Abstract
Despite extensive efforts in tuberculosis (TB) drug research, very few novel inhibitors have been discovered. This issue emphasizes the need for innovative methods to discover new anti-TB drugs. In this study, we established a new high-throughput screen (HTS) platform technology that differs from traditional TB drug screens because it utilizes Mycobacterial–Protein Fragment Complementation (M-PFC) to identify small molecule inhibitors of protein–protein interactions in mycobacteria. Several examples of protein–protein interactions were tested with M-PFC to highlight the diversity of selectable drug targets that could be used for screening. These included interactions of essential regulators (IdeR dimerization), enzymatic complexes (LeuCD), secretory antigens (Cfp10-Esat6), and signaling pathways (DevR dimerization). The feasibility of M-PFC in a HTS platform setting was tested by performing a proof-of-concept quantitative HTS of 3,600 small molecule compounds on DevR–DevR interaction, which was chosen because of its strong implications in Mycobacterium tuberculosis persistence and the need for effective drugs against latent TB. The calculated Z′-factor was consistently ≥0.8, indicating a robust and reproducible assay. Completion of the proof-of-concept screen allowed for the identification of advantages and disadvantages in the current assay design, where improvements made will further pioneer M-PFC-based applications in a large-scale HTS format.
Introduction
Mycobacterium tuberculosis (Mtb) is an infectious organism that causes the development and progression of tuberculosis (TB) in humans and claims ∼2 million lives per year.1 The impact of this disease is devastating considering that TB is also a leading cause of death among people infected with human immunodeficiency virus.2 Despite the availability of the Bacillus-Calmette Guerin (BCG) vaccine and current chemotherapeutic drugs, neither of these approaches has proven completely effective in the prevention or treatment of TB.3–5 This problem is a direct consequence of the sharp rise of multidrug-resistant/extensively drug-resistant strains, as well as the known limitations of current anti-TB drugs and their inability to act on dormant bacilli. These barriers underscore the need to discover new drug targets and develop new drugs for combating both active and latent TB.
Conventional methods for TB drug discovery are hampered by bottlenecks that point toward a lack of effective, innovative, and rational approaches in efforts to develop new anti-TB drugs. Most rely on the old paradigm of phenotypic screening, where drug-like molecules are often serendipitously identified based on antitubercular activity.6 Results obtained from these empirical screens offer little information on the drug target and lack mechanistic detail.7 On the other hand, target-based screens are often molecularly based and performed in vitro, but face substantial drawbacks because many compounds identified from these screens show no antibacterial activity when tested against whole cells.8–10 Therefore, novel techniques that screen for inhibitors that are specific for their target and are tested against cells offer a great advantage for the discovery of new TB drugs.
The search for inhibitors against protein–protein interactions has garnered much attention in the study of new drug development technologies and represents an emerging paradigm in TB drug discovery. Since biological functions involve a tightly regulated and balanced interplay between the association and dissociation of proteins, the selective disruption of these essential processes can be exploited for the discovery of small molecule inhibitors. However, this approach has been met with significant conceptual and physical challenges.
Skepticism on identifying inhibitors via this method stems from the observation that protein contact surfaces are large (typically ≈1,600 Å2), flat, and buried. These attributes may prevent a small molecule from binding and exerting its effect.11–13 Despite these challenges, the discovery of hot spots and hot regions has pioneered the search for small molecule inhibitors of protein–protein interactions. These hot spots and regions account for the majority of the binding energy between two proteins and consist of conserved, hydrophobic residues that define the protein contact interface.12,14–18 As a result of this research, a diverse set of small molecule inhibitors that target protein–protein interactions has been discovered for both mammalian and bacterial protein complexes in recent years.11,19,20 While novel inhibitors have been found against the RNA polymerase assembly machinery,21,22 ZipA-FtsZ interaction of Escherichia coli,23 and ToxT dimerization of Vibrio cholerae,24,25 none have been discovered against protein–protein interactions of Mtb in a whole-cell-based assay.
Previous applications of protein fragment complementation assays (PCAs) were developed in various organisms and have been a great advancement to the study of protein–protein interactions in biological systems.26–29 However, until now it has been impossible to find inhibitors against protein–protein interactions in mycobacteria due to the lack of tools necessary to detect disruption of protein associations in vivo. This deficiency gave us the rationale to exploit Mycobacterial–Protein Fragment Complementation (M-PFC)30 as a tool for the discovery of drugs against protein–protein interactions in their native environment. It is known that certain interactions like Pup and FabD require mycobacterial-specific host factors31 and thus cannot be investigated fully with other systems. For all M-PFC-related studies, Mycobacterium smegmatis (Msm) is used as a surrogate host for the evaluation of Mtb protein interactions. Two particularly attractive attributes of working with Msm are that it has a faster replication time than Mtb and can be safely handled in BSL2 laboratory settings. In addition, Msm shares many biosynthetic and signaling pathways with Mtb,32,33 and has been shown to properly express, secrete, and post-translationally modify Mtb proteins.32,34–36 Therefore, Msm is a suitable and popular model system for the screening of drugs against Mtb proteins.37,38
Another benefit is that M-PFC technology is optimally designed for detecting cytoplasmic and membrane-bound protein associations within mycobacterial cells through the functional reconstitution of the murine dihydrofolate reductase (DHFR) reporter fragments [F(1,2)) and F(3)],29 and the subsequent survival in trimethoprim (TRIM)-containing media (Fig. 1).30 TRIM is a specific inhibitor of both murine and bacterial DHFR, but an important difference between the two is that murine DHFR has ∼12,000 times lower affinity for TRIM.29 This distinction allows for the selection of positive interactions from the M-PFC expressed proteins that are fused to the murine DHFR fragments.30 The Alamar Blue-TRIM detection assay (AB-TRIM) is an extension of M-PFC in which strength of protein–protein interaction is quantified based on the ability of cells to survive in the presence of TRIM. These metabolically active cells reduce AB from a blue to a fluorescent pink color and then fluorescence is measured.
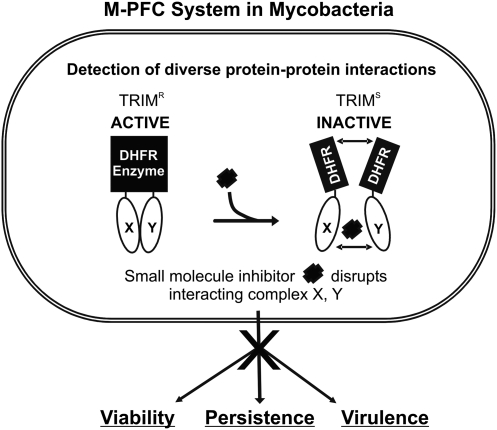
Fig. 1.
M-PFC in mycobacteria. Conceptual diagram of using M-PFC to search for inhibitors that can disrupt key mycobacterial protein interactions involved in essential, persistence, and virulence pathways. Black rectangles represent DHFR F(1,2) and F(3) fragments. X and Y are the translationally fused mycobacterial proteins selected for protein interaction studies with M-PFC and can include interactions involved in cell wall synthesis, two-component signaling, virulence, persistence, secretion, etc. A HTS platform based on M-PFC can detect small molecule inhibitors that disrupt the interaction between X and Y, which leads to an abolishment of downstream effects on viability, persistence, and virulence. HTS, high-throughput screen. DHFR, dihydrofolate reductase; M-PFC, Mycobacterial–Protein Fragment Complementation.
As proof of concept, we developed and optimized a whole-cell-based assay fit for high-throughput screen (HTS) by exploiting M-PFC technology to detect small molecule inhibitors of mycobacterial interactions. The targeted disruption of interactions X and Y can lead to downstream effects on viability, persistence, and virulence (Fig. 1). Current chemotherapy targets only actively replicating Mtb,39,40 so the development of drugs for nonreplicating Mtb represents an area of intense focus in TB drug discovery programs.
DevR is the response regulator in the Mtb DevR/S/T dormancy pathway, which when activated by DevS or DevT, regulates ∼48 genes that are thought to aid Mtb in its transition to the persistent state.41–46 Because DevR has been implicated in promoting mycobacterial persistence and reactivation,41,47,48 we chose to demonstrate the utility of M-PFC technology against DevR dimerization as the initial drug screen target. Completion of this proof-of-concept screen establishes, for the first time, the feasibility of this whole-cell-based approach as a novel technique for antitubercular drug discovery.
Materials and Methods
Strains and Culture Conditions
E. coli strains were grown in LB and supplemented with either kanamycin (Kan; 50 μg/mL; Fisher Scientific, Fairlawn, NJ) or hygromycin (Hyg; 150 μg/mL; Invitrogen™, Carlsbad, CA). Msm strains were grown in 7H9 complete media, which is 7H9 (Difco™ BD™, Sparks, MD) supplemented with 0.5% glycerol, 0.5% glucose, and 0.2% Tween80. When necessary, Kan (25 μg/mL), Hyg (50 μg/mL), and TRIM (6.25–100 μg/mL; Sigma-Aldrich®, St. Louise, MO) were added.
Plasmid Constructs for M-PFC
Plasmid constructs for M-PFC were made by replacing the GCN4 domains from pUAB100-400 with target DNA sequences. Both devR (Rv3133c) and ideR (Rv2711) were PCR amplified with complimentary oligonucleotides (Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/adt) and cloned into pUAB100 at BamHI/ClaI and pUAB200 at MunI/ClaI restriction sites. pUAB300 and pUAB400 were used as backbone vectors for leuC (Rv2988c) and leuD (Rv2987c), and were digested with BamHI/PstI and MunI/PvuII enzymes, respectively, then ligated, and transformed into E. coli. After verifying sequences of all constructs, plasmids were transformed into Msm and plated on 7H11 (BBL™ BD™, Sparks, MD) KanHyg agar to generate the DevR–DevR, IdeR-IdeR, and LeuD-LeuC interacting strains. Protein interaction was detected by streaking each strain and its respective negative control onto 7H11 KanHyg agar plates containing varying concentrations of TRIM. Msm cells expressing the negative control M-PFC constructs contained an unrelated protein fused to one of the DHFR fragments and the target protein fused to the other corresponding DHFR fragment. The Cfp10-Esat6 control strain was made previously.30
AB-TRIM Assay to Determine Strength of Interaction
AB-TRIM assays were performed as described.30 Briefly, Msm cells containing interacting plasmids were first cultured in 7H9 complete KanHyg media to an OD600 ∼1.0 and immediately frozen in 20% glycerol to generate individual glycerol stocks. All experiments were subsequently performed in μClear® 96-well flat-bottom, polystyrene plates (Greiner®, Frickenhausen, Germany). Each well was prefilled with 100 μL of 7H9 complete KanHyg media before adding increasing concentrations of TRIM. Cells were thawed, washed with 1 × PBS, and recovered for 1 h in 7H9 complete broth without any antibiotics. They were then diluted in 7H9 complete KanHyg media and loaded at a final concentration of 105 cells/well. The total volume per well was 200 μL. Plates were incubated for 8–12 h at 37°C before adding 20 μL of Alamar Blue® (AB; AbD Serotec Ltd., Kidlington, Oxford, United Kingdom) and incubated further for 24 h. Fluorescence intensity was measured at 530 nm (ex)/590 nm (em) using a Biotek® Synergy™ HT microplate reader (Biotek®, Winooski, VT) in bottom-read mode, with a sensitivity value of 41. For all AB-TRIM experiments, fluorescence intensity is represented in arbitrary units. Samples were loaded in triplicate wells and experiments were performed at least twice. Data points are represented as mean ± standard deviation (SD).
Rapamycin-Induced Interaction and FK506 Inhibition
The open reading frames of FKBP12 and FRB were amplified with complimentary oligonucleotides and cloned into pUAB100 and pUAB200, respectively, with the same restriction sites as cited above for pUAB100 and pUAB200. Basic protocols for the AB-TRIM assay were then followed, with the addition of increasing concentrations (0–50 nM) of Rapamycin (RAP; LC Laboratories®, Woburn, MC) to induce FRB-FKBP12 interaction under a set TRIM concentration, usually 20 μg/mL. For FK506 (Cell Signaling Technology®, Danvers, MA) inhibition studies, 100 μL of 7H9 complete KanHyg media was supplemented with 20 μg/mL TRIM and 12.5 nM RAP before preloading into wells of a 96-well plate. Increasing concentrations of FK506 were then added to wells before cell loading. Cells were recovered in 7H9 complete media with the addition of 12.5 nM RAP for 1–1.5 h before diluting to 105 cells/well and loading into respective wells. IdeR-IdeR (see Materials and Methods section) was used as control to evaluate the effects of RAP or FK506 on: (1) viability of Msm cells and (2) interaction of an un-related M-PFC interacting strain. Percent interaction was calculated as the [(fluorescent value at a given FK506 concentration)/(untreated FK506 cells)] × 100, where untreated FK506 cells in the presence of RAP and TRIM were representative of 100% interaction of FRB-FKBP12 or IdeR-IdeR. Samples were loaded in triplicate wells and experiments were performed at least twice. Data points are represented as mean ± SD.
Optimization of Drug Screen Conditions
Optimization experiments were performed in 384-well black, clear-bottom plates (Nunc™, Copenhagen, Denmark). For determining optimal cell number, Msm cells were grown to an OD600 of ∼1.0 and then diluted to where wells contained 102 to 106 cells/well in an 80 μL total volume. Cells were allowed to incubate for 8 h at 37°C before addition of 10% AB, or 8 μL. Time points were taken every hour for 30 h with the BioTek® Synergy™ HT, where the plate was internally incubated in the machine. For dimethylsulfoxide (DMSO; Fisher Scientific) and rifampicin (RIF; Sigma-Aldrich) studies, cells at 105 cells/well were incubated in the presence of increasing DMSO or RIF concentrations. In the case of the DMSO experiment, 0–8 μL of solvent was added to respective wells to achieve increasing percent concentrations of DMSO. Samples were loaded in triplicate wells and experiments were performed at least twice. The Z′-factor49 was calculated based on the formula: 1 − [(3 × SDhigh) + (3 × SDlow)/(Avghigh − Avglow)], where high and low values are generated from the DMSO only and RIF-containing wells, respectively.
HTS Protocol
The 3,600 compounds tested in the primary screen were randomly selected from a nonbiased, 50,000 compound library purchased from ChemBridge Corp. (ChemBridge™, San Diego, CA). This library contained synthetic drug-like compounds with an average molecular weight of 400 Da, but ranged from 250 to 500 Da and had high chemical structural diversity. Individual compounds were arranged in 96-well compound source plates as 10 mM solutions in DMSO, which were further diluted 1:5 with DMSO to 2 mM working stocks. Before screening, each source plate was thawed to room temperature and spun down at 900 rpm for 5 min to collect condensation droplets that formed on the lids. Fifteen source plates, or 1,200 compounds, were screened at a time in two separate conditions: No TRIM (to control for viability) and +TRIM at 6.25 μg/mL (to assess compound effects on protein–protein interaction). All procedures involving transfer of compounds to test plates used the iLinkPro® integration software, version 1.1.45.1 (Caliper Life Sciences, Hopkinton, MA), to control the core instruments. These included the Caliper Sciclone ALH 3000 workstation (Caliper Life Sciences) version 4.1.7, as well as the Twister® II Plate Handler (Caliper Life Sciences).
Before primary screening, glycerol stocks of Msm expressing DevR–DevR were thawed, washed twice with 1 × PBS, resuspended in 7H9 complete media, and then recovered for 1 h at 37°C. Forty microliters of 7H9 complete media (+antibiotics) were predispensed into all wells in the 384-well plates, except for the media-only control wells, which contained 80 μL of media. Compounds were delivered to two sets of plates: control plates and test plates. Specifically, 1.5 μL of each compound was added to position one of the 384-well plates, and then serially diluted sixfold to positions 2–4, sequentially. RIF (10 μg/mL) controls were independently added to each plate. After compound loading, cells were diluted and 40 μL of prepared cells was added to each plate so that the final concentration reached 105 cell/well, with a total volume of ∼80 μL per well. Plates were sealed with TempPlate® film (USA Scientific®, Ocala, FL) to prevent evaporation. All prepared plates were then incubated at 37°C for the duration of the entire screen. Plates were first incubated for 8 h before adding 10% AB to each well and then re-sealed. Automatic readings were then taken with the BioTek Synergy HT every 3 h for up to 36 h at 530 nm (ex)/590 nm (em). Data were analyzed at the 24 h time point. Data from each set were analyzed based upon the percentage of inhibition compared to control for each concentration, which was calculated as: 100 × (compound/avg. DMSO control)control − (compound/avg. DMSO control)test. Compounds that were active in two or more concentrations and inhibited interaction by at least ± 4 × SD were selected for secondary screening.
Dose–Response Curves of Potential Hits
Msm expressing Cfp10-Esat6 and DevR–DevR were used in the AB-TRIM assay to test for the effect of the compounds on viability and interaction. Briefly, methods for the AB-TRIM assay were followed for each strain at a set TRIM concentration of 6.25 μg/mL, with the addition of increasing concentrations of compound. For observing effects of the compounds on viability, no TRIM was added to the media. Samples were loaded in triplicate wells; experiments were performed at least twice. Data points are represented as mean ± SD.
Results
The ability of the F(1,2) and F(3) DHFR fragments to associate and dissociate depends on the interaction status of the fused interacting proteins. For some PCA reporter enzymes like green fluorescent protein or yellow fluorescent protein, they remain locked in a stable, folded state once they are assembled.50,51 Therefore, a prerequisite in determining if M-PFC can be used to identify small molecule inhibitors of protein–protein interactions is to test whether the DHFR reporter fragments are reversible through the sequential and forced separation of previously reconstituted DHFR enzyme fragments.
Small Molecule-Based Induction or Inhibition of Protein–Protein Interactions Using M-PFC
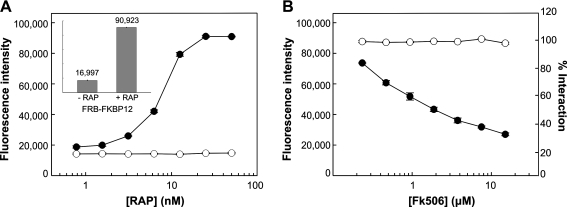
A M-PFC rapamycin (RAP) inducible system was developed to monitor induction and disruption of the entire M-PFC DHFR complex. FRB and FKBP12 were independently fused to F(3) and F(1,2) fragments of DHFR, respectively. Since FRB and FKBP12 interact in the presence of RAP,52,53 Msm cells overexpressing FRB-(F3) and FKBP12-(F1,2) fusions were grown in the presence of RAP and protein association was analyzed in 20 μg/mL TRIM media. Complete association of the FRB-FKBP12 complex was detected in cells at 50 nM RAP (Fig. 2A, inset) and minimally at 3.2 nM RAP, whereas the control expressing an unrelated protein with FKBP12 did not interact at any concentration of RAP. This result suggests that inducible association of FRB-FKBP12 was specific (Fig. 2A). To demonstrate that addition of FK506 (a competitive inhibitor of RAP binding to FKBP12)54,55 disrupts and inhibits the formation of the FRB-FKBP12 complex, Msm cells were cultured in RAP-containing media and exposed to increasing concentrations of FK506. Loss of protein interaction was examined with the AB-TRIM assay. As shown in Figure 2B, 15 μM FK506 was sufficient to achieve a 70% inhibition of complex formation, whereas interaction and viability of the negative control remained unaffected (Fig. 2B). With this result, we demonstrate that FK506 is an effective and specific inhibitor of FRB-FKBP12 interaction, and that disruption of the entire M-PFC DHFR complex could be detected with the AB-TRIM assay. Similarly, a recent study demonstrated that the humanized form of Gaussia princeps luciferase (hGluc), a well-known PCA reporter enzyme, was induced and inhibited through the use of an eukaryotic RAP-inducible system.56 In sum, these results demonstrate that M-PFC can be adapted as a whole-cell-based screening system to identify small molecules that bridge or disrupt protein–protein interactions.
Fig. 2.
Chemically induced association and disruption of protein interactions with the M-PFC AB-TRIM assay. (A) Use of AB-TRIM assay to demonstrate (•) FRB-FKBP12 and (○) neg-FKBP12 (negative control) interaction in the presence RAP (0.78, 1.56, 3.13, 6.25, 12.5, 25.0, and 50.0 nM). Positive interaction of FRB-FKBP12 is shown by survival in TRIM 20 μg/mL media, as indicated by increasing fluorescence intensity of AB. Inset shows bar graph of FRB-FKBP12 interaction at 0 and 50 nM RAP, where relative fluorescence intensities are indicated. (B) Cells were pre-exposed to 12.5 nM RAP to facilitate FRB-FKBP12 interaction before treating them with FK506 (0.23, 0.47, 0.94, 1.88, 3.75, 7.50, and 15.0 μM). Step-wise inhibition of (•) FRB-FKBP12 interaction was observed with increasing FK506 concentration, where 100% interaction was defined as the fluorescence intensity at 0 μM FK506. Msm expressing the M-PFC construct (○) IdeR-IdeR was used as a viability and specificity control against FK506. Each data point represents the mean of triplicate wells ± SD. AB-TRIM, Alamar Blue–trimethoprim; Msm, Mycobacterium smegmatis; RAP, rapamycin; SD, standard deviation.
Identifying Protein Complexes for HTS
To demonstrate the versatility of M-PFC in detecting a diverse set of protein associations in mycobacteria, several protein–protein interactions were selected as key examples for the study and included those involved in essential complexes (IdeR-IdeR, Rv2711),57 enzymatic complexes (LeuC-LeuD, Rv2988c-Rv2987c),58,59 virulence pathways/secretion (Cfp10-Esat6, Rv374-Rv3875),60 and persistence (DevR–DevR, Rv3133c).41 Consistent with current literature, the results show a strong association between IdeR-IdeR, Cfp10-Esat6, and DevR–DevR, and are evident by the growth of Msm on plates containing 30 μg/mL TRIM (Fig. 3A). Alternatively, LeuD-LeuC expressing cells grew at 15 μg/ml of TRIM, suggesting a weak interaction (Fig. 3B). This result also represents the first report of this interacting complex in vivo. Controls expressing unrelated proteins with the test proteins showed no growth on either 30 or 15 μg/mL TRIM plates, confirming that the observed interactions were specific (Fig. 3A, B). These results, as well as those from previously published studies,30,61 highlight the versatility of M-PFC to test different interactions as potential drug targets in an M-PFC-based HTS.
Fig. 3.
Mycobacterial protein–protein interactions with M-PFC. (A) Cells expressing strong interacting pairs and their corresponding negative controls were streaked onto TRIM 30 μg/mL plates and incubated at 37°C for 4 days. (1) neg-DevR; (2) DevR–DevR; (3) Cfp10-neg; (4) Cfp10-Esat6; (5) neg-IdeR; (6) IdeR-IdeR. (B) A weak interacting pair and its negative control were streaked onto TRIM 15 μg/mL plates and incubated at 37°C for 4 days. (1) neg-LeuC; (2) LeuD-LeuC. (C) The AB-TRIM assay (see Materials and Methods section) was performed to assess the strength of DevR–DevR interaction. Strong and weak interactions are determined by the ability to survive in high and low concentrations of TRIM, respectively. Msm cells expressing (•) DevR–DevR were exposed to increasing concentrations of TRIM and maximum interaction was detected between 1.6 and 6.3 μg/mL TRIM. Red arrows point to the concentration of TRIM selected for DevR–DevR in HTS. Inset shows a colorimetric representation of AB being reduced at specified TRIM concentrations. Each data point represents the mean of triplicate wells ± SD.
Optimizing Parameters to Screen for Small Molecule Inhibitors Against DevR–DevR Interaction Using M-PFC
For every protein interaction target, it is critical to determine the strength of interaction with the AB-TRIM assay. As a prerequisite for this proof-of-concept screen, the strength of DevR–DevR interaction was determined (Fig. 3C). Using a 96-well format, the DevR–DevR expressing strain was exposed to increasing concentrations of TRIM for 12 h before addition of AB. Cells with AB were incubated for an additional 24 h to allow AB conversion from blue to pink, and fluorescence was measured at 530 nm (ex)/590 nm (em). We found that DevR–DevR interaction was indeed present. A high level of interaction was observed during growth in 6.25 μg/mL of TRIM, followed by a progressive decline in the strength of interaction at higher concentrations of TRIM, as indicated by the blue wells (Fig. 3C, inset). Thus, TRIM at 6.25 μg/mL was used for detecting DevR–DevR interaction in the proof-of-concept HTS. It was determined that lower TRIM concentrations resulted in nonspecific interactions (data not shown) and higher concentrations of TRIM led to sub-optimal fluorescence intensity and strength in interaction (Fig. 3C).
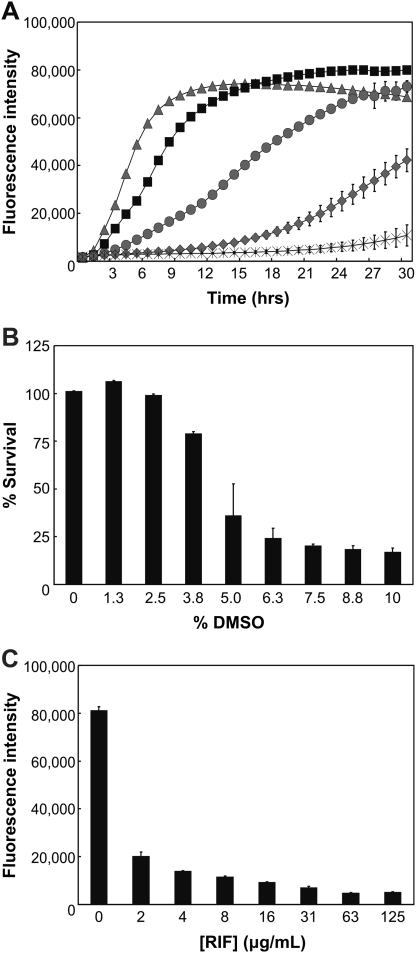
To prepare for HTS, a series of optimization experiments in 384-well plates were performed to evaluate the optimal cell number and effect of DMSO on the HTS assay. First, Msm cells (106 to 102 cells per well) were incubated for 8 h at 37°C. AB was then added and a change in fluorescence was measured for a total of 30 h in 1 h intervals. The data were analyzed by plotting the averages of triplicate wells (Fig. 4A). It was determined that 105 cells changed AB fluorescence in a time-dependent manner and exhibited a growth curve that was neither too fast (106 cells) nor too slow (104 cells). On the other hand, the AB fluorescence from other cell numbers, such as those from 103 or 102 cells, was variable and inconsistent. In addition, we observed that cells at 24 h post-AB (Fig. 4A) reached maximum fluorescence before they entered complete stationary phase, which resulted in no significant change in AB fluorescence. Thus, a concentration of 105 cells/well was used in the proof-of-concept screen, and initial analysis of the data was performed at 24 h post-AB.
Fig. 4.
Optimizing conditions for HTS. (A) Optimal cell concentration was determined by observing growth curves of Msm cells from wells containing ( ) 106, (▪) 105, (
) 106, (▪) 105, ( ) 104, (
) 104, ( ) 103, and (X) 102 cells over the course of 30 h. (B) The tolerance of up to 10% DMSO (8 μL) was determined in Msm cells as described in the Materials and Methods section. Viability of Msm cells was not dramatically affected at a concentration below 2.5% DMSO, and thus a threshold of 2% DMSO was selected for HTS. (C) RIF susceptibility was assessed for Msm cells to determine optimal RIF concentrations for HTS. Cells were susceptible to ≥2 μg/mL RIF by at least 75%, and 10 μg/mL RIF was chosen for HTS to produce low values of ∼8,000 arbitrary units. The data are means ± SD from triplicate wells. Each experiment was performed at least twice. DMSO, dimethylsulfoxide; RIF, rifampicin.
) 103, and (X) 102 cells over the course of 30 h. (B) The tolerance of up to 10% DMSO (8 μL) was determined in Msm cells as described in the Materials and Methods section. Viability of Msm cells was not dramatically affected at a concentration below 2.5% DMSO, and thus a threshold of 2% DMSO was selected for HTS. (C) RIF susceptibility was assessed for Msm cells to determine optimal RIF concentrations for HTS. Cells were susceptible to ≥2 μg/mL RIF by at least 75%, and 10 μg/mL RIF was chosen for HTS to produce low values of ∼8,000 arbitrary units. The data are means ± SD from triplicate wells. Each experiment was performed at least twice. DMSO, dimethylsulfoxide; RIF, rifampicin.
Since the ChemBridge source plates used DMSO to dissolve individual chemical compounds, the effects of this organic solvent were monitored on Msm cells to determine the smallest percentage (or maximum tolerable dose) of DMSO allowed for screening. Three-hundred-eighty-four-well plates were filled with 105 Msm cells in 7H9 complete media and treated with 0–8 μL of DMSO, representing increasing concentrations of this solvent to 10% DMSO. The results indicate that addition of >2.5% DMSO, or 2 μL, lead to a decrease in Msm viability by >10% in the total assay mixture. We chose to include no >2% DMSO as a solvent for the HTS (Fig. 4B).
Since there are no known examples of TB drugs that dissociate mycobacterial protein–protein interactions, RIF was used as an additional control in the study. RIF affects overall viability of Msm and functions to provide baseline fluorescence values for the screen. We expected a compound that affected 100% inhibition on DevR–DevR interaction to have the same fluorescence intensity value as the RIF control wells. A susceptibility test was performed in 384-well plates to select the appropriate RIF concentration for HTS. The results indicate that Msm viability is immediately affected by 75% at 2 μg/mL RIF. This effect is otherwise represented as a drop in fluorescence intensity to ∼20,000 units from untreated cells (∼80,000 units) (Fig. 4C). In the proof-of-concept screen, an RIF concentration of 10 μg/mL was used because it produced baseline fluorescence values of ∼8,000 arbitrary units and allowed for a 10-fold dynamic range between DMSO control wells (high) and RIF control values (low).
Finally, inter- and intra-plate variations were tested by analyzing high (DMSO only wells) and low (RIF wells) values to calculate the Z′-factor and dynamic range as an assessment of the quality of the screen.49 Values were obtained from different quadrants within plates, as well as from corresponding wells among various plates. The calculated Z′-factor for these assay plates was consistently >0.8, and the dynamic range between high and low wells was at least 10-fold. Taken together, optimal conditions for the HTS were determined and implemented, which led to the establishment of a robust and precise screen.
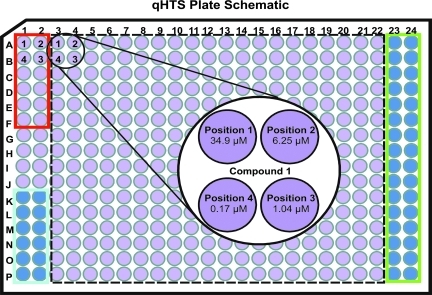
Plate Schematic
The design of the preliminary drug screen included two sets of 384-well plates for every compound source plate. One plate contained 7H9 complete media, which allowed us to examine the effects of inhibitors on cell viability. The second plate contained 6.25 μg/mL TRIM to score for inhibitors that affected protein–protein interaction. These conditions provided an important distinction between a hit that killed the cell nonspecifically versus a hit that modulated interaction of the target proteins. Eighty small chemical compounds randomly derived from a 50,000 ChemBridge library were tested for every set, followed by successive dilutions of these compounds. Our approach was based on quantitative HTS (qHTS).62 This application allowed us to determine the bioactivity of each compound by covering a range of concentrations from 34.9 to 0.17 μM (Fig. 5). Each plate had three internal control areas, as indicated by the colored boxes (Fig. 5). The wells in the red box were for the DMSO control. The volume of DMSO in each position was identical to the amount of DMSO + inhibitor distributed in the compound wells, and did not affect viability, as suggested by Figure 4B. RIF was manually added to the wells in the blue box at a final concentration of 10 μg/mL. Wells in the green box represented the media plus AB only control, where no cells were added to exclude any possibility of media contamination.
Fig. 5.
Design of qHTS: 384-well-plate schematic. Each test plate contains 80 different test compounds (within dotted black line). Each compound is serially diluted sixfold, in a clockwise pattern, as indicated by positions 1–4 within the black circle. (1) 34.9 μM; (2) 6.25 μM; (3) 1.04 μM; (4) 0.17 μM. DMSO vehicle control wells (red box), RIF at 10 μg/mL control wells (blue box) and negative control wells, without cells (green box), are as indicated. qHTS, quantitative HTS.
Adoption of the Assay Protocol on a HTS Platform
We adopted the AB-TRIM assay for rapid assessment with HTS. The final assay protocol depended on media loading (40 μL per well) with a multichannel dispenser and then drug loading using a robotic platform. The 96-well source plates contained 80 small molecules each. About 1.5 μL of compound solution was aseptically transferred to designated assay plates and then subsequently diluted approximately sixfold in a clockwise manner for a total of four dilutions. RIF was then manually added to negative control wells using a multichannel pipettor at 10 μg/mL. After compound loading, 40 μL of cells was dispensed at regular 5.5 min intervals to synchronize the time it took for one plate to be read with the microplate reader. The total volume per well was ∼80 μL except for those in position 1 of the dilution series. All wells in this position contained ∼74.8 μL of total media due to the initial media uptake for subsequent dilutions. Therefore, the concentration of compounds in position 1 was appropriately adjusted to 34.9 μM. The remaining steps were performed at 37°C. First, plates were manually sealed and incubated for 8 h. AB was then dispensed in 5.5 min intervals to a final concentration of 10% (or 8 μL). Plates were resealed and then placed on a stacker that was connected to a microplate reader for data acquisition.
Automatic readings were taken every 3 h for a total of 36 h. One run of a stack of 30 plates, 15 control plates (7H9 only) and 15 test plates (7H9 + TRIM), took ∼2 h 55 min to complete (Table 1). As a whole, the outlined protocol emphasizes the simplicity of the HTS platform, with results easily obtained after 2 days, and can be summarized in four major steps: compound and cell loading, AB addition and plate reading.
Table 1.
Example of HTS Assay Protocol Table
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Media | 40 μL | 7H9 complete media; Hyg, Km, −/+ TRIM: (control vs. test) |
| 2 | Library compounds | 1.5 μLa | 4 concentrations: 34.9 to 0.17 μM |
| 3 | Cells | 40 μL | Mycobacterium smegmatis expressing DevR–DevR |
| 4 | Incubation time | 8 h | 37°C |
| 5 | Alamar Blue | 8 μL | Redox indicator dye; 10% final solution |
| 6 | Incubation time | 36 h | 37°C |
| 7 | Assay readout | 530 nm/590 nm | Biotek microplate reader; fluorescent mode |
1. Black 384-well plate, clear bottom; 1,200 compounds per round; μFill dispenser used to load media into all wells
2. Chembridge source plates in 100% DMSO, 2 mM; 1.5 μL transfer from source plates,a 6.7 μL for dilutions
3. Prepared cells; μFill dispenser used to load cells into designated wells at 105 cells/well
4. Plates sealed to prevent evaporation
5. μFill dispenser used to load Alamar Blue
6. Plates sealed to prevent evaporation; total 30 plates (2 sets of 15); Placed on stacker, fluorescence read every 3 h
7. Biotek microplate reader; data analyzed at 24 h postaddition of Alamar Blue; compared control vs. test plates
For future screens, it is recommended that compound dilutions are performed in DMSO first, before transferring to test plates.
DMSO, dimethylsulfoxide.
Proof-of-Concept Screening of a 3,600-Compound Chemical Library
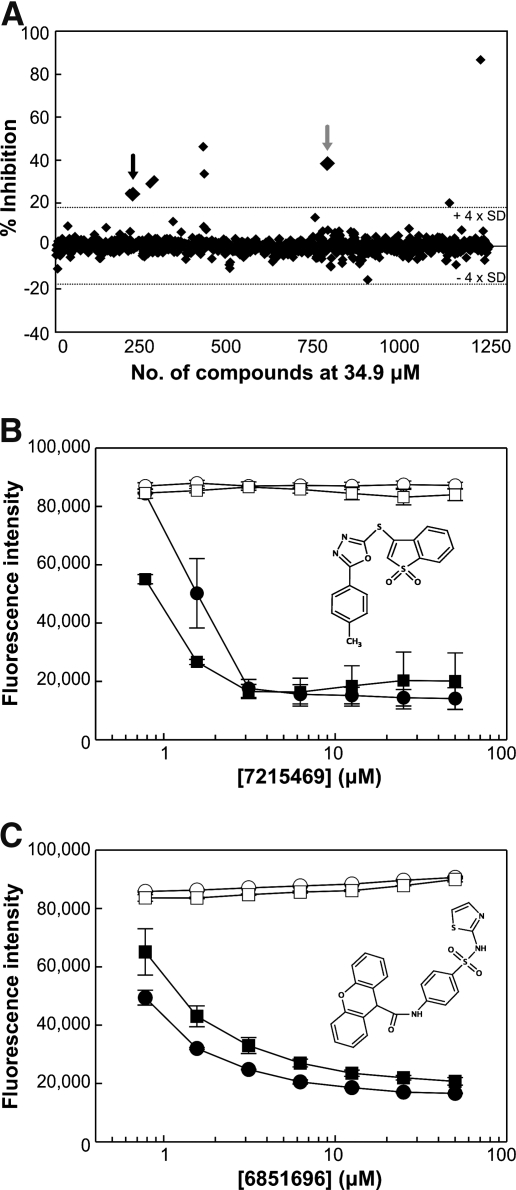
Once parameters were firmly established, a limited, semi-automated proof-of-concept screen involving 3,600 small chemical molecules was performed. These compounds averaged a molecular weight of 400 Da and were part of a nonbiased, chemically diverse library from ChemBridge. After the primary screen, the compounds selected for secondary screening were limited to a cutoff of greater than ± 4 × SD and demonstrated inhibitory activity in at least two or more concentrations. A subset of results at 34.9 μM obtained from the proof-of-concept screen was presented as an example of potential hits that disrupted DevR–DevR interaction, whereby the two compounds of interest were highlighted by the black and gray arrows (Fig. 6A). The first had a 24.5% and 36.3% inhibition in two concentration ranges, 34.9 and 6.25 μM, respectively. The second was active in three concentrations ranges at 34.9, 6.25, and 1.04 μM and inhibited the interaction by 38.4%, 35.9%, and 13.7% (data not shown). All other hits that exhibited high inhibitory activity were representative of those compounds active in single concentration ranges, and were excluded from future validation screens.
Fig. 6.
Example of HTS hits. (A) Representative scatter plot of 1,200 compounds at 34.9 μM analyzed at 24 h. Data points are represented as percent inhibition of DevR–DevR interaction in 6.25 μg/mL TRIM as compared to control, without TRIM. The dotted line represents the 4 × SD threshold (±17.4%). The black and gray arrows represent the two potential hits, 7215469 and 6851696, respectively, identified from the proof-of-concept screen. All other compounds above 4 × SD were representative of those that exhibited inhibitory activity at only 34.9 μM, and were excluded from secondary screens. (B and C) Titration curves of compounds (B) 7215469 and (C) 6851696 on Cfp10-Esat6 (counter screen control) and DevR–DevR (target interaction). Concentrations used are 0.78, 1.56, 3.13, 6.25, 12.5, 25.0, and 50.0 μM. (○) Cfp10-Esat6 and (□) DevR–DevR are assayed without TRIM to look for effects of compound on viability. (•) Cfp10-Esat6 and (▪) DevR–DevR are assayed in media containing 6.25 μg/mL TRIM to assess effects of compound on interaction. Chemical structures of 7215469 and 6851696 are as shown. Each data point for B and C represent the mean of triplicate wells ± SD.
Results from the proof-of-concept screen indicated that the assay could identify potential hits against viability vs. interaction. The two compounds that were identified were subsequently purchased from ChemBridge for secondary screening with a different M-PFC expressing strain as control. The validation assay included side-by-side AB-TRIM assays between the DevR–DevR expressing test and the Cfp10-Esat6 expressing control strains to identify and eliminate nonspecific hits. Those that affected the DHFR reporter enzyme and/or viability in the presence of TRIM were grouped into the nonspecific category and were excluded from future use. Dose–response curves were generated for compounds 7215469 (black arrow) and 6851696 (gray arrow) in no TRIM and 6.25 μg/mL TRIM media. The compounds were subjected to two-fold dilutions from 50 μM to include the concentrations used in the proof-of-concept screen. In the absence of TRIM, compounds 7215469 and 6851696 had no effect on the viability of Cfp10-Esat6 and DevR–DevR expressing cells for up to 50 μM (Fig. 6B, C). However, in its presence, the compounds seemed to affect Cfp10-Esat6 and DevR–DevR interaction in a similar manner, as indicated by the drop in AB fluorescence (Fig. 6B, C). These results suggest that both compounds were not specific in disrupting DevR–DevR interaction because Cfp10-Esat6 interaction was similarly affected. Due to the limited number of compounds tested in the proof-of-concept screen, we did not expect to identify compounds for lead development. Instead, the goal was to demonstrate that this screen could identify potential hits from a primary qHTS and that the validation assay could discriminate against nonspecific hits. All together, these results demonstrate that the HTS platform is fit for future large-scale studies.
Discussion
In this study, we assessed the potential of utilizing M-PFC to screen for inhibitors against protein–protein interactions in mycobacteria. Our goal was two-fold: to establish whether M-PFC technology could be used to facilitate and dissociate protein interactions, and to develop and optimize M-PFC in a HTS platform setting for inhibitors against mycobacterial protein–protein interactions.
The M-PFC RAP-inducible dimerization system was a novel tool initially developed for detecting inducers of protein interactions in mycobacteria, but could also be exploited for future use in ligand-dependent technologies in the mycobacterial field. It was subsequently shown that dissociation of the DHFR reporter fragments through the forced disruption of FRB-FKBP12 could be detected with the AB-TRIM assay (Fig. 2B), although at higher FK506 concentrations than in previously published studies.56 The difference in sensitivity between the M-PFC system and hGluc56 can be explained by several factors. First, the eukaryotic hGluc system uses luciferase as a reporter enzyme with a luminescence signal as its readout.56 It is inherently more sensitive than M-PFC because our DHFR-based PCA depends on fluorescence emissions from AB.30 Second, mycobacteria are already intrinsically resistant to many drugs due to the low permeability barrier of the extraordinarily thick cell wall, as well as porins, multidrug efflux pumps and drug-inactivating enzymes.63–65 Collectively, these characteristics can also lead to decreased sensitivity of FK506 and emphasize the importance of screening for small molecule inhibitors of protein–protein interactions in mycobacterial cells. Finally, Mtb proteins are over-expressed under the control of a constitutive promoter (hsp60) with M-PFC. Over-expression of target genes allows for the study of protein interactions without the dependence of signal-induced expression. However, it can also lead to a saturated system66 that results in decreased sensitivity of FK506 to FKBP12, thereby requiring higher concentrations of drug to disrupt the FRB-FKBP12 complex. As expected, overexpression of certain mycobacterial genes is also toxic (data not shown). In light of these discoveries, efforts are currently underway to develop an inducible promoter system for controlling target protein expression and represent the focus of another study.
Nevertheless, the current M-PFC system is designed to detect a wide array of physiologically relevant protein interaction complexes in mycobacteria. Along with the capability to detect protein sub-domain interfaces (data not shown), these examples provide limitless possibilities for a selectable target in a HTS platform setting. It is important to note that while ideR57 and leuD58,67 are essential in Mtb, M-PFC has the flexibility to accommodate these proteins as potential targets for HTS. This opportunity is possible because ideR is not essential in Msm,68 and exogenous leucine can be supplemented in media when searching for inhibitors against LeuC-LeuD interaction.67
We chose to address the problem of mycobacterial persistence and the lack of drugs against dormant bacilli by performing a proof-of-concept screen for inhibitors against DevR dimerization, which occurs mostly in the N′ terminal domains at the α5 and α6 helices.69 For most response regulators, oligomerization occurs and is necessary for DNA binding.70–72 Therefore, disruption of any part of this process may influence downstream expression of regulated genes. In support of this theory, recent structure-based development strategies led to the discovery of a potential compound that was capable of binding and locking DevR in a conformation that inhibited DNA binding.73
We implemented qHTS62 to assay a chemical library for the generation of a comprehensive data set. As determined from Figure 4A, data are evaluated as an end-point analysis at 24 h post-AB addition. However, it is well known that current TB drugs target bacilli during active growth39 and that some require activation by components expressed only during bacterial replication.39,74 Because the physiology and metabolic state of Msm cells vary at different growth phases, there is a possibility that some compounds may show better activity at certain growth states. This example is readily observed with other compounds that are growth-phase dependent.75 Unfortunately, the two potential hits identified from this proof-of-concept screen were deemed not specific to DevR–DevR, and thus time-course data for these compounds were not assessed for growth-phase dependency. Nevertheless, the option to analyze time-course data generated from a single screen increases the versatility of the M-PFC HTS platform and allows for the collection of additional information on individual compounds (Supplementary Fig. S1).
Of the 3,600 compounds tested, two potential hits were identified from the proof-of-concept screen, resulting in a 0.06% hit rate. By comparison, HTS of small molecule inhibitors against the whole bacterium consistently produce hit rates of 1%–3%.76,77 Therefore, for a single target among all 3,959 genes in the Mtb genome, the hit rate is expected to be significantly lower. This expectation encourages future efforts in performing an extensively larger HTS to accommodate for the specificity of the M-PFC-based screen. Compounds 7215469 and 6851696 inhibited Cfp10-Esat6 interaction with doses similar to DevR–DevR interaction, suggesting a mode of action that was not specific to DevR–DevR (Fig. 6B, C). This behavior was observed only in the presence of TRIM, and suggests one of three possible outcomes: targeted disruption of the DHFR enzyme, adverse effects on components of endogenous essential pathways, or artifacts as a result of the screen.
Some compounds, like 7215469, slightly precipitated out of solution, but still exhibited similar activities for Cfp10-Esat6 and DevR–DevR in TRIM-containing media. The current protocols for the qHTS and the validation assay involve the initial transfer of compound(s) to the wells in the first position, before subsequent dilutions of compound in 7H9. We reasoned that serial dilution in media could have facilitated compound aggregation to influence compound behavior. However, re-evaluation of both compounds under conditions where primary dilutions were first performed in DMSO showed similar nonspecific activity in TRIM containing media (Supplementary Fig. S2).
While examination of these potential outcomes is complex, the validation assay was sufficient in identifying nonspecific hits and allowed us to exclude these compounds from future experiments. Upon further investigation, we observed the presence of several sulfonamide compounds from within a group of hits that was subjected to a more lenient cut-off (data not shown). We hypothesized that this specific group of sulfa drugs, like 6851696 (Fig. 6C, inset), acted synergistically with TRIM to affect the endogenous Msm tetrahydrofolic acid biosynthesis pathway. The clinical significance of sulfa drugs (e.g., sulfamethoxazole78) is well documented in that they are often used in conjunction with TRIM as a potent combination to treat TB, as well as a significant number of other microbial infections.79–85 These two components work together to inhibit specific steps in the synthesis of tetrahydrofolic acid. Blocking this pathway ultimately results in inhibition of DNA, RNA, and protein synthesis. Therefore, we believe that the combination of 6851696 and TRIM blocked essential cellular processes of Msm, which led to a decrease in AB fluorescence.
To overcome this problem, we plan to generate a customized library that will exclude chemical structures showing similarity to sulfa drugs as well as derivatives against mycobacterial DHFR.86,87 A potential caveat to the current assay design is that the screened compounds undergo several dilutions in media, which may increase the number of false-positive hits due to compound precipitation events. With access to a fully automated HTS facility, we plan to minimize these common solubility issues88 by prediluting all initial compounds in DMSO first, before placing them into test plates. The qHTS platform will also be expanded to screen two interacting targets in parallel. One target will act as a direct counter screen for the other, thereby making use of the validation assay in the actual screen and eliminating the need to include a no-TRIM control. We expect that this will be a novel and time-saving approach in the search for new anti-TB drugs against two protein interaction targets.
By optimizing assay conditions for HTS, we were able to complete a proof-of-concept screen against DevR–DevR interaction, where advantages and disadvantages of the M-PFC HTS platform were identified. This identification allowed us to address potential pitfalls and propose solutions for the establishment of more efficient screens. We believe that the data presented in this study demonstrate the novelty and versatility of M-PFC in searching for inhibitors against mycobacterial protein–protein interactions. We conclude that the M-PFC-based HTS is suitable, robust and compatible with the automation necessary for an upscale HTS platform format, where current efforts are underway to screen for 100,000 compounds against multiple protein–protein interactions in mycobacteria.
Supplementary Material
Abbreviations
- AB
Alamar Blue
- AB-TRIM
Alamar Blue–trimethoprim
- BCG
Bacillus-Calmette Guerin
- DHFR
dihydrofolate reductase
- DMSO
dimethylsulfoxide
- Hyg
hygromycin
- HTS
high-throughput screen
- Kan
kanamycin
- M-PFC
Mycobacterial–Protein Fragment Complementation
- Msm
Mycobacterium smegmatis
- Mtb
Mycobacterium tuberculosis
- PCA
protein fragment complementation assay
- qHTS
quantitative high-throughput screen
- RAP
rapamycin
- RIF
rifampicin
- SD
standard deviation
- TB
tuberculosis
- TRIM
trimethoprim
Acknowledgments
The authors would like to thank Amit Singh for critical review of this article, as well as other members of the Steyn, Kutsch, and Hartman laboratories for excellent technical assistance. This work was supported by National Institutes of Health Grant AI058131 (to A.J.C. Steyn). Additional funding was provided by the University of Alabama at Birmingham (UAB) Center for AIDS Research, UAB Center for Free Radical Biology, and UAB Center for Emerging Infections and Emergency Preparedness (CEIEP).
Disclosure Statement
No competing financial interests exist.
References
- 1.TB Fact Sheet: WHO. 2009. www.who.int/tb/publications/2009/tbfactsheet_2009update_one_page.pdf www.who.int/tb/publications/2009/tbfactsheet_2009update_one_page.pdf
- 2.TB/HIV Fact Sheet: WHO. 2009. www.who.int/tb/challenges/hiv/factsheet_hivtv_2009update.pdf www.who.int/tb/challenges/hiv/factsheet_hivtv_2009update.pdf
- 3.Franco-Paredes C. Rouphael N. del Rio C. Santos-Preciado JI. Vaccination strategies to prevent tuberculosis in the new millennium: from BCG to new vaccine candidates. Int J Infect Dis. 2006;10:93–102. doi: 10.1016/j.ijid.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Duncan K. Progress in TB drug development and what is still needed. Tuberculosis (Edinb) 2003;83:201–207. doi: 10.1016/s1472-9792(02)00076-8. [DOI] [PubMed] [Google Scholar]
- 5.Brewer TF. Preventing tuberculosis with Bacillus Calmette-Guerin vaccine: a meta-analysis of the literature. Clin Infect Dis. 2000;31(Suppl 3):S64–S67. doi: 10.1086/314072. [DOI] [PubMed] [Google Scholar]
- 6.Casenghi M. Schoen-Angerer T. Development of new drugs for TB chemotherapy: analysis of the current drug pipeline. MSF. 2006:1–48. [Google Scholar]
- 7.Rubin EJ. Toward a new therapy for tuberculosis. N Engl J Med. 2005;352:933–934. doi: 10.1056/NEJMcibr044438. [DOI] [PubMed] [Google Scholar]
- 8.Payne DJ. Gwynn MN. Holmes DJ. Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 9.Alksne LE. Dunman PM. Target-based antimicrobial drug discovery. Methods Mol Biol. 2008;431:271–283. doi: 10.1007/978-1-60327-032-8_21. [DOI] [PubMed] [Google Scholar]
- 10.Projan SJ. New (and not so new) antibacterial targets—from where and when will the novel drugs come? Curr Opin Pharmacol. 2002;2:513–522. doi: 10.1016/s1471-4892(02)00197-2. [DOI] [PubMed] [Google Scholar]
- 11.Wells JA. McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 12.Bogan AA. Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 13.Jones S. Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clackson T. Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 15.Lo Conte L. Chothia C. Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 16.Ma B. Elkayam T. Wolfson H. Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci U S A. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma B. Nussinov R. Trp/Met/Phe hot spots in protein-protein interactions: potential targets in drug design. Curr Top Med Chem. 2007;7:999–1005. doi: 10.2174/156802607780906717. [DOI] [PubMed] [Google Scholar]
- 18.Keskin O. Ma B. Nussinov R. Hot regions in protein—protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 19.Arkin MR. Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 20.Loregian A. Palu G. Disruption of protein-protein interactions: towards new targets for chemotherapy. J Cell Physiol. 2005;204:750–762. doi: 10.1002/jcp.20356. [DOI] [PubMed] [Google Scholar]
- 21.Andre E. Bastide L. Villain-Guillot P. Latouche J. Rouby J. Leonetti JP. A multiwell assay to isolate compounds inhibiting the assembly of the prokaryotic RNA polymerase. Assay Drug Dev Technol. 2004;2:629–635. doi: 10.1089/adt.2004.2.629. [DOI] [PubMed] [Google Scholar]
- 22.Andre E. Bastide L. Michaux-Charachon S. Gouby A. Villain-Guillot P. Latouche J. Bouchet A. Gualtieri M. Leonetti JP. Novel synthetic molecules targeting the bacterial RNA polymerase assembly. J Antimicrob Chemother. 2006;57:245–251. doi: 10.1093/jac/dki426. [DOI] [PubMed] [Google Scholar]
- 23.Tsao DH. Sutherland AG. Jennings LD. Li Y. Rush TS., 3rd Alvarez JC. Ding W. Dushin EG. Dushin RG. Haney SA. Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorg Med Chem. 2006;14:7953–7961. doi: 10.1016/j.bmc.2006.07.050. [DOI] [PubMed] [Google Scholar]
- 24.Hung DT. Shakhnovich EA. Pierson E. Mekalanos JJ. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 25.Shakhnovich EA. Hung DT. Pierson E. Lee K. Mekalanos JJ. Virstatin inhibits dimerization of the transcriptional activator ToxT. Proc Natl Acad Sci U S A. 2007;104:2372–2377. doi: 10.1073/pnas.0611643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michnick SW. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr Opin Struct Biol. 2001;11:472–477. doi: 10.1016/s0959-440x(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 27.Michnick SW. Ear PH. Manderson EN. Remy I. Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 28.Remy I. Ghaddar G. Michnick SW. Using the beta-lactamase protein-fragment complementation assay to probe dynamic protein-protein interactions. Nat Protoc. 2007;2:2302–2306. doi: 10.1038/nprot.2007.356. [DOI] [PubMed] [Google Scholar]
- 29.Pelletier JN. Campbell-Valois FX. Michnick SW. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc Natl Acad Sci U S A. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh A. Mai D. Kumar A. Steyn AJ. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc Natl Acad Sci U S A. 2006;103:11346–11351. doi: 10.1073/pnas.0602817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearce MJ. Mintseris J. Ferreyra J. Gygi SP. Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs WRJ. Mycobacterium tuberculosis: a once genetically intractable organism. In: Hatfull GF, editor; Jacobs WRJ, editor. Molecular Genetics of Mycobacteria. ASM Press; Washington, DC: 2000. pp. 1–18. [Google Scholar]
- 33.Lee JM. Cho HY. Cho HJ. Ko IJ. Park SW. Baik HS. Oh JH. Eom CY. Kim YM. Kang BS. O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis. J Bacteriol. 2008;190:6795–6804. doi: 10.1128/JB.00401-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garbe T. Harris D. Vordermeier M. Lathigra R. Ivanyi J. Young D. Expression of the Mycobacterium tuberculosis 19-kilodalton antigen in Mycobacterium smegmatis: immunological analysis and evidence of glycosylation. Infect Immun. 1993;61:260–267. doi: 10.1128/iai.61.1.260-267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagchi G. Mayuri Tyagi JS. Hypoxia-responsive expression of Mycobacterium tuberculosis Rv3134c and devR promoters in Mycobacterium smegmatis. Microbiology. 2003;149:2303–2305. doi: 10.1099/mic.0.C0120-0. [DOI] [PubMed] [Google Scholar]
- 36.Harth G. Lee BY. Horwitz MA. High-level heterologous expression and secretion in rapidly growing nonpathogenic mycobacteria of four major Mycobacterium tuberculosis extracellular proteins considered to be leading vaccine candidates and drug targets. Infect Immun. 1997;65:2321–2328. doi: 10.1128/iai.65.6.2321-2328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaturvedi V. Dwivedi N. Tripathi RP. Sinha S. Evaluation of Mycobacterium smegmatis as a possible surrogate screen for selecting molecules active against multi-drug resistant Mycobacterium tuberculosis. J Gen Appl Microbiol. 2007;53:333–337. doi: 10.2323/jgam.53.333. [DOI] [PubMed] [Google Scholar]
- 38.Taneja NK. Tyagi JS. Resazurin reduction assays for screening of anti-tubercular compounds against dormant and actively growing Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. J Antimicrob Chemother. 2007;60:288–293. doi: 10.1093/jac/dkm207. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y. The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 40.Wayne LG. Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park HD. Guinn KM. Harrell MI. Liao R. Voskuil MI. Tompa M. Schoolnik GK. Sherman DR. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dasgupta N. Kapur V. Singh KK. Das TK. Sachdeva S. Jyothisri K. Tyagi JS. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber Lung Dis. 2000;80:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 43.Roberts DM. Liao RP. Wisedchaisri G. Hol WG. Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J Biol Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sousa EH. Tuckerman JR. Gonzalez G. Gilles-Gonzalez MA. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 2007;16:1708–1719. doi: 10.1110/ps.072897707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar A. Toledo JC. Patel RP. Lancaster JR., Jr. Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A. Deshane JS. Crossman DK. Bolisetty S. Yan BS. Kramnik I. Agarwal A. Steyn AJ. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J Biol Chem. 2008;283:18032–18039. doi: 10.1074/jbc.M802274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honer zu Bentrup K. Russell DG. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 2001;9:597–605. doi: 10.1016/s0966-842x(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 48.Malhotra V. Sharma D. Ramanathan VD. Shakila H. Saini DK. Chakravorty S. Das TK. Li Q. Silver RF. Narayanan PR. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol Lett. 2004;231:237–245. doi: 10.1016/S0378-1097(04)00002-3. [DOI] [PubMed] [Google Scholar]
- 49.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 50.Hu CD. Chinenov Y. Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 51.Magliery TJ. Wilson CG. Pan W. Mishler D. Ghosh I. Hamilton AD. Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 52.Chen J. Zheng XF. Brown EJ. Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang J. Choi J. Clardy J. Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:736–744. doi: 10.1107/s0907444998014747. [DOI] [PubMed] [Google Scholar]
- 54.Van Duyne GD. Standaert RF. Karplus PA. Schreiber SL. Clardy J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- 55.Choi J. Chen J. Schreiber SL. Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 56.Remy I. Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez GM. Voskuil MI. Gold B. Schoolnik GK. Smith I. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bange FC. Brown AM. Jacobs WR., Jr. Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect Immun. 1996;64:1794–1799. doi: 10.1128/iai.64.5.1794-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McAdam RA. Weisbrod TR. Martin J. Scuderi JD. Brown AM. Cirillo JD. Bloom BR. Jacobs WR., Jr. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect Immun. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiGiuseppe Champion PA. Cox JS. Protein secretion systems in Mycobacteria. Cell Microbiol. 2007;9:1376–1384. doi: 10.1111/j.1462-5822.2007.00943.x. [DOI] [PubMed] [Google Scholar]
- 61.Callahan B. Nguyen K. Collins A. Valdes K. Caplow M. Crossman DK. Steyn AJ. Eisele L. Derbyshire KM. Conservation of structure and protein-protein interactions mediated by the secreted mycobacterial proteins EsxA, EsxB, and EspA. J Bacteriol. 2009;192:326–335. doi: 10.1128/JB.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inglese J. Auld DS. Jadhav A. Johnson RL. Simeonov A. Yasgar A. Zheng W. Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danilchanka O. Pavlenok M. Niederweis M. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob Agents Chemother. 2008;52:3127–3134. doi: 10.1128/AAC.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen L. Thompson CJ. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 2006;14:304–312. doi: 10.1016/j.tim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 65.Brennan PJ. Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 66.Joshi PB. Hirst M. Malcolm T. Parent J. Mitchell D. Lund K. Sadowski I. Identification of protein interaction antagonists using the repressed trans-activator two-hybrid system. Biotechniques. 2007;42:635–644. doi: 10.2144/000112434. [DOI] [PubMed] [Google Scholar]
- 67.Hondalus MK. Bardarov S. Russell R. Chan J. Jacobs WR., Jr. Bloom BR. Attenuation of and protection induced by a leucine auxotroph of Mycobac-terium tuberculosis. Infect Immun. 2000;68:2888–2898. doi: 10.1128/iai.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dussurget O. Rodriguez M. Smith I. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol. 1996;22:535–544. doi: 10.1046/j.1365-2958.1996.1461511.x. [DOI] [PubMed] [Google Scholar]
- 69.Wisedchaisri G. Wu M. Sherman DR. Hol WG. Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J Mol Biol. 2008;378:227–242. doi: 10.1016/j.jmb.2008.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao R. Mack TR. Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao R. Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maris AE. Sawaya MR. Kaczor-Grzeskowiak M. Jarvis MR. Bearson SM. Kopka ML. Schroder I. Gunsalus RP. Dickerson RE. Dimerization allows DNA target site recognition by the NarL response regulator. Nat Struct Biol. 2002;9:771–778. doi: 10.1038/nsb845. [DOI] [PubMed] [Google Scholar]
- 73.Gupta RK. Thakur TS. Desiraju GR. Tyagi JS. Structure-based design of DevR inhibitor active against nonreplicating Mycobacterium tuberculosis. J Med Chem. 2009;52:6324–6334. doi: 10.1021/jm900358q. [DOI] [PubMed] [Google Scholar]
- 74.Vilcheze C. Jacobs WR., Jr. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu Rev Microbiol. 2007;61:35–50. doi: 10.1146/annurev.micro.61.111606.122346. [DOI] [PubMed] [Google Scholar]
- 75.Beggs WH. Influence of growth phase on the susceptibility of Candida albicans to butoconazole, oxiconazole, and sulconazole. J Antimicrob Chemother. 1985;16:397. doi: 10.1093/jac/16.3.397. [DOI] [PubMed] [Google Scholar]
- 76.Maddry JA. Ananthan S. Goldman RC. Hobrath JV. Kwong CD. Maddox C. Rasmussen L. Reynolds RC. Secrist JA., 3rd Sosa MI. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis (Edinb) 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ananthan S. Faaleolea ER. Goldman RC. Hobrath JV. Kwong CD. Laughon BE. Maddry JA. Mehta A. Rasmussen L. Reynolds RC. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalkut G. Sulfonamides and trimethoprim. Cancer Invest. 1998;16:612–615. doi: 10.3109/07357909809032892. [DOI] [PubMed] [Google Scholar]
- 79.Weidner Wells MA. Macielag MJ. Antibacterial Agents, Sulfonamides. 2003. onlinelibrary.wiley.com/doi/10.1002/0471238961.1921120606152505.a01.pub2/full onlinelibrary.wiley.com/doi/10.1002/0471238961.1921120606152505.a01.pub2/full Wiley Online Library.
- 80.Bush K. Freudenberger JS. Slusarchyk DS. Sykes RB. Meyers E. Activity of sulfa drugs and dihydrofolate reductase inhibitors against Candida albicans. Cell Mol Life Sci. 1982;38:436–437. doi: 10.1007/BF01952625. [DOI] [PubMed] [Google Scholar]
- 81.Suling WJ. Seitz LE. Reynolds RC. Barrow WW. New Mycobacterium avium antifolate shows synergistic effect when used in combination with dihydropteroate synthase inhibitors. Antimicrob Agents Chemother. 2005;49:4801–4803. doi: 10.1128/AAC.49.11.4801-4803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith CL. Powell KR. Review of the sulfonamides and trimethoprim. Pediatr Rev. 2000;21:368. [PubMed] [Google Scholar]
- 83.Porras MC. Lecumberri JN. Castrillon JL. Trimethoprim/sulfamethoxazole and metabolic acidosis in HIV-infected patients. Ann Pharmacother. 1998;32:185–189. doi: 10.1345/aph.17042. [DOI] [PubMed] [Google Scholar]
- 84.Wallace RJ., Jr. Wiss K. Bushby MB. Hollowell DC. In vitro activity of trimethoprim and sulfamethoxazole against the nontuberculous mycobacteria. Rev Infect Dis. 1982;4:326–331. doi: 10.1093/clinids/4.2.326. [DOI] [PubMed] [Google Scholar]
- 85.Wormser GP. Keusch GT. Heel RC. Co-trimoxazole (trimethoprim-sulfamethoxazole) Drugs. 1982;24:459–518. doi: 10.2165/00003495-198224060-00002. [DOI] [PubMed] [Google Scholar]
- 86.Suling WJ. Reynolds RC. Barrow EW. Wilson LN. Piper JR. Barrow WW. Susceptibilities of Mycobacterium tuberculosis and Mycobacterium avium complex to lipophilic deazapteridine derivatives, inhibitors of dihydrofolate reductase. J Antimicrob Chemother. 1998;42:811–815. doi: 10.1093/jac/42.6.811. [DOI] [PubMed] [Google Scholar]
- 87.Suling WJ. Seitz LE. Pathak V. Westbrook L. Barrow EW. Zywno-Van-Ginkel S. Reynolds RC. Piper JR. Barrow WW. Antimycobacterial activities of 2,4-diamino-5-deazapteridine derivatives and effects on mycobacterial dihydrofolate reductase. Antimicrob Agents Chemother. 2000;44:2784–2793. doi: 10.1128/aac.44.10.2784-2793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McGovern SL. Caselli E. Grigorieff N. Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.