Abstract
Purpose
Picoplatin is a new generation platinum designed to overcome platinum resistance. The goal of this study was to assess picoplatin anti-tumor activity and measure various cellular parameters in small-cell lung cancer (SCLC) cells resistant to cell killing by cisplatin and carboplatin.
Methods
We developed several platinum-resistant SCLC cell lines to evaluate picoplatin activity and drug resistance mechanisms in vitro. Drug cytotoxicity was measured by MTS assay. Total cellular platinum accumulation was measured by inductively coupled plasma mass spectrometry (ICP-MS). Whole genome gene expression profiling was carried out by microarray analysis.
Results
Picoplatin retained significant cytotoxic activity in platinum-resistant SCLC lines compared to cisplatin and carboplatin. Cellular picoplatin accumulation in platinum-resistant and parental cells was high relative to levels of cellular platinum found in the same cell lines after cisplatin or carboplatin treatment. Gene expression analyses revealed substantial differences in gene expression and highlighted specific annotation clusters in carboplatin-resistant cells. In addition, a similar gene expression pattern was observed in picoplatin-treated carboplatin-resistant and parental cells.
Conclusions
Our study demonstrates that picoplatin can overcome carboplatin and cisplatin resistance. The results suggest decreased platinum accumulation as a potential mechanism of platinum resistance in SCLC cells, provide candidate markers (e.g. several genes in the Hox, glutathione biosynthetic process, and MAGE families) that may serve as signatures for platinum resistance, support distinct effects of picoplatin on SCLC cells compared to other platinums, and provide a rationale to develop picoplatin for the treatment of recurrent SCLC following initial therapy with cisplatin or carboplatin.
Electronic supplementary material
The online version of this article (doi:10.1007/s00280-010-1435-5) contains supplementary material, which is available to authorized users.
Keywords: Picoplatin (AMD473, JM473, ZD0473); Cisplatin; Carboplatin; Small-cell lung cancer; Drug resistance; Platinum analog
Introduction
Platinum-based chemotherapy has been the primary treatment for cancer patients diagnosed with small-cell lung cancer (SCLC) treatment following initial diagnosis. In the United States, 82% of SCLC patients were treated with either carboplatin or cisplatin in combination with etoposide (IntrisiQ, 2008). Despite high initial response rates of 40–90% [1], the majority of patients develop treatment resistance. Effective second-line treatment for recurrent SCLC is a major unmet medical need. There is no standard chemotherapy for second-line platinum-refractory (no response to initial therapy) or platinum-resistant SCLC (early relapse after initial therapy).
Currently, the understanding of platinum resistance mechanisms is largely based on studies of cisplatin. Resistance is multi-factorial and can vary between cell lines and types and this finding is consistent with variable responses to platinum therapies between patients with similar tumor types. Several mechanisms of resistance have been hypothesized and are related directly or indirectly to the platinum DNA binding. Potential mechanisms of resistance include processes that alter the net intracellular accumulation of platinum due to influx and efflux transporters, the inactivation of platinum drugs once inside the cell by thiol-containing proteins such as glutathione or metallothionine, the removal of platinum adducts from DNA by nucleotide excision repair, the status of DNA mismatch repair and bypass of DNA adducts by DNA polymerase, and the mechanisms that control apoptosis (for review, see [2]).
Gene expression analysis has been used recently to identify individual genes and pathways whose transcriptional regulation contributes to platinum resistance. To enable physicians to provide the most appropriate care for individual patients, biomarkers for patient stratification for treatment as well as real-time monitoring of response are needed; gene expression signatures may provide this much-needed tool. A small number of recent publications report gene expression analysis of platinum resistance in ovarian cancer, osteosarcoma, esophageal cancer, and Hodgkin’s lymphoma. For ovarian cancer, potential gene signatures for resistance or correlation have been identified by profiling tumor samples from platinum responder and non-responder patients [3] and by profiling the response to carboplatin in an ovarian cell line [4]. Despite the potential clinical benefit, to date, no studies have been reported characterizing gene expression in SCLC cell lines (resistant or responsive to platinum agents) or SCLC clinical samples.
Picoplatin (AMD473, JM473, ZD0473) is a new generation platinum-based analog designed to overcome platinum resistance [5]. In vitro preclinical studies confirmed the ability of picoplatin to retain activity in the presence of elevated levels of glutathione and also demonstrated that picoplatin can overcome mechanisms which decrease accumulation of cisplatin [6, 7]. The suggestive in vitro findings were substantiated by in vivo preclinical observations: picoplatin has a greater anti-tumor effect on cisplatin-resistant ovarian tumors relative to other platinum agents [8].
Limited preclinical data are available regarding the activity of picoplatin in SCLC. The only publication documents picoplatin cytotoxicity against two SCLC lines, H69 and SBC-3, and against cisplatin-resistant derivatives of these two lines [7]. The following report describes the development of novel SCLC cell lines resistant to cisplatin and carboplatin followed by their characterization and the pharmacologic evaluation of picoplatin compared to other platinums in these tumor cells.
Materials and methods
Cell culture and reagents
Small-cell lung cancer lines, DMS53 and DMS114, were obtained from American Type Culture Collection (Manassas, VA). DMS53 and DMS114 are cultured in Waymouth’s MB 752/1 medium supplemented with 10% FBS. Injection grade cisplatin (1 mg/ml) and carboplatin (10 mg/ml) were obtained from Sicor (Irvine, CA) and oxaliplatin (5 mg/ml) was obtained from Sanofi-Aventis (Bridgewater, NJ). Picoplatin (0.5 mg/ml) was provided by Poniard Pharmaceuticals (South San Francisco, CA).
Drug selection
Drug-resistant sub-cultures were generated by repeated cycling in increasing selection drug concentration according to the following schedule: cells were treated in supplemented media with selection drug for 7 days. Cells were then gently washed and allowed to recover for 21 days in fresh supplemented media without drug. During this time, cells were tested for drug resistance. Drug concentrations used for selection were increased incrementally until cell lines acquired resistance. For DMS114, selection was initiated at 2 μM cisplatin and increased to 5 μM in 1 μM increments, and at 4 μM carboplatin and increased to 11 μM in 1 μM increments. Similarly in DMS53, cisplatin selection was from 3 to 6 μM, carboplatin from 5 to 11 μM, and oxaliplatin from 1 to 5 μM. These cell lines were then cycled at maintenance drug concentrations to retain resistance. In some cases, cell lines acquired durable resistance to the selection drug after 10–12 rounds of selection and no longer required cycling to maintain resistance. In cell lines where continued cycling is necessary to maintain drug resistance, cells in the recovery period (7–21 days after removal of selection drug) were used for experiments.
Cytotoxicity assay
An MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay was used to assess cytotoxicity. The CellTiter 96 AQueous Cell Proliferation Assay (Promega, Madison, WI) was performed essentially according to the manufacturer’s protocol. Briefly, 104 cells per well were seeded in flat bottom 96-well plates in supplemented media and allowed to adhere overnight. Following the addition of a serial dilution of drug, cells were incubated for 4 days. 20 μl of MTS reagent was added to each well, and plates were incubated for an additional 1–2 h at 37°C before spectrometric measurement of absorbance at 490 nm. Media-only background was subtracted from all samples. Data were analyzed using GraphPad Prism 5. Fifty percent inhibitory concentration (IC50) values were calculated based on normalized log transformed data using a non-linear regression algorithm. The resistance factor (RF) serves as a measure of drug resistance in resistant cells and is defined as the IC50 drug concentration of the resistant line divided by that of the parental line.
Cellular platinum accumulation
Total cellular platinum accumulation was determined by ICP-MS essentially as described by Kitada et al. [9]. Briefly, 106 cells were seeded in supplemented media in 6-well plates and allowed to adhere overnight, washed with PBS, and treated with drug in serum-free media for 24 h. Cells were then washed three times with PBS, lysed and sonicated in 1 ml H2O. A volume of 0.5 ml of sample was then reacted with aqua regia, digested at 95°C, and diluted to a final volume of 5 ml with H2O. Samples were analyzed by conventional ICP-MS and quantified against a certified standard using multi-point weighted linear regression (CanTest, Burnaby, Canada).
Gene expression profiling by microarray
In supplemented Waymouth’s medium, 106 carboplatin-resistant (7–21 days into the drug cycling recovery period) and parental DMS53 cells were allowed to adhere overnight and treated with drug for 24 h. RNA was extracted using the RNeasy total RNA purification kit (Qiagen, Valencia, CA) according to manufacturer protocol. Total RNA quality was determined using a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Four independent batches of RNA were prepared for gene expression profiling to allow for adequate statistical power. Sample preparation, labeling, and array hybridizations were performed according to standard protocols by the UCSF Shared Microarray Core Facilities and Agilent Technologies (http://www.arrays.ucsf.edu and http://www.agilent.com). Equal amounts of Cy3-labeled cRNA target were hybridized to Agilent human whole genome 4 × 44 K Ink-jet arrays (Agilent). Arrays were scanned using the Agilent microarray scanner (Agilent) and raw signal intensities were extracted with Feature Extraction 10.1.1 software (Agilent). Coefficients were included to adjust for batch specific differences. Subsequent gene ontology and functional clustering analyses were performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID 2008; National Institute of Allergy and Infectious Disease (NIAID), NIH; [10, 11]).
Results
Generation of drug-resistant SCLC cell lines
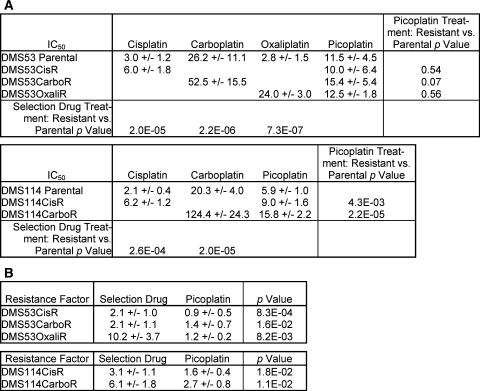
As a part of our effort to characterize the mechanism of action of the new generation platinum analog picoplatin in SCLC cell lines, we also characterized the activities of cisplatin and carboplatin, which are platinum-based drugs used in the first-line treatment of SCLC. Sensitivity to cisplatin, carboplatin and picoplatin in DMS53 and DMS114 was determined by measuring the inhibition of proliferation at various test concentrations using an MTS-based cytotoxicity assay. IC50 values were obtained by non-linear regression of normalized transformed data. Although oxaliplatin is not currently used in the treatment of SCLC, it is we included oxaliplatin to compare picoplatin effectiveness to the current marketed platinum agents. Relative cell cytotoxicity profiles were generated for picoplatin, cisplatin, carboplatin, and oxaliplatin. Picoplatin IC50 values were slightly higher than those of cisplatin and oxaliplatin but substantially lower than that of carboplatin (Fig. 1a). This ranking in these SCLC cell lines is consistent with previous observations in ovarian cell lines [6].
Fig. 1.
IC50 and RF values for DMS53 and DMS114 parental and resistant cell lines. a 50% inhibitory concentration (IC50) values (μM) measured in by the MTS assay (18 point serial titration) after treatment of drug for 96 h. b Resistance factors (RFs) for resistant cells were calculated by dividing the IC50 value of the resistant line by that of the corresponding parental line within a single experiment. Data are the average for at least 6 independent experiments. The standard deviation is reported for each IC50 and RF value. Student’s t-test (two-tailed) was used to determine p values. p < 0.05 is considered significant
We generated cisplatin-, carboplatin-, and oxaliplatin-resistant SCLC lines in order to study the mechanisms of acquired drug resistance. Drug-resistant lines were generated by repeat cycling of cells in increasing concentrations of selection drug over time (see “Materials and Methods”). Simultaneously, we also attempted to generate picoplatin-resistant SCLC lines with little success (unpublished observations). Kelland et al. had similar findings in their attempts to generate picoplatin-resistant ovarian cell lines (personal communication). Resistance to oxaliplatin in DMS53 cells was obtained more quickly (after 4 rounds) and was more stable (no longer required cycling in drug to retain resistance) than cisplatin or carboplatin, which is consistent with a study by Stordal et al. [12] with H69 cells. Cisplatin and carboplatin-resistant DMS53 and DMS114 lines became more stably resistant to selection drug after 10–12 rounds of selection.
To assess whether picoplatin retains activity in platinum-resistant cells, we performed cytotoxicity assays comparing IC50 drug concentrations in parental and resistant cells (Fig. 1a). The resistance factor (RF) values with the selection drug were 2–10, indicating that the cell lines that had undergone successive rounds of drug selection were resistant to the selection drug (Fig. 1b). The RF values with picoplatin treatment in cisplatin, carboplatin, or oxaliplatin-resistant cells remained at or near 1, with exception of carboplatin-resistant DMS114 cells, which showed some resistance to picoplatin. However, the same cells showed markedly higher resistance to carboplatin. These data demonstrate that picoplatin has the capacity to overcome at least some mechanisms of resistance to cisplatin and carboplatin (and oxaliplatin) in SCLC cells.
Cellular platinum accumulation after picoplatin treatment
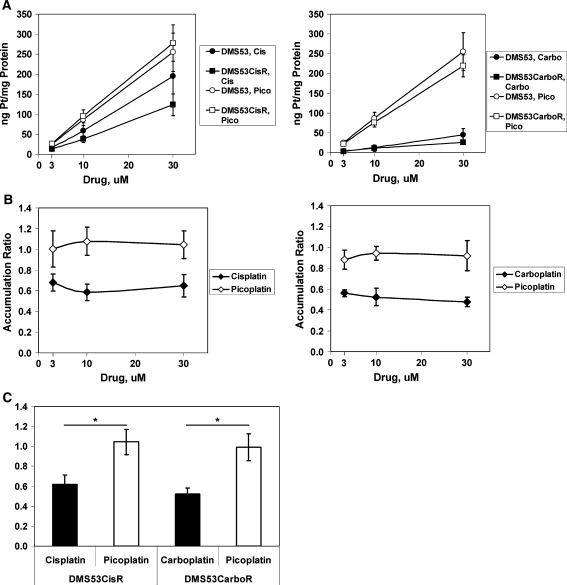
Multiple drug influx and efflux mechanisms are known to regulate intracellular drug levels and in turn play a role in determining drug sensitivity [13]. Cellular drug accumulation is a major topic of study in resistance to platinum drugs. In order to ascertain whether cellular platinum accumulation varies between picoplatin and other platinum drugs, lysates from cells treated with drug were analyzed by ICP-MS for platinum. In all cases, platinum accumulation was essentially linear through the drug titration range for all drugs tested (Fig. 2a). In parental DMS53 cells, carboplatin treatment resulted in the lowest level of cellular platinum accumulation whereas picoplatin treatment resulted in the highest level. Platinum accumulation was not significantly different in parental DMS53 after treatment with cisplatin or picoplatin (P > 0.05). However, cisplatin-resistant DMS53 accumulated significantly less platinum after cisplatin treatment compared to picoplatin treatment (P < 0.0003). In both parental and carboplatin-resistant DMS53, picoplatin treatment resulted in strikingly higher cellular platinum levels than carboplatin treatment (P < 0.00005). Similar results were observed in DMS114 cells (Supplementary Fig. S1). Because platinum accumulation was linear over the treatment concentrations used, it was possible to express the data as an accumulation ratio, which is defined as the platinum accumulation in resistant cells divided by that of parental cells for each drug concentration. We found that this ratio was essentially constant over the concentration ranges tested, which is expected given the linear relationship between platinum accumulation and drug concentration (Fig. 2b). A number less than 1 indicates reduced drug accumulation in resistant cells relative to parental cells. Therefore, it was possible to calculate an average accumulation ratio (Fig. 2c), which represents the fraction of platinum accumulation in drug-resistant cells relative to its corresponding parental cells. Both cisplatin- and carboplatin-resistant DMS53 cells accumulated a lower level of their respective selection drug when compared with parental cells (62 and 52%, respectively; Fig. 2b), suggesting that influx/efflux mechanisms may be involved in drug resistance in these cells. Cisplatin-resistant DMS114 cells accumulated 71% of the selection drug relative to parental cells, whereas carboplatin-resistant DMS114 cells did not accumulate less selection drug (91%, Supplementary Fig. S1). Strikingly, platinum accumulation in cisplatin- and carboplatin-resistant DMS53 cells treated with picoplatin remained unchanged when compared with parental cells (104 and 99%, respectively, Fig. 2a–c). Together, these data suggest that the mechanisms involved in reducing platinum accumulation in cisplatin- and carboplatin-resistant SCLC cells do not affect platinum accumulation after picoplatin treatment.
Fig. 2.
Total cellular platinum accumulation in parental and drug-resistant DMS53 cells. a Cellular platinum levels versus drug concentration. Attached cells were treated with drug (3, 10, and 30 μM) for 24 h, and lysates were harvested and analyzed by ICP-MS. Platinum levels were normalized to protein concentration for each sample. b Accumulation ratio is calculated by dividing the amount of platinum per mg of protein for the resistant cells by that of the parental cells. A ratio of one indicates that there is no difference in platinum accumulation between resistant and parental cells whereas a value of less than one represents reduced platinum accumulation in resistant cells. c Average accumulation ratio was obtained by averaging the ratios of the 3 concentrations tested for each drug. This was possible because the accumulation did not change over the drug concentration range used. Filled symbols = selection drug; Open symbols = picoplatin. CisR = cisplatin-resistant line; CarboR = carboplatin-resistant line. Data are the average of 3 independent experiments. Error bars represent the standard deviation for each data point. *p < 0.000025, Student’s t-test, two-tailed
Whole genome gene expression profiling
Drug resistance pathways such as drug transport, neutralization, metabolism, decreased apoptosis, and improved DNA repair have been suggested to be involved with drug resistance in various cancer cell types. However, there has been little done to discern the pathways of platinum resistance in SCLC. Therefore, we sought to identify genes and pathways that are differentially expressed between carboplatin-resistant and parental SCLC cells. In addition, gene expression changes after picoplatin or carboplatin treatment were assessed.
Unsupervised hierarchical cluster analysis
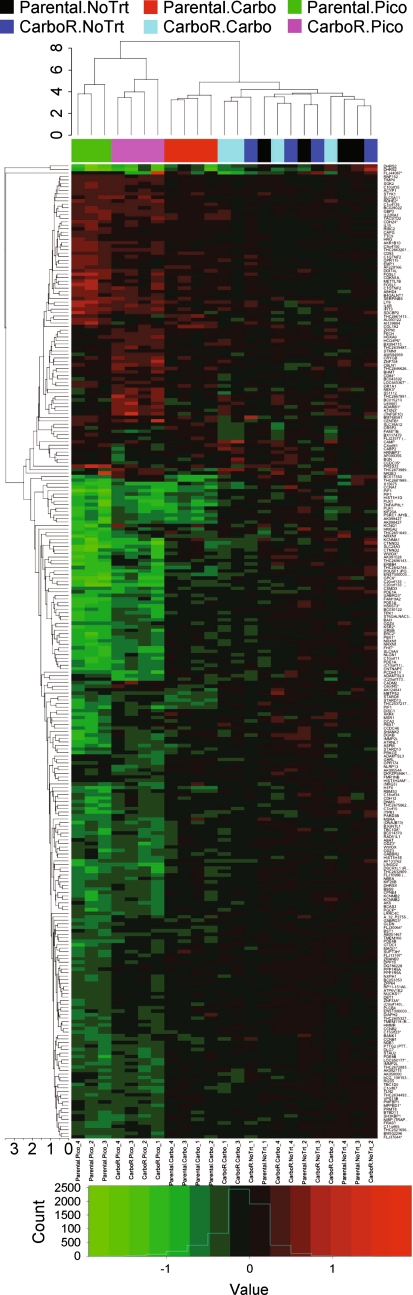
In order to evaluate the gene expression changes between treatments, an unsupervised hierarchical clustering analysis was performed (Fig. 3). Because of the substantial basal differences between carboplatin-resistant and parent cells, treatment-induced fold changes were calculated relative to the no treatment expression levels from their respective cell lines. As expected, treatment replicates generally clustered well (Fig. 3, x-axis dendrogram). Interestingly, clustering of treatment-induced differential expression also resulted in clustering of picoplatin-treated parental and carboplatin-resistant samples, suggesting that picoplatin elicits a similar response in both parental and carboplatin-resistant cells but is distinct from that of carboplatin. Carboplatin-treated parental samples clustered together whereas carboplatin-treated carboplatin-resistant replicates clustered loosely with no treatment replicates of both lines, suggesting that gene expression was not substantially affected by selection drug treatment in carboplatin-resistant cells. This is likely explained by the relative lack of differential gene expression after carboplatin treatment of carboplatin-resistant cells.
Fig. 3.
Dendrogram of two-dimensional hierarchical clustering analysis of differential expression by treatment. The heatmap indicates log2 fold change of all sample replicates relative to the average across arrays (aveA) for genes that were differentially expressed in any of the treatment contrasts; red upregulated and green downregulated. Due to the large basal gene expression differences between carboplatin-resistant and parental cells, fold changes were calculated relative to the no treatment group of the respective cell line to discern treatment-induced changes in each line. The line height of dendrograms indicates the Euclidean average distance between clusters. Treatment groups are represented by color below the treatment dendrogram on the x-axis (black parental, no treat; red parental, carbo; green parental, pico; dark blue carboR, no treat; light blue carboR, carbo; pink carboR, pico)
Analysis of individual genes most highly upregulated in carboplatin-resistant DMS53 cells revealed a number of genes linked to cancer (Table 1, Adj P < 0.01, fold change >5). For instance, phosphorylated HCLS1, known for its function in immune synapse formation and chemotaxis, is associated with poor prognosis in chronic lymphocytic leukemia [34]. Angiogenin is well known for its pro-angiogenic properties [35] and may be involved in cell proliferation and angiogenesis in lung adenocarcinoma [36]. HOXB9 overexpression was frequently seen in lung cancer [26], and its upregulation was found in Hodgkin lymphoma and shown to be involved in proliferation and apoptosis [37]. STMN2 was found to be upregulated in hepatoma via beta-catenin/TCF, a key component of the Wnt signaling pathway [38]. Interestingly, five genes in the variable charge, X/Y-linked (VCX/Y) family and four genes in the paraneoplastic antigen like (PNMA) family—whose functions are not known—were upregulated more than fivefold in carboplatin-resistant cells.
Table 1.
Highest differentially upregulated genes: no treatment, carboplatin-resistant (CarboR) versus parental
| Gene name | Description | GenBank accession | Expression ratio (CarboR/Parental) | Adjusted P Value |
|---|---|---|---|---|
| HCLS1 | Hematopoietic cell-specific Lyn substrate 1 | NM_005335 | 62.4 | 1.72E-16 |
| UNQ2541 | MSFL2541 | NM_203347 | 33.5 | 5.23E-13 |
| KLHL1 | Kelch-like 1 (Drosophila) | NM_020866 | 21.6 | 2.06E-10 |
| SEPT1 | Septin 1 | NM_052838 | 18.4 | 6.34E-12 |
| FUT3 | Fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) | NM_000149 | 13.3 | 1.10E-10 |
| TPM4 | Tropomyosin 4 | NM_003290 | 12.8 | 1.26E-13 |
| ANG | Angiogenin, ribonuclease, RNase A family, 5 | NM_001145 | 12.3 | 9.98E-08 |
| RNASE4 | Ribonuclease, RNase A family, 4 | NM_194430 | 11.2 | 9.89E-08 |
| CLCA1 | Chloride channel, calcium activated, family member 1 | NM_001285 | 9.8 | 4.06E-06 |
| LOC199725 | Hypothetical protein LOC199725 | XM_086001 | 9.4 | 2.11E-05 |
| PNMA3 | Paraneoplastic antigen MA3 | NM_013364 | 9.1 | 1.34E-11 |
| VCX3A | Variable charge, X-linked 3A | NM_016379 | 9.0 | 1.64E-06 |
| PNMA5 | Paraneoplastic antigen like 5 | NM_052926 | 8.7 | 5.35E-13 |
| LOC340061 | Hypothetical protein LOC340061 | NM_198282 | 8.6 | 3.86E-07 |
| SUCNR1 | Succinate receptor 1 | NM_033050 | 8.5 | 8.00E-08 |
| VCX | Variable charge, X-linked | NM_013452 | 8.3 | 1.84E-05 |
| VCX2 | Variable charge, X-linked 2 | NM_016378 | 8.2 | 1.29E-06 |
| PNMA6A | Paraneoplastic antigen like 6A | NM_032882 | 8.1 | 1.57E-06 |
| FBXL13 | F-box and leucine-rich repeat protein 13 | NM_145032 | 8.1 | 1.20E-08 |
| VCY | Variable charge, Y-linked | NM_004679 | 8.0 | 5.85E-09 |
| VCX2 | Variable charge, X-linked 2 | NM_016378 | 8.0 | 5.98E-08 |
| TMEM71 | Transmembrane protein 71 | NM_144649 | 7.8 | 4.59E-09 |
| STMN2 | Stathmin-like 2 | NM_007029 | 7.4 | 1.00E-06 |
| HOXB9 | Homeobox B9 | NM_024017 | 7.3 | 2.09E-12 |
| ZNF179 | Zinc finger protein 179 | NM_007148 | 7.0 | 1.02E-10 |
| TSPAN1 | Tetraspanin 1 | NM_005727 | 6.7 | 2.68E-09 |
| FER1L3 | Fer-1-like 3, myoferlin (C. elegans) | NM_013451 | 6.6 | 1.13E-07 |
| SEZ6 | Seizure related 6 homolog (mouse) | NM_178860 | 6.2 | 0.000643 |
| COL8A1 | Collagen, type VIII, alpha 1 | NM_001850 | 6.0 | 1.74E-05 |
| FER1L3 | Fer-1-like 3, myoferlin (C. elegans) | NM_013451 | 5.9 | 4.15E-08 |
| ANXA3 | Annexin A3 | NM_005139 | 5.9 | 3.19E-08 |
| LECT1 | Leukocyte cell derived chemotaxin 1 | NM_007015 | 5.8 | 0.000185 |
| SHC2 | SHC (Src homology 2 domain containing) transforming protein 2 (SHC2) | XM_939572 (NM_012435) | 5.8 | 0.001418 |
| ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | NM_001175 | 5.7 | 0.002958 |
| PRSS16 | Protease, serine, 16 (thymus) | NM_005865 | 5.7 | 1.33E-06 |
| BRDG1 | BCR downstream signaling 1 | NM_012108 | 5.5 | 0.001209 |
| HOXC9 | Homeobox C9 | NM_006897 | 5.3 | 6.11E-12 |
| FST | Follistatin | NM_013409 | 5.1 | 0.002365 |
| HGD | Homogentisate 1,2-dioxygenase (homogentisate oxidase) | NM_000187 | 5.0 | 2.08E-05 |
Adjusted p value <0.01, greater than fivefold differential expression
Carboplatin-resistant versus parental, no treatment
A comparison of carboplatin-resistant and parental DMS53 cells revealed significant baseline differences in gene expression. Functional annotation cluster analyses (DAVID) highlighted overrepresentation of genes in homeodomain, glutathione biosynthetic/metabolic process, MAGE, homophilic cell adhesion, and regulation of phosphorylation functional groups in carboplatin-resistant cells (Table 2, Adjusted P < 0.05, fold change >2). Underrepresented functional groups in carboplatin-resistant cells include genes in blood coagulation/homeostasis/wound healing, neuron development and differentiation, EGF/EGF-like, and protease inhibitor functional groups.
Table 2.
Functional annotation cluster analysis: no treatment, carboplatin-resistant versus parental
| Functional group | Category | Fold enrichment | Genes | P Value | |
|---|---|---|---|---|---|
| 1 | Homeobox; Homeodomain related | U, I, S, SP | 4.9–6.4 | LBX1, ARX, DLX1, DLX2, DLX4, TLX2, HOXA7, HOXA9, HOXA13, HOXB2, HOXB5, HOXB6, HOXB7, HOXB8, HOXB9, HOXC9, SIX6, PAX3 | 1.4E-7 to 2.3E-9 Median: 8.4E-8 |
| 2 | Acyltransferase activity; glutathione biosynthetic process | SP, GM, I, GB, K | 1.6–37.4 | GGT1, GGT3, GSTT1, GGTL4, GGTLA4, GLYATL1 | 1.1E-4 to 3.9E-1 Median: 6.3E-3 |
| 3 | Tumor associated protein MAGE | U, I, P | 7.3–7.8 | MAGEA8, MAGEA9, MAGEA11, MAGEB2 | 1.5E-2 to 5.5E-2 Median: 1.7E-2 |
| 4 | Cadherin; Homophilic cell adhesion | GB, I, U, S | 3.0–4.5 | CDH3, PCDH8, PCDH12, PCDHB8, PCDHB16, CLSTN2, FRAS1 | 2.5E-2 to 1.0E-1 Median: 4.6E-2 |
| 5 | Regulation of phosphorylation | GB | 1.8–3.9 | ANG, HCLS1, IGFBP3, SYK, TNF | 4.6E-2 to 4.8E-1 Median: 8.1E-2 |
Fold upregulated >2, adjusted p value <0.05
Annotation database/category abbreviations: U UP seq feature, I interpro, S smart, SP SP PIR keywords, GM GOTerm MF ALL, GB GOTerm BP ALL, P PIR superfamily, K KEGG pathway
Picoplatin versus no treatment, parental, and carboplatin-resistant cells
In addition, we assessed differential gene expression after picoplatin treatment versus no treatment in either parental or carboplatin-resistant cells. Treatment with picoplatin resulted in relatively few upregulated genes in both cell lines, and consequently no striking functional clustering patterns were found in either cell line (data not shown). Downregulated functional gene clusters in parental cells after picoplatin treatment include genes involved in M phase of cell cycle and cyclins, phosphodiesterase, phorbol ester binding, EGF/EGF like, and ion transport. Similarly, picoplatin treatment in carboplatin-resistant cells led to decreased expression of EGF/EGF-like and cell cycle groups (data not shown). Notable among cell cycle genes downregulated in both cell lines after picoplatin treatment are cyclin A1, Polo-like kinase 1, and RAD51-like 1.
Picoplatin versus carboplatin treatment in parental cells
We also compared gene expression changes after picoplatin versus carboplatin treatment. The most striking observation from functional clustering analyses was that picoplatin treatment of parental cells reduced expression of genes in ion transporter activity (ion/transmembrane, metal cation, potassium ion, alkali metal ion) compared to carboplatin treatment. The majority of genes downregulated by picoplatin in these clusters are related to neuronal cell type or processes.
Discussion
Picoplatin is an investigational agent that has been evaluated in >1100 cancer patients to date. Picoplatin has been and continues to be evaluated as an anti-tumor agent as a single agent or in combination with other cytotoxics in a wide variety of tumor types. Two independent Phase II monotherapy studies of picoplatin in SCLC demonstrated a median overall survival of at least 27 weeks and were comparable to other therapeutic options [43, 44]. In the SPEAR (Study of Picoplatin Efficacy After Relapse) pivotal Phase III trial conducted by Poniard Pharmaceuticals, the efficacy and safety of picoplatin in combination with best supportive care (BSC) vs BSC alone was evaluated in a total of 401 SCLC patients who were refractory to or who progressed within 6 months of first-line platinum-based therapy. Picoplatin was well-tolerated with manageable hematologic toxicity. Although the analysis of overall survival (321 events) did not achieve statistical significance (P = 0.0895) in the intent-to-treat population, a statistically significant imbalanced use of post-study chemotherapy in favor of the BSC arm may have impacted this result (P = 0.012). In 273 patients who did not receive post-study chemotherapy, there is statistically significant difference in overall survival in favor of the picoplatin arm (P = 0.0345). More importantly, picoplatin demonstrated significant survival improvement in 294 refractory patients who never responded or relapsed within 45 days (P = 0.0173). Picoplatin addresses a major unmet need in the treatment of second-line small-cell lung cancer as there is currently no FDA approved therapy for patients who are refractory or relapse within 45 days of first-line platinum-based therapy [22].
In this study, we demonstrated the ability of picoplatin to overcome platinum drug resistance in SCLC using newly generated cisplatin- and carboplatin-resistant SCLC cell lines. These observations were consistent with a study by Kawamura et al. [7], which showed continued sensitivity to picoplatin in two cisplatin-resistant SCLC and two cisplatin-resistant NSCLC lines. A number of resistance mechanisms to platinum drugs have been postulated [2]. However, no reports of studies related to platinum resistance have been done in small-cell lung cancer. In this study, we identified decreased cellular platinum accumulation after drug treatment as a potential mechanism that could contribute to cisplatin and carboplatin resistance in these cell lines. In both platinum-resistant cell lines, picoplatin treatment resulted in high levels of cellular platinum that were indistinguishable from those of parental cells. Similar results were observed in human ovarian cancer lines [6]. In addition, picoplatin appears to retain cytotoxic activity against both these platinum-resistant lines.
Gene expression microarray analysis has been useful to identify signatures of drug resistance and provide potential clinical biomarkers. A comprehensive study of the carboplatin-resistant cell line using gene expression profiling revealed additional pathways that may potentially contribute to carboplatin resistance. Functional annotation clustering showed significant upregulation of HOX-containing genes, genes involved in glutathione biosynthesis, and MAGE genes, many of which have been implicated in cancer.
Our study shows that homeodomain (HOX)-containing gene functional clusters are highly overrepresented in carboplatin-resistant DMS53 cells (Table 2, Cluster 1). HOX genes are well known for their role in development and hematopoiesis [14, 15]. Their involvement in cancer was initially identified in hematologic malignancies such as myeloid lymphocytic leukemia where chromosomal translocation of HOX with Nup98 is thought to contribute to tumorigenesis [16]. More recently, a significant number of studies suggest that the expression of HOX genes is dysregulated in multiple solid tumor types including SCLC [17–19] and is involved in progression, invasion, metastasis, and DNA repair [20–22]. For instance, the disruption of HOX interaction with PBX leads to increased apoptosis in NSCLC, RCC, and melanoma cells [23–25]. HOXA9 and HOXB9, both upregulated in carboplatin-resistant DMS53 cells, were found to be frequently overexpressed in lung cancer [26]. Interestingly, HOXB7, also upregulated in carboplatin-resistant DMS53, is believed to be involved in DNA repair [22]. Activation of HOX gene clusters has been linked to poor survival and drug resistance in glioblastoma and resembles a self-renewal signature in leukemia [27]. Little else is known regarding the role of HOX genes in drug resistance.
Increased glutathione levels and glutathione-S-transferase activity have frequently been implicated in platinum neutralization and resistance, and the correlation between increased glutathione levels and drug resistance has been well documented in multiple tumor types [28–33]. However, a causal relationship between the two has thus far not been convincingly demonstrated to date. We found that glutathione biosynthetic and metabolic processes were overrepresented in carboplatin-resistant DMS53 relative to parental DMS53 cells. Consistent with these data, total cellular glutathione level was shown to be significantly higher in carboplatin-resistant DMS53 cells in a separate set of experiments (Supplementary Fig. S2). Together, these data show that upregulation of the glutathione pathway is associated with carboplatin resistance and that drug neutralization by glutathione may contribute to drug resistance in SCLC. Although the results of our study do not establish a causal relationship between increased glutathione and drug resistance, they do suggest that the glutathione biosynthetic pathway may be regulated by the development of drug resistance in SCLC. Interestingly, picoplatin retained cytotoxic activity against carboplatin-resistant DMS53 cells, and cellular accumulation remained high in both drug-resistant and parental cells, which suggests that picoplatin is able to circumvent glutathione mediated inactivation.
Expression of MAGE genes has been linked to SCLC and suggested as possible biomarkers for detection as well as potential targets for immune-based therapeutic modalities [42, 43]. The first melanoma-associated antigen (MAGE-A1) was isolated from a melanoma cell line and found to be recognized by cytotoxic T lymphocytes [44]. In subsequent years, numerous additional MAGE genes were identified. MAGE genes are subclassified into four groups (A-D) based on sequence homology and divided into two subgroups (I, containing groups A-C, and II, containing group D) based on expression pattern. Subgroup I, also called cancer/testis antigens, are expressed in testis and frequently in tumor cells and have become the subject of substantial interest in tumor immunotherapy. Although the functions of MAGE proteins have remained largely elusive, they are believed to be involved in embryonic development and aspects of tumor transformation and progression. Our data suggest that upregulation of several MAGE genes (MAGE A8, A9, A11, and B2) may be related to the acquisition of resistance to carboplatin in SCLC cells. If confirmed, these observations could potentially have utility in predicting SCLC resistance to platinum and guiding patient treatment decisions.
Interestingly, our results also point to the upregulation of a number of genes in the VCX/Y and PNMA families, the functions of which are currently unknown. Further studies will be required to assess whether these differentially regulated genes play a crucial role in conferring resistance to carboplatin.
Our gene expression data suggest a possible mechanism for resistance to carboplatin in SCLC cells and begin to reveal a distinct mode of action for picoplatin. Further studies will be required to confirm our findings in additional platinum-resistant SCLC cell lines and establish a potential signature for platinum resistance. Since the kinetics of DNA binding of picoplatin and cell cycle/death response to picoplatin appear to be somewhat slow relative to other platinums [5], gene expression profiling of platinum-treated tumor cells over time may prove to be useful. Based on our finding that picoplatin treatment can overcome resistance to other platinum drugs and the observation that picoplatin elicits a gene expression profile that is distinct from that of carboplatin in SCLC, we believe that additional studies along this line of inquiry could help to determine the mechanism of action for picoplatin and potentially have clinical application by the establishment of gene expression signature of carboplatin resistance in SCLC.
The primary objective of this work was to determine whether picoplatin could overcome resistance to treatment by cisplatin and carboplatin in small-cell lung cancer. We found that picoplatin retains cytotoxic activity against cisplatin and carboplatin-resistant SCLC lines. In addition, our results demonstrated that platinum-resistant cells accumulated less platinum and differential gene expression. Picoplatin treatment resulted in high levels of platinum accumulation in all resistant and parental lines tested, suggesting that mechanisms controlling cellular cisplatin and carboplatin levels were not effective in controlling cellular picoplatin levels. Finally, we showed by gene expression profiling that picoplatin treatment triggers a distinct gene expression pattern relative to treatment with carboplatin and that this pattern is similar in parental and carboplatin-resistant SCLC cell line. Together our data suggest that picoplatin action is distinct from those of the other platinums tested and could have clinical benefit in the treatment of small-cell lung cancer resistant to cisplatin and carboplatin therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Giaccone G. Clinical perspectives on platinum resistance. Drugs. 2000;59(Suppl 4):9–17. doi: 10.2165/00003495-200059004-00002. [DOI] [PubMed] [Google Scholar]
- 2.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 3.Helleman J, Jansen MP, Span PN, van Staveren IL, Massuger LF, Meijer-van Gelder ME, Sweep FC, Ewing PC, van der Burg ME, Stoter G, Nooter K, Berns EM. Molecular profiling of platinum resistant ovarian cancer. Int J Cancer. 2006;118:1963–1971. doi: 10.1002/ijc.21599. [DOI] [PubMed] [Google Scholar]
- 4.Konstantinopoulos PA, Fountzilas E, Pillay K, Zerbini LF, Libermann TA, Cannistra SA, Spentzos D. Carboplatin-induced gene expression changes in vitro are prognostic of survival in epithelial ovarian cancer. BMC Med Genomics. 2008;1:59. doi: 10.1186/1755-8794-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holford J, Raynaud F, Murrer BA, Grimaldi K, Hartley JA, Abrams M, Kelland LR. Chemical, biochemical and pharmacological activity of the novel sterically hindered platinum co-ordination complex, cis-[amminedichloro(2-methylpyridine)] platinum(II) (AMD473) Anticancer Drug Des. 1998;13:1–18. [PubMed] [Google Scholar]
- 6.Holford J, Sharp SY, Murrer BA, Abrams M, Kelland LR. In vitro circumvention of cisplatin resistance by the novel sterically hindered platinum complex AMD473. Br J Cancer. 1998;77:366–373. doi: 10.1038/bjc.1998.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamura-Akiyama Y, Kusaba H, Kanzawa F, Tamura T, Saijo N, Nishio K. Non-cross resistance of ZD0473 in acquired cisplatin-resistant lung cancer cell lines. Lung Cancer. 2002;38:43–50. doi: 10.1016/S0169-5002(02)00175-7. [DOI] [PubMed] [Google Scholar]
- 8.Raynaud FI, Boxall FE, Goddard PM, Valenti M, Jones M, Murrer BA, Abrams M, Kelland LR. cis-Amminedichloro(2-methylpyridine) platinum(II) (AMD473), a novel sterically hindered platinum complex: in vivo activity, toxicology, and pharmacokinetics in mice. Clin Cancer Res. 1997;3:2063–2074. [PubMed] [Google Scholar]
- 9.Kitada N, Takara K, Minegaki T, Itoh C, Tsujimoto M, Sakaeda T, Yokoyama T. Factors affecting sensitivity to antitumor platinum derivatives of human colorectal tumor cell lines. Cancer Chemother Pharmacol. 2008;62:577–584. doi: 10.1007/s00280-007-0640-3. [DOI] [PubMed] [Google Scholar]
- 10.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 11.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 12.Stordal BK, Davey MW, Davey RA. Oxaliplatin induces drug resistance more rapidly than cisplatin in H69 small cell lung cancer cells. Cancer Chemother Pharmacol. 2006;58:256–265. doi: 10.1007/s00280-005-0148-7. [DOI] [PubMed] [Google Scholar]
- 13.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 14.Scielzo C, Ghia P, Conti A, Bachi A, Guida G, Geuna M, Alessio M, Caligaris-Cappio F. HS1 protein is differentially expressed in chronic lymphocytic leukemia patient subsets with good or poor prognoses. J Clin Invest. 2005;115:1644–1650. doi: 10.1172/JCI24276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tello-Montoliu A, Patel JV, Lip GY. Angiogenin: a review of the pathophysiology and potential clinical applications. J Thromb Haemost. 2006;4:1864–1874. doi: 10.1111/j.1538-7836.2006.01995.x. [DOI] [PubMed] [Google Scholar]
- 16.Yuan Y, Wang F, Liu XH, Gong DJ, Cheng HZ, Huang SD. Angiogenin is involved in lung adenocarcinoma cell proliferation and angiogenesis. Lung Cancer. 2009;66:28–36. doi: 10.1016/j.lungcan.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E, Rosell R, Gemmill RM, Drabkin HA. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci USA. 2000;97:12776–12781. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagel S, Burek C, Venturini L, Scherr M, Quentmeier H, Meyer C, Rosenwald A, Drexler HG, MacLeod RA. Comprehensive analysis of homeobox genes in Hodgkin lymphoma cell lines identifies dysregulated expression of HOXB9 mediated via ERK5 signaling and BMI1. Blood. 2007;109:3015–3023. doi: 10.1182/blood-2006-08-044347. [DOI] [PubMed] [Google Scholar]
- 19.Lee HS, Lee DC, Park MH, Yang SJ, Lee JJ, Kim DM, Jang Y, Lee JH, Choi JY, Kang YK, Kim DI, Park KC, Kim SY, Yoo HS, Choi EJ, Yeom YI. STMN2 is a novel target of beta-catenin/TCF-mediated transcription in human hepatoma cells. Biochem Biophys Res Commun. 2006;345:1059–1067. doi: 10.1016/j.bbrc.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Eckardt JR, Bentsion DL, Lipatov ON, Polyakov IS, Mackintosh FR, Karlin DA, Baker GS, Breitz HB. Phase II study of picoplatin as second-line therapy for patients with small-cell lung cancer. J Clin Oncol. 2009;27:2046–2051. doi: 10.1200/JCO.2008.19.3235. [DOI] [PubMed] [Google Scholar]
- 21.Treat J, Schiller J, Quoix E, Mauer A, Edelman M, Modiano M, Bonomi P, Ramlau R, Lemarie E. ZD0473 treatment in lung cancer: an overview of the clinical trial results. Eur J Cancer. 2002;38( Suppl 8):S13–S18. doi: 10.1016/S0959-8049(02)80016-8. [DOI] [PubMed] [Google Scholar]
- 22.Ciuleanu T (2010) Randomized phase III study (SPEAR) of picoplatin plus best supportive care (BSC) or BSC alone in patients with SCLC refractory or progressive within 6 months after first-line platinum-based chemotherapy. Oral Presentation, Abstract No: 7002. ASCO Annual Meeting, Chicago
- 23.Chen H, Sukumar S. Role of homeobox genes in normal mammary gland development and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:159–175. doi: 10.1023/A:1025996707117. [DOI] [PubMed] [Google Scholar]
- 24.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 25.Borrow J, Shearman AM, Stanton VP, Jr, Becher R, Collins T, Williams AJ, Dube I, Katz F, Kwong YL, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman DE. The t(7;11)(p15; p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 26.Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, Tamoto E, Shindo G, Matsunaga A, Teramoto K, Komuro K, Kondo S, Katoh H, Koike T, Moriuchi T. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep. 2005;13:673–679. [PubMed] [Google Scholar]
- 27.Abe M, Hamada J, Takahashi O, Takahashi Y, Tada M, Miyamoto M, Morikawa T, Kondo S, Moriuchi T. Disordered expression of HOX genes in human non-small cell lung cancer. Oncol Rep. 2006;15:797–802. [PubMed] [Google Scholar]
- 28.Cillo C. HOX genes in human cancers. Invasion Metastasis. 1994;14:38–49. [PubMed] [Google Scholar]
- 29.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, Shevde LA, Li W, Eschrich S, Daud A, Ju J, Matta J. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashita T, Tazawa S, Yawei Z, Katayama H, Kato Y, Nishiwaki K, Yokohama Y, Ishikawa M. Suppression of invasive characteristics by antisense introduction of overexpressed HOX genes in ovarian cancer cells. Int J Oncol. 2006;28:931–938. [PubMed] [Google Scholar]
- 31.Rubin E, Wu X, Zhu T, Cheung JC, Chen H, Lorincz A, Pandita RK, Sharma GG, Ha HC, Gasson J, Hanakahi LA, Pandita TK, Sukumar S. A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res. 2007;67:1527–1535. doi: 10.1158/0008-5472.CAN-06-4283. [DOI] [PubMed] [Google Scholar]
- 32.Plowright L, Harrington KJ, Pandha HS, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer) Br J Cancer. 2009;100:470–475. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shears L, Plowright L, Harrington K, Pandha HS, Morgan R. Disrupting the interaction between HOX and PBX causes necrotic and apoptotic cell death in the renal cancer lines CaKi-2 and 769-P. J Urol. 2008;180:2196–2201. doi: 10.1016/j.juro.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Morgan R, Pirard PM, Shears L, Sohal J, Pettengell R, Pandha HS. Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of melanoma. Cancer Res. 2007;67:5806–5813. doi: 10.1158/0008-5472.CAN-06-4231. [DOI] [PubMed] [Google Scholar]
- 35.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, Zimmer Y, Cairncross JG, Janzer RC, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 36.Mellish KJ, Kelland LR, Harrap KR. In vitro platinum drug chemosensitivity of human cervical squamous cell carcinoma cell lines with intrinsic and acquired resistance to cisplatin. Br J Cancer. 1993;68:240–250. doi: 10.1038/bjc.1993.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boubakari, Bracht K, Neumann C, Grunert R, Bednarski PJ. No correlation between GSH levels in human cancer cell lines and the cell growth inhibitory activities of platinum diamine complexes. Arch Pharm (Weinheim) 2004;337:668–671. doi: 10.1002/ardp.200400620. [DOI] [PubMed] [Google Scholar]
- 38.Tanner B, Hengstler JG, Dietrich B, Henrich M, Steinberg P, Weikel W, Meinert R, Kaina B, Oesch F, Knapstein PG. Glutathione, glutathione S-transferase alpha and pi, and aldehyde dehydrogenase content in relationship to drug resistance in ovarian cancer. Gynecol Oncol. 1997;65:54–62. doi: 10.1006/gyno.1996.4593. [DOI] [PubMed] [Google Scholar]
- 39.Goddard P, Valenti M, Kelland LR. The role of glutathione (GSH) in determining sensitivity to platinum drugs in vivo in platinum-sensitive and -resistant murine leukaemia and plasmacytoma and human ovarian carcinoma xenografts. Anticancer Res. 1994;14:1065–1070. [PubMed] [Google Scholar]
- 40.Bracht K, Boubakari, Grunert R, Bednarski PJ. Correlations between the activities of 19 anti-tumor agents and the intracellular glutathione concentrations in a panel of 14 human cancer cell lines: comparisons with the National Cancer Institute data. Anticancer Drugs. 2006;17:41–51. doi: 10.1097/01.cad.0000190280.60005.05. [DOI] [PubMed] [Google Scholar]
- 41.Hamberger J, Liebeke M, Kaiser M, Bracht K, Olszewski U, Zeillinger R, Hamilton G, Braun D, Bednarski PJ. Characterization of chemosensitivity and resistance of human cancer cell lines to platinum(II) versus platinum(IV) anticancer agents. Anticancer Drugs. 2009;20:559–572. doi: 10.1097/CAD.0b013e32832d513b. [DOI] [PubMed] [Google Scholar]
- 42.Sugita M, Geraci M, Gao B, Powell RL, Hirsch FR, Johnson G, Lapadat R, Gabrielson E, Bremnes R, Bunn PA, Franklin WA. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62:3971–3979. [PubMed] [Google Scholar]
- 43.Tajima K, Obata Y, Tamaki H, Yoshida M, Chen YT, Scanlan MJ, Old LJ, Kuwano H, Takahashi T, Takahashi T, Mitsudomi T. Expression of cancer/testis (CT) antigens in lung cancer. Lung Cancer. 2003;42:23–33. doi: 10.1016/S0169-5002(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 44.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.