Abstract
The C–terminal domain of the Escherichia coli RNA polymerase α subunit (αCTD) plays a key role in transcription initiation at many activator-dependent promoters. This domain is connected to the N–terminal domain by an unstructured linker, which is proposed to confer a high degree of mobility on αCTD. To investigate the role of this linker in transcription activation we tested the effect of altering the linker length on promoters dependent on the cyclic AMP receptor protein (CRP). Short deletions within the α linker decrease CRP-dependent transcription at a Class I promoter while increasing the activity of a Class II promoter. Linker extension impairs CRP-dependent transcription from both promoters, with short extensions exerting a more marked effect on the Class II promoter. Activation at both classes of promoter was shown to remain dependent upon activating region 1 of CRP. These results show that the response to CRP of RNA polymerase containing linker-modified α subunits is class specific. These observations have important implications for the architecture of transcription initiation complexes at CRP-dependent promoters.
Keywords: α subunit/cyclic AMP receptor protein/inter-domain linker/RNA polymerase/transcription activation
Introduction
Escherichia coli holo RNA polymerase (RNAP) is a multi-subunit complex consisting of two identical α subunits, single β and β′ subunits, and one of several σ subunit species. During transcription initiation at many promoters, RNAP containing the major σ factor, σ70, recognizes three promoter elements: the –10 hexamer, the –35 hexamer and the UP element. In addition, interactions between RNAP and one or more transcription activators are frequently required, often involving the α subunit. Each α subunit consists of 329 amino acids organized in two independently folding domains (Blatter et al., 1994; Negishi et al., 1995). The N–terminal domain (αNTD; residues 8–231) contains determinants for dimerization and assembly into RNAP (Igarashi et al., 1991; Kimura et al., 1994; Kimura and Ishihama, 1995a,b; Zhang and Darst, 1998) and plays a role in transcription activation at some promoters (see below). The C–terminal domain (αCTD; residues 249–329) plays multiple roles in transcription (Igarashi and Ishihama, 1991; Ross et al., 1993; Jeon et al., 1995; Gaal et al., 1996; Kainz and Gourse, 1998). During transcription initiation at some promoters, αCTD recognizes the A+T-rich UP element (Ross et al., 1993; Giladi et al., 1996; Tagami and Aiba, 1999). At the rrnB P1 promoter, this interaction is responsible for a 30- to 70-fold increase in promoter activity (Ross et al., 1993; Rao et al., 1994; Estrem et al., 1998). In addition, αCTD is also a target for a variety of transcription activator proteins at positively regulated promoters, including the well studied cyclic AMP receptor protein (CRP; also known as the catabolite activator protein, CAP) (reviewed by Ishihama, 1993; Ebright and Busby, 1995; Busby and Ebright, 1999).
CRP can activate transcription at >100 different promoters in E.coli in response to the intracellular concentration of its allosteric effector, cAMP (reviewed by Kolb et al., 1993a; Busby and Kolb, 1996). The cAMP–CRP complex binds as a dimer to a 22 bp inverted repeat, possessing a consensus of aaaTGTGAtctagaTCACAttt (Berg and von Hippel, 1988). The binding of the CRP dimer to its DNA target results in a sharp bend in the DNA (Wu and Crothers, 1984) estimated to be 80–90° (Schultz et al., 1991; Busby and Ebright, 1999). Promoters that are dependent upon CRP as sole activator can be divided into two classes according to the location of the bound CRP (Ushida and Aiba, 1990; Ebright, 1993). At Class I CRP-dependent promoters, the CRP target site is located upstream of the –35 region (at sites centred near positions –61.5, –71.5, –81.5 or –91.5 with respect to the transcription start point), whereas at Class II CRP-dependent promoters the CRP-binding site is centred near position –41.5, and therefore overlaps the –35 region of the target promoter (Gaston et al., 1990). At both classes of promoter, αCTD makes contact with activating region 1 (AR1) of CRP (Zhou et al., 1994a; Niu et al., 1996; Rhodius et al., 1997) and is thereby recruited to DNA sequences adjacent to CRP. The interaction of CRP with αCTD is a requirement for CRP-dependent activation at these promoters. AR1 is a surface-exposed loop (residues 156–164), which is located adjacent to the helix–turn–helix motif responsible for DNA target site recognition by CRP (Zhou et al., 1993a; Niu et al., 1994). At Class I CRP-dependent promoters, recruitment occurs through the ‘downstream’ subunit of the bound CRP dimer so that one or both αCTDs occupies a position on the template between CRP and the rest of the RNAP holoenzyme (Kolb et al., 1993b; Zhou et al., 1993b, 1994b; Law et al., 1999), while at Class II CRP-dependent promoters αCTD is recruited by the ‘upstream’ CRP subunit and therefore binds to the DNA upstream of bound CRP (Attey et al., 1994; Belyaeva et al., 1996, 1998; Murakami et al., 1997). The determinant on αCTD required for the interaction with AR1 at both classes of promoter is proposed to comprise the side chains of amino acids 285–289, 315 and 317–318 (Savery et al., 1998; Busby and Ebright, 1999; N.J.Savery, G.S.Lloyd, S.J.W.Busby, M.S.Thomas, R.H.Ebright and R.L.Gourse, in preparation). In addition, the DNA-binding determinant on αCTD is involved in interactions with upstream promoter sequences during transcription activation by CRP at both classes of promoter. Transcription activation at Class II CRP-dependent promoters also requires a second activating region on CRP, AR2, located in the N–terminal cAMP-binding domain (Niu et al., 1996; Rhodius et al., 1997). AR2 of the downstream CRP subunit makes functional contacts with residues 162–165 of αNTD at this class of promoter (Niu et al., 1996).
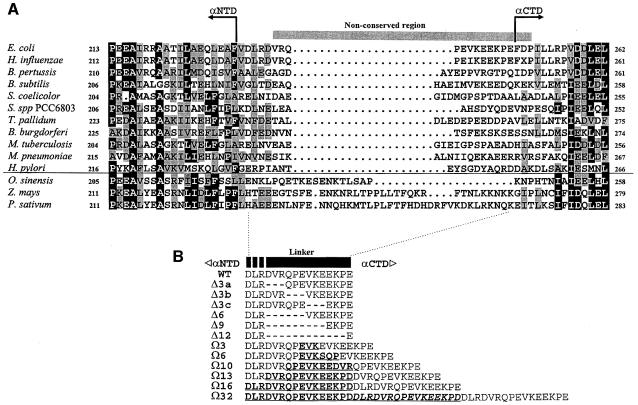
The ability of αCTD to contact CRP bound at different upstream positions at target promoters suggests that this domain possesses a degree of motional freedom. Several lines of evidence indicate the presence of a flexible linker connecting the mobile αCTD to a fixed αNTD. First, limited proteolysis studies pointed to an accessible region between amino acids 234 and 249 (Blatter et al., 1994; Negishi et al., 1995). Secondly, NMR analysis of an isolated C–terminal fragment of α (amino acids 233–329) showed that a region of at least 13 amino acids, extending from D236 (but possibly from D233) to E248, exhibits a high degree of flexibility (Jeon et al., 1997). The more N–terminal limit for the inter-domain linker is consistent with the crystallographic data for αNTD, where the C–terminal limit of helix 3 is assigned to F231 (Zhang and Darst, 1998). Finally, amino acid sequence alignments of the α subunits of eubacteria and chloroplasts reveal a non-conserved sequence corresponding to amino acids V237–P251 of the E.coli α subunit (Figure 1A). Despite the sequence variation, the length of this region is fairly well conserved in the bacterial α subunits (14–18 residues), but exhibits considerable length variation in the chloroplast α homologues (18–38 residues).
Fig. 1. (A) Amino acid sequence alignment of putative inter-domain regions of RNAP α subunits from representative bacteria and chloroplasts. Thirty sequences (14 from bacteria and 16 from chloroplasts) obtained from the Swiss Protein sequence database were aligned, although for clarity only 14 (11 from bacteria and three from chloroplasts) are shown. The amino acid denoted by the letter X in the Haemophilus influenzae α sequence is aspartate or glutamate (GAA/T). Amino acid residues at any one position that are identical in ≥50% of sequences throughout the alignment are highlighted in black boxes, and residues exhibiting similarity in ≥50% of more sequences are shown in grey boxes. The region where sequence conservation is absent is indicated by a horizontal bar over the E.coli sequence. The C–terminal limit of αNTD and the N–terminal limit of αCTD are indicated on the E.coli sequence and are inferred from Zhang and Darst (1998) and Jeon et al. (1997). Chloroplast sequences are separated from bacterial sequences by a horizontal line. (B) Amino acid sequences of wild-type and mutant inter-domain linkers of the E.coli RNAP α subunit used in this work. Amino acids 233–248 of the wild-type linker are shown. Deletions are represented by dashes, and insertions (duplicated sequences) are in bold and underlined. The second linker repeat in the Ω32 mutant derivative is also italicized. Deletions are confined to residues 236–247, insertions involve duplication of parts of or all of the region shown, with the exception that αΩ6 has a serine rather than an arginine in the duplicated linker segment, and E248 is replaced by an aspartate residue at each position where it is replicated in the Ω13, Ω16 and Ω32 derivatives.
As Class I and Class II CRP-dependent promoters require the interaction of αCTD with CRP (and adjacent DNA sequences) at a variety of distances upstream from the core promoter elements, they constitute an ideal system to study the role of the α inter-domain linker in transcription activation. In this work, we have constructed a series of deletions and insertions within the inter-domain linker of the E.coli RNAP α subunit. These linker-modified α derivatives were then either reconstituted in vitro or incorporated in vivo into RNAP and the length requirement of the linker at different classes of CRP-dependent promoter was investigated. Our results indicate that the linker can tolerate gross changes without affecting transcription from the CRP-independent, UP element-independent lacUV5 promoter, whereas length is important for CRP-activated transcription at both classes of CRP-dependent promoter.
Results
Construction of RNAP mutants containing α subunits with inter-domain linkers of different lengths
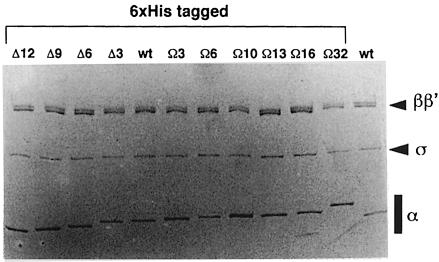
To test the requirements for the RNAP α subunit inter-domain linker at CRP-dependent promoters, a series of 12 rpoA mutants was constructed by PCR and introduced into the rpoA expression plasmid pHTT7f1NHα (Figure 1B; Table I). Six of the variants contain sequences encoding shorter linkers, giving rise to α derivatives harbouring deletions of 3, 6, 9 and 12 amino acids (Δ3a–c, Δ6, Δ9 and Δ12, respectively; three different Δ3 derivatives were designed) within the 13-amino-acid flexible region (D236–E248) defined by NMR as constituting the minimum limits of the α linker (Jeon et al., 1997). Additionally, six rpoA variants were constructed in which the linker encoding sequence was extended (Figure 1B). For the constructs encoding 3, 6, 10 and 13 additional amino acids in the α linker (Ω3, Ω6, Ω10 and Ω13), extension was achieved by duplicating segments within the 13-amino-acid flexible region. For the Ω16 α mutant, the DLR motif (residues 233–235 of α) was also duplicated in addition to residues 236–248. This 16-amino-acid sequence is present in three tandem copies in the Ω32 variant. All the mutant α subunits, as well as the wild-type subunit, were overproduced as N–terminal His-tagged fusions under the control of a phage T7 promoter in E.coli, purified to homogeneity and then reconstituted into holo RNAP with wild-type β, β′ and σ70 subunits (Figure 2).
Table I. Linker coding sequences of mutant rpoA expression plasmids.
| α derivative | Nucleotide sequence of modified linker coding regionsa | pHTT7f1-NHα/pLAW2 derivatives |
|---|---|---|
| Δ12 | GACCTAAGG…GAG | pMGM34/pMGM35 |
| Δ9 | GACCTAAGG…GAGAAACCAGAG | pMGM6/pMGM17 |
| Δ6 | GACCTAAGG…GTGAAAGAAGAGAAACCAGAG | pMGM5/pMGM16 |
| Δ3c | GACCTAAGGGATGTACGTCAGCCTGAA…GAGAAACCAGAG | pMGM4/pMGM15 |
| Δ3b | GACCTAAGGGATGTACGT…GTGAAAGAAGAGAAACCAGAG | pMGM3/pMGM14 |
| Δ3a | GACCTAAGG…CAGCCTGAAGTGAAAGAAGAGAAACCAGAG | pMGM2/pMGM13 |
| Wild-type (native) | GACTTACGTGATGTACGTCAGCCTGAAGTGAAAGAAGAGAAACCAGAG | pHTT7f1-NHα/pLAW2 |
| Wild-type (Bsu36I) | GACCTAAGGGATGTACGTCAGCCTGAAGTGAAAGAAGAGAAACCAGAG | pMGM1/pMGM12 |
| Ω3 | GACCTAAGGGATGTACGTCAGCCTGAGGTCAAGGAAGTGAAAGAAGAGAAACCAGAG | pMGM7/pMGM18 |
| Ω6 | GACCTAAGGGATGTACGTCAGCCTGAGGTCAAGAGTCAACCAGAAGTGAAAGAAGAGAAACCAGAG | pMGM8/pMGM19 |
| Ω10 | GACTTACGTGATGTACGTCAGCCTGAAGTGAAAGAGGAAGATGTACGTCAGCCTGAAGTGAAAGAAGAGAAACCAGAG | pMGM31/pMGM32 |
| Ω13 | GACTTACGTGATGTACGTCAGCCTGAAGTGAAAGAGGAAAAGCCTGACGATGTACGTCAGCCTGAAGTGAAAGAAGAGAAACCAGAG | pMGM10/pMGM21 |
| Ω16 | GACTTACGTGATGTACGTCAGCCTGAAGTGAAAGAGGAAAAGCCTGACGACCTAAGGGATGTACGTCAGCCTGAAGTGAAAGAAGAGAAACCAGAG | pMGM9/pMGM20 |
| Ω32 | GACCTAAGGGATGTACGTCAGCCTGAAGTGAAAGAGGAAAAGCCTGACGACCTAAGGGATGTACGTCAGCCTGAAGTGAAAGAGGAAAAGCCTGACGACCTAAGGGATGTACGTCAGCCTGAAGTGAAAGAGAAACCAGAG | pMGM11/pMGM22 |
aCodons 233–248 of wild-type rpoA and the corresponding codons in the mutant sequences are shown. Locations of deleted linker codons are indicated by a dotted line. Codons inserted into the linker are in bold. Bsu36I sites introduced into the α coding sequence to create pMGM1 and subsequent linker-modified derivatives are underlined (there are no Bsu36I sites in the Ω10 and Ω13 derivatives). Bases changed in the wild-type α coding sequence in order to introduce the Bsu36I site, without changing the encoded amino acids, are also underlined.
Fig. 2. SDS–PAGE of reconstituted E.coli RNAP carrying wild-type or linker-modified α subunits. The proteins were analysed on an 8–15% polyacrylamide gradient gel. Each lane contains reconstituted RNAP carrying linker-modified α subunits (all N–terminal His-tagged) as indicated, or wild-type (wt) RNAP (Boehringer). Positions of α, β, β′ and σ subunits are indicated.
Effect of α linker length on CRP-dependent transcription in vitro
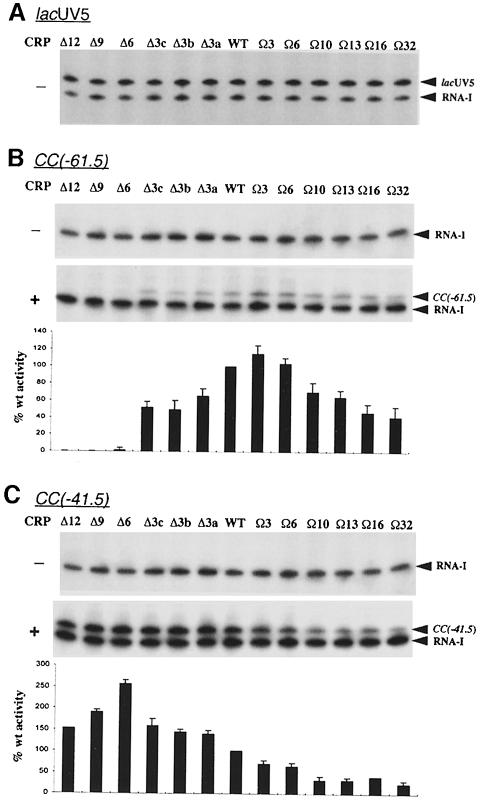
The effects of α subunit linker length on CRP-dependent transcription were assessed in vitro by performing multiple round transcription assays with template DNA carrying either the Class I CRP-dependent promoter, CC(–61.5), or the Class II CRP-dependent promoter, CC(–41.5) (Gaston et al., 1990). As a control, transcription from the CRP-independent lacUV5 promoter was also examined. As expected, transcription from this promoter occurred in the absence of CRP, and was not appreciably affected by alterations in linker length (⩽10% variation; Figure 3A), whereas under the same conditions (without CRP) no transcription was detectable from either CRP-dependent promoter by any of the RNAPs (Figure 3B and C). When CRP was included in the transcription reactions, transcription occurred from both CRP-dependent promoters in the presence of wild-type RNAP. Shortening the α inter-domain linker caused a sharp decrease in CRP-dependent transcriptional activity at the CC(–61.5) promoter. Thus, deletion of three amino acids resulted in a 35–50% decrease in CRP-dependent transcription from this promoter, depending upon which amino acid triplet was removed, while deletion of a further three amino acids almost completely abolished transcription (Figure 3B). Insertion of three amino acids in the inter-domain linker caused a small but reproducible enhancement in the response of RNAP to CRP at the Class I promoter (∼20% increase), whereas further extension of the α linker resulted in a gradual decrease in CRP-dependent activity, with RNAP containing the Ω32 α derivative initiating transcription ∼40–50% as efficiently as the wild-type enzyme.
Fig. 3. In vitro transcription from CRP-dependent and CRP-independent promoters by RNAP reconstituted with α subunits containing wild-type or altered inter-domain linkers. Transcription was carried out at the lacUV5 promoter in the absence of CRP (A), and at the CC(–61.5) and CC(–41.5) promoters (B and C, respectively) in the absence (–) or presence (+) of wild-type CRP. The mutant α subunits present in each reconstituted RNAP are indicated above the gel in each panel. Concentrations of RNAP were: Δ12α RNAP, 15.0 nM; Δ9α RNAP, 14.6 nM; Δ6α RNAP, 14.6 nM; Δ3cα RNAP, 13.6 nM; Δ3bα RNAP, 14.6 nM; Δ3aα RNAP, 13.2 nM; WTα RNAP, 10.5 nM; Ω3α RNAP, 13.7 nM; Ω6α RNAP, 13.2 nM; Ω10α RNAP, 12.3 nM; Ω13α RNAP, 13.6 nM; Ω16α RNAP, 12.9 nM; Ω32α RNAP, 11.9 nM. The identities of specific transcripts are indicated by arrows (the vector-encoded replication repressor, RNA-I, 108 nucleotides; CC promoters, 123 nucleotides; lacUV5, 131 nucleotides). The abundance of transcripts originating from the CC(–61.5) and CC(–41.5) promoters in the presence of CRP was quantified from three experiments and plotted. The values were calculated as a percentage of transcript obtained with wild-type RNAP (with standard deviations) and are presented below the appropriate transcription gel, aligned with the corresponding gel lane.
With the CC(–41.5) promoter, in contrast to the behaviour of linker-modified RNAPs at CC(–61.5), all RNAPs produced transcripts, regardless of α linker length. However, the level of transcription activity in response to CRP varied markedly. Deletion of the α linker caused an increase in the CRP-dependent activity of RNAP, reaching a maximum with RNAP reconstituted with the Δ6 α derivative (2.5- to 3-fold increase over wild type) (Figure 3C). Further deletion reversed this trend, but even for the Δ12 α derivative the activity remained greater than that conferred by wild-type α. On the other hand, insertions within the linker caused a reduction in transcriptional activity (∼30–75% decrease in activity as the linker is increased by 3–32 amino acids). Thus, the effect of alterations in linker length on CRP-dependent transcription is different at the CC(–41.5) and CC(–61.5) promoters.
Effect of α linker length on the interaction between AR1 of CRP and αCTD
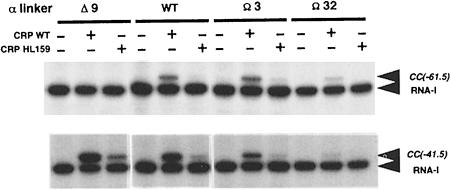
Both Class I and Class II CRP-dependent promoters share a requirement for AR1 of CRP, which is involved in direct interactions with αCTD. The CRP–αCTD interaction can be disrupted by single amino acid changes in AR1, such as the HL159 substitution (Bell et al., 1990; Zhou et al., 1993a). Thus, to determine whether the AR1–αCTD contact is still required for the response of the linker-modified RNAPs to CRP, the ability of three reconstituted mutant RNAPs (containing the Δ9, Ω3 and Ω32 mutant α subunits) to respond to HL159 CRP was examined in a multiple round in vitro transcription assay. The results show that the observed CRP-dependent activity of the three mutant RNAP preparations at CC(–61.5) is severely impaired by the HL159 substitution in AR1 (Figure 4). Similarly, the HL159 substitution also exerted a negative effect on CRP-dependent transcription from the CC(–41.5) promoter (Figure 4). This indicates that the αCTD–AR1 contact remains a requirement for efficient transcription initiation by the linker-modified RNAPs at both classes of CRP-dependent promoter.
Fig. 4. Requirement for CRP AR1 at the CC(–61.5) and CC(–41.5) promoters. Transcription was carried out with RNAP reconstituted with wild-type (WT), Δ9, Ω3 and Ω32 α subunits in the presence of wild-type or HL159 CRP (20 nM, where indicated). Concentrations of RNAP were: Δ9α RNAP, 14.6 nM; WTα RNAP, 10.5 nM; Ω3α RNAP, 13.7 nM; Ω32α RNAP, 11.9 nM. The identities of specific transcripts are indicated by arrows.
Effect of α linker length on interactions between αCTD and upstream DNA sequences at Class II CRP-dependent promoters
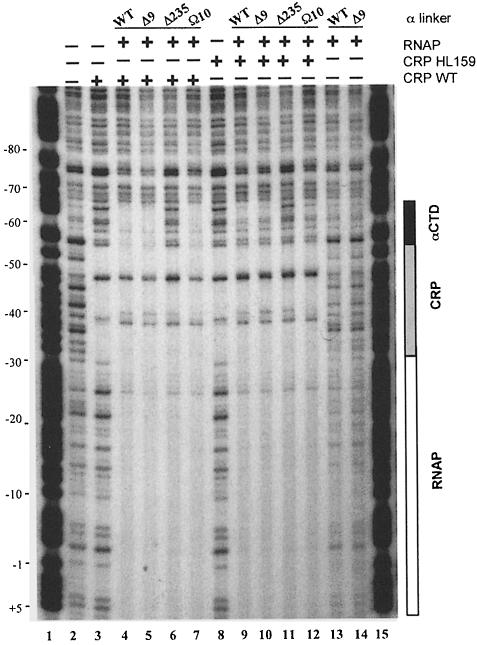
At Class II CRP-dependent promoters, CRP recruits αCTD to the DNA segment upstream of the CRP-binding site (Attey et al., 1994; Savery et al., 1995; Belyaeva et al., 1996, 1998). The requirement of AR1 for CRP-dependent activation of transcription from the CC(–41.5) promoter following deletion of nine amino acids from the α subunit inter-domain linker suggests that αCTD may still be recruited to upstream promoter sequences (despite the decrease in linker length). To explore the location of αCTD in RNAP–CRP–DNA ternary complexes at CC(–41.5), DNase I footprinting was performed with RNAP preparations carrying either the Δ9 or Ω10 α variants (Figure 5). The results showed that wild-type RNAP or RNAP reconstituted with the Δ9 α subunit affords only weak protection of the core promoter sequences in the absence of CRP (Figure 5, lanes 13 and 14). CRP alone binds to and protects the DNA site for CRP centred at –41.5 (Figure 5, lane 3). In the presence of wild-type CRP, the protection afforded by wild-type RNAP and RNAP reconstituted with the Δ9 or Ω10 α subunits was both enhanced and extended upstream of the CRP-binding site (Figure 5, lanes 4, 5 and 7). As expected, the protection upstream of CRP was not evident with RNAP reconstituted with the C–terminal-deleted Δ235 α mutant (Figure 5, lane 6; Attey et al., 1994). When wild-type CRP was replaced by HL159 CRP in the footprinting assay, the extension of upstream protection by RNAPs containing wild-type, Δ9 and Ω10 α was less pronounced (Figure 5, lanes 9, 10 and 12). However, the enhancement of core promoter protection was retained, presumably due to the CRP AR2–αNTD interaction (Busby and Ebright, 1999). These results indicate that αCTD, tethered to αNTD by a linker shortened by nine amino acids, can still be recruited to DNA sequences upstream of the CRP-binding site at Class II CRP-dependent promoters, and this recruitment remains dependent upon AR1. In addition, extension of the linker does not compromise the ability of αCTD to be recruited by CRP at this promoter.
Fig. 5. DNase I footprinting analysis of complexes formed by linker-modified RNAPs at the CC(–41.5) promoter. Protection of the CC(–41.5) promoter by CRP alone (wild-type and HL159) and RNAP reconstituted with wild-type (WT), Δ9, Ω10 and Δ235 α subunits (with or without wild-type or HL159 CRP) is shown. Protection by αCTD, CRP and RNAP is indicated by a filled box, stippled box and open box, respectively. Lanes 1 and 15, A+G ladder; lane 2, no protein; lane 3, WT CRP; lanes 4–7, WT CRP plus RNAP reconstituted with the α derivative indicated; lane 8, HL159 CRP; lanes 9–12, HL159 CRP plus the RNAP indicated; lanes 13 and 14, the RNAP indicated in the absence of CRP. Base co-ordinates are numbered relative to the transcription startpoint (+1) for CC(–41.5).
Effect of α linker length on open complex formation at CRP-dependent promoters
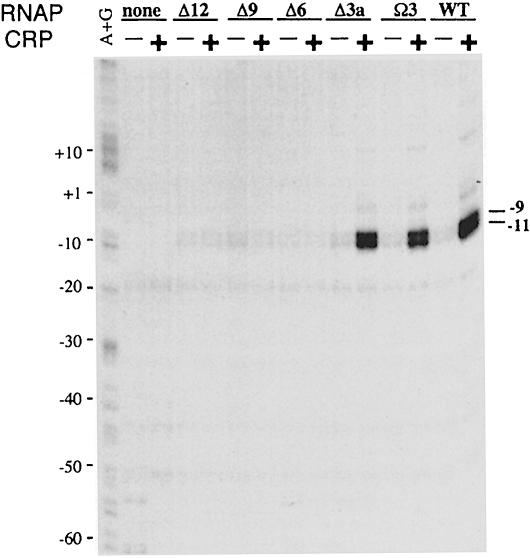
The inhibition of transcription initiation at the Class I CRP-dependent promoter due to removal of amino acids from the α linker could occur either prior to open complex formation or after open complex formation. To discriminate between these two possibilities, we used permanganate to detect open complex formation in the absence of transcript formation. The footprinting was performed at the CC(–61.5) promoter with RNAP preparations carrying linker-deleted α variants, in the presence and absence of CRP. The results showed that, in the presence of CRP, RNAP reconstituted with the Δ3, Ω3 or wild-type α subunits, which respond to CRP in the transcription assay, gave rise to a pair of bands indicative of enhanced reactivity of T residues at positions –9 and –11 in the template strand (Figure 6). In contrast, RNAP containing the Δ6, Δ9 or Δ12 α subunits, which fail to produce transcripts in response to CRP, did not give rise to these bands. As expected, these bands were also absent when CRP was omitted from the assay. The absence of DNA melting in the –10 region catalysed by RNAP reconstituted with the Δ6, Δ9 or Δ12 mutant α subunits, in the presence of CRP, indicates that the impairment of CRP-dependent transcription from the CC(–61.5) promoter caused by shortening the α linker occurs at a step prior to open complex formation.
Fig. 6. Effect of α subunit linker length on open complex formation at the CC(–61.5) promoter. Complexes formed by RNAP reconstituted with the α subunits indicated in the presence or absence of wild-type CRP at the CC(–61.5) promoter were reacted with potassium permanganate. The location of the reactive thymidine residues was determined by PCR with a labelled primer followed by gel electrophoresis and autoradiography. Base co-ordinates are numbered relative to the transcription startpoint.
Incorporation of linker-modified α variants into RNAP in vivo
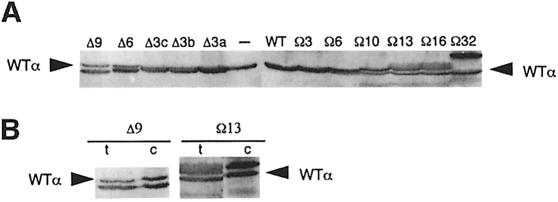
To assess the effect of linker modifications on CRP-dependent transcription in vivo, the mutant rpoA alleles were transferred to the rpoA expression plasmid pLAW2 (Zou et al., 1992). In this plasmid, rpoA is under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lpp/lac dual promoter system and the resultant α derivative is synthesized without a His tag. The level of production of mutant α subunits and the efficiency of their incorporation into RNAP were ascertained by immunoblotting following induction of the lpp/lac promoter for 3–4 generations. Figure 7A shows an immunoblot of SDS–PAGE-fractionated whole-cell extracts using polyclonal anti-α antibody. The experiment showed that each plasmid-encoded α variant was overproduced 2- to 3-fold relative to chromosome-encoded α. To investigate the efficiency with which the plasmid-encoded α subunits are incorporated into RNAP core and holoenzyme, RNAP complexes were immunoprecipitated with a polyclonal anti-β′ antibody, subjected to SDS–PAGE and immunoblotted with anti-α antibody. The blot showed that the ratio of plasmid-encoded mutant α to chromosome-encoded wild-type α was 2.0–2.5:1 (Figure 7B), which is similar to the ratio observed in total cell protein. This result indicates that the different plasmid-encoded α variants accumulate to similar levels in this system and are incorporated into RNAP complexes equally as efficiently as the chromosome-encoded wild-type α. Therefore, in cells expressing the mutant alleles from a pLAW2 derivative, ∼90% of RNAP molecules in the cell would contain at least one plasmid-encoded α, while ∼50% of RNAP molecules would contain two plasmid-encoded α subunits (cf. Tang et al., 1994).
Fig. 7. Relative abundance of chromosomal and plasmid-encoded α subunits in total cell protein and RNAP complexes. (A) Western blot of SDS–PAGE-fractionated cell extracts from RLG4649 harbouring pMGM12-pMGM22 and pMGM32 grown in the presence of IPTG (1 mM). Blots were probed with a polyclonal anti-α antibody. Plasmid-encoded α subunits present in the cell extracts are indicated at the top of each lane (–, extracts from cells not harbouring an rpoA expression plasmid). Wild-type (WT) α is indicated by an arrow. (B) Comparison of the relative abundance of chromosome-encoded wild-type α and plasmid-encoded Δ9 and Ω32 α subunits in total protein (t) and RNAP complexes (c). RNAP complexes were prepared by immunoprecipitation of cell extracts with a polyclonal anti-β′ antibody. Cell extracts (t) and immunoprecipitates (c) were subjected to western blotting as in (A). The chromosome-encoded wild-type α subunit is indicated by an arrow.
Effect of α linker length on CRP-dependent transcription in vivo
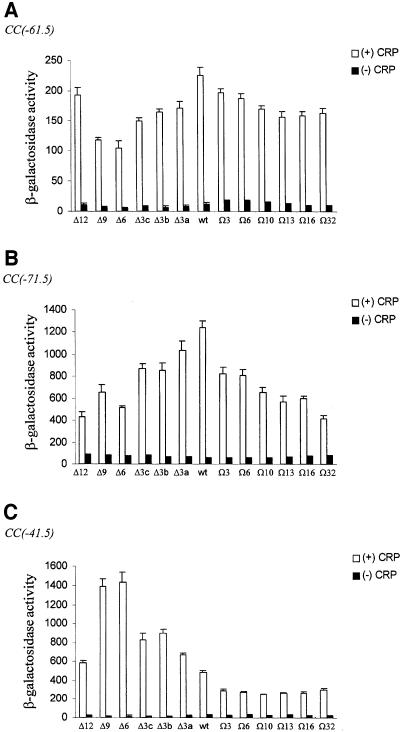
To investigate the effects of altering the length of the α inter-domain linker on CRP-dependent transcription in vivo, the pLAW2 derivatives expressing the mutant rpoA alleles were transformed into M182 or M182Δcrp cells carrying various CRP-dependent promoter–lacZ fusions in single copy. β-galactosidase activities were assayed following induction of mutant α synthesis for 3–4 generations. In general agreement with our observations in vitro, increasing the number of amino acids removed from or inserted into the α inter-domain linker exerted a progressively stronger negative effect on transcription from the Class I CRP-dependent promoter, CC(–61.5), in the crp+ background (Figure 8A). The exception to this trend occurred in cells containing the Δ12 α subunit, where transcriptional activity was only slightly impaired with respect to cells containing solely wild-type RNAP. The CC(–61.5) promoter appeared to be most active in cells containing only wild-type α, rather than in cells also containing the Ω3 α derivative, which confers the highest activity on RNAP at this promoter in vitro. The in vivo activity of the mutant RNAP derivatives at the natural Class I CRP-dependent promoter, lacP1, exhibited the same trend as that observed at CC(–61.5) (data not shown). Qualitatively, the other Class I CRP-dependent promoter tested, CC(–71.5), behaved similarly to CC(–61.5) except for the fact that deletion of 12 amino acids from the α linker resulted in a further decrease in transcriptional activity, consistent with the results obtained at CC(–61.5) in vitro (Figure 8B). Linker extension exerted a more severe effect on CRP-dependent transcription from CC(–71.5) than CC(–61.5).
Fig. 8. Effect of deletions and insertions within the RNAP α subunit inter-domain linker on the activity of CRP-dependent promoters in vivo. β-galactosidase activities were measured in M182 (open bars) or M182Δcrp (filled bars) harbouring a single-copy lacZ fusion to the CC(–61.5) promoter (RLG4650 and NJS1011, respectively) (A), the CC(–71.5) promoter (NJS1020 and WAM137, respectively) (B), the CC(–41.5) promoter (RLG4649 and NJS1001, respectively) (C), and a pLAW2 derivative (pMGM12-pMGM22, pMGM32 or pMGM35) expressing different mutant rpoA alleles as indicated. The activities shown are the mean (with standard deviation) of three or more independent assays and are given in Miller units.
For the Class II CRP-dependent promoter, CC(–41.5), our in vivo observations were also consistent with the transcription analysis in vitro: deletions in the inter-domain linker increase RNAP activity at this promoter relative to cells containing only wild-type RNAP (Figure 8C). The most active RNAP population was present in cells producing the Δ6 α mutant (∼3-fold increase relative to wild-type RNAP). Amino acid insertions within the linker exerted a negative effect on CRP-dependent transcription from this promoter, with the longest linker reducing the activity to ∼60% of the wild-type level. The effect of altering linker length at CC(–41.5) in vivo was also reproduced with the natural Class II CRP-dependent promoter, galP1 (data not shown). The contrast between the effect of linker modification on CRP-dependent activation at the two classes of promoter, as revealed by the in vitro and in vivo assays, is summarized in Figure 9. Comparison of the two panels in Figure 9 also highlights the strong correspondence between the results obtained from the in vitro and in vivo assays at each promoter.
Fig. 9. Comparison of the effect of deletions and insertions within the α inter-domain linker on transcription from CRP-dependent promoters in vitro and in vivo. (A) Amount of transcripts produced by RNAP reconstituted with each mutant α subunit in the presence of CRP at the CC(–41.5) (▴) and CC(–61.5) (○) promoters in vitro. Data are taken from Figure 3. (B) Activity of the CC(–41.5) (▴) and CC(–61.5) (○) promoters in cells expressing chromosome-encoded wild-type α and plasmid-encoded mutant or wild-type α. Data are taken from Figure 8. Values in both panels are expressed as a percentage of the promoter activity in the presence of RNAP containing only wild-type α.
Discussion
During transcription initiation at many natural and artificial promoters, the RNAP αCTD has been shown to bind DNA at various upstream locations (Belyaeva et al., 1996; Murakami et al., 1997). In some cases, binding is dependent upon recruitment of αCTD to the DNA by transcription activator proteins. Previous studies have indicated that the inter-domain linker of the α subunit may provide the necessary flexibility responsible for the mobility of αCTD (Blatter et al., 1994; Jeon et al., 1997). In this work we investigated the length requirements of the α inter-domain linker at two classes of CRP-dependent promoter. Our study shows that the α subunit inter-domain linker can tolerate gross changes in length without impairing assembly of the α subunit into RNAP and without affecting the efficiency of transcription at the activator-independent lacUV5 promoter. These observations are consistent with previous studies which demonstrate that the determinants for assembly of the α subunit into RNAP reside within αNTD and do not include the proposed linker region (Igarashi et al., 1991; Kimura et al., 1994; Kimura and Ishihama, 1995a,b). They also confirm the minimal role played by αCTD in transcription initiation at the lacUV5 promoter (Igarashi and Ishihama, 1991). In contrast, our results establish that inter-domain linker length is important at both classes of CRP-dependent promoter and that the optimum length requirement at each class of promoter differs.
Deletions within the α linker have different effects on transcription at the two classes of CRP-dependent promoter. At the Class I CRP-dependent promoter we tested, deletion of only three residues in the linker exerts a strong negative effect on CRP-dependent transcription in vitro, while deletion of a further three residues almost completely inhibits CRP-dependent transcription. Qualitatively similar results were obtained with the lacP1 promoter (Fujita et al., 2000). It is known that the role of CRP at Class I promoters is to increase the affinity of RNAP for promoter DNA via the CRP AR1–αCTD 287 determinant interaction, thus resulting in an increase in the binding constant, KB, for the formation of the closed complex (Malan et al., 1984; Busby and Ebright, 1999). Our results indicate that at Class I CRP-dependent promoters, linker deletion affects a step in the transcription initiation pathway that precedes open complex formation. Thus, it is very likely that the deletions prevent contact between the 287 determinant on αCTD and AR1 of the downstream CRP subunit (Figure 10).
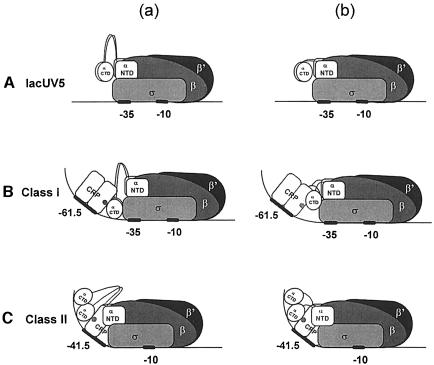
Fig. 10. Models illustrating the effect of decreased α linker length on CRP-dependent and -independent transcription. (A) At activator-independent promoters such as lacUV5, αCTD makes no specific interactions with activators or upstream DNA sequences (a) and is therefore unaffected by alterations in linker length (b). (B) At Class I CRP-dependent promoters, αCTD is recruited by the downstream subunit of the CRP dimer to the promoter region immediately upstream of the –35 hexamer and additionally makes contacts with the σ subunit (Busby and Ebright, 1999) (a). Decreasing linker length prevents αCTD from making contact with AR1 of CRP and/or the template (b). (C) At Class II CRP-dependent promoters, αCTD makes contacts with the upstream subunit of CRP and DNA upstream of the CRP dimer (a). Owing to the proximity of this region of the DNA to αNTD, as a result of CRP-induced bending, RNAP is able to tolerate substantial reductions in linker length (b). Adapted from Busby and Ebright (1999).
At the Class II CRP-dependent promoter, CC(–41.5), deletion of up to 12 amino acids does not impair CRP-dependent transcription. To explain this we suggest that αCTD recruited to upstream DNA sequences by CRP at a Class II promoter is in closer proximity to αNTD than αCTD bound at a Class I promoter (Figure 10). Thus, although αCTD interacts with sequences nearer the –35 region at Class I CRP-dependent promoters than is the case at Class II CRP-dependent promoters [i.e. αCTD contacts the –50 to –41 region at the lac promoter in the presence of CRP, whereas αCTD is recruited to the –86 to –58 region at galP1 (Kolb et al., 1993b; Belyaeva et al., 1996)], the architecture of the ternary CRP–RNAP–promoter DNA complex at the Class II promoter may bring the DNA-binding site for αCTD into closer proximity to the fixed αNTD. However, why should shortening the α inter-domain linker result in an increased response of RNAP to CRP at Class II promoters? The simplest explanation is that deletions within the linker suppress an inhibitory role of αCTD at Class II CRP-dependent promoters. This inhibition arises from the energetic cost of displacing αCTD from the –42 region, its preferred location in the absence of CRP (Belyaeva et al., 1996; Busby and Ebright, 1999). αCTD tethered to RNAP by shorter inter-domain linkers is less able to make stable contacts with DNA immediately upstream of the –35 region (see Figure 10A, b) and will therefore not compete with CRP for this region of the DNA. Thus, the CRP AR1–αCTD interaction, which contributes to KB (Niu et al., 1996; Rhodius et al., 1997) and compensates for the energetic cost of displacing αCTD from its preferred location at Class II CRP-dependent promoters (‘anti-inhibition’; Busby and Ebright, 1997), would make a greater contribution to KB where displacement of αCTD from the –42 region is unnecessary.
As the extent of linker deletion is increased to 9–12 amino acids, the response to CRP at the Class II promoter decreases. It is probable that, with deletions as long as these, linker length somewhat restricts the ability of αCTD to access the upstream CRP monomer and adjacent DNA sequences. Nevertheless, transcription remains AR1 dependent and αCTD can still bind sequences upstream of CRP. It is possible that the loss of mobility conferred on αCTD by decreasing the linker length can be compensated for by increased DNA bending.
Insertions within the linker of more than six amino acids impair transcription from both Class I and Class II CRP-dependent promoters. However, none of these alterations abolish the response to CRP completely. Particularly striking was the observation that transcriptional activity at the CC(–61.5) promoter did not fall below 40% of the level obtained with RNAP containing wild-type α. This suggests that, providing the amino acid sequence organization of the extended linker is maintained, even large insertions within the inter-domain region can be accommodated without preventing efficient recruitment of αCTD by CRP at Class I promoters. The decrease in CRP-dependent transcription caused by linker extension at both classes of promoter may be explained by an increased probability of αCTD making non-productive contacts with more distant sites on the upstream DNA, and/or by conformational constraints imposed by the architecture of the initiation complexes at these classes of promoter.
Our analysis suggests that a different length of inter-domain linker in the RNAP α subunit is required for most efficient transcription at different classes of CRP-dependent promoter. The length of the natural linker appears to be close to optimal for Class I CRP-dependent promoters, but not for Class II promoters. In E.coli, there is only one kind of α subunit, thus the choice of natural linker must be that which offers most overall benefit to the cell. In this respect, it is interesting to note that the E.coli RNAP α subunit linker appears to be optimized for UP-dependent transcription from rRNA P1 promoters (W.Meng, N.Savery, R.Gourse, S.Busby and M.Thomas, unpublished observations), which at fastest growth rates account for ≥50% of total transcription (Bremer and Dennis, 1987).
Materials and methods
Strains and plasmids
The E.coli strains and plasmids used in this work are listed in Table II. Standard molecular biology methods for plasmid isolation and DNA manipulation were used throughout (Sambrook et al., 1989). The plasmids pSR/CC(–41.5) and pSR/CC(–61.5) were constructed by cloning EcoRI–HindIII fragments carrying the CC(–41.5) and CC(–61.5) promoters (Gaston et al., 1990), respectively, into plasmid pSR (Kolb et al., 1995). These promoters are derivatives of the melR promoter with CRP sites centred at position –41.5 and position –61.5 with respect to the transcription startpoint. Plasmid pSR/lacUV5 is described in Savery et al. (1998). The location of the downstream HindIII site used to transfer these promoters to pSR is at position +36 at both CRP-dependent promoters and at +43 at the lacUV5 promoter (Lloyd et al., 1997; Savery et al., 1998). pSR carries the bacteriophage λ oop RNA transcription terminator (to) downstream of the HindIII site, which gives rise to discrete transcripts of predicted length (transcribed vector sequences, including the HindIII site, contribute 88 nucleotides to the resultant transcripts).
Table II. Bacterial strains and plasmids.
| Designation | Genotype/relevant characteristics | Source/reference |
|---|---|---|
| E.coli strains | ||
| BL21(DE3) | F–ompT hsdS (rB–mB–) gal λDE3 lysogen | Studier and Moffatt (1986) |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 (rK–mK+) supE44 relA1 lac λ– [F′ lacIqZΔM15 proAB+ Tn10] | Bullock et al. (1987) |
| DH5α | F– recA1 endA1 gyrA96 thi-1 hsdR17 (rK–mK+) supE44 relA1 Δ(lacZYA-argF)U169 deoR phoA λ– φ80dlacZΔM15 | Jessee (1986) |
| M182 | Δ(lacIPOZYA)X74 galU galK rpsL150 (strA) mel | Casadaban and Cohen (1980) |
| M182 Δcrp | Δcrp derivative of M182 | Busby et al. (1983) |
| DM0068 | MG1655Δcrp::Cm | Kim et al. (1992) |
| RLG4649 | M182 carrying chromosomal λCC(–41.5)-lacZ prophage | Savery et al. (1998) |
| RLG4650 | M182 carrying chromosomal λCC(–61.5)-lacZ prophage | Savery et al. (in preparation)c |
| NJS1020 | M182 carrying chromosomal λCC(–71.5)-lacZ prophage | Savery et al. (in preparation)c |
| NJS1001 | Δcrp::Cm derivative of RLG4649 | this work |
| NJS1011 | Δcrp::Cm derivative of RLG4650 | this work |
| WAM137 | Δcrp::Cm derivative of NJS1020 | this work |
| Plasmidsa | ||
| pHTT7f1NHαb | T7 φ10 promoter directing expression of rpoA on pET21a. His6-tag coding sequence present between the first two codons of rpoA | Tang et al. (1995) |
| pHTT7f1NHαΔ235 | pHTT7f1NHα deleted for the C–terminal 94 codons of rpoA | Tang et al. (1995) |
| pMGM1b | pHTT7f1NHα with Bsu36I site overlapping codons 233–235 of rpoA | this work |
| pMGM2-pMGM11, pMGM31, pMGM34b | pMGM1 derivatives containing deletions or insertions within the linker coding region of rpoA | this work |
| pMKSe2 | pUC19 derivative carrying rpoB (encoding RNA polymerase β subunit) under control of the lac promoter | Severinov et al. (1993) |
| pT7β′ | pT76 derivative carrying rpoC (encoding RNA polymerase β′ subunit) under control of the T7 φ10 promoter | Zalenskaya et al. (1990) |
| pLHN12σ | pBR322 derivative carrying rpoD (encoding RNA polymerase σ subunit) under control of the T7 φ10 promoter | Savery et al. (1998) |
| pSR/lacUV5 | pBR322 derivative carrying lacUV5 promoter cloned upstream of transcription terminator | Savery et al. (1998) |
| pSR/CC(–41.5) | pBR322 derivative carrying CC(–41.5) promoter cloned upstream of transcription terminator | Savery et al. (1998) |
| pSR/CC(–61.5) | pBR322 derivative carrying CC(–61.5) promoter cloned upstream of transcription terminator | Savery et al. (in preparation)c |
| pLAW2b | pBR322 derivative carrying rpoA under lpp/lacUV5 promoter control | Zou et al. (1992) |
| pMGM12-pMGM22, pMGM32, pMGM35b | pLAW2 derivatives in which rpoA contains the modified linker coding sequences from pMGM1-pMGM11, pMGM31 and pMGM34 | this work |
| pBR322 | general purpose cloning vector | Bolivar et al. (1977) |
Two sets of rpoA expression plasmids were used. The pHTT7f1NHα derivatives, overexpressing rpoA alleles encoding His-tagged α subunits with modified inter-domain linkers, were constructed by PCR using different combinations of synthetic oligodeoxyribonucleotide primers. PCR fragments were amplified as HindIII–BamHI fragments, Bsu36I–BamHI fragments or HindIII–Bsu36I fragments from pMGM1, a pHTT7f1NHα derivative containing a unique Bsu36I site overlapping codons 233–235 of rpoA, and substituted for the corresponding fragment of pMGM1 (the HindIII site overlaps codons 229–231 of rpoA and the BamHI site is located immediately downstream of the rpoA stop codon in pHTT7f1NHα and its derivatives). All deletions and insertions were confirmed by DNA sequencing and are listed in Table I. For measurement of trancription activities in vivo, the mutant rpoA alleles were expressed from the tandem lpp-lacUV5 promoter by replacing the EcoRI–SacI fragment of pLAW2, containing the wild-type rpoA linker coding region, with the corresponding fragment from the pHTT7f1NHα derivatives. The λ lysogens RLG4649, RLG4650 and NJS1020, each carrying a single-copy transcriptional fusion of the CC(–41.5), CC(–61.5) and CC(–71.5) promoters to the lacZ gene, respectively, were constructed in strain M182 by the method of Simons et al. (1987), as described in Rao et al. (1994). [The CC(–71.5) promoter is derived from the FF(–71.5) promoter (Wing et al., 1995) and comprises the melR promoter with a DNA-binding site for CRP centred at position –71.5 relative to the transcription startpoint (S.Busby, unpublished).] Δcrp derivatives of these strains were made by introduction of the Δcrp::Cm allele by P1 transduction from DM0068.
Subunit purification and reconstitution of RNAP
The His-tagged linker-modified RNAP α subunits were overproduced in strain BL21(DE3) containing the pHTT7f1NHα derivatives following 3 h of induction of the plasmid-borne T7 promoter and purified by Ni2+-affinity chromatography as described in Tang et al. (1995) and Gaal et al. (1996). Preparation of inclusion bodies of RNAP β, β′ and σ subunits from strains XL1-Blue (pMKSe2), BL21(DE3) (pT7β′) and BL21(DE3) (pLHN12σ), respectively, and reconstitution of RNAP were performed as described in Tang et al. (1995). Wild-type CRP and CRP HL159 were purified from M182Δcrp containing pDCRP or pDCRP HL159 according to Ghosaini et al. (1988).
In vitro transcription
Multiple-round transcription reactions were performed at 37°C for 15 min in a volume of 25 μl containing 100 mM KCl, 40 mM Tris–acetate pH 7.9, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 100 μg/ml bovine serum albumin (BSA), 200 μM ATP, 200 μM CTP, 200 μM GTP, 10 μM UTP and 5 μCi of [α-32P]UTP (DuPont/NEN). Each reaction also contained 0.2 nM supercoiled template DNA [pSR/lacUV5, pSR/CC(–41.5) or pSR/CC(–61.5)] and, where appropriate, 20 nM wild-type or HL159 CRP plus 200 μM cAMP. Template plasmid DNA was prepared using the Qiagen plasmid midi kit and purified further with an Elutip-D column (Schleicher & Schuell). Reactions were initiated by addition of reconstituted RNAP and terminated with 25 μl of stop solution (7 M urea, 1% SDS, 10 mM EDTA, 0.04% bromophenol blue). Samples were fractionated on a 6% acrylamide gel containing 7 M urea and transcript abundance was quantified using a PhosphorImager (Bio-Rad). The activities of the reconstituted RNAPs were titrated at the constitutive RNA-I promoter present on pSR/CC(–41.5) in the absence of CRP, and amounts giving rise to equivalent activity (between 10 and 15 nM each reconstituted RNAP) were used to measure RNAP activity at the test promoters.
DNase I footprinting
DNase I footprinting was performed according to Belyaeva et al. (1998) and Savery et al. (1996). The PstI–HindIII fragment carrying the CC(–41.5) promoter from pSR/CC(–41.5), in which the downstream HindIII site is located at position +36 with respect to the transcription initiation site (Lloyd et al., 1997), was purified on a 7.5% acrylamide gel and labelled with [γ-32P]ATP (DuPont/NEN) at the HindIII end on the non-template strand using T4 polynucleotide kinase. The labelled fragment (0.5–1.0 nM) was incubated at 37°C for 30 min in a total volume of 20 μl of buffer containing 5 mM MgCl2, 50 mM potassium glutamate, 1 mM DTT, 500 μg/ml BSA, 20 mM HEPES pH 8.0 and, where applicable, reconstituted RNAP (100 nM RNAP reconstituted with His-tagged wild-type α, 150 nM RNAP reconstituted with Δ9 α, 170 nM RNAP reconstituted with Δ235 α and 100 nM RNAP reconstituted with Ω10 α) and 100 nM wild-type or HL159 CRP plus 200 μM cAMP. Digestion was allowed to occur at room temperature for 30–60 s following addition of 3 μl of an appropriate DNase I dilution. Reactions were terminated by addition of 200 μl of stop solution (0.3 M sodium acetate, 10 mM EDTA). Following phenol extraction and ethanol precipitation, samples were fractionated on a 6% acrylamide–7 M urea gel and visualized by autoradiography. Footprinting gels were calibrated with Maxam–Gilbert A+G sequencing reactions carried out on the same DNA fragment.
Permanganate footprinting
Permanganate footprinting was performed according to Savery et al. (1996). Plasmid DNA (10 nM) was incubated with ∼100 nM reconstituted RNAP, with or without 20 nM CRP plus 200 μM cAMP, in a total volume of 20 μl of binding buffer (5% glycerol, 0.5 M NaCl, 25 mM MgCl2, 0.5 mM EDTA, 5 mM DTT, 0.25 mg/ml BSA, 100 mM Tris–HCl pH 8.0) at 37°C for 30 min. One microlitre of freshly made 200 mM KMnO4 was added to the binding mixture and the incubation was continued for 4 min. The reaction was terminated by adding 50 μl of stop buffer (3 M ammonium acetate, 0.1 mM EDTA, 1.5 M β–mercaptoethanol), whereupon the DNA was purified by phenol extraction and used as a PCR template. The PCR was carried out with Dynazyme II (FMC) and a single primer, P18 (5′-GGCCCTTTCGTCTTCAAGAATTC-3′), 32P-labelled at the 5′ end with T4 polynucleotide kinase. PCR products were purified by phenol extraction and analysed on a 6% polyacrylamide–7 M urea gel alongside a Sanger G+A DNA sequencing ladder.
Immunoprecipitation and Western blotting
Determination of the ratio of mutant to wild-type α subunits in whole cells and in RNAP complexes was performed according to Zou et al. (1997). Cultures of RLG4649 containing pLAW2 derivatives were grown to mid-logarithmic phase and induced as described for the β-galactosidase assay (see below). For determination of the ratio of mutant to wild-type α in total cellular protein, cells were lysed in denaturing buffer (50 mM Tris–HCl pH 6.8, 100 mM DTT, 2% SDS, 10% glycerol, 0.1% bromophenol blue), whereas for the determination of the ratio in RNAP complexes, cells were disrupted by sonication in non-denaturing buffer (32 mM Tris–HCl pH 8.0, 0.8 mM MgCl2, 16 mM KCl, 0.8 mM EDTA, 0.4% Brij-58, 0.8 mM phenylmethylsulfonyl fluoride, 0.8 mM benzamidine, 0.8 mM DTT, 0.4 mg/ml lysozyme, 20% sucrose), following which RNAP complexes (α2ββ′ and α2ββ′σ) were immunoprecipitated with anti-β′ antibodies. Both whole cell protein and immunoprecipitates were subjected to SDS–PAGE (10% gel) and western blot analysis with anti-α antibodies. Immunopositive bands due to wild-type and linker-modified α, if well separated, were analysed densitometrically.
Measurement of β-galactosidase activity
Overnight cultures of lacZ fusion strains transformed with pLAW2 derivatives were diluted 1:50 in fresh pre-warmed LB broth containing ampicillin (100 μg/ml) and 1 mM IPTG, and grown at 37°C with vigorous shaking for 3–4 generations (OD600 ∼0.3–0.5). β-galactosidase activity was determined by the method of Miller (1972) using the chloroform–SDS procedure and the results are expressed in Miller units. Results presented are the averages from three or more independent assays.
Acknowledgments
Acknowledgements
We would like to thank Akira Ishihama for providing RNAP α subunit and β′ subunit antibodies and Nobuyuki Fujita for communicating unpublished results. Also, thanks are due to Georgina S.Lloyd and Tamara Belyaeva for their excellent technical assistance. This work was supported by project grant 050794 from The Wellcome Trust to M.S.T. and S.J.W.B.
References
- Attey A., Belyaeva, T., Savery, N., Hoggett, J., Fujita, N., Ishihama, A. and Busby, S. (1994) Interaction between cyclic AMP receptor protein and the α subunit of RNA polymerase at the E.coli galP1 promoter. Nucleic Acids Res., 22, 4375–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A., Gaston, K., Williams, R., Chapman, K., Kolb, A., Buc, H., Minchin, S., Williams, J. and Busby, S. (1990) Mutations that alter the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res., 18, 7243–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva T., Bown, J., Fujita, N., Ishihama, A. and Busby, S. (1996) Location of the C–terminal domain of the RNA polymerase α subunit in different open complexes at the E.coli galactose operon regulatory region. Nucleic Acids Res., 24, 2243–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva T., Rhodius, V., Webster, C. and Busby, S. (1998) Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organization of the RNA polymerase α subunits. J. Mol. Biol., 277, 789–804. [DOI] [PubMed] [Google Scholar]
- Berg O. and von Hippel, P. (1988) Selection of DNA binding sites by regulatory proteins. The binding specificity of cyclic AMP receptor protein to recognition sites. J. Mol. Biol., 200, 709–723. [DOI] [PubMed] [Google Scholar]
- Blatter E.E., Ross, W., Tang, H., Gourse, R.L. and Ebright, R.H. (1994) Domain organization of RNA polymerase α subunit: C–terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell, 78, 889–896. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez, R.L., Greene, P.J., Betlach, M.C., Heynecker, H.L., Boyer, H.W., Crosa, J.H. and Falkow, S. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113. [PubMed] [Google Scholar]
- Bremer H. and Dennis,P.P. (1987) Modulation of chemical composition and other parameters of the cell by growth rate. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1527–1542. [Google Scholar]
- Bullock W.O., Fernandez, J.M. and Short, J.M. (1987) XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. Biotechniques, 5, 376–378. [Google Scholar]
- Busby S. and Ebright, R.H. (1997) Transcription activation at Class II CAP-dependent promoters. Mol. Microbiol., 23, 853–859. [DOI] [PubMed] [Google Scholar]
- Busby S. and Ebright, R.H. (1999) Transcription activation by catabolite activator protein (CAP). J. Mol. Biol., 293, 199–213. [DOI] [PubMed] [Google Scholar]
- Busby S. and Kolb,A. (1996) The CAP modulon. In Lin,E.C.C. and Lynch,A.S. (eds), Regulation of Gene Expression in Escherichia coli. R.G.Landes Co, Austin, TX, pp. 255–279. [Google Scholar]
- Busby S., Kotlarz, D. and Buc, H. (1983) Deletion mutagenesis of the Escherichia coli galactose operon promoter region. J. Mol. Biol., 167, 259–274. [DOI] [PubMed] [Google Scholar]
- Casadaban M.J. and Cohen, S.N. (1980) Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol., 138, 179–207. [DOI] [PubMed] [Google Scholar]
- Ebright R.H. (1993) Transcription activation at class I CAP-dependent promoters. Mol. Microbiol., 8, 797–802. [DOI] [PubMed] [Google Scholar]
- Ebright R.H. and Busby, S. (1995) The Escherichia coli RNA polymerase α subunit: structure and function. Curr. Opin. Genet. Dev., 5, 197–203. [DOI] [PubMed] [Google Scholar]
- Estrem S.T., Gaal, T., Ross, W. and Gourse, R.L. (1998) Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl Acad. Sci. USA, 95, 9761–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Endo,S. and Ishihama,A. (2000) Structural requirements for the interdomain linker of α subunit of Escherichia coli RNA polymerase. Biochemistry, in press. [DOI] [PubMed] [Google Scholar]
- Gaal T., Ross, W., Blatter, E.E., Tang, H., Jia, X., Krishnan, V.V., Assa-Munt, N., Ebright, R.H. and Gourse, R.L. (1996) DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev., 10, 16–26. [DOI] [PubMed] [Google Scholar]
- Gaston K., Bell, A., Kolb, A., Buc, H. and Busby, S. (1990) Stringent spacing requirements for transcription activation by CRP. Cell, 62, 733–740. [DOI] [PubMed] [Google Scholar]
- Ghosaini L.R., Brown, A.M. and Sturtevant, J.M. (1988) Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry, 27, 5257–5261. [DOI] [PubMed] [Google Scholar]
- Giladi H., Murakami, K., Ishihama, A. and Oppenheim, A.B. (1996) Identification of an UP element within the IHF binding site at the PL1–PL2 tandem promoter of bacteriophage λ. J. Mol. Biol., 260, 484–491. [DOI] [PubMed] [Google Scholar]
- Igarashi K. and Ishihama, A. (1991) Bipartite functional map of the E.coli RNA polymerase α subunit: involvement of the C–terminal region in transcription activation by cAMP-CRP. Cell, 65, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Fujita, N. and Ishihama, A. (1991) Identification of a subunit assembly domain in the α subunit of Escherichia coli RNA polymerase. J. Mol. Biol., 218, 1–6. [PubMed] [Google Scholar]
- Ishihama A. (1993) Protein–protein communication within the transcription apparatus. J. Bacteriol., 175, 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y.H., Negishi, T., Shirakawa, M., Yamazaki, T., Fujita, N., Ishihama, A. and Kyogoku, Y. (1995) Solution structure of the activator contact domain of the RNA polymerase α subunit. Science, 270, 1495–1497. [DOI] [PubMed] [Google Scholar]
- Jeon Y.H., Yamazaki, T., Otomo, T., Ishihama, A. and Kyogoku, Y. (1997) Flexible linker in the RNA polymerase α subunit facilitates the independent motion of the C–terminal activator contact domain. J. Mol. Biol., 267, 953–962. [DOI] [PubMed] [Google Scholar]
- Jessee J. (1986) New subcloning efficiency competent cells: >1 × 106 transformants/μg. Focus (BRL), 8, 9–10. [Google Scholar]
- Kainz M. and Gourse, R.L. (1998) The C–terminal domain of the α subunit of Escherichia coli RNA polymerase is required for Rho-dependent transcription termination. J. Mol. Biol., 284, 1379–1390. [DOI] [PubMed] [Google Scholar]
- Kim J., Adhya, S. and Garges, S. (1992) Allosteric changes in the cAMP receptor protein of Escherichia coli–hinge reorientation. Proc. Natl Acad. Sci. USA, 89, 9700–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. and Ishihama, A. (1995a) Functional map of the α subunit of Escherichia coli RNA polymerase: insertion analysis of the amino–terminal assembly domain. J. Mol. Biol., 248, 756–767. [DOI] [PubMed] [Google Scholar]
- Kimura M. and Ishihama, A. (1995b) Functional map of the α subunit of Escherichia coli RNA polymerase: amino acid substitution within the amino–terminal assembly domain. J. Mol. Biol., 254, 342–349. [DOI] [PubMed] [Google Scholar]
- Kimura M., Fujita, N. and Ishihama, A. (1994) Functional map of the α subunit of Escherichia coli RNA polymerase. Deletion analysis of the amino–terminal assembly domain. J. Mol. Biol., 242, 107–115. [DOI] [PubMed] [Google Scholar]
- Kolb A., Busby, S., Buc, H., Garges, S. and Adhya, S. (1993a) Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem., 62, 749–795. [DOI] [PubMed] [Google Scholar]
- Kolb A., Igarashi, K., Ishihama, A., Lavigne, M., Buckle, M. and Buc, H. (1993b) E.coli RNA polymerase, deleted in the C–terminal part of its α-subunit, interacts differently with the cAMP–CRP complex at the lacP1 and at the galP1 promoter. Nucleic Acids Res., 21, 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Kotlarz, D., Kusan, S. and Ishihama, A. (1995) Selectivity of the Escherichia coli RNA polymerase Eσ38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res., 23, 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law E., Savery, N. and Busby, S. (1999) Interactions between the Escherichia coli cyclic AMP receptor protein and the C–terminal domain of the α subunit of RNA polymerase at Class I promoters. Biochem. J., 337, 415–423. [PMC free article] [PubMed] [Google Scholar]
- Lloyd G.S., Busby, S.J.W. and Savery, N.J. (1997) Spacing requirements for interactions between the C–terminal domain of the α subunit of Escherichia coli RNA polymerase and the cAMP receptor protein. Biochem. J., 330, 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan T.P., Kolb, A., Buc, H., McClure, W.R. (1984) Mechanism of CRP-cAMP activation at the lac operon: transcription initiation activation of the P1 promoter. J. Mol. Biol., 180, 881–909. [DOI] [PubMed] [Google Scholar]
- Miller J. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Murakami K., Owens, J.T., Belyaeva, T., Meares, C.F., Busby, S.J.W. and Ishihama, A. (1997) Positioning of two α subunit carboxy–terminal domains of RNA polymerase along promoter DNA by two transcription factors. Proc. Natl Acad. Sci. USA, 94, 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi T., Fujita, N. and Ishihama, A. (1995) Structural map of the α subunit of Escherichia coli RNA polymerase: structural domains identified by proteolytic cleavage. J. Mol. Biol., 248, 723–728. [DOI] [PubMed] [Google Scholar]
- Niu W., Zhou, Y., Dong, Q., Ebright, Y. and Ebright, R.H. (1994) Characterization of the activation region of Escherichia coli catabolite gene activator protein (CAP). I. Saturation and alanine scanning mutagenesis. J. Mol. Biol., 243, 595–602. [DOI] [PubMed] [Google Scholar]
- Niu W., Kim, Y., Tau, G., Heyduk, T. and Ebright, R.H. (1996) Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell, 87, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L., Ross, W., Appleman, J.A., Gaal, T., Leirmo, S., Schlax, P.J., Record, M.T., Jr and Gourse, R.L. (1994) Factor-independent activation of rrnB P1. An ‘extended’ promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol., 235, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Rhodius V.A., West, D.M., Webster, C.L., Busby, S.J.W. and Savery, N.J. (1997) Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res., 25, 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Gosink, K.K., Salomon, J., Igarishi, K., Zou, C., Ishihama, A., Severinov, K. and Gourse, R.L. (1993) A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science, 262, 1407–1413. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Savery N.J., Rhodius, V.A., Wing, H.J. and Busby, S.J.W. (1995) Transcription activation at Escherichia coli promoters dependent on the cyclic AMP receptor protein: effects of binding sequences for the RNA polymerase α subunit. Biochem. J., 309, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery N.J., Belyaeva,T. and Busby,S.J.W. (1996) Protein–DNA interactions. In Docherty,K. (ed.), Gene Transcription: DNA Binding Proteins, Essential Techniques. Wiley/BIOS Press, Chichester, UK, pp. 1–43. [Google Scholar]
- Savery N.J., Lloyd, G.S., Kainz, M., Gaal, T., Ross, W., Ebright, R.H., Gourse, R.L. and Busby, S.J.W. (1998) Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C–terminal domain of the RNA polymerase α subunit. EMBO J., 12, 3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S., Shields, G. and Steitz, T. (1991) Crystal structure of a CAP–DNA complex: the DNA is bent by 90 degrees. Science, 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Severinov K., Soushko, M., Goldfarb, A. and Nikiforov, V. (1993) Rifampicin region revisited–new rifampicin-resistant and streptolydigin-resistant mutants in the β-subunit of Escherichia coli RNA-polymerase. J. Biol. Chem., 268, 14820–14825. [PubMed] [Google Scholar]
- Simons R.W., Houman, F. and Kleckner, N. (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene, 53, 85–96. [DOI] [PubMed] [Google Scholar]
- Studier F.W. and Moffatt, B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Tagami H. and Aiba, H. (1999) An inactive open complex mediated by an UP element at Escherichia coli promoters. Proc. Natl Acad. Sci. USA, 96, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Severinov, K., Goldfarb, A., Fenyo, D., Chait, B. and Ebright, R.H. (1994) Location, structure and function of the target of a transcriptional activator protein. Genes Dev., 8, 3058–3067. [DOI] [PubMed] [Google Scholar]
- Tang H., Severinov, K., Goldfarb, A. and Ebright, R.H. (1995) Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA, 92, 4902–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushida C. and Aiba, H. (1990) Helical phase dependent action of CRP: effect of the distance between the CRP site and the –35 region on promoter activity. Nucleic Acids Res., 18, 6325–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing H., Williams, S. and Busby, S. (1995) Spacing requirements for transcription activation by Escherichia coli FNR protein. J. Bacteriol., 177, 6704–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.M. and Crothers, D.M. (1984) The locus of sequence-directed and protein-induced DNA bending. Nature, 308, 509–513. [DOI] [PubMed] [Google Scholar]
- Zalenskaya K., Lee, J.Y., Gujuluva, C.N., Shin, Y.K., Slutsky, M. and Goldfarb, A. (1990) Recombinant RNA polymerase-inducible overexpression, purification and assembly of Escherichia coli rpo gene-products. Gene, 89, 7–12. [DOI] [PubMed] [Google Scholar]
- Zhang G. and Darst, S. (1998) Structure of the Escherichia coli RNA polymerase α subunit amino–terminal domain. Science, 281, 262–266. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhang, X. and Ebright, R.H. (1993a) Identification of the activating region of CAP: isolation and characterization of mutants of CAP specifically defective in transcription activation. Proc. Natl Acad. Sci. USA, 90, 6081–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Busby, S. and Ebright, R.H. (1993b) Identification of the functional subunit of a dimeric transcription activator protein by use of ‘oriented heterodimers’. Cell, 73, 375–379. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Merkel, T.J. and Ebright, R.H. (1994a) Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP) II. Role at class I and class II CAP-dependent promoters. J. Mol. Biol., 243, 603–610. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Pendergrast, P.S., Bell, A., Williams, R., Busby, S. and Ebright, R.H. (1994b) The functional subunit of a dimeric transcription activator protein depends on promoter architecture. EMBO J., 13, 4549–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Fujita, N., Igarishi, K. and Ishihama, A. (1992) Mapping the cAMP receptor protein contact site on the α subunit of Escherichia coli RNA polymerase. Mol. Microbiol., 6, 2599–2605. [DOI] [PubMed] [Google Scholar]
- Zou C., Thomas, M.S., Keen, J. and Glass, R. (1997) A nested set of C–terminal deletions of the α subunit of Escherichia coli RNA polymerase define regions concerned with assembly, proteolysis, stability and transcriptional activation in vivo. Genes Cells, 2, 81–94. [DOI] [PubMed] [Google Scholar]