Abstract
Cardiac myocytes are known to be influenced by the rigidity and topography of their physical microenvironment. It was hypothesized that 3D heterogeneity introduced by purely physical microdomains regulates cardiac myocyte size and contraction. This was tested in vitro using polymeric microstructures (G′=1.66 GPa) suspended with random orientation in 3D by a soft Matrigel matrix (G′=22.9 Pa). After 10 days of culture, the presence of 100 μm-long microstructures in 3D gels induced fold increases in neonatal rat ventricular myocyte size (1.61±0.06, p<0.01) and total protein/cell ratios (1.43± 0.08, p<0.05) that were comparable to those induced chemically by 50 μM phenylephrine treatment. Upon attachment to microstructures, individual myocytes also had larger cross-sectional areas (1.57±0.05, p<0.01) and higher average rates of spontaneous contraction (2.01±0.08, p<0.01) than unattached myocytes. Furthermore, the inclusion of microstructures in myocyte-seeded gels caused significant increases in the expression of beta-1 adrenergic receptor (β1-AR, 1.19±0.01), cardiac ankyrin repeat protein (CARP, 1.26±0.02), and sarcoplasmic reticulum calcium-ATPase (SERCA2, 1.59±0.12, p<0.05), genes implicated in hypertrophy and contractile activity. Together, the results demonstrate that cardiac myocyte behavior can be controlled through local 3D microdomains alone. This approach of defining physical cues as independent features may help to advance the elemental design considerations for scaffolds in cardiac tissue engineering and therapeutic microdevices.
Keywords: Cardiomyocyte, Beat frequency, Cell remodeling, Focal adhesion, Mechanotransduction, Microstructure, Microenvironment, Three dimensions, Hypertrophy, Spontaneous contraction
1 Introduction
The emergence of three dimensional (3D) scaffolds for cell and tissue engineering has focused the need to understand how cells interact with cues from the physical environment of supporting substrates. It is clear that physical variables regulate numerous processes of many cell types, including motility, growth, differentiation, apoptosis, and signal transduction (Brown 1982; Pelham and Wang 1997; Huang et al. 2004; Peyton and Putnam 2005; Ruiz and Chen 2008). For instance, both the shape and size of a target-activated surface affect cell morphology and viability (Chen et al. 1997; Wang and Ho 2004; Lee et al. 2004). The effective stiffness or rigidity of a flat substrate has influence over the intracellular cytoskeletal stability (Engler et al. 2006; Saez et al. 2007). In addition, behavior is altered when cells are grown on surfaces with microtopography or in uniform 3D matrices (Pedersen and Swartz 2005; Lee et al. 2007; Thakar et al. 2008). Nonetheless, more needs to be learned about the nature of the interaction of specific cell types with a tightly-controlled microenvironment that mimics the more natural state a cell might find in a scaffold in vivo. Therefore, this article explores how cardiac muscle cells (myocytes) respond to microscale heterogeneity in a 3D matrix.
It seems likely that the recognition of physical cues by cardiac myocytes occurs through a contact-dependent interaction of cells with the extracellular matrix (ECM) and features of external topography. Myocytes develop force internally and bear loads through cell attachments. It is the balance of these extrinsic and intrinsic loads that regulate protein synthesis, sarcomere assembly, cell size, and contractile activity (Katz 2002). Cardiac myocytes possess specialized forms of focal adhesions at their ends and encircling the peripheral myofibrils at Z-discs. These junctions link the transmembrane integrin receptors to the actin cytoskeleton via adaptor proteins such as vinculin, paxillin, and α-actinin (Samarel 2005). Mechanical information detected by focal adhesions and other internal protein complexes are then signaled downstream to mediate pathways involved in key transcriptional and cell remodeling events (Durieux et al. 2007; Ingber 2008; Russell et al. 2010). Thus, the formation and stabilization of anchorage in cardiac myocytes is the critical step for mechanotransduction which, in turn, converts physical perturbations into cell-wide structural changes (Wang et al. 1993; Hoshijima 2006; Senyo et al. 2007). These signaling pathways ultimately control myocyte hypertrophy and optimize contractile work to pump blood effectively through the body (Ogawa et al. 2000; Russell et al. 2000; Raskin et al. 2009).
Specific responses of isolated cardiac myocytes to scaffolds have been explored with single interventions of rigidity, microtopography, or the third dimension. Upon modification of extracellular rigidity, cardiac myocytes grown on 10–17 kPa substrates yield more highly-ordered sarcomeres, fewer stress cables, and greater mechanical forces than myocytes on stiffer substrates (Jacot et al. 2008; Engler et al. 2008). In the transition between 2D and 3D environments, the introduction of 5 μm-high vertical textures to a planar substrate affects cardiac myocyte shape, gene expression, and localization of attachment proteins (Motlagh et al. 2003b). Neonatal cardiac myocytes have been shown to align and spontaneously contract in 5 μm grooves (Deutsch et al. 2000; Motlagh et al. 2003a) or upon flat surfaces stamped with protein bands of 10 μm width (Gopalan et al. 2003). In a full 3D collagen matrix, neonatal cardiac myocytes display more mature morphological qualities, such as parallel-arranged sarcomeres and well-developed T-tubules (Zimmermann et al. 2002). Furthermore, varying the stiffness of a uniform 3D fibrinogen gel is known to alter the rate and amplitude of myocyte contraction (Shapira-Schweitzer and Seliktar 2007).
In this study, a combination of microscale rigidity and topography in 3D for cardiac myocytes is assessed for the first time. Such physical cues are presented to single cells in vitro as polymeric microstructures in a supporting 3D gel. Inclusion of rigid microstructures at low concentrations in a soft 3D matrix greatly reduces cardiac fibroblast proliferation even though the bulk stiffness of the gel remains unchanged (Norman et al. 2008). Neonatal rat ventricular myocytes are shown here to recognize the local heterogeneity created by microstructure addition, forming cell contacts with the discrete features in 3D. Microstructures significantly affect the spontaneous beating rate, gene expression, and cross-sectional area of myocytes, which suggests that cellular processes in cardiac muscle are sensitive to physical microdomains in 3D.
2 Materials and methods
2.1 Fabrication of microstructures
Polymeric microstructures measuring 50×15×15 or 100× 15×15 μm3 (L×W×H) were fabricated using basic photo-lithographic methods as described previously (Norman et al. 2008). Briefly, negative photoresist SU-8 2010 (Microchem, Newton, MA) was spun onto a clean 3-inch silicon wafer to a depth of 15 μm. After a short pre-baking step, the wafer was centered under a Karl Suss MJB3 mask aligner (Karl Suss, Munich, Germany) and a 3-inch square transparency mask patterned with an array of 50 or 100 μm-long microstructure shapes (Infinite Graphics, Minneapolis, MN). The wafer was then exposed to a 365 nm light source (13 mW/cm2) for 13 s, followed by a 3 min post-bake. Gentle submersion in SU-8 developer (Microchem) removed all non-crosslinked photoresist, yielding the final, fully-formed microstructures. The dimensions of microstructures were confirmed by measurements from a Tencor P-1 profilometer (KLA Tencor, Milpitas, CA) and a Nikon interferometric microscope (Nikon, Melville, NY). Microstructures were collected from wafers, sterilized in 70% ethanol, counted, and stored in serum-free culture media at 4°C until use.
Microstructures made from poly(ethylene glycol) dimethacrylate (PEGDMA) were fabricated by a similar protocol. A photoinitiator solution containing 75 mg 2,2-dimethyloxy-2-phenylacetophenone (DMPA) per 1 mL vinyl-2-pyrrolidone was vortexed and added as 1.4% of an 83.3%/15.3% PEGDMA/PBS solution (Sigma, St. Louis, MO). After sonication for 30 min, the mixed solution was spun onto a silicon wafer to a 15 μm thickness and exposed to 365 nm light with no pre- or post-baking steps. Non-crosslinked PEGDMA was washed with water and isopropyl alcohol, and the resulting microstructures were collected as before.
2.2 Suspension of cardiac myocytes and microstructures in 3D
Primary ventricular myocytes were enzymatically isolated from 1–2 day old Sprague-Dawley rat hearts by established methods (Motlagh et al. 2003a). Myocytes and microstructures were suspended in a 3D gel matrix using protocols originally described for studies with fibroblasts; the provided mechanical modulus of the microstructures and gel matrix components are values reported previously (Norman et al. 2008; Ayala et al. 2010). Isolated myocytes and sterile microstructures (SU-8 G′=1.66 GPa) were uniformly mixed on ice in Matrigel (BD Biosciences, San Jose, CA), a commercially available ECM protein-rich (e.g.—collagen IV, laminin) extract that approximates a physiologic cell-supporting environment. The unpolymerized composite (G′=22.9 Pa), containing randomly dispersed myocytes (4×106/mL) and microstructures (2×104/mL, 4×104/mL, or 8×104/mL corresponding to 0.045%, 0.09%, or 0.18% total volume) in diluted Matrigel (4.0 mg/mL), was pipetted as 100 μL droplets onto 35 mm glass-bottom dishes (MatTek, Ashland, MA) and allowed to polymerize. These concentrations of cells and microstructures attempted to ensure that myocytes, which are largely non-migratory, were positioned no greater than 100–150 μm from a microstructure, thereby maximizing potential cell-microstructure interactions without promoting clustering of either component. Control groups of similar amounts of myocytes and Matrigel but with no added microstructures were mixed and plated in the same manner. The resulting gel layers (0.5–1.0 mm in depth) were immersed in 2 mL complete medium consisting of DMEM F-12 without L-glutamine (Sigma), 5% fetal bovine serum, palmitic (2.56 mg/L) and linoleic (0.84 mg/L) fatty acids, penicillin G/streptomyocin (1 mg/mL), and 8 μM AraC (Sigma). Myocytes were grown for up to 10 days at 37°C with 5% CO2.
2.3 Fluorescent staining of cardiac myocytes
To image live myocytes in 3D settings, the fluorescent stains calcein AM and Hoechst 34580 (Invitrogen, Carlsbad, CA) were used to visualize cell cytoplasm and nuclei, respectively. Culture media was supplemented with 5 μM calcein AM and 10 μM Hoechst 34580 and incubated with myocyte-seeded gels for 20 min at 37°C. Fluorescent signals from cells and microstructures (which autofluoresce with 405 nm excitation) were captured using a Zeiss LSM 510 META laser confocal microscope (Zeiss, Peabody, MA). Acquired images (1024×1024 pixels) were processed with LSM Image Browser software (Zeiss).
For immunofluorescent staining of myocytes, seeded gels were fixed in 3% paraformaldehyde for 5 min, followed by a 0.3 M glycine/PBS rinse. After soaking gels in 0.5% Triton X-100 (Sigma) for 5 min and 10% goat serum/PBS for 20 min, cells were labeled for 1 h with antibodies for sarcomeric α-actinin (monoclonal ab9465, Abcam, Cambridge, MA), paxillin (monoclonal ab32084, Abcam), or N-cadherin (polyclonal ab12221, Abcam) diluted 1/200 in 1% BSA/PBS. After 3 PBS washes, species-compatible Alexa Fluor IgG secondary antibodies (Invitrogen) diluted 1/1000 were added for 45 min. Gels were then rinsed with PBS and stored in 1 nM DAPI/PBS (Vector Labs, Burlingame, CA) for imaging. Fluorescence was captured as single or a series of 1 μm optical slices using a laser confocal microscope (Zeiss) as detailed above.
2.4 Evaluation of cardiac myocyte size
An assessment of the morphological or size changes caused by microstructures after 10 days was carried out by several analogous methods. The simplest approach involved an analysis of images captured from live myocytes stained with calcein AM and Hoechst 34580. Measurement of the fluorescence-based contours of myocytes was performed with ImageJ software (NIH, Bethesda, MA) to produce scaled values of cell perimeter and area (in μm and μm2, respectively). Such measurements were averaged from a pool of at least 8 images acquired for separate gel conditions with no, 50, or 100 μm-long microstructures.
A second, more robust image analysis technique was used to compare microstructure-induced cellular changes to a pharmacological mediator of myocyte hypertrophy. This positive control was established by treating a cell-seeded gel lacking microstructures with 50 μM phenylephrine for 48 h beginning on day 2. After fluorescent staining for α-actinin and nuclei, images were collected from gel layers as described using a 25× objective. A minimum of 8 blindly-selected images were taken for each experimental group and repeated over 3 independent cultures. For each image, a stereological average of sarcomeric content, based on α-actinin signals, was measured using ImageJ software. This value was divided by the number of contained nuclei present to obtain an index of myocyte size. Numerical results from groups with an added stimulus were normalized over each culture to the negative control group.
An assessment of myocyte size for the whole cell population in 3D gels was determined using protein/cell number ratios. With phenylephrine-treated myocytes again serving as a positive control, viable cells were removed from gels by incubation with 400 μL Dispase (BD Biosciences) at 37°C for 2 h. The resulting cell suspensions were collected in centrifuge tubes and spun for 5 min at 500×g. Resuspended pellet volumes were then split, with half used for 5 separate cell counts via trypan blue staining and a hemocytometer. The remaining cells were lysed with 100 μL 1% SDS buffer. Protein levels in each group were measured with a Qubit fluorometer and Quant-iT protein assay (Invitrogen). Ratios of average protein readings and respective cell counts were normalized over 3 cultures to those from negative control gels, as before.
2.5 Calculation of cardiac myocyte cross-sectional area
A comparison of cross-sectional area for myocytes in 3D gels with or without local microstructures was made by analyzing stacks of 1 μm-thick image slices captured from α-actinin and nuclei signals. Scaling tools within ImageJ software were used to measure both the total cell thickness and the maximum transverse width at the linear midpoints along reconstructed myocytes. Using these values and an ellipsoid shape assumption, the cross-sectional area was calculated. The process was repeated over 20 image stacks for each condition of anchorage, as pooled from 3 separate cultures.
2.6 Counting of myocyte clusters in 3D
To gauge whether the presence of microstructures contributed to an overall change in myocyte aggregation, a random sampling of cell cluster sizes were tallied for each condition. Myocytes were fixed and co-immunostained with α-actinin and N-cadherin and viewed under a laser confocal microscope (Zeiss) with high-powered objective. Cells in clusters (containing more than a single isolated myocyte) were verified by the appearance of N-cadherin at cell-cell junctions. At least 50 single cells or clusters were counted in control and microstructure-containing gels and averaged over 3 separate cultures. Final averages were plotted in terms of the relative frequency of each cluster size.
2.7 Analysis of cardiac myocyte contraction rate
Two methods were used to survey the rate of contraction for live cardiac myocytes in 3D gels with or without microstructure contact. The first made use of a Nikon TMS inverted microscope (Nikon) to identify single myocytes visually. The number of spontaneous beats for each myocyte was noted over a 60 s interval, and a minimum of 30 cells were observed for each anchored condition over 5 cultures. The second method used the same confocal microscope detailed earlier to capture line scans of isolated myocytes. Briefly, cells in gels were incubated with 5 μM calcein AM and 10 μM Hoechst 34580 (Invitrogen) at 37°C for 20 min to confirm cell viability. Individual beating myocytes oriented with a long axis parallel to the XY plane were focused at their widest region. A time series was then started to collect transmitted light (from DIC channel) over a pixel-wide line along the cell length every 25 ms for a total of 5 s. The ability for this 3D system to assess contraction rate was validated by recapturing line scans on the same cell positions upon switching of culture media containing 10 μM dobutamine or 10 mM 2,3-butanedione monoxime (BDM, Sigma).
2.8 Gene expression analysis: microarray and real-time PCR
Global gene expression analysis of cardiac myocytes made use of the Affymetrix GeneChip platform, with all steps of microarray hybridization carried out compliant to manufacturer recommendations (Affymetrix, Santa Clara, CA). Briefly, RNA was extracted directly from myocytes in gels with or without microstructures at day 10 by TRIzol extraction (Invitrogen) and purified with DNase I treatment (Invitrogen). Isolated RNA was pooled equally by mass over 3 separate cultures and submitted to the University of Illinois Core Genomics Facility (Chicago, IL) for sample viability, reverse-transcription, cDNA preparation, cRNA labeling, hybridization, and scanning protocols. Using the Rat Expression Array 230 2.0 (Affymetrix), a total of 6 chips were scanned, with 3 replicates from each experimental group.
Expression data analysis was performed with the dChip software package (Li and Hung Wong 2001). Scanned results were normalized on median intensity profiles across all replicates to generate raw gene expression data. From here, model-based expression data was calculated using the Perfect Match/Mismatch (PM/MM) Difference Model (Li and Hung Wong 2001). Within the software, sorting filters were then applied to over 28,000 representative genes to identify those with significant differences (p<0.05) between control and microstructure-containing replicates. These genes were grouped by biological process from gene ontology annotations of corresponding microarray probes. The gene list was further filtered manually for biological relevance to cardiac muscle, with respect to calcium cycling, metabolism, cytoskeleton remodeling, and mechanical signaling. A final subset of 10 genes was plotted with heat maps of normalized expression data.
Confirmation of microstructure-induced fold changes for several members of this gene subset was achieved by real-time PCR analysis. Here, RNA was isolated from cardiac myocytes in gels by TRIzol extraction (Invitrogen) and reverse transcribed with the enzyme M-MLV (Invitrogen). The resulting cDNA and primers specific to adrenergic receptor beta-1 (β1-AR, 5′-ATTAGTTGGAAGGACC AGGCG and 5′-GCAAAATGGCCTTTCAACCAC), sarcoplasmic reticulum calcium-ATPase (SERCA2, 5′-TCTGCCAGTGGGACTCTTCT and 5′-GTTGAGCAT CCTGTCCTTCC), cardiac ankyrin repeat protein (CARP, 5′-CACACCGTTTCCAGTGTCAT and 5′-CCAGCCTTCAATGGGTTAAG), ankyrin-1 (ANK1, 5′-TCCCGTTGCTAGTAGGAGCTGT and 5′-GAGCATGT GAGAGAGAGCCAAG), cytochrome c oxidase subunit VIII heart/muscle (COX8H, 5′-GAGAATCATGCC AAGGCTTC and 5′-GCCAGCGATTATGACTGACA) and the housekeeping control beta-2-microglobulin (β2M, 5′-CAGTTCCACCCACCTCAGAT and 5′-TTTTGGGCTCCTTCAGAGTG) were set up with a SYBR Green assay and run in an ABI 7900HT thermocycler (Applied Biosystems, Warrington, WA). The β2M control was selected based on the overall stability and transcript levels of the gene previously confirmed within the heart (Pérez et al. 2007). Gene products were compared for myocytes grown with or without microstructures using the relative standard curve method. The analysis was repeated over 4 separate cultures.
2.9 Statistics
Statistical analysis was performed using Excel software (Microsoft, Redmond, CA). Differences in quantitative variables were determined by a two-tailed Student’s t-test or one-way ANOVA, with significance marked as p<0.05. All data is expressed as mean ± SEM.
3 Results
3.1 Microstructure design and effectiveness
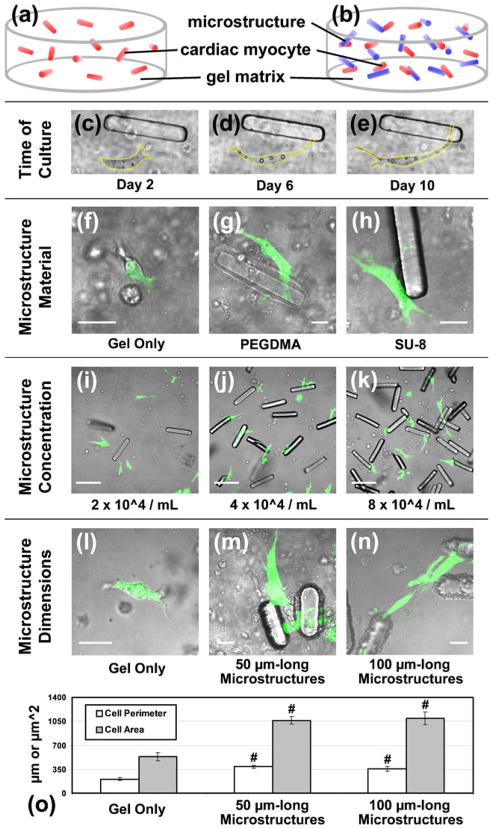
Several optimization experiments were first required in order to study the responses of cardiac muscle cells to microscale domains. All experiments used a constant seeding density of neonatal rat ventricular myocytes (4×106/mL) to yield unsynchronized, isolated cells in 3D gel matrices. This concentration of myocytes suspended in Matrigel (Fig. 1(a)) served as a control for comparison to gels containing the same number of cells with engineered microstructures (Fig. 1(b)). Both culture conditions produced independently-beating myocytes that remained viable beyond 10 days. In this time frame, single myocytes demonstrated an ability to make contact with other neighboring features, whether another cell or a locally-positioned microstructure (Fig. 1(c–e)). Myocyte-microstructure contact occurred similarly when microstructures were comprised of either poly (ethylene glycol) dimethacrylate (PEGDMA) or SU-8 photo-resist (Fig. 1(f–h)). Next, the minimum microstructure concentration required to permit contact with the majority of myocytes was determined (Fig. 1(i–k)). At a microstructure concentration of 4×104/mL (0.09% of total volume), most myocytes were within a cell length (<100 μm) of a discrete microstructure (Fig. 1(j)). This randomly-oriented but uniform distribution of microstructures was found to cause changes in live myocyte morphology after 10 days. Cell perimeter and area, as quantified through cytoplasmic staining with calcein AM (Fig. 1(l–n)), both increased significantly with microstructure presence (Fig. 1(o)). However, no average morphological differences were detected when microstructures of two distinct dimensions were contrasted (50×15×15 or 100×15×15 μm3 at fixed volume percentages). As a result, only 100 μm-long microstructures were used in additional testing to more closely approximate the scale of single myocytes (Fraticelli et al. 1989).
Fig. 1.
Microstructure optimization. Isolated neonatal rat ventricular myocytes were suspended with random orientation in a supporting 3D gel matrix (Matrigel) (a) without or (b) with engineered 100 μm-long microstructures (c–e) and cultured for 10 days while observed with phase microscopy (yellow line = cell boundary). (f) Live myocytes (green with calcein AM) in gel only or making contact with microstructures fabricated from either (g) poly (ethylene glycol) dimethacrylate (PEGDMA) or (h) SU-8 photo-resist (scale = 20 μm). (i–k) The concentration of microstructures in 3D gels was varied, with 4×104/mL yielding a single microstructure within approximately 100 μm of each cell (scale = 100 μm). Myocyte morphology in (l) gel alone or with (m) 50× 15×15 or (n) 100×15×15 μm3 microstructures in 3D (scale = 20 μm). (o) The average perimeter and area of live myocytes is significantly lower in control gels than with 50 and 100 μm-long microstructures, but cell morphology across the two microstructure groups is similar (n≥8). Mean ± SEM. #p<0.01
3.2 Immunostaining of microstructure-interfacing myocytes
Upon immunofluorescent staining at day 10, paxillin was found to accumulate at regions of myocyte-microstructure contact, indicative of active focal adhesion complexes (Fig. 2(b)). In control gels with no microstructures, paxillin staining in myocytes was speckled and punctate in appearance (Fig. 2(a)). However, both 3D-suspended groups exhibited focal adhesions that were less prominent than those typically found in myocytes firmly anchored to conventional 2D substrates (not shown). The interaction between cells and microstructures was evident even in myocytes showing stable attachments to adjacent cells, as evaluated by N-cadherin staining of adherens junctions (Fig. 2(c),(d)). The angles single myocytes made with microstructures were random in 3D, with contact occurring anywhere along each microstructure (Fig. 2(e–g)). In all such cases, sarcomeric α-actinin staining showed that myocytes attached to microstructures with terminal striations, as the first sarcomeres at cell ends were adjacent to microstructure surfaces.
Fig. 2.
Immunostaining of myocyte interfaces with microstructures. (a, b) Staining of cardiac myocytes for paxillin (green), sarcomeric α-actinin (red), and nuclei (blue) show punctate focal adhesions in control gels but accumulation at regions of microstructure contact (scale = 20 μm). Note that microstructures also fluoresce blue. White arrowheads label focal adhesions along the microstructure. (c, d) Antibody labeling of N-cadherin (green) and α-actinin (red) reveal that small clusters of myocytes (2 cells) also interact with microstructures. Arrowheads mark regions of cell-cell contact. (e) In gel alone, myocytes (α-actinin, red) exhibit a common spindle shape. Striations are visible in the inset and are identified with arrowheads. (f) Myocytes form perpendicular, oblique, or parallel associations with randomly-oriented microstructures in 3D, with (g) showing a series of confocal image slices over an 8 μm thickness
3.3 Indices of cardiac myocyte hypertrophy
Microstructure-related changes in the general characteristics of cardiac myocytes were examined more thoroughly with assays of cell growth. By 10 days in culture with microstructures, myocytes were visibly larger and displayed a greater area of muscle striations (Fig. 3(a),(c)). At this time point, a stereology-based assessment of area in 3D-suspended myocytes (via sarcomeric α-actinin staining) revealed that microstructures caused a 1.61-fold increase in muscle cell size (Fig. 3(d)). In a separate analysis, myocyte populations recovered from depolymerized gels were counted and subsequently lysed to quantify protein levels; the determined protein/cell ratios, when normalized, showed a 1.43-fold difference between microstructure conditions (Fig. 3(e)). The physiological context of microstructure cues was evaluated by comparison to a pharmacologic stimulus of myocyte hypertrophy, phenylephrine (Eble et al. 1998), which caused increases in cell area and protein/cell ratios that were similar to those induced by microstructures (Fig. 3(b),(d),(e)). Furthermore, an increased myocyte cross-sectional size correlated with microstructure presence; measurements of α-actinin-based 3D reconstructions for microstructure-interfacing myocytes consistently yielded larger cross-sectional areas (60.63± 2.26 μm2) than myocytes in gel alone (38.39±1.98 μm2) (Fig. 3(f)). Microstructures had no effect on whole cell cluster sizes, and the prevalence of isolated cells did not differ in gels with or without microstructures (Fig. 3(g)).
Fig. 3.
Microstructure-induced myocyte hypertrophy. Images captured using laser confocal microscopy reveal differences in cardiac myocyte size after 10 days between (a) myocytes in 3D gels, (b) myocytes in gels treated with 50 μM phenylephrine, and (c) myocyte-seeded gels containing 100 μm-long SU-8 microstructures. Cells were stained for muscle-specific α-actinin (red) and nuclei (blue) (scale =50 μm). (d) Image analysis (n=3 cultures, 24 total images per culture) confirms similar fold increases in myocyte size (1.61±0.06) due to microstructures or phenylephrine (1.68±0.02). (e) Protein levels per total cell count in each dish (n=3), microstructure-containing gels and phenylephrine-treated gels both show similar fold increases (1.43± 0.08 and 1.46±0.03, respectively) relative to control myocytes. (f) Myocyte cross-sectional area from α-actinin-based 3D reconstructions (n=3 cultures, 7 stacks per culture) also increases between conditions without (38.39±1.98 μm2) or with microstructures (60.63±2.26 μm2). (g) Cell clustering does not vary with microstructures, and the majority of myocytes exist as single cells in both experimental groups (n=3 cultures, >50 cell samplings per culture). Mean ± SEM. #p< 0.01, *p<0.05
3.4 Spontaneous contraction of microstructure-supported cardiac myocytes
Microstructure contact also affected the beat-to-beat character of individual cardiac myocytes. Specifically, myocytes situated near microstructures in 3D exhibited 2-fold higher average rates of spontaneous contraction (33.0±1.4 BPM) at day 10 than cells with no adjacent microstructures (16.4±1.1 BPM) (Fig. 4(a)). Since only single, isolated cells were used in the analysis, such chronotropic differences were not the result of an altered clustering effect of the myocytes themselves. Examples of the varied beating rates due to microstructures were captured using live cell imaging and high-speed line scanning (Fig. 4(b–e)). Apart from these baseline differences, myocytes with or without microstructure contact similarly increased beating rates when exposed to dobutamine, a selective adrenergic agonist (Fig. 4(a),(f),(g)). In addition, BDM, an inhibitor of actin-myosin cross-bridge cycling, blocked all beating as expected both in the presence or absence of microstructures (Fig. 4(a),(h),(i)).
Fig. 4.
Myocyte beating with microstructures. (a) Myocytes near microstructures have higher spontaneous beating rates (33.0±1.4 BPM) than myocytes in gels without microstructures (16.4±1.1 BPM) (n=5 cultures, >10 samplings per culture). Following addition of 10 μM dobutamine (DOB), myocytes exhibit significant increases in beating rates with microstructures (68.8±2.6 BPM) and in gel alone (39.4±3.3 BPM). 10 mM 2,3-butanedione monoxime (BDM) arrests contraction in both groups. (b, c) Confocal line scans (positions shown in red) are captured for live myocytes, as identified by calcein AM (green) and Hoechst 34580 (blue) staining of cytoplasm and nuclei, respectively. (d, e) Myocytes in conditions with or without microstructure contact show differences in baseline beating rates. White arrowheads mark each recorded beat; note that green and blue signals are artificially overlapped with the collected grayscale channel to help represent cell dimensions. (f, g) Upon addition of DOB, each myocyte shows a similar increase in beating rate. (h, i) Contraction in the same cells is inhibited after switching to media with BDM
3.5 Microstructure-induced changes in gene expression
The effect of 10 day microstructure inclusion on global gene profiles in myocytes was carried out by microarray scanning. Comparison of the resulting normalized expression values revealed 276 distinct genes that differed significantly upon microstructure addition in 3D gels. When grouped by biological process, many genes occupied broad ontological classifications such as cell localization, metabolism, and development (Fig. 5(a)). Other common categories—including the regulation of cell processes, signal transduction, and the response to a stimulus—were of particular interest to cardiac muscle. Such genes were filtered by annotation and relevance to produce a small, focused subset of genes associated with membrane-cytoskeletal linkage and muscle regulation (Fig. 5(b)). Several of these microstructure-affected genes had significant fold changes that were confirmed through real-time PCR assays (Fig. 5(c)). Increases in expression were identified for adrenergic receptor beta-1 (β1-AR, 1.19± 0.01), sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2, 1.6±0.12), cardiac ankyrin repeat protein (CARP, 1.26±0.02), ankyrin-1 (ANK1, 1.42±0.05), and cytochrome c oxidase subunit VIII heart/muscle (COX8H, 2.05±0.2) as relative to a beta-2 microglobulin (β2M) housekeeping control gene.
Fig. 5.
Gene expression altered by microstructures. Global microarray gene expression analysis was performed using RNA isolated from 10 day cultures of myocytes in 3D gels with or without microstructures (n=3). (a) Over 250 unique genes significantly altered by microstructures (p<0.05) were grouped by biological process. Several ontology categories with relevance to cardiac muscle (highlighted in yellow) were explored to yield a list of 10 genes of interest. (b) The filtered genes are listed and shown with corresponding chip-level expression values (red = higher normalized expression). (c) Expression changes were confirmed for several genes by independent real-time PCR analysis, showing significant microstructure-related increases in adrenergic receptor beta-1 (β1-AR, 1.19±0.01), sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2, 1.6± 0.12), cardiac ankyrin repeat protein (CARP, 1.26±0.02), ankyrin-1 (ANK1, 1.42±0.05), and cytochrome c oxidase subunit VIII heart/muscle (COX8H, 2.05±0.2) when normalized to a beta-2 micro-globulin (β2M) housekeeping control gene (n=4). Mean ± SEM. #p< 0.01, *p<0.05
4 Discussion
Results show that the addition of a small quantity of microstructures in 3D gels introduces heterogeneity in stiffness and topography, which is sufficient to alter the hypertrophy, spontaneous contraction, and gene expression of neonatal rat ventricular myocytes. Myocytes interact with microstructures and are sensitive to such local heterogeneity over time. Direct contact of an individual myocyte with a microstructure thus provides an effective stimulus for augmenting muscle cell function.
4.1 Microstructures promote hypertrophy in myocytes
Ventricular myocytes undergo rapid changes in functional size and intracellular organization in response to under-worked or overworked conditions both in vivo and in vitro (Thompson et al. 1984; Cooper et al. 1986; Nishimura et al. 2004). Similar changes are evident in the current study, as total sarcomeric area and protein levels per myocyte show significant increases by day 10 in 3D culture with microstructures (Fig. 3). Indeed, the magnitude of the observed myocyte growth due to microstructures approaches that of phenylephrine, a standard chemical stimulus of muscle cell hypertrophy (Eble et al. 1998). This physically-induced hypertrophic response leads to the simple interpretation that microstructures affect the adaptive behaviors of nearby myocytes. Interestingly, other studies have demonstrated that modifications in micropatterned surfaces (Geisse et al. 2009) and micropegged and/or microgrooved substrata (Motlagh et al. 2003b) are not sufficient to elicit differences in total myocyte size. Yet the firm anchorage of cells on these substrates may prevent such microscale geometric or topographical variables from influencing the active shortening and contractile effort of myocytes. In full 3D, however, myocytes contract against the discrete 100×15×15 μm3 microstructures and thus cause hypertrophy in a loaded manner comparable to that in the body (Russell et al. 2000).
Real-time PCR analysis identified several genes affected by the inclusion of microstructures (Fig. 5) that may contribute to the ongoing hypertrophic processes in cardiac myocytes. The gene coding adrenergic receptor beta-1 is significantly increased in microstructure groups; the G-protein-coupled receptor is a known marker of pathological and physiological hypertrophy, with expression levels increased in both pressure-overloaded and chronically-exercised rat hearts (Iemitsu et al. 2001). Cytochrome c oxidase subunit VIII heart/muscle (COX8H) is also affected by microstructures; the terminal enzyme of the mitochondria electron transport chain is a key metabolic gene with altered expression during hypertrophy (Strøm et al. 2004). Ankyrin-1 and cardiac ankyrin repeat protein (CARP) share some sequence homology but assume distinct structural and signaling functions in myocytes. An alternatively spliced ankyrin-1 product is concentrated in the sarcoplasmic reticulum and helps to connect the organelle to the contractile apparatus (Bagnato et al. 2003). CARP acts as a transcription factor that operates with the myofibrillar protein titin in the muscle stretch-sensor system (Zolk et al. 2002; Witt et al. 2005). CARP expression has also been found to be upregulated rapidly in response to hypertrophic stimuli (Aihara et al. 2000; Nagueh et al. 2004). Taken together, the activation of these genes at day 10 in culture suggests that microstructures have a prolonged effect on myocyte-specific growth mechanisms.
4.2 Microstructures and contractile activity
The significant differences in cardiac myocyte beating rates due to microstructure contact imply that additional regulatory pathways are affected (Fig. 4). Yet myocytes with or without local microstructures both display normal responses to the chronotropic drugs dobutamine and BDM, affirming that basic physiological activity is not impaired in either 3D-cultured group. Myocyte beating frequency is known to be influenced by the physical environment, as a correlation has been described between 2D substrate stiffness and spontaneous beating rate in embryonic cardiac myocytes (Engler et al. 2008). The spontaneous contraction of neonatal cardiac myocytes is also dependent on the uniform stiffnesses of 3D gels (Shapira-Schweitzer and Seliktar 2007; Lee et al. 2008). Current data shows that this bulk property can be overridden (based on cellular responses) by the inclusion of rigid domains that account for less than 1% of the total volume.
Beating rate changes in myocytes from microstructure-containing gels may be related to the increased gene expression of SERCA2, a protein integral to intracellular calcium cycling and thus the overall rate of contraction (Weisser-Thomas et al. 2005). Myocyte SERCA2 expression varies with 2D substrate stiffness (Jacot et al. 2008), which parallels the study relating the same variable to myocyte beating rate (Engler et al. 2008). Other external loading stimuli have shown contrasting effects SERCA2 expression, with evidence of both upregulation and downregulation of the gene through different regimes of cyclic stretch (Cadre et al. 1998; Kögler et al. 2006). SERCA2 may therefore be a key player in the apparent feedback between microstructure-myocyte contact and contractile cell dynamics.
4.3 Recognition of a microscale domain
Certain physical attributes of microstructures play a greater role than others in eliciting cardiac myocyte responses. The polymer itself (PEGDMA or SU-8) does not appear to be relevant in this in vitro system, as myocyte interactions occur regardless of microstructure material. This is surprising since both PEGDMA and SU-8 are known to resist protein or cell attachment when used as unmodified 2D substrates (Kim et al. 2005; Tao et al. 2008). However, when PEGDMA or SU-8 microstructures are surrounded by the rich extracellular matrix (Matrigel) during seeding, cells seem to sustain physiologically meaningful contact with the 3D features.
It is clear that the heterogeneity introduced by microstructures into the uniform gel matrix in 3D is an effective biophysical cue. Cardiac myocytes recognize such local heterogeneity, forming randomly-oriented perpendicular, oblique, or parallel associations with microstructures by day 10 (Fig. 2(e–g)). The underlying cause of this interaction may be template guidance, as demonstrated with the alignment of myocytes in the direction of micro-textured grooves (Motlagh et al. 2003b). Other cell types such as cardiac fibroblasts (Norman et al. 2008) and mesenchymal stem cells (Collins et al. 2010) have also been shown to detect heterogeneity in 3D culture.
The addition of microstructures in 3D gels alters not only the spatial heterogeneity around cells, but the mechanical heterogeneity as well. Myocytes interact similarly with PEGDMA and SU-8 microstructures, which have considerably different scales of elastic moduli (approximately 20 kPa and 4 GPa, respectively) (Norman et al. 2008; Ayala et al. 2010). These observations suggest that the differential stiffness provided by relatively rigid microstructures in the very soft 3D gel (G′=22.9 Pa) is a contributing factor in affecting myocytes. From the level of single myocytes, the uniformly soft control gels may thus lack a resistive hub that is otherwise provided by the microstructures.
Cardiac myocytes also display a limited response to microstructure size. Both 50 and 100 μm-long stiff microstructures (at the same volume percentage) cause a significant change in myocyte morphology relative to control cells without microstructures (Fig. 1(l–o)). However, the nearly equivalent results between 50 and 100 μm groups indicate that myocytes do not distinguish a 3D surface with a scale above that of a single cell.
5 Conclusions
As a whole, the collected data shows that microscale physical domains in a 3D environment control many aspects of cardiac myocyte function, including cell size, contractile beating rate, and gene expression. The ability for myocytes to be regulated by the heterogeneity provided by discrete 3D structures thus poses important considerations for various cardiac tissue engineering strategies. In exploring the physical requirements of single myocytes, the results also provide new insight for the understanding of cardiac muscle cell adaptation and remodeling.
Acknowledgments
Many thanks to Perla Ayala at the University of California at San Francisco for her ongoing help in fabricating microstructures. This work was supported by the National Institutes of Health grants T32 HL007692, PO1 HL62426, and RO1 HL090523.
Abbreviations
- 3D
three dimensions
- AraC
cytosine β-D-arabino-furanoside
- β1-AR
beta-1 adrenergic receptor
- β2M
beta-2 microglobulin
- BDM
2,3-butanedione monoxime
- BPM
beats per minute
- BSA
bovine serum albumin
- CARP
cardiac ankyrin repeat protein
- COX8H
cytochrome c oxidase subunit VIII heart/muscle
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DMEM
Dulbecco’s modified Eagle’s medium
- DOB
dobutamine
- ECM
extracellular matrix
- PBS
phosphate buffered saline
- PE
phenylephrine
- PEGDMA
poly(ethylene glycol) dimethacrylate
- SDS
sodium dodecyl sulfate
- SEM
standard error of measurement
- SERCA2
sarcoplasmic reticulum calcium-ATPase
Contributor Information
Matthew W. Curtis, Department of Bioengineering, University of Illinois at Chicago, Chicago, IL, USA
Sadhana Sharma, Department of Physiology and Biophysics (MC 901), University of Illinois at Chicago, 835 S. Wolcott Avenue, Chicago, IL 60612-7342, USA.
Tejal A. Desai, Department of Physiology and Division of Bioengineering, University of California at San Francisco, San Francisco, CA, USA
Brenda Russell, Email: russell@uic.edu, Department of Physiology and Biophysics (MC 901), University of Illinois at Chicago, 835 S. Wolcott Avenue, Chicago, IL 60612-7342, USA.
References
- Aihara Y, Kurabayashi M, Saito Y, Ohyama Y, Tanaka T, Takeda S, Tomaru K, Sekiguchi K, Arai M, Nakamura T, Nagai R. Cardiac ankyrin repeat protein is a novel marker of cardiac hypertrophy: role of M-CAT element within the promoter. Hypertension. 2000;36:48–53. doi: 10.1161/01.hyp.36.1.48. [DOI] [PubMed] [Google Scholar]
- Ayala P, Lopez JI, Desai TA. Microtopographical cues in 3D attenuate fibrotic phenotype and extracellular matrix deposition: implications for tissue regeneration. Tissue Eng Part A. 2010 doi: 10.1089/ten.tea.2009.0815. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AF. Neutrophil granulocytes: adhesion and locomotion on collagen substrata and in collagen matrices. J Cell Sci. 1982;58:455–467. doi: 10.1242/jcs.58.1.455. [DOI] [PubMed] [Google Scholar]
- Cadre BM, Qi M, Eble DM, Shannon TR, Bers DM, Samarel AM. Cyclic stretch down-regulates calcium transporter gene expression in neonatal rat ventricular myocytes. J Mol Cell Cardiol. 1998;30:2247–2259. doi: 10.1006/jmcc.1998.0788. [DOI] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Collins JM, Ayala P, Desai TA, Russell B. Three-dimensional culture with stiff microstructures increases proliferation and slows osteogenic differentiation of human mesenchymal stem cells. Small. 2010;6:355–360. doi: 10.1002/smll.200901757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G, 4th, Mercer WE, Hoober JK, Gordon PR, Kent RL, Lauva IK, Marino TA. Load regulation of the properties of adult feline cardiocytes. The role of substrate adhesion. Circ Res. 1986;58:692–705. doi: 10.1161/01.res.58.5.692. [DOI] [PubMed] [Google Scholar]
- Deutsch J, Motlagh D, Russell B, Desai TA. Fabrication of microtextured membranes for cardiac myocyte attachment and orientation. J Biomed Mater Res. 2000;53:267–275. doi: 10.1002/(sici)1097-4636(2000)53:3<267::aid-jbm12>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Durieux AC, Desplanches D, Freyssenet D, Flück M. Mechanotransduction in striated muscle via focal adhesion kinase. Biochem Soc Trans. 2007;35:1312–1313. doi: 10.1042/BST0351312. [DOI] [PubMed] [Google Scholar]
- Eble DM, Qi M, Waldschmidt S, Lucchesi PA, Byron KL, Samarel AM. Contractile activity is required for sarcomeric assembly in phenylephrine-induced cardiac myocyte hypertrophy. Am J Physiol. 1998;274:C1226–C1237. doi: 10.1152/ajpcell.1998.274.5.C1226. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, Speicher DW, Sanger JW, Sanger JM, Discher DE. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraticelli A, Josephson R, Danziger R, Lakatta E, Spurgeon H. Morphological and contractile characteristics of rat cardiac myocytes from maturation to senescence. Am J Physiol. 1989;257:H259–H265. doi: 10.1152/ajpheart.1989.257.1.H259. [DOI] [PubMed] [Google Scholar]
- Geisse NA, Sheehy SP, Parker KK. Control of myocyte remodeling in vitro with engineered substrates, In Vitro. Cell Dev Biol Anim. 2009;45:343–350. doi: 10.1007/s11626-009-9182-9. [DOI] [PubMed] [Google Scholar]
- Gopalan SM, Flaim C, Bhatia SN, Hoshijima M, Knoell R, Chien KR, Omens JH, McCulloch AD. Anisotropic stretch-induced hypertrophy in neonatal ventricular myocytes micro-patterned on deformable elastomers. Biotechnol Bioeng. 2003;81:578–587. doi: 10.1002/bit.10506. [DOI] [PubMed] [Google Scholar]
- Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290:H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Kamm RD, Lee RT. Cell mechanics and mechanotransduction: pathways, probes, and physiology. Am J Physiol Cell Physiol. 2004;287:C1–C11. doi: 10.1152/ajpcell.00559.2003. [DOI] [PubMed] [Google Scholar]
- Iemitsu M, Miyauchi T, Maeda S, Sakai S, Kobayashi T, Fujii N, Miyazaki H, Matsuda M, Yamaguchi I. Physiological and pathological cardiac hypertrophy induce different molecular phenotypes in the rat. Am J Physiol Regul Integr Comp Physiol. 2001;281:R2029–R2036. doi: 10.1152/ajpregu.2001.281.6.R2029. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Tensegrity-based mechanosensing from macro to micro. Prog Biophys Mol Biol. 2008;97:163–179. doi: 10.1016/j.pbiomolbio.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot JG, McCulloch AD, Omens JH. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys J. 2008;95:3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz AM. Maladaptive growth in the failing heart: the cardiomyopathy of overload. Cardiovasc Drugs Ther. 2002;16:245–249. doi: 10.1023/a:1020604623427. [DOI] [PubMed] [Google Scholar]
- Kim P, Kim DH, Kim B, Choi SK, Lee SH, Khademhosseini A, Langer R, Suh KY. Fabrication of nanostructures of polyethylene glycol for applications to protein adsorption and cell adhesion. Nanotechnology. 2005;16:1–7. doi: 10.1088/0957-4484/16/10/072. [DOI] [PubMed] [Google Scholar]
- Kögler H, Schott P, Toischer K, Milting H, Van PN, Kohlhaas M, Grebe C, Kassner A, Domeier E, Teucher N, Seidler T, Knöll R, Maier LS, El-Banayosy A, Körfer R, Hasenfuss G. Relevance of brain natriuretic peptide in preload-dependent regulation of cardiac sarcoplasmic reticulum Ca2+ ATPase expression. Circulation. 2006;113:2724–2732. doi: 10.1161/CIRCULATIONAHA.105.608828. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Blumenkranz MS, Fishman HA, Bent SF. Controlling cell adhesion on human tissue by soft lithography. Langmuir. 2004;20:4155–4161. doi: 10.1021/la035467c. [DOI] [PubMed] [Google Scholar]
- Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Kim do E, Azeloglu EU, Costa KD. Engineered cardiac organoid chambers: toward a functional biological model ventricle. Tissue Eng Part A. 2008;14:215–225. doi: 10.1089/tea.2007.0351. [DOI] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:1–11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh D, Hartman TJ, Desai TA, Russell B. Microfabricated grooves recapitulate neonatal myocyte connexin43 and N-cadherin expression and localization. J Biomed Mater Res A. 2003a;67:148–157. doi: 10.1002/jbm.a.10083. [DOI] [PubMed] [Google Scholar]
- Motlagh D, Senyo SE, Desai TA, Russell B. Microtextured substrata alter gene expression, protein localization and the shape of cardiac myocytes. Biomaterials. 2003b;24:2463–2476. doi: 10.1016/s0142-9612(02)00644-0. [DOI] [PubMed] [Google Scholar]
- Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- Nishimura S, Yasuda S, Katoh M, Yamada KP, Yamashita H, Saeki Y, Sunagawa K, Nagai R, Hisada T, Sugiura S. Single cell mechanics of rat cardiomyocytes under isometric, unloaded, and physiologically loaded conditions. Am J Physiol Heart Circ Physiol. 2004;287:H196–H202. doi: 10.1152/ajpheart.00948.2003. [DOI] [PubMed] [Google Scholar]
- Norman JJ, Collins JM, Sharma S, Russell B, Desai TA. Microstructures in 3D biological gels affect cell proliferation. Tissue Eng A. 2008;14:379–390. doi: 10.1089/tea.2007.0077. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Saito Y, Harada M, Kamitani S, Kuwahara K, Miyamoto Y, Ishikawa M, Hamanaka I, Kajiyama N, Takahashi N, Nakagawa O, Masuda I, Kishimoto I, Nakao K. Outside-in signalling of fibronectin stimulates cardiomyocyte hypertrophy in cultured neonatal rat ventricular myocytes. J Mol Cell Cardiol. 2000;32:765–776. doi: 10.1006/jmcc.2000.1119. [DOI] [PubMed] [Google Scholar]
- Pedersen JA, Swartz MA. Mechanobiology in the third dimension. Ann Biomed Eng. 2005;33:1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez S, Royo LJ, Astudillo A, Escudero D, Alvarez F, Rodríguez A, Gómez E, Otero J. Identifying the most suitable endogenous control for determining gene expression in hearts from organ donors. BMC Mol Biol. 2007;8:114. doi: 10.1186/1471-2199-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- Raskin AM, Hoshijima M, Swanson E, McCulloch AD, Omens JH. Hypertrophic gene expression induced by chronic stretch of excised mouse heart muscle. Mol Cell Biomech. 2009;6:145–159. [PMC free article] [PubMed] [Google Scholar]
- Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B, Motlagh D, Ashley WW. Form follows function: how muscle shape is regulated by work. J Appl Physiol. 2000;88:1127–1132. doi: 10.1152/jappl.2000.88.3.1127. [DOI] [PubMed] [Google Scholar]
- Russell B, Curtis MW, Koshman YE, Samarel AM. Mechanical stress-induced sarcomere assembly for cardiac muscle growth in length and width. J Mol Cell Cardiol. 2010;48:817–823. doi: 10.1016/j.yjmcc.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Ghibaudo M, Buguin A, Silberzan P, Ladoux B. Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates. Proc Natl Acad Sci U S A. 2007;104:8281–8286. doi: 10.1073/pnas.0702259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarel AM. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am J Physiol Heart Circ Physiol. 2005;289:H2291–H2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- Senyo SE, Koshman YE, Russell B. Stimulus interval, rate and direction differentially regulate phosphorylation for mechanotransduction in neonatal cardiac myocytes. FEBS Lett. 2007;581:4241–4247. doi: 10.1016/j.febslet.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira-Schweitzer K, Seliktar D. Matrix stiffness affects spontaneous contraction of cardiomyocytes cultured within a PEGylated fibrinogen biomaterial. Acta Biomater. 2007;3:33–41. doi: 10.1016/j.actbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Strøm CC, Kruhøffer M, Knudsen S, Stensgaard-Hansen F, Jonassen TE, Orntoft TF, Haunsø S, Sheikh SP. Identification of a core set of genes that signifies pathways underlying cardiac hypertrophy. Comp Funct Genomics. 2004;5:459–470. doi: 10.1002/cfg.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao SL, Popat KC, Norman JJ, Desai TA. Surface modification of SU-8 for enhanced biofunctionality and nonfouling properties. Langmuir. 2008;24:2631–2636. doi: 10.1021/la703066z. [DOI] [PubMed] [Google Scholar]
- Thakar RG, Chown MG, Patel A, Peng L, Kumar S, Desai TA. Contractility-dependent modulation of cell proliferation and adhesion by microscale topographical cues. Small. 2008;4:1416–1424. doi: 10.1002/smll.200701302. [DOI] [PubMed] [Google Scholar]
- Thompson EW, Marino TA, Uboh CE, Kent RL, Cooper G., 4th Atrophy reversal and cardiocyte redifferentiation in reloaded cat myocardium. Circ Res. 1984;54:367–377. doi: 10.1161/01.res.54.4.367. [DOI] [PubMed] [Google Scholar]
- Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- Wang YC, Ho CC. Micropatterning of proteins and mammalian cells on biomaterials. FASEB J. 2004;18:525–527. doi: 10.1096/fj.03-0490fje. [DOI] [PubMed] [Google Scholar]
- Weisser-Thomas J, Kubo H, Hefner CA, Gaughan JP, McGowan BS, Ross R, Meyer M, Dillmann W, Houser SR. The Na+/Ca2+ exchanger/SR Ca2+ ATPase transport capacity regulates the contractility of normal and hypertrophied feline ventricular myocytes. J Card Fail. 2005;11:380–387. doi: 10.1016/j.cardfail.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Witt SH, Labeit D, Granzier H, Labeit S, Witt CC. Dimerization of the cardiac ankyrin protein CARP: implications for MARP titin-based signaling. J Muscle Res Cell Motil. 2005;26:401–408. doi: 10.1007/s10974-005-9022-9. [DOI] [PubMed] [Google Scholar]
- Zimmermann WH, Schneiderbanger K, Schubert P, Didié M, Münzel F, Heubach JF, Kostin S, Neuhuber WL, Eschenhagen T. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 2002;90:223–230. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- Zolk O, Frohme M, Maurer A, Kluxen FW, Hentsch B, Zubakov D, Hoheisel JD, Zucker IH, Pepe S, Eschenhagen T. Cardiac ankyrin repeat protein, a negative regulator of cardiac gene expression, is augmented in human heart failure. Biochem Biophys Res Commun. 2002;293:1377–1382. doi: 10.1016/S0006-291X(02)00387-X. [DOI] [PubMed] [Google Scholar]