chain of Ser-1 at the base of the turn in the Nak peptide interacts primarily with those of the hydrophobic residues Cys198 and Leu199 of the protein (Figure 3C). The close proximity of Ser-1 to Cys198 and Leu199 of the Numb PTB domain explains why its mutation to alanine, which is expected to promote hydrophobic interactions, results in a significant increase in binding affinity (Table I). The C–terminal residues of the peptide, including Phe0, Glu+1, Asp+2, Phe+3 and Pro+4, interact with residues Ile144, Glu145, Lys146, Val147 from the β4–β5 loop and Arg165 from the β6 strand of the protein (Figure 3C).
It is interesting to compare the role of pTyr of the NPXpY motif in binding to the IRS-1 PTB domain with that of Phe0 of the NMSF motif to the Numb PTB domain. While the pTyr residue was shown to be crucial for binding to the IRS-1 and Shc PTB domains by a mechanism partly involving charge–charge interactions mediated through the phosphate group, the phenylalanine residue within the NMSF sequence of the Nak peptide appears to utilize a different mechanism for binding. While residues Glu145 and Ser148 of the Numb PTB domain are within interaction range of Phe0, Phe0 appears also to have a structural role in stabilizing the N–terminal β–turn of the Nak peptide through a potential aromatic amino interaction with the side chain of residue Asn-3 (Figure 3C). In addition, the ring protons of Phe0 showed detectable NOEs to β–protons of Asp+2 and the ring protons of Phe+3 (data not shown).
Comparison with other PTB domains
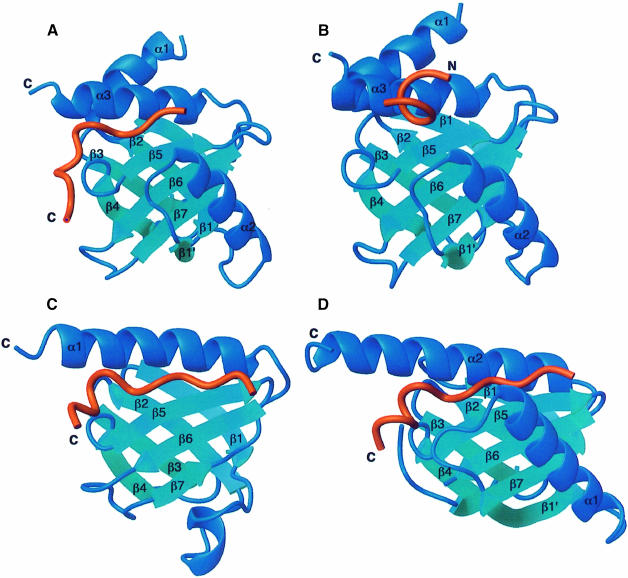
The Numb PTB domain–Nak peptide complex displays many features of the Shc and IRS-1 PTB domain–peptide complexes (Zhou et al., 1995, 1996; Eck et al., 1996; Zhang et al., 1997). Apart from similarities in the PTB domain structure, the Nak peptide adopts a β–turn conformation initiated at the asparagine residue of the NMSF sequence, similar to those observed for the more conventional NPXpY motif. Moreover, all these peptides appear to be localized to the same region on the corresponding PTB domain surface (Figure 4). Nevertheless, there are significant differences among these complexes. Most notably, the Nak peptide is not phosphorylated and, indeed, does not contain a tyrosine residue in its sequence. In contrast to the Shc and the IRS-1 PTB domains, which possess positively charged sites at the protein interaction surface for phosphate recognition, the Numb PTB domain lacks such a site and is thus unlikely to display a strong preference for phosphorylated peptides.
Fig. 4. Ribbon representations of (A) the Numb PTB domain–Nak peptide complex, (B) the Numb PTB domain–GPpY peptide complex, (C) the IRS-1 PTB domain–IL-4R peptide complex (Zhou et al., 1996) and (D) the X11 PTB domain–βAPP peptide complex (Zhang et al., 1997) shown in the same orientation. Proteins are in blue and peptides in red.
In addition, the Nak peptide differs from canonical PTB ligands in the function of residues at the N–terminus. For instance, the IRS-1 PTB domain-binding peptide derived from the interleukin–4 receptor (IL-4R) is characterized by an NPApY motif preceded by an N–terminal extension enriched in hydrophobic residues. Recognition of this peptide by the IRS-1 PTB domain is mediated by both the NPApY motif and the N–terminal residues, which make extensive hydrogen bonding interactions with the β5 strand of the protein, adding essentially another strand to the protein's β–sheet. Favorable hydrophobic interactions are also present between the N–terminal residues and hydrophobic residues of the protein (Eck et al., 1996; Zhou et al., 1996). In contrast, the Nak peptide has a shortened N–terminus and hence lacks extensive hydrogen bonding interactions with the β5 strand of the protein. Indeed, the length of the β5 strand of the Numb PTB domain is significantly shorter than the corresponding strands of other PTB domains, and is therefore unable to accommodate a longer N–terminal peptide segment (Figure 4). These observations explain the binding data showing that N–terminal extension of the Nak–c peptide did not enhance affinity for the Numb PTB domain.
Instead of using an elongated N–terminus to promote binding, the Nak peptide appears to employ residues C–terminal to the β–turn motif to enhance interactions with the Numb PTB domain. The turn immediately C–terminal to the NMSF β–turn leads to a compact structure for the Nak peptide, and allows the side chains of the C–terminal residues to interact with residues from the β4–β5 loop of the protein (Figure 3C). This provides a structural basis for the observation that the C–terminal residues of the Nak peptide are indispensable for binding (Table I).
It is interesting to compare the Numb PTB domain–Nak peptide complex with that between the X11 PTB domain and the βAPP peptide (Zhang et al., 1997). Both the Nak and βAPP peptides are unphosphorylated, and indeed neither one requires the presence of tyrosine for high affinity binding (Zhang et al., 1997). The C–terminal residues of the βAPP peptide are shown to be in a 310-helix conformation and interact with a defined hydrophobic pocket on the surface of the protein in the X11 PTB domain–βAPP peptide complex (Zhang et al., 1997). However, the conformation of the βAPP peptide, which consists of a long, extended N–terminus followed sequentially by a β–turn and a turn of 310-helix, is noticeably different from that of the Nak peptide, which features a short N–terminus followed by two β–turns (Figure 3). Apparently, the N–terminal residues of the βAPP peptide contribute significantly more binding energy than the corresponding residues of the Nak peptide to their respective PTB domains. In addition, many of the residues involved in binding the C–terminal turn of Nak are not hydrophobic in nature, in contrast to those of the X11 PTB domain shown to contact the C–terminus of the βAPP peptide (Zhang et al., 1997). Furthermore, the fact that the Numb PTB domain possesses two modes of binding, one (with the Nak peptide) comparable to that of the X11 PTB–βAPP complex and a second quite distinct mode (with the GPpY peptide), shows the versatile nature of this protein module.
Structure-based mutagenesis identifies residues critical for Numb PTB–peptide interactions
To probe further the molecular mechanism for specific peptide recognition, we conducted mutagenesis on selected residues of the Numb PTB domain based on structural criteria. As shown in Table III, mutation of residues found in the hydrophobic core of the peptide–protein interface, such as Phe149, Cys150 and Phe195, resulted in a drastic loss of binding affinity for both peptides, especially for the F149V and F195V PTB domain variants. Similar mutations in the full-length Drosophila Numb protein compromise its function in transgenic flies (Yaich et al., 1998). Although these mutations maintain the hydrophobic nature of the side chains, they remove their aromatic character, suggesting that aromatic interactions play important roles in the ligand-binding site.
Table III. Relative affinitiesa of dNumb PTB domain mutants.
| Numb PTB | Nak–c | GPpY |
|---|---|---|
| Wild type | 100 ± 8 | 100 ± 15 |
| F149V | <1 | <1 |
| C150A | 30 ± 3 | 25 ± 6 |
| F195V | <1 | <1 |
| S148A | 103 ± 8 | 35 ± 3 |
| R170M | 105 ± 24 | 36 ± 24 |
| R171Q | 144 ± 24 | 90 ± 11 |
| I144A | <5 | 34 ± 6 |
| E145A | 215 ± 35 | 250 ± 28 |
| K146A | 39 ± 7 | 30 ± 4 |
| K147A | <2 | 15 ± 5 |
| R165A | 44 ± 9 | 16 ± 3 |
| H189A | 47 ± 5 | 45 ± 10 |
| C198A | 27 ± 4 | 22 ± 2 |
aThe relative affinity of a mutant protein was calculated by comparing its dissociation constant (Kd) for either the Nak–c or the GPpY peptide with that of the wild-type protein measured against the same peptide. Note that the Kd value for the wild-type PTB domain for peptide Nak–c is 1.70 ± 0.52 μM, whereas the Kd value of the PTB–GPpY complex is 0.58 ± 0.10 μM.
A major difference in the binding properties of the Shc and Numb PTB domains is in the effect of phosphorylation on peptide binding. The Shc PTB domain possesses a relatively well defined, positively charged pocket formed by side chains from residues Arg67, Ser151, Lys169 and Arg175. Mutations of these residues, such as Ser151 to alanine and Arg175 to glutamine, either decrease or eliminate the binding of the Shc PTB domain to its ligand, p145 SHIP (Zhou et al., 1995). In contrast, the Numb PTB domain lacks a positively charged pocket to accommodate a phosphate group on its peptide-binding surface. Consequently, mutations of residues Ser148 and Arg171 in the Numb PTB domain, which correspond to Ser151 and Arg175 in the Shc PTB domain, to alanine and glutamine had no apparent effect on Nak peptide binding (Table III). Interestingly, mutation of another neighboring residue, Arg170 to methionine, increased the affinity of the mutant PTB domain for the Nak peptide by an unknown mechanism. However, these same mutations, especially R171M and S148A, decreased the affinity of the mutant protein for the GPpY peptide appreciably, indicating that differences exist between the Numb PTB domain interaction with the Nak and the GPpY peptides. It should be noted that residues Arg170 and Arg171 are located outside the peptide-binding groove of the Numb PTB domain, and are thus unlikely to be involved directly in peptide recognition (Figure 3A).
The differences between the binding modes of the two peptides were explored further by mutagenesis of residues that show contacts with one peptide, but not the other. To this end, we generated Numb PTB mutants bearing alanine substitution for residues Ile144, Glu145, Lys146, Val147, Arg165, His189 or Cys198. These residues interact with C–terminal residues of the Nak peptide but not residues of the GPpY peptide. As shown in Table III, four of the seven Numb PTB variants, namely E145A, K146A, H189A and C198A, displayed comparable relative affinities for both the Nak and GPpY peptides. Mutation of Arg165 to alanine had a more negative effect on binding to the GPpY peptide than to the Nak peptide, although this residue is not involved in recognizing the former peptide. It is possible that the smaller binding area of the GPpY peptide on the Numb PTB domain renders it more sensitive than the Nak peptide to perturbations of local environment. Interestingly, mutations of Ile144 and Val147 to alanine caused a much greater reduction in affinity of the PTB domain for the Nak peptide than the GPpY peptide, suggesting that these two residues play a crucial role in binding the Nak peptide, but a less important role in binding the GPpY peptide. These mutants also provide a tool to selectively disrupt one mode of peptide interaction with the Numb PTB domain while leaving the other mode relatively intact.
Discussion
Modular domains that mediate specific protein–protein and protein–lipid interactions are involved centrally both in the targeting of polypeptides to specific subcellular locations and in controlling the activation of intracellular signaling pathways in response to external cues (Pawson and Scott, 1997). Some of these modules are also used to coordinate the cellular response to intrinsic determinants of cell fate. This is exemplified by proteins that control polarity and asymmetric cell division during invertebrate embryonic development. Numb is one of several Drosophila proteins, including Inscuteable, Miranda and PON, that can form a complex network of protein–protein interactions during cell division, which in turn dictates the specific localization of these polypeptides relative to the mitotic spindle (Jan and Jan, 1998). Depending on the cell, the components of this network determine the plane of division and the identities of the daughter cells. We wish to understand at a molecular level how a protein module, the PTB domain, originally identified as an important component of signaling from receptor tyrosine kinases, might be used to generate cell diversity by regulating asymmetric cell division.
The Numb PTB domain potentially mediates multiple protein complexes
Genetic and biochemical data indicate that the invertebrate and mammalian Numb PTB domains may have multiple potential functions and interacting partners, including PON, Nak, Lnx, Notch and Mdm2. The Numb PTB domain binds preferentially to both phosphorylated and non-phosphorylated peptides containing a YIGP(p)YL sequence, identified through an in vitro library screening (Li et al., 1997), and to peptide motifs such as GFSNMSFEDFP from the Nak protein. Comparison of these Numb PTB-binding sequences reveals the following. First, they have rather divergent amino acid sequences, indicating that the Numb PTB domain is promiscuous in its recognition of peptide sequences. Secondly, they all contain residues known to promote β–turn structures in proteins such as asparagine and/or proline (Chou and Fasman, 1977). Also, they all contain one or more aromatic residues, which appear critical in mediating peptide–protein interactions. Thirdly, tyrosine phosphorylation and, in some instances, even a tyrosine residue is not required for their interaction with the Numb PTB domain. The latter two characteristics are also shared by other PTB domain ligands, notably those for the X11 and Fe65 PTB domains.
The Numb PTB domain peptide ligands studied to date displayed moderate binding affinities, with Kd values ranging from 0.5 to 1.8 μM (Li et al., 1998). We examined the interaction between the Numb PTB domain and the Nak peptide by performing alanine scanning substitutions, and found that the Nak peptide sequence is not optimized for binding to the Numb PTB domain. In fact, a peptide bearing a double alanine substitution (Nak-AA) bound to Numb 15 times more strongly than the wild-type peptide. The observation that a PTB-binding protein has a suboptimal binding affinity may have functional relevance. The PTB domain forms multiple complexes with other proteins in asymmetric cell division. Furthermore, the formation and dissociation of these complexes have to be orchestrated with the cell cycle (Knoblich et al., 1997; Jan and Jan, 1998). It is therefore conceivable that a ligand with very high affinity for the Numb PTB domain would trap Numb in a particular complex, and thus prevent it from being recruited by other functionally important binding partners. Consistent with this possibility, a Nak mutant bearing the affinity-enhancing, double alanine substitution displays a stronger overexpression phenotype in Drosophila than wild-type Nak (C.-T.Chien and S.–C.Li, unpublished data).
Structural basis for diverse peptide recognition by the Numb PTB domain
We determined the structure of a Numb PTB domain–Nak peptide complex using NMR spectroscopy. The structure explains the results obtained from the binding studies on the Nak peptide, as well as from mutagenesis studies on the PTB domain. It is informative to compare the present structure with that of the Numb PTB domain–GPpY complex determined previously (Li et al., 1998). The Nak peptide bound to the Numb PTB domain adopts a conformation characterized by consecutive β-turns, whereas the GPpY peptide forms a single helical turn. Although both peptides bind to the same groove on the PTB domain surface, they differ significantly in their contact areas. The GPpY peptide interacts with a patch of hydrophobic residues. The Nak peptide, however, not only utilizes this patch but also engages residues outside of it to maximize its tertiary contacts with the Numb PTB domain. The two related, yet different, modes of peptide recognition by the Numb PTB domain provide a structural basis for the promiscuous nature of its peptide recognition. These results demonstrate that the ability of the Numb PTB domain to recognize peptide motifs is not governed solely by the primary sequence motifs or secondary structure of the ligand, but rather is dictated by tertiary contacts between the PTB domain and the peptide. Since surface compatibility has long been recognized as being critical for protein–protein and protein–peptide interactions, it may be an oversimplification to characterize PTB domain ligands rigidly by sequence motifs. Our results show that this simplification, which had been extremely useful in identifying potential PTB domain ligands, could hinder the identification of potential new targets for the Numb and possibly other PTB domains. In support of this notion, the PTB domain of the docking protein FRS2 was recently shown to engage not only a conventional phosphorylated NPXpY motif in the juxtamembrane region of the TrkA receptor tyrosine kinase, but also a completely different non-phosphorylated motif in the fibroblast growth factor receptor (Ong et al., 2000). Our data indicate at a structural level how a single PTB domain can recognize diverse ligands.
The modes of peptide recognition by the Numb PTB domain are significantly different from those used by the Shc, IRS-1 and X11 PTB domains. Apart from the distinct conformations adopted by the Numb PTB ligands, the Shc and IRS-1 PTB domains each contain a site on the protein enriched in positively charged residues to interact with the pTyr phosphate group of their NPXpY-containing ligands. Moreover, ligands for all these three PTB domains generally contain long N–terminal sequences that can interact extensively with the β5 strand of the protein. In contrast, the Nak peptide contains no tyrosine residue in its sequence and the Numb PTB domain does not possess a positively charged site for phosphate recognition. Also, the N–terminal residues in Nak play a less important role in PTB domain binding, apparently due to the shortened β5 strand of the Numb PTB domain compared with those of other PTB domains.
Conclusions
Our data suggest that the Numb PTB domain recognizes multiple ligands through its ability to engage different amounts of surface area dictated by tertiary contacts rather than primary sequences. This may allow Numb to interact with a diverse set of proteins during the process of asymmetric division and specification of cell fate. These results reinforce the view that the backbone fold found in PTB domains can serve as a molecular scaffold for a broad range of intermolecular interactions. PH domains, which commonly bind phosphoinositides (Harlan et al., 1994), EVH1 domains, which recognize proline-rich sequences (Prehoda et al., 1999), and the Ran-binding domain of RBP1, which associates with the activated Ran-GTPase (Vetter et al., 1999), all have the same topology as PTB domains but use different structural elements to engage their ligands. Together with previous genetic and biochemical data, our study provides insights into how this flexible module may control multiple molecular events leading to the determination of cell identity during embryonic development.
Materials and methods
Peptide synthesis and binding studies
Peptides were synthesized on an Applied Biosystem 431A peptide synthesizer using standard 9-fluorenyl methoxycarbonyl (Fmoc) solid phase chemistry and purified following published procedures (Li et al., 1996). The identities of the peptides were confirmed by mass spectrometry and the concentrations of peptide stock solutions were determined by quantitative amino acid analysis. For NMR studies, the Nak peptide was also labeled selectively for the proline, glycine and phenylalanine residues starting from the corresponding amino acid uniformly enriched with the 15N and 13C isotopes (Cambridge Isotope Laboratories, Andover, USA).
Fluorescence polarization experiments used to examine peptide–protein interactions were conducted on a Beacon Fluorescence Polarization System (PanVera Co., USA) equipped with a 100 μl sample chamber. A C–terminal GGK extension was added to the Nak peptide for fluorescence labeling via the lysine side chain amino group (Li et al., 1997). For fluorescence polarization studies, the labeled peptides were dissolved in a phosphate buffer, pH 7.4, containing 0.1 mM EDTA and 1.0 mM dithiothreitol (DTT). The GST fusion PTB protein used in the study was prepared according to published procedures (Li et al., 1997). For competition studies, various concentrations of unlabeled peptides were mixed with 4.0 μM protein in the phosphate buffer in the presence of labeled peptides and the mixture was allowed to stand at room temperature for 5 min before measurement. All measurements were carried out at 25°C.
NMR sample preparation
Preparation of isolated dNumb PTB protein from Escherichia coli was carried out following published procedures (Li et al., 1997, 1998). Four protein samples with different labeling schemes were prepared. Sample A was formed from a uniformly 15N–labeled PTB domain in a 1:1 complex with natural abundance Nak peptide. Sample B, used for backbone and side chain assignment, was the uniformly 15N/13C–labeled dNumb PTB domain in complex with the natural abundance Nak peptide. Sample C consisted of a uniformly 15N/10% 13C–labeled PTB domain complexed to natural abundance peptide, used for stereo-specific assignments of valine and leucine methyl groups. Sample D consisted of natural abundance PTB domain in complex with a Nak peptide that was 15N and 13C labeled at residues Gly-1, Phe-2, Phe-7, Phe-10 and Pro-11. This sample was used to aid peptide assignment.
NMR spectroscopy
NMR experiments were carried out at 30°C on Varian Inova 500 and 600 MHz spectrometers equipped with z-axis pulsed-field gradients. Data were processed using NMRPipe (Delaglio et al., 1995) and analyzed with the NMRView (Johnson and Blevins, 1994) software packages. 1HN, 15N, 13Cα, 13Cβ and 13C–carbonyl assignments were obtained from (HB)CBCA(CO)-NNH (Grzesiek and Bax, 1992), HNCACB (Wittekind and Muller, 1993) and HNCO (Kay et al., 1994) spectra recorded using enhanced sensitivity pulsed-field gradient methodology (Kay et al., 1992) (sample B). Side chain resonances were assigned from HCCH-TOCSY (Kay et al., 1993), CCC–TOCSY(CO)NNH/HCC–TOCSY(CO)NNH (Montelione et al., 1992; Grzesiek et al., 1993) and HAHB (CACBCO)NNH (Grzesiek and Bax, 1993) spectra recorded on sample B. Chemical shifts have been deposited at BioMagResBank (BMRB) under accession number 2482.
Structural restraints were obtained from a set of four NOESY experiments, both 15N–edited HSQC–NOESY (Zhang et al., 1994) (sample A) and simultaneous 13C, 15N–edited NOESY-HSQC (Pascal et al., 1994) (sample B), recorded with mixing times of 40 and 90 ms. Restraints for the peptide were derived from a 150 ms double-filtered NOESY (Ikura and Bax, 1992) on sample B. Intermolecular restraints were derived from a 150 ms half-filtered NOESY (Zwahlen et al., 1997) on sample B.
Structure calculation
Structures were calculated using torsion angle dynamics simulated annealing protocols implemented in the program CNS 0.3 (Brünger et al., 1998). Distance restraints for the PTB domain were assigned automatically based on a previously determined structure of the dNumb PTB domain (Li et al., 1998). Constraints for the peptide were assigned manually from the half- and the double-filtered NOESY spectra. Converged structures were then used as input to ARIA (Nilges et al., 1998), which was utilized for iterative, automatic NOE assignment of all the spectra and for the refinement of structures generated from CNS 0.3. Tolerance for automatic assignment was 0.05 p.p.m. in the proton dimension and 0.2 p.p.m. in the heteronucleus dimensions. Seven iterations were performed to reduce the cut-off for ambiguous assignments from 0.99 to 0.85. Twenty structures were calculated for each iteration, from which the best eight were used for interactive NOE assignment of the next iteration. NOE assignments that satisfied at least 50% of the eight structures were included in the next iteration. Hydrogen bonds that were present in 80% of the eight structures of the previous iteration and could be assigned to regular secondary structure elements were introduced progressively. Dihedral restraints were defined according to the Cα and Cβ chemical shifts, and were used at the start of the iterative procedure. The final structures were evaluated using AQUA and Procheck (Laskowski et al., 1996). Coordinates have been deposited with the Brookhaven Data Bank, accession code 1DDM.
Site-directed mutagenesis
A pGEX4T2 vector containing the dNumb PTB domain (residues 58–205) (Li et al., 1997) was used as a template for the PCR-based, Quickchange site-directed mutagenesis (Stratagene, La Jolla, CA). Specifically, primers containing the desired mutations were used to amplify the plasmid. Plasmids containing the mutant proteins were obtained after enzymatic digestion of the plasmid DNA containing the wild-type protein. All mutations were verified by DNA sequencing. Production and purification of the mutant proteins following transformation into E.coli were the same as for the wild-type protein (Li et al., 1997).
Acknowledgments
Acknowledgements
S.-C.L. is the recipient of a Centennial Fellowship from the Medical Research Council of Canada (MRC). C.Z. is the recipient of a Swiss Science National Fund fellowship and a Human Frontier Science Program post-doctoral fellowship. This work was supported by grants from the Human Frontier Science Program, Asashi Chemical Company, National Cancer Institute of Canada (NCIC) and the MRC. T.P. is a Distinguished Scientist of the MRC. T.P. and L.E.K. are Howard Hughes International Research Scholars.
References
- Blaikie P., Immanuel, D., Wu, J., Li, N., Yajnik, V. and Margolis, B. (1994) A region in Shc distinct from the SH2 domain can bind tyrosine-phosphorylated growth factor receptors. J. Biol. Chem., 269, 32031–32034. [PubMed] [Google Scholar]
- Borg J.-P., Ooi, J., Levy, E. and Margolis, B. (1996) The phosphotyrosine interaction domains of X11 and Fe65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol. Cell. Biol., 16, 6229–6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. and Margolis, B. (1995) A phosphotyrosine interaction domain. Cell, 80, 693–694. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998)Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chien C.-T., Wang, S., Rothenberg, M., Jan, L.Y. and Jan, Y.N. (1998) Numb-associated kinase interacts with the phosphotyrosine binding domain of Numb and antagonizes the function of Numb in vivo. Mol. Cell. Biol., 18, 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P.Y. and Fasman, G.D. (1977) Beta-turns in proteins. J. Mol. Biol., 115, 135–175. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J. and Bax, A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Dho S.E., Jacob, S., Wolting, C.D., French, M.B., Rohrschneider, L.R., McGlade, C.J. (1998) The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J. Biol. Chem., 273, 9179–9187. [DOI] [PubMed] [Google Scholar]
- Eck M.J., Dhe-Paganon, S., Trub, T., Nolte, R.T. and Shoelson, S.E. (1996) Structure of the IRS–1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell, 85, 695–705. [DOI] [PubMed] [Google Scholar]
- Grzesiek S. and Bax, A. (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc., 114, 6291–6293. [Google Scholar]
- Grzesiek S., Anglister, J. and Bax, A. (1993) Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B, 101, 114–119. [Google Scholar]
- Guo M., Jan, L.Y. and Jan, Y.N. (1996) Control of daughter cell fates during asymmetric cell division: interaction of Numb and Notch. Neuron, 17, 27–41. [DOI] [PubMed] [Google Scholar]
- Gustafson T.A, He, W., Craparo, A., Schaub, C.D., O'Neil, T.J. (1995) Phosphotyrosine-dependent interaction of SHC and insulin receptor substrate 1 with the NPEY motif of the insulin receptor via a novel non-SH2 domain. Mol. Cell. Biol., 15, 2500–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan J.E., Hajduk, P.J., Yoon, H.S. and Fesik, S.W. (1994) Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature, 371, 168–170. [DOI] [PubMed] [Google Scholar]
- Howell B.W., Lanier, L.M., Frank, R., Gertler, F.B. and Cooper, J.A. (1999) The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol. Cell. Biol., 19, 5179–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M. and Bax, A. (1992) Isotopic-filtered 2D NMR of a protein–peptide complex: study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J. Am. Chem. Soc., 114, 2433–2440. [Google Scholar]
- Jan Y.N. and Jan, L.Y. (1998) Asymmetric cell division. Nature, 392, 775–778. [DOI] [PubMed] [Google Scholar]
- Johnson B.A. and Blevins, R.A. (1994) NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR, 4, 603–614. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T., Shifman, O., Unger, T., Elkeles, A., Haupt, Y. and Oren, M. (1998) The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol. Cell. Biol., 18, 3974–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh W.M. and Williams, L.T. (1994) An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science, 266, 1862–1865. [DOI] [PubMed] [Google Scholar]
- Kay L.E., Keifer, P. and Saarinen, T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc., 114, 10663–10665. [Google Scholar]
- Kay L.E., Xu, G.-Y., Singer, A.U., Muhandiram, D.R., Forman-Kay, J.D. (1993) A gradient enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlation in H2O samples of proteins. J. Magn. Reson. B, 101, 333–337. [Google Scholar]
- Kay L.E., Xu, G.-Y. and Yamazaki, T. (1994) Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. A, 109, 129–133. [Google Scholar]
- Knoblich J.A., Jan, L.Y. and Jan, Y.N. (1995) Asymmetric segregation of Numb and Prospero during cell division. Nature, 377, 624–627. [DOI] [PubMed] [Google Scholar]
- Knoblich J.A., Jan, L.Y. and Jan, Y.N. (1997) The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc. Natl Acad. Sci. USA, 94, 13005–13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R., Billeter, M. and Wuthrich, K. (1996) MOLMOL, a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Laminet A.A., Apell, G., Conroy, L. and Kavanaugh, W.M. (1996) Affinity, specificity and kinetics of the interaction of the SHC phosphotyrosine binding domain with asparagine-X-X-phosphotyrosine motifs of growth factor receptors. J. Biol. Chem., 271, 264–269. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann, J.A.C., MacArthur, M.W., Kaptein, R. and Thornton, J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Li S.-C., Lai, K.M.V., Gish, G.D., Parris, W.E., van der Geer, P., Forman-Kay, J. and Pawson, T. (1996) Characterization of the phosphotyrosine-binding domain of the Drosophila Shc protein. J. Biol. Chem., 271, 31855–31862. [DOI] [PubMed] [Google Scholar]
- Li S.-C., Songyang, Z., Vincent, S.J., Zwahlen, C., Wiley, S., Cantley, L., Kay, L.E., Forman-Kay, J. and Pawson, T. (1997) High affinity binding of the Drosophila numb phosphotyrosine-binding domain to peptides containing a Gly-Pro-(p)Tyr motif. Proc. Natl Acad. Sci. USA, 94, 7204–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-C., Zwahlen, C., Vincent, S.J., McGlade, C.J., Kay, L.E., Pawson, T., Forman-Kay, J.D. (1998) Structure of a Numb PTB domain–peptide complex suggests a basis for diverse binding specificity. Nature Struct. Biol., 5, 1075–1083. [DOI] [PubMed] [Google Scholar]
- Lu B., Rothenberg, M., Jan, L.Y. and Jan, Y.N. (1998) Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell, 95, 225–235. [DOI] [PubMed] [Google Scholar]
- Montelione G.T., Lyons, B.A., Emerson, S.D. and Tashiro, M. (1992) An efficient triple resonance experiment using carbon 13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J. Am. Chem. Soc., 114, 10974–10975. [Google Scholar]
- Nilges M., O'Donoghue, S.I. (1998) Ambiguous NOEs and automated NOE assignment. Prog. NMR Spectrosc., 32, 107–139. [Google Scholar]
- Ong S.H., Hadari, Y.R., Laks, S., Gotoh, N., Schlessinger, J. and Lax, I. (2000) FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell. Biol., 20, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal S.M., Muhandiran, D.R., Yamazaki, T., Forman-Kay, J.D. and Kay, L.E. (1994) Simultaneous acquisition of 15N– and 13C–edited NOE spectra of proteins dissolved in H2O. J. Magn. Reson. B., 103, 197–201. [Google Scholar]
- Pawson T. (1995) Protein modules and signaling networks. Nature, 373, 573–580. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Scott, J.D. (1997) Signaling through scaffold, anchoring and adaptor proteins. Science, 278, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Prehoda K.E., Lee, D.J. and Lim, W.A. (1999) Structure of the Enabled/VASP homology 1 domain–peptide complex: a key component in the spatial control of actin assembly. Cell, 97, 471–480. [DOI] [PubMed] [Google Scholar]
- Salcini A.E., Confalonierri, S., Doria, M., Santolini, E., Tassi, E., Minenkova, O., Cesareni, G., Pelicci, P.G., Di Fiore, P.P. (1997) Binding specificity and in vivo targets of the EH domain, a novel protein–protein interaction module. Genes Dev., 11, 2239–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z. et al. (1993)SH2 domains recognize specific phospho– peptide sequences. Cell, 72, 767–778. [DOI] [PubMed] [Google Scholar]
- Trüb T., Choi, W.E., Wolf, G., Ottinger, E., Chen, Y., Weiss, M. and Shoelson, S.E. (1995) Specificity of the PTB domain of Shc for β turn-forming pentapeptide motifs amino-terminal to phosphotyrosine. J. Biol. Chem., 270, 18205–18208. [DOI] [PubMed] [Google Scholar]
- Uemura T., Shepherd, S., Ackerman, L., Jan, L.Y. and Jan, Y.N. (1989) Numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell, 58, 349–360. [DOI] [PubMed] [Google Scholar]
- van der Geer P., Wiley, S., Lai, V.K., Olivier, J.P., Gish, G.D., Stephens, R., Kaplan, D., Shoelson, S. and Pawson, T. (1995) A conserved amino-terminal Shc domain binds to phosphotyrosine motifs in activated receptors and phosphopeptides. Curr. Biol., 5, 404–412. [DOI] [PubMed] [Google Scholar]
- van der Geer P., Wiley, S., Gish, G.D., Lai, V.K., Stephens, R., White, M.F., Kaplan, D. and Pawson, T. (1996) Identification of residues that control specific binding of the Shc phosphotyrosine-binding domain to phosphotyrosine sites. Proc. Natl Acad. Sci. USA, 93, 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdi J.M., Schmandt, R., Bashirullah, A., Jacob, S., Salvino, R., Craig, C.G., Program, A.E., Lipshitz, H.D., McGlade, C.J. (1996) Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol., 6, 1134–1145. [DOI] [PubMed] [Google Scholar]
- Vetter I.R., Arndt, A., Kutay, U., Görlich, D. and Wittinghofer, A. (1999) Structural view of the Ran–importin β interaction at 2.3 Å resolution. Cell, 97, 635–646. [DOI] [PubMed] [Google Scholar]
- Wittekind M. and Muller, L. (1993) HNCACB, a high sensitivity 3D NMR experiment to correlate amide proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. B, 101, 201–205. [Google Scholar]
- Wolf G., Trub, T., Ottinger, E., Groninga, L., Lynch, A., White, M.F., Miyazaki, M., Lee, J. and Shoelson, S.E. (1995) PTB domains of IRS–1 and Shc have distinct but overlapping binding specificities. J. Biol. Chem., 270, 27407–27410. [DOI] [PubMed] [Google Scholar]
- Yaich L., Ooi, J., Park, M., Borg, J.P., Landry, C., Bodmer, R. and Margolis, B. (1998) Functional analysis of the Numb phosphotyrosine-binding domain using site-directed mutagenesis. J. Biol. Chem., 273, 10381–10388. [DOI] [PubMed] [Google Scholar]
- Zambrano N. et al. (1997)Interaction of the phosphotyrosine interaction/phosphotyrosine binding-related domains of Fe65 with wild-type and mutant Alzheimer's β-amyloid precursor proteins. J. Biol. Chem., 272, 6399–6405. [DOI] [PubMed] [Google Scholar]
- Zhang O., Kay, L.E., Olivier, J.P., Forman-Kay, J.D. (1994) Backbone 1H and 15N resonance assigments of the N–terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR, 4, 845–858. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lee, C.-H., Mandiyan, V., Borg, J.-P., Margolis, B., Schlessinger, J. and Kuriyan, J. (1997) Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain. EMBO J., 16, 6141–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Feder, J.N., Jiang, M.M., Jan, L.Y. and Jan, Y.N. (1996) Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron, 17, 43–53. [DOI] [PubMed] [Google Scholar]
- Zhou M.-M. et al. (1995)Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature, 378, 584–592. [DOI] [PubMed] [Google Scholar]
- Zhou M.-M., Olejniczak, E.T., Meadows, R.P., Shuker, S.B., Miyazaki, M., Trub, T., Shoelson, S.E. and Fesik, S.W. (1996) Structural basis for IL–4 receptor phosphopeptide recognition by the IRS-1 PTB domain. Nature Struct. Biol., 3, 388–393. [DOI] [PubMed] [Google Scholar]
- Zwahlen C., Legault, P., Vincent, S.J.F., Greenblatt, J., Konrat, R. and Kay, L.E. (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage λ N–peptide/boxB RNA complex. J. Am. Chem. Soc., 119, 6711–6721. [Google Scholar]