Abstract
Cytochrome P450 (CYP or P450)-mediated drug metabolism requires the interaction of P450s with their redox partner, cytochrome P450 reductase (CPR). In this work, we have investigated the role of P450 hydrophobic residues in complex formation with CPR and uncovered novel roles for the surface-exposed residues V267 and L270 of CYP2B4 in mediating CYP2B4-CPR interactions. Using a combination of fluorescence labeling and stopped-flow spectroscopy we have investigated the basis for these interactions. Specifically, in order to study P450-CPR interactions, a single reactive cysteine was introduced in to a genetically engineered variant of CYP2B4 (C79SC152S) at each of 7 strategically selected surface-exposed positions. Each of these cysteine residues was modified by reaction with fluorescein-5-maleimide (FM) and the CYP2B4-FM variants were then used to determine the Kd of the complex by monitoring fluorescence enhancement in the presence of CPR. Furthermore, the intrinsic Km values of the CYP2B4 variants for CPR were measured and stopped-flow spectroscopy was used to determine the intrinsic kinetics and the extent of reduction of the ferric P450 mutants to the ferrous P450-CO adduct by CPR. A comparison of the results from these three approaches reveals that the sites on P450 exhibiting the greatest changes in fluorescence intensity upon binding CPR are associated with the greatest increases in the Km values of the P450 variants for CPR and with the greatest decreases in the rates and extents of reduced P450-CO formation.
The cytochromes P450 comprise a superfamily of heme-containing mono-oxygenases that play central roles in the metabolism of a wide variety of endogenous compounds including drugs and carcinogens. The oxidation of their substrates occurs through a series of steps that collectively constitute the P450 catalytic cycle. An essential component of the cycle is the delivery of two electrons from NADPH-CYP450 reductase (CPR). The first electron reduces the ferric heme to the ferrous state that binds molecular oxygen to produce an oxyferrous complex. The second electron leads to the formation of the peroxo intermediate, which is protonated twice: the first proton serves to yield the hydroperoxo intermediate, and the second to cleave the oxygen-oxygen bond. Finally, oxidation of the substrate by an oxyferryl intermediate leads to product release and regeneration of the resting state of the enzyme(1,2).
The crystal structure of rat CPR reveals that an FMN-binding domain, an NADPH/FAD-binding domain and a “linker” domain compose the three catalytic regions of the enzyme(3–5). The FAD serves as an electron acceptor from NADPH, whereas the FMN-binding domain interacts with P450 to transfer electrons. Although we have known for some time that the P450-CPR interaction is required for P450 catalysis, details are lacking with respect to the fundamental principles that govern redox partner recognition. What is the molecular basis for the rapid association and dissociation of the P450-CPR complex that allows for efficient electron transfer? Since the ratio of CPR to P450 in the endoplasmic reticulum is estimated to be 1:20(6,7), how does CPR orchestrate the recognition and reduction of not only multiple P450 isoforms but cytochrome b5(8), cytochrome c (9), heme oxygenase(10) and squalene monoxygenase(11) as well?
Currently, it is thought that the association of P450s with CPR is largely driven by charge-pairing interactions between a small cluster of positively charged amino acid residues located on the surface of the side at which the prosthetic heme is ligated to the P450 (proximal), and another cluster of negatively charged amino acid residues located on the FMN domain of CPR(3,12,13). For example, Shimizu et al. mutated a series of Lys and Arg residues in order to identify the binding site of CPR on 1A2. Their studies showed that the mutated residues may form charge interactions with CPR and/or orient the two proteins for functional interaction(14). Some of these surface-exposed residues were on the proximal side of CYP2B4 and correspond to residues Arg-422, Lys-433, and Arg-443 in CYP2B4, which were investigated extensively by Bridges et al. using alanine-scanning mutagenesis(15). In addition to these three basic residues, Bridges et al.’s work demonstrated that CYP2B4 R122A, R126A, R133A, and K139A variants exhibited dramatically decreased binding to CPR. The use of chemical cross-linking reagents coupled with mass spectrometry to investigate CYP interaction with its redox partner has also been recently evaluated(16,17). These studies have shown that key lysine residues on the proximal side of P450 are involved in complex formation. For instance, it has recently been demonstrated through chemical cross-linking studies that residues presumed to be located in the C-helix of CYP2B6 form contacts with residues located in the linker region between the FAD and FMN domains(17). These same residues in the C-helix of CYP2B4 have been shown by mutagenesis to be involved in complex formation.
Although it is generally accepted that the site at which CPR interacts with P450s involves the proximal region of the P450, much of our knowledge regarding the specific residues that mediate this interaction stems from a very limited number of mutagenesis studies. Furthermore, most of these experimental studies have focused on mutating charged residues to neutral ones and measuring changes in activity of the CPR-P450 complex in attempts to identify these residues. A central assumption in these studies is that the mutation does not alter electron transfer and/or the heme’s catalytic integrity. However, since CPR affinity is measured indirectly through substrate turnover, it is not possible to distinguish the effect of mutagenesis on protein activity from CPR-P450 complex formation by relying exclusively on steady-state measurements. Additionally, the significant biochemical and crystallographic work performed on the P450cam and P450BM3 redox complexes suggests the likelihood that ionic interactions do not exclusively dominate protein-protein contact regions in all P450s(18). The structure between the heme domain of P450BM3 and its FMN domain is the only available P450-redox partner complex structure and reveals that only a single intermolecular ionic bond mediates domain interaction(19). The shared role that hydrophobic and ionic interactions play in redox proteins was also demonstrated in the recent crystal structure of the Pdx-Pdr complex which showed that the interface includes two potential ionic interactions but is predominantly hydrophobic(20).Work from the Schenkman lab has questioned the universality of electrostatic pairing interactions and the extent that such interactions contribute to P450-CPR complex formation(21,22). Their results have shown that increasing ionic strength actually stimulated P450 reduction in an isoform dependent manner, thus suggesting that electrostatic forces inhibit the P450-CPR interaction. These observations, in part, inspired us to investigate the role of hydrophobic residues in P450-CPR interactions.
In an effort to experimentally identify the CPR-P450 interface and the residues that dictate functional interaction at this interface, it is essential that the changes measured by such investigations result directly from protein-protein interactions. Fluorescent probes that can be chemically attached to various sites on P450 and that exhibit varying fluorescence intensities as a function of their proximity to the CPR-P450 binding site are powerful tools for mapping protein-protein interfaces(23–25). Additionally, measurements of the kinetics of the reduction of ferric P450 by CPR using a stopped-flow spectrophotometer can be used to determine whether the effect of site-directed mutagenesis is on protein binding or electron transfer. Since the rate of reduction of P450 by CPR can be measured by mixing a preformed P450-CPR complex with NADPH in the presence of CO, then the mutation of residues on P450 that mediate P450-CPR complex formation should decrease the rate of reduction and the extent of P450-CO complex formation.
The results presented in this work demonstrate that when fluorescein-5-maleimide (FM) is attached to the proximal side of CYP2B4 and incubated with excess CPR, this region shows the largest fluorescence enhancement compared to studies where FM was adducted to residues located in regions distal and perpendicular to the axis of the heme-thiolate ligation. The mutation of hydrophobic residues to cys on the proximal side of 2B4 reduced the extent and rate of P450-CO complex formation to virtually the same extent as a positively-charged proximal residue (R133) that was also mutated to cys and was previously reported to be the single largest contributor to P450-CPR interaction(15).
To our knowledge, this is the first study that circumvents the limitations inherent in identifying residues at protein-protein interfaces exclusively by using mutagenesis and steady-state measurements by complementing those studies with stopped-flow and fluorescence spectroscopy. These results identify a novel role of hydrophobic residues V267 and L270 of CYP2B4 in CPR recognition and suggest that our approach will be informative for acquiring a detailed molecular understanding of protein-protein interactions.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals used are of ACS grade unless otherwise specified and obtained from commercial vendors. Benzphetamine, NADPH, sodium dithionite and tert-butyl hydroperoxide were purchased from Sigma. Trifluoroacetic acid and fluorescein-5-maleimide were purchased from Pierce Chemicals. Dilauroylphosphatidylcholine (DLPC) was purchased from Doosan Serdary Research Laboratory (Toronto, Canada). Carbon monoxide gas (purity >99.5%) was purchased from Cryogenic Gases (Detroit, MI).
Construction of CYP2B4 variants
Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit according to the manufacturer’s protocol (Stratagene). The forward and reverse mutagenic primers for C79S, C79SC152S, and the other 7 variants are listed in Supporting Information Table S1. These other 7 variants used C79SC152S as a template plasmid with a single additional mutation: V267C, L270C, L420C, R133C, Y484C, H226C or E60C. Each site-specific mutation was confirmed by DNA sequencing at the University of Michigan DNA Sequencing Core.
Overexpression and Purification of CYP2B4 WT and variants
CYP2B4, its variants and CPR were expressed and purified from Escherichia coli as described previously(26). The concentrations of CYP2B4 and its variants were determined using an extinction coefficient of Δε450–490 nm of 91 mM cm−1 as described by Omura and Sato(27). The concentration of CPR was determined using an extinction coefficient of 21 mM cm−1 at 456 nm for the oxidized enzyme(28).
Labeling of CYP2B4 variants with FM
FM was dissolved in 5% DMF and prepared as a 10 mM stock in 0.1M potassium phosphate buffer, pH 7.1. To determine the optimal concentration for labeling, aliquots of the FM stock solution were added to solutions of CYP2B4 (0.83 µM) in opaque Eppendorf tubes (to protect from light) such that the final concentration of FM was 50, 100, 200, 500 and 1000 fold over the concentration of CYP2B4 and the total volume was 400 µL. The protein samples were either allowed to sit at 4 °C overnight or for 2 h at room temperature. Any precipitated FM was then removed by centrifuging at 13.2k rpm for 5 min and the molecular masses of the labeled and unlabeled samples were then analyzed using an LCQ ion trap mass spectrometer (ThermoFinnigan, Inc.). Once the optimal concentrations of FM required for labeling each variant were identified, the above steps were repeated on larger samples and the proteins were subsequently dialyzed extensively to remove any free FM from solution. The FM-labeled proteins were dialyzed for 4–5 hr at a time for a number of times and after each dialysis, the fluorescence emission of the dialysis solution was measured to determine the presence of unreacted FM. The dialysis was considered complete when FM could no longer be detected in the dialysis solution
ESI-LC-MS Characterization of the FM Labeled CYP2B4 variants
The CYP2B4 samples (50 µL) were injected onto an Agilent Zorbax 300SB-C3 column (3.0 × 150 mm; Agilent, Santa Clara, CA). ESI-LC-MS was carried out using a ThermoFinnigan LCQ ion trap mass spectrometer interfaced with a Hewlett Packard 1100 series HPLC system (Hewlett Packard, Palo Alto, CA). The sheath gas was set at 90 (arbitrary units) and the auxiliary gas was set at 30 (arbitrary units). The spray voltage was 3.5 kV and the capillary temperature was 200 °C. The flow rate was 0.2 ml/min and the initial conditions were 70% 0.1% TFA (v/v) in water (solvent A) and 30% of 0.1% TFA (v/v) in acetonitrile (solvent B). The percentage of B was maintained at 30% for 5 min followed by a linear gradient to 90% B from 5 min to 35 min and maintained at 90% B for another 10 min. The gradient was then lowered from 90% to 30%B in 5 min and held for 15 min to equilibrate the column.
Characterization of CYP2B4FM Variants
The Kd reported for the CPR-P450 complex is in the range from 0.02 – 0.1 µM (15,29–31). Thus, to ensure that under our experimental conditions >95% of the CYP2B4FM variants were complexed with CPR, final concentrations of 3 µM CPR, 0.1 µM CYP2B4FM and 0.1 mg/ml DLPC were first reconstituted overnight at 4°C and then diluted to a final volume of 600 µL in 50 mM potassium phosphate, pH 7.4. Fluorescence measurements of the CYP2B4FM variants were performed on a Shimadzu RF – 5301PC spectrofluorophotometer (Columbia, MD) using 10 nm excitation and 10 nm emission bandwidth slits. The excitation wavelength was set at 490 nm and the emission spectrum was obtained by scanning from 495 nm to 600 nm. The emission intensity of the blank (in the absence of CPR) was subtracted from the intensity in the presence of 3 µM CPR and that difference was divided by the fluorescence intensity in the absence of CPR to give a percent intensity enhancement for each CYP2B4FM – CPR variant complex. The percent fluorescence enhancement of each CYP2B4FM – CPR variant complex was then normalized to the percentage fluorescence enhancement recorded for CYP2B4FM (L420C) to give a relative measure of the fluorescence change as result of reconstituting with CPR.
Determination of the Apparent Kd Values of the CYP2B4FM Variants for CPR
The apparent dissociation constants of the CYP2B4FM variants for CPR were determined by measuring the difference in fluorescence intensity of the CYP2B4FM variants in the presence and absence of varying concentrations of CPR. These data were normalized to the fluorescence intensity obtained from the concentration of CPR that yielded a plateau in fluorescence enhancement. The fluorescence of the 0.05 µM CYP2B4FM variants was individually recorded in the presence of 0.1 mg/ml DLPC and 50 mM potassium phosphate pH 7.4 at 22 °C to obtain initial intensity (Ii) at the emission maximum. Increasing concentrations of CPR were reconstituted overnight at 4°C in the presence of 0.05 µM of the CYP2B4FM variant and 0.1 mg/ml DLPC. These samples were diluted in 50 mM potassium phosphate, pH 7.4, to 600 µL final volume and the emission of the mixture was recorded to obtain (I). The fluorescence intensity obtained from the concentration of CPR that produced the highest fluorescence intensity was recorded to obtain maximum intensity (Imax).
Assuming the formation of a 1:1 complex, the equilibrium dissociation between CYP2B4FM and CPR can be described by the equation:
The equilibrium dissociation constant, or Kd is given by the equation:
| Eq.1 |
where [CYP2B4FM] and [CPR] are the free concentrations of CYP2B4FM and CPR, respectively. This can be rewritten as:
| Eq.2 |
where F is obtained from the equation:
| Eq.3 |
Kd can be obtained by plotting F vs [CPR] and fitting the data to a non-linear regression using GraphPad Prism 5.0 from GraphPad software.
Determination of the Apparent Km and kcat Values for CYP2B4 WT and Variants for CPR Using the N-demethylation of Benzphetamine in the Absence of FM
The Km and kcat values for CYP2B4 WT and its variants for CPR were determined at 30°C by measuring the rate of formaldehyde formation as a result of the N-demethylation of benzphetamine at a constant P450 concentration with increasing concentrations of CPR. CYP2B4 WT and its variants (0.2 µM) were reconstituted in triplicate with varying concentrations of CPR (0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 1.0 and 1.2 µM) and 0.1 mg/ml DLPC on ice for 1 h. The reconstituted mixtures were then added to 50 mM potassium phosphate buffer, pH 7.4, and 1 mM benzphetamine. After the samples were equilibrated at 30°C for 15 min, the reactions were initiated by adding 7.5 µL of 20 mM NADPH for a final reaction volume of 500 µL. The reactions were allowed to proceed for 5 min at 30 °C before they were quenched by the addition of 25 µL of 50% TFA. The protein was precipitated by centrifugation at 13.2k rpm for 5 min and a 500 µL aliquot of the supernatant was assayed for formaldehyde using the Nash reaction(32). The kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using GraphPad Prism 5.0 from GraphPad software (La Jolla, CA).
Characterization of the tert-butyl hydroperoxide-supported Metabolism of Benzphetamine by CYP2B4 WT and its Variants in the Absence of FM
To determine the rates for the tert-butyl hydroperoxide–supported metabolism of benzphetamine, final concentrations of 0.25 µM CYP2B4 WT and its variants were each incubated with 0.1 mg/ml DLPC, 50 mM potassium phosphate buffer, pH 7.4, and 1 mM benzphetamine at 30 °C for 15 min. 52.5 µL of 1 M tert-butyl hydroperoxide were added to give a final volume of 500 µL and the reactions were allowed to proceed for 5 min after which they were terminated by the addition of 25 µL of 50% TFA. The samples were centrifuged at 13.2k rpm for 5 min and 500 µL aliquots of the supernatants were assayed for formaldehyde using the Nash reaction(32).
Determination of the Rate of Electron Transfer from CPR to CYP2B4 WT and its Variants in the Absence of FM
In order to investigate the effects of the mutations on the interactions between the CYP2B4 variants and CPR, the rates of the first electron transfer from CPR to the CYP2B4 WT and its variants were measured. The rates of reduction were determined using a stopped-flow spectrophotometer (Hi-Tech SF61DX2, TgK Scientific, Bradford-on-Avon, UK) to monitor the increase in the absorbance at 450 nm as a result of the formation of the ferrous CYP2B4-CO adduct following reduction of ferric CYP2B4 by CPR at 30 °C. One syringe contained CYP2B4 WT or a variant (3 µM), CPR (3 µM) and DLPC (0.15 mg/ml) that had been reconstituted on ice for 1 hr before being diluted in 0.1 M potassium phosphate buffer, pH 7.4, to a final volume of 1.5 mL. This mixture was then rapidly mixed with an equal volume of 0.1 M potassium phosphate buffer, pH 7.4, and 0.1 mM NADPH from another syringe. Both solutions had been saturated with CO by passing a gentle stream of CO gas over the sample solutions. The increase in absorbance at 450 nm was monitored over time for 100 sec. The data were then analyzed to determine the apparent rate constants and amplitudes for the rates of electron transfer from CPR to CYP2B4 WT and its variants by fitting the absorbance changes at 450 nm to a three-exponential equation using the KinetAsyst software (Bradford-on-Avon, UK).
RESULTS
Rationale for the Selection of Solvent Exposed Residues for FM Labeling
CYP2B4 is the paradigmatic mammalian P450. It can be expressed and purified in relatively large quantities(15) and has been studied extensively from a structure-function point of view(33). Furthermore, a crystal structure is readily available from the Brookhaven National Laboratory Protein Data Bank site (PDB ID 1suo) (34) which, along with the work of Bridges et al.(15), guided the selection of solvent exposed residues for mutagenesis.
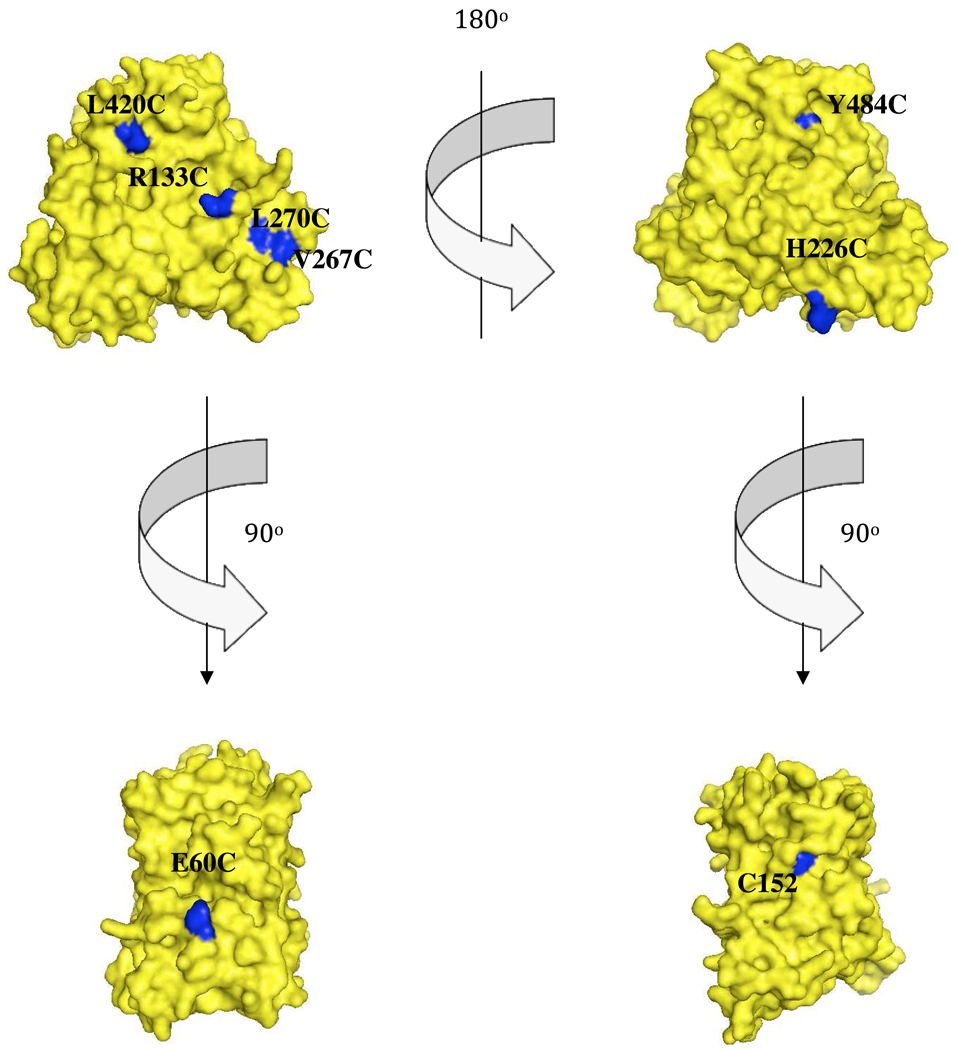
CYP2B4 WT possesses four cys residues, two of which (C79 and C152) are solvent exposed. When the WT enzyme was incubated with FM and analyzed using ESI-LC/MS, an increase in molecular mass corresponding to the adduct formation by reaction of two FM molecules was observed. Since C152, but not C79, was in a position of interest for our current study, C79 was mutated to a Ser in order to block maleimide labeling, thus leaving C152 available for investigation (Figure 2). The additional residues that were selected for FM labeling following cys mutagenesis were E60, H226, Y484, V267, R133, L270 and L420 (Figure 2). A double cysteine to serine mutation (C79S/C152S) was generated to serve as a starting point for studying the mutation of residues E60, H226, Y484, V267, R133, L270 and L420 to cys. For simplicity, a specific CYP2B4 such as C79S/C152S/E60C is denoted as CYP2B4 (E60C), while the single C79S mutation is simply denoted as CYP2B4 C79S. All variants containing the FM abbreviation (eg. (V267C)FM) are P450s that are chemically labeled by fluorescein-5-maleimide.
Figure 2.
Selection of solvent-exposed residues for FM labeling. van der Waal surface representations of CYP2B4 (PDB 1SUO) in the upper left and right corners display the proximal and distal regions of CYP2B4 respectively. The lower left and right structures represent regions perpendicular to the axis of the heme-thiolate ligation. The residues that were selected for cysteine mutagenesis and subsequent FM labeling are shown in blue, while the rest of the structure is shown in maize. Since WT CYP2B4 possesses two surface-exposed cysteines (C79 and C152) almost all our studies required generating a C79S/C152S template to which cysteine residues were introduced at sites of interest. However, residue C152 (lower right) was already in a position of interest and studying this site simply required replacing C79 with serine.
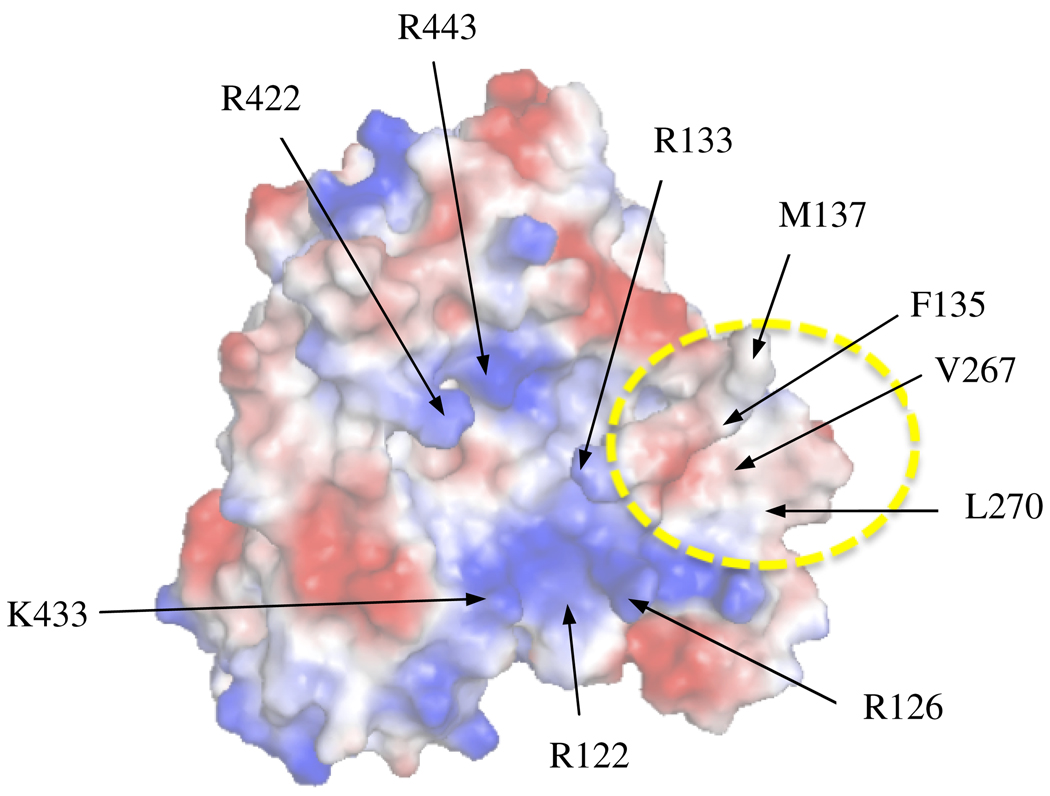
In order to determine which hydrophobic residues would be selected for site-directed mutagenesis studies on CYP2B4’s proximal region, we performed an APBS surface electrostatic plot of CYP2B4’s crystal structure (PDB code 1SUO). This analysis revealed that the solvent-exposed proximal region is composed of a variety of charged and neutral residues (Figure 1). Specifically, the center of the proximal region is mostly composed of hydrophobic residues that are surrounded by a series of Arg and Lys. Most notably, we observed a relatively large and concentrated patch of hydrophobic residues near R126, R133 and K274. The hydrophobic patch consists of the following residues: A123, F127, F135, G136, M137, P258, P261, F264, V267, L270 and F283. Of these residues, L270 and V267 are not within van der Waal contact with other amino acids and their roles in CPR binding have yet to be characterized. Therefore, the absence of significant intermolecular interactions between V267 and L270 and their surrounding residues made them good candidates for mutagenesis.
Figure 1.
Surface electrostatic plot of the proximal region of CYP2B4. Electrostatic surface of CYP2B4 as determined by APBS which shows the central basic residues proposed to be involved in CPR binding and a hydrophobic patch that includes V267 and L270 indicated by the dashed yellow circle. Red represents acidic residues while blue represents basic residues on the surface of CYP2B4.
In contrast, Bridges et al studied the role of the basic proximal residue R133 in mediating protein-protein interaction extensively(15). Their study showed that mutating this residue to Ala had the largest impact on increasing the apparent Kd for the CYP2B4-CPR complex. Thus R133 was also chosen for mutation to cys due to its previously demonstrated role in complex formation. Furthermore, two more residues, H226 and Y484 were chosen for cys mutagenesis to investigate the possibility that the distal side of CYP2B4 might serve as a docking site for CPR(35).
Generation of CYP2B4 Variants
The site-directed mutations were produced as described in Experimental Procedures. As evidenced by a reduced CO difference spectrum, all of the mutants absorbed predominantly at 450 nm with minimal absorption at 420 nm (~10%). Chemical labeling of the C79S and triple mutants by FM followed by ESI-LC/MS analysis showed that the MW of the variants increased by a mass which corresponded to the adduction of a single FM label and thus suggested they both had a single solvent exposed cysteine. The WT incorporated 2 FM labels while C79SC152S was completely resistant to labeling.
Labeling of the CYP2B4 Mutants with FM
In order to study directly the binding interface of the CYP2B4-CPR complex, we chose FM because it combines an environmentally sensitive fluorescent moiety (fluorescein) (24) with the labeling specificity of the maleimide group (36). Monitoring the degree of labeling using ESI-LC/MS allowed us to optimize our reaction conditions. Typically, this involved incubating CYP2B4 variants (0.83 µM) each with 41.5, 83, 166, 415, and 830 µM of FM overnight at 4°C, as described in Experimental Procedures. Each CYP2B4-FM was dialyzed against 0.1M potassium phosphate buffer, pH 7.4, until all free flouresceine was removed from solution. The crystal structure of CYP2B4 in the closed conformation shows that there are two native solvent-exposed cysteine residues (C79 and C152) and incubation of the WT with FM confirms this observation by incorporating two FM labels. As expected, when C79S and the triple mutants were incubated separately with FM they each reacted with the FM to give a single adduct to the protein, as measured by the increase in mass, while C79SC152S showed no increase in mass This is evidenced by the presence of a peak that corresponds to the mass of a singly labeled CYP2B4-FM variant (± 50 Da) and the absence of a peak that corresponds to an unlabeled CYP2B4 variant (data not shown). The lowest concentration of FM that produced the highest degree of labeling in the given amount of time was chosen for large scale labeling.
Fluorescence Characterization of the CYP2B4FM variants with CPR
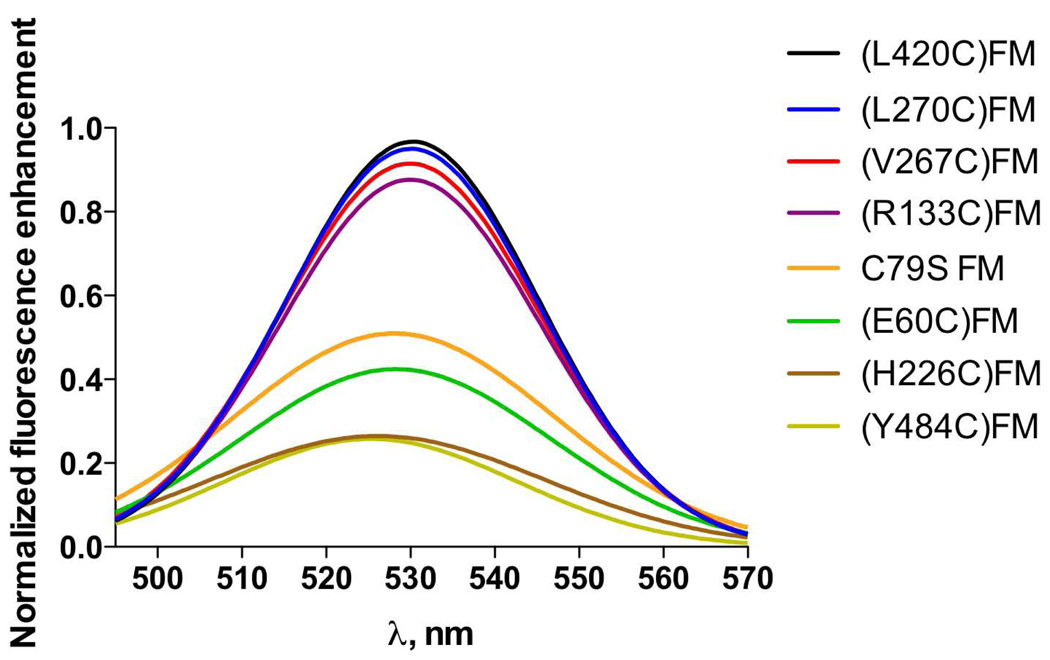
In an attempt to study the protein-protein interactions at the binding interface of the CYP2B4-CPR complex, a fluorescent probe that has previously been shown to be sensitive to changes in its local environment was attached to solvent exposed cys residues on the surface of CYP2B4. When reconstituted with DLPC and CPR, we observed changes in the fluorescence emission intensity of the CYP2B4FM variants which were dependent on the position of the label. For example, the variants of CYP2B4 labeled on the (L420C), (L270C), (V267C) and (R133C) showed the largest increases in fluorescence intensity in the presence of CPR (Figure 3). In contrast, the variants of CYP2B4 labeled on the distal side, (H226C) and (Y484C), showed virtually no fluorescence enhancement, while CYP2B4 (E60C) and C79S showed changes that were intermediate between those observed for the proximal and distal variants. To test whether the CYP2B4-FM fluorescence enhancement is the result of specific binding of CPR to CYP2B4 and not due to random protein-protein interactions, we tested whether Bovine Serum Albumin (BSA) could, under the same conditions, reproduce the results observed with CPR. The addition of up to a fifty-fold excess of BSA only enhanced the fluorescence emission by 5%, thus demonstrating that the results observed in Figure 3 are specific to the interactions of CPR with CYP2B4FM to form a complex (data not shown). To further investigate the specificity of these results, we performed titration studies with CPR to see if the increase in the fluorescence enhancement with increasing CPR would allow us to determine dissociation constants for the complex.
Figure 3.
Fluorescence emission spectra for all the FM-labeled CYP2B4 variants (0.05 µM) in the presence of 3 µM CPR, 0.1 mg/ml DLPC at 23°C, pH 7.4. When reconstituted with excess CPR, the proximal variants showed the greatest increases in fluorescence enhancement, the distal variants showed the lowest enhancement, while the variants on regions perpendicular to the heme-thiolate ligand exhibited enhancements that were intermediate between the proximal and distal variants.
Determination of the Kd Value for the CPR-CYP2B4-FM Complex using Fluorescence Titration
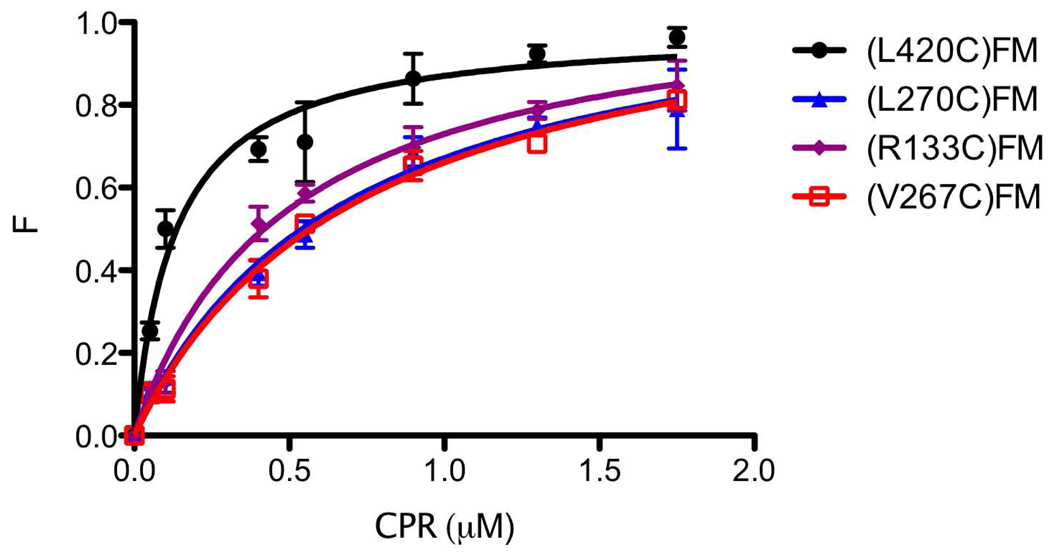
Previous studies have shown that fluorescent compounds can be used as probes to understand the quantitative basis for protein-protein interactions(31). Since (L420C), (L270C), (V267C) and (R133C) variants of CYP2B4FM showed the largest enhancements in fluorescence emission, they were used to determine the dissociation constants for the complex. The results are presented in Figure 4 and Table 1. (R133C)FM, (L270C)FM and (V267C)FM showed 3.8, 5.2 and 5.6 fold increases respectively in their apparent Kd for CPR compared to (L420C)FM. This difference is indicative of weaker binding of CPR to these variants. The results obtained with (R133C)FM in this assay parallel those for the Ala mutagenesis study reported by Bridges et al (15). That is, R133 appears to be an important residue mediating CPR binding. Thus, this residue serves as an excellent positive control for our approach and the titration data suggests that L270 and V267 may also be implicated in the interaction of CPR with CYP2B4. If, like R133, L270 and V267 constitute part of the binding interface, we hypothesize that the Km’s of these variants for CPR should also be higher than those for all other variants and WT.
Figure 4.
Plot of F vs. [CPR] for the CYP2B4-FM proximal variants used to determine the Kd values for CPR. Measurements were carried out in 0.1M potassium phosphate buffer, pH 7.4, with 0.1 mg/ml DLPC at 23°C as described under “Experimental Procedures”. Error bars are the standard deviations for three measurements.
Table 1. Values for the Kd’s determined for the interactions between the CYP2B4-FM variants on the proximal side of the protein and CPR by fluorescence titration.
| CYP2B4-FM variant | Kd |
|---|---|
| (L420C)FM | 0.13±0.02 |
| (L270C)FM | 0.68±0.09 |
| (R133C)FM | 0.49±0.05 |
| (V267C)FM | 0.73±0.07 |
The fluorescence emission of the 0.05 µM CYP2B4FM proximal variants were recorded in the presence of 0.1 mg/ml DLPC and 50 mM potassium phosphate, pH 7.4, at 22 ° C. The CYP2B4FM samples were then reconstituted with increasing concentrations of CPR. The resulting titration curves were fit to the Michaelis-Menten equation from which Kd’s were determined.
Determination of the Km and kcat Values for the Interactions of CYP2B4 WT and its Variants with CPR in the Absence of FM
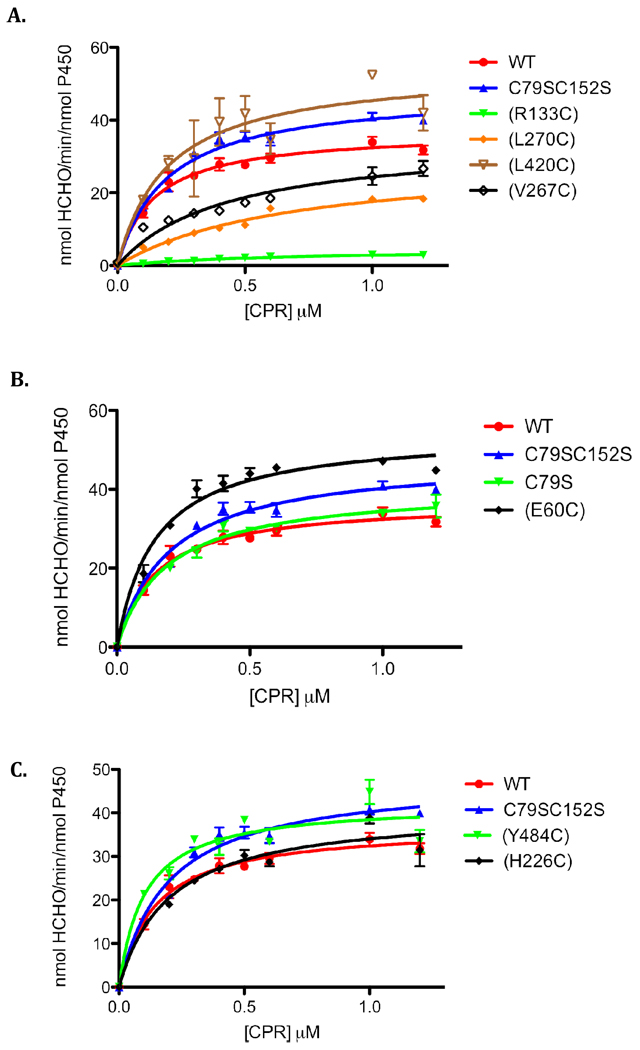
Complex formation between CPR and CYP is an essential step in the transfer of electrons to the heme that is required for substrate oxidation. It has been proposed that the rate of substrate oxidation is directly proportional to the concentration of the CPR-CYP2B4 complex(15,29). Thus, by measuring the rate of benzphetamine oxidation in the presence of a fixed concentration of CYP2B4 and increasing concentrations of CPR, Km and kcat values can be determined. By comparing these values with the Kd values that were determined by fluorescence spectroscopy, we can investigate the effect of the mutations on CPR affinity in the absence of FM. The mutation of a residue involved in redox partner recognition should disrupt complex formation and increase the concentration of uncomplexed CYP2B4 which is not able to oxidize substrate. As shown in Figure 5 and Table 2, the rate of benzphetamine oxidation as a function of varying CPR concentrations follows a rectangular hyperbolic relationship, which can be fit to the Michaelis-Menten equation.
Figure 5.
Determination of the apparent Km and kcat values of the CYP2B4 (A) proximal, (B) side and (C) distal variants.The N-demethylation of benzphetamine to produce formaldehyde (HCHO) was measured at a constant concentration of CYP2B4 (0.2 µM) with increasing concentrations of CPR as described under “Experimental Procedures”. Error bars are the standard deviations from three measurements.
Table 2. Km’s and kcat’s determined for CYP2B4 WT and its variants and CPR.
| 2B4 | Km (µM) | kcat (min−1) |
|---|---|---|
| WT | 0.14±0.022 | 37±1.5 |
| C79SC152S | 0.19±0.023 | 48±1.7 |
| C79S | 0.18±0.025 | 41±1.6 |
| (E60C) | 0.14±0.019 | 54±1.9 |
| (H226C) | 0.20±0.036 | 41±2.3 |
| (Y484C) | 0.10±0.024 | 42±2.1 |
| (V267C) | 0.43±0.10 | 34±3.1 |
| (R133C) | 0.65±0.11 | 4.7±0.40 |
| (L270C) | 0.69±0.12 | 30±2.5 |
| (L420C) | 0.19±0.064 | 54±5.2 |
As described under “Experimental Procedures”, the kinetic parameters were determined by measuring the rate of formaldehyde formation as a result of CYP2B4-mediated N-demethylation of benzphetamine under fixed concentrations of CYP2B4 and excess benzphetamine with increasing concentrations of CPR.
It is apparent that the majority of the mutations did not significantly affect Km or kcat. Most notable, however were the significant increases in the Km values observed for CYP2B4 (R133C), (L270C) and (V267C) as well as a decrease in the kcat value for (R133C). As expected, the Kd’s observed in Table 1 agree with the increases in the Km values and this agreement suggests that hydrophobic residues V267 and L270, in addition to charged residues, may also mediate CPR-CYP2B4 interactions. Since the results obtained from this assay rely on substrate metabolism, it might also be argued that a decrease in benzphetamine oxidation may be attributed to an alteration in the P450 active site instead of decreases in the concentrations of the CPR-CYP2B4 variants complex. Such alterations can be assessed by determining the tert-butyl hydroperoxide-supported metabolism of benzphetamine by the CYP2B4 WT enzyme and its variants.
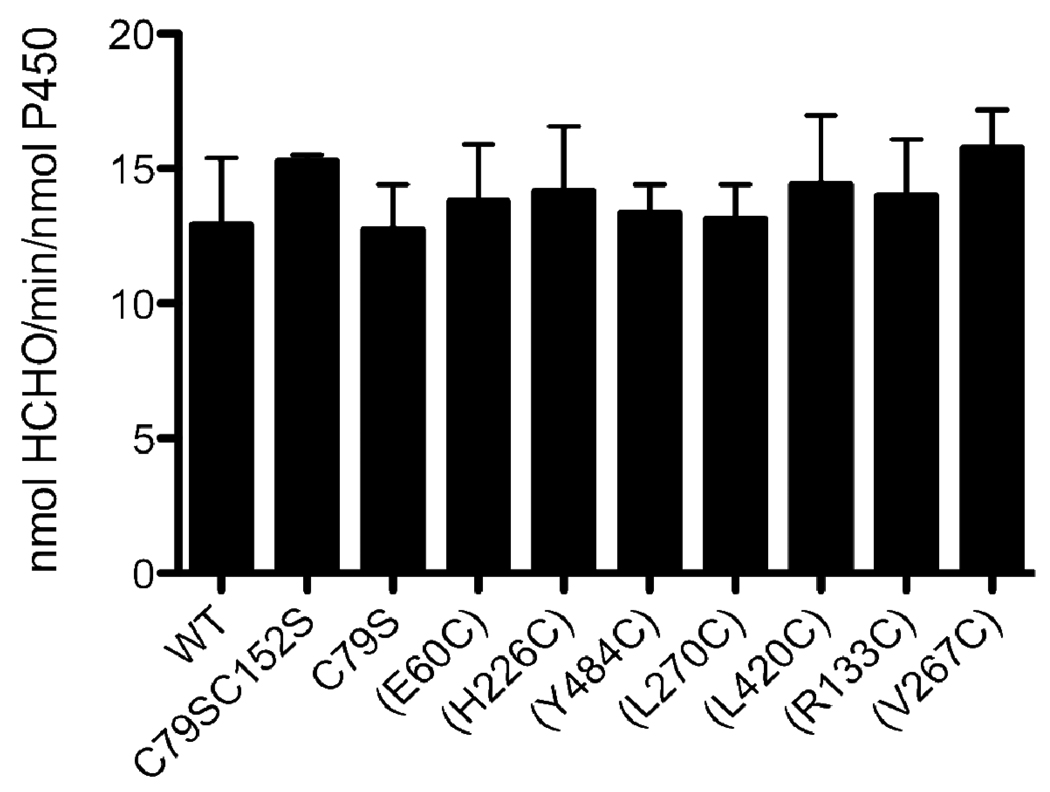
Characterization of the tert-butyl hydroperoxide-Supported Metabolism of Benzphetamine by the CYP2B4 WT Enzyme and its Variants in the Absence of FM
In order to determine whether the increase in Km is due to disruption of the interaction of CYP2B4 with CPR, or another step in the catalytic cycle, the benzphetamine catalytic activity supported by tBHP was characterized and compared to that supported by CPR. As shown in Figure 5 and table 2, CYP2B4 (R133C), (L270C) and (V267C) exhibited increases in the Km values when titrated with CPR. However when the reaction was performed in the presence of tBHP none of the variants displayed any significant changes in benzphetamine activity (Figure 6). Since hydroperoxides and other artificial oxygen donors are able to support the catalytic turnover of P450s in the absence of redox partners such as CPR, the decrease in the CPR supported activity suggests that the increase in Km occurs as a result of mutations impairing the binding of CYP2B4 (R133C), (L270C) and (V267C) variants with CPR or electron transfer pathways. To distinguish between the two we measured the rate and extent of the reduction of CYP2B4 variants by CPR using stopped-flow spectrophotometry.
Figure 6.
Comparison of the tBHP-supported metabolism of benzphetamine by CYP2B4 WT and variants. CYP2B4 WT and its variants were incubated with DLPC, potassium phosphate buffer, pH 7.4, and excess benzphetamine as described under “Experimental Procedures”. The reactions were initiated by the addition of a 0.1 M final concentration of tBHP and the rate of formaldehyde formation was assessed spectrometrically after reaction with Nash’s reagent. The data are an average of two experiments with the error bars representing standard error mean.
Determination of the Rates of Electron Transfer from CPR to CYP2B4 WT and its Variants in the Absence of FM
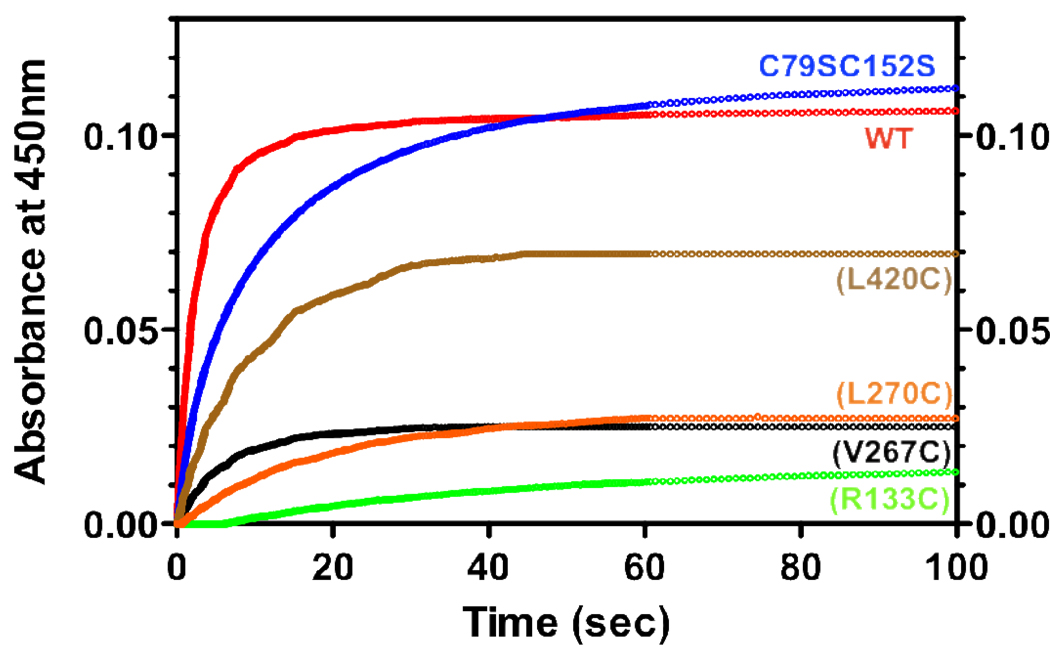
To test our hypothesis that residues L270 and V267, like R133 are involved in the binding of CPR, we measured the rate and extent of reduction of ferric CYP2B4 WT and its variants by CPR under pre-steady state conditions. Using stopped-flow spectrophotometry, we mixed preformed CYP2B4 WT or variant – CPR complexes with NADPH in the presence of CO and monitored the increase in absorbance at 450 nm with time. The production of the reduced-CO complex of P450 absorbing at 450 nm is the result of a series of steps. First NADPH must reduce CPR, which then delivers an electron to P450. This electron reduces the ferric heme to its ferrous state that can bind CO and absorbs at 450 nm. The binding of CO is very rapid compared to the reduction reaction and has no effect on the rate observed. When the CPR-P450 complex is not pre-formed by lengthy reconstitution, the association reaction and the subsequent reduction of P450 by CPR are very slow(37). For this reason, the rate of first electron transfer from CPR to P450 is typically measured by mixing a preformed P450-CPR complex with NADPH. Since the formation of a P450-CO complex depends on the amount of ferrous heme, which in turn depends on the concentration of the CPR-P450 complex, the total amplitude of the fast and slow phases is a reflection of the affinity of P450 for CPR. By deriving the concentration of the P450-CO complex from the total amplitude of the fast and slow phases, and examining the rate constants of those phases we can distinguish the effect of our mutations on protein-protein interaction from studies on P450 reduction. Figure 7 shows the absorbance increase for the proximal variants, C79SC152S and WT. As expected the reduction of all variants and WT followed biphasic kinetics. The rate constants of the fast and slow phases for the reduction of CYP2B4 WT in the absence of benzphetamine are 0.70 and 0.16 s−1 respectively and the amplitude of the fast phase is 62%. The final concentration of the CYP2B4WT-CO complex was determined from the total amplitude of the fast and slow phases and for the WT this was 1.22 µM (81% of the theoretical concentration). Variants C79SC152S, C79S, (E60C) and (Y484C) were all reduced to virtually the same levels and rate as the WT, while (H226C) and (L420C) displayed slightly lower total amplitudes (by 44% and 36% respectively) and rates compared to WT. As reflected in the total concentration of the CYP2B4-CO complex, variants (V267C), (L270C) and (R133C) had the most significant effect on CPR binding. These variants reduced the total amount of CYP2B4-CO complex by an average of 77% and their kinetic constants for the fast phase of reduction were similarly affected. It is interesting to note that when the kinetics of the reduction of these mutants using equimolar ratios of P450 and CPR were compared to the kinetics of reduction with a 3-fold excess of CPR over P450, no significant increase in the rate of the fast phase was observed, however the total amplitude increased significantly (Figure 1S.). Therefore, these data clearly demonstrate that V267 and L270 in CYP2B4 are involved in binding CPR.
Figure 7.
Kinetics for the reduction of the CYP2B4 proximal variants by CPR in the presence of NADPH. CYP2B4, CPR and DLPC were reconstituted for 1 hr on ice and mixed with NADPH in a stopped-flow apparatus. After mixing, the final concentrations of CYP2B4 and CPR were 1.5 µM and NADPH was 50 µM. Experiments were conducted as described under “Experimental Procedures” with a saturated solution of CO in both syringes. For convenience, all the absorbance data are offset to the same baseline.
DISCUSSION
The results presented here demonstrate that the site for the binding of CPR to CYP2B4 is on the proximal side of CYP2B4 and that mutation of residues L270 and V267 to cys disrupts the CYP2B4-CPR interaction. To investigate the residues involved in binding we mutated strategically selected residues on the surface of CYP2B4 to cys for site-specific labeling with FM and observed that when these P450s were reconstituted with excess CPR and DLPC, the largest fluorescence enhancement was recorded for those variants with labels on their proximal side. With the goal of uncovering whether hydrophobic residues might be involved in recognition and binding of CPR, we investigated whether the V267C and L270C mutations could inhibit complex formation by studying the Km’s of the variants for CPR and monitoring the rate and extent of P450 reduction by CPR. We observed a significant increase in Km and a decrease in the rates and extents of P450 reduction for the proximal variants (L270C), (V267C) and (R133C), and virtually no change for (L420C) compared to WT.
Bridges et al. (15) have previously demonstrated that R133, when mutated to ala, exhibited the single largest perturbation on the apparent Kd of CYP2B4 for CPR when compared to WT. Therefore, we used this residue as a positive control to evaluate the results obtained using our strategy. Experiments where the CYP2B4FM variants were incubated with CPR revealed that fluorescence enhancement occurs to some extent for each site of FM attachment with the highest extent of fluorescence enhancement occurring with variants (V267C), (L270C), (R133C), and (L420C). It is likely that interactions of the amino acid residues from CPR with the FM label on the P450 increases the probe’s quantum yield by restricting its conformational flexibility and reducing the amount of energy dissipated as non-radiative decay. This would increase the emission intensity of the probe and explain the observed results. Another, although highly unlikely explanation for the increase in fluorescence emission, is that the binding of CPR to CYP2B4 induces a conformational change in the protein that results in the burial of the probe further into the core of CYP2B4. Thus, the changes observed could possibly be due to some type of allosteric effect rather than direct binding. Although P450s are known to undergo relatively large conformational changes in the presence of substrate(34,38,39), conformational changes of the magnitude required to internalize FM in CYP2B4 have not been reported. Moreover, the possibility of multiple docking sites for CPR on CYP2B4 is not supported by the data obtained from this study. Instead our findings support the hypothesis that only a single, proximally located site on CYP2B4 is involved in complex formation.
The spectral results shown in Figure 3 led us to investigate the quantitative basis for the CYP2B4-CPR interaction by titrating fixed concentrations of the CYP2B4-FM variants with increasing concentrations of CPR. By plotting the increase in fluorescence intensity with increasing concentrations of CPR and fitting this to Eq. 2, we were able to determine the affinity of the various FM-modified proximal mutants for CPR. As shown in Figure 4 and Table 1 CYP2B4 (L420C)-FM had the lowest Kd while (V267C)-, (L270C)-, and (R133C)-FM all displayed similarly lower affinities for CPR. Although the addition of two FM labels to the WT P450 prevented us from obtaining fluorescence titration data for an unmutagenized CYP2B4, the Kd obtained for CYP2B4 (L420C)-FM corresponds closely to previously reported Kd’s for WT CYP2B4-CPR. Since this variant also exhibited similar Km and kcat values for CPR as WT, it not only serves as a reasonable reference to the WT but also suggests that the FM-labeled L420C residue is close enough to the CPR docking site to exhibit changes in fluorescence enhancement in the presence of CPR but is not involved in mediating CPR binding.
To characterize the effect of mutating the selected residues to cysteines in the absence of the fluorescent probe, we opted to characterize the Km and kcat values of the mutants for CPR. This approach would not only provide information regarding the possible roles of V267 and L270 in CPR binding in the absence of FM but would also reveal whether the lack of the fluorescence enhancement observed for the distal and side mutants was due to either a lack of interaction with the reversibly bound CPR or an obliteration of the interaction as a consequence of mutagenesis. Although we expected it was due to the former we needed a definitive way to investigate the latter possibility. By studying the rate of benzphetamine oxidation as a function of increasing concentrations of CPR, an apparent Km can be determined to assess the affinity of the unmodified variants for CPR. Although this approach has been used in the past, the results obtained from steady-state kinetic analyses of P450 variants do not necessarily directly determine which residues mediate CPR-P450 interaction. For example, electron transfer from CPR to P450 is thought to be mediated by amino acid residues that facilitate electron tunneling through proteins(40). The identity of these particular residues is currently unknown, but when mutated to non-functional ones, their effect on protein activity could be indistinguishable from that of residues involved in CPR-P450 interactions. This limitation highlights the need to employ a holistic approach that combines steady-state turnover kinetics with fluorescence and stopped-flow spectroscopy to the study of residues involved in redox partner recognition and forms the basis on which the experiments presented in this study were conceived.
The hypothesis that residues L270 and V267, like R133, occupy the binding interface is supported by our Km data and further strengthened by stopped-flow experiments. In the kinetic experiments for the determination of the Km, all of the proximal mutants with the exception of (L420C) showed increases in Km values that, based on our studies on the tBHP supported catalytic activity could not be attributed to an altered P450 active site. Furthermore, the possibility that no fluorescence enhancement was observed when the distal FM-labeled variants were incubated with CPR because of complete disruption of complex formation can be conclusively ruled out by the observation that all of the other variants exhibited Km’s that were virtually identical to WT.
Data from measurements of the rate and extent of P450 reduction by CPR argue in favor of the conclusion that the higher Km and Kd values for (V267C), (L270C) and (R133C) for CPR are due to impaired protein-protein interactions. Disruption of CYP2B4-CPR binding as a result of proximal residue mutagenesis can be inferred from the total amplitude of the slow and fast phases and is critical to our understanding of the roles of L270 and V267 in complex formation. In theory, the binding of CO to ferrous heme should promote further CYP2B4-CPR complex formation by depleting the concentration of ferric heme and shifting the equilibrium between ferric and ferrous forms in the direction of the ferrous state. However, experimentally, we, and others, find that the association of P450 with CPR is very slow when the complex between the two proteins is not pre-formed by extensive reconstitution(37). Therefore, in this study the concentration of the CYP2B4-CO complex is limited by the concentration of the CYP2B4-CPR complex and the total amplitude of the fast and slow phases is a reflection of the affinity of the protein partners for each other. In the case of residues V267, L270 and R133, only 24, 26 and 19% respectively of the P450 variants was reduced by CPR compared to WT and strongly supports the conclusion that these residues are involved in CPR binding.
Since protein-protein interaction between CPR and CYP2B4 appears to involve both complementary charge pairing and hydrophobic interactions, how do the two different intermolecular forces coordinate protein recognition? Jones and Thornton have surveyed the interfaces of protein heterocomplexes with the goal to identify the types of residues that distinguish those found at the interface from those present on the surface of the protein(41). They have noted that large hydrophobic and uncharged polar residues were present more often at the binding interface compared to the rest of the solvent exposed surface. Similarly, work on the human growth hormone receptor has demonstrated that when hydrophobic contacts are distributed over a relatively large area they can contribute substantially to the binding energy (42). Shielding these hydrophobic contacts from the influence of water are charged residues that frame the binding interface and are postulated to dictate specificity in the interaction. The notion that hydrophobic residues can dominate the interfaces of redox-partners was recently reinforced by the crystal structure of a chemically cross-linked putidaredoxin reductase (Pdr) and putidaredoxin (Pdx) complex that is essential in the P450cam monooxygenase system. These studies revealed that the Pdr-Pdx interface is predominantly hydrophobic with a central salt bridge assisting docking and orientation(20).
Since the structure of CYP102A1 (P450BM3) (PDB code 1BVY) represents the only known crystal structure of a P450 complexed with its redox partner (19), it is interesting to examine whether the residues currently believed to contribute to the CYP2B4-CPR interaction (15) are conserved in the heme domain of P450BM3. The amino acid sequences of CYP2B4 and P450BM3 were subjected to analysis by the EMBOSS Pairwise Alignment Algorithm (43) which revealed that the charge of approximately half of the CYP2B4 residues believed to mediate CPR recognition are conserved between these two P450s (Figure 2S). Specifically, we find that the charges of only R122, F135, M137, L270 and K433 of CYP2B4 are conserved in P450BM3. Interestingly, the P450BM3 counterparts to the CYP2B4 basic amino acids identified by Bridges et al (15) to be involved in binding CPR are almost all neutral residues while V267 on CYP2B4 is H237 in P450BM3. Of all the P450BM3 residues presented in Table 2S only K98 and Q398 are within the FMN domain binding region. However, only Q396 of BM3’s heme domain could potentially form a hydrogen bond with E494 of the FMN domain (although the two are 6.2 Å apart) since K98 of the heme domain likely contributes to the koff of the complex as it forms a charge repulsion interaction with R498 of BM3’s FMN domain. Thus, although the P450BM3 structure is the only available crystal structure of a P450-redox partner complex, it is difficult to extend our understanding of specific CYP2B4’s residues involved in binding CPR to P450BM3.
In the CPR-P450 interaction field roles for hydrophobic residues and charged residues in mediating complex formation have been identified in the past(14,15,17,21,44–46). However most studies relied on steady state turnover kinetics to determine the binding interface and thus could not distinguish perturbations in electron transfer pathways from binding affinity. To our knowledge, this study is the first to identify roles for L270 and V267 in the interaction between CYP2B4 and CPR using a combination of kinetic and non-kinetic methods.
The residues present at protein-protein interaction sites not only dictate the kon of the association but the koff of the dissociation as well. This is usually achieved either through steric clashing or electrostatic repulsion. Knowledge regarding these residues can be harnessed to create P450 mutants with residues that contribute to the koff re-engineered to promote the kon (47,48). As a result of the change, we would anticipate a tighter binding CPR to P450 and thus higher P450 catalytic activity that could be exploited industrially in the P450-mediated biosynthesis of chemicals(49). Therefore further studies are under way to determine the effect of mutating V267 and L270 to charged residues on the Kd of the CYP2B4-CPR complex.
The insights gained into the roles of selected amino acid residues in the interactions between CYP2B4 and CPR by combining stopped-flow and fluorescence spectroscopy, site-specific mutagenesis and steady-state kinetics are invaluable. They demonstrate that FM can be used to site-specifically label surface exposed residues on 2B4 and, when reconstituted with CPR, the CYP2B4-FM variants with labels on their proximal side showed the largest fluorescence enhancement compared to other residues on the surface of CYP2B4. These residues were L420C, L270C, V267C and R133C. With the exception of L420C, all the proximal variants showed increased Kd and Km values for CPR and were reduced by CPR at rates and to extents significantly less than WT. Thus we propose an expanded role for hydrophobic residues in mediating complex formation between CYP2B4 and CPR.
Supplementary Material
Table 3. The rate constants and amplitudes observed for the reduction of CYP2B4 and its variants by CPR at 23°C.
The apparent rate constants for the two kinetic phases are k1 and k2, respectively, and the relative amplitudes for each phase are expressed as A1% and A2%. The total absolute amplitude for the overall reduction is AT.
| 2B4 | Kinetic parameters | ||||
|---|---|---|---|---|---|
| AT | A1% | k1,S−1 | A2% | k2,S−1 | |
| WT | 0.110 | 62 | 0.70 | 39 | 0.16 |
| C79SC152S | 0.110 | 28 | 0.51 | 72 | 0.067 |
| C79S | 0.079 | 18 | 0.205 | 82 | 0.052 |
| (E60C) | 0.120 | 28 | 0.678 | 72 | 0.077 |
| (H226C) | 0.061 | 33 | 0.15 | 67 | 0.043 |
| (Y484C) | 0.079 | 21 | 0.212 | 79 | 0.049 |
| (V267C) | 0.027 | 64 | 0.057 | 36 | 0.0138 |
| (R133C) | 0.021 | 85 | 0.019 | 15 | 0.0031 |
| (L270C) | 0.029 | 10 | 0.032 | 90 | 0.0110 |
| (L420C) | 0.070 | 39 | 0.360 | 61 | 0.0035 |
ACKNOWLEDGEMENTS
We thank Drs. Dave Ballou and Tatyana Spolitak for help with stopped-flow experiments, Hsia-Lien Lin for the expression and purification of CPR and Dr. Djemel Hamdane for help with APBS.
The abbreviations used are
- CYP or P450
cytochrome P450
- CPR
NADPH-dependent cytochrome P450 reductase
- WT
wild-type
- Pdx
Putidaredoxin
- Pdr
Putidaredoxin reductase
- FM
fluorescein-5-maleimide
- CO
carbon monoxide
- ESI-LC-MS
electrospray ionization liquid chromatography mass spectrometry
- FMN
flavin mononucleotide
- FAD
flavin adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- DLPC
dilauroylphosphatidylcholine
- HCHO
formaldehyde
- tBHP
tert-butyl hydroperoxide
- PDB
Protein Data Bank
- APBS
Adaptive Poisson-Boltzmann Solver
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant CA16954 (to P.F.H), a Horace H. Rackham Graduate Student Research Grant (to C.K) and an ASPET Institutional Summer Undergraduate Research Fellowship (to E.V.S.).
SUPPORTING INFORMATION AVAILABLE
A figure displaying the kinetics for the reduction of the CYP2B4 V267C and L270C proximal variants under saturating and subsaturating amounts of CPR in the presence of NADPH and BNZ. A figure showing the amino acid sequence alignment of CYP2B4 and P450BM3. A table containing the CYP2B4 proximal residues, identified by Bridges et al. and this study, to be involved in CPR binding and their corresponding residues in P450BM3. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and Chemistry of Cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 2.Rittle J, Green MT. Cytochrome P450 Compound I: Capture, Characterization, and C-H Bond Activation Kinetics. Science. 2010;330:933–937. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 3.Hamdane D, Xia C, Im SC, Zhang H, Kim JJ, Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J Biol Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubbard PA, Shen AL, Paschke R, Kasper CB, Kim JJ. NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer. J Biol Chem. 2001;276:29163–29170. doi: 10.1074/jbc.M101731200. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cawley GF, Batie CJ, Backes WL. Substrate-dependent competition of different P450 isozymes for limiting NADPH-cytochrome P450 reductase. Biochemistry. 1995;34:1244–1247. doi: 10.1021/bi00004a018. [DOI] [PubMed] [Google Scholar]
- 7.Backes WL, Kelley RW. Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol Ther. 2003;98:221–233. doi: 10.1016/s0163-7258(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 8.Enoch HG, Strittmatter P. Cytochrome b5 reduction by NADPH-cytochrome P-450 reductase. J Biol Chem. 1979;254:8976–8981. [PubMed] [Google Scholar]
- 9.Williams CH, Jr., Kamin H. Microsomal triphosphopyridine nucleotide-cytochrome c reductase of liver. J. Biol. Chem. 1962;237:587–595. [PubMed] [Google Scholar]
- 10.Kikuchi G, Yoshida T, Noguchi M. Heme oxygenase and heme degradation. Biochem Biophys Res Commun. 2005;338:558–567. doi: 10.1016/j.bbrc.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Ono T, Bloch K. Solubilization and partial characterization of rat liver squalene epoxidase. J. Biol. Chem. 1975;250:1571–1579. [PubMed] [Google Scholar]
- 12.Jang HH, Jamakhandi AP, Sullivan SZ, Yun CH, Hollenberg PF, Miller GP. Beta sheet 2-alpha helix C loop of cytochrome P450 reductase serves as a docking site for redox partners. Biochim Biophys Acta. 2010;1804:1285–1293. doi: 10.1016/j.bbapap.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen AL, Kasper CB. Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J Biol Chem. 1995;270:27475–27480. doi: 10.1074/jbc.270.46.27475. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu T, Tateishi T, Hatano M, Fujii-Kuriyama Y. Probing the role of lysines and arginines in the catalytic function of cytochrome P450d by site-directed mutagenesis. Interaction with NADPH-cytochrome P450 reductase. J Biol Chem. 1991;266:3372–3375. [PubMed] [Google Scholar]
- 15.Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 16.Gao Q, Doneanu CE, Shaffer SA, Adman ET, Goodlett DR, Nelson SD. Identification of the interactions between cytochrome P450 2E1 and cytochrome b5 by mass spectrometry and site-directed mutagenesis. J. Biol. Chem. 2006;281:20404–20417. doi: 10.1074/jbc.M601785200. [DOI] [PubMed] [Google Scholar]
- 17.Bumpus NN, Hollenberg PF. Cross-linking of human cytochrome P450 2B6 to NADPH-cytochrome P450 reductase: Identification of a potential site of interaction. J Inorg Biochem. 2010;104:485–488. doi: 10.1016/j.jinorgbio.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sevrioukova IF, Poulos TL. Structural biology of redox partner interactions in P450cam monooxygenase: A fresh look at an old system. Arch Biochem Biophys. 2011;501:66–74. doi: 10.1016/j.abb.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci U S A. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sevrioukova IF, Poulos TL, Churbanova IY. Crystal structure of the putidaredoxin reductase x putidaredoxin electron transfer complex. J Biol Chem. 2010;285:13616–13620. doi: 10.1074/jbc.M110.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voznesensky AI, Schenkman JB. The cytochrome P450 2B4-NADPH cytochrome P450 reductase electron transfer complex is not formed by charge-pairing. J Biol Chem. 1992;267:14669–14676. [PubMed] [Google Scholar]
- 22.Voznesensky AI, Schenkman JB. Quantitative analyses of electrostatic interactions between NADPH-cytochrome P450 reductase and cytochrome P450 enzymes. J Biol Chem. 1994;269:15724–15731. [PubMed] [Google Scholar]
- 23.Hassan AQ, Wang Y, Plate L, Stubbe J. Methodology to probe subunit interactions in ribonucleotide reductases. Biochemistry. 2008;47:13046–13055. doi: 10.1021/bi8012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drees BL, Rye HS, Glazer AN, Nelson HC. Environment-sensitive labels in multiplex fluorescence analyses of protein-DNA complexes. J Biol Chem. 1996;271:32168–32173. doi: 10.1074/jbc.271.50.32168. [DOI] [PubMed] [Google Scholar]
- 25.Sevrioukova IF, Hazzard JT, Tollin G, Poulos TL. The FMN to heme electron transfer in cytochrome P450BM-3. Effect of chemical modification of cysteines engineered at the FMN-heme domain interaction site. J Biol Chem. 1999;274:36097–36106. doi: 10.1074/jbc.274.51.36097. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Im SC, Waskell L. Cytochrome b5 increases the rate of product formation by cytochrome P450 2B4 and competes with cytochrome P450 reductase for a binding site on cytochrome P450 2B4. J Biol Chem. 2007;282:29766–29776. doi: 10.1074/jbc.M703845200. [DOI] [PubMed] [Google Scholar]
- 27.Omura T, Sato R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. II. Solubilization, Purification and Properties. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 28.Vermilion JL, Coon MJ. Purified liver microsomal NADPH-cytochrome P-450 reductase. Spectral characterization of oxidation-reduction states. J Biol Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- 29.Miwa GT, West SB, Huang MT, Lu AY. Studies on the association of cytochrome P-450 and NADPH-cytochrome c reductase during catalysis in a reconstituted hydroxylating system. J Biol Chem. 1979;254:5695–5700. [PubMed] [Google Scholar]
- 30.French JS, Guengerich FP, Coon MJ. Interactions of cytochrome P-450, NADPH-cytochrome P-450 reductase, phospholipid, and substrate in the reconstituted liver microsomal enzyme system. J Biol Chem. 1980;255:4112–4119. [PubMed] [Google Scholar]
- 31.Davydov DR, Knyushko TV, Kanaeva IP, Koen YM, Samenkova NF, Archakov AI, Hui Bon Hoa G. Interactions of cytochrome P450 2B4 with NADPH-cytochrome P450 reductase studied by fluorescent probe. Biochimie. 1996;78:734–743. doi: 10.1016/s0300-9084(97)82531-x. [DOI] [PubMed] [Google Scholar]
- 32.Nash T. The colorimetric estimation of formaldehyde by means of the Hantzech reaction. Biochem. J. 1953;55:416–421. doi: 10.1042/bj0550416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coon MJ, Hoeven TA, Kaschnitz RM, Strobel HW. Biochemical studies on cytochrome P-450 solubilized from liver microsomes: partial purification and mechanism of catalysis. Ann N Y Acad Sci. 1973;212:449–457. doi: 10.1111/j.1749-6632.1973.tb47613.x. [DOI] [PubMed] [Google Scholar]
- 34.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 35.Lehnerer M, Schulze J, Achterhold K, Lewis DF, Hlavica P. Identification of key residues in rabbit liver microsomal cytochrome P450 2B4: importance in interactions with NADPH-cytochrome P450 reductase. J Biochem. 2000;127:163–169. doi: 10.1093/oxfordjournals.jbchem.a022578. [DOI] [PubMed] [Google Scholar]
- 36.Hermanson GT. Bioconjugate Techniques. San Diego: Academic Press; 1996. [Google Scholar]
- 37.Backes WL, Eyer CS. Cytochrome P-450 LM2 reduction. Substrate effects on the rate of reductase-LM2 association. J Biol Chem. 1989;264:6252–6259. [PubMed] [Google Scholar]
- 38.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci U S A. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Kenaan C, Hamdane D, Hoa GH, Hollenberg PF. Effect of conformational dynamics on substrate recognition and specificity as probed by the introduction of a de novo disulfide bond into cytochrome P450 2B1. J Biol Chem. 2009;284:25678–25686. doi: 10.1074/jbc.M109.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray HB, Winkler JR. Electron transfer in proteins. Annu Rev Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 41.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clackson T, Ultsch MH, Wells JA, de Vos AM. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J Mol Biol. 1998;277:1111–1128. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- 43. http://www.ebi.ac.uk/Tools/emboss/align/index.html.
- 44.Rodgers KK, Sligar SG. Mapping electrostatic interactions in macromolecular associations. J Mol Biol. 1991;221:1453–1460. doi: 10.1016/0022-2836(91)90945-3. [DOI] [PubMed] [Google Scholar]
- 45.Black SD, French JS, Williams CH, Jr., Coon MJ. Role of a hydrophobic polypeptide in the N-terminal region of NADPH-cytochrome P-450 reductase in complex formation with P-450LM. Biochem Biophys Res Commun. 1979;91:1528–1535. doi: 10.1016/0006-291x(79)91238-5. [DOI] [PubMed] [Google Scholar]
- 46.Stayton PS, Sligar SG. The cytochrome P-450cam binding surface as defined by site-directed mutagenesis and electrostatic modeling. Biochemistry. 1990;29:7381–7386. doi: 10.1021/bi00484a005. [DOI] [PubMed] [Google Scholar]
- 47.Selzer T, Albeck S, Schreiber G. Rational design of faster associating and tighter binding protein complexes. Nat Struct Biol. 2000;7:537–541. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- 48.Xiong P, Nocek JM, Griffin AK, Wang J, Hoffman BM. Electrostatic redesign of the [myoglobin, cytochrome b5] interface to create a well-defined docked complex with rapid interprotein electron transfer. J Am Chem Soc. 2009;131:6938–6939. doi: 10.1021/ja902131d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guengerich FP. Cytochrome P450 enzymes in the generation of commercial products. Nat Rev Drug Discov. 2002;1:359–366. doi: 10.1038/nrd792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.