Abstract
Adipogenesis is tightly controlled by a complex network of transcription factors acting at different stages of differentiation. Peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer-binding protein (C/EBP) family members are key regulators of this process. We have employed DNase I hypersensitive site analysis to investigate the genome-wide changes in chromatin structure that accompany the binding of adipogenic transcription factors. These analyses revealed a dramatic and dynamic modulation of the chromatin landscape during the first hours of adipocyte differentiation that coincides with cooperative binding of multiple early transcription factors (including glucocorticoid receptor, retinoid X receptor, Stat5a, C/EBPβ and -δ) to transcription factor ‘hotspots’. Our results demonstrate that C/EBPβ marks a large number of these transcription factor ‘hotspots’ before induction of differentiation and chromatin remodelling and is required for their establishment. Furthermore, a subset of early remodelled C/EBP-binding sites persists throughout differentiation and is later occupied by PPARγ, indicating that early C/EBP family members, in addition to their well-established role in activation of PPARγ transcription, may act as pioneering factors for PPARγ binding.
Keywords: adipocyte differentiation, C/EBPβ, ChIP-seq, chromatin remodelling, DNase I hypersensitivity
Introduction
Adipogenesis is a tightly controlled differentiation process regulated by a complex transcriptional network involving both pro-adipogenic and anti-adipogenic signals (MacDougald and Mandrup, 2002; Farmer, 2006; Tontonoz and Spiegelman, 2008; Lefterova and Lazar, 2009). The murine 3T3-L1 cell line (Green and Kehinde, 1974) is the most extensively used adipogenic model system, and the ability of this cell line to undergo a rather synchronous differentiation process upon addition of an adipogenic cocktail has been instrumental for the ability to decipher the complex regulatory network of adipocyte differentiation. Ex vivo as well as in vivo investigations have clearly established that peroxisome proliferator-activated receptor γ (PPARγ) is obligate for adipocyte differentiation (Barak et al, 1999; Rosen et al, 1999). In addition, three members of the CCAAT/enhancer-binding protein (C/EBP) family (α, β, and δ) (Wang et al, 1995; Tanaka et al, 1997; Linhart et al, 2001), along with several other transcription factors acting at different stages of differentiation (Kim and Spiegelman, 1996; Nanbu-Wakao et al, 2002; Chen et al, 2005; Mori et al, 2005; Oishi et al, 2005; Birsoy et al, 2008; Steger et al, 2010), have been demonstrated to have key roles. Immediately following addition of the adipogenic cocktail the expression of C/EBPβ and -δ is induced. These members of the C/EBP family constitute important factors in a transcriptional network involved in the induction of the key adipocyte transcription factors, PPARγ and C/EBPα.
Recently, we have used chromatin immunoprecipitation in combination with deep sequencing to profile genome-wide PPARγ and RXR binding during 3T3-L1 adipocyte differentiation (Nielsen et al, 2008). Similarly, others have characterized PPARγ and C/EBPα-binding sites in mature 3T3-L1 adipocytes using ChIP-chip (Lefterova et al, 2008). These analyses show that both PPARγ and C/EBPα-binding sites are enriched in the vicinity of most genes induced during adipogenesis. Furthermore, the data sets reveal an extensive overlap between the binding sites of PPARγ and C/EBPα in mature adipocytes. These genome-wide studies have greatly increased our understanding of the transcriptional networks controlling adipocyte differentiation and revealed a hitherto unrecognized cross talk between key transcriptional regulators of adipogenesis (Lefterova et al, 2008; Nielsen et al, 2008; Siersbæk et al, 2010). In addition, a few recent studies have identified the changes in histone marks that accompany adipocyte differentiation (Wakabayashi et al, 2009; Lefterova et al, 2010; Mikkelsen et al, 2010; Steger et al, 2010).
The ability of transcription factors to gain access to DNA is regulated in part by the local chromatin configuration, as well as the exact positioning of nucleosomes. ATP-dependent chromatin remodelling complexes, such as SWI/SNF and ISWI, reorganize local nucleosome structures and thereby modulate access of transcription factors to their binding elements (Felsenfeld and Groudine, 2003; John et al, 2008; Wu et al, 2009). These remodelling complexes are targeted to DNA in part through interactions with transcription factors, which thereby direct remodelling to specific sites on DNA (Kowenz-Leutz and Leutz, 1999; Pedersen et al, 2001; Kadam and Emerson, 2003). DNase I HyperSensitive (DHS) site analysis is a well-established technique that has been widely used to identify DNA residing in open chromatin regions (Dorschner et al, 2004; John et al, 2008). This analysis identifies perturbations in the chromatin structure as a result of chromatin remodelling, and can consequently be used to locate sites of protein binding with no a priori knowledge about the identity of the protein (Hager, 2009). Recently, DHS site analysis has been combined with high-throughput techniques such as microarray (DHS-chip) (Sabo et al, 2006; Crawford et al, 2006a; Boyle et al, 2008) or deep sequencing (DHS-seq) (Crawford et al, 2006b; Boyle et al, 2008; Hesselberth et al, 2009; John et al, 2011) to study alterations in chromatin structure on a global scale.
Little is known about the chromatin remodelling events that are required for the development of adipocytes. A few studies have investigated the development of open chromatin regions at specific loci during adipogenesis (Salma et al, 2004; Eguchi et al, 2008; Xiao et al, 2011); however, no studies have addressed these remodelling events at a genome-wide scale. Here, we have used DHS site analysis in combination with deep sequencing to obtain a temporal genome-wide map of the chromatin structure during 3T3-L1 adipocyte differentiation. We describe a dramatic increase in the number of discrete, localized DHS sites within a few hours after addition of the adipogenic cocktail. Most of these sites are only transiently open, whereas others remain open later in adipogenesis. Notably, 33% of PPARγ target sites are remodelled within 4 h after induction well before PPARγ binding. The majority of these sites are occupied by members of the C/EBP family, indicating that these transcription factors prime regulatory sites for subsequent PPARγ binding. Furthermore, we identify many transcription factor ‘hotspots’ where multiple transcription factors bind in a cooperative manner and remodel the chromatin structure within hours after induction of adipogenesis. C/EBPβ marks a large fraction of these ‘hotspots’ before induction of differentiation and chromatin remodelling and is required for efficient recruitment of the other factors. Our results indicate that C/EBPβ is a key player acting in concert with multiple other early transcription factors to initiate chromatin remodelling at early stages of adipogenesis and prime the chromatin template for subsequent binding of late-acting adipogenic factors, including PPARγ. This study provides the first genome-wide map of the structural transitions of chromatin through a differentiation process and reveals a novel direct cross talk between early and late events during adipocyte differentiation.
Results
Genome-wide mapping of open chromatin using DHS-seq during 3T3-L1 adipogenesis
To investigate the temporal genome-wide changes in chromatin structure during adipocyte differentiation, we mapped regions of DNase I hypersensitivity using DHS-seq at different time points during 3T3-L1 adipocyte differentiation. Two days post confluency, the cells were induced to differentiate by addition of a standard cocktail of adipogenic inducers consisting of dexamethasone, a cyclic-AMP elevating agent, insulin, and fetal calf serum. Nuclei were isolated and subjected to a limited DNase I digestion at time points 0 h, 2 h, 4 h, 1 day, 2 days, and 6 days after induction, and DNA was size fractionated on a sucrose gradient (Figure 1A). Small DNA fragments (100 bp–1 kb), representing open chromatin regions where DNase I had cut twice within a short distance were isolated. This pool of small DNA fragments liberated by DNase I digestion has previously been shown to represent true DHS sites with a high sensitivity and very low false positive rate (Sabo et al, 2006). DHS samples were subsequently subjected to deep sequencing using the Illumina platform, and sequencing data for the different time points were normalized to equal number of tags for proper comparison. The time points for DHS-seq were chosen to represent different transcriptional stages of adipocyte differentiation, i.e., pre-adipocytes (day 0), early initiation events (2 and 4 h), intermediate phase of differentiation (days 1 and 2), and mature adipocytes (day 6).
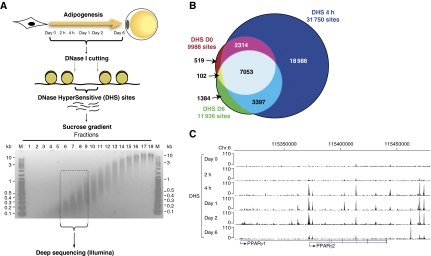
Figure 1.
DNase I HyperSensitive (DHS) site analysis during 3T3-L1 adipogenesis. (A) Experimental outline. Nuclei from 3T3-L1 cells were isolated at the indicated time points of adipocyte differentiation and subsequently subjected to a limited DNase I digestion. Small DNA fragments where DNase I cut twice (i.e., DHS sites) were purified over a sucrose gradient and subsequently subjected to deep sequencing using the Illumina platform. (B) Venn diagram representing the overlap between DHS sites in pre-adipocytes immediately before induction of differentiation (day 0; red), in adipocytes (day 6; green), and in cells stimulated for 4 h with the differentiation cocktail (blue). Sizes of the circles are proportional to the number of sites. (C) DHS-seq data at the PPARγ locus viewed in the UCSC genome browser.
Tag density profiles show robust and distinct DHS sites for all time points (day 0: 9988; 2 h: 9054; 4 h: 31 750; day 1: 35 389; day 2: 25 097; and day 6: 11 936 sites). The notable increase in DHS sites at 4 h includes 22 383 novel DHS sites, the majority of which are transiently open (Figure 1B), indicating that these represent regulatory sites that have a role early but not late in the differentiation process. Importantly however, most of the DHS sites identified in mature adipocytes overlap with a DHS site at the 4-h time point (Figure 1B). Thus, for most sites that are open and likely to represent regulatory sites in mature adipocytes, the transition from closed to open chromatin occurs already within a few hours after induction of adipogenesis, suggesting that different transcription factors prime these regulatory sites during early differentiation. As an example, tag density profiles at the PPARγ locus are shown in Figure 1C. Interestingly, DHS sites develop at the PPARγ2 promoter and distal sites already within 4 h after induction of differentiation, indicating that the locus is primed by remodelling of the chromatin template much before the PPARγ gene is induced at days 1–2 (Nielsen et al, 2008).
Cluster analysis of DHS sites during adipogenesis reveals four distinct temporal profiles of chromatin remodelling
To determine the major representative patterns of changes in intensity of DHS sites during adipogenesis, the DNase I hypersensitivity data were subjected to cluster analysis using a K-means algorithm. This analysis identified four clusters of DHS sites with distinct temporal profiles (Figure 2). Cluster 1 contains DHS sites that are induced early in adipogenesis (4 h after induction) and persist throughout differentiation (Figure 2A). A subset of cluster 1 DHS sites has a high tag density already at day 0 and remains open throughout differentiation. Clusters 2 and 3 contain DHS sites that are transiently induced 4 and 24 h after induction of adipogenesis, respectively (Figure 2B and C). Cluster 4 contains DHS sites that increase in intensity throughout adipogenesis (Figure 2D). Representative examples of the temporal development of DHS sites in each cluster are shown (Figure 2).
Figure 2.
Cluster analysis of DHS sites reveals four clusters with distinct temporal profiles (A–D). A K-means cluster analysis was performed on DHS sites to classify the temporal profiles of the magnitudes of their DNase I hypersensitivity. Cluster plots show the relative tag densities of DHS sites for each time point during differentiation (left). Yellow line represents the mean and red line represents the medoid for each cluster. Representative examples of DHS sites in each cluster are indicated by red arrows in screen shots from the UCSC genome browser (middle). Genes were grouped based on their RNA Pol II ChIP-seq tag counts during 3T3-L1 differentiation (Nielsen et al, 2008). A group of induced genes (>3-fold increase from day 0 to day 4 and not repressed at 4 h; 429 genes), repressed genes (>3-fold decrease from day 0 to day 4 and not induced at 4 h; 640 genes), transient genes (>3-fold increase from day 0 to day 1 and <2-fold increase from day 0 to day 4; 104 genes), and constitutive genes (expression >50th percentile at day 0 and <1.5-fold change from day 0 to day 1 and day 4, and neither transiently repressed nor induced at 4 h; 2692 genes) was defined. The number of DHS sites within 100 kb (divided into 10 kb bins) of the TSS of genes in each group was determined and the number of DHS sites per gene was visualized in a bar plot (right).
We have previously used RNA Pol II ChIP-seq to monitor transcriptional changes during 3T3-L1 adipogenesis (Nielsen et al, 2008). To determine if changes in the chromatin landscape are correlated with transcriptional changes, we counted the number of DHS sites in each cluster within 100 kb of the TSS of induced, repressed, transiently induced, and constitutively transcribed genes (Figure 2) (see Supplementary Table S1 for GO analysis (Dennis et al, 2003) of gene groups). These analyses revealed that DHS sites found in cluster 1 are enriched in the vicinity of induced and transiently induced but not repressed genes. This suggests that DHS sites in this cluster include sites where chromatin remodelling precedes transcriptional activation of nearby genes, such as the example above for the sites in the PPARγ locus (Figure 1C). In addition, cluster 1 DHS sites are specifically enriched in the promoter region of constitutively transcribed genes. DHS cluster 3, and to a lesser extent cluster 2, are enriched in the vicinity of transiently induced genes, suggesting that transcription factors regulate adipocyte differentiation by associating with these regulatory sites specifically at the early stages of adipogenesis. As expected, DHS sites in cluster 4 are highly enriched in the vicinity of induced genes, suggesting that these regulatory sites are occupied by late adipogenic factors (e.g., PPARγ and C/EBPα) that induce the mature adipocyte phenotype. In addition, surprisingly cluster 4 sites are also enriched in the vicinity of transiently induced genes. This may be explained by the heterogeneous nature of cluster 4, which also contains a subset of DHS sites that are induced early. Taken together, we find a high correlation between chromatin remodelling of regulatory sites and transcriptional activation of nearby genes; however, there are multiple sites where remodelling of the chromatin structure precedes transcriptional activation of nearby genes, or where a DHS site persists although the expression of the nearby gene is reduced.
Two recent studies have characterized epigenetic changes during 3T3-L1 adipogenesis on a global scale (Mikkelsen et al, 2010; Steger et al, 2010). Consistent with previous findings (Barski et al, 2007; Heintzman et al, 2007; The ENCODE Project Consortium, 2007; Wang et al, 2008), we find that distal and promoter DHS sites at day 2 are located in regions enriched in well-characterized active distal (H3K4me1 and H3K27ac) and promoter (H3K4me3 and H3K27ac) epigenetic marks, respectively. By contrast, there is little overlap with the repressive chromatin mark H3K27me3 (Supplementary Figure S1A). Similarly, we observed a good correlation between transient DHS sites and regions having transient H3K9ac at day 1 reported by Lazar and coworkers (Supplementary Figure S2). Notably, however, there are a large number ‘active chromatin marks’ outside DHS regions (e.g., 25 170 regions enriched in H3K27Ac do not overlap with a DHS at day 2). Furthermore, there is only limited correlation between the development of chromatin marks and DHS sites (Supplementary Figure S1B–E), clearly demonstrating that DHS sites and active histone marks represent two different chromatin signatures.
Early development of DNase I hypersensitivity at PPARγ target sites in mature adipocytes
Given the importance of PPARγ for adipocyte differentiation, a key question is whether the chromatin structure at target sites is remodelled before or coincident with PPARγ binding. To investigate this, we performed PPARγ ChIP-seq on chromatin from day 6 of adipogenesis and assigned identified PPARγ-binding sites to the four DHS clusters based on positional overlap with sites in the respective clusters (Figure 3A). A large fraction (41%) of the PPARγ sites that can be assigned to a DHS cluster are found in cluster 4, indicating that the development of an open chromatin structure at these sites correlates with PPARγ binding, which increases throughout differentiation (Nielsen et al, 2008). Interestingly however, a high fraction of cluster-associated PPARγ sites identified on day 6 are found in clusters 1 through 3—i.e., these sites develop an open chromatin structure early in adipogenesis before PPARγ binding. In total, 33% of all PPARγ-binding sites detected in mature adipocytes (1801 sites) overlap with a DHS site 4 h after induction of differentiation. Thus, a significant fraction of PPARγ-binding sites develop an open chromatin configuration very early in differentiation well before PPARγ is induced and able to bind to chromatin.
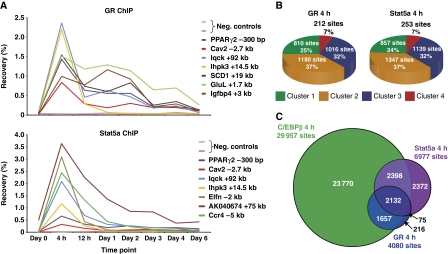
Figure 3.
PPARγ target sites that are remodelled early in differentiation are occupied by C/EBPβ. (A) A subset of PPARγ target sites is open before PPARγ binding. PPARγ-binding sites in mature adipocytes (day 6) displaying a positional overlap with a DHS site at any given time point were assigned to the corresponding DHS cluster (Figure 2). Pie charts indicate the percentage of PPARγ-binding sites in a given cluster out of all the target sites that could be assigned to the four clusters. (B) PPARγ, C/EBPβ, and C/EBPδ ChIP-PCR for selected sites. Results are representative of at least three independent experiments. (C) Early binding of C/EBPβ and -δ at several PPARγ sites at the caveolin 1 (Cav1) and Cav2 locus viewed in the UCSC genome browser. (D) Classification of PPARγ-binding sites at day 6 based on their status at 4 h. PPARγ target sites in mature adipocytes were divided based on their positional overlap with DHS sites at 4 h of differentiation. The PPARγ sites overlapping with open chromatin at 4 h were further subdivided based on the presence of C/EBPβ at these sites at the 4-h time point.
Early RXR binding shows limited overlap with PPARγ binding in mature adipocytes
We have previously shown that RXR associates mainly with PPARγ target sites in mature adipocytes presumably as part of a PPARγ:RXR heterodimer (Nielsen et al, 2008). Interestingly, we also found that RXR is recruited to >3000 sites at day 1 of adipogenesis, where very little PPARγ binding was detected. Furthermore, we showed that some of these sites later become occupied by PPARγ in mature adipocytes, suggesting that RXR could act early in adipogenesis as a heterodimer with other partners than PPARγ (e.g., PPARδ) or as a homodimer to prime regulatory sites for subsequent PPARγ binding (Nielsen et al, 2008). Investigation of RXR binding during adipogenesis to a selected number of early RXR sites revealed that RXR is recruited already within the first 4 h after induction of differentiation to these sites (Supplementary Figure S3A). To identify early RXR-binding sites on a genome-wide scale, we performed RXR ChIP-seq at the 4-h time point. This analysis identified 2076 RXR-binding sites, which are primarily associated with DHS clusters 2 and 3 containing early transient DHS sites (Supplementary Figure S3B). Consistently, few 4 h RXR sites are also occupied by RXR at day 1 and later in adipogenesis (data not shown), indicating that RXR is involved in different transcriptional programs at 4 h, compared with later time points. Consistent with this observation, the overlap between RXR binding at 4 h and early remodelled PPARγ sites is limited to 354 sites (Supplementary Figure S3C), suggesting that RXR binding at this stage of adipogenesis is not a major priming factor for PPARγ.
C/EBPβ and -δ bind at PPARγ target sites before PPARγ
To identify transcription factors binding to the DHS sites that develop during the first 4 h of adipogenesis, we conducted de novo motif analysis using the MEME algorithm on the 6000 most intense peaks induced at 4 h. The analysis revealed significant enrichment of a C/EBP consensus sequence (Supplementary Figure S4A). We and others have previously shown that C/EBPβ binds to several PPARγ target sites (i.e., to regions containing a PPARγ ChIP-seq peak) early in adipogenesis (Lefterova et al, 2008; Nielsen et al, 2008). Interestingly, investigation of the early time course of binding of C/EBPs to selected PPARγ target sites revealed that both C/EBPβ and -δ are recruited very early to these sites following addition of the adipogenic cocktail well before PPARγ binding (Figure 3B). To investigate the prevalence of this finding, we performed ChIP-seq on chromatin from early time points of 3T3-L1 adipogenesis using C/EBPβ and -δ antibodies. Tag density profiles for C/EBPβ, -δ, and PPARγ at the caveolin 1 (Cav1) and Cav2 locus (Figure 3C) clearly show how C/EBPβ and -δ binding is induced within 4 h after induction of differentiation at several PPARγ-binding sites in this locus. In total, we identified 29 957 C/EBPβ and 18 327 C/EBPδ target sites at the 4-h time point. Interestingly, 1324 (24%) and 1032 (19%) of the sites identified as PPARγ-binding sites in mature adipocytes (day 6) are occupied by C/EBPβ and -δ, respectively, and have an open chromatin structure 4 h after induction of differentiation (Figure 3D; Supplementary Figure S4B). Importantly, 94% of the PPARγ sites in mature adipocytes that are open and bound by C/EBPβ at the 4-h time point maintained an open chromatin structure until day 2, where PPARγ is first expressed and recruited to these sites, and the majority of these sites were also retained at day 6 (data not shown). This suggests that C/EBPβ and -δ may regulate PPARγ activity at a subset of PPARγ target sites by regulating chromatin accessibility early in adipogenesis. Furthermore, most day 6 PPARγ sites overlapping a DHS site at 4 h are also occupied by C/EBPβ (Figure 3D) and -δ (Supplementary Figure S4B), respectively, suggesting that these transcription factors are involved in most early remodelling events at PPARγ target sites.
C/EBPs, GR, and Stat5a bind to shared sites early in adipogenesis
In addition to the C/EBP motifs, de novo motif analysis of the sequences within the DHS sites that develop during the first 4 h of differentiation, revealed enrichment of binding motifs for the glucocorticoid receptor (GR) and signal transducer and activator of transcription (Stat) (Supplementary Figure S5A and B). The synthetic glucocorticoid dexamethasone is an important component of the adipogenic cocktail (Pantoja et al, 2008), and recently it was shown that GR binding to chromatin is induced in the early phase of differentiation, and that GR is required for 3T3-L1 differentiation (Steger et al, 2010). Stat5A/B are the only members of the Stat family that have been shown to regulate adipocyte differentiation (Teglund et al, 1998; Nanbu-Wakao et al, 2002; Kawai et al, 2007), and they are specifically activated during the early phase of differentiation by tyrosine phosphorylation (Nanbu-Wakao et al, 2002; Baugh et al, 2007). Consistently, GR and Stat5a ChIP-PCR at a few selected sites showed that these two transcription factors associate with the investigated binding sites mainly at the 4-h time point (Figure 4A).
Figure 4.
Extensive overlap between C/EBPβ, GR, and Stat5a binding during early adipocyte differentiation. (A) GR and Stat5a ChIP-PCR for a few selected sites. Results are representative of three independent experiments. (B) GR and Stat5a-binding sites at the 4-h time point displaying a positional overlap with a DHS site at any given time point were assigned to the corresponding DHS cluster (Figure 2). Pie charts indicate the percentage of GR and Stat5a-binding sites in a given cluster out of all the target sites that could be assigned to the four clusters. (C) Venn diagram representing the overlap between the C/EBPβ, GR, and Stat5a-binding profiles 4 h after induction of adipogenesis. Circles are proportional to the number of sites.
To get a global picture of GR and Stat5a binding, we performed ChIP-seq at the 4-h time point, which revealed 4080 GR and 6977 Stat5a-binding sites, respectively. Assigning these binding sites to the four DHS clusters showed that GR and Stat5a mainly bind to DHS sites that develop early in differentiation (Figure 4B). Most of these DHS sites are transient in nature, whereas others persist throughout differentiation, suggesting that GR and Stat5a may also be involved in priming regulatory sites for subsequent binding by other transcription factors. However, only a limited number of early remodelled PPARγ target sites are occupied by GR and Stat5a 4 h after induction of differentiation (Supplementary Figure S5C and D), indicating that neither GR nor Stat5a are predominant priming factors for PPARγ. Instead, GR and Stat5a target sites that maintain hypersensitivity to DNase I throughout differentiation may be occupied by other late adipogenic factors than PPARγ (e.g., C/EBPα and Krüppel-like factor 15).
Given that C/EBPβ binding is most prominent 4 h after induction of differentiation, we determined the overlap between the C/EBPβ, GR, and Stat5a-binding profiles at this time point to determine if these transcription factors bind to the same sites early in adipogenesis. Remarkably, 3789 GR sites (93%) and 4530 Stat5a sites (65%) overlap with C/EBPβ binding, and 2132 sites are occupied by all three factors (Figure 4C). C/EBPδ was found to have a similar overlap with GR and Stat5a binding (Supplementary Figure S5E). This extensive overlap between the binding profiles suggests that C/EBPβ, -δ, GR, and Stat5a coordinate early transcriptional events from shared regulatory sites early in adipogenesis.
Adipogenic pathways converge at transcription factor ‘hotspots’ early in adipogenesis
Interestingly, we find that 1849 (89%) RXR-binding sites overlap with both C/EBPβ and -δ binding at the 4-h time point (Supplementary Figure S6). Together with the finding that C/EBPβ, -δ, GR, and Stat5a have highly overlapping profiles, this led us to determine the co-occurrence of all five factors (Figure 5A). This analysis revealed a remarkable overlap between the binding profiles of the five adipogenic transcription factors with C/EBPβ being the central transcription factor in this network. Interestingly, we identified as many as 938 transcription factor ‘hotspots’ associated with all five factors. Thus, this indicates a direct cross talk on the chromatin template between multiple adipogenic pathways early in adipogenesis. Given the importance of PPARγ in adipocyte differentiation, it is interesting to note that several transcription factor ‘hotspots’ are located within and immediately adjacent to the PPARγ locus (Supplementary Figure S7A), indicating that multiple early transcriptional pathways converge on the transcriptional regulation of this receptor.
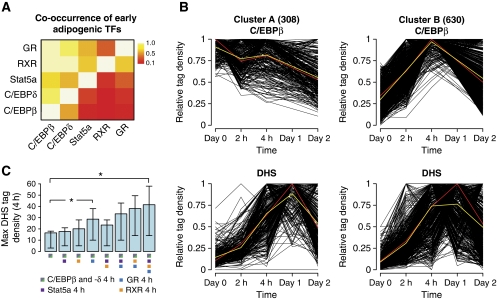
Figure 5.
C/EBPβ binds to transcription factor ‘hotspots’ before induction of differentiation and chromatin remodelling. (A) Co-occurrence of early adipogenic transcription factors (TFs). The proportion of binding sites for the transcription factors on the y axis that overlaps with a binding site for the TFs on the x axis at the 4-h time point was calculated for each TF pair and visualized as a heat map. (B) Classification of transcription factor ‘hotspots’ by K-means cluster analysis based on C/EBPβ ChIP-seq data. The corresponding DHS profiles are shown below. Cluster plots show the relative tag densities for each time point during differentiation. Yellow line represents the mean and red line represents the medoid for each cluster. (C) Maximum DHS tag density at transcription factor-binding sites 4 h after induction of adipogenesis. Error bars represent the 25th and 75th quantiles, respectively. * denotes a statistically significant difference with a P-value <2.2E-16 as determined using a Wilcoxon's test.
To delineate the temporal binding pattern of C/EBPβ at the identified ‘hotspots’, we subjected C/EBPβ ChIP-seq data from time points day 0, 2 h, 4 h, and day 2 to K-means cluster analysis (Figure 5B). This analysis revealed that the ‘hotspots’ can be subdivided into two clusters with distinct temporal C/EBPβ-binding profiles. Cluster A contains ‘hotspots’ that have stable C/EBPβ binding during the first 2 days of differentiation, whereas cluster B contains ‘hotspots’ where C/EBPβ binding is induced and peaks at 4 h. Remarkably, DNase I hypersensitivity is induced at ‘hotspots’ found in both clusters during the first 4 h of differentiation (see Supplementary Figure S7B and C for examples). This indicates that binding of C/EBPβ per se is insufficient to initiate remodelling of the chromatin template. Rather, C/EBPβ appears to mark a subset of ‘hotspots’ (33%) before induction of differentiation, but only upon activation and recruitment of additional transcription factors is the chromatin template remodelled resulting in the development of a DHS site. Consistently, C/EBP-binding sites at 4 h that are co-occupied by one or more of the other transcription factors investigated generally have a more open chromatin structure than sites only occupied by the C/EBPs (Figure 5C). Notably, especially GR binding seems to be more highly correlated with hypersensitivity to DNase I attack than binding of the other early transcription factors, indicating that an open chromatin structure may be particularly important for GR binding, and/or that GR is an important inducer of early chromatin remodelling. The preferred association of GR with open chromatin regions is consistent with previous findings (John et al, 2008, 2011). Taken together, this suggests that C/EBPβ marks a subset of transcription factor ‘hotspots’ before induction of differentiation and chromatin remodelling.
‘Hotspots’ are enriched in the vicinity of early induced genes
To investigate if binding of the early transcription factors to chromatin is correlated with early transcriptional changes, we performed RNA Pol II ChIP-seq for day 0 and the 4-h time point and counted the number of transcription factor binding sites within 50 kb of early induced, repressed, and constitutively transcribed genes (Figure 6) (see Supplementary Table S2 for GO analysis (Dennis et al, 2003) of gene groups). Shared sites occupied by RXR and especially GR are highly correlated with transcriptional induction of nearby genes, whereas none of the investigated transcription factors are enriched in the vicinity of repressed genes. Furthermore, sites co-occupied by all five investigated transcription factors (‘hotspots’) seem to be most highly correlated with transcriptional induction. Taken together, these analyses suggest that the cross talk between multiple transcription factors at ‘hotspots’ is important for the transcriptional induction of nearby genes during the early phase of differentiation with GR binding being particularly highly correlated with induction of early adipogenic transcription.
Figure 6.
Transcription factor ‘hotspots’ are enriched in the vicinity of early induced genes. RNA Pol II ChIP-seq profiles were determined for day 0 and 4 h as previously described (Nielsen et al, 2008) and genes were grouped as induced (795 genes), repressed (776), or constitutive (2414 genes) as described in Figure 2. The number of transcription factor-binding sites per 10 000 binding sites within 50 kb of the TSS of genes in each group was determined.
C/EBPβ is required for efficient transcription factor binding to ‘hotspots’ early in adipogenesis
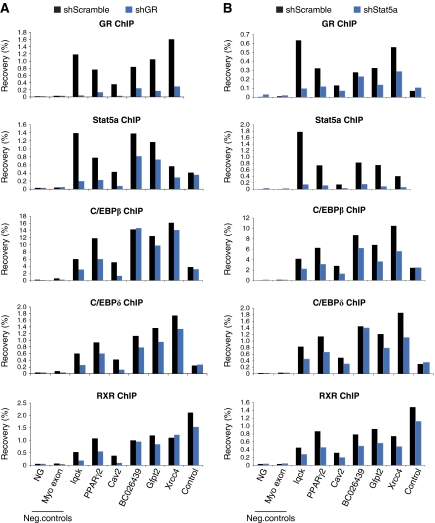
The finding that C/EBPβ marks transcription factor ‘hotspots’ before induction of adipogenesis led us to investigate the requirement for C/EBPβ for the recruitment of other early transcription factors to shared sites. Using lentiviral-mediated shRNA delivery, we obtained a robust knockdown of C/EBPβ at the level of total cellular protein (Figure 7A) as well as at the level of binding to chromatin (Figure 7B). Interestingly, knockdown of C/EBPβ results in a significant reduction in the binding of GR, Stat5a, and RXR to most shared sites, whereas recruitment of GR, Stat5a, and RXR to control sites that are not co-occupied by C/EBPβ is unaffected by the C/EBPβ knockdown (Figure 7C–E). This indicates that C/EBPβ is required for efficient binding of these transcription factors to shared sites early in adipocyte differentiation. Notably, C/EBPδ binding to shared C/EBPβ and -δ sites is unaffected or slightly increased by the C/EBPβ knockdown (Figure 7F). This suggests that C/EBPδ can bind to these sites independent of C/EBPβ and that for a few sites (especially BC026439, Gfpt2, and Xrcc4) C/EBPδ may even replace C/EBPβ, which may explain the less dramatic reduction in binding of GR, Stat5a, and RXR to these sites. Taken together, C/EBPβ is required for efficient binding of GR, Stat5a, and RXR, but not C/EBPδ, to most shared target sites early in adipogenesis.
Figure 7.
C/EBPβ is required for efficient binding of GR, Stat5a, and RXR, but not C/EBPδ, to shared target sites. (A) Lentiviral-mediated knockdown of C/EBPβ expression. 3T3-L1 cells were transduced with lentivirus expressing shRNA targeting LacZ (control) or C/EBPβ, respectively, and cells were induced to differentiate for 4 h. The three different isoforms of C/EBPβ (liver inhibitory protein (LIP), liver activating protein (LAP), and LAP*) are indicated. TFIIB was used as a loading control. C/EBPβ (B), Stat5a (C), GR (D), RXR (E), and C/EBPδ (F) ChIP-PCR for a few selected shared sites 4 h after induction of differentiation of cells expressing shRNA targeting LacZ (control) or C/EBPβ, respectively. Control denotes sites that are occupied by the respective factors but not co-occupied by C/EBPβ. Results are representative of three independent experiments. A full-colour version of this figure is available at The EMBO Journal Online.
Formaldehyde-assisted isolation of regulatory elements (FAIRE) is a simple method to determine the relative nucleosome occupancy at selected sites (Nagy et al, 2003; Hogan et al, 2006). To investigate if knockdown of C/EBPβ affects early chromatin remodelling at selected sites, we performed FAIRE on chromatin from cells expressing shRNA against LacZ (control) and C/EBPβ, respectively (Supplementary Figure S8). Interestingly, FAIRE analysis revealed that C/EBPβ is required for efficient chromatin remodelling at most of the investigated shared sites, which is highly concordant with the observed reduced binding of GR, RXR, and Stat5a upon C/EBPβ depletion (Figure 7C–E).
Cooperative binding of factors to transcription factor ‘hotspots’
To investigate the importance of the other early transcription factors for transcription factor recruitment to ‘hotspots’, we performed lentiviral-mediated knockdown of GR, Stat5a, RXR, and C/EBPδ in 3T3-L1 cells. This approach resulted in robust knockdown of GR and Stat5a (>80%; Supplementary Figure S9) without affecting cell growth, whereas knockdown of RXR and C/EBPδ with several different constructs resulted in significantly reduced cell growth impeding further analyses. Interestingly, both GR and Stat5a knockdown results in an overall decrease in binding of the other early transcription factors to selected ‘hotspots’ 4 h after standard hormone induction of differentiation (Figure 8). In particular, GR is important for Stat5a binding and vice versa. Taken together, this suggests that transcription factors bind cooperatively to transcription factor ‘hotspots’ early in adipogenesis.
Figure 8.
GR and Stat5a are required for efficient binding of transcription factors to selected ‘hotspots’. 3T3-L1 cells were transduced with lentivirus expressing scrambled shRNA or shRNA targeting GR (A) or Stat5a (B), and cells were induced to differentiate using a standard hormone induction of adipogenesis. GR, Stat5a, C/EBPβ, C/EBPδ, and RXR ChIP-PCR for selected ‘hotspots’ 4 h after induction of differentiation. Control denotes sites that are occupied by the respective factors but not co-occupied by GR (A) or Stat5a (B). Results are representative of two independent experiments. A full-colour version of this figure is available at The EMBO Journal Online.
Based on the findings presented here, we propose the model depicted in Figure 9, where C/EBPβ marks specific closed chromatin sites before induction of adipogenesis. Upon induction of differentiation, additional transcription factors (e.g., GR, Stat5a, and RXR) are activated and bind in a cooperative manner to these sites to remodel the chromatin and form transcription factor ‘hotspots’. Most of these are only transiently open, whereas others maintain an open chromatin configuration and are subsequently occupied by PPARγ and other late adipogenic factors (e.g., C/EBPα) in the later stages of adipogenesis.
Figure 9.
Model illustrating C/EBPβ as a pioneering factor for adipogenic transcription factors and chromatin remodelling. C/EBPβ binds to closed chromatin in pre-adipocytes. Upon induction of differentiation, several other transcription factors are activated and recruited to C/EBPβ sites, resulting in remodelling of the chromatin structure and formation of transcription factor ‘hotspots’. Some of these are transient in nature, whereas others persist throughout differentiation and are later occupied by PPARγ and C/EBPα, which induce the mature adipocyte phenotype.
Discussion
In this study, we have mapped the development of open chromatin structures during 3T3-L1 adipocyte differentiation using DHS site analysis. We identify four clusters of DHS sites with distinct temporal profiles that correlate well with the regulation of nearby genes. Two clusters contain DHS sites that increase dramatically in intensity already within 4 h after induction of differentiation coincident with re-entry into the cell cycle and initiation of the adipogenic program. Interestingly, although these sites are enriched in the vicinity of early induced genes, they are also found in the vicinity of late induced genes indicating that remodelling precedes gene activation.
The number of DHS sites identified in pre-adipocytes (day 0) and adipocytes (day 6) is surprisingly low (9988 and 11 936 DHS sites, respectively) compared with what is found for the 4-h time point (31 750 DHS sites) and what has previously been reported for other cell types (The ENCODE Project Consortium, 2007; Boyle et al, 2008). It should be noted that we used very conservative criteria for identification of DHS sites (see Materials and methods) to focus our analyses on robustly enriched sites. However, to specifically validate the low number of sites at days 0 and 6, we have performed a biological replicate for these days, which is highly concordant with the data presented here (>70% overlap between the data sets).
Given the importance of PPARγ for adipocyte differentiation, a key question is whether the chromatin structure is remodelled before or concomitantly with PPARγ binding. Interestingly, one third of all PPARγ target sites overlap with a DHS site 4 h after induction of differentiation, indicating that these sites are remodelled and primed for subsequent PPARγ binding by pioneering factors early in adipogenesis. Motif discovery analysis revealed enrichment of motifs for members of the C/EBP family at DHS sites that develop during the first 4 h of differentiation. C/EBPβ and -δ are known to have important and partially redundant roles in adipogenesis (Tanaka et al, 1997; Tang et al, 2003) in part by directly activating the expression of PPARγ (Wu et al, 1996). Both C/EBP subtypes are transcriptionally (Clarkson et al, 1995; Yeh et al, 1995) and post-translationally (Tang et al, 2005) activated by the adipogenic cocktail. Interestingly, we found that the early remodelled PPARγ-binding sites display a high degree of overlap with early binding of C/EBPβ, and to some extent C/EBPδ, but none of the other early transcription factors investigated. This indicates that C/EBPβ and -δ, in addition to their well-established role in induction of PPARγ expression, also directly affect PPARγ binding by regulating chromatin accessibility at specific genomic regions.
In addition to the binding motif for C/EBP family members, motif discovery analysis of DHS sites that develop during the first 4 h of adipogenesis also revealed enrichment of GR and Stat motifs. We found that on a genome-wide scale GR and Stat5a associate mainly with transiently open chromatin regions, suggesting that these factors regulate adipogenesis through transient action at regulatory sites. Interestingly, we identified multiple transcription factor ‘hotspots’ at 4 h that are occupied by all five early factors (GR, Stat5a, C/EBPβ, -δ, and RXR). Thus, several different adipogenic pathways converge at specific genomic sites during early adipogenesis, indicating a hitherto unrecognized direct cross talk between different adipogenic pathways on the chromatin template. Notably, such early ‘hotspots’ are highly enriched in the vicinity of genes induced during the first 4 h of adipocyte differentiation, and in particular GR binding is highly correlated with induction of nearby genes. Interestingly, there are multiple transcription factor ‘hotspots’ within and immediately adjacent to the PPARγ locus consistent with the importance of this receptor for adipocyte differentiation. However, in this case the locus is not transcriptionally activated until later in adipogenesis, indicating that additional factors/signals are required for activation.
Intriguingly, cluster analysis of the temporal binding pattern of C/EBPβ revealed that C/EBPβ binds to a subset of transcription factor ‘hotspots’ before induction of differentiation and chromatin remodelling. This indicates that C/EBPβ is able to associate with closed chromatin in pre-adipocytes. Similarly, we identify 16 304 C/EBPβ-binding sites at 4 h that are found in closed chromatin. Consistent with our findings, a truncated version of C/EBPβ has previously been shown to associate with the mim-1 enhancer in fibroblasts without inducing chromatin remodelling (Plachetka et al, 2008). Similarly, FoxA1, which is considered to be a pioneering factor for ER (Carroll et al, 2005; Hurtado et al, 2011), is able to associate with closed chromatin (Eeckhoute et al, 2009). Importantly, we show that C/EBPβ is required for binding of GR, Stat5a, and RXR, but not C/EBPδ, to shared sites including ‘hotspots’ and for efficient chromatin remodelling at some of these sites. Similarly, we show that GR and Stat5a are also required for efficient binding of transcription factors to some ‘hotspots’. Thus, collectively these data indicate that transcription factors bind in a cooperative manner to ‘hotspots’, and that C/EBPβ marks many of these ‘hotspots’ before induction of differentiation and functions as a pioneering factor for the assembly of transcription factor complexes at these sites.
In conclusion, we have used DHS-seq to map the development of open chromatin structures on a genome-wide scale during 3T3-L1 adipocyte differentiation, and we show that the major chromatin remodelling events occur within the first 4 h of adipogenesis. We identified multiple transcription factor ‘hotspots’ at this early stage of differentiation, where all investigated transcription factors bind to open chromatin regions in a cooperative manner. C/EBPβ acts as a pioneering factor and cooperating transcription factor for the assembly of this network of early adipogenic transcription factors. In addition, C/EBPβ may also be involved in early remodelling of PPARγ target sites before PPARγ binding, thereby indicating a novel direct cross talk between early and late events during adipogenesis.
Materials and methods
3T3-L1 cell culture
3T3-L1 fibroblasts were differentiated to adipocytes by stimulation with 3-isobutyl-1-methylxanthine, dexamethasone, and insulin as described previously (Helledie et al, 2002).
DHS-seq
DNase I digestions of nuclei were performed essentially as described previously (Sabo et al, 2006). Briefly, 3T3-L1 cells were induced to differentiate and cells were harvested by trypsination at various time points of differentiation. Cells were washed twice in cold PBS and resuspended in Buffer A (15 mM Tris–HCl pH=8, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, and protease inhibitors (Complete; Roche)) at 2 × 106 cells/ml. One volume of Buffer A containing 0.04% NP-40 was gently added and the suspension was incubated for 10 min on ice to release nuclei. Nuclei were washed twice in cold Buffer A and 2 × 107 nuclei were pelleted at 1200 g for 7 min. The pellet was incubated at 37°C for 1 min to equilibrate the nuclei. Nuclei were subsequently resuspended in 2 ml DNase I digestion buffer (Buffer A without spermidine and protease inhibitors but with the addition of 6 mM CaCl2 and 75 mM NaCl) containing 60 U/ml DNase I (Roche, cat# 11 284 932 001) and incubated for 3 min at 37°C. We have previously used this concentration of DNase I to map DHS sites at selected genomic regions using qPCR (John et al, 2008). Digestion was stopped by the addition of 2 ml Stop buffer (50 mM Tris–HCl pH=8, 100 mM NaCl, 0.1% SDS, 100 mM EDTA, and 0.5 mM spermidine) containing 10 μg/ml RNase (Roche, cat# 11 119 915 001). Samples were subsequently incubated for 4–6 h at 55°C to remove RNA. Samples were then treated over night with 50 μg/ml proteinase K at 55°C. Cell debris was removed by very gentle phenol/chloroform extraction and DNA fragments were separated by ultracentrifugation through a sucrose gradient. Fractions of 0.5 ml were collected and run on a 2% agarose gel, which was subsequently stained with SYBR green (Invitrogen). Fractions containing small DNA fragments representing two-hit DNase I cutting events were precipitated, pooled, and prepared for sequencing according to the manufacturer's instructions (Illumina).
ChIP and ChIP-seq
ChIP experiments were performed according to a standard protocol as described previously (Nielsen et al, 2006). Antibodies used: PPARγ antibody (H-100, sc-7196, Santa Cruz), RXR antibody (Δ197, sc-774, Santa Cruz), C/EBPβ antibody (C-19, sc-150, Santa Cruz), C/EBPδ antibody (M-17, sc-636, Santa Cruz), Stat5a antibody (L-20, sc-1081, Santa Cruz), and GR antibodies (PA1-511A and MA1-510, Affinity Bioreagents). ChIP'ed DNA was analysed by qPCR or prepared for sequencing according to the manufacturer's instructions (Illumina).
The hotspot algorithm for detection of significantly enriched regions from DHS-seq or ChIP-seq data
Hotspots. We identified regions of local enrichment of 36-mer short-read tags mapped to the reference genome mm9 by a procedure similar to the previously employed algorithm for DHS (Hesselberth et al, 2009). Enrichment of tags in a 250-bp target window relative to a 200-kb surrounding window (local background) is gauged by the model based on the binomial distribution. Each tag is extended to the 150-bp length into its strand and assigned a z-score (explained below) using the target window and the background window both centered on the tag. An unthresholded hotspot is defined as a contiguous union of 250 bp windows whose z-scores are >2. Once a hotspot is called, the z-score of the hotspot is given by the maximum z-score of the constituent tags.
Mappability-adjusted z-scores. If there are n observed tags and N tags that overlap the target window and the local background window, respectively, then the probability p of a tag in the background window overlapping the target window is given by the ratio (# of uniquely mappable tags for the 250-bp window)/(# of uniquely mappable tags for the 200-kb window). Because not all 36-mers in a window can be aligned uniquely to the reference genome, p differs greatly from genomic region to region. The expected number of tags overlapping the target window is then μ=Np after this important step of adjusting for the differential mappability of short reads. The standard deviation of the expectation is  . The z-score for the observed number of tags in the target window is defined by z=(n−μ)/σ.
. The z-score for the observed number of tags in the target window is defined by z=(n−μ)/σ.
Peak detection and input adjustment for ChIP-seq. The above algorithm can be applied to DHS tag data without modification. However, the sequencing data from matching input samples are used for the processing of ChIP data, as a measure of background signal which might be significant. After normalizing the input data to match the number of tags in the ChIP data, the number of input tags is subtracted from the number of ChIP tags in the target window before calculating its z-score. In addition, we refine ChIP hotspots into 150 bp peaks using a peak-finding procedure. For each hotspot, we compute a new z-score ‘peak z’ similarly as for hotspot detection, but instead using a 150-bp window within the hotspot and the hotspot as the target window and local background window, respectively. We compute the peak z-score scanning through the hotspot with bp resolution for all possible 150 bp windows. A putative peak is defined as a 150-bp window whose peak z-score is >30. If nearby putative peaks overlap within a hotspot, the 150 bp window with the highest peak z-score is selected to be the peak among the cluster of overlapping putative peaks. Therefore, a hotspot may not have a peak or may have multiple peaks.
Repeat sequence filtering. In both DHS and ChIP sequencing reads, artifacts are inevitably found and typically concentrated on relatively small regions. We remove these artifacts by filtering out sequence reads which overlap satellites, LINE (long interpersed repetitive elements), and STR (single tandem repeats) after extension of 150 bp. Repeat-masking data were downloaded from the UCSC genome browser site (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/database/simpleRepeat.txt.gz).
False discovery rate calculations. We imposed a final z-score threshold on hotspots for each data set based on false discovery rate (FDR). To calculate FDR, we performed random sampling of uniquely mappable tags and generated a random data set with the same number of tags as for the observed data set. Hotspots are called for the random data set and their z-scores are calculated. Then, the FDR for the observed hotspots at a given z-score threshold z0 is estimated as (# of hotspots with z>z0 in the random data)/(# of hotspots with z>z0 in the observed data). Hotspots are selected by setting a z-score threshold that is just above the highest z-score encountered in the random set. We considered hotspots defined at this stringency level as significant enrichment at FDR=1/N, where N is the number of observed hotspots (i.e., FDR<1.1E-4 in this study).
Tag density thresholds. As a stringent filtering step for robustly enriched regions, the following thresholds were used to require that DHS hotspots or ChIP peaks have their (maximum) tag density exceed these values. To adjust for different depths of sequencing and facilitate cross-data set comparison, the tag density for each data set was normalized to match 10 million uniquely aligned tags. Because ChIP efficiency and specificity are affected by the antibodies used, we chose data-specific tag density thresholds based on visual inspection of a large number of low signal peaks. DHS: 12.5, PPARγ: 32.5, C/EBPβ: 16.5, C/EBPδ: 19.5, GR: 36, Stat5a: 19, RXR: 17. All subsequent analyses used these tag density thresholded DHS hotspots or ChIP peaks.
Cluster analysis
We performed K-means cluster analysis on DHS hotspots to classify the temporal profiles of their tag density. To begin with a master set of hotspots, we merged hotspots from different time points if they overlapped with each other. Redefining hotspot boundaries this way resulted in a non-redundant set of 34 269 DHS hotspots. These represent all DHS hotspots that are present in one or more time points. The magnitude of DNase I hypersensitivity of a hotspot at a given time point was used for cluster analysis and defined as the relative density: For hotspot i and time point j, the relative density is given by Mij/maxjMij where Mij is the tag density of the hotspot. In effect, this amounted to rescaling of all hotspot density values to range from zero to one over the time course. We employed the function ‘clara’ (Kaufman and Rousseeuw, 1990) of ‘cluster’ package of R statistical language. The number of clusters, K, was varied and the final K was selected based on cluster information from the silhouette plots.
Motif discovery analysis
We performed a motif discovery analysis on selected DNA sequences using MEME (Bailey and Gribskov, 1998) on parallel clusters at the NIH Biowulf supercomputing facility. DNA sequences for MEME input were from the top 6000 (by tag density) DHS hotspots among all 4 h DHS hotspots not overlapping day 0 hotspots. To limit the computational load, only the 400-bp region with the highest tag density was used instead of the entire width of a hotspot in cases where the hotspot spanned >400 bp. The width of motifs to discover was set to 6 and 20 for minimum and maximum, respectively. To identify motifs for known transcription factor binding, we queried individual position-specific probability matrices against the Transfac database using the Tomtom software (http://meme.nbcr.net/meme/cgi-bin/tomtom.cgi). We retrieved statistically significant matches that share the majority of specific nucleotides in the sequence motifs.
Lentiviral-mediated knockdown
For knockdown of C/EBPβ, shRNA oligo DNA targeting LacZ (control) and CEBPβ was cloned into pSicoR PGK puro vectors (Addgene) and lentiviral particles were produced in human embryonic kidney 293T cells as described (Ventura et al, 2004). For knockdown of GR and Stat5a, pLKO.1 vectors expressing scrambled shRNA (control) and shRNA against GR and Stat5a were purchased from Sigma-Aldrich. Proliferating 3T3-L1 cells were transduced with lentivirus and subsequently induced to differentiate using a standard hormone cocktail. Cells were harvested 4 h after induction of differentiation, and protein and chromatin were prepared and analysed by western blotting and ChIP as described previously (Nielsen et al, 2006).
Quantitative real-time PCR
Real-time qPCR was performed using SYBR Green mix (Sigma) with an MX3000 machine (Stratagene) as previously described (Nielsen et al, 2006). Primer sequences used for real-time qPCR are available upon request.
Accession numbers
DHS-seq and ChIP-seq raw data are deposited into GEO (accession number GSE27826).
Supplementary Material
Acknowledgments
We thank members of the Mandrup and Hager laboratories for fruitful discussions. This work was supported by grants from the Danish Natural Science Research Council, the Novo Nordisk Foundation, and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bailey TL, Gribskov M (1998) Combining evidence using P-values: application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM (1999) PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell 4: 585–595 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui KR, Roh TY, Schones DE, Wang ZB, Wei G, Chepelev I, Zhao KJ (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Baugh JE, Floyd ZE, Stephens JM (2007) The modulation of STAT5A/GR Complexes during fat cell differentiation and in mature adipocytes. Obesity 15: 583–590 [DOI] [PubMed] [Google Scholar]
- Birsoy KI, Chen Z, Friedman J (2008) Transcriptional regulation of adipogenesis by KLF4. Cell Metabol 7: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao WL, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM (2005) Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metabol 1: 93–106 [DOI] [PubMed] [Google Scholar]
- Clarkson RWE, Chen CM, Harrison S, Wells C, Muscat GEO, Waters MJ (1995) Early responses of transactivating factors to growth-hormone in preadipocytes—differential regulation of Ccaat Enhancer-Binding Protein-Beta (C/Ebp-Beta) and C/Ebp-delta. Mol Endocrinol 9: 108–120 [DOI] [PubMed] [Google Scholar]
- Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS (2006a) DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat Methods 3: 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, Zhou D, Luo S, Vasicek TJ, Daly MJ, Wolfsberg TG, Collins FS (2006b) Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res 16: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane HC, Lempicki R (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- Dorschner M, Hawrylycz M, Humbert R, Wallace JC, Shafer A, Kawamoto J, Mack J, Hall R, Goldy J, Sabo PJ, Kohli A, Li QL, McArthur M, Stamatoyannopoulos JA (2004) High-throughput localization of functional elements by quantitative chromatin profiting. Nat Methods 1: 219–225 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Lupien M, Meyer CA, Verzi MP, Shivdasani RA, Liu XS, Brown M (2009) Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res 19: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Yan QW, Schones DE, Karnal M, Hsu CH, Zhang MQ, Crawford GE, Rosen ED (2008) Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metabol 7: 86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR (2006) Transcriptional control of adipocyte formation. Cell Metabol 4: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M (2003) Controlling the double helix. Nature 421: 448–453 [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O (1974) Sublines of mouse 3T3 cells that accumulate lipid. Cell 1: 113–116 [Google Scholar]
- Hager G (2009) Footprints by deep sequencing. Nat Methods 6: 254–256 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu YT, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu CX, Ching KA, Wang W, Weng ZP, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Helledie T, Grontved L, Jensen SS, Kiilerich P, Rietveld L, Albrektsen T, Boysen MS, Nohr J, Larsen LK, Fleckner J, Stunnenberg HG, Kristiansen K, Mandrup S (2002) The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J Biol Chem 277: 26821–26830 [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA (2009) Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD (2006) Cell cycle specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet 2: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS (2011) FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL (2008) Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 29: 611–624 [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM (2003) Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389 [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ (1990) Finding Groups in Data: An Introduction to Cluster Analysis. Wiley: New York [Google Scholar]
- Kawai M, Namba N, Mushiake S, Etani Y, Nishimura R, Makishima M, Ozono K (2007) Growth hormone stimulates adipogenesis of 3T3-L1 cells through activation of the Stat5A/5B-PPARgamma pathway. J Mol Endocrinol 38: 19–34 [DOI] [PubMed] [Google Scholar]
- Kim JB, Spiegelman BM (1996) ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev 10: 1096–1107 [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A (1999) A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell 4: 735–743 [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Lazar MA (2009) New developments in adipogenesis. Trends Endocrinol Metabol 20: 107–114 [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA (2010) Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol 30: 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Liu XS, Lazar MA (2008) PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 22: 2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B, Bick RJ, Darlington GJ (2001) C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc Natl Acad Sci USA 98: 12532–12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougald OA, Mandrup S (2002) Adipogenesis: forces that tip the scales. Trends Endocrinol Metabol 13: 5–11 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143: 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M (2005) Role of Krüppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem 280: 12867–12875 [DOI] [PubMed] [Google Scholar]
- Nagy PL, Cleary ML, Brown PO, Lieb JD (2003) Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci USA 100: 6364–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbu-Wakao R, Morikawa Y, Matsumura I, Masuho Y, Muramatsu MA, Senba E, Wakao H (2002) Stimulation of 3T3-L1 adipogenesis by signal transducer and activator of transcription 5. Mol Endocrinol 16: 1565–1576 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Grontved L, Stunnenberg HG, Mandrup S (2006) Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Mol Cell Biol 26: 5698–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TÅ, Hagenbeek D, Moulos P, Siersbæk R, Megens E, Denissov S, Børgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG (2008) Genome-wide profiling of PPARgamma: RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev 22: 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R (2005) Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metabol 1: 27–39 [DOI] [PubMed] [Google Scholar]
- Pantoja C, Huff JT, Yamamoto KR (2008) Glucocorticoid signaling defines a novel commitment state during adipogenesis in vitro. Mol Biol Cell 19: 4032–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C (2001) Cooperation between C/EBP alpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev 15: 3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetka A, Chayka O, Wilczek C, Melnik S, Bonifer C, Klempnauer KH (2008) C/EBPbeta induces chromatin opening at a cell-type-specific enhancer. Mol Cell Biol 28: 2102–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM (1999) PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 4: 611–617 [DOI] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M et al. (2006) Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods 3: 511–518 [DOI] [PubMed] [Google Scholar]
- Salma N, Xiao HG, Mueller E, Imbalzano AN (2004) Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol Cell Biol 24: 4651–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R, Nielsen R, Mandrup S (2010) PPARgamma in adipocyte differentiation and metabolism—novel insights from genome-wide studies. FEBS Lett 584: 3242–3249 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Lazar MA (2010) Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 24: 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yoshida N, Kishimoto T, Akira S (1997) Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 16: 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Grønborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD (2005) Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci USA 102: 9766–9771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD (2003) CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci USA 100: 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teglund S, Mckay C, Schuetz E, van Deursen JM, Stravopodis D, Wang DM, Brown M, Bodner S, Grosveld G, Ihle JN (1998) Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93: 841–850 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM (2008) Fat and beyond: the diverse biology of PPAR gamma. Annu Rev Biochem 77: 289–312 [DOI] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T (2004) Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA 101: 10380–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi KI, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, Kitakami JI, Ihara S, Hashimoto Y, Hamakubo T, Kodama T, Aburatani H, Sakai J (2009) The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 29: 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Milton JF, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ (1995) Impaired energy homeostasis in C/EBPalpha knockout mice. Science 269: 1108–1112 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR (2009) Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Bucher NL, Farmer SR (1996) Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol Cell Biol 16: 4128–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, LeBlanc SE, Wu Q, Konda S, Salma N, Marfella CGA, Ohkawa Y, Imbalzano AN (2011) Chromatin accessibility and transcription factor binding at the PPARgamma2 promoter during adipogenesis is protein kinase A-dependent. J Cell Physiol 226: 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WC, Cao ZD, Classon M, Mcknight SL (1995) Cascade regulation of terminal adipocyte differentiation by 3 members of the C/Ebp family of leucine-zipper proteins. Genes Dev 9: 168–181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.