Abstract
Polymorphic variants of DNA repair genes can increase the carcinogenic potential of exposure to ionizing radiation. Two single nucleotide polymorphisms (SNPs) in Prkdc, the gene encoding the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), have been identified in BALB/c mice and linked to reduced DNA-PKcs activity and mammary cancer susceptibility. We examined three additional mouse strains to better define the roles of the BALB/c Prkdc SNPs (R2140C and M3844V). One is a congenic strain (C.B6) that has the C57BL/6 Prkdc allele on a BALB/c background, and the other is a congenic strain (B6.C) that has the BALB/c variant Prkdc allele on a C57BL/6 background. We also examined the LEWES mouse strain, which possesses only one of the BALB/c Prkdc SNPs (M3844V). Our results demonstrate that both Prkdc SNPs are responsible for deficient DNA-PKcs protein expression, DNA repair and telomere function, while the LEWES SNP affects only DNA-PKcs expression and repair capacity. These studies provide insight into the separation of function between the two BALB/c SNPs as well as direct evidence that SNPs positioned within Prkdc can significantly influence DNA-PKcs function involving DNA repair capacity, telomere end-capping, and potentially cancer susceptibility.
INTRODUCTION
It is now well documented that genes involved in DNA repair pathways can play important roles in cancer predisposition (1). However, mutations in known cancer predisposition genes like BRCA1 and BRCA2 account for only a small fraction of the total breast cancers diagnosed. Evidence suggests that a large portion (20–40%) of the remaining “spontaneous” breast cancers may be due to currently unidentified mutations in DNA repair genes (2). In many cases, the consequences of such alterations do not become apparent until DNA repair is enlisted after damage incurred by an exogenous factor, e.g. exposure to ionizing radiation. In support of this concept, an association between polymorphisms in the non-homologous end-joining (NHEJ) DNA-dependent protein kinase catalytic subunit (DNA-PKcs) gene, Prkdc, and risk of breast cancer was recently reported in the U.S. Radiologic Technologist cohort (3). Decreased DNA-PKcs kinase activity has also been linked to individuals with lung cancer (4). Most recently, a Prkdc mutation was found in a human patient that exhibited a severe combined immune-deficient (SCID) phenotype (5). These and other studies suggest that SNPs in DNA repair genes such as Prkdc have the potential to affect cancer predisposition in the human population.
The relationship between SNPs in Prkdc, DNA-PKcs deficiency and breast carcinogenesis has been examined previously using the BALB/c mouse model. A significant increase in the incidence of both lung and mammary adenocarcinomas has been demonstrated in female BALB/c mice after exposure to 137Cs γ radiation (6). Cytogenetic analysis of mammary epithelial cells revealed significantly elevated frequencies of chromatid-type aberrations several population doublings postirradiation in BALB/c but not C57BL/6 mice (7). Due to the nature of chromatid-type aberrations forming only in the cell cycle of collection, this observation suggested that BALB/c but not C57BL/6 mice have an increased risk of radiation-induced delayed genomic instability. Subsequent investigations examined radiation-induced DNA repair capacity and found a significant deficiency in DSB repair activity and repair time in BALB/c mice compared to other mouse strains (8). Subsequent studies also demonstrated reduced DNA-PKcs protein expression and kinase activity in both SCID and BALB/c mice as compared to C57BL/6 mice while expression of Ku70/Ku80 remained unchanged. Additional analysis led to the discovery of two SNPs located in the coding region of Prkdc that may be responsible for the decrease in DNA-PKcs function observed in BALB/c mice (9). One SNP (R2140C), located in exon 48, resides in a highly conserved region located between a leucine zipper domain and an autophosphorylation cluster, a position that could potentially impact autophosphorylation ability, DNA-protein and/or protein-protein interactions and three-dimensional conformation (10, 11). The other BALB/c SNP (M3844V), located near the C-terminal domain in exon 81, is not well conserved among species. However, due to its location within the kinase domain, it could potentially affect enzymatic activity.
In addition to its function in DNA repair, an unanticipated role for DNA-PKcs in mammalian telomeric end-capping function has been identified (12, 13). Initial studies involving primary cells derived from null DNA-PKcs SCID and DNA-PKcs knockout mice displayed significantly elevated frequencies of telomere-telomere fusion, which is not readily observed in BALB/c mice. The predominant telomere fusion event found in BALB/c mice is radiation-induced telomere-DSB fusion (14). Mechanistic links between uncapped/dysfunctional telomeres in DNA-PKcs-deficient backgrounds, radiation-induced instability and breast cancer susceptibility have recently been provided (15). DNA-PKcs has also been shown to be involved in cell communication via the bystander effect in that it is required to generate a bystander signal but is not necessary to receive one (16).
As discussed above, genetic studies suggest a link between the BALB/c variant of Prkdc and DNA repair, kinase activity and genomic instability (9). To directly examine this relationship, two congenic mouse strains were generated, one having the BALB/c Prkdc allele on a C57BL/6 background (B6.C) and the other having the C57BL/6 Prkdc allele on a BALB/c background (C.B6). This approach eliminates most of the potential problems arising from the various mouse genetic backgrounds, which can complicate interpretation and evaluation of a role for the Prkdc allele. In the present series of studies, we examined DNA-PKcs expression, DSB repair, and telomere function as measures of radiosensitivity in the two congenic mouse strains. Additionally, we examined the LEWES/EiJ mouse, an inbred strain recently derived from wild mice that possesses only one of the BALB/c SNPs, M3844V, located in the kinase domain of the Prkdc allele.
MATERIALS AND METHODS
Cell Irradiation
The isolation and culture of mouse kidney fibroblasts have been described previously (8, 9). Log-phase primary or low-passage kidney fibroblasts were plated 24 h prior to radiation exposure and maintained following specific protocols for each end point as described below. All irradiations were performed at a dose rate of 3.9 Gy/min using a Mark I 137Cs γ irradiator (J. L. Shepherd and Associates).
Mice
Female LEWES/EiJ, C57BL/6ByJ and BALB/cByJ mice were obtained from the Jackson Laboratory and female CB17SCRF-FC.B-Igh-1b/IcrTac-Prkdcscid (SCID) mice were obtained from Taconic. Mice were maintained in specific-pathogen-free conditions at the AAALAC-approved Laboratory Animal Resource facility at Colorado State University and used at 2–3 months of age. All animal work was approved by the Institutional Animal Care and Use Committee at Colorado State University.
Generation of C.B6 and B6.C Congenic Strains by Marker-Assisted “Speed” Congenics
Two novel strains of congenic mice were generated by a combination of conventional and marker-assisted backcrossing (17). For the C.B6-PrkdcB6 congenic strain (C.B6), the common allele of Prkdc (PrkdcB6) was transferred by repeated backcrosses (introgression) onto a BALB/c background. For the B6.C-PrkdcBALB congenic strain (B6.C), the BALB/c allele (PrkdcBALB) was introgressed onto a resistant background strain (C57BL/6). A detailed description of the generation of these congenic strains can be found in the Supplementary Methods and Results.
DNA/RNA/cDNA Isolation and Purification
DNA and RNA were extracted from primary mouse kidney fibroblasts and purified using the DNeasy blood and tissue kit and RNeasy kit from Qiagen. The cDNA was synthesized from RNA using random primers and the iScript cDNA synthesis kit (BioRad). All PCR products were purified using a PCR purification kit or by agarose gel electrophoresis and gel extraction (QIAquick PCR purification kit and QIAquick gel extraction kit, Qiagen).
Genotyping
The PCR restriction fragment length polymorphism (PCR/RFLP) method was used to genotype all mouse strains as described previously (9). The R2140C SNP, located downstream of the leucine zipper, abolishes a BsmB1 site, the M3844V SNP, in the kinase domain, creates a novel Hph1 site. PCR primers were designed to flank the SNP loci and sequences were amplified using Taq polymerase (Invitrogen; Taq DNA polymerase, Recombinant 10342-020). Primer sequences used are: exon 48, PKF-GCCTAAGGTAAGGTGCTGTA and PKR-GCCATGATCCTTAGCAAGTG; exon 81, 81F-ATGTTCTTTGCCATGCAGT and 81R-TTCTTCCCTCCCTTCTCAGTA. The PCR products were digested with BsmB1 or Hph1 and were compared against known size samples by electrophoresis through a 2% or 3% agarose gel.
Sequencing
The entire coding region of the LEWES mouse Prkdc gene was sequenced and compared to the sequences for the C57BL/6 and BALB/c mouse genes obtained from the Ensemble and NCBI databases (http://www.ensembl.org/index.html and http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccoreandid=124517705). A total of 19 PCR primer sets were designed using the Primer3 website (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) (primer sequences in Supplementary Methods). Segments of the gene were amplified using PfuUltra high-fidelity polymerase (PfuUltra High-Fidelity Polymerase, Stratagene) according to the manufacturer’s recommended protocol; amplified products were purified and sequenced.
Reverse Transcriptase Quantitative Real-Time PCR
Quantitative real-time PCR was used to determine Prkdc mRNA levels in C57BL/6, BALB/c and LEWES mice. RNA was isolated and converted into cDNA as described previously. Samples were added in triplicate to 96-well plates containing a SYBR green mix (QPCR SYBR Green Fluorescein mix, Thermo Scientific). The standard real-time protocol was followed using the BioRad Icycler thermal cycler iQ5. Data were then retrieved and analyzed using Icycler-iQ software version 3.1, and relative expression in the three strains was compared. HPRT (primer set: F-GCTGACCTGCTGGATTACAT and R-TTGGGGCTGTACTGCTTAAC) was used for the internal control. Prkdc primers (PRKDC4491F-GCCTGCAGTCTTTGGACCC and PRKDC4564R-TCCTCCAAAACCAAAGGCTAATT) were used for these experiments.
Immunoblotting
Mouse primary kidney fibroblasts were lysed and 60 µg of lysate was loaded onto a 4–15% gradient SDS-PAGE gel (BioRad). Electrophoresis was carried out at 125 V for 1–1.5 h; then samples were transferred onto nitrocellulose membranes (Amersham Biosciences) for 20 h at 4°C. Membranes were incubated in blocking buffer (1× TBST with 5% dry milk) for 1 h at room temperature. Primary antibody for DNA-PKcs (DNA-PKcs Ab-4 cocktail, Neomarkers) was added at a 1:500 dilution and incubated at room temperature for 1 h. Secondary antibody (Amersham: ECL Mouse IgG) was added at a 1:1000 dilution and incubated for 1 h at room temperature. Membranes were washed with 1× TBST and incubated with an ECL detection kit (ECL plus Western Blotting Detection Reagents, Amersham) for 2 min. Detection of signal was done using a STORM 860 Fluor Imager system (Molecular Dynamics), and comparison of protein levels was done using Microsoft Image Quant version 5.1 as described previously (8, 9).
DNA DSB Repair/γ-H2AX Foci
Immunofluorescence was used to detect γ-H2AX foci in mouse primary kidney fibroblasts. Cells were plated in a 4-well chamber slide at an approximate density of 15,000 cells per chamber. Leucine-deprived medium was added to synchronize the cells in the G1 phase of the cell cycle (18). Primary antibody (Upstate) was added at a 1:500 dilution and incubated at room temperature for 1 h. Secondary antibody (Mouse IgG Alexafluor 488, Invitrogen) was added at a 1:200 dilution and incubated for 1 h at room temperature. DAPI was then added to stain nuclei and mount the slide. The γ-H2AX focus images were analyzed and captured using a Zeiss Axioskop2 plus microscope equipped with a Photometrics Coolsnap ES2 camera and Metavue 7.1 software. At least 50 interphase cells were scored, and experiments were repeated to provide a minimum of 100 scored cells per sample.
Chromosome-Orientation Fluorescence In Situ Hybridization (CO-FISH)
After irradiation, cell cultures were incubated for various times, trypsinized and subcultured with 5′-bromo-2′-deoxyuridine (BrdU) for one cell cycle; Colcemid (0.1 µg/ml; Gibco) was added during the final 3–4 h to accumulate mitotic cells, which were collected and processed for telomere CO-FISH as described previously with some modifications (19, 20). Briefly, samples were fixed and then slides were dried and treated with RNase A (100 µg/ml; Sigma, 10 min at 37°C), fixed in 1% formaldehyde (10 min at room temperature), then dehydrated through a cold ethanol series (75%, 85% and 100%). Slides were stained with Hoechst 33258 (0.5 ng/µl; Fisher) for 15 min and exposed to 365 nm UV light (Stratalinker 2400) for 25 min. After UV irradiation, strands that had incorporated BrdU were digested with exonuclease III (2 U/µl in provided reaction buffer; Promega) at room temperature for 10 min. Slides were rinsed and samples denatured in 70% formamide at 75°C for 1 min and 15 s. After an additional ethanol dehydration and drying, a Cy-3-conjugated (TTAGGG)3 PNA telomere probe (0.2 µg/ml; Applied Biosystems) was hybridized to the samples at 37°C for 1.5 h. Slides were rinsed in 70% formamide at 32°C for 10 min and dehydrated in an ethanol series before reprobing at 37°C for 2 h. After the second hybridization, slides were rinsed with 70% formamide at 32°C for 15 min followed by a 5-min rinse in PN Buffer. Chromosomes were counterstained with DAPI (Vectashield with DAPI, Vector Laboratories).
Radiation-Induced Bystander Effect/ SCE
The bystander studies used to detect the generation of a bystander signal and the reception of radiation-induced bystander signals as measured by sister chromatid exchange (SCE) frequencies were performed as described previously (16). Both donor and recipient cells were counted using a Coulter Counter (Beckman Coulter, Fullerton, CA), and 4 × 105 human 5C fibroblasts were plated into T-75 flasks, then co-cultured with a 1:100 dilution of either unirradiated or 1-Gy γ-irradiated exponentially growing donor mouse primary kidney fibroblasts. BrdU was added to cultures at a final concentration of 2 × 1025 M and cells were allowed to grow for two rounds of cell division. Colcemid (Invitrogen) was added at a final concentration of 0.2 µg/ml and cells were harvested approximately 2–3 h later. Cells were trypsinized, centrifuged, resuspended in 0.075 M KCl for 15 min at room temperature, and then fixed with 3:1 methanol:acetic acid. After fluorescence plus Giemsa staining of slides, second-cycle cells with characteristic harlequin staining patterns were scored for SCE; a total of 50 cells (two experiments with 25 metaphases scored per experiment) were counted. Images were analyzed and captured using a Zeiss Axioskop2 Plus microscope equipped with a Photometrics Coolsnap ES2 camera and Metavue 7.1 software.
RESULTS
C.B6-PrkdcB6 (C.B6) and B6.C-PrkdcBALB (B6.C) Mice are Congenic for the Common and BALB Variant Alleles of Prkdc
Using conventional and marker-assisted backcrossing, we generated two congenic mouse strains with minimal donor contamination (17). C.B6 mice have the common Prkdc allele (PrkdcB6) associated with resistance to radiation-induced mammary cancer on a BALB/c background, whereas B6.C mice have the susceptible allele (PrkdcBALB) on a resistant strain background (C57BL/6). (A detailed description of the generation and genotyping of the congenic mouse strains can be found in the Supplementary Methods and Results.)
BALB/c mice also have two SNPs in Cdk2na that are responsible for altered cell cycle control and susceptibility to plasmacytomas (21). Genotyping of the two Cdkn2a SNPs by SNP resequencing confirmed that each congenic strain contained its appropriate background Cdkn2a allele (data not shown).
Genome-wide SNP genotyping revealed minimal donor haplotype contamination in the congenic strains C.B6 and B6.C (see Supplementary Fig. 1A and B). Therefore, phenotypic differences between the congenic strains and their corresponding background strains were interpreted to be a consequence of the introgressed Prkdc allele.
LEWES Mouse Strain Possesses the BALB/c SNP in the Kinase Domain
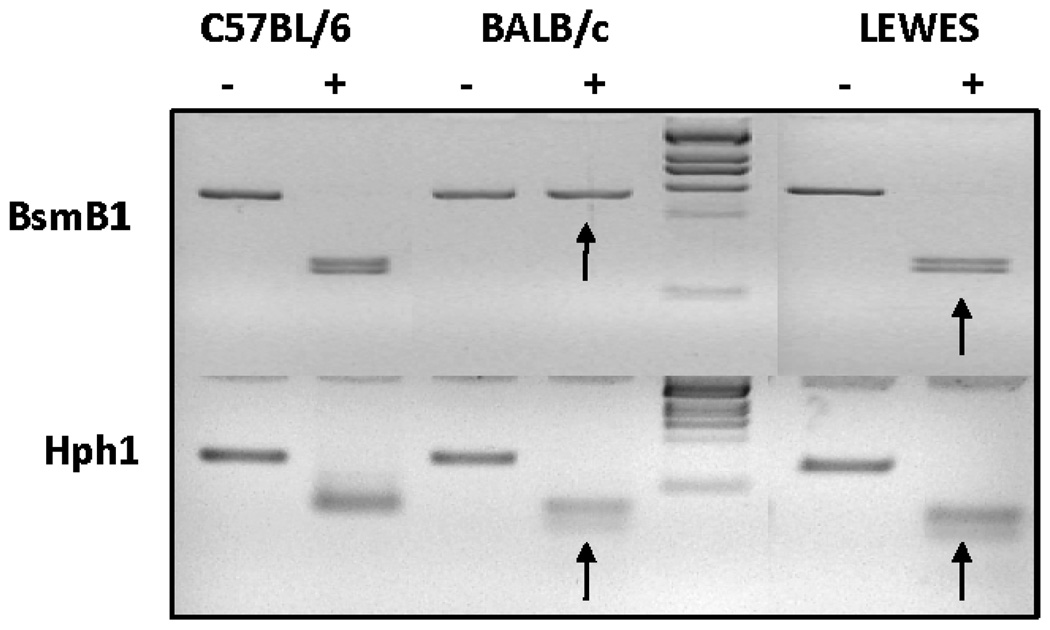
Genotyping and sequencing were performed on the LEWES mouse strain to ensure the presence of the kinase SNP and the absence of other additional changes in Prkdc that could potentially affect DNA-PKcs function. Primary mouse kidney fibroblasts were isolated from C57BL/6ByJ, BALB/cByJ and LEWES/EiJ female mice. PCR products were purified and digested with either BsmB1 or Hph1 (for the R2140C or M3844V SNP, respectively). The first BALB/c SNP abolishes a BsmB1 site; consequently, amplified DNA from C57BL/6 mice was digested but amplified DNA from BALB/c mice was not (Fig. 1). Digestion of amplified LEWES mouse DNA with BsmB1 yielded fragments similar to those seen in C57BL/6 mouse DNA. The second SNP in BALB/c mouse Prkdc creates a novel Hph1 site that yields an additional fragment after digestion. As expected, two fragments were generated by Hph1 digestion of the C57BL/6-generated amplicon, whereas the BALB/c and LEWES mouse amplicons yielded three fragments when digested (Fig. 1). These results confirmed the presence of the BALB/c SNP in the kinase domain and the common nucleotide at the conserved locus downstream of the leucine zipper in LEWES mice. Further confirmation of these results was obtained by sequencing PCR products derived from all three strains.
FIG. 1.
Genotyping of the LEWES strain. The R2140C SNP located downstream of the leucine zipper abolishes a BsmB1 site while the M3844V SNP in the kinase domain creates a novel Hph1 site. PCR products from both SNP loci were purified and digested with BsmB1 or Hph1. Both C57BL/6 and LEWES mouse products were cleaved with BsmB1, while the BALB/c mouse product containing R2140C was not (arrow). After Hph1 digestion only one cut occurred in the C57BL/6 mouse products, but BALB/c and LEWES mouse products containing the M3844V SNP yielded an extra digested fragment (arrows).
The entire coding region of the LEWES mouse Prkdc was sequenced to ensure that there were no additional sequence variations that could affect function. Nineteen PCR primer sets were designed to amplify overlapping portions of the Prkdc cDNA. PCR products were purified, sequenced and compared to the C57BL/6 Prkdc cDNA sequence. Our data showed no differences in Prkdc sequence between LEWES and C57BL/6 mice with the exception of the SNP in the kinase domain (M3844V) (data not shown).
DNA-PKcs Protein Expression in the Congenic and LEWES Mouse Strains
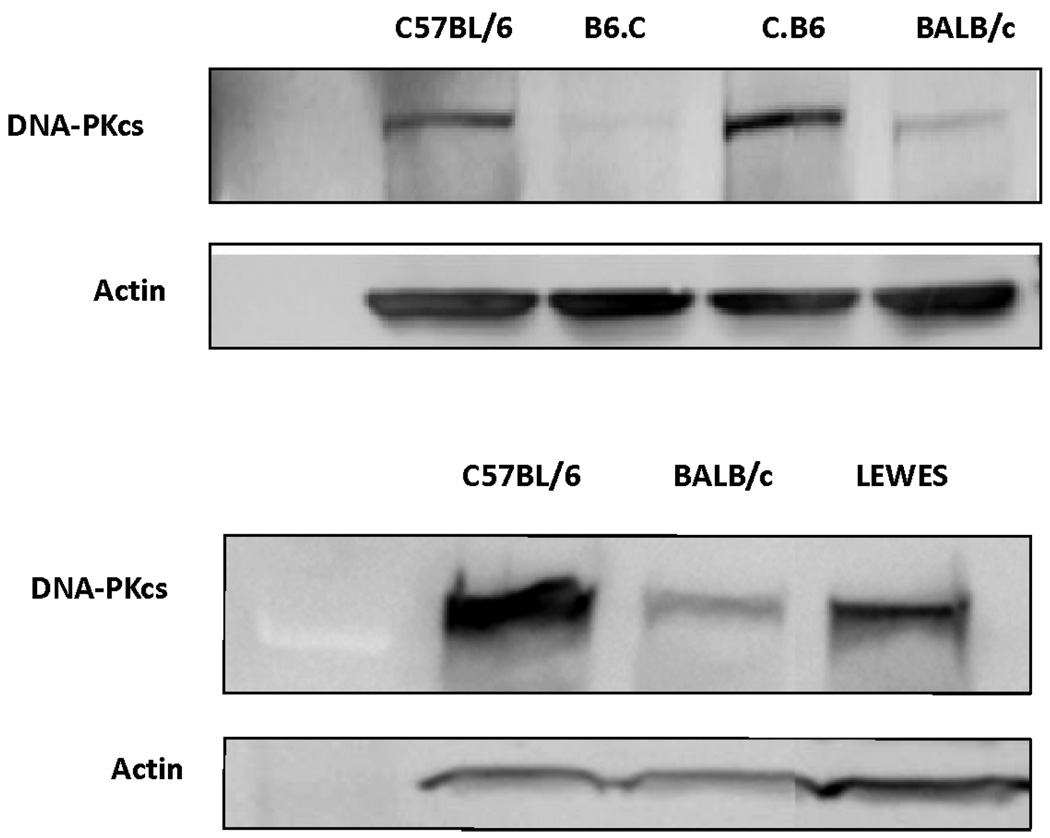
DNA-PKcs protein levels are significantly lower in BALB/c mice than in C57BL/6 mice in several tissues, including the mammary gland (8). In all mouse strains, expression of DNA-PKcs was lowest in mammary tissue compared with other tissues, with expression in the BALB/c mouse mammary gland being lower still. In the present study, we compared protein levels in the congenic and LEWES strains to the C57BL/6 and BALB/c strains to directly determine the role of the BALB/c SNPs on expression of DNA-PKcs. Because expression of DNA-PKcs is significantly lower in mammary tissue than in other tissues, we used primary kidney fibroblasts to facilitate the strain comparisons. The results clearly demonstrated that expression of DNA-PKcs was significantly reduced in mice having the BALB/c variant of Prkdc, independent of the genetic background. DNA-PKcs protein levels in C.B6 mice were similar to those in C57BL/6 mice, while DNA-PKcs levels in the B6.C strain were similar to those in the BALB/c strain (Fig. 2). Protein expression in LEWES mice was intermediate between that in C57BL/6 and BALB/c mice, suggesting that both SNPs play a role in expression. Consistent with previous observations, quantitative real-time PCR showed no difference in mRNA levels between the various strains, suggesting that post-translational events may be responsible for the reduced DNA-PKcs protein levels (8).
FIG. 2.
DNA-PKcs protein expression as a function of genotype. The C.B6 congenic strain has DNA-PKcs expression similar to the C57BL/6 strain while the B6.C strain has decreased amounts of DNA-PKcs, comparable to the BALB/c strain (top panel). The DNA-PKcs protein expression of the LEWES strain is intermediate between those of the C57BL/6 and BALB/c strains (bottom panel).
DNA DSB Repair in the Congenic and LEWES Mouse Strains
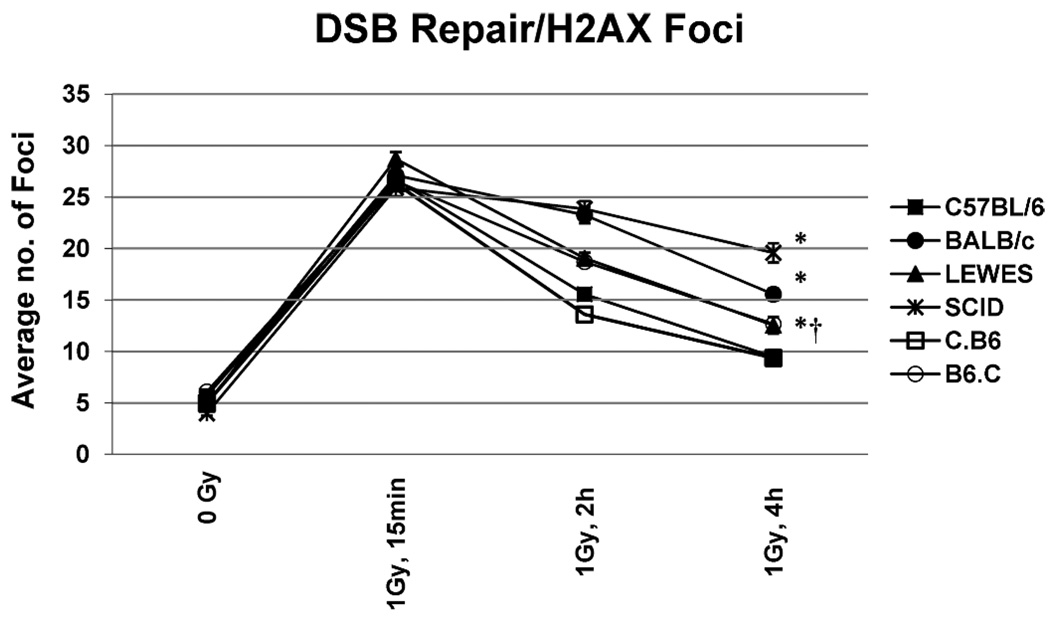
Previous work using the fraction of activity released (FAR) assay demonstrated that BALB/c mice are deficient in DSB repair after exposure to low-LET γ radiation compared to other mouse strains (8) and suggested that this deficiency was correlated with the BALB/c variant form of Prkdc. To directly examine the impact of the BALB/c polymorphisms, we evaluated DSB repair kinetics in the parental, congenic and LEWES mouse strains with the FAR and the γ-H2AX assays. The γ-H2AX assay involves detection of large tracks of the phosphorylated histone variant H2AX around DSB sites that dissipate once repair has occurred. Therefore, counting the number of phosphorylated H2AX (γ-H2AX) foci over time provides a measure of initial DSB induction and the kinetics of their repair (22). For the γ-H2AX studies, primary kidney fibroblasts from C57BL/6, BALB/c, C.B6, B6.C and LEWES mice were used. SCID mice possessing a truncation mutation in DNA-PKcs were used as a positive control. Cells were irradiated with 1 Gy of γ radiation, and the frequency of γ-H2AX foci was determined at 15 min, 2 h and 4 h postirradiation. Because previous studies indicated that most DSB repair occurs by 4 h, we did not include times beyond 4 h. All strains had similar background levels of γ-H2AX foci and showed similar increases at the 15 min, indicating that there was no difference in the frequency of DSBs initially induced in these strains (Fig. 3). By comparing the number of foci at 15 min and 4 h postirradiation, we determined that 82% of DSBs were repaired in C57BL/6 mice, 79% in C.B6 mice, 56% in LEWES mice, 58% in B6.C mice, 48% in BALB/c mice, and only 29% in SCID mice over this period. Clearly, the introgression of the common allele of Prkdc (C.B6) restored the DNA repair capacity to that observed in the C57BL/6 mice. Conversely, introgression of the BALB/c variant onto the C57BL/6 background (B6.C) reduced DSB repair capacity to a level near that of BALB/c mice. Repair in the LEWES strain, which harbors the M3844V SNP in the kinase region, was similar to that observed in B6.C mice, suggesting that reduced DNA-PKcs expression and DSB repair in these strains are largely a result of the SNP in the kinase region of Prkdc. Similar trends were observed using the FAR assay (see Supplementary Methods and Results). Clonogenic survival studies (see Supplementary Methods and Results) provided further support for these conclusions, in that similar levels of survival were observed in C57B/6 and C.B6 mice, while the surviving fraction as a function of dose was significantly lower in BALB/c and B6.C mice containing the variant allele.
FIG. 3.
DSB repair γ-H2AX assay as a function of genotype. As shown previously (8), the C57BL/6 strain is efficient in DNA repair, with SCID mice being substantially deficient in repair and the BALB/c mouse showing an intermediate level of repair. The C.B6 strain containing the common allele of Prkdc was comparable to the C57BL/6 strain, while in the B6.C strain containing the BALB variant allele and the LEWES strain containing only the M3844V SNP, the level of repair was significantly reduced compared to the C57BL/6 strain (*P < 0.0001 for C57BL/6, BALB/c and SCID; †P < 0.005 for LEWES). Bars are means ± SEM.
Telomeric End-Capping Function and the Prkdc SNPs
Cytogenetic analysis was performed using Chromosome Orientation-Fluorescence In Situ Hybridization (CO-FISH) to determine the impact of DNA-PKcs variants/SNPs on telomeric end-capping function and accurate DSB repair via NHEJ. Uncapped telomeres can be recognized as unrepaired DSB ends and so are prone to fusion with radiation-induced DSBs (15). The strand-specific nature of CO-FISH allows detection of telomere-DSB fusions, which are operationally observed as single-sided telomere signals along the length of the chromosome (one interstitial telomere signal), as opposed to telomere-telomere fusions, which appear as CO-FISH telomere signals on both chromatids (two opposing interstitial telomere signals).
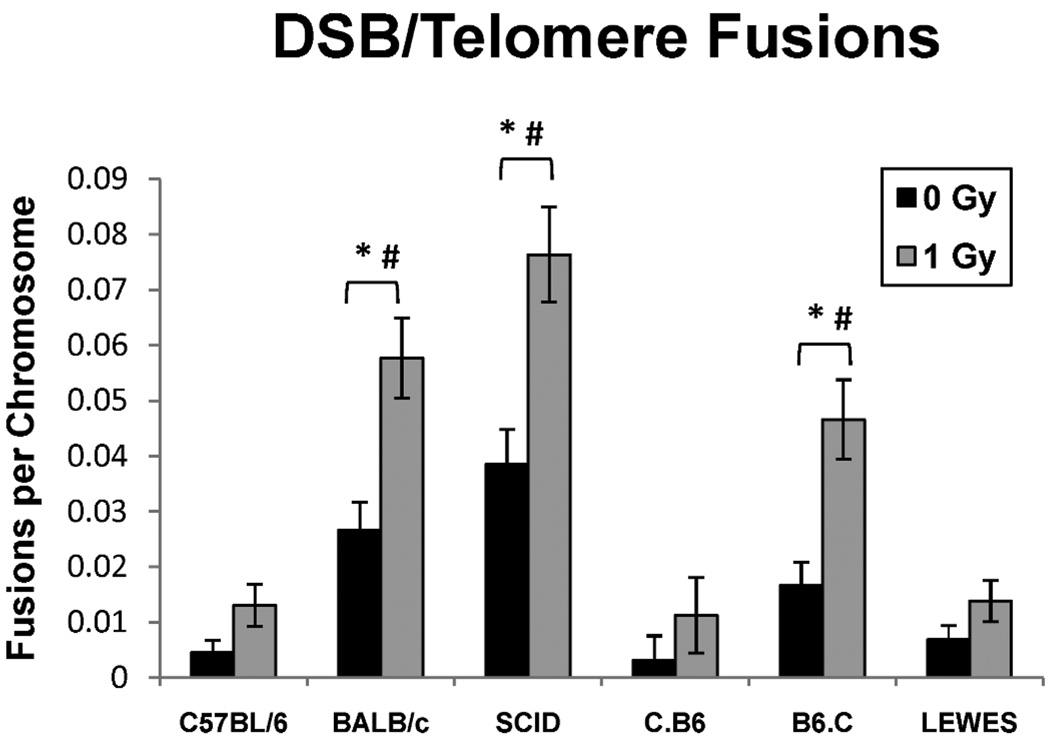
The C57BL/6 mouse strain served as the negative control for telomere-DSB fusion analysis. No significant difference was observed (P > 0.05) in the telomere-DSB fusion frequencies in the LEWES, C.B6 and C57BL/6 control mice in either irradiated or nonirradiated populations. In contrast, telomere-DSB fusion frequencies in both BALB/c and B6.C mice differed significantly from those in both irradiated and nonirradiated populations of C57/BL6 mice (P < 0.05) (Fig. 4). Furthermore, BALB/c and B6.C mice showed no significant difference from one another in telomere-DSB fusion frequencies in either irradiated or nonirradiated populations. These data demonstrate that the variant DNA-PKcs allele containing both SNPs (the leucine zipper and kinase domain), as exists in BALB/c and B6.C mice, results in extensive telomere uncapping and telomere-based fusion events. The frequency of telomere-DSB fusions in the LEWES mouse cells (containing only the SNP within the PIKK-like kinase domain, M3844V) resembles that of the C57/BL6 and C.B6 mouse cells. Taken together, these data suggest that the SNP located downstreamof the leucine zipper near the autophosphorylation cluster (R2140C) is critical for proper telomere end-capping and M3844V SNP is required for optimal function of DNA-PKcs in accurate DSB repair via NHEJ.
FIG. 4.
Frequency of telomere-DSB fusions in the 6 mouse strains. The C.B6 and LEWES strains showed low levels of telomere/DSB fusions after exposure to 1 Gy γ rays, similar to C57BL/6, and consistent with wild-type DNA-PKcs. The SCID, BALB/c and B6.C strains had significantly higher levels of telomere/DSB fusions after irradiation (*P < 0.0005, #P < 0.0001, 0 Gy compared to 1 Gy), consistent with mutant DNA-PKcs.
LEWES Allele of Prkdc Generates a Radiation-Induced Bystander Effect
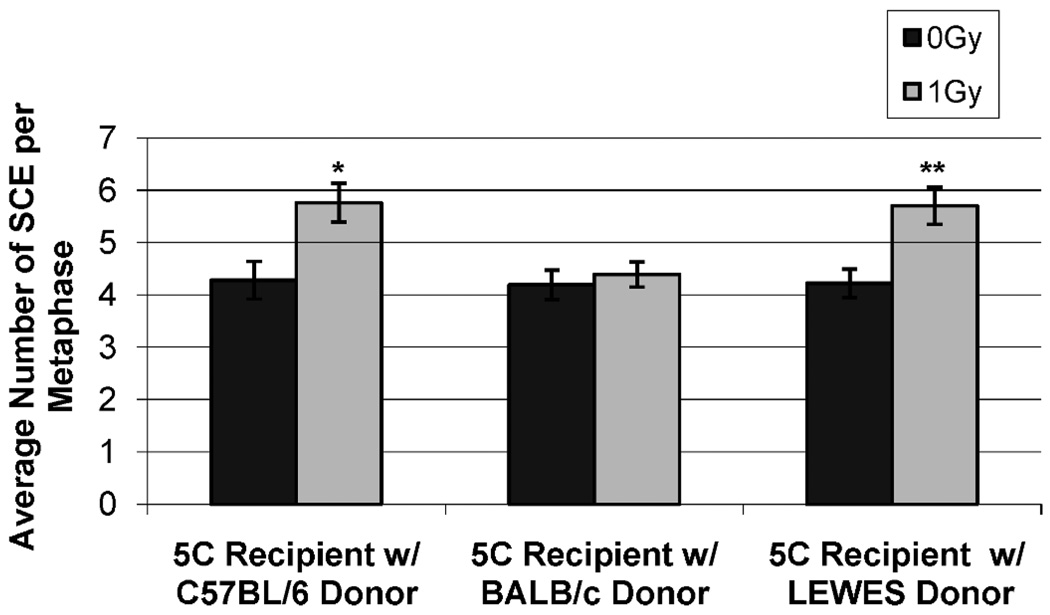
We have shown previously that there are differences in C57BL/6, BALB/c and SCID mice (also DNA-PKcs deficient) and in the congenic strains regarding the bystander effect (16). In these earlier studies, irradiated C57BL/6 and C.B6 mouse cells generated a bystander response measured by increased sister chromatid exchanges (SCE) in nonirradiated human 5C human dermal fibroblast (HDF) cells. Conversely, cells from BALB/c, SCID and B6.C mice were not capable of generating such a bystander response. Since data from the present studies for DNA-PKcs expression and DSB repair suggested that the SNP in the kinase region was largely responsible for reduced expression and repair, we next determined whether cells from the LEWES strain containing only the SNP in the kinase region could produce a bystander response. As demonstrated previously, there was a significant elevation in SCE frequency in human 5C cells co-cultured with cells from C57BL/6 mice, but no increase was observed in cells co-cultured with BALB/c mouse cells. A clear bystander effect was seen in the human recipient cells co-cultured with irradiated donor LEWES cells (5.70 SCE/metaphase) compared to co-cultures of 5C cells with unirradiated control or BALB/c cells (4.22 and 4.19 SCE/metaphase, respectively) (Fig. 5). These results provide additional support for our conclusion that the ability to generate a radiation-induced bystander effect is dependent upon DNA-PKcs. However, because LEWES cells were fully capable of generating a bystander response, the present data reveal a role for the R2140C, but not the M3844V, SNP in the generation of such a response.
FIG. 5.
The bystander effect in the LEWES strain. Both the C57BL/6 and LEWES strains (wild-type DNA-PKcs) were capable of generating a bystander effect as determined by an increased frequency of SCE in recipient cells, but the BALB/c strain (mutant DNA-PKcs) was not (0 Gy compared to 1 Gy *P < 0.0030; 1 Gy C57BL/6 compared to BALB/c, SCID and B6.C **P < 0.0001).
DISCUSSION
It has been well documented that defects in DNA damage response proteins like p53, ATM or BRCA1/2 strongly correlate with increased risk for the development of a number of different cancer types. However, such highly penetrant mutations can account for only a small percentage of total cancer cases. Research now suggests that more subtle mutations or polymorphisms may play a significant role in carcinogenesis and potentially could account for a larger number of “spontaneous” breast cancers (23). Several epidemiological studies have reported associations between polymorphisms in DNA repair genes and increased cancer risk (24–26). Three different SNPs in the Prkdc gene have been shown to be associated with elevated breast cancer incidences in the U.S. Radiologic Technologist cohort (3). Most recently, a SCID mutation in Prkdc, which produced a hypomorphic version of DNA-PKcs, was observed in a human cancer patient for the first time (5). While these associations are suggestive, clear mechanistic links cannot be established from such studies. SNPs in Prkdc can also be observed in mice, making mice attractive models for studying the impact of SNPs on cancer predisposition. The purpose of the current studies was to directly examine the impact of such SNPs on the function of the NHEJ protein DNA-PKcs.
The BALB/c mouse strain is susceptible to spontaneous and radiation-induced lung and mammary adenocarcinomas (27). This strain also possesses a hypomorphic variant of DNA-PKcs that has been linked to two SNPs (designated R2140C and M3844V) in the coding sequence of the Prkdc gene (9) that appear, based on genetic studies using F1 hybrids, to be associated with reduced expression of DNA-PKcs, reduced repair capacity, altered telomere function, and sensitivity to radiation-induced genomic instability (9, 15). While these genetic studies strongly support the role of Prkdc polymorphisms in such phenotypes, evidence that is more direct is required to serve as the basis for understanding the molecular mechanisms involved and the potential impacts on cancer susceptibility. Here, we directly analyzed the impact of the BALB/c mouse Prkdc SNPs on a variety of end points using two unique congenic strains: C.B6 (BALB/c background with the C57BL/6 Prkdc wild-type allele) and B6.C (C57BL/6 background with the BALB/c Prkdc variant allele). The LEWES mouse strain, which only has the M3844V SNP in the kinase domain, was also characterized using identical end points to provide insight into the potential impact of this polymorphism on DNA-PKcs function.
The congenic and LEWES mouse strains were characterized with respect to DNA-PKcs protein levels, Prkdc mRNA expression, DNA DSB repair kinetics, and telomere function to further elucidate the impact of the Prkdc SNPs to radiosensitivity phenotypes in vivo. The C.B6 congenic strain was comparable to C57BL/6 with respect to all of these end points (wild-type Prkdc), while the phenotypes of B6.C mice were similar to those of BALB/c mice (variant Prkdc). Our results provide the first direct evidence that these particular polymorphisms are mechanistically responsible, necessary and sufficient for the reduced DSB repair capacity and altered telomere function observed in BALB/c mice. Data from the studies using the congenic strains also suggested that the genetic background plays only a minor role with respect to DNA-PKcs expression and functional activity. Although the DSB repair capacity of LEWES mouse cells was slightly better than that BALB/c mouse cells (approximately 40% vs. 30% reduction), we found reduced DNA-PKcs expression levels and repair capacity compared to C57BL/6 mouse cells, suggesting an important role for M3844V in these functional end points. LEWES telomere function was similar to that of C57BL/6 and C.B6 cells, which have the common Prkdc allele. While genetic background cannot be completely excluded as a contributing factor to the phenotypic differences we observed between the two Prkdc SNPs, the results for the congenic strains suggest that genetic background plays only a minor role in modifying the effects of these polymorphisms on the DNA-PKcs function associated with either DSB repair or telomere function. The potential separation of these phenotypes offers an important opportunity to dissect the role of DNA-PKcs in telomere function and its potential link to cancer susceptibility. Clearly, additional studies are warranted.
Because of its location in the kinase domain, it is easy to envision how the M3844V SNP might affect DNA-PKcs functions, particularly those associated with DNA repair. Previous studies using a pull-down assay have found that both expression and kinase activity of DNA-PKcs are lower (8). However, it remains unclear whether this SNP affects the specific activity of the enzyme or whether kinase activity is lower simply because of the lower levels of DNA-PKcs expression, or both. The stability of the BALB/c mouse variant of DNA-PKcs is also important when considering potential mechanisms. However, studies designed to quantify specific kinase activity and to determine the half-life of the variant form of DNA-PKcs using standard biochemical approaches have been problematic because of the extremely low levels of expression. Regardless, whether this SNP affects the specific activity of the protein or is simply a result of reduced expression, it is clear that kinase activity is reduced. Numerous investigations have demonstrated the important role of DNA-PKcs phosphorylation of various substrates, including DNA-PKcs itself (autophosphorylation), in the processes involved in DNA repair [reviewed in (28)]. Additionally, we have previously shown that DNA-PKcs autophosphorylation at the Thr-2609 cluster is a critical event for proper telomeric end-capping function and for prevention of ligase IV-mediated telomere fusion (15).
Here, we shed light on separating DNA-PKcs function in end joining (DSB repair) vs. end capping (protecting telomeres). Our results suggest that telomeric end capping requires the DNA-PKcs leucine zipper DNA binding domain (R2140C), which we speculate may be important for the conformational changes required for site-specific DNA-PKcs autophosphorylation. Previous studies in our laboratory have suggested that some level of DNA-PKcs kinase activity is required for this occurrence at telomeres (29). It has recently been proposed that DNA-PKcs acts at telomeres to block backup pathways of NHEJ (30); our results suggest that this protective function at telomeres may well require the DNA-PKcs leucine zipper DNA binding domain.
In summary, we have directly demonstrated that the Prkdc SNPs (R2140C and M3844V) contained in the BALB/c mouse strain (PrkdcBALB) are both necessary and sufficient to result in decreased expression of DNA-PKcs, deficient repair capacity, and dysfunctional telomeres. Further, the data suggest separation of function between the two PrkdcBALB SNPs, in that M3844V (located in the kinase domain) has its primary effect on protein expression and repair capacity of DNA-PKcs, while R2140C (located near leucine zipper DNA binding domain) is required for normal telomere end-capping function.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge support from the NIH (CA09236; CA043322) and NASA (NNX09AM08G; NNJ04HD83G).
Footnotes
SUPPLEMENTARY INFORMATION
Supplementary methods, results and figure legends. http://dx.doi.org/10.1667/RR2431.1.S1
Supplementary Figs. 1–3. http://dx.doi.org/10.1667/RR2431.1.S2
REFERENCES
- 1.Madhusudan S, Middleton MR. The emerging role of DNA repair proteins as predictive, prognostic and therapeutic targets in cancer. Cancer Treat. Rev. 2005;31:603–617. doi: 10.1016/j.ctrv.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Scott D. Chromosomal radiosensitivity and low penetrance predisposition to cancer. Cytogenet. Genome Res. 2004;104:365–370. doi: 10.1159/000077517. [DOI] [PubMed] [Google Scholar]
- 3.Bhatti P, Struewing JP, Alexander BH, Hauptmann M, Bowen L, Mateus-Pereira LH, Pineda MA, Simon SL, Weinstock RM, Sigurdson AJ. Polymorphisms in DNA repair genes, ionizing radiation exposure and risk of breast cancer in U.S. radiologic technologists. Int. J. Cancer. 2008;122:177–182. doi: 10.1002/ijc.23066. [DOI] [PubMed] [Google Scholar]
- 4.Auckley DH, Crowell RE, Heaphy ER, Stidley CA, Lechner JF, Gilliland FD, Belinsky SA. Reduced DNA-dependent protein kinase activity is associated with lung cancer. Carcinogenesis. 2001;22:723–727. doi: 10.1093/carcin/22.5.723. [DOI] [PubMed] [Google Scholar]
- 5.van der Burg M, van Dongen JJ, van Gent DC. DNA-PKcs deficiency in human: long predicted, finally found. Curr. Opin. Allergy Clin. Immunol. 2009;9:503–509. doi: 10.1097/ACI.0b013e3283327e41. [DOI] [PubMed] [Google Scholar]
- 6.Ullrich RL. Tumor induction in BALB/c female mice after fission neutron or gamma irradiation. Radiat. Res. 1983;93:506–515. [PubMed] [Google Scholar]
- 7.Ponnaiya B, Cornforth MN, Ullrich RL. Radiation-induced chromosomal instability in BALB/c and C57BL/6 mice: the difference is as clear as black and white. Radiat. Res. 1997;147:121–125. [PubMed] [Google Scholar]
- 8.Okayasu R, Suetomi K, Yu Y, Silver A, Bedford JS, Cox R, Ullrich RL. A deficiency in DNA repair and DNA-PKcs expression in the radiosensitive BALB/c mouse. Cancer Res. 2000;60:4342–4345. [PubMed] [Google Scholar]
- 9.Yu Y, Okayasu R, Weil MM, Silver A, McCarthy M, Zabriskie R, Long S, Cox R, Ullrich RL. Elevated breast cancer risk in irradiated BALB/c mice associates with unique functional polymorphism of the Prkdc (DNA-dependent protein kinase catalytic subunit) gene. Cancer Res. 2001;61:1820–1824. [PubMed] [Google Scholar]
- 10.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, Tainer JA. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 14.Bailey SM, Cornforth MN, Ullrich RL, Goodwin EH. Dysfunctional mammalian telomeres join with DNA double-strand breaks. DNA Repair (Amst.) 2004;3:349–357. doi: 10.1016/j.dnarep.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Williams ES, Klingler R, Ponnaiya B, Hardt T, Schrock E, Lees-Miller SP, Meek K, Ullrich RL, Bailey SM. Telomere dysfunction and DNA-PKcs deficiency: characterization and consequence. Cancer Res. 2009;69:2100–2107. doi: 10.1158/0008-5472.CAN-08-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagelstrom RT, Askin KF, Williams AJ, Ramaiah L, Desaintes C, Goodwin EH, Ullrich RL, Bailey SM. DNA-PKcs and ATM influence generation of ionizing radiation-induced bystander signals. Oncogene. 2008;27:6761–6769. doi: 10.1038/onc.2008.276. [DOI] [PubMed] [Google Scholar]
- 17.Weil MM, Brown BW, Serachitopol DM. Genotype selection to rapidly breed congenic strains. Genetics. 1997;146:1061–1069. doi: 10.1093/genetics/146.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato TA, Nagasawa H, Weil MM, Little JB, Bedford JS. Levels of gamma-H2AX foci after low-dose-rate irradiation reveal a DNA DSB rejoining defect in cells from human ATM heterozygotes in two at families and in another apparently normal individual. Radiat. Res. 2006;166:443–453. doi: 10.1667/RR3604.1. [DOI] [PubMed] [Google Scholar]
- 19.Bailey SM, Goodwin EH, Cornforth MN. Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet. Genome Res. 2004;107:14–17. doi: 10.1159/000079565. [DOI] [PubMed] [Google Scholar]
- 20.Bailey SM, Williams ES, Cornforth MN, Goodwin EH. Chromosome orientation fluorescence in situ hybridization or strand-specific FISH. Methods Mol. Biol. 659:173–183. doi: 10.1007/978-1-60761-789-1_12. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Ramsay ES, Mock BA. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc. Natl. Acad. Sci. USA. 1998;95:2429–2434. doi: 10.1073/pnas.95.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato TA, Nagasawa H, Weil MM, Genik PC, Little JB, Bedford JS. γ-H2AX foci after low-dose-rate irradiation reveal Atm haploinsufficiency in mice. Radiat. Res. 2006;166:47–54. doi: 10.1667/RR3587.1. [DOI] [PubMed] [Google Scholar]
- 23.Tempfer CB, Hefler LA, Schneeberger C, Huber JC. How valid is single nucleotide polymorphism (SNP) diagnosis for the individual risk assessment of breast cancer? Gynecol. Endocrinol. 2006;22:155–159. doi: 10.1080/09513590600629175. [DOI] [PubMed] [Google Scholar]
- 24.Mandal RK, Gangwar R, Mandhani A, Mittal RD. DNA repair gene x-ray repair cross-complementing group 1 and xeroderma pigmentosum group D polymorphisms and risk of prostate cancer: a study from north India. DNA Cell Biol. 2010;29:183–190. doi: 10.1089/dna.2009.0956. [DOI] [PubMed] [Google Scholar]
- 25.Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Suzuki T, Kato T, Hirai T, Kanemitsu Y, Tajima K. A gene-gene interaction between ALDH2 Glu487Lys and ADH2 His47Arg polymorphisms regarding the risk of colorectal cancer in Japan. Carcinogenesis. 2006;27:1018–1023. doi: 10.1093/carcin/bgi282. [DOI] [PubMed] [Google Scholar]
- 26.Conde J, Silva SN, Azevedo AP, Teixeira V, Pina JE, Rueff J, Gaspar JF. Association of common variants in mismatch repair genes and breast cancer susceptibility: a multigene study. BMC Cancer. 2009;9:344. doi: 10.1186/1471-2407-9-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullrich RL. Cellular and molecular changes in mammary epithelial cells following irradiation. Radiat. Res. 1991;128 Suppl:S136–S140. [PubMed] [Google Scholar]
- 28.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey SM, Brenneman MA, Halbrook J, Nickoloff JA, Ullrich RL, Goodwin EH. The kinase activity of DNA-PK is required to protect mammalian telomeres. DNA Repair (Amst.) 2004;3:225–233. doi: 10.1016/j.dnarep.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, Salles B, Calsou P. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 2010;29:1573–1584. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.