SUMMARY
HIV infection is characterized by gradual immune system collapse and hematopoietic dysfunction. We recently showed that HIV enters multipotent hematopoietic progenitor cells and establishes both active cytotoxic and latent infections that can be reactivated by myeloid differentiation. However, whether these multipotent progenitors include long-lived hematopoietic stem cells (HSCs) that could establish viral reservoirs for the life of the infected person remains unknown. Here we provide direct evidence that HIV targets long-lived HSCs and show that infected HSCs yield stable, multilineage engraftment in a xenograft model. Furthermore, we establish that the capacity to use the chemokine receptor CXCR4 for entry determines whether a virus will enter multipotent versus differentiated progenitor cells. Because HSCs live for the lifespan of the infected person and are crucial for hematopoietic health, these data may explain the poor prognosis associated with CXCR4-tropic HIV infection and suggest HSCs as long-lived cellular reservoirs of latent HIV.
INTRODUCTION
The natural course of HIV disease is characterized by progressive destruction of the host immune system, manifested as a decline in CD4+ T cell counts over several years. Depletion of CD4+ T cells invariably causes an immunocompromised state in the host, leading to the onset of AIDS and ultimately death from opportunistic infections. Despite extensive study, the exact mechanisms triggering the progression to AIDS remain unclear.
HIV entry into permissive cells is mediated by interactions of the HIV envelope (Env) protein with CD4 and a chemokine coreceptor (CCR5 or CXCR4 (Alkhatib et al., 1996; Deng et al., 1996; Dragic et al., 1996; Feng et al., 1996)). Initial transmission is mediated primarily by CCR5-utilizing (R5-tropic) HIV (Lathey et al., 1999; van't Wout et al., 1994) and R5-tropic isolates are more commonly detected early in disease (reviewed in (Margolis and Shattock, 2006)), but eventually, X4-tropic isolates predominate in most infected individuals (Richman and Bozzette, 1994; Shankarappa et al., 1999). The conversion of HIV Env from R5-tropic to X4-tropic requires only a small number of changes in the Env V3 region. This conversion has been associated with more rapid disease progression manifested as reduced CD4+ T cell counts and a poor clinical prognosis (Connor et al., 1997; Daar et al., 2007; Karlsson et al., 1994; Scarlatti et al., 1997; Schuitemaker et al., 1992; Shepherd et al., 2008; Waters et al., 2008; Weiser et al., 2008; Yu et al., 1998; Zhou et al., 2008). Furthermore, in the rare instances when infection is initiated by dual (R5X4) or X4-tropic HIV, CD4 counts decline rapidly and disease progression is sometimes accelerated (Sheppard et al., 2002; Yu et al., 1998). It is not clear whether the conversion to CXCR4-tropic virus plays a causal role in disease progression or whether other factors account for this association.
CD4+ T cells, myeloid cells and subsets of hematopoietic stem and progenitor cells (HSPCs) express HIV receptors (CD4 (Morrison and Weissman, 1994) and CCR5 or CXCR4 (Carter et al., 2010; Ishii et al., 1999; Majka et al., 1999; Peled et al., 1999; Shen et al., 1999; Viardot et al., 1998)), but whether HSPCs can be infected has been controversial in the literature (Folks et al., 1988; Redd et al., 2007; Shen et al., 1999; Stanley et al., 1992; Zhang et al., 2007) and there is evidence that these cells may be relatively resistant to infection (Shen et al., 1999; Zhang et al., 2007). Recent reports indicate that low-level infection of multi-potent HSPCs occurs in vivo and in vitro (Carter et al., 2010; Redd et al., 2007) but active infection is cytotoxic and hard to detect in long term culture (Carter et al., 2010). Importantly, the assays used in these studies could not distinguish whether infected cells were long-lived hematopoietic stem cells (HSCs) or short lived common myeloid progenitor cells. Thus, it is still unknown whether HIV infects HSCs, a subset of HSPCs defined by their ability to stably engraft and generate multiple lineages upon transplantation into immunocompromised mice. The distinction between HSCs and other multipotent hematopoietic progenitor cells (HPCs) is of key importance, as infection of the long-lived HSC population would have a greater impact on hematopoiesis and this population would have greater potential to serve as a long-term reservoir of HIV in infected people.
In this study, we provide evidence that HIV Envs can target HSCs and that integration can occur within these cells. Moreover, we show that HIV Env tropism influences which subset(s) of HSPCs are infected: only CXCR4-tropic envelopes permit entry into multipotent HSPCs, including HSCs. These findings suggest not only that HSCs can become infected by HIV and thus have the potential to serve as a long-term reservoir of virus, but also that the association between the emergence of CXCR4-tropic isolates and declining CD4+ T cell counts could be related to infection of multipotent HSPCs.
RESULTS
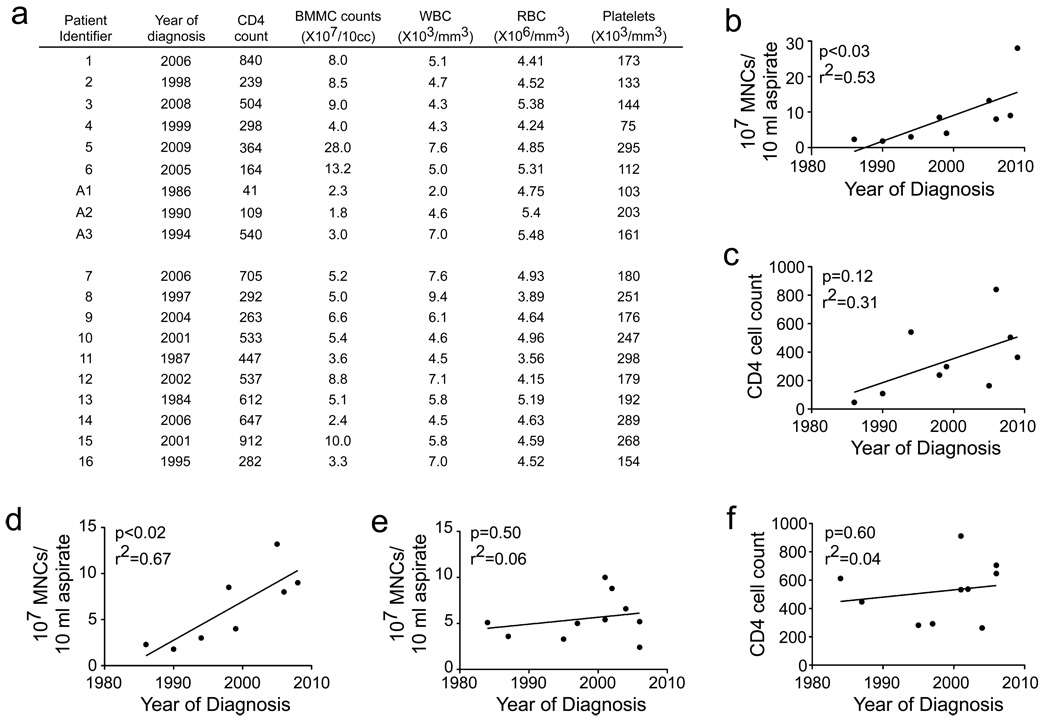
Recent work has indicated that bone marrow CD34+ HSPCs from HIV+ donors are targets of HIV infection in vivo (Carter et al., 2010). In this study, three of six donors with high viral loads had evidence of active HIV infection of bone marrow CD34+ cells. In the remaining three donors, active infection could be induced by culturing the cells in GMCSF and TNFα (Carter et al., 2010). However, CD34+ cells are a heterogeneous population and it is not known whether stem cells or multipotent progenitor cells are infected in HIV+ people. Healthy stem cells and multipotent progenitor cells are needed to maintain all hematopoietic lineages as well as normal bone marrow cellularity. Thus, it is expected that infection of primitive HSPCs by HIV would eventually be reflected in a loss of total bone marrow mononuclear cells. To examine this, we quantified the bone marrow cellularity of high viral load (>50,000 copies/ml) donors (Carter et al., 2010) who had relatively normal complete white blood cell counts (Figure 1a). Interestingly, we found a striking correlation between the number of mononuclear cells isolated from 10 ml of aspirate and the year of diagnosis (Figure 1b). This correlation was more significant in our cohort (p<0.03) than the correlation between CD4 cell count and year of diagnosis (p=0.12, Figure 1c) and was significant even when an outlier with a very high cell count was excluded (p<0.02, Figure 1d). A second cohort of patients with undetectable (<48 copies/ml) viral loads on highly active antiviral therapy (HAART) was also studied (Figure 1a). In this group, HIV genomes could be detected within CD34+ cells from 40% of donors but Gag expression was only detectable after culturing the cells in GMCSF and TNFα, consistent with latent infection of this cell type (Carter et al., 2010). In this group, we observed no correlation between bone marrow cellularity and year of diagnosis (Figure 1e) or between CD4 cell count and year of diagnosis (Figure 1f). These data provide in vivo support for a potent effect of HIV on the bone marrow that requires active viral replication. While this effect may be due to the chronic inflammation associated with unsuppressed HIV replication, it is also consistent with direct infection of multipotent HSPCs by HIV.
Figure 1. Total number of bone marrow mononuclear cells is correlated with year of diagnosis.
(a) Complete blood count of HIV+ donors with high viral loads (1–6 and A1–A3, 61,000 to 202,000 copies/ml) and low viral loads (7–16, <48 copies/ml). (b) The number of purified bone marrow cells versus year of diagnosis for high viral load donors. (c) CD4 counts of high viral load donors at the time of bone marrow aspiration plotted as a function of year of diagnosis. (d) As (b), but with an outlier with a high mononuclear cell yield (Donor 5) removed. (e) The number of purified bone marrow cells versus year of diagnosis for low viral load donors. (f) CD4 counts of low viral load donors at the time of bone marrow aspiration plotted as a function of year of diagnosis.
X4-tropic Envs infect multipotent HSPCs
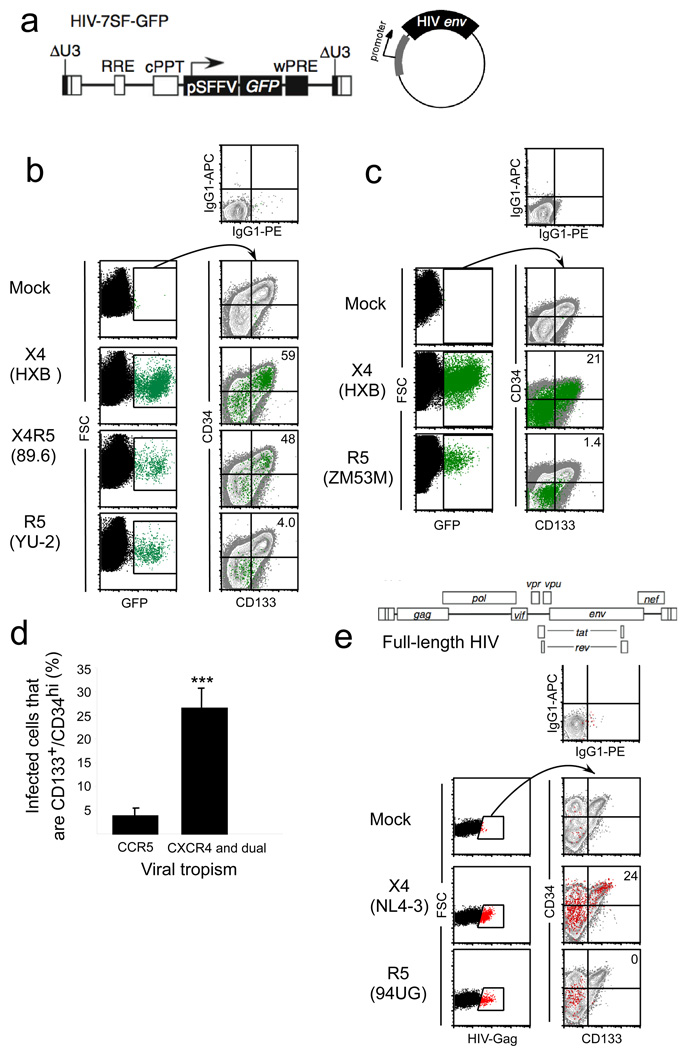
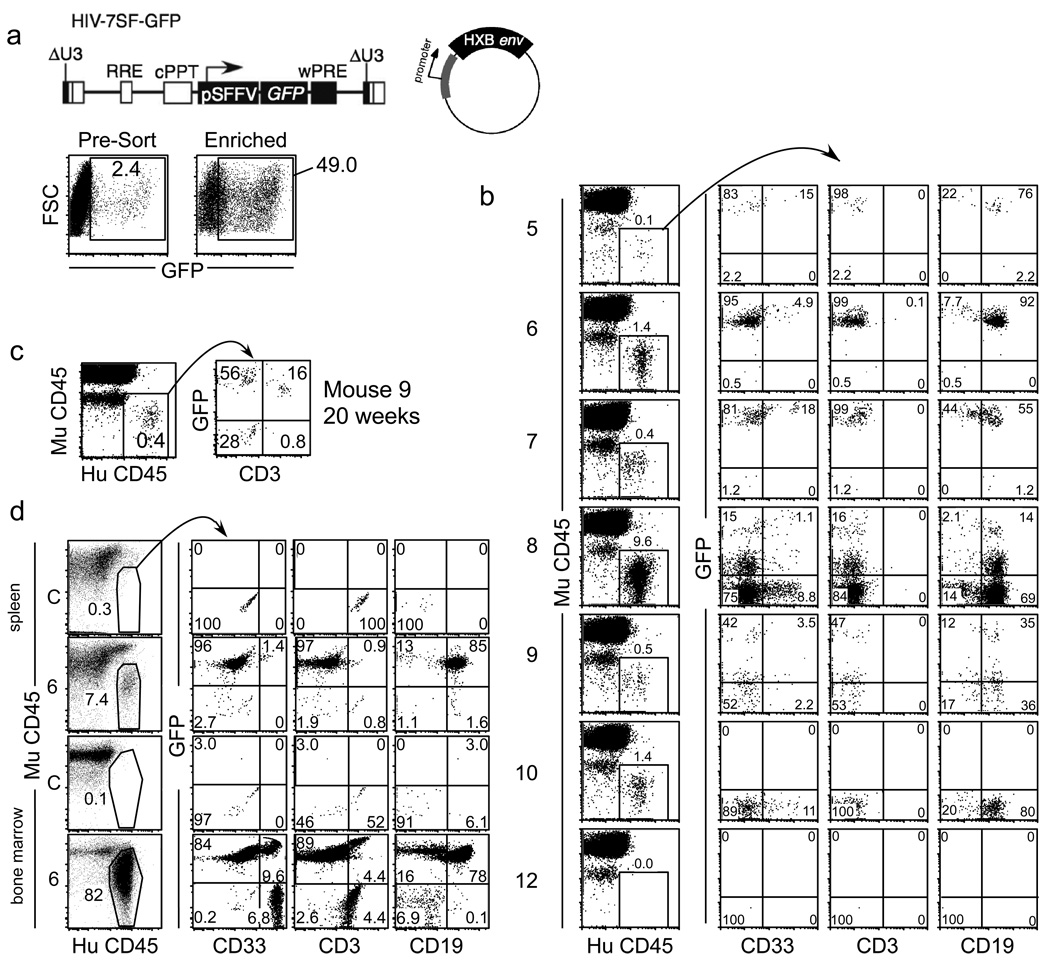
The conversion of HIV Env from R5-tropic to X4-tropic is associated with more rapid disease progression manifested as reduced CD4+ T cell counts and a poor clinical prognosis (Connor et al., 1997; Daar et al., 2007; Karlsson et al., 1994; Scarlatti et al., 1997; Schuitemaker et al., 1992; Shepherd et al., 2008; Waters et al., 2008; Weiser et al., 2008; Yu et al., 1998; Zhou et al., 2008). To determine whether chemokine receptor expression influences the type of HSPC infected by HIV, we tested a panel of HIVs differing only in the Env they contained. These virus particles were generated by co-transfecting a GFP-expressing minimal HIV construct (HIV-7SF-GFP, Figure 2a (Yam et al., 2002)), which expresses GFP but no HIV proteins, along with a packaging-null/env-null HIV genome and an HIV env-expressing plasmid (Carter et al., 2010). The resulting virus particles thus contain unmodified HIV proteins, including Env, integrase, and reverse transcriptase. Once the HIV genome is integrated into the genome of the target cell, however, new HIV proteins are not transcribed and instead GFP is expressed from the constitutively active SFFV promoter. These viral supernatants were used to infect HSPCs isolated by magnetic sorting. After infection, the cell surface phenotype of GFP-positive cells was determined by flow cytometry.
Figure 2. CXCR4-tropic HIV Envs infect CD133+, CD34+ HSPCs.
(a) Schematic of HIV-7SF-GFP construct and generic HIV envelope plasmid used to construct viruses used in b–d. (b and c) Flow cytometric analysis of cord-blood derived CD133+ cells infected with a minimal HIV (HIV-7SF-GFP (Yam et al., 2002)) that lacked expression of HIV gene products and was pseudotyped with the indicated HIV Env. In this construct, GFP is expressed from a heterologous promoter (Yam et al., 2002). Cells were analyzed three days post-infection. The left panels show the GFP+ gating. These events were overlaid in green on the right plots that also show staining of the total cell population in grey. Isotype control staining is shown in the top panel. The percentage of GFP+ cells that were CD34HighCD133+ is shown in the upper right hand corner (d), Summary plot of HIV-1 Env data. Results are compiled from ten experiments with CCR5-tropic Env proteins (94UG114.1.6, 8 replicates; YU-2, one experiment; ZM53M.PB12, one experiment) and sixteen experiments with CXCR4 and dual tropic Env proteins (HXB2, 8 replicates; 89.6, 7 replicates; 92HT593, 1 experiment); error bars are standard error of the mean. ***p<0.0001. (e) Upper panel, schematic of full-length HIV used in lower panel. Lower panel, flow cytometric analysis of cord-blood derived CD133+ cells three days post infection with full length, wild type HIVs. NL4-3 has an X4-tropic Env and 94UG114.1.6 has an R5-tropic Env. Gag+ cells were gated on in the left panels, and these events (red dots) were overlaid on CD34 vs. CD133 plots in the right panels to determine the immunophenotype of the infected cells compared with the total cell population (grey dots). Isotype control staining is shown in the top panel. The percentage of Gag+ cells that were CD34+CD133+ is shown in the upper right hand corner. See also Figure S1.
Remarkably, we found that both the X4-tropic Env HXB and the R5X4-tropic Env 89.6 were able to target various cell types, including cells with a surface phenotype consistent with multipotent HSPCs (CD34HighCD133+) (Figure 2b, 2c). Infection occurred both when the cells were infected by spin infection and when cells were pulsed with virus for 2h at 37°C (Figure S1). In contrast, the R5 tropic Envs YU-2 (Figure 2b, lower panel) and ZM53M.PB12 (Figure 2c, lower panel) infected CD34HighCD133+ cells inefficiently; most infection was seen in cells with a surface phenotype consistent with less primitive cells (CD34LowCD133−). Similar results were observed in ten replicate experiments using three different CCR5-tropic Envs and sixteen replicates using three different CXCR4- or dual-tropic Envs (summarized in Fig 2d).
We performed additional experiments using full length HIVs encoding CXCR4-tropic Env (NL4-3) or CCR5-tropic Env (94UG114.1.6), except that in this case we detected infection by expression of intracellular Gag. As shown in Figure 2e, we again found that CXCR4-tropic NL4-3 was able to infect cells with a surface phenotype consistent with HSPCs (CD34HighCD133+). In contrast, CCR5-tropic 94UG114.1.6 only infected cells with a surface phenotype consistent with less primitive cells (CD34LowCD133−) (Figure 2e). Collectively, these findings indicate that wild type X4- and R5-tropic HIVs target distinct subsets of HSPCs and that X4-tropic viruses have an increased capacity to target cells with surface markers characteristic of multipotent HSPCs.
X4-tropic HIVs infect multipotent HSPC
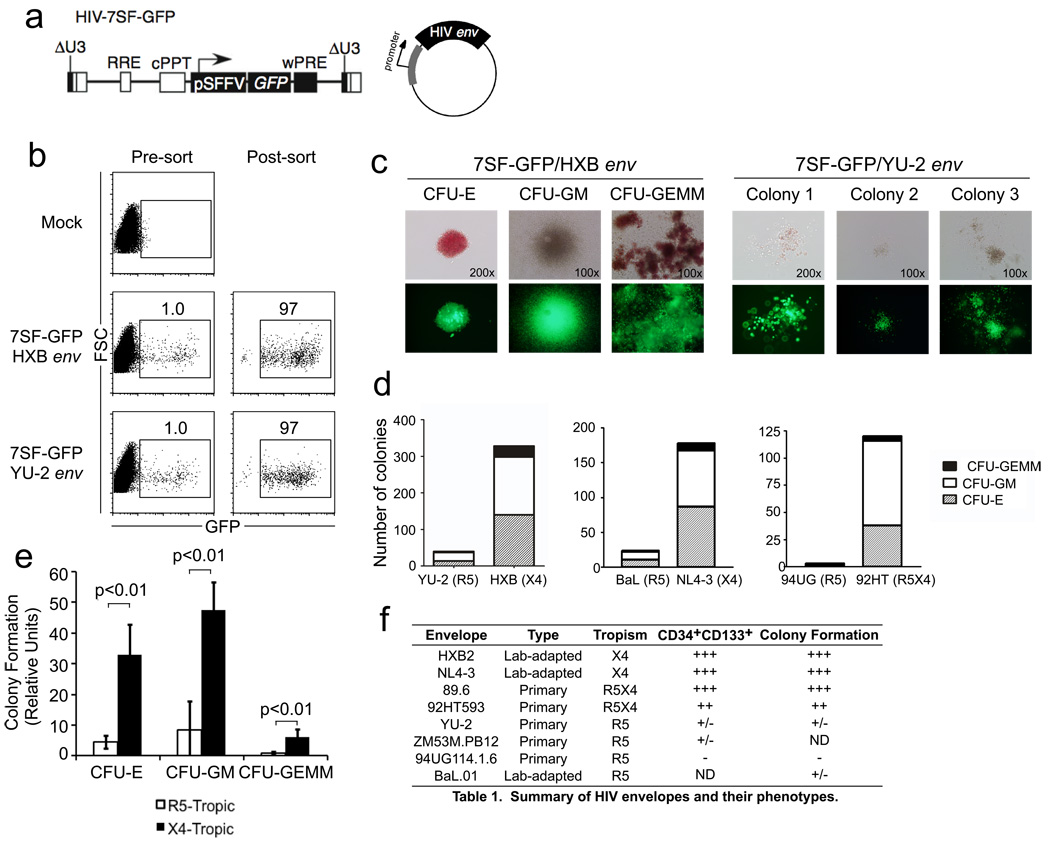
To test whether X4-tropic HIVs were capable of infecting multipotent HSPCs, we asked whether the targeted cells formed multilineage colonies in culture. For these experiments, cord-blood derived HSPCs were infected with the minimal HIV construct HIV-7SF-GFP pseudotyped with HXB Env (X4-tropic) or YU-2 Env (R5-tropic) (Figure 3a). The use of this construct was critical for these experiments because wild type HIV kills actively infected HSPCs within a few days (Carter et al., 2010), making it difficult to determine the developmental capacity of the targeted cell type by colony formation as this assay takes weeks.
Figure 3. CXCR4-tropic HIV Envs infect HSPCs with the capacity to form multilineage colonies.
(a) Schematic of HIV-7SF-GFP construct and generic HIV envelope plasmid used to construct viruses used in b–e. (b) Flow cytometric analysis of cord-blood derived CD133+ cells infected with HIV-7SF-GFP pseudotyped with HXB (X4-tropic) or YU2 (R5-tropic) Env proteins, and purified by flow sorting. (c) Example colonies identified after culturing cells isolated as shown in part (b) for 14 days. Phase contrast and epifluorescence microscopy are shown (erythroid, CFU-E; myeloid, CFU-GM; or multilineage, CFU-GEMM). (d) Quantification of colony formation for the experiment shown in part (c) and for two similar experiments using other HIV Envs as indicated (erythroid, CFU-E; myeloid, CFU-GM; or multilineage, GFU-GEMM). The total number of colonies from 6 replicate wells is displayed. (e) Summary of colony formation results. Data were compiled from five independent experiments using six different Env proteins. The average normalized number of colonies observed with CXCR4- or dual-tropic Env versus CCR5-tropic Env is depicted. Error bars represent standard deviation and p-values were determined using the two-tailed Student’s T test (erythroid, CFU-E; myeloid, CFU-GM; or multilineage, GFU-GEMM). (f) Summary table of the ability of HIV-1 Envs of different tropism to infection CD133+CD34High and multipotent HSPCs.
Three days after infection with HIV-7SF-GFP, the GFP+ cells were purified (>95% pure, Figure 3b), and 6000 GFP+ cells from each infection were plated in methylcellulose medium. After two weeks, colonies were analyzed for morphology and GFP expression. We found 140 erythroid (CFU-E), 158 myeloid (CFU-GM), and 30 multilineage (CFU-GEMM) colonies generated from cells infected with HXB enveloped virus (Figure 3c and d). CFU-GEMM colonies form only from hematopoietic stem cells (HSCs), multipotent progenitor cells and common myeloid progenitors, indicating that the HXB envelope permitted entry into one of these immature HSPC types. In contrast, HSPCs infected with YU-2 enveloped virus gave rise to about ten-fold fewer colonies, primarily small granulocyte/macrophage colonies (Total of 14 CFU-E, 23 CFU-GM, and 3 CFU-GEMM; Figure 3c and d).
To determine whether the capacity to infect multipotent HSPCs was consistently associated with chemokine receptor use, we analyzed additional Envs (Figure 3d). In each case, we found X4-tropism or dual-tropism (NL4-3 and 92HT593) associated with infection of cells able to generate multilineage colonies. In contrast, cells infected using R5-tropic Env (BaL and 94UG114.1.6) largely lacked this capacity; we observed only 2 CFU-GEMM colonies generated from cells infected with HIV-7SF-GFP pseudotyped with BaL Env and none from cells infected using 94UG114.1.6 Env (Figure 3d–f). These findings demonstrate that X4-tropic Envs have the capacity to infect multipotent HSPCs, whereas R5-tropic Envs primarily infect more mature HSPCs.
CD4 and CXCR4 receptor use is required for infection of primitive HSPCs
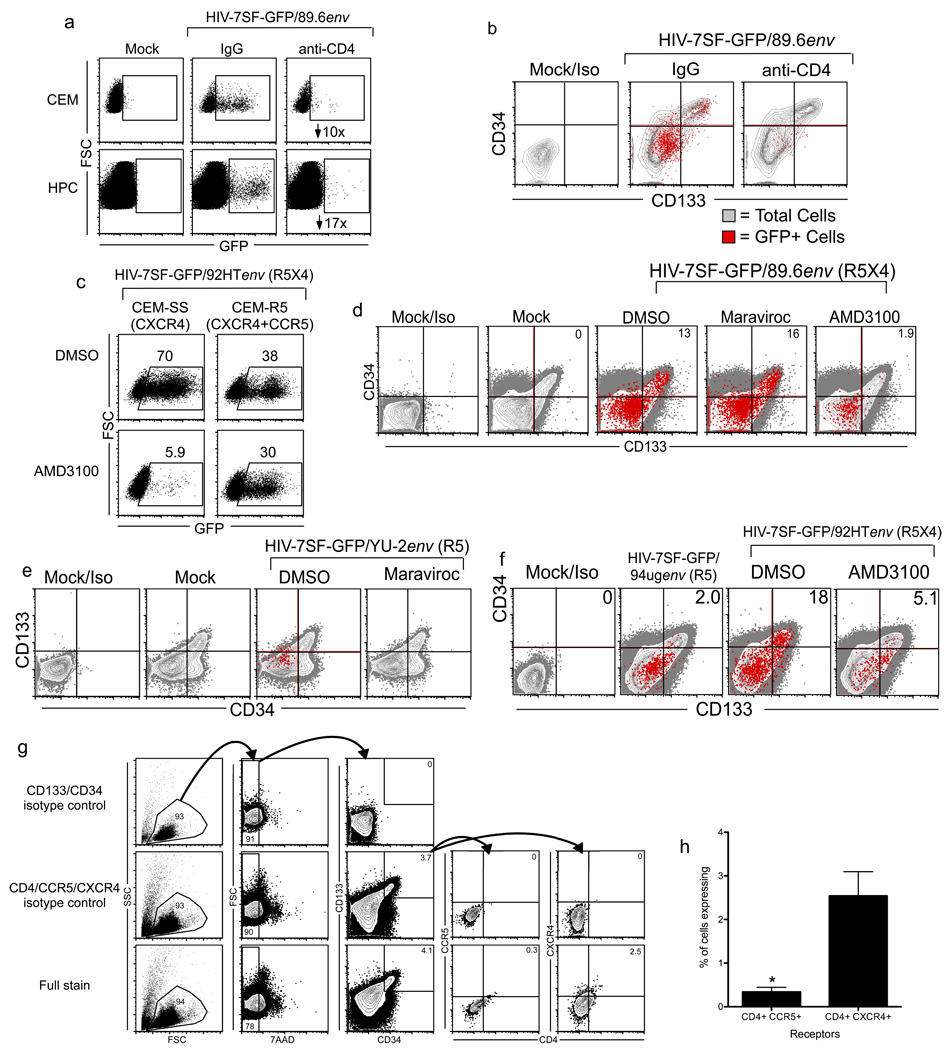
We hypothesized that infection of primitive HSPCs with X4-tropic Envs occurred by the canonical mechanism, wherein HIV Env triggers membrane fusion after binding CD4 and CXCR4 on the target cell. While this is the most common scenario, numerous reports have documented HIV infection by CD4-independent mechanisms, usually involving the use of CXCR4 alone to facilitate entry (Endres et al., 1996; Hoxie et al., 1998; Liu et al., 2004; Saha et al., 2005; Zerhouni et al., 2004). To explore this possibility, we treated cord-blood derived HSPCs and CEM T cells with the CD4-blocking antibody L3T4 before infecting the cells with HIV-7SF-GFP pseudotyped with the dual tropic 89.6 Env. As expected, pre-treatment with CD4-blocking antibody substantially reduced infection of CEM T cells (Figure 4a). Pretreatment of HSPCs with CD4-blocking antibody inhibited infection even more robustly, causing a near-complete block in infection (Figure 4a). These data demonstrate that infection of HSPCs is strictly dependent on CD4. Moreover, CD4 antibody treatment blocked infection of all cells tested, those with a surface phenotype consistent with primitive HSPCs (CD34HighCD133+) as well as more mature progenitors, suggesting that infection of all HSPCs is CD4-dependent (Figure 4b).
Figure 4. Infection of CD34HighCD133+ HSPCs is dependent on CD4 and CXCR4.
(a) Flow cytometric analysis of cord blood-derived CD133+ HSPCs or CEM T cells pre-incubated with anti-CD4 antibody (L3T4, 20µg/mL) or control antibody and then infected with HIV-7SF-GFP pseudotyped with 89.6 Env. The cells were analyzed three days post-infection. The numbers in the right panels are fold inhibition of infection. (b) Flow cytometric analysis of cord blood-derived CD133+ HSPCs pre-incubated with control antibody or antibody to CD4 (L3T4, 20µg/mL) and then infected with HIV-7SF-GFP pseudotyped with 89.6 Env. The cells were analyzed three days after infection. GFP+ (red) cells are overlaid on the total population (grey). (c) Flow cytometric analysis of CEM-SS (CXCR4-expressing) or CEM-R5 (CXCR4 and CCR5 expressing) cells treated with 10µg/mL AMD3100 or an equal volume of DMSO and then infected with HIV-7SF-GFP pseudotyped with the dual tropic HIV Env 92HT593. The cells were analyzed three days post-infection. The percent GFP+ cells are shown in the numbers above each gate. Results are representative of two independent experiments. (d–f) Flow cytometric analysis of cord-blood derived CD133+ cells infected with HIV-7SF-GFP pseudotyped with the dual-tropic 89.6 (d) or 92HT (f) HIV Envs or the R5-tropic YU-2 HIV env (e) in the presence and absence of 20µM maraviroc (R5-blocking) or 10µg/mL AMD3100 (X4-blocking). The cells were analyzed three days after infection. GFP+ cells (red) are overlaid on the total population (grey). (g) Flow cytometric analysis of CD133+ UCB cells expanded 4–7 days in STIF media and then stained for the indicated cell surface markers. Samples stained with isotype control antibodies for CD133 and CD34 (top) or with antibodies to CD133 and CD34 but isotype control antibodies for CD4, CXCR4, and CCR5 (middle) were included to determine gating. Numbers indicate the percent of cells falling within each gate. (h) Summary graph showing percent of total CD133+CD34High cells that express both of the indicated receptors for three independent experiments; mean and standard deviation are shown. *p<0.03, paired t test. See also Figure S2.
As described above, infection of multipotent HSPCs by various HIVs correlates with their capacity to use CXCR4 for entry (Figure 3). These data likely reflect the expression pattern of chemokine receptors on multipotent HSPCs (i.e. that multipotent HSPCs express CXCR4 more widely than CCR5) (Carter et al., 2010). A less likely possibility was that some multipotent HSPCs express both chemokine receptors but that the X4-tropic Env is required for another function other than entry, or that signaling through CXCR4 is required for productive infection. To distinguish these possibilities, we asked whether blockade of CXCR4 would reduce infection of multipotent HSPCs by dual-tropic HIV Envs. We tested this approach using two related T cell lines: CEM-SS, which expresses only CXCR4, and CEM-R5, which expresses both CXCR4 and CCR5. These cells were treated with the small molecule CXCR4 antagonist AMD3100 (Donzella et al., 1998) and infected with HIV-7SF-GFP pseudotyped with the dual-tropic Env 92HT593. As expected, AMD3100 blocked infection of CEM-SS cells almost completely but only partially blocked infection of CEM-R5 cells, demonstrating use of CCR5 by 92HT593 Env in these cells (Figure 4c).
We then treated cord blood-derived HSPCs with AMD3100 or maraviroc (CCR5-blocking) and infected the cells with HIV-7SF-GFP pseudotyped with 89.6 Env. We found that not only did AMD3100 reduce the rate of infection in total HSPCs, it nearly eliminated infection of CD34High/CD133+ cells (Figure 4d). By contrast, maraviroc did not impede infection of CD34High/CD133+ cells by 89.6-pseudotyped HIV-7SF-GFP (Figure 4d) but did eliminate infection of HSPCs when we used the R5-tropic Env YU-2 (Figure 4e). We further observed that when we infected HSPCs with HIV-7SF-GFP pseudotyped with 92HT593 Env, treatment with AMD3100 reduced overall infection rates in CD34HighCD133+ cells to rates comparable to those observed with the R5-tropic 94UG Env (Figure 4f). Consistent with these results, we observed that CD34HighCD133+ HSPCs express CXCR4 at higher levels than CCR5 (Figure 4g and h). We confirmed the low levels of CCR5 on these cells functionally with a calcium flux assay to assess response to CXCR4 and CCR5 ligands (Figure S2a and b) and with an acid wash to confirm that bound ligand was not masking CCR5 on these cells (Figure S2c). Finally, to assess whether crosslinking of CXCR4 could permit entry by CCR5-tropic virus, we infected CD133+ HSPCs with HIV-7SF-GFP pseudotyped with YU2 Env in the presence or absence of full-length NL4-3. We then examined GFP expression in the infected populations to determine which cell types were infected by the R5-tropic virus and found that NL4-3 did not permit the CCR5-tropic virus to enter CD133+ cells (Figure S2d).
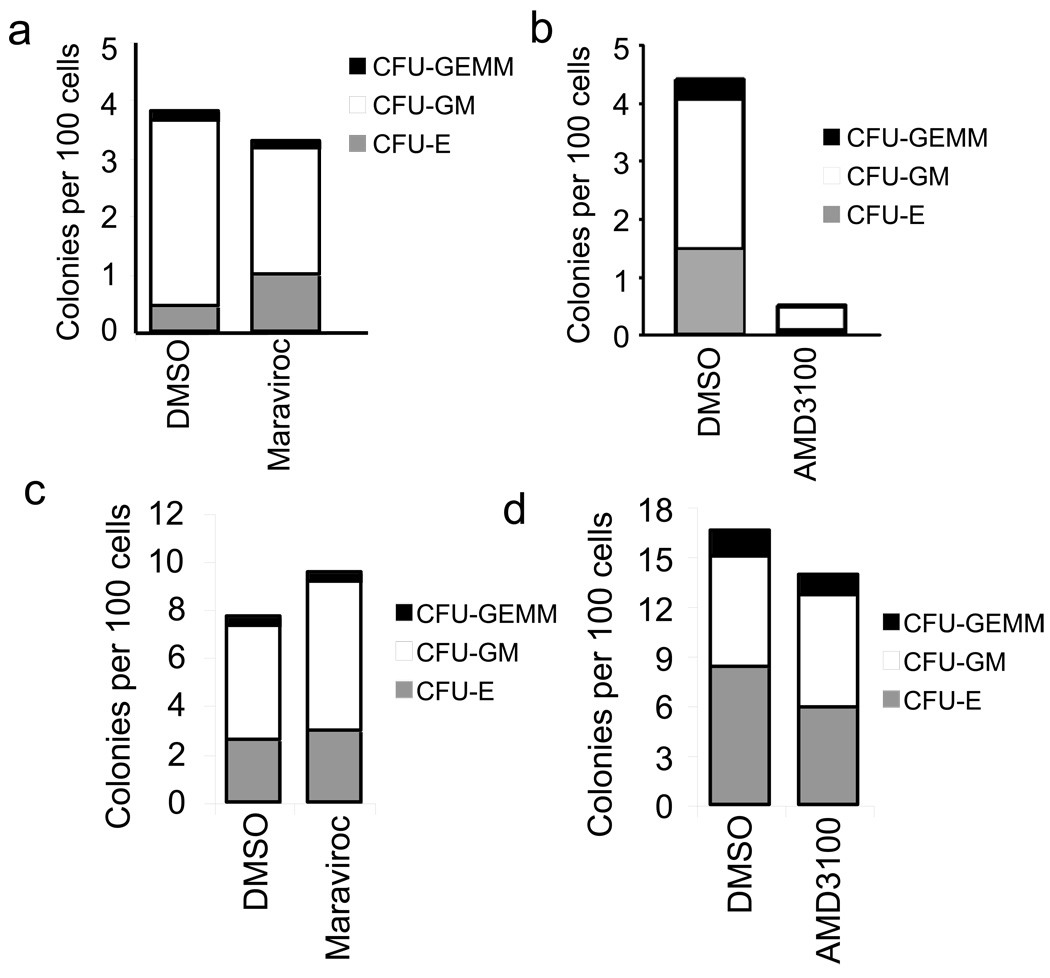
Next, we treated cord-blood derived HSPCs with AMD3100 or maraviroc and infected with HIV-7SF-GFP pseudotyped with dual-tropic 89.6 Env. We sorted GFP+ cells from AMD3100-treated, maraviroc-treated, and control-treated cultures and plated the cells in methylcellulose medium. Infected control-treated and maraviroc-treated cells formed numerous erythroid, myeloid and multilineage colonies (Figure 5a), whereas infection of AMD3100-treated colony-forming cells was dramatically reduced (Figure 5b). Treatment with maraviroc (Figure 5c) or AMD3100 (Figure 5d) had no effect on colony formation by uninfected cells. These data show that CXCR4 usage is necessary for the infection of multipotent HSPCs by HIV and that CCR5 cannot substitute.
Figure 5. Infection of multipotent cells requires CXCR4.
(a and b) Colony formation by cord blood-derived HSPCs treated with 20µM maraviroc (a) 10µg/mL AMD3100 (b) or an equal volume of DMSO, infected with HIV-7SF-GFP pseudotyped with the dual tropic HIV Env 89.6 and purified by FACS. Two blinded counters analyzed the colonies 14 to 18 days after plating in methylcellulose medium. Data are represented as number of colonies per 100 cells plated. The mean of the two scorers’ counts is shown. (c and d) Colony formation by CD133+ UCB mock-infected in the presence of 20µM maraviroc (c) or 10µg/mL AMD3100 (d) or an equal volume of DMSO and plated in methylcellulose three days after exposure. Colonies were scored after two weeks by two blinded counters. The mean of the two scorers’ counts is shown. (Erythroid, CFU-E; myeloid, CFU-GM; or multilineage, CFU-GEMM.)
X4-tropic HIVs infect HSCs capable of multilineage reconstitution of immunocompromised mice
Having demonstrated that HIV can infect multipotent HSPCs, we next asked whether HIV could infect hematopoietic stem cells (HSCs) capable of stably engrafting irradiated NOD/SCID IL-2Rγnull mice. Stable multilineage reconstitution in immunocompromised mice can be accomplished only by engraftment of hematopoietic stem cells, and thus this is a definitive assay for infection of HSCs (Christensen and Weissman 2001, Jones et al. 1990, Osawa et al. 1996, Uchida and Weissman 1992). Because we observed little infection of multipotent HSPCs with R5-tropic HIV Envs, we used only X4-tropic Env for these experiments. We infected purified CD133+ cells with replication defective, minimal HIV (HIV-7SF-GFP) pseudotyped with X4-tropic HXB Env (Figure 6a, upper panel). Three days after infection, GFP+ cells were enriched to 40–70% purity (Figure 6a, lower panel) and intrafemorally injected into sublethally irradiated NOD/SCID IL-2Rγnull mice. In all, 13 animals were injected with infected CD133+ cells and 4 were injected with mock-infected, unsorted HSPCs. We used a population of mixed GFP+ and GFP− cells for two reasons: first, to minimize loss of infected cells that would occur with more stringent purification; and second, to enable us to distinguish between mice that specifically failed to engraft infected (GFP+) cells and those that failed to engraft at all due to technical error.
Figure 6. CXCR4-tropic HIV can infect HSCs that stably engraft and generate multiple lineages in NOD/SCID IL2γnull mice.
(a) Upper panel, schematic of HIV-7SF-GFP and HXB envelope plasmid used to construct virus used in lower panel. Lower panel, flow cytometric analysis of cord blood derived CD133+ HPCs infected with HIV-7SF-GFP pseudotyped with HXB Env after three days in culture. Cells in the right panel were sorted for GFP positivity. (b) Flow cytometric analysis of peripheral blood from transplanted mice 16–18 weeks post transplant. Leukocytes that were positive for human CD45 and negative for mouse CD45 were gated in the left panels. The right panels show staining of the indicated markers within this subpopulation. CD33, myeloid; CD3, T cell; CD19, B cell (c) Flow cytometric analysis of peripheral blood cells harvested 20 weeks post transplantation in animal 9. (d) Flow cytometric analysis of bone marrow and spleen from animal 6 after 26 weeks. (C: untreated control animal). See also Figure S3.
Because the HIV genome we used was not cytotoxic and expressed GFP from a constitutively active promoter, we were able to detect infected, mature peripheral blood cells that were the progeny of the originally infected HSPCs. The use of this construct thus enabled us to evaluate the developmental potential of all infected HSPCs, whereas we have previously shown that when HSPCs are infected with replication-competent HIV, the actively infected cells die rapidly (Carter et al., 2010).
Beginning 4 weeks after transplantation and continuing monthly for 20 weeks after transplantation, peripheral blood was collected from the mice and analyzed for GFP expression by flow cytometry. Over time, we detected human cells (HuCD45+MuCD45−) in the periphery of all four mice that received mock-infected transplants (animals 1–4 in Table 1 and Figure S3 and animals 1–2 in Figure S4). In addition, we detected human cells in 7 of 13 mice that received infected HPC transplants (animals 5–11 in Table 1 and Figure S4, animals 5–10 in Figure 6b, and animal 11 in Figure S2). Of the mice that engrafted, 71% (5/7) had GFP+ cells, indicating successful engraftment of cells infected with an HXB-Env bearing virus (animals 5–9 in Table 1, Figure 6b, and Figure S4). Two of the seven mice engrafted human cells that were all GFP-negative (animals 10 and 11 in Table 1, Figure 6b, Figure S3, and Figure S4). An example of one of the six mice that failed to engraft human cells is also shown (animal 12 in Figure 6b, animals 12–17 in Table 1). Although animals 5–11 received both GFP+ and GFP− human cells, several animals (animals 5–7, 10–11) engrafted only GFP+ or only GFP− human cells. This is because only a small fraction of the transplanted cells have the capacity to engraft in the mouse, and these few cells account for all of the human cells found in the peripheral blood. Because so few cells actually engraft, it is not surprising that in some mice, all of the engrafting cells were GFP+ or all were GFP−.
Table 1. Summary of murine xenotransplantation results.
Animals 1–4 were injected with mock-infected HPCs and successfully engrafted. Animals 5–11 were injected with virus-treated HPCs and were successfully engrafted with human cells. Animals 12–17 were injected with virus-treated HPCs but did not successfully engraft. Percents of human and GFP+ cells are percents of total peripheral blood leukocytes at 14–18 weeks after transplant.
| Animal | Number of cells transplanted |
Condition | % Human Leukocytes |
% of Human Leukocytes that are GFP+ |
% of GFP+ Human Leukocytes that are: |
||
|---|---|---|---|---|---|---|---|
| Myeloid | B cells | T cells | |||||
| 1 | 50,000 | Mock | 0.6 | 0.0 | - | - | - |
| 2 | 50,000 | Mock | 0.1 | 0.0 | - | - | - |
| 3 | 100,000 | Mock | 0.1 | 0.0 | - | - | - |
| 4 | 150,000 | Mock | 0.1 | 0.0 | - | - | - |
| 5 | 50,000 | 7SF-GFP/HXB | 0.1 | 98 | 16 | 78 | 0.0 |
| 6 | 50,000 | 7SF-GFP/HXB | 1.4 | 100 | 4.9 | 92 | 0.1* |
| 7 | 100,000 | 7SF-GFP/HXB | 0.4 | 99 | 19 | 56 | 0.0 |
| 8 | 150,000 | 7SF-GFP/HXB | 9.6 | 16 | 6.9 | 88 | 0.0 |
| 9 | 150,000 | 7SF-GFP/HXB | 0.5 | 46 | 7.7 | 77 | 0.0** |
| 10 | 100,000 | 7SF-GFP/HXB | 1.4 | 0.0 | - | - | - |
| 11 | 100,000 | 7SF-GFP/HXB | 0.6 | 0.0 | - | - | - |
| 12–17 | 150,000 | 7SF-GFP/HXB (No engraftment) | 0.0 – 0.03 | 0.0 | - | - | - |
Although the frequency of human cells varied widely in the engrafted mice (0.1–9.6% human leukocytes, Table 1, Figure 6b and Figure S3), all mice that engrafted with infected human cells had GFP+ lymphoid (CD3+ and/or CD19+) and GFP+ myeloid (CD33+) cells in the periphery for at least 20 weeks after transplantation. In all cases, the frequency of GFP+ human cells in the peripheral blood increased after 8–10 weeks post-transplant (Figure S4). As only HSCs can maintain multilineage reconstitution for more than 4–6 weeks in vivo, the increasing frequency of GFP+ cells at later time points clearly demonstrates that infected HSCs have engrafted.
For both GFP+ and GFP− engraftments, T lymphocytes (CD3+) were slow to appear in peripheral blood, consistent with prior reports that human thymopoiesis is inefficient in NOD/SCID/IL-2Rγnull mice after stem-cell transplantation (Lan et al., 2006). For example, by 20 weeks animal 9 clearly had GFP+CD3+ cells, whereas these cells were not apparent at an early time point (compare Figure 6c 20 week time point for animal 9 with 16 week time point shown in Figure 6b). Additionally, animal 6 was sacrificed 26 weeks after transplantation and the tissues were examined for T cell chimerism. As shown in Figure 6d, human CD3+ cells were present in both bone marrow and spleen. Thus, T cells were clearly present in at least 2 of 5 mice that stably engrafted stem cells targeted by HIV.
DISCUSSION
The identification and eradication of long-lived cellular reservoirs is necessary to cure HIV and eliminate the need for lifelong therapy. We have previously demonstrated that HIV can infect multipotent HSPCs, establishing both active and latent infections (Carter et al., 2010). Here we demonstrate that, similar to infection of T cells, infection of multipotent HSPCs depends on CD4. However, based on the panel of Envs we tested, robust infection of primitive HSPCs capable of generating multilineage colonies in soft agar only occurs with CXCR4- or dual-tropic viruses. Blockade of CXCR4 dramatically reduced infection of multipotent hematopoietic cells by dual-tropic HIVs. In contrast, the R5-tropic HIVs we tested had only minimal infectivity in multipotent HSPCs and blockade of CCR5 had no effect on infection of multipotent cells by dual-tropic HIVs.
The simplest explanation for the inability of CCR5-bearing viruses to infect multipotent HSPCs is that CCR5 is not expressed at high enough levels to support infection. An alternative hypothesis is that engagement of R5-tropic Envs with the CCR5 chemokine receptor affects the ability of multipotent HSPC to form colonies or is toxic to the cells. This hypothesis is less likely because dual-tropic HIVs able to bind both CXCR4 and CCR5 can infect cells with a multipotent phenotype. In addition, we have detected minimal CCR5 expression and signaling in response to CCR5 ligands on human CD34+CD133+ cells.
HIV infection of multipotent HSPCs could lead to the presence of viral genomes in multiple hematopoietic lineages. However, HIV is primarily detected in myeloid and T cells, but not in B cells. This apparent enigma may be explained by the fact that active infection of HSPCs leads to the upregulation of markers of apoptosis and rapid depletion of infected cells from the culture (Carter et al., 2010). Thus, the lack of evidence for HIV genomes in B cells in infected people may be due to the fact that active HIV infection kills early HSPCs, preventing the development of infected cells in some lineages (Carter et al., 2010). Latent infection can also occur in HSPCs, but induction of differentiation may induce viral activation and subsequent cell death (Carter et al., 2010).
We have also determined that HIV infects HSCs that are capable of stable, multilineage engraftment in irradiated NOD/SCID IL-2Rγnull mice. All of the mice that were successfully transplanted with infected HSPCs demonstrated multilineage engraftment of infected, GFP+ human cells. These results have significant implications for viral persistence because HSCs are capable of long-term self-renewal in vivo. Thus, latently infected HSCs would persist indefinitely, forming a long-term viral reservoir. Additionally, based on our prior results (Carter et al., 2010), active infection can trigger cell death in multipotent HSPCs. If a sufficient number of HSPCs were infected, the subsequent death of these cells could disrupt the entire hematopoietic cascade. Over many years, this disruption could impact the function of the bone marrow. Unfortunately, the toxicity of wild type HIV precluded us from generating sufficient numbers of infected cells to test whether wild type virus from patient samples infects cells in vivo that have the capacity to engraft. The experiments presented here were only possible with the use of replication defective HIV constructs that do not express additional cytotoxic HIV proteins following integration.
The findings in this study appear to conflict with previous reports indicating that HSCs are resistant to infection with HIV-1 as well as with lentiviral constructs pseudotyped with HIV Envs (Shen et al., 1999; Weichold et al., 1998; Zhang et al., 2007). The mechanism by which HIV-1 infection of HSCs is purported to be blocked has been inconsistent: one study found that the block was solely at the level of entry due to insufficient expression of viral receptors and that VSVG-pseudotyped viral particles could efficiently infect HSCs (Shen et al. 1999), whereas another group found that there was a post-entry block to infection mediated by p21 (Zhang et al. 2007). The apparent difference between our findings and those of previous groups can be explained by low rates of infection (typically less than 2%) that rapidly decline over time because of the cytoxitcity of the virus. Such infection rates are too low to yield detectable infection with many of the non-flow cytometric assays used in previous studies, especially those that require that the cells be cultured for more than a couple days. The use of recently optimized culture conditions for HSPCs (Zhang et al., 2008) has allowed us to increase infection rates in these cells due to improved health of the cells. The previously described blocks to infection likely contribute to the low infection rates that we observe in HSPCs, but importantly, we show that these blocks are not absolute and that X4-tropic HIVs can infect HSCs at a low but significant rate.
In sum, we have shown that multipotent HSPCs and HSCs can be infected by HIV and that this infection is primarily accomplished by CXCR4-tropic HIVs. The infection and destruction of multipotent HSPCs may contribute to the more rapid decline in CD4 counts associated with CXCR4-tropic HIV isolate emergence. Alternatively, as infected HSCs could create an extremely long-lived reservoir of virus, preferential infection of these cells by CXCR4-tropic virus could provide a reservoir for the emergence of CXCR4-tropic isolates late in disease: as other viral reservoirs are depleted, CXCR4-tropic virus from the HSC and HSPC reservoir could begin to predominate. In addition, our demonstration that HIV can infect cells capable of stably engrafting for months in the xenograft model indicates that HIV can infect HSCs that are capable of self-renewal and, if the integrated viral genome is latent, that it can be maintained and even expanded by cell division.
Based on these data, there should be a renewed focus on primitive hematopoietic progenitors as an important reservoir for HIV that will require eradication to improve the treatment of HIV-infected people. Our data suggesting that a subset of HSCs and other primitive hematopoietic progenitors could function as a latent reservoir for HIV raise the possibility that combining HIV therapies with approaches to activate HSCs might deplete this reservoir by triggering the apoptosis of infected HSCs.
EXPERIMENTAL PROCEDURES
Antibodies and reagents
Antibodies to the following proteins were used for flow cytometry: CD34 (FITC-conjugated, BD biosciences), CD34 (APC-conjugated, Caltag), CD34 (647-conjugated, eBioscience), CD133 (PE-conjugated, Miltenyi Biotech), CD133 (biotin-conjugated with streptavidin-APC/Cy7 (eBioscience)), Gag (FITC-conjugated, Coulter), Gag (PE-conjugated, Coulter), CD45 (APC/Cy7-conjugated, BD Biosciences), CD33 (PE-conjugated, BD Biosciences), CD3 (PE/Cy5-conjugated, BD Biosciences), CD19 (APC-conjugated BD Biosciences), CD4 (BD Biosciences), CXCR4 (PE/Cy7–conjugated, eBioscience), CCR5 (PE-conjugated, eBioscience), mouse IgG (FITC-conjugated, Invitrogen). For receptor blocking experiments, functional grade antibody against CD4 was used (clone L3T4, eBioscience).
The env gene expression plasmid pcDNA-89.6env was created as described (Carter et al., 2010). pcDNA-94UGenv was created by digesting p94UG114.1.6 with XbaI and SmaI. The resulting 3021-nucleotide fragment was then ligated to pcDNA3.1 (+), which had been digested with ApaI, blunted by Klenow treatment and digested with XbaI. We created the env gene expression plasmid pEBB-NLenv by digesting pNL4-3 with SalI and NotI and blunting the resulting fragment by Klenow treatment. The resulting fragment was then ligated into pEBB that had been digested with NotI and blunted by Klenow treatment.
Cell culture
We obtained pre-existing umbilical cord blood lacking subject identifiers after scheduled cesarean section procedures. We obtained fresh whole bone marrow aspirates from a commercial source (AllCells Ltd.). We prepared BMMCs and UCB mononuclear cells by Ficoll-Paque density separation (GE Healthcare) according to the manufacturer's instructions. UCB-MNC were frequently cryopreserved in 10% DMSO in FBS. BM-MNCs were always used fresh. We prepared CD34+ or CD133+ cells from adherence-depleted mononuclear cells with commercially available kits (positive selection MACS, Miltenyi Biotech). After isolation we maintained HSPCs in STIF medium (StemSpan or Stemline II medium supplemented with 50ng/ml SCF, 50ng/ml TPO, 100ng/ml IGFBP-2, and 50ng/ml Flt3-L) 40.
We conducted methylcellulose colony-forming assays according to the manufacturer's recommendation (Methocult H4034, StemCell Technologies). Colonies were scored based on morphology using an inverted brightfield microscope at 40× or 100× magnification. CFU-GEMM morphology was verified at high power (200×). GFP expression was analyzed on an inverted epifluorescent microscope.
HIV preparation
We prepared infectious supernatants by transfection of proviral plasmids into 293T cells using polyethylenimine. For pseudotyped viruses, we concentrated supernatants with high-molecular-weight polyethylene glycol precipitation 41. Pellets were resuspended in 1/5th to 1/10th the original volume of StemSpan medium and stored at −80°C. Virus infectivity was determined by infection of CEM-SS or CEM-R5 cells under identical conditions. MOI were calculated by applying the percent of infected CEM-SS cells to the formula MOI = −Ln (1−p) where p is the proportion of cells infected. We conducted HIV infections with a standard spin infection technique for primary cells (1048.6 × g for two hours at room temperature) or by incubating the cells with virus at 37°C for two hours.
Flow cytometry
We stained cells in FACS buffer (2% FBS, 1% human serum, 2mM HEPES, 0.025% NaN3/PBS) for 10–20 minutes on ice, washed and fixed them in 2% paraformaldehyde/PBS. For intracellular Gag staining, we then incubated the cells for 5 min in 0.1% Triton X-100 in PBS at 25 °C. We incubated washed cells with anti-Gag antibody in FACS buffer for 30 minutes on ice. We analyzed the cells on a FACScan or FACSCanto flow cytometer. We excluded dead cells using 7AAD.
For analysis of murine peripheral blood, we lysed erythrocytes with IO Test 3 lysis buffer (Beckman-Coulter) and we stained leukocytes as described above. We analyzed cells on a FacsCantoII analyzer. We excluded dead cells from analysis by DAPI uptake and we excluded cell doublets using FSC-A/FSC-H ratio.
We sorted cells with a FACSVantage SE or FACSAria cytometer (Becton Dickinson). For high-purity sorting, we used normal-R mode with a 1.0 sorted drop envelope. For cell enrichment, we sorted cells in enrich mode with a 1.0 sorted drop envelope.
For the calcium flux assay, cells were suspended in cell loading media (RPMI with 2% FBS and 25mM HEPES) and loaded with 1.5µM Indo-1 AM. Cells were incubated at 37°C for 45 minutes, then washed twice and resuspended in FACS buffer. Surface staining was conducted as described above. Following surface stain, cells were resuspended in cell loading media and equilibrated at 37°C for 30–60 minutes prior to analysis. EGTA was added 1 hour prior to analysis. Samples were analyzed using a FACSDiVa cyotmeter (BD). For the acid wash assay, cells were incubated in pH 2.7 glycine buffer for 1 minute, then washed with 9 ml PBS and stained as above except that serum-free buffer (PBS + 0.1% BSA was used).
Mice
Nonobese diabetic severe combined immunodeficiency mice lacking the interleukin-2 gamma receptor (NOD/SCID IL-2Rnull mice, strain NOD.CB17-Prkdcscid Il2rgtm1Wjl/Szj, (Jackson Laboratory)) were maintained at the University of Michigan by the Unit for Laboratory Animal Medicine. All experiments were conducted in accordance to with research protocols approved by the University Committee on the Use and Care of Animals.
Mouse transplantation
We cultured cord blood-derived CD133+ cells for 4 days in STIF medium to expand HSCs prior to infection. We then infected the cells with HIV-7SF-GFP pseudotyped with HXB2 Env. After three days, we sorted GFP-positive cells, rested them overnight in STIF medium and then transplanted them into sublethally irradiated mice (340cGy). We used a Hamilton syringe fitted with a 27Ga needle to inject 25µL of cells in PBS into the femur. We gave the transplanted mice antibiotic water (1.1gl neomycin and 0.121gl polymyxin B).
Isolation of CD34+ cells from HIV-infected donors
HIV+ individuals were recruited from the University of Michigan HIV/AIDS Treatment Program Outpatient Clinic. The human subjects protocol was approved by the Institutional Review Board and General Clinical Research Center and, as outlined in the protocol, all subjects signed informed consent documents. Using sterile procedure, a Jamshidi needle was used to aspirate one mL of marrow aspirate from the posterior iliac crest. The sample was evaluated for spicules to ensure adequate quality, and then ten ml of marrow aspirate was obtained in preservative free heparin. The subjects experienced no adverse events from the procedure. The bone marrow mononuclear cells were prepared by density separation using Ficoll-Paque (GE healthcare) according to the manufacturer’s instructions, and total mononuclear cells were counted.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by US National Institutes of Health grant RO1 AI051192, MO1-RR000042, the Burroughs Wellcome Foundation, the Rackham Predoctoral Fellowship (C.C.C.), Medical Scientist Training Grant to the University of Michigan (C.C.C) the Molecular Mechanisms in Microbial Pathogenesis Training Grant to the University of Michigan (C.C.C) a US National Science Foundation Predoctoral Fellowship (L.A.M.) and a Bernard Maas Fellowship (L.A.M.). Mark Shackleton was supported by the Australian National Health and Medical Research Council, the Human Frontiers Science Program. Sean Morrison is an Investigator of the Howard Hughes Medical Institute. We are grateful to the University of Michigan flow cytometry core and the University of Michigan DNA sequencing core for their services. We thank Cosmos van de Ven and the University of Michigan Department of Obstetrics surgical staff for umbilical cord blood. We are indebted to Carole McIntyre-Ramm for assistance with recruitment of donors to our study and for help with human subjects regulatory documentation. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, US National Institutes of Health: pNL4-3 from M. Martin (Viral Pathogenesis and Vaccine Section at the US National Institute of Allergy and Infectious Diseases); p89.6 from R.G. Collman (University of Pennsylvania); p94UG–114.1, pCRII-92HT593.1 and pYU-2 from B. Hahn (University of Alabama at Birmingham); pHXB2-env from Dr. Kathleen Page and Dr. Dan Littman; HIV-1 clone BaL.01 (Cat#11445) from Dr. Mascola; CEM.NKR-CCR5 from Dr. Alexandra Trkola; bicyclam JM-2987 (hydrobromide salt of AMD-3100) and maraviroc (Cat #11580). pCMV-HIV-1 and pHIV-7/SF-GFP were gifts of S.-J.-K. Yee (City of Hope National Medical Center). The YU2 env expression plasmid was a kind gift of Joseph Sodroski.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Carter CC, Onafuwa-Nuga A, McNamara LA, Riddell J, Bixby D, Savona MR, Collins KL. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med. 2010;16:446–451. doi: 10.1038/nm.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc Natl Acad Sci. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar ES, Kesler KL, Petropoulos CJ, Huang W, Bates M, Lail AE, Coakley EP, Gomperts ED, Donfield SM. Baseline HIV type 1 coreceptor tropism predicts disease progression. Clin Infect Dis. 2007;45:643–649. doi: 10.1086/520650. [DOI] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Donzella GA, Schols D, Lin SW, Este JA, Nagashima KA, Maddon PJ, Allaway GP, Sakmar TP, Henson G, De Clercq E, et al. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Endres MJ, Clapham PR, Marsh M, Ahuja M, Turner JD, McKnight A, Thomas JF, Stoebenau-Haggarty B, Choe S, Vance PJ, et al. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Folks TM, Kessler SW, Orenstein JM, Justement JS, Jaffe ES, Fauci AS. Infection and replication of HIV-1 in purified progenitor cells of normal human bone marrow. Science. 1988;242:919–922. doi: 10.1126/science.2460922. [DOI] [PubMed] [Google Scholar]
- Hoxie JA, LaBranche CC, Endres MJ, Turner JD, Berson JF, Doms RW, Matthews TJ. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J Reprod Immunol. 1998;41:197–211. doi: 10.1016/s0165-0378(98)00059-x. [DOI] [PubMed] [Google Scholar]
- Ishii T, Nishihara M, Ma F, Ebihara Y, Tsuji K, Asano S, Nakahata T, Maekawa T. Expression of stromal cell-derived factor-1/pre-B cell growth-stimulating factor receptor, CXC chemokine receptor 4, on CD34+ human bone marrow cells is a phenotypic alteration for committed lymphoid progenitors. J Immunol. 1999;163:3612–3620. [PubMed] [Google Scholar]
- Jones RJ, Wagner JE, Celano P, Zicha MS, Sharkis SJ. Separation of pluripotent haematopoietic stem cells from spleen colony-forming cells. Nature. 1990;347:188–189. doi: 10.1038/347188a0. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Parsmyr K, Sandstrom E, Fenyo EM, Albert J. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J Clin Microbiol. 1994;32:364–370. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Tonomura N, Shimizu A, Wang S, Yang YG. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood. 2006;108:487–492. doi: 10.1182/blood-2005-11-4388. [DOI] [PubMed] [Google Scholar]
- Lathey JL, Tsou J, Brinker K, Hsia K, Meyer WA, 3rd, Spector SA. Lack of autologous neutralizing antibody to human immunodeficiency virus type 1 (HIV-1) and macrophage tropism are associated with mother-to-infant transmission. J Infect Dis. 1999;180:344–350. doi: 10.1086/314886. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu H, Kim BO, Gattone VH, Li J, Nath A, Blum J, He JJ. CD4-independent infection of astrocytes by human immunodeficiency virus type 1: requirement for the human mannose receptor. J Virol. 2004;78:4120–4133. doi: 10.1128/JVI.78.8.4120-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka M, Rozmyslowicz T, Lee B, Murphy SL, Pietrzkowski Z, Gaulton GN, Silberstein L, Ratajczak MZ. Bone marrow CD34(+) cells and megakaryoblasts secrete beta-chemokines that block infection of hematopoietic cells by M-tropic R5 HIV. J Clin Invest. 1999;104:1739–1749. doi: 10.1172/JCI7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis L, Shattock R. Selective transmission of CCR5-utilizing HIV-1: the 'gatekeeper' problem resolved? Nat Rev Microbiol. 2006;4:312–317. doi: 10.1038/nrmicro1387. [DOI] [PubMed] [Google Scholar]
- Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors--central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-Term Lymphohematopoietic Reconstitution by a Single CD34-Low/Negative Hematopoietic Stem Cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, Nagler A, Ben-Hur H, Many A, Shultz L, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- Redd AD, Avalos A, Essex M. Infection of hematopoietic progenitor cells by HIV-1 subtype C, and its association with anemia in southern Africa. Blood. 2007;110:3143–3149. doi: 10.1182/blood-2007-04-086314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- Saha K, Yan H, Nelson JA, Zerhouni-Layachi B. Infection of human and non-human cells by a highly fusogenic primary CD4-independent HIV-1 isolate with a truncated envelope cytoplasmic tail. Virology. 2005;337:30–44. doi: 10.1016/j.virol.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- Schuitemaker H, Koot M, Kootstra NA, Dercksen MW, de Goede RE, van Steenwijk RP, Lange JM, Schattenkerk JK, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Cheng T, Preffer FI, Dombkowski D, Tomasson MH, Golan DE, Yang O, Hofmann W, Sodroski JG, Luster AD, et al. Intrinsic human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J Virol. 1999;73:728–737. doi: 10.1128/jvi.73.1.728-737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JC, Jacobson LP, Qiao W, Jamieson BD, Phair JP, Piazza P, Quinn TC, Margolick JB. Emergence and persistence of CXCR4-tropic HIV-1 in a population of men from the multicenter AIDS cohort study. J Infect Dis. 2008;198:1104–1112. doi: 10.1086/591623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard HW, Celum C, Michael NL, O'Brien S, Dean M, Carrington M, Dondero D, Buchbinder SP. HIV-1 infection in individuals with the CCR5-Delta32/Delta32 genotype: acquisition of syncytium-inducing virus at seroconversion. J Acquir Immune Defic Syndr. 2002;29:307–313. doi: 10.1097/00126334-200203010-00013. [DOI] [PubMed] [Google Scholar]
- Stanley SK, Kessler SW, Justement JS, Schnittman SM, Greenhouse JJ, Brown CC, Musongela L, Musey K, Kapita B, Fauci AS. CD34+ bone marrow cells are infected with HIV in a subset of seropositive individuals. J Immunol. 1992;149:689–697. [PubMed] [Google Scholar]
- Uchida N, Weissman IL. Searching for Hematopoietic Stem Cells: Evidence That Thy-1.1lo Lin− Sca-1+ Cells Are the Only Stem Cells in C57BL/Ka-Thy-1.1 Bone Marrow. J Exp Med. 1992;175:175–184. doi: 10.1084/jem.175.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Wout AB, Kootstra NA, Mulder-Kampinga GA, Albrecht-van Lent N, Scherpbier HJ, Veenstra J, Boer K, Coutinho RA, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viardot A, Kronenwett R, Deichmann M, Haas R. The human immunodeficiency virus (HIV)-type 1 coreceptor CXCR-4 (fusin) is preferentially expressed on the more immature CD34+ hematopoietic stem cells. Ann Hematol. 1998;77:193–197. doi: 10.1007/s002770050442. [DOI] [PubMed] [Google Scholar]
- Waters L, Mandalia S, Randell P, Wildfire A, Gazzard B, Moyle G. The impact of HIV tropism on decreases in CD4 cell count, clinical progression, and subsequent response to a first antiretroviral therapy regimen. Clin Infect Dis. 2008;46:1617–1623. doi: 10.1086/587660. [DOI] [PubMed] [Google Scholar]
- Weichold FF, Zella D, Barabitskaja O, Maciejewski JP, Dunn DE, Sloand EM, Young NS. Neither human immunodeficiency virus-1 (HIV-1) nor HIV-2 infects most-primitive human hematopoietic stem cells as assessed in long-term bone marrow cultures. Blood. 1998;91:907–915. [PubMed] [Google Scholar]
- Weiser B, Philpott S, Klimkait T, Burger H, Kitchen C, Burgisser P, Gorgievski M, Perrin L, Piffaretti JC, Ledergerber B. HIV-1 coreceptor usage and CXCR4-specific viral load predict clinical disease progression during combination antiretroviral therapy. AIDS. 2008;22:469–479. doi: 10.1097/QAD.0b013e3282f4196c. [DOI] [PubMed] [Google Scholar]
- Yam PY, Li S, Wu J, Hu J, Zaia JA, Yee JK. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther. 2002;5:479–484. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
- Yu XF, Wang Z, Vlahov D, Markham RB, Farzadegan H, Margolick JB. Infection with dual-tropic human immunodeficiency virus type 1 variants associated with rapid total T cell decline and disease progression in injection drug users. J Infect Dis. 1998;178:388–396. doi: 10.1086/515646. [DOI] [PubMed] [Google Scholar]
- Zerhouni B, Nelson JA, Saha K. Isolation of CD4-independent primary human immunodeficiency virus type 1 isolates that are syncytium inducing and acutely cytopathic for CD8+ lymphocytes. J Virol. 2004;78:1243–1255. doi: 10.1128/JVI.78.3.1243-1255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC, Kaba M, Iizuka S, Huynh H, Lodish HF. Angiopoietin-like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood. 2008;111:3415–3423. doi: 10.1182/blood-2007-11-122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Scadden DT, Crumpacker CS. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Shen L, Yang HC, Siliciano RF. Preferential cytolysis of peripheral memory CD4+ T cells by in vitro X4-tropic human immunodeficiency virus type 1 infection before the completion of reverse transcription. J Virol. 2008;82:9154–9163. doi: 10.1128/JVI.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.