SUMMARY
Modular polyketide synthases (PKS) make novel natural products through a series of pre-programmed chemical steps catalyzed by an assembly line of multi-domain modules. Each assembly line step involves unique extension and modification reactions, resulting in tremendous diversity of polyketide products. Dehydratase domains catalyze formation of an α,β-double bond in the nascent polyketide intermediate. We present crystal structures of the four dehydratase domains from the curacin A PKS. The catalytic residues and substrate binding site reside in a tunnel within a single monomer. The positions of the catalytic residues and shape of the substrate tunnel explain how chirality of the substrate hydroxyl group may determine the configuration of the product double bond. Access to the active site may require opening the substrate tunnel, forming an open trench. The arrangement of monomers within the dimer is consistent among PKS dehydratases and differs from that seen in the related mammalian fatty acid synthases.
INTRODUCTION
Polyketide and non-ribosomal peptide natural products are important secondary metabolites of microbial systems. These small molecules display a rich diversity of biosynthetic chemistry and are the basis of many therapeutic compounds in clinical use, including antibiotics, immunosuppressants and anti-tumor agents. The biosynthesis of polyketides and non-ribosomal peptides occurs in a coordinated step-wise fashion by enzyme assembly lines, the polyketide synthases (PKS), non-ribosomal peptide synthetases (NRPS), or NRPS/PKS hybrid systems. There is a significant interest in producing novel non-natural polyketides by exploiting the natural organization of the PKS pathways into modules (Weissman and Muller, 2008). Each acetate-derived two-carbon extension in a PKS pathway, catalyzed by one module, can differ in character at the β-position (keto, R-hydroxyl, S-hydroxyl or fully reduced) and at the α-position (R-methyl, S-methyl, no modification, cis α,β-double bond or trans α,β-double bond). The modifications at any two-carbon unit are independent of those at all other positions in the finished product, and the deliberate engineering of modules can result in many unique products not yet discovered or synthesized. Toward this end, understanding substrate specificity of PKS enzymes and the detailed molecular interactions of multi-domain modules is essential to guide the design of novel PKS pathways (Sherman and Smith, 2006).
Modular PKS systems are thought to share a common ancestor with mammalian fatty acid synthases (mFAS) (Smith and Sherman, 2008). Both have multifunctional polypeptide chains with catalytic domains for elongation and modification of intermediate products, which are covalently attached to an acyl carrier protein (ACP) domain via a thioester linkage. In the mFAS and modular PKS pathways, intermediate compounds are extended by the action of two elongation domains: the acyltransferase (AT) and ketosynthase (KS). The β-keto product of the elongation reactions may be altered by the sequential actions of ketoreductase (KR), dehydratase (DH), and enoylreductase (ER) modifying domains, which form a hydroxyl group, double bond and single bond, respectively. While fatty acids are synthesized in an iterative fashion, with the intermediate chain extended and modified within the same polypeptide, modular PKS systems operate in an assembly-line fashion, in which intermediates are passed to successive modules in a megacomplex (Smith and Sherman, 2008). The assembly-line architecture prescribes incorporation of a unique building block at each step and a unique set of modifying reactions, leading to the tremendous diversity that is a hallmark of polyketide natural products.
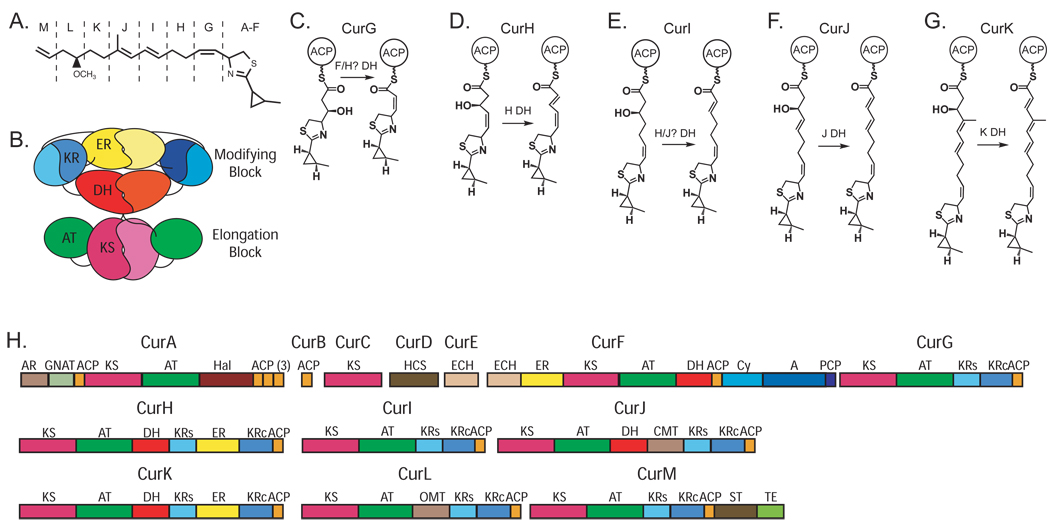
Curacin A is an excellent example of the chemical diversity of natural products with a polyketide skeleton (Figure 1A). It is an important anti-cancer lead agent that blocks the cell cycle by inhibition of microtubule polymerization (Blokhin et al., 1995; Verdier-Pinard et al., 1998). The biosynthetic pathway includes at least 41 catalytic domains in 13 polypeptides. A cyclopropyl ring, cis-double bond, thiazoline ring and terminal alkene make curacin A a chemical stand-out even among the diverse polyketides (Figure 1). Formation of the cyclopropyl ring is catalyzed by the actions of a hydroxymethyl glutarate (HMG)-ACP synthase and modification domains in CurA—CurF (Gu et al., 2009a). The cysteine-derived thiazoline ring is generated by an NRPS module within CurF. The terminal alkene is formed by an unusual sulfotransferase-thioesterase didomain in CurM (Gu et al., 2009b). CurG—CurL appear to be more prototypical PKS monomodules, albeit with additional methyltransferase domains within CurJ and CurL (Chang et al., 2004). The cyclopropyl ring, thiazoline ring and cis-double bond formation are early steps in curacin biosynthesis, thus all subsequent functional modules must accommodate these relatively bulky substituents within the growing polyketide chain.
Figure 1. Curacin A synthase.
(A) Curacin A. Unusual features include a thiazoline ring, a cyclopropane ring, a cis-double bond and a terminal double bond. Single letter codes refer to the module for extension and modification of the functional groups outlined with dashed lines, and correspond to modules outlined in part (H).
(B) Architectural model of a complete dimeric PKS module. The N-terminal elongation block comprises the KS (magenta) and AT (green) domains, while the C-terminal modifying block comprises the DH (red), ER (yellow) and KR (blue) domains. The ER domain is inserted between the structural (KRs) and catalytic (KRc) halves of the functional KR domain. A C-terminal ACP domain is not shown. The model is based on structures of mFAS (Maier et al., 2008) and PKS fragments (Keatinge-Clay, 2008; Keatinge-Clay and Stroud, 2006; Tang et al., 2006).
(C—G) Proposed dehydratase-catalyzed reactions occurring on intermediates of the CurG (C), CurH (D), CurI (E), CurJ (F) and CurK (G) modules. Dehydratase reactions are assumed based on the mature curacin A product. The CurG and CurI polypeptides lack DH domains. In contrast to DH substrates of other modules, the CurG intermediate (C) is proposed to have inverted stereochemistry at the β–C atom based both on the sequence of the CurG KR domain and the cis configuration of the product (see text). It is unknown whether methylation at the α position of the CurJ intermediate (F) occurs before or after the dehydration reaction, thus the CurJ DH substrate may contain an α-imethyl substituent.
(H) Curacin A biosynthetic proteins. Complete domain assignment for polypeptides (CurA – CurM) is colored as in (B) for the PKS domains with addition of ACP. Other domains include the AR/GNAT loading module, Hal halogenase, hydroxymethyl-glutarate-ACP synthase (HCS), dehydratase and decarboxylase (ECHs), C-methyl transferase (CMT), O-methyl transferase (OMT), sulfotransferase (ST), thioesterase (TE), and the NRPS-derived adenylation (A), condensation/cyclization (Cy) and peptidyl carrier (PCP) domains. Box sizes are proportional to domain sizes.
The structure of the mammalian fatty acid synthase (mFAS) reveals much detail about the architecture of homologous mFAS and PKS systems (Maier et al., 2008). In particular they share an overall dimeric structure in which the elongation domains (KS and AT) form a discrete structural unit known as the “elongation block”, and the modifying domains (KR, DH and ER) form a separate structural unit, the “modifying block” (Figure 1B). In the shared mFAS/PKS architecture, the dimer interface is formed by three domains: the KS domain of the elongation block, and the DH and ER domains of the modifying block. The elongation blocks of mFAS and PKS have remarkably similar structures (Maier et al., 2008; Tang et al., 2006). In the modifying block, the ER domain is inserted between the N-terminal structural half (KRs) and the C-terminal catalytic half (KRc) of the KR (Castonguay et al., 2008; Keatinge-Clay and Stroud, 2006). The excised DH domain from module 4 of the erythromycin PKS is dimeric (Keatinge-Clay, 2008), as in the intact mFAS (Maier et al., 2008). However, the orientation of the Ery4 DH monomers differs from that seen in the mFAS complex: the Ery4 DH forms an extended dimer, while mFAS DH forms a V-shaped dimer. Other differences among the mFAS and PKS modifying blocks are certain to arise as a result of the absence of either the ER or both the DH and ER domains in many PKS modules. The ACP domains, which are fused to the C-terminus of the modifying block, also differ in PKS and mFAS systems. The mFAS ACPs are loosely tethered and monomeric when excised, and thus the motions of the two ACPs in the dimer are presumed to be independent of one another. In contrast, the domains immediately downstream of the PKS ACPs are generally dimeric and thus may restrict the motion of the PKS ACP domains (Broadhurst et al., 2003; Buchholz et al., 2009; Tang et al., 2007; Tsai et al., 2002; Tsai et al., 2001).
The PKS DH domain catalyzes reversible dehydration of the β-hydroxyl group, resulting in formation of an α,β-double bond in a cis or trans configuration. The mFAS and PKS dehydratase domains are evident products of gene duplication and fusion, and share a common ancestor with the bacterial FAS DH enzymes. Bacterial FAS DH proteins (FabA and FabZ) are standalone dimers of a structure known as the “hotdog fold”, and contain two bipartite active sites comprising a histidine from one monomer, and aspartate from the second monomer (Kostrewa et al., 2005; Leesong et al., 1996). For a time, it was thought that in dimeric PKSs and mFAS, the DH domains would have a similar inter-subunit active site, but it is now clear that these DH domains are double hotdogs in which the active site contains a histidine from the N-terminal hotdog and an aspartate from the C-terminal hotdog (Keatinge-Clay, 2008; Maier et al., 2006). Our efforts were motivated by open questions regarding the mechanism of action of DH domains including how substrates and products are loaded and unloaded, how cis- vs. trans- stereochemistry is determined and what, if any, structural features define substrate specificity.
Here we present the crystal structures of the four DH domains from the curacin A biosynthetic pathway, including those from the CurF, CurH, CurJ and CurK polypeptides. The DH domains were assigned based on a re-annotation of the curacin PKS sequence in which one DH is newly identified and two modules lack DH where it is anticipated. Our analysis of the structures, combined with insights from previously reported structures, suggests that uncovering of a substrate tunnel may be essential for loading substrates and unloading products, and that the orientation of DH monomers differs in PKS modules from that observed for mammalian fatty acid synthase.
RESULTS AND DISCUSSION
DH domain excision
The curacin PKS is expected to catalyze five dehydration reactions involving β-OH groups of pathway chain elongation intermediates. Based on the biosynthetic pathway deduced from the sequence of the Cur gene cluster and the chemical structure of curacin A (Chang et al., 2004), dehydratase domains are expected within the CurG, CurH, CurI, CurJ and CurK polypeptides (Figure 1C–G). However, sequence analysis revealed a striking deviation from expectation (Figure 1H). We identified DH domains within CurH, CurJ and CurK, as expected, but not within CurG or CurI. We also identified a DH domain within CurF, although dehydration is not among the expected collinear reactions catalyzed by the PKS module within CurF. Further, the DH domain within CurJ precedes the predicted methyltransferase domain. These changes to the original domain assignments reflect the growing understanding of DH as gleaned from the mFAS-derived structure (Maier et al., 2006; Maier et al., 2008) and from predictions (Keatinge-Clay and Stroud, 2006), which were unavailable when the curacin A gene cluster was characterized. The four Cur DH domains have moderate levels of sequence identity (21%–28%), with the exception of the CurF and CurK DHs, which are 68% identical.
Based on sequence similarity with other PKS DHs, we defined minimal Cur DH domains to include residues 1687–1968 of CurF, 939–1218 of CurH, 946–1239 of CurJ, and 963–1245 of CurK, with N- and C-termini precisely at the boundaries of strong sequence conservation. These conserved regions are fused to well defined adjacent domains by rather short linkers of 11–21 residues, with the exception of CurJ, which appears to have a full domain inserted between its DH and MT domains (144-residue linker). In initial experiments, the minimal constructs of CurF DH, CurH DH and CurJ DH were well behaved in solution and eluted from a gel filtration column with apparent molecular weight of ~81 kDa, consistent with a dimeric association in solution. All three proteins also crystallized, however the CurH DH and CurJ DH crystals diffracted poorly. Therefore, we expressed a total of 24 constructs encoding 0, 5 or 10 additional N-terminal residues and 0 or 5 additional C-terminal residues for all four proteins. We chose to limit extensions to 10 residues for the N-terminus and 5 residues for the C-terminus because our analysis of DH sequences indicated that longer extensions would include structurally ordered residues from either the preceding AT (Tang et al., 2006) or subsequent KRs domain (Keatinge-Clay and Stroud, 2006). The C-terminal extensions were particularly beneficial for crystallization. The C-terminal extended CurH DH structure was solved by multiwavelength anomalous diffraction from the selenomethionyl protein, and the other three DH structures were solved by molecular replacement (Table 1). The crystal structures present two crystallographically independent views of each of the CurF and CurH DH dimers, and one view of the CurJ and CurK dimers.
Table 1.
Crystallographic Summary
| CurH DH (SeMet) | CurF DH | CurJ DH | CurK DH | ||
|---|---|---|---|---|---|
| Diffraction Data | |||||
| Space group | P21 | C2221 | P212121 | P212121 | |
| a, b, c (Å) | 82.2 74.5 103.5 | 87.5 198.5 146.8 | 47.4 70.4 174.8 | 38.1 94.3 151.0 | |
| α, β, γ (°) | 90 108.84 90 | 90 90 90 | 90 90 90 | 90 90 90 | |
| Peak | Inflection | ||||
| Wavelength (Å) | 0.97945 | 0.97962 | 0.97942 | 0.97945 | 1.0332 |
| Resolutiona (Å) | 2.80 (2.91-2.80) | 2.90 (3.00-2.90) | 2.70 (2.80-2.70) | 2.45 (2.54-2.45) | 1.70 (1.76-1.70) |
| <I/σI> | 19.7 (2.68) | 14.2 (1.5) | 18.1 (2.4) | 19.7 (2.5) | 20.1 (2.3) |
| Rsymm | 0.107 (0.813)b | 0.096 (0.874)b | 0.109 (0.792) | 0.092 (0.799) | 0.072 (0.656) |
| Completeness | 99.9 (100.0) | 99.9 (100.0) | 100.0 (100.0) | 97.4 (91.9) | 98.0 (85.8) |
| Avg. Redundancy | 3.9 (3.9) | 3.9 (3.9) | 7.4 (6.4) | 3.6 (3.4) | 3.6 (2.8) |
| Unique Reflections | 29969 | 26831 | 35542 | 22090 | 60671 |
| Refinement | |||||
| Data range (Å) | 50.0—2.80 | 50.0—2.70 | 50.0—2.45 | 50.0—1.70 | |
| No. Reflections | 27848 | 33290 | 20616 | 56370 | |
| Rwork / Rfreec | 0.208 / 0.269 | 0.212 / 0.264 | 0.195 / 0.259 | 0.182 / 0.217 | |
| RMS Deviations | |||||
| Bonds (Å) | 0.008 | 0.006 | 0.009 | 0.008 | |
| Angles (°) | 1.085 | 0.916 | 1.166 | 1.123 | |
| No. atoms | |||||
| Protein | 9031 | 8796 | 4508 | 4441 | |
| Water | 45 | 52 | 74 | 513 | |
| B-factors (Å2) | |||||
| Protein | 93.0 | 73.8 | 56.7 | 27.6 | |
| Water | 65.6 | 52.0 | 48.8 | 38.6 | |
| Ramachandran | |||||
| Favored | 97.3% | 97.7% | 96.9% | 97.6% | |
| Allowed | 2.6% | 2.2% | 3.1% | 2.4% | |
| Outliers | 0.1% | 0.1% | 0% | 0% | |
Highest resolution shell shown in parentheses.
Anomalous data were included in calculation of Rsymm values.
5% percent of data were withheld from refinement for Rfree data set.
Dehydratase fold
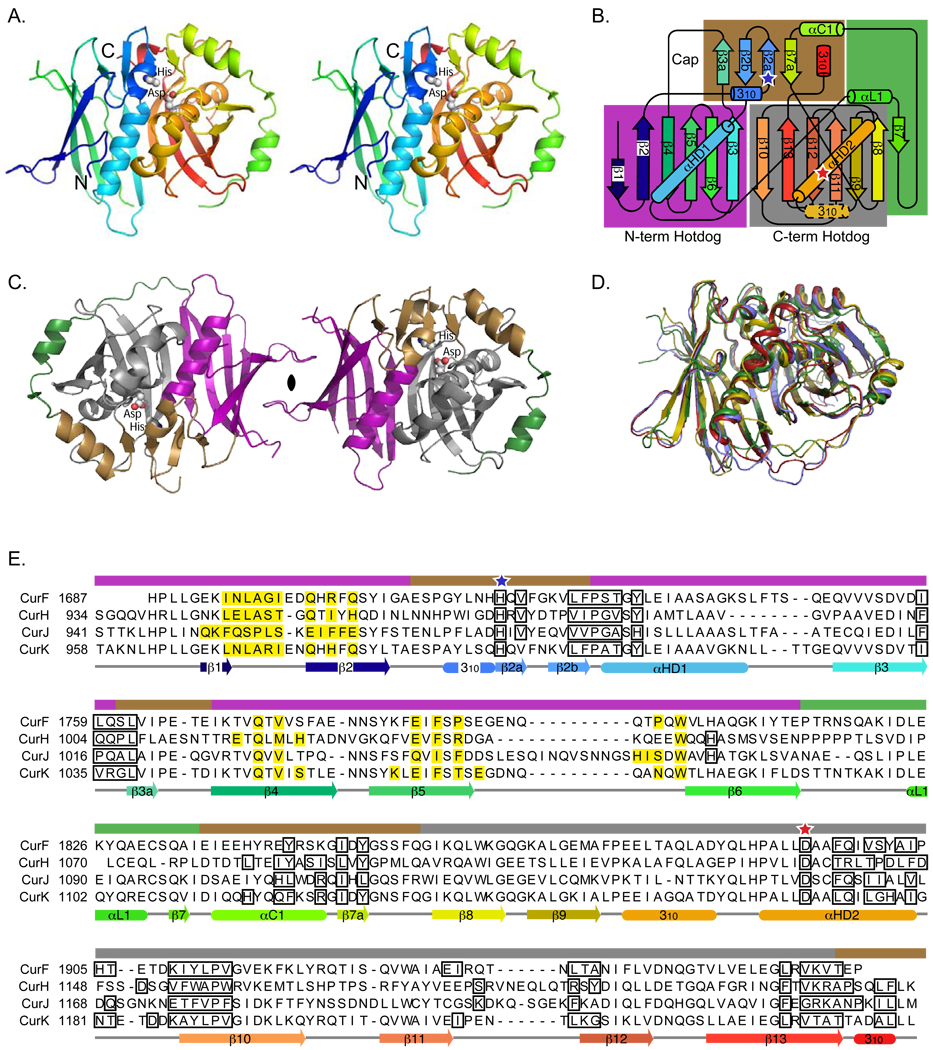
The four Cur DHs are dimeric, with monomers having essentially identical folds (core region Cα rmsd values of 1.0–2.0 Å, Figure 2A–D, Figures S1, Table 2). As expected, the monomers comprise two repeats of the hotdog fold. The ~125-residue N-terminal hotdog is slightly shorter than the ~140-residue C-terminal hotdog (residues 1687–1811 and 1832–1968 of CurF, 939–1055 and 1076–1223 of CurH, 946–1078 and 1097–1244 of CurJ, and 963–1087 and 1108–1245 of CurK). The intervening ~20 residues connect the two hotdog domains with an extended linker and a short helix (αL1). The double-hotdog fold forms a continuous antiparallel β-sheet in which the β-strands curve around the “hotdog” helices (αHD1 from the N-terminal and αHD2 from the C-terminal hotdog folds). In all four Cur DHs, the double-hotdog core is topped by a cap motif comprising a 310 helix, a 4-stranded β-sheet (β2a, β2b, β3a and β7a), and a single α-helix (αC1), and additionally, a short C-terminal 310 helix can be considered part of the cap (Figure 2). The CurF and CurK DHs contain a short 310 helix in the second hotdog motif, which is topologically analogous to the first 310 helix of the cap motif and presumably is a remnant from the ancestral hotdog fold as observed in the prokaryotic FAS DH proteins (Kostrewa et al., 2005; Leesong et al., 1996). The five C-terminal residues of the excised domains of the CurH, CurJ and CurK DHs form the short 310 helix that packs against the small cap β-sheet, accounting for the importance of these residues to protein integrity and crystal diffraction quality. Structural landmarks include the primary β-sheet, the hotdog helices, the cap β-sheet, and the short C-terminal helix.
Figure 2. Structure and sequence of curacin A dehydratases.
(A) Backbone trace of the CurK DH double-hotdog monomer. The active-site histidine and aspartate side chains are shown as spheres and labeled. The stereo diagram is colored from blue at the N-terminus to red at the C-terminus.
(B) Topology diagram. The curacin DH domains have identical topologies with the exception of the 310 helix with a dashed outline, which is observed only in the CurK and CurJ DHs. Tertiary structural motifs (N- and C- terminal hotdogs, cap and linker) are highlighted with purple, grey, brown and green boxes, respectively. Active site residues are marked with blue (histidine) and red (aspartate) stars.
(C) CurK DH dimer. Structural motifs are colored purple, grey, brown and green as for the boxes of the topology diagram in (B). The monomer on the right is viewed in the same orientation as in (A).
(D) Superposition of the CurF (red) CurH (gold), CurJ (green) and CurK (blue) DH monomers, showing the overall similarity of the structures. Figure S1 is a stereo view of this figure.
(E) Structure-based sequence alignment of CurF, CurH, CurJ, and CurK DH domains. Active site residues are indicated with blue (histidine) and red (aspartate) stars. Residues involved in the dimer interface are highlighted yellow. Residues lining the active site tunnel are outlined in black. The helices and β-strands of each protein are shown below the sequence alignment colored blue to red, N- to C- termini, as for parts (A) and (B). Colored bars above the alignment indicate the tertiary structure motifs as in parts (B) and (C). Secondary structure assignments for other DH domains, including DEBS module 4 DH and mFAS DH domains are given in Figure S3.
Table 2.
Structure and Sequence Similarity of DH Domains
| CurF | CurH | CurJ | CurK | Ery4 DH | mFAS DH | |
|---|---|---|---|---|---|---|
| CurF | ||||||
| RMSDa | 2.0 Å | 1.5 Å | 1.0 Å | 1.9 Å | 1.9 Å | |
| # Cαb | 247 | 246 | 257 | 233 | 203 | |
| % identityc | 23% | 32% | 68% | 20% | 23% | |
| CurH | 1.8 Å | 1.9 Å | 2.1 Å | 2.1 Å | ||
| 256 | 248 | 235 | 200 | |||
| 25% | 21% | 22% | 18% | |||
| CurJ | 1.7 Å | 1.8 Å | 1.8 Å | |||
| 252 | 232 | 201 | ||||
| 31% | 23% | 17% | ||||
| CurK | 1.6 Å | 2.1 Å | ||||
| 226 | 208 | |||||
| 21% | 21% | |||||
| FabA | 2.2 Å | 3.2 Å | 2.2 Å | 2.3 Å | ||
| (to N term) | 105 | 104 | 104 | 105 | ||
| 11% | 12% | 12% | 11% | |||
| FabA | 2.2 Å | 3.0 Å | 2.8 Å | 2.8 Å | ||
| (to C term) | 113 | 117 | 128 | 129 | ||
| 12% | 9% | 7% | 11% | |||
RMSD of superimposed Cα atoms
number of residues superimposed
sequence identity for superimposed residues only
Structure based alignments were calculated using SSM (Krissinel and Henrick, 2004).
Active site
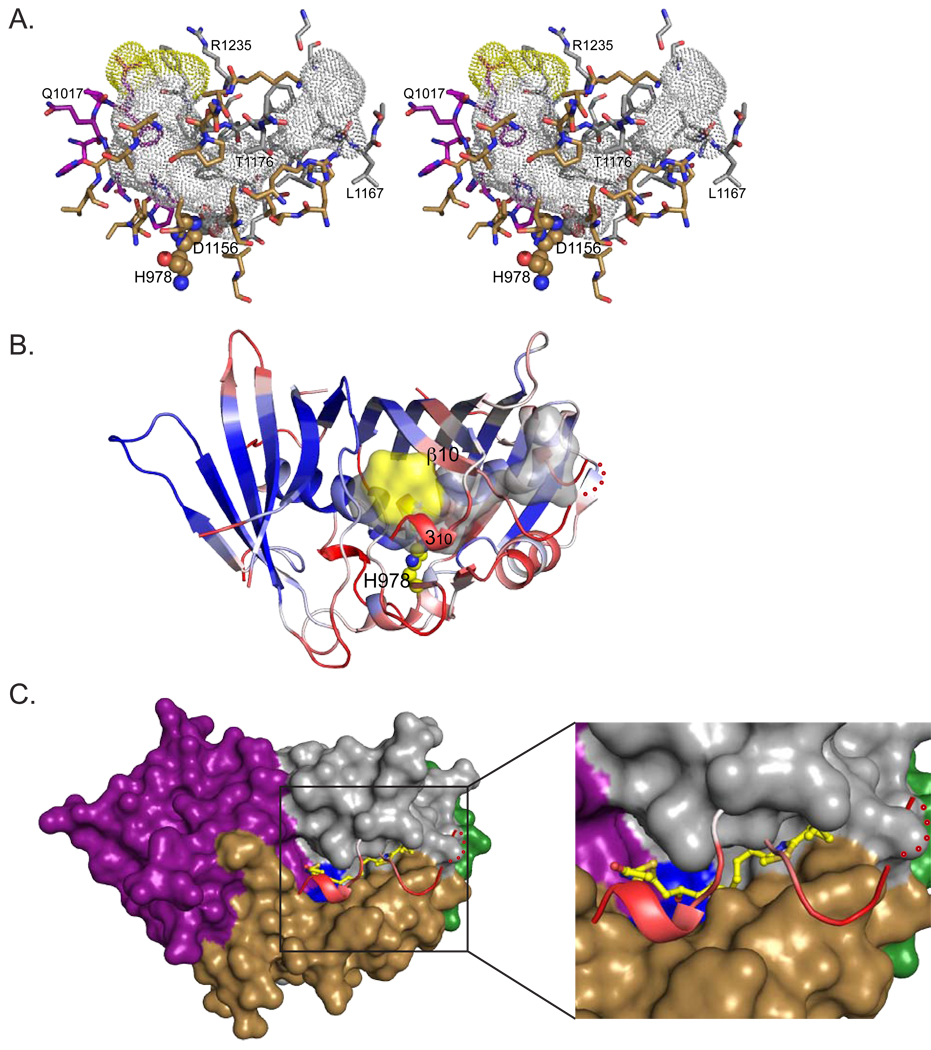
The DH active site is a His-Asp “catalytic dyad”, with the histidine from the N-terminal hotdog and the aspartate from the C-terminal hotdog (CurF His1720 and Asp1893, CurH His971 and Asp1136, CurJ His978 and Asp1156, CurK His996 and Asp1169). All PKS DHs have similarly arranged active sites, based on sequence conservation. The histidine is on the small cap strand β2a, and the aspartate is on the second hotdog helix (αHD2). The active site is located at the bottom of a deep V within a tunnel formed by elements from the N- and C-terminal hotdog folds and closed off by the cap motif (Figure 3A, Figure S2). The catalytic dyad is at the most constricted region of the substrate tunnel. The tunnel extends into the C-terminal hotdog fold between the hotdog helix and the primary β-sheet. The tunnel entrance lies at the junction of the N- and C-terminal hotdog folds (yellow surface in Figure 3A,B). The entrance is bounded by β3 from the N-terminal hotdog, β10 from the C-terminal hotdog, and the C-terminal 310 helix (Figure 3B). The catalytic residues, substrate tunnel and tunnel entrance are similar in the four Cur DHs (Figure 3A, Figure S2) and also in Ery DH, mFAS DH and the bacterial fatty acid DHs, FabA and FabZ (Keatinge-Clay, 2008; Kostrewa et al., 2005; Leesong et al., 1996; Maier et al., 2008).
Figure 3. Active site tunnel and substrate access.
(A) Stereodiagram of the active site tunnel for CurJ DH. The tunnel is shown as a dotted surface with gray interior and yellow entrance. Residues lining the tunnel (outlined in Figure 2E) are shown as sticks with C atoms colored by structural motif (purple N-terminal hotdog, gray C-terminal hotdog, brown cap) as for Figure 2B,C. Active site residues (His and Asp) are shown as spheres. The tunnel surface was calculated using HOLLOW (Ho and Gruswitz, 2008). Active site tunnels for the other three Cur DHs are shown in Figure S2.
(B) CurJ monomer colored according to average atomic temperature (B) factor per residue. This view is rotated ~90° along the horizontal axis with respect to (A). The lowest B-factors (blue) occur in the N-terminal hotdog fold, which forms the dimer interface. The highest B-factors are in residues covering the active site tunnel (the loop connecting helix αHD2 and strand β10, and the C-terminal 310 helix) and also in the linker connecting the two hotdog motifs. The substrate tunnel is shown as a translucent surface colored as in (A), and the active site histidine is shown as spheres with yellow C.
(C) Surface diagram of CurJ DH monomer without the tunnel cover. Removal of residues with high B-factors (αHD2—β10 loop and C-terminal 310 helix, shown in ribbon form) allows unfettered access to the active site (His-Asp dyad in blue surface), which now resides in the bottom of a trench. The surface is colored by structural motif (as in Figure 2C). The CurJ DH substrate is modeled as sticks with yellow C (more detail inset). These views are in the same orientation as (B).
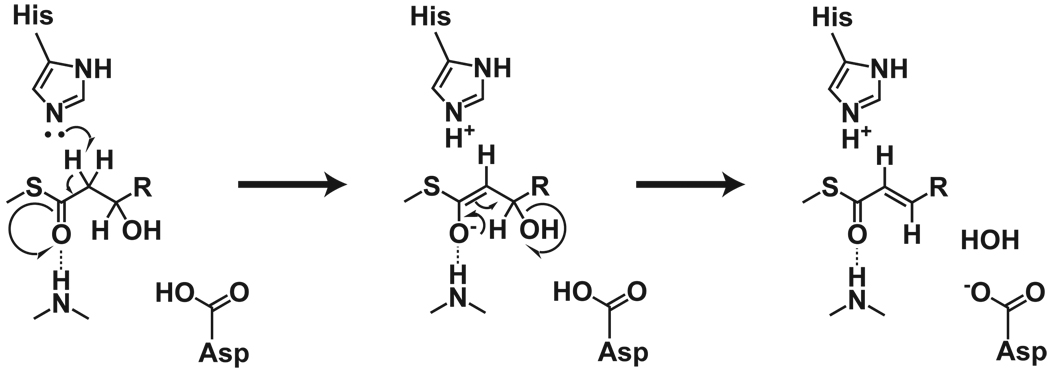
Catalysis is thought to proceed by histidine abstraction of a proton from the α position (Helmkamp and Bloch, 1969) and aspartic acid abstraction of a hydroxyl from the β position (Leesong et al., 1996) (Scheme 1). The structure of bacterial FabA with a covalent label at the catalytic histidine (Leesong et al., 1996) guided our modeling of substrates and products into the substrate chambers of the Cur DHs (Figure 4). Two water molecules within the active site guided the placement of the substrate β-OH and thioester carbonyl groups (Figure 4A). This modeling placed the substrate thioester carbonyl in position to form a hydrogen bond with an amide at the N-terminus of αHD1 (a conserved Pro-Gly/Ala/Ser), and the substrate β-OH in contact with the catalytic aspartate. These locations are likely important as the catalytic aspartate is almost certainly responsible for removal of the β-OH and the positive dipole of the N-terminus of αHD1 may play a role in stabilizing a proposed enolate intermediate (Scheme 1) (Schwab et al., 1986). While positioning of the α-C and β-OH is critical, exquisite substrate recognition of the polyketide portion of the chain is probably unnecessary, as the substrate is always delivered on a linked ACP domain (referred to as ‘channeling’). Consistent with this idea, the substrate tunnels are largely hydrophobic, generally lack hydrogen-bonding groups, and do not have shapes complementary to their presumed substrates (Figure 3A, Figure S2).
Scheme 1.
Figure 4. Active site and substrate chirality.
(A) Stereodiagram of CurK DH active site with electron density (1.7-Å 2m| Fo|-| Fc| contoured at 1σ). Active site residues (His996 and Asp1169) are shown with yellow C. Two waters, W1 hydrogen bonded to the N-terminus of αHD1 (Ala1006 amide) and W2 to active site Asp1169, were used when modeling substrate.
(B) Stereodiagram of CurK DH substrate (yellow ball and stick) and product (transparent yellow) modeled in the active site. The thioester carbonyl and β-OH oxygen are in the positions of W1 and W2 from (A). The CurK DH product has four conjugated double bonds (Figure 1G) resulting in a long rigid region. The orientation of the CurK DH monomer is similar to that of the CurJ DH monomer in Figure 3A.
(C) Stereodiagram of CurF DH active site with CurG intermediates (DH substrate and product) modeled as in (B). Relative to the CurK DH substrate in (B), the β-OH of the CurG DH substrate has inverted chirality, which is hypothesized to lead to formation of the cis double bond.
Each substrate/product combination presents a unique set of restrictions that must be accommodated within the active site. The products of all DH reactions contain α,β-double bonds conjugated with the thioester carbonyl. Additionally, the γ,δ-double bond of the CurH and CurJ substrates, and the γ,δ- and ε,ζ-double bonds of the CurK substrate result in multiply conjugated products (Figure 1C–G). Our substrate modeling did not predict which Cur DH acts on the CurG and CurI β-hydroxy intermediates, as these potential substrates would fit into any of the Cur DH tunnels. All DH substrates contain the thiazoline and cyclopropyl rings of curacin A (Figure 1C–G). While there is sufficient space to accommodate both rings within the substrate tunnels, there may not be enough space to thread them into the tunnel and past the active site constriction (Figure 3A, Figure S2).
Substrate modeling also provides insight into the relationship between substrate and product stereochemistry (Figure 4B,C). It has been proposed that the chirality of the β-OH determines the outcome of the dehydratase reaction (cis or trans) and the β-OH stereochemistry in turn is determined by the preceding action of the ketoreductase (Caffrey, 2003; Reid et al., 2003). Caffrey designated KR domains as A-type or B-type depending on the sequences of two motifs that were predicted to determine the chirality of the β-OH product (Caffrey, 2003). Products of A-type KRs are predicted to be dehydrated to cis double-bond products and B-type products to trans double-bonds (Reid et al., 2003). Consistent with these predictions, the CurH, CurI, CurJ and CurK KR domains are of the B-type and the modules yield trans double bonds whereas the CurG KR is an A-type and the presumed CurG intermediate has a corresponding cis double bond.
The structures of the four Cur DH domains are consistent with the hypothesis that the chirality of the substrate β-OH determines the configuration of the product α,β-double bond. In models of the products of A-type KRs (e.g. the CurG intermediate) into DH active sites, the substrate tunnel binds the polyketide tail in a cis-like conformation, thereby favoring the cis product (Figure 4C). In contrast, products of B-type KRs (e.g. the CurK intermediate) model into the substrate tunnels in a trans-like conformation, favoring the trans product (Figure 4B). The critical difference in substrate conformation results from the modeling constraint that the substrate β-OH should form a hydrogen bond with the catalytic aspartate, analogous to a highly ordered water molecule in the crystal structures (Figure 4A: W2). Intriguingly, the models predict that the catalytic histidine abstracts different α-protons in formation of cis and trans products: the pro-S proton in forming cis double bonds and the pro-R proton for trans double bonds, as demonstrated for the bacterial enzyme FabA (Schwab et al., 1986). The hypothesis that substrate identity determines product configuration is analogous to the mechanism proposed for the pikromycin thioesterase in which conjugation of double bonds in the substrate is believed to be a critical determinant between cyclization or hydrolysis of the final product (Akey et al., 2006). Our assessment of chiral determinants is similar to that proposed in the evaluation of the Ery4 DH structure (Keatinge-Clay, 2008).
Access to and Egress from the Active Site
Entrances to the tunnels of all four Cur DHs are near the C-terminus of the domain (Figures 3A and S2). The substrate tunnel is formed in part by three structural elements that cover the core of the C-terminal hotdog fold and are among the least ordered elements of the structures, with high temperature factors and poor electron density (Figure 3B). They include a small helix (αC1) leading to the cap β-sheet, the loop connecting the C-terminal hotdog helix to the β-sheet (αHD2—β10), and the small C-terminal 310 helix with its connecting loop. Elevated temperature factors and disorder were also observed in the homologous regions of the Ery4 DH and the prokaryotic FabZ DH structures (Keatinge-Clay, 2008; Kostrewa et al., 2005). We propose that these parts of the proteins are even more disordered in solution where the tunnel may be transiently uncovered, transforming it into an elongated trench with the active site at the bottom (Fig. 3F). If the tunnels can open, then a PKS intermediate may access the active site in two ways: either the substrate may be threaded through the tunnel entrance until the β-OH is proximal to the catalytic dyad, or the tunnel may open, allowing the substrate to settle onto the active site. Threading may be a topologically difficult maneuver given that many DH substrates are quite long and can contain a variety of sterically demanding and rigid functional groups, including formation of an alkene group from the DH reaction (e.g. the quadruple conjugated double bond system of the CurK DH product; Figure 1G, 4B). In contrast, opening of the tunnel (i.e. formation of a cleft) provides a simple and general mechanism for loading and unloading of substrates. Opening of the tunnel may be facilitated by specific interaction with ACP domains, providing a level of control on the activity of the DH domains. This mechanism of access does not affect stereochemical outcome of the DH reaction, as the restraints with regard to the cis and trans bond formation are determined by the bend at the active site and are not affected by the open or closed state of the lid.
Subunit interface
The PKS DHs form elongated dimers using residues in the two N-terminal β-strands (β1 and β2) and the β1-β2 connecting loop, as well as residues in strands β4 and β5 and the turn immediately preceding β6 (Figures 2, 5A). The interface between the two DH monomers is remarkably small, with buried surface areas of 560 Å2 for CurF DH, 645 Å2 for CurH DH, 765 Å2 for CurJ DH and 717 Å2 for CurK DH, accounting for only 4–6% of the total surface area. Nevertheless, the interface is formed by highly complementary and hydrophobic surfaces in all four DHs (Figure 2E, Figure 5A). The hydrophobic surfaces of the interface residues are completely buried in the subunit interfaces of the four proteins. All DH domains in PKS pathways are likely to form a similar dimer because the hydrophobic character of these positions is conserved. While relatively small, the subunit interaction is stable enough that the excised domains are dimeric in solution. The elongated shape of the DH dimer, with an axial ratio of nearly 3:1, accounts for the greater apparent molecular weight (~81 kDa) of all Cur DHs on gel filtration compared to the calculated molecular weights (~66 kDa).
Figure 5. DH dimer formation.
(A) Hydrophobic dimer interface. The backbone is rendered as a silver ribbon in this stereo diagram. Residues in the interface are shown in stick form with C atoms colored green for one monomer and yellow for the other. The view is along the dimer axis, similar to Figure 2C.
(B) PKS DH dimer. The dimer is fully extended with an angle of ~180° between the monomers. The N-termini are marked with blue spheres and the C-termini with red spheres. The substrate tunnel is shown for one monomer as a gray surface with yellow marking the entrance. The dimer axis is vertical in this view, and rotated ~90° about the horizontal axis with respect to Figures 2C and 5A.
(C) mFAS DH dimer. The dimer has an overall V-shape with an angle of ~90° between the monomers. The N- and C-termini and substrate tunnel are marked as in (B) with the tunnel entrance in orange. The dimer axis is vertical, as for the PKS DH dimer in (B).
(D) Superposition of the PKS and mFAS DH dimers. The dimer two-fold axes were superimposed and then rotated to match N-termini (blue spheres), C-termini (red spheres) and substrate tunnel entrances (yellow and orange surfaces). These key features of the domains can be superimposed despite the substantial difference in dimer shape. Figure S4 is a stereo view of this figure.
(E) Superposition of the PKS and mFAS DH dimers viewed along the dimer axis, perpendicular to (D). The PKS and mFAS DH dimers have different shapes. However, their dimer axes, N-termini, C-termini and tunnel entrances can be superimposed, indicating that the observed dimers can readily fit into multi-domain dimers of similar architecture.
Comparison to other dehydratases
DH monomers from PKS and mFAS pathways share a high degree of structural similarity. All the structures have the double-hotdog fold, similarly situated active sites, dimerization through N-terminal β-strands, and N-termini close to the dimer interface. The structural identity in so many details is remarkable given the low sequence identity of the proteins. As expected, structural similarity within the PKS DHs is reasonably high, with Cα rmsd values less than 2.0 Å for ~250 aligned residues (Table 2). Structural similarity correlates with sequence identity among the core aligned residues. Structure-based sequence alignments to the mFAS DH have 17% – 23% identity within the shorter regions (~200 residues) that can be aligned (Table 2). Nevertheless, the structural similarity is high within these alignable regions, with Cα rmsd values less than 2.2 Å (Table 2). The percent identities are lower outside the aligned regions. While the divergence from the prokaryotic fatty-acid DH, FabA, is higher, with 104 –105 aligned residues (11% – 12% sequence identity) to the N-terminal hotdogs and 113 – 119 residues (7% – 12% identity) to the C-terminal hotdog, the folds are well conserved with Cα rmsd values 2.2 – 3.2 Å.
Dimeric dehydratase within the PKS module
How does the DH dimer fit into a PKS module, comprising fused KS-AT-DH-KRs-ER-KRc-ACP domains? No structure of an intact PKS module, with domains homologous to the mFAS, is available. However, the elongation blocks of PKS modules and mFAS have nearly identical architectures (Maier et al., 2006; Maier et al., 2008; Tang et al., 2006), and thus PKS modules are also likely to be comprised of an elongation block and a modifying block, perhaps with a domain organization similar to the mFAS modifying block. It is particularly notable that the PKS DH dimers and the mFAS DH dimer differ substantially in the angle between subunits (Figure 5B,C). The angle between DH monomers is similar in the independent views of the DH dimers in the crystal forms presented here. Together with the Ery DH domain (Keatinge-Clay, 2008), all these PKS DH dimers have similarly extended conformations (Figure 5B). This extended conformation differs by ~90° from the V-shaped DH dimer observed within the mFAS megacomplex (Figure 5C). The V-shape of the mFAS DH domains seems to be important in properly orienting the downstream ER and KR domains (Figure 1B). While it is plausible that hinge motion in the dimer interface could result in a similar V-shape for the PKS DH domains, several observations suggest that the observed extended conformations of the excised domains may properly reflect the conformation in the PKS megacomplex. The extended conformation has been observed in all crystal structures reported here, as well as in the Ery4 DH domain. If the dimer interface is highly flexible, it is unlikely that eight crystal environments of five different DH domains would, fortuitously, display non-physiological interaction angles that are identical within a few degrees. In addition, the structures and sequences of the subunit interfaces of PKS and mFAS DH dimers differ substantially in detail (Figure S3). The dimer symmetry axes, domain N-termini, domain C-termini, and active-site tunnel entrances of PKS DH dimers can be superimposed on the corresponding structures of the mFAS DH dimer with no alteration of inter-subunit hinge (Figure 5D,E, Figure S4). This demonstrates that the relationship among key links to other domains and the access to ACP-delivered substrates can be identical with very different overall dimer shapes. Thus an extended DH dimer, as observed in the crystal structures, would not impede an mFAS-like domain architecture for the modifying block of the PKS module. Domain content differs among PKS modules and also between PKS and mFAS modules. The mFAS modifying domains include, in order, the DH domain, a cryptic methyltransferase-like (ΨMT) domain, a KRs, an ER and a KRc domain (Maier et al., 2008). PKS modules that contain a DH generally lack an MT domain, and frequently lack the ER domain. In addition, the sequences of peptides that link domains are substantially different in PKS and mFAS polypeptides, in which some linkers contribute secondary structures to catalytic domains. For example, the mFAS ΨMT domain is packed against the KRs domain, which is substantially smaller than the PKS KRs domain, and the DH-ΨMT linker contributes β-strands to the KRs domain (Maier et al., 2008). As PKS module composition is variable, sharing of secondary structural elements between domains cannot be conserved. Most PKS modules lack an MT domain, and the linkers are generally too short to contribute secondary structures to catalytic domains. Thus it should not be surprising that the orientation of DH domains may differ between PKS and mFAS systems.
Catalysis of “Orphan” Double Bonds
Given the new annotation of the curacin A biosynthetic pathway, we observe that double bonds formed in the extensions added by the CurG and CurI modules must be formed by DH domains in different modules. The structures of the four Cur DHs do not provide clues about which DHs form the “orphan” double bonds. The likely candidates for these reactions would be the DH domains from adjoining modules: the CurF or CurH DH for the CurG product and the CurH or CurJ DH for the CurI product. As the CurF polypeptide is not expected to catalyze a DH reaction, the CurF DH may play a role in the CurG extension. In contrast, an adjoining module whose DH domain forms the double bond within the CurI extension would be responsible for two DH reactions. Dehydrations catalyzed by DH domains within downstream modules should precede the downstream extension reactions if an enolate intermediate facilitates dehydration, as expected (Scheme 1). Visits of ACP domains to non-canonical active sites may have precedent elsewhere in the curacin PKS. The minor curacin products of the Curaçao strain of L. majuscula, curacin B and curacin C, differ from curacin A only in having cis double bonds at positions catalyzed by the CurJ and CurI modules, respectively (Yoo and Gerwick, 1995), raising the possibility that the CurI and CurJ ACPs occasionally visit the A-type CurG KR where they are primed for cis double bond formation. To the best of our knowledge, modification of extensions across module boundaries within PKS systems has not been detected previously.
Similarities between the four structures presented here and the reported structure of the Ery4 DH domain suggests that loading and unloading of intermediates is likely facilitated by reordering of mobile elements opening the active site tunnel. This feature may be shared by the mFAS DH domains. The dimer interface is consistent among PKS DHs and results in a shared architecture that differs from the mFAS DH in the angle between monomers. Lastly, modeling of substrates reveals that the deep bend at the active site and the positions of the catalytic histidine and aspartate side chains are likely responsible for the dependency of double bond conformation on the chirality of the β−OH substrate.
EXPERIMENTAL PROCEDURES
Cloning and expression
Plasmids pJRR038 (CurF: residues 1687–1968), pJRR187 (CurH: residues 929–1223), pJRR192 (CurJ: residues 936–1244), and pJRR198 (CurK: residues 958–1250) were generated by PCR amplification of the designated coding sequences from the pLM9 cosmid and insertion into pMCSG7 (Donnelly et al., 2006). Insert identities were confirmed by DNA sequencing. Expression plasmids were transformed into E.coli BL21(DE3) and plated onto LB agar plates containing 100 µg/mL ampicillin. Colonies were picked and used to grow 5-mL starter cultures overnight at 37°C. 2 mL of the starter cultures were added to 2-L baffled flasks containing 0.5 L of 4% glycerol TB media and grown at 37°C to an A600 of 0.9–1.2. The cultures were adjusted to 18°C for 1 hr with shaking, then augmented with IPTG at a final concentration of 0.1 mM, and grown 16–20 hr. Cells were harvested by centrifugation and the cell pellets were frozen immediately at −20°C. Selenomethionine-labeled CurH DH protein was expressed using the above protocol modified according to Guerrero et al. (Guerrero et al., 2001) in which cells from a 100-mL overnight culture were added to SeMet-supplemented minimal media prior to induction.
Protein purification
All purification steps, except the His-tag removal step, were performed at 4°C. Frozen cell pellets containing a given Cur DH were resuspended in 35 mL Buffer A (50mM Tris pH 7.5, 300mM NaCl, 10% glycerol) and lysed by sonication. Lysates were cleared by 45-min centrifugation at 28,600 g. Supernatants were filtered through 0.45-µm filters, loaded onto a 5-mL His Trap column (GE Healthcare) and washed with 10 column volumes of Buffer A. The proteins eluted at about 200 mM imidazole using a linear gradient of Buffer B (Buffer A plus 400 mM imidazole). Peak fractions were pooled. His-tags were removed from CurF DH, CurH DH and CurJ DH by addition of His-tagged tobacco etch virus protease (1 mg His-tagged TEV: 50 mg His-tagged DH domain) and 200 mM DTT with the reaction mixes incubated 2–4 hr at room temperature then dialyzed at 4°C overnight in Buffer C (Buffer A plus 200 mM DTT). The dialyzed mixtures were re-loaded onto the 5-mL His-trap column and flow-through fractions were pooled and concentrated. All proteins were further purified on a High Prep 16/60, Sephacryl, S200 High Res (GE Healthcare) column pre-equilibrated with Buffer C. The fractions were combined, concentrated to about 10 mg/mL, and frozen in liquid N2 or stored at 4°C.
Crystallization
Crystals were grown by vapor diffusion using the hanging-drop method at 20°C. Prior to crystallization, constructs were dialyzed against 10 mM HEPES pH 7.0, 2 mM DTT. Selenomethionyl CurH DH crystals were grown by equilibration of a 1:1 or 1:1.5 mix of protein stock (~8 mg/mL) and reservoir solution (15–20% PEG 4000, 10% PEG 400, 2 mM DTT, 100 mM bis-Tris propane pH 6.5) against the reservoir solution. Crystals usually formed in 2–3 days. CurJ DH crystals were grown by equilibration of 1:1 or 1:1.5 mix of protein stock (~8 mg/mL) and reservoir solution (18–23% PEG 3350, 2–10% 1,4 butanediol, 250 mM NaCl, 100 mM bis-Tris propane pH 6.5) against reservoir solution. CurJ DH crystal grew overnight, and multiple crystal forms were often present in the same drops. CurF DH crystals were grown by equilibration of 1:1 mix of protein stock (~10 mg/mL) and reservoir solution containing 14% (w/v) PEG 4000, 200 mM CaCl2, 10% (v/v) glycerol, 2 mM DTT and Tris pH 8.0. CurK crystals were grown by equilibration of 1:2 mix of protein stock (~8 mg/mL) and reservoir solution containing 1.3 M tri-sodium citrate, 30 mM (D)+sucrose, 100 mM tris pH 8.5. Crystals of all proteins were harvested directly from the drops and flash frozen in liquid nitrogen.
Data collection and structure determination
Data were collected at the GM/CA beamline 23-ID-D at the Advanced Photon Source (APS, Argonne National Laboratory). CurH DH crystals diffracted to 2.8 Å (Table 1) and were primitive monoclinic with four polypeptides in the asymmetric unit. The Wilson B factor for the CurH DH data is 81 Å2. The structure of SeMet CurH DH was solved from a two-wavelength MAD data set using SOLVE/RESOLVE (Terwilliger, 2003; Terwilliger and Berendzen, 1999) as implemented in the PHENIX software suite (Adams et al., 2002). All 20 expected Se sites were identified and the resultant phases had a figure-of-merit of 0.45. Density modification in RESOLVE using 4-fold non-crystallographic symmetry averaging resulted in phases with a figure-of-merit of 0.64. The CurF DH, CurJ DH and CurK DH structures were solved by molecular replacement using PHASER as implemented in PHENIX. Wilson B factors for CurF, CurJ and CurK data are 70 Å2 , 54 Å2 , and 24 Å2 , respectively. The CurF DH and CurK DH crystals contained four and two polypeptides, respectively, in the asymmetric unit. Two crystal forms for CurJ DH were solved, a primitive orthorhombic form with two chains in the asymmetric unit and a C-centered orthorhombic form with one chain in the asymmetric unit. The two crystal forms have identical CurJ DH dimers; only the primitive orthorhombic form is reported here. Disordered loops between β5 and β6 from the first hotdog fold (CurF DH chains A and D; CurJ DH both chains; CurK DH both chains), between β6 and αL1 in the linker (CurK DH chain B; CurJ DH both chains), between αHD2 and β10 in the second hotdog fold (CurF DH chains A, C and D; CurJ DH both chains) and between β11 and β12 (CurF DH chain A) were not modeled. All model refinement used PHENIX and REFMAC5 with TLS modeling of molecular motion, and model building was done with Coot (Adams et al., 2002; CCP4, 1994; Emsley and Cowtan, 2004; Murshudov et al., 1997; Winn et al., 2003). Four TLS groups per monomer, corresponding to the N- and C- terminal hotdogs, linker, and cap region, were defined for refinement. Structures were validated with MolProbity (Davis et al., 2007).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants DK-42303 to JLS and CA-108874 to DHS and WHG. GM/CA CAT is supported by the NIH National Institute of General Medical Sciences and the National Cancer Institute at the APS, which is supported by the US Dept. of Energy Office of Science. The authors thank Clay Brown and Jim DelProposto at the High-Throughput Protein Lab in the Center for Structural Biology at the University of Michigan for assistance with construct design and production.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structures factors have been deposited in the Protein Data Bank (http://rcsb/pdb) under the accession codes 3KG6 for CurF DH, 3KG7 for CurH DH, 3KG8 for CurJ DH, and 3KG9 for CurK DH.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Akey DL, Kittendorf JD, Giraldes JW, Fecik RA, Sherman DH, Smith JL. Structural basis for macrolactonization by the pikromycin thioesterase. Nat Chem Biol. 2006;2:537–542. doi: 10.1038/nchembio824. [DOI] [PubMed] [Google Scholar]
- Blokhin AV, Yoo HD, Geralds RS, Nagle DG, Gerwick WH, Hamel E. Characterization of the interaction of the marine cyanobacterial natural product curacin A with the colchicine site of tubulin and initial structure-activity studies with analogues. Mol Pharmacol. 1995;48:523–531. [PubMed] [Google Scholar]
- Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem Biol. 2003;10:723–731. doi: 10.1016/s1074-5521(03)00156-x. [DOI] [PubMed] [Google Scholar]
- Buchholz TJ, Geders TW, Bartley FE, 3rd, Reynolds KA, Smith JL, Sherman DH. Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains. ACS Chem Biol. 2009;4:41–52. doi: 10.1021/cb8002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- Castonguay R, Valenzano CR, Chen AY, Keatinge-Clay A, Khosla C, Cane DE. Stereospecificity of ketoreductase domains 1 and 2 of the tylactone modular polyketide synthase. J Am Chem Soc. 2008;130:11598–11599. doi: 10.1021/ja804453p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. D. 1994:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009a;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Wang B, Kulkarni A, Gehret JJ, Lloyd KR, Gerwick L, Gerwick WH, Wipf P, Hakansson K, Smith JL, Sherman DH. Polyketide Decarboxylative Chain Termination Preceded by O-Sulfonation in Curacin A Biosynthesis. J Am Chem Soc. 2009b doi: 10.1021/ja9071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero SA, Hecht HJ, Hofmann B, Biebl H, Singh M. Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl Microbiol Biotechnol. 2001;56:718–723. doi: 10.1007/s002530100690. [DOI] [PubMed] [Google Scholar]
- Helmkamp GM, Jr, Bloch K. Beta-hydroxydecanoyl thioester dehydrase. Studies on molecular structure and active site. J Biol Chem. 1969;244:6014–6022. [PubMed] [Google Scholar]
- Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay A. Crystal structure of the erythromycin polyketide synthase dehydratase. J Mol Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Winkler FK, Folkers G, Scapozza L, Perozzo R. The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum. Protein Sci. 2005;14:1570–1580. doi: 10.1110/ps.051373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure. 1996;4:253–264. doi: 10.1016/s0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- Schwab JM, Habib A, Klassen JB. A thorough study of the stereochemical consequences of the hydration/dehydration reaction catalyzed by β-hydroxydecanoyl thioester dehydrase. J. Am. Chem. Soc. 1986;108:5304–5308. [Google Scholar]
- Sherman DH, Smith JL. Clearing the skies over modular polyketide synthases. ACS Chem Biol. 2006;1:505–509. doi: 10.1021/cb600376r. [DOI] [PubMed] [Google Scholar]
- Smith JL, Sherman DH. Biochemistry. An enzyme assembly line. Science. 2008;321:1304–1305. doi: 10.1126/science.1163785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U S A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SC, Lu H, Cane DE, Khosla C, Stroud RM. Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases. Biochemistry. 2002;41:12598–12606. doi: 10.1021/bi0260177. [DOI] [PubMed] [Google Scholar]
- Tsai SC, Miercke LJ, Krucinski J, Gokhale R, Chen JC, Foster PG, Cane DE, Khosla C, Stroud RM. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc Natl Acad Sci U S A. 2001;98:14808–14813. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier-Pinard P, Lai JY, Yoo HD, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, et al. Structure-activity analysis of the interaction of curacin A, the potent colchicine site antimitotic agent, with tubulin and effects of analogs on the growth of MCF-7 breast cancer cells. Mol Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- Weissman KJ, Muller R. Protein-protein interactions in multienzyme megasynthetases. Chembiochem. 2008;9:826–848. doi: 10.1002/cbic.200700751. [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Yoo HD, Gerwick WH. Curacins B and C, New antimitotic natural products from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 1995;58:1961–1965. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.