Abstract
The metazoan cyclin-dependent kinase Cdk7 was purified originally as part of a biochemical activity called CAK (Cdk-activating kinase) capable of phosphoryla-ting and activating in vitro the Cdks that promote the different cell cycle transitions. Cdk7 is also found in the transcription factor complex TFIIH, suggesting that it participates in vivo in the control of RNA polymerase II. We have examined the physiological role of Cdk7 during the course of Drosophila development. By expressing dominant-negative forms of the kinase, we were able to alter Cdk7 function at given developmental stages. Expression of Cdk7 mutants severely delayed the onset of zygotic transcription in the early embryo, but did not alter the timing of the first 13 embryonic nuclear cycles. These results implicate Cdk7 in the control of transcriptional machinery in vivo. While cell cycle regulation is not sensitive to our manipulations of Cdk7 activity, it suggests that a distinct pool of CAK activity that is unaffected by expression of the cdk7DN mutants is present in these embryos.
Keywords: CAK/Cdk7/Drosophila/mid-blastula transition/transcription
Introduction
The cyclin-dependent kinases (Cdks) constitute a conserved family of molecules involved in the control of cell cycle and transcriptional machineries. Analysis of their function in controlling cell division has pointed to different regulatory mechanisms that modulate their activities: Cdks are themselves controlled by association with either regulatory subunits called cyclins, or inhibitory molecules called CKIs, and by a complex series of phosphorylations and dephosphorylations (reviewed in Morgan, 1995). Three phosphorylation sites have been identified that control the activity of Cdks. Thr14 and Tyr15 (on human Cdc2) are phosphorylated by the Myt1 and Wee1 kinases and keep Cdc2 inactive until dephosphorylated by the Cdc25 phosphatases (reviewed in Solomon and Kaldis, 1998). The third phosphorylation site (Thr169 on human Cdc2) is in the ‘T–loop’; it was first shown to be essential for Cdc2 activity and for viability in Schizosaccharomyces pombe (Gould et al., 1991). An activity called CAK (Cdk-activating kinase) is responsible for phosphorylation of this conserved threonine in all eukaryotes examined so far. Based on a biochemical assay, a putative CAK complex has been purified from starfish and frog oocytes as well as from human cell extracts (Fesquet et al., 1993; Poon et al., 1993; Solomon et al., 1993), which consist of a stoichiometric association between Cdk7/p40MO15, cyclin H (Fisher and Morgan, 1994; Makela et al., 1994; Tassan et al., 1994) and Mat1 (Devault et al., 1995; Fisher et al., 1995; Tassan et al., 1995; Adamczewski et al., 1996). Initially known to phosphorylate and activate the Cdc2–cyclin B complex in vitro, CAK was also shown to phosphorylate the conserved threonine of other Cdks (Cdk2, Cdk3, Cdk4 and Cdk6) (reviewed in Nigg, 1996).
It came as a major surprise more recently that the three CAK subunits are also found in the transcription factor TFIIH, one of the basal transcription factors involved in the formation of the pre-initiation complex with RNA polymerase II (polII) (Roy et al., 1994; Fisher et al., 1995; Serizawa et al., 1995; Shiekhattar et al., 1995; Adamczewski et al., 1996). TFIIH has a kinase activity that is directed towards the C-terminal domain (CTD) of the large subunit of polII whose phosphorylation coincides with the transition from pre-initiation to elongation (Dahmus, 1996). The CAK complex is responsible for the kinase activity of TFIIH and is capable of phosphorylating the CTD of polII, suggesting that Cdk7 is a potential CTD kinase in vivo (Roy et al., 1994; Makela et al., 1995). Even though, in some cases, inactivated CAK-containing TFIIH complex can allow transcription in vitro (Makela et al., 1995), it has been shown that, under conditions where the CTD is required, the Cdk7 kinase is also required for transcription in vitro (Akoulitchev et al., 1995). These results strongly suggest that Cdk7 plays a role in transcription through its participation in the TFIIH complex, although no direct evidence in vivo has been reported so far in higher eukaryotes.
Studies of yeast homologs of Cdk7 give new hints about the biological function of the complex. In S.pombe, Mcs6 and Mcs2, homologs of Cdk7 and cyclin H, respectively, display both CAK and CTD kinase activities in vitro (Molz and Beach, 1993; Buck et al., 1995; Damagnez et al., 1995). mcs2 is an essential gene but its loss does not result in a phenotype consistent with a CAK function. In contrast, Csk1, a putative regulator of Mcs6, is a monomeric kinase that phosphorylates and activates Mcs6/Mcs2 in vitro and in vivo (Hermand et al., 1998), as well as Cdc2/Cdc13 in vitro (Lee et al., 1999). Furthermore, an mcs6ts/csk1Δ strain is deficient in CAK function, suggesting that both kinases are required for CAK activity in S.pombe (Lee et al., 1999). In Saccharomyces cerevisiae, the Cdk7 homolog, Kin28, is devoid of CAK activity in vitro as well as in vivo (Cismowski et al., 1995; Valay et al., 1995; Holstege et al., 1998). Its role in transcription in vivo has been established by the observation that polII transcription is drastically reduced in a kin28ts mutant at restrictive temperature, and this correlates with a decreased phosphorylation of the polII CTD. A search for CAK activity in S.cerevisiae has led to the biochemical characterization of a monomeric kinase termed Cak1 or Civ1, which is only partially related to the Cdk family (Espinoza et al., 1996; Kaldis et al., 1996; Thuret et al., 1996), and mutational analysis provided an in vivo demonstration that Cak1 is required for Cdc28/Cdc2 phosphorylation and activation. Thus, in S.cerevisiae, the CAK and TFIIH kinase functions are distinct and carried out by two separate enzymes.
A more recent report suggested that Cdk7 could be required in vivo for CAK function in flies. Embryos laid from cdk7ts mothers showed decreased Cdc2–T170 phosphorylation after their mothers were kept at restrictive temperature for increasing times (Larochelle et al., 1998). In contrast, Cdk2 phosphorylation is not affected, suggesting that a different function can activate this enzyme separately. Although this is the strongest data so far linking Cdk7 function to Cdc2 activation, one cannot exclude the possibility that an impaired transcriptional activity of Cdk7 could contribute indirectly to such effects.
Here, we address the physiological role of Cdk7 in a developmental system, using ectopic expression of dominant-negative forms of the kinase (Cdk7DN) to alter specifically the function of the kinase in a given tissue and at a precise period of development. In early Drosophila pre-blastoderm embryos, transcription is absent and nuclear division cycles run strictly on maternal information, thus providing a system in which the possible cell cycle function of Cdk7 can be studied in the absence of any interference due to its transcriptional function. At the end of this period, zygotic trancription takes place, allowing testing of the transcriptional role of Cdk7. In these conditions, we showed that alteration of Cdk7 activity induced marked defects during the onset of zygotic transcription. This is strong in vivo evidence for the involvement of a metazoan Cdk7 in the control of the transcriptional machinery. In contrast, the early embryonic division cycles are not affected, thus suggesting that a distinct pool of CAK activity that is unaffected by expression of the cdk7DN mutants can fulfill a cell cycle function in these embryos.
Results
Generating dominant-negative forms of the Cdk7 kinase
In order to analyze in vivo the function of the Cdk7 kinase, we generated modified cdk7 cDNAs with mutations known to produce a dominant-negative effect in several Cdks (van den Heuvel and Harlow, 1993). Two mutants were used, each producing proteins with a single amino acid change (D137R or T170A) and tagged with the hemagglutinin (HA) sequence at the C-terminal end. D137R corresponds to a mutation in the central core of the kinase domain shown previously to inactivate the catalytic function of the kinase. T170A is a mutation of the conserved threonine in the T–loop, whose phosphorylation has been shown to be necessary for full Cdk7 and Kin28 activity in vivo (Labbé et al., 1994; Espinoza et al., 1998; Kimmelman et al., 1999). The two mutants and a wild-type cdk7 cDNA, driven by a UASGAL4 enhancer, were introduced into flies and expressed after crossing with Gal4-expressing lines.
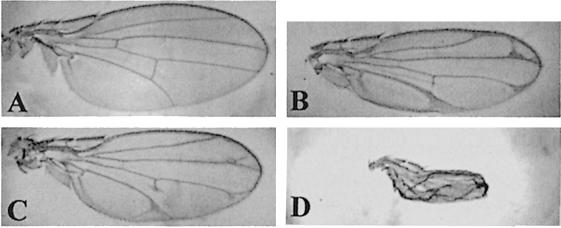
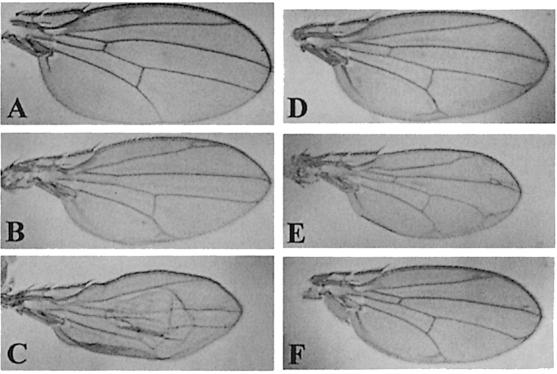
In order to test their effect on the endogenous Cdk7 function, we first expressed the mutant proteins in the larval wing imaginal disc, a sensitive tissue where alterations of both transcription and proliferation controls provoke easily scorable and non-lethal adult phenotypes. Indeed, when expressed in the wing disc using the MS1096 GAL4 driver, D137R and T170A cdk7 mutants induced malformations ranging from alteration of the vein pattern to severe wing blade reduction, depending on the transgene copy number and the strength of the UAS line used (Figure 1). In order to test whether these phenotypes were due to a specific alteration of Cdk7 function, we tried either to rescue them by co-expressing a wild-type allele of cdk7, or to enhance them by reducing the dosage of the endogenous cdk7 gene. The intermediate phenotypes obtained with two copies of weak cdk7 dominant constructs were enhanced in a cdk7null#3 heterozygous background (Figure 2A–C). Conversely, overexpression of a wild-type cdk7 allele did not produce a wing phenotype by itself, but rescued the phenotypes induced by expression of both cdk7 dominant alleles (Figure 2D–F). We could conclude from these experiments that expression of the two dominant cdk7 alleles D137R and T170A specifically interfered with endogenous Cdk7 function. We then tested the capacity of the DN mutants to rescue the null cdk7null#3 mutation under basal expression levels. In these experiments, expression of the transgenes was driven by the HSP70 basal promoter and resulted in an accumulation level of the various Cdk7 proteins ∼20–fold lower than normal endogenous Cdk7 expression (not shown). Despite this low expression level, cdk7wt constructs were able to rescue the cdk7null#3 mutant phenotypes when neither the cdk7T170A nor the cdk7D137R construct did, indicating that the two mutants were defective in cdk7 function.
Fig. 1. Effects of mutant Cdk7 expression on the wing morphology. (A) Wild-type wing. (B–D) Alteration of the wing morphology after expression of dominant-negative forms of Cdk7 using the MS1096 driver. (B) MS1096/+; UAS-Cdk7D137R/UAS-Cdk7D137R line. (C) MS1096/+; UAS-Cdk7T170A/UAS-Cdk7T170A. (D) MS1096/MS1096; UAS-Cdk7T170A/UAS-Cdk7T170A (same line as in C).
Fig. 2. The mutant forms of Cdk7 are dominant over endogenous Cdk7 function. Wild-type and mutant Cdk7 proteins are expressed using the MS1096 driver. (A) cdk7null#3/+ wing. (B) MS1096/+; UAS-cdk7T170A/UAS-cdk7T170A wing. (C) MS1096/cdk7null#3; UAS-cdk7T170A/UAS-cdk7T170A wing. (D) MS1096/MS1096; UAS-cdk7wt/UAS-cdk7wt wing. (E) MS1096/MS1096; UAS-cdk7T170A/UAS- cdkT170A wing. (F) MS1096/MS1096; UAS-cdk7wt/UAS-cdk7wt; UAS-cdk7T170A/UAS-cdk7T170A wing. Comparable results are obtained with the cdk7D137R mutant.
Altering Cdk7 function in the early embryo
Altering Cdk7 function in the wing disc did not allow us to distinguish between effects on transcription or cell cycle control, since in imaginal disc cells these two mechanisms are intimately linked. Indeed, alterations of the transcriptional machinery could induce cell cycle defects and, conversely, modification of the cell cycle program could alter general transcription. In order to ask specific questions about the proposed transcription or cell cycle functions of Cdk7, we took advantage of the special features of the early Drosophila embryo, which runs through the first 13 nuclear divisions without requiring transcription from the zygote genome. Zygotic transcription starts progressively at around cycle 9 and increases strongly during interphase of cell cycle 14. By driving expression of the dominant-negative cdk7 mutants through maternal expression, before the onset of zygotic transcription, we could therefore test the effect of altering Cdk7 function on the control of the early division program. In the same conditions, we could also easily detect an interference with the transcriptional machinery during the onset of zygotic transcription.
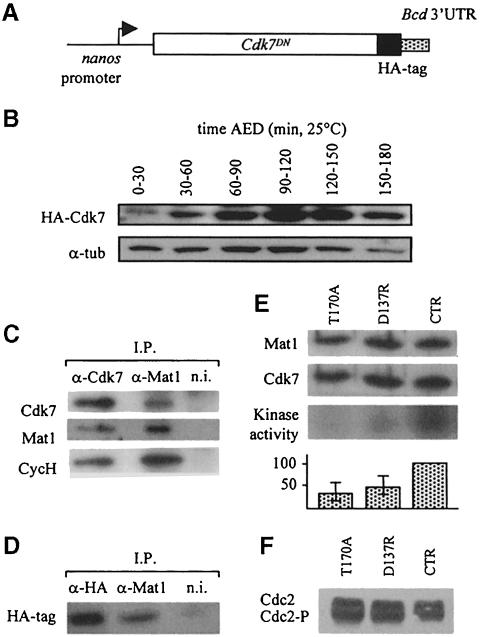
We first used a genetic approach that allowed high maternal accumulation of the transgene mRNA and a regionalized accumulation of the corresponding protein in the early embryo. In this system, the nanos (nos) promoter drives expression of the transgene in the maternal germline, and the 3′–untranslated region (3′–UTR) sequences from the bicoid gene allow translation and accumulation of the protein in the anterior part of the embryo after fertilization (pnos>cdk7D137R>bcd3′–UTR transgene; Figure 3A). Using this expression system, the mutant proteins could be detected within 30 min after egg deposition (AED) and stably accumulated during pre-blastoderm and blastoderm stages (Figure 3B) at a level corresponding to ∼20–fold endogenous Cdk7 levels for our strongest lines, estimated by comparing relative sensitivities of anti-HA and anti-Cdk7 antibodies. In order to follow the behavior of exogenous Cdk7DN, we performed immunoprecipitation experiments with anti-Mat1 antibodies on total extracts from pre-blastoderm and blastoderm embryos laid from pnos>cdk7D137R>bcd3′–UTR homozygous mothers. Using anti-HA antibodies, we could specifically detect Cdk7DN proteins in Mat1 immunoprecipitates (Figure 3C and D). Since Mat1 can only associate with Cdk7–cyclin H heterodimers (Devault et al., 1995; Tassan et al., 1995), this indicated that ectopically produced Cdk7DN formed trimeric CAK complexes with endogenous partners. We then asked whether ectopic expression of the dom– inant-negative mutants could alter Cdk7 kinase activity in vivo. For this purpose, wild-type Cdk7 and the two mutants were expressed maternally and Cdk7 activity was then measured in anti-Mat1 immunoprecipitates on exogenous GST–Cdk2–cyclin A substrate using a standard CAK assay (Labbé et al., 1994). Up to 80% of the overall Cdk7 activity could be suppressed in 0–2 h pnos>cdk7DN>bcd3′–UTR embryos [Figure 3E; three independent experiments, each on 100 embryos: 33, 59 and 63% inhibition for D137R (average 52%); 53, 64 and 81% inhibition for T170A (average 66%)]. Given that this system allowed localized accumulation of the protein products from an anterior source of mRNAs, this suggested that the inhibition rate was much higher at the anterior pole than the average value measured on whole embryos.
Fig. 3. Expression of Cdk7DN in the anterior part of the early embryo. (A) To induce expression of Cdk7DN in the anterior part of the early embryo, transgenic animals expressing different HA-tagged Cdk7 proteins under the control of the nanos promoter and the 3′-UTR sequences of bicoid were used. (B) Western blot on embryos laid from pnanos>cdk7D137R>bcd3′–UTR homozygous mothers. Cdk7DN protein is detected using anti-HA antibodies; control, anti-α–tubulin (for additional information, wild-type Cdk7 protein accumulation during that period is presented in Figure 7B). (C) Co-immunoprecipitation of Cdk7, Mat1 and cyclin H in wild-type embryos. Immunoprecipitations were performed on pre-blastoderm and blastoderm embryonic extracts using anti-Cdk7, anti-Mat1 or non-immune (n.i.) antibodies. Immunoprecipitates were analyzed by Western blotting using anti-Cdk7, anti-Mat1 and anti-CycH antibodies. (D) Co-immuno- precipitation of Cdk7DN–HA with Mat1 in mutant-expressing embryos. Immunoprecipitations were performed on pre-blastoderm embryos laid from pnos>cdk7D137R> bcd3′–UTR mothers using anti-HA tag, anti-Mat1 or non-immune (n.i.) antibodies and analyzed by Western blotting with anti-HA tag antibodies. Similar results were obtained with Cdk7T170A-expressing embryos (not shown). (E) CAK activity in 0–2 h embryos laid from homozygous pnos>cdk7DN> bcd3′–UTR or control mothers, after anti-Mat1 immunoprecipitation. D137R, pnos>cdk7D137R>bcd3′–UTR; T170A, pnos>cdk7T170A>bcd3′–UTR; CTR, pnos>GAL4>bcd3′–UTR. Anti-Mat1 and anti-Cdk7 Western blotting as well as kinase assay are shown (graphs present an average of three independent experiments). (F) Mobility shifts of Cdc2 in embryos laid from homozygous pnos>cdk7DN>bcd3′–UTR or control mothers. D137R, pnos>cdk7D137R>bcd3′–UTR; T170A, pnos>cdk7T170A>bcd3′–UTR; CTR, pnos>GAL4>bcd3′–UTR.
Cdk7DN proteins interfere with the transcription program in the embryo
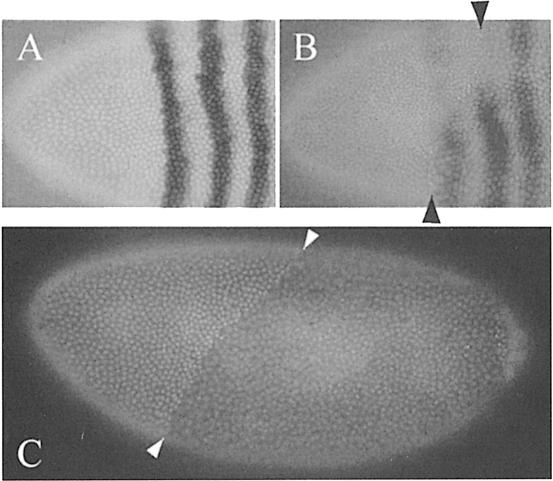
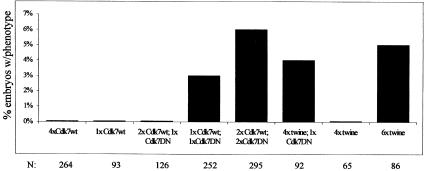
High expression of the Cdk7DN proteins in the anterior part of the pnos>cdk7DN>bcd3′–UTR embryos repeatedly resulted in a defect in transcript accumulation of the fushi tarazu (ftz) gene, one of the earliest zygotic genes transcribed in the blastoderm embryo (Figure 4B, compare with control in A). Furthermore, at the end of the maternal division program, a corresponding zone with twice the normal density of interphase 14 nuclei was observed, resulting from the appearance of an extra synchronous division in the domain of expression of the Cdk7DN proteins (Figure 4B and C). In order to characterize this effect better, we used an early zygotic GAL4 driver (krüppel-GAL4) allowing a strong central expression of UAS constructs in the blastoderm embryo. When crossed with UAS-cdk7 D137R and T170A lines, we could again observe a delay in early zygotic transcription of the ftz gene occurring in the domain of expression of the Cdk7DN proteins (Figure 5D and E). The defect in ftz transcription was also accompanied by a delay in cellularization and an extra division of the blastoderm nuclei, supporting the idea of a general delay in the onset of zygotic transcription (Figure 5B, D and F). It has been described previously that termination of the maternal division program in Drosophila embryos is under the control of zygotic transcription, which induces an abrupt degradation of the maternal transcripts coding for the two Cdc25 mitotic inducers twine and string. Blocking the onset of zygotic transcription generates an extra round of maternal division by stabilizing maternal twine and string messages (Edgar and Datar, 1996). To confirm that the Cdk7DN effect on the division program was a consequence of a stabilization of the maternal cdc25 transcripts, we increased the maternal dose of the cdc25twine gene in an embryo expressing low levels of Cdk7DN. Although neither one copy of the cdk7DNtransgene nor four copies of maternal cdc25twine could induce extra divisions on their own, together they promoted an extra mitosis in the Cdk7DN expression domain (Figure 6). Thus, increasing gene dosage of cdc25twine enhanced the cdk7DN effect. This indicated that the primary effect of Cdk7DN was to delay zygotic transcription, therefore stabilizing maternal twine transcripts and preventing exit from syncytial divisions. Extra divisions were observed with similar frequencies in embryos obtained from 6×twine mothers by us and others (Figure 6 and Edgar and Datar, 1996). This low penetrance can probably be explained by the fact that even small amounts of transcription might allow activation of the zygotic degradation pathway, and that this pathway is only needed to complete a degradation that has already started under maternal control (Edgar and Datar, 1996; Bashirullah et al., 1999).
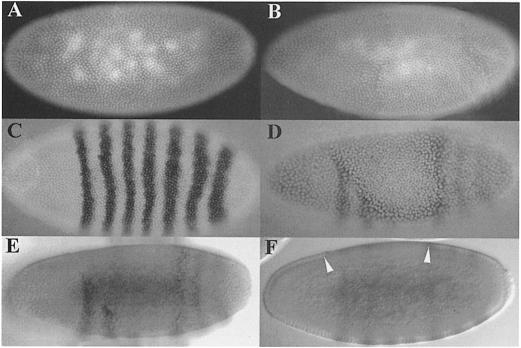
Fig. 4. Effects of Cdk7DN expression in the anterior part of early embryos. (A and B) The anterior part of embryos labeled with a ftz probe and Hoechst 33258. (A) Control embryo showing the normal striped pattern of the ftz gene. (B) An embryo laid from a pnos>cdk7D137R>bcd3′–UTR homozygous mother; note the disappearance of the two first ftz-expressing bands in the zone of increased nuclear density. (C) Embryos laid from pnos>cdk7D137R>bcd3′–UTR homozygous mothers stained with Hoechst 33258. Arrows indicate the limit between normal nuclear density at interphase 14 and a zone of increased nuclear density in the anterior.
Fig. 5. Expression of Cdk7DN delays the onset of zygotic transcription. A krüppel-GAL4 driver was used to direct early zygotic expression of the cdk7 transgenes in the middle part of the embryo. (A) A wild-type interphase 14 embryo stained with Hoechst. (B) A kr-GAL4/+; UAS-cdk7T170A/UAS-cdk7T170A embryo, at the same stage; note the zone of double nuclear density in the central region of the embryo. (C) An interphase 14 wild-type embryo labeled with a ftz probe. (D–F) A kr-GAL4/+; UAS-cdk7T170A/UAS-cdk7T170A embryo. (D) Staining with Hoechst and ftz probe; (E) upper view of the same embryo with Nomarski optics, showing the absence of ftz expression in the zone of extra maternal division; and (F) a section view of the same embryo showing a delayed cellularization in the zone of extra maternal division (between arrows).
Fig. 6. Increasing the dose of maternal twine enhances the Cdk7DN phenotype. Overexpression of cdk7wt or cdk7T170A resulted from expression with one copy of the kr-GAL4 driver. Embryos with a zone of double nuclear density at the mid-blastula transition are counted as positive. N, total number of embryos analyzed. 1×cdk7wt, embryos heterozygous for the cdk7null#3 null mutation; 2×Cdk7wt, wild-type embryos; 4×Cdk7wt, wild-type embryos carrying two copies of the UAS-cdk7wt transgene. The y-axis shows the percentage of embryos presenting an extra maternal division.
Cdk7DN do not affect the program of maternal nuclear cycles
Two different strategies were used to express the dominant mutants in the early embryo before the start of transcription. In both cases, the division cycle program was followed by time lapse video microscopy, using an Ncd–green fluorescent protein (GFP) marker to record spindle formation and the timing of each maternal mitosis (Endow and Komma, 1996). We first injected mRNA coding for either Cdk7wt, Cdk7D137R or Cdk7T170A. Using anti-HA antibodies, we could readily detect accumulation of the mutant proteins during the first hour following injection and at levels comparable to those obtained in the larval wing disc using the GAL4/UAS system (data not shown). Nevertheless, no modification or delay was observed in the timing of the first 13 nuclear divisions. In particular, the timing of cycles 11–13 was comparable to that of control embryos (cycle 11, 13 ± 2 min, cycle 12, 16 ± 2 min, cycle 13, 24 ± 2 min). To confirm these observations, we then used the maternal nanos expression system, which allowed a robust localized expression in the anterior part of the embryo. Although this expression was shown locally to disrupt most of the Cdk7 kinase activity and provoke severe transcription defects, no modification of the division cycle program was observed in the early embryos. Mobility shift experiments did not reveal any differences in the level of phosphorylation of Cdc2 between control and pnos>cdk7DN >bcd3′–UTR embryos during this period (Figure 3F). This set of data suggested either that the cell cycle function of Cdk7 required sensibly less activity than the transcription function, or that another enzymatic activity that was unaffected by Cdk7DN expression could fulfill a cell cycle function in the absence of Cdk7 in these embryos.
Cdk7 transcription, protein accumulation and polII phosphorylation during the onset of transcription in the embryo
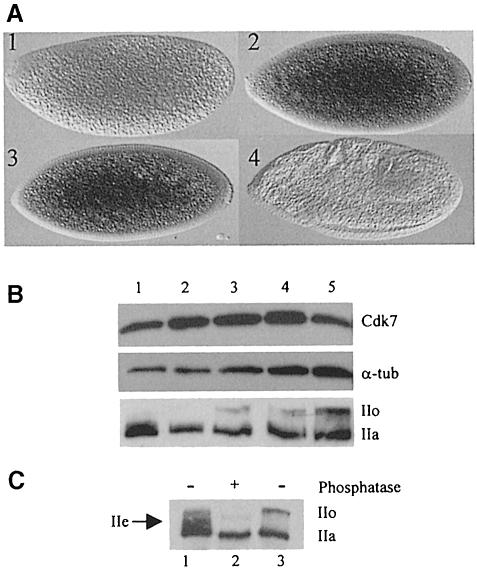
In an effort to correlate the timing of accumulation of Cdk7 with its function during early embryogenesis, we performed an analysis of RNA and protein levels during the first period of development. We found that Cdk7 is maternally provided, although early embryos are only faintly labeled using a cdk7 antisense probe (Figure 7A). Surprisingly, embryos at the time of pole cell formation (cycles 9–11) repeatedly presented a much stronger labeling, suggesting that zygotic transcription of the gene might be starting at that period. A weaker signal was present in older embryos. Protein level analysis on time-sorted embryos showed only a modest increase of the Cdk7 protein during early embryogenesis (Figure 7B). Western blot analysis of polII indicated that the transcriptionally active, phosphorylated form of the CTD (IIo) is not present during the pre-blastoderm period and appears only after pole cell formation to accumulate progressively during later stages (Figure 7B). Careful analysis of polII accumulation revealed that an intermediate phosphorylated form is present transiently during the pre-blastoderm period (Figure 7C), which could correspond to the previously described polIIe in mammalian embryos (Bellier et al., 1997). This phosphorylation intermediate disappears at the time of pole cell formation and is replaced by the typical IIo form, which involves Cdk7 kinase activity (Figure 7B). Thus, it appears that although Cdk7 protein is present during early embryonic stages, its physiological target is not phosphorylated in vivo.
Fig. 7. Expression of cdk7 in the early embryo. (A) Expression of cdk7 mRNA visualized by in situ hybridization: 1, pre-blastoderm; 2, cycle 9 (pole cells bud at the posterior); 3, syncytial blastoderm; 4, germ band extension. (B) Western blot on embryonic extracts using anti-Cdk7, anti-α–tubulin (DM1a) and anti-RNA polII (ARNA3). Lane 1, pre-blastoderm; lane 2, pole cell formation; lane 3, cellularization; lane 4, gastrulation; lane 5, germ band elongation. (C) Western blot on embryonic extracts using anti-RNA polII (ARNA3) antibodies. Lane 1, pre-blastoderm embryonic extracts; lane 2, pre-blastoderm embryonic extracts treated with calf intestine phosphatase; lane 3, gastrulation stage embryonic extracts.
Discussion
Cdk7 is suspected to have a dual role in the cell. Because of its co-purification with the TFIIH complex and its in vitro CTD kinase activity, it is a very good candidate as a direct regulator of polII activity. By phosphorylating the conserved threonine of Cdc2 and of other Cdks in vitro, it is also a good candidate as an activator of the cell cycle machinery. It has been remarkably difficult to assess a direct role for Cdk7 in the control of the cell cycle machinery in many model systems, due to the fact that altering its activity would have an effect on its transcriptional function that could affect the control of the cell cycle indirectly.
We have analyzed the physiological role of the Cdk7 kinase by expressing dominant-negative forms of the kinase at different stages of Drosophila development. By first expressing them in the wing disc, we could easily detect a dominant effect on both the morphology and growth of the adult wing. These dominant phenotypes were either enhanced or suppressed by decreasing or increasing the dose of cdk7wt, thus confirming that in these conditions we could alter Cdk7 function specifically. Both DN mutations were able to disrupt Cdk7 function. D137R is a kinase-dead mutation and expression of this mutant could compete with endogenous Cdk7 for cyclin H and Mat1 partners or substrates in the cells. T170A is a mutation in the T–loop of Cdk7 on a phosphorylation site equivalent to the activating phosphorylation site of most Cdks. The functional significance of T170 phosphorylation (or equivalent T in other species) on Cdk7 is still controversial as it seems to be required for full Cdk7 and Kin28 activity in vivo (Labbé et al., 1994; Espinoza et al., 1998; Kimmelman et al., 1999), but dispensable in vitro as long as the Mat1 assembly factor is present (Devault et al., 1995; Fisher et al., 1995; Tassan et al., 1995; Martinez et al., 1997). Our in vivo results show that ectopic expression of the T170A mutant creates a dominant phenotype in Drosophila, associated with an inhibition of the endogenous Cdk7 activity, thus inferring that T170 phosphorylation is necessary for the normal function of the kinase in a cellular context in this organism. Furthermore, basal constitutive expression of T170A and D137R mutants did not complement the cdk7null#3 null mutation in conditions where expression of cdk7wt did, confirming that these modifications altered normal in vivo function of the kinase.
In order to test a direct cell cycle function for Cdk7 without interfering with transcription, we focused on the early embryonic divisions that run without zygotic transcription. In these conditions, expression of Cdk7DN proteins did not alter the program of maternal nuclear cycles as measured by time-lapse video microscopy. Further experiments using zygotic Gal4 expression that allowed disruption of Cdk7 function at mid-blastula transition showed that the dominant Cdk7 effects appeared within 20 min after induction of the Cdk7DN proteins in the embryo. This ruled out the possibility that the absence of effect on early cycles could be due to the appearance of a necessary lag before the action of dominant proteins. It has been proposed previously that two distinct Cdk7 complexes found in the cell could participate in distinct functions: one, corresponding to the Cdk7–cyclin H–Mat1 ternary complex (‘free CAK’), would allow activation of Cdc2 kinase, while the other, corresponding to complete TFIIH, would participate in transcriptional function. Our results show that exogenous HA-tagged Cdk7DN proteins associate with endogenous Mat1 in early embryos, indicating that Cdk7DN proteins compete with endogenous Cdk7 to form trimeric CAK, acting indifferently on free CAK and TFIIH-associated activities. We further showed that expression of both Cdk7DN proteins alters endogenous CAK activity. Reduction of Cdk7 activity reached 80% in early embryos using the nanos maternal constructs. This system allows a very strongly regionalized expression at the anterior pole of the embryo, therefore suggesting that, in these conditions, inhibition at the pole might be nearly complete. Under these conditions, although transcriptional activity of the embryos could be perturbed strongly (see below), we did not observe any effect on Cdc2 phosphorylation and the early division program.
We had previously analyzed the phosphorylation state of Cdc2 during early embryonic cycles (Edgar et al., 1994). Our results had established that, starting from cell cycle 8, Cdc2 was dephosphorylated periodically on T161 during interphase, thus requiring a CAK activity to re-enter mitosis. In this context, our present data suggest two possible explanations. Different levels of activity could be required for the two possible functions of Cdk7 in vivo, transcription control requiring a higher level than cell cycle control. This would allow DN mutant expression to induce transcription defects in conditions where cell cycle control is not affected. Alternatively, a different pool of CAK activity distinct from Cdk7 complexes might be present to allow re-activation of the Cdc2 kinase prior to each mitosis in the absence of Cdk7 function. In S.cerevisiae, the TFIIH-associated CTD kinase and CAK activities are carried out by separate enzymes, Kin28/Ccl1/Tfb3 and Cak1, respectively (for a review, see Kaldis, 1999). In S.pombe, both Cdk7 (or Mcs6) and Csk1, a monomeric kinase, are involved in CAK function in vivo (Lee et al., 1999). Analysis of a cdk7 null mutation in Drosophila had suggested that Cdk7 might be responsible only for Cdc2 phosphorylation and that another CAK activity could be responsible at least for the activation of Cdk2 (Larochelle et al., 1998). Very recently, Nagahara and colleagues have described a distinct CAK activity in human HepG2 cells that does not fractionate with Cdk7 complexes, reacts with anti-yeast Cak1 antibodies and could be the target of transforming growth factor–β (TGF–β) leading to G1 arrest in these cells (Nagahara et al., 1999). A similar activity, distinct from the Cdk7 complex and reacting with antibodies against yeast Cak1, has recently been partially purified from HeLa cells (P.Kaldis and M.Solomon, in preparation). We also repeatedly found in Drosophila embryonic extracts a 42 kDa protein species that specifically cross-reacts with anti-yeast Cak1 antibodies (not shown). These and our present data strongly suggest that a distinct enzyme might be able to re-activate Cdc2 kinase periodically in the absence of Cdk7 during early embryonic cycles.
Expression of the Cdk7DN proteins repeatedly induced a delay in early zygotic transcription, manifested by the absence of pair-ruled ftz gene transcription, a delay in cellularization and a failure to terminate syncytial divisions following interphase 13. Several lines of evidence have shown that a defect in early transcription in embryos is associated with a failure to undergo cellularization and to end the program of maternal divisions. These two events are the hallmark of mid-blastula transition as they are the earliest developmental events in the embryo known to require zygotic transcription. Blastoderm cellularization is under the control of a number of zygotic genes (bottleneck, serendipity–α and nullo), whose expression is required early in interphase 14 (Merrill et al., 1988; Wieschaus and Sweeton, 1988; Schweisguth et al., 1990; Simpson and Wieschaus, 1990; Schejter and Wieschaus, 1993). The termination of blastoderm mitoses depends on the degradation of maternal cdc25twine and cdc25string transcripts. Recent work has pointed to two different pathways leading to the degradation of maternal mRNAs (Bashirullah et al., 1999). One is maternally encoded and responsible for a progressive disappearance of maternal transcripts. The second is zygotically activated 2 h after fertilization and results in a more abrupt RNA degradation. The function of both pathways is necessary for the complete elimination of transcripts at mid-blastula transition. Indeed, embryos injected with α–amanitin do not cellularize and display a delay in degradation of both maternal cdc25 RNAs, resulting in a 14th synchronous syncytial nuclear division (Edgar and Datar, 1996). Our experiments confirmed that the Cdk7DN effect on the program of syncytial divisions is a consequence of a transcriptional effect on mRNA degradation, since increasing the cdc25twine maternal dosage results in a strong enhancement of the cdk7DN phenotype.
Despite the effects of cdk7DN expression on the transcription program, we have not been able to observe consistent variations of the phosphorylation levels of polII CTD, one of the known targets of the TFIIH kinase (data not shown). This is probably due to the fact that the expression system we use was too localized to induce a general decrease of polII phosphorylation levels that could be detected. Additionally, during this early period, polII phosphorylation evolves rapidly (see Figure 7B), which makes the staging of the embryos used for analysis particularly crucial.
The transcriptional role of the S.cerevisae Cdk7 homolog, Kin28, has been studied recently by genome-wide expression analysis (Holstege et al., 1998). The results reveal that Kin28 is required for the general expression of most protein-coding genes, as widely as polII itself. In Cdk7DN-expressing embryos, the variety of phenotypes also suggests that transcription of many early genes might be affected.
Taken together, these experiments implicate Cdk7 function in the onset of transcription at mid-blastula transition and constitute the first in vivo evidence that Cdk7 function is essential for the regulation of the transcriptional machinery in higher eukaryotes.
Interestingly, two recent reports suggest that TFIIH could be the target of a mechanism of transcriptional repression during mitosis (Akoulitchev and Reinberg, 1998; Long et al., 1998). In these studies, it was demonstrated that upon entry into mitosis, TFIIH is phosphorylated on its p36 (Cdk7) subunit and that this phosphorylation results in inhibition of the Cdk7-associated CTD kinase and, consequently, of the transcriptional activity of the TFIIH complex. Furthermore, this inhibitory phosphorylation can be carried out in vitro by mitotic kinase Cdc2–cyclin B, suggesting that there might be a direct interaction between Cdc2 and Cdk7 that leads to inhibition of the CTD kinase activity of Cdk7. Mitotic inhibition of transcription can be related through many points to pre-blastoderm inhibition of transcription. In early embryos, it has been shown that the mitotic kinase Cdc2 is constitutively active through the first nuclear cycles, until cell cycle 9, when inactivation of Cdc2 can be detected first in interphase (Edgar et al., 1994). This coincides precisely with the moment when earliest traces of transcription can be detected using ftz probes (Pritchard and Schubiger, 1996). Inhibition of transcription by the Cdc2 kinase is one of the mechanisms that has been proposed for delaying the onset of zygotic transcription (Hartl et al., 1993). More recently, analysis of the grapes and mei–41 mutations in flies has shown that the progressive lengthening of interphases 11, 12 and 13 is also essential for proper accumulation of zygotic transcripts (Sibon et al., 1997), but suggests that additional mechanisms repress transcription during blastoderm cycles (Sibon et al., 1999). We propose that, as in mitotic cells, the transcription machinery in the pre-blastoderm embryo could be inhibited by a specific mechanism involving the mitotic kinase Cdc2, with Cdk7 as one of its possible targets. Our results reinforce this hypothesis, since we find no polIIo accumulating at pre-blastoderm stages, suggesting a possible early inhibition of the in vivo CTD kinase activity of Cdk7. We also noticed the presence of a modified IIe-like form of polII in pre-blastoderm embryos. Alternatively, absence of CTD phosphorylation by Cdk7 at this stage could be explained by a transient cytosolic localization of polII, as has been described for the IIe form in rabbit embryos (Bellier et al., 1997). Although mostly speculative, these models should serve as a framework for further analyses of the regulation of polII machinery during early embryogenesis.
Materials and methods
Constructs and germline transformation
pUAST transformation plasmids encoding Cdk7, Cdk7D137R and Cdk7T170A tagged at the C-terminus with an HA tag were constructed as follows. Point mutations were induced by PCR in the cdk7 cDNA subcloned in pBluescript. The resulting plasmids pKScdk7D137R, pKScdk7T170A and pKScdk7 were subjected to another PCR with a sense primer made of the EcoRI site and nucleotides –52 to –27 of cdk7 and an antisense primer containing an EcoRI site, a stop codon, an HA tag and codons 353–348 of cdk7. The PCR products were subcloned in the EcoRI site of the pUAST vector. The inserts of the resulting plasmids pUAScdk7D137R, pUAScdk7T170A and pUAScdk7 were sequenced entirely to ensure that no unintended mutations were introduced. The different pnos>cdk7>bcd3′–UTR constructs were made by subcloning the previous cdk7 inserts into pKSGal4VP16, which contains a Gal4–VP16 cassette under the control of the nanos promoter (pnos) and the 3′-UTR of bicoid (N.Dostatni, Marseille, France), after removal of the Gal4–VP16 cassette. Germline transformation was performed using standard procedures.
Fly strains
Fly cultures and crosses were performed at 25°C except for Gal4/UAS crosses, which were performed at 29°C. Gal4 was expressed in the wing disc using the MS1096 line (Capdevila and Guerrero, 1994) and in the syncytial blastoderm embryo using the kr-GAL4 line (Castelli-Gair et al., 1994). The following lines have been described previously: ncd–GFP (Endow and Komma, 1996), 6×twine (Edgar and Datar, 1996) and the cdk7null#3/FM7 line (Larochelle et al., 1998). To produce 4×twine/1×pnos>gal4-GCN4>bcd3′–UTR/1× UAS-cdk7 embryos, 6×twine females (w P[w+ twn3.7]/w P[w+ twn3.7]; +; P[w+ twn10.0]/w P[w+ twn10.0]) were crossed with GAL4-GCN4 43B (w/Y; +; P[w+ pnos>GAL4-GCN4>bcd3′–UTR]/ +; P[w+ pnos>GAL4-GCN4> bcd3′-UTR]). Males and the resulting females were crossed with UAS-cdk7, UAS-cdk7D137R or UAS-cdk7T170A homozygous males.
RNA injection experiments
For mRNA injection experiments, cdk7, cdk7D137R and cdk7T170A mRNAs were produced from pSP64T(R) vector (M.Dorée, Montpellier, France). Embryos were collected 0–30 min AED and injected at the posterior pole. The timing of nuclear divisions was followed by time-lapse video microscopy on a Zeiss inverted fluorescent microscope using a Princeton CCD camera and a Metamorph acquisition system. Images were collected at 30 s intervals and processed with Photoshop (Adobe). For Western blot analyses, collected embryos were crushed directly into SDS sample buffer and boiled.
Biochemistry
Affinity-purified rabbit polyclonal antibodies directed against a synthetic Cdk7 C–terminal peptide were used at 1/300 for immunobloting experiments. Anti-cyclin H and anti-Mat1 antibodies were kindly provided by M.Dorée (Montpellier). Characterization of DmCyclin H and DmMat1 will be described elsewhere (C.Lehner, V.Leclerc and P.Léopold, unpublished data). Mouse monoclonal anti-polII antibodies ARNA3 (Kramer et al., 1980) were a gift from E.K.Bautz. For immunoblots, hand-selected, dechorionated embryos were crushed directly either into SDS sample buffer or into lysis buffer [50 mM HEPES–K+, 150 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium bisulfite, 1 mM benzamidine, 10 mM β–glycerophosphate, 50 mM NaF, 4 mM sodium orthovanadate, 0.5% Triton X–100 pH 7.6]. DM1a (Sigma; 1/10 000) and 12CA5 (Roche; 1/1000) mouse monoclonal antibodies were used to detect α–tubulin and HA tag, respectively. For CAK assays, 100 embryos were selected under the microscope and crushed in lysis buffer. Anti-Mat1 immunoprecipitations were carried out for 2 h at 4°C essentially as described (Leclerc et al., 1996) and precipitates were divided into two aliquots. One was processed for Western blotting, the other was used for a two-step CAK assay as described (Labbé et al., 1994). Mobility shifts on Cdc2 were observed using the procedure described in Edgar et al. (1994): 20 dechorionated embryos (60–120 min AED) were selected under the microscope and crushed in 2× Laemmli buffer including phosphatase inhibitors, then boiled for 4 min. Cdc2 was detected using anti-PSTAIR antibodies (Sigma) at 1/5000.
Phenotypic analysis
Wings were dehydrated and dissected in ethanol before mounting in DPX (BDH). Embryos were fixed with formaldehyde and stained with Hoechst 33258 directly or after in situ hybridization with a ftz DIG-labeled DNA probe. Embryos were mounted in Vectastain (Vector) or in Mowiol mounting medium to reduce UV background.
Acknowledgments
Acknowledgements
We thank M.Dorée for initially providing us with early sequence data from starfish MO15 protein and for very stimulating discussions, J.C.Cavadore for helping with anti-Cdk7 peptide antibodies with unequalled efficiency and kindness, J.P.Vincent for providing us with MS1096 and kr-GAL4 strains as well as always valuable advice, B.Edgar for 6×twine flies, S.Endow for ncd–GFP flies, N.Dostatni for the pKSGal4VP16 plasmid, and B.Suter for cdk7null#3 flies. This work was initiated in the Research Unit of C.Sardet (URA 671 CNRS, Villefranche-sur-mer). His support and special help with imaging was much appreciated. We thank P.Dru and N.Scrima for technical assistance, and Evelyn Houliston and Ellen Van Obberghen-Schilling for careful reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer and the Fondation pour la Recherche Médicale.
References
- Adamczewski J.P., Rossignol, M., Tassan, J.P., Nigg, E.A., Moncollin, V. and Egly, J.M. (1996) MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J., 15, 1877–1884. [PMC free article] [PubMed] [Google Scholar]
- Akoulitchev S. and Reinberg, D. (1998) The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev., 12, 3541–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoulitchev S., Makela, T.P., Weinberg, R.A. and Reinberg, D. (1995) Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature, 377, 557–560. [DOI] [PubMed] [Google Scholar]
- Bashirullah A. et al. (1999)Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J., 18, 2610–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier S., Chastant, S., Adenot, P., Vincent, M., Renard, J.P. and Bensaude, O. (1997) Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J., 16, 6250–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck V., Russell, P. and Millar, J.B. (1995) Identification of a cdk-activating kinase in fission yeast. EMBO J., 14, 6173–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J. and Guerrero, I. (1994) Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J., 13, 4459–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli-Gair J., Greig, S., Micklem, G. and Akam, M. (1994) Dissecting the temporal requirements for homeotic gene function. Development, 120, 1983–1995. [DOI] [PubMed] [Google Scholar]
- Cismowski M.J., Laff, G.M., Solomon, M.J. and Reed, S.I. (1995) KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol., 15, 2983–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M.E. (1996) Phosphorylation of mammalian RNA polymerase II. Methods Enzymol., 273, 185–193. [DOI] [PubMed] [Google Scholar]
- Damagnez V., Makela, T.P. and Cottarel, G. (1995) Schizosaccharomyces pombe Mop1-Mcs2 is related to mammalian CAK. EMBO J., 14, 6164–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devault A., Martinez, A.M., Fesquet, D., Labbe, J.C., Morin, N., Tassan, J.P., Nigg, E.A., Cavadore, J.C. and Dorée, M. (1995) MAT1 (‘menage a trois’) a new RING finger protein subunit stabilizing cyclin H–cdk7 complexes in starfish and Xenopus CAK. EMBO J., 14, 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A. and Datar, S.A. (1996) Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes Dev., 10, 1966–1977. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Sprenger, F., Duronio, R.J., Leopold, P. and O'Farrell, P.H. (1994) Distinct molecular mechanisms regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev., 8, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A. and Komma, D.J. (1996) Centrosome and spindle function of the Drosophila Ncd microtubule motor visualized in live embryos using Ncd–GFP fusion proteins. J. Cell Sci., 109, 2429–2442. [DOI] [PubMed] [Google Scholar]
- Espinoza F.H., Farrell, A., Erdjument, B.H., Tempst, P. and Morgan, D.O. (1996) A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science, 273, 1714–1717. [DOI] [PubMed] [Google Scholar]
- Fesquet D. et al. (1993)The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J., 12, 3111–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesquet D., Morin, N., Dorée, M. and Devault, A. (1997) Is Cdk7/cyclin H/MAT1 the genuine cdk activating kinase in cycling Xenopus egg extracts? Oncogene, 15, 1303–1307. [DOI] [PubMed] [Google Scholar]
- Fisher R.P. and Morgan, D.O. (1994) A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell, 78, 713–724. [DOI] [PubMed] [Google Scholar]
- Fisher R.P., Jin, P., Chamberlin, H.M. and Morgan, D.O. (1995) Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell, 83, 47–57. [DOI] [PubMed] [Google Scholar]
- Gould K.L., Moreno, S., Owen, D.J., Sazer, S. and Nurse, P. (1991) Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J., 10, 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl P., Gottesfeld, J. and Forbes, D.J. (1993) Mitotic repression of transcription in vitro. J. Cell Biol., 120, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermand D., Pihlak, A., Westerling, T., Damagnez, V., Vandenhaute, J., Cottarel, G. and Makela, T.P. (1998) Fission yeast csk1 is a CAK-activating kinase (CAKAK). EMBO J., 17, 7230–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S. and Young, R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Kaldis P. (1999) The cdk-activating kinase (CAK): from yeast to mammals. Cell. Mol. Life Sci., 55, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis P., Sutton, A. and Solomon, M.J. (1996) The Cdk-activating kinase (CAK) from budding yeast. Cell, 86, 553–564. [DOI] [PubMed] [Google Scholar]
- Kimmelman J., Kaldis, P., Hengartner, C.J., Laff, G.M., Koh, S.S., Young, R.A. and Solomon, M.J. (1999) Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol., 19, 4774–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A., Haars, R., Kabisch, R., Will, H., Bautz, F.A. and Bautz, E.K. (1980) Monoclonal antibody directed against RNA polymerase II of Drosophila melanogaster. Mol. Gen. Genet., 180, 193–199. [DOI] [PubMed] [Google Scholar]
- Labbe J.C. et al. (1994)p40MO15 associates with a p36 subunit and requires both nuclear translocation and Thr176 phosphorylation to generate cdk-activating kinase activity in Xenopus oocytes. EMBO J., 13, 5155–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle S., Pandur, J., Fisher, R.P., Salz, H.K. and Suter, B. (1998) Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev., 12, 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc V., Tassan, J.P., O'Farrell, P.H., Nigg, E.A. and Leopold, P. (1996) Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol. Biol. Cell, 7, 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.M., Saiz, J.E., Barton, W.A. and Fisher, R.P. (1999) Cdc2 activation in fission yeast depends on mcs6 and csk1, two partially redundant cdk-activating kinases (CAKs). Curr. Biol., 9, 441–444. [DOI] [PubMed] [Google Scholar]
- Long J.J., Leresche, A., Kriwacki, R.W. and Gottesfeld, J.M. (1998) Repression of TFIIH transcriptional activity and TFIIH-associated cdk7 kinase activity at mitosis. Mol. Cell. Biol., 18, 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela T.P., Tassan, J.P., Nigg, E.A., Frutiger, S., Hughes, G.J. and Weinberg, R.A. (1994) A cyclin associated with the CDK-activating kinase MO15. Nature, 371, 254–257. [DOI] [PubMed] [Google Scholar]
- Makela T.P., Parvin, J.D., Kim, J., Huber, L.J., Sharp, P.A. and Weinberg, R.A. (1995) A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc. Natl Acad. Sci. USA, 92, 5174–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A.M., Afshar, M., Martin, F., Cavadore, J.C., Labbe, J.C. and Dorée, M. (1997) Dual phosphorylation of the T-loop in cdk7: its role in controlling cyclin H binding and CAK activity. EMBO J., 16, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill P.T., Sweeton, D. and Wieschaus, E. (1988) Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development, 104, 495–509. [DOI] [PubMed] [Google Scholar]
- Molz L. and Beach, D. (1993) Characterization of the fission yeast mcs2 cyclin and its associated protein kinase activity. EMBO J., 12, 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. (1995) Principles of CDK regulation. Nature, 374, 131–134. [DOI] [PubMed] [Google Scholar]
- Nagahara H., Ezhevsky, S.A., Vocero-Akbani, A.M., Kaldis, P., Solomon, M.J. and Dowdy, S.F. (1999) Transforming growth factor β targeted inactivation of cyclin e:cyclin-dependent kinase 2 (Cdk2) complexes by inhibition of cdk2 activating kinase activity. Proc. Natl Acad. Sci. USA, 96, 14961–14966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. (1996) Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control? Curr. Opin. Cell Biol., 8, 312–317. [DOI] [PubMed] [Google Scholar]
- Poon R.Y., Yamashita, K., Adamczewski, J.P., Hunt, T. and Shuttleworth, J. (1993) The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J., 12, 3123–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard D.K. and Schubiger, G. (1996) Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev., 10, 1131–1142. [DOI] [PubMed] [Google Scholar]
- Roy R., Adamczewski, J.P., Seroz, T., Vermeulen, W., Tassan, J.P., Schaeffer, L., Nigg, E.A., Hoeijmakers, J.H. and Egly, J.M. (1994) The MO15 cell cycle kinase is associated with the TFIIH transcription–DNA repair factor. Cell, 79, 1093–1101. [DOI] [PubMed] [Google Scholar]
- Schejter E.D. and Wieschaus, E. (1993) bottleneck acts as a regulator of the microfilament network governing cellularization of the Drosophila embryo. Cell, 75, 373–385. [DOI] [PubMed] [Google Scholar]
- Schweisguth F., Lepesant, J.A. and Vincent, A. (1990) The serendipity α gene encodes a membrane-associated protein required for the cellularization of the Drosophila embryo. Genes Dev., 4, 922–931. [DOI] [PubMed] [Google Scholar]
- Serizawa H., Makela, T.P., Conaway, J.W., Conaway, R.C., Weinberg, R.A. and Young, R.A. (1995) Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature, 374, 280–282. [DOI] [PubMed] [Google Scholar]
- Shiekhattar R., Mermelstein, F., Fisher, R.P., Drapkin, R., Dynlacht, B., Wessling, H.C., Morgan, D.O. and Reinberg, D. (1995) Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature, 374, 283–287. [DOI] [PubMed] [Google Scholar]
- Sibon O.C., Stevenson, V.A. and Theurkauf, W.E. (1997) DNA-replication checkpoint control at the Drosophila midblastula transition. Nature, 388, 93–97. [DOI] [PubMed] [Google Scholar]
- Sibon O.C., Laurencon, A., Scott Hawley, W.E. and Theurkauf, W.E. (1999) The Drosophila ATM homologue mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol., 9, 302–312. [DOI] [PubMed] [Google Scholar]
- Simpson L. and Wieschaus, E. (1990) Zygotic activity of the nullo locus is required to stabilize the actin–myosin network during cellularization in Drosophila. Development, 110, 851–863. [DOI] [PubMed] [Google Scholar]
- Solomon M.J. and Kaldis, P. (1998) Regulation of CDKs by phosphorylation. Results Probl. Cell Differ., 22, 79–109. [DOI] [PubMed] [Google Scholar]
- Solomon M.J., Harper, J.W. and Shuttleworth, J. (1993) CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J., 12, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan J.P., Schultz, S.J., Bartek, J. and Nigg, E.A. (1994) Cell cycle analysis of the activity, subcellular localization and subunit composition of human CAK (CDK-activating kinase). J. Cell Biol., 127, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan J.P., Jaquenoud, M., Fry, A.M., Frutiger, S., Hughes, G.J. and Nigg, E.A. (1995) In vitro assembly of a functional human CDK7–cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J., 14, 5608–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuret J.Y., Valay, J.G., Faye, G. and Mann, C. (1996) Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell, 86, 565–576. [DOI] [PubMed] [Google Scholar]
- Valay J.G., Simon, M., Dubois, M.F., Bensaude, O., Facca, C. and Faye, G. (1995) The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol., 249, 535–544. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S. and Harlow, E. (1993) Distinct roles for cyclin-dependent kinases in cell cycle control. Science, 262, 2050–2054. [DOI] [PubMed] [Google Scholar]
- Wieschaus E. and Sweeton, D. (1988) Requirements for X-linked zygotic gene activity during cellularization of early Drosophila embryos. Development, 104, 483–493. [DOI] [PubMed] [Google Scholar]