Abstract
The oculocerebrorenal syndrome of Lowe (OCRL), also called Lowe syndrome, is characterized by defects of the nervous system, the eye and the kidney. Lowe syndrome is a monogenetic X-linked disease caused by mutations of the inositol-5-phosphatase OCRL1. OCRL1 is a membrane-bound protein recruited to membranes via interaction with a variety of Rab proteins. The structural and kinetic basis of OCRL1 for the recognition of several Rab proteins is unknown. In this study, we report the crystal structure of the Rab-binding domain (RBD) of OCRL1 in complex with Rab8a and the kinetic binding analysis of OCRL1 with several Rab GTPases (Rab1b, Rab5a, Rab6a and Rab8a). In contrast to other effectors that bind their respective Rab predominantly via α-helical structure elements, the Rab-binding interface of OCRL1 consists mainly of the IgG-like β-strand structure of the ASPM-SPD-2-Hydin domain as well as one α-helix. Our results give a deeper structural understanding of disease-causing mutations of OCRL1 affecting Rab binding.
Keywords: Dent disease, Lowe syndrome, OCRL, Rab, Rab8
Introduction
Membrane trafficking is a fundamental biological process of eukaryotic cells. Vectorial membrane trafficking is crucial for the establishment and maintenance of cell polarity and is strictly regulated by the concerted action of Rab GTPases and phosphatidylinositols (Mellman and Nelson, 2008; Stenmark, 2009). A hallmark of Rab proteins and phosphatidylinositols is their distinct subcellular localization to membrane compartments (Di Paolo and De Camilli, 2006; Stenmark, 2009). For example, Rab5 is mainly localized at early endosomes, whereas Rab6 is localized preferentially at the Golgi apparatus. Likewise, phosphatidylinositol-4,5-bisphosphate is mainly present in the plasma membrane, whereas phosphatidylinositol-4-phosphate is largely confined to the Golgi apparatus. Rab proteins and phosphatidylinositols are both integral parts of the molecular recognition machinery of subcellular membrane compartments, which regulate targeted membrane trafficking between organelles. Rab proteins function as molecular switches by changing from the active GTP-bound to the inactive GDP-bound form and vice versa. Rab GTPases regulate membrane trafficking through the recruitment of effector proteins, such as sorting adaptors, tethering factors, kinases, phosphatases and motors (Segev, 2001; Stenmark, 2009). The Rab-effector proteins specifically interact with the active GTP-bound state of the Rab GTPase. One of these effectors is oculocerebrorenal syndrome of Lowe protein (OCRL1). It shows a broad range of Rab-binding activity and interacts with several Golgi-associated (e.g. Rab1, Rab6 and Rab8) and endosomal (Rab5) Rab proteins (Hyvola et al, 2006; Fukuda et al, 2008). OCRL1 is a member of the type II family of inositol polyphosphate 5-phosphatases and preferentially dephosphorylates phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) and phosphatidylinositol-3,4,5-trisphosphate (PI(3,4,5)P3) at the 5-position (Lowe, 2005; Ooms et al, 2009). OCRL1 is a peripheral membrane protein mainly localized at the Golgi apparatus and early endosomes (Faucherre et al, 2003; Ungewickell et al, 2004; Choudhury et al, 2005). In addition, growth factor stimulation leads to translocation of OCRL1 to plasma membrane ruffles (Faucherre et al, 2003). Mutations of OCRL1 are responsible for the oculocerebrorenal syndrome of Lowe (Lowe syndrome) (Attree et al, 1992). Additionally, OCRL1 mutations were recently also identified in a subset of Dent disease patients, which is like Lowe syndrome an X-linked disorder (Hoopes et al, 2005). Lowe syndrome is characterized by congenital cataracts, mental retardation and kidney readsorption defects caused by proximal tubule dysfunction (Delleman et al, 1977; Kenworthy and Charnas, 1995; Kleta, 2008), whereas Dent disease (type 2) shows a phenotype largely confined to the kidney (Hoopes et al, 2005; Utsch et al, 2006; Shrimpton et al, 2009). OCRL1 has been implicated in several membrane trafficking steps, such as the transport between the early endosome and the Golgi apparatus or endocytosis (Choudhury et al, 2005; Erdmann et al, 2007). Moreover, recently it was reported that OCRL1 regulates migration and spreading of human dermal fibroblasts (Coon et al, 2009). However, the precise function of OCRL1 and its role in disease is not clear.
OCRL1 is a multi-domain protein comprising an N-terminal pleckstrin homology (PH) domain followed by a central inositol-5-phosphatase domain, an ASPM-SPD-2-Hydin (ASH) domain, and a C-terminal, catalytically inactive RhoGAP-like domain (Figure 1) (Erdmann et al, 2007; Mao et al, 2009). In addition to binding to multiple Rab proteins, OCRL1 also interacts with clathrin heavy chain, the clathrin adaptor AP2, Rac, Cdc42, the Rab5 effector APPL1 (adaptor protein containing PH domain, PTB domain and leucine zipper motif 1), and the endocytic proteins Ses1/2 (Faucherre et al, 2003; Ungewickell et al, 2004; Choudhury et al, 2005, 2009; Erdmann et al, 2007; Swan et al, 2010). Interestingly, Lowe syndrome cannot be reproduced in OCRL1-knockout mice, probably because of the existence of the close OCRL1 relative INPP5B. INPP5B has similar domain architecture and substrate specificity, but no clathrin- or AP2-binding sites (Jefferson and Majerus, 1995). In line with at least partially redundant functions of OCRL1 and INPP5B is the fact that OCRL1/INPP5B double-knockout mice are not viable and appear to die early in embryogenesis, whereas OCRL1-knockout mice show no obvious and INPP5B-knockout mice only a minor phenotype (male sterility) (Janne et al, 1998).
Figure 1.
Schematic representation of the domain architecture of OCRL1.
Recent work by the De Camilli group has given first insights into the detailed molecular structure of OCRL1. The X-ray structure of the ASH-RhoGAP-like tandem domain revealed the localization of the clathrin-binding site within an extended loop in the RhoGAP-like domain. Subsequent NMR-structural analysis of the N-terminus revealed the presence of a PH domain and the presence of a second clathrin-binding site. Together with the previously solved X-ray structure of an inositol-5-phosphatase domain of Schizosaccharomyces pombe, a first model for membrane-bound OCRL1 was proposed taking into account recruitment of OCRL1 to the Golgi apparatus and to endosomal membranes via Rab proteins (Erdmann et al, 2007; Mao et al, 2009). Rab proteins (e.g. Rab1, Rab6, Rab8 and Rab14) may also mediate the localization of OCRL1 to chlamydial inclusions and OCRL1 depletion from host cells impairs chlamydial infection (Moorhead et al, 2010). Moreover, active Rab5 and active Rab6 stimulate the 5-phosphatase activity of OCRL1 in hydrolyzing PI(4,5)P2 (Hyvola et al, 2006) and several Lowe syndrome causing amino-acid substitutions are localized in the Rab-binding region of OCRL1 (Hichri et al, 2010). Thus, investigation of the structural and kinetic basis of the OCRL1–Rab interaction is pivotal for the understanding of the molecular details of OCRL1 function. Here, we present the crystal structure of the Rab-binding domain (RBD) of OCRL1 in complex with active Rab8a (Fukuda et al, 2008; Rodriguez-Gabin et al, 2010). We have also analysed the kinetics and affinities of OCRL1 binding to the Golgi-associated and endosomal Rab proteins Rab1b, Rab5a, Rab6a and Rab8a. Our results reveal a novel Rab-effector binding mode for OCRL1. In addition, our data contribute valuable information for a structural model of membrane-bound OCRL1. Finally, our results explain the structural effects of disease-causing mutations in OCRL1 leading to a better understanding of Lowe syndrome.
Results and Discussion
Interactions of OCRL1 with Rab proteins
Many Lowe syndrome causing amino-acid substitutions are localized in the 5-phosphatase domain of OCRL1, suggesting that impairment of catalytic activity is part of the OCRL1 disease mechanism (Hichri et al, 2010). Major substrates for OCRL1 are the membrane localized phosphatidylinositols (PI(4,5)P2) and (PI(3,4,5)P3). Thus, understanding the precise mechanism of membrane recruitment of OCRL1 is crucial for the understanding of OCRL1 function. Two mechanisms of membrane recruitment have been described for OCRL1. Recruitment to plasma membrane localized membrane ruffles after growth factor stimulation has been attributed to interaction with activated Cdc42, while localization of OCRL1 to the Golgi apparatus and endosomal membranes has been shown to be mediated via Rab proteins (Faucherre et al, 2003; Hyvola et al, 2006).
The Rab-binding site of OCRL1 was mapped to a region from amino acids 540–725 in a previous report (Hyvola et al, 2006) (for a schematic depiction of the domain architecture of OCRL1 see Figure 1). Various data indicate binding of OCRL1 to a number of different Rab proteins in vitro (Hyvola et al, 2006; Fukuda et al, 2008). The interactions were also further supported by co-localization in cells for some OCRL1/Rab pairs; however, the affinities for their interactions have not been determined. Using a fluorescence-based approach, we were able to monitor the binding of OCRL1539–901 (OCRL1 amino acids 539–901) to mantGppNHp (a non-hydrolyzable and fluorescent GTP analogue)-loaded Rabs (Rab1b, Rab3a, Rab5a, Rab6a, Rab7, Rab8a, Rab13, Rab14 and Rab31) by exploiting the change in fluorescence polarization (Supplementary Figure S1). The fluorescence polarization increased upon Rab binding to OCRL1539–901, indicating complex formation. The reaction was fully reversed by displacement of the mantGppNHp-loaded Rab from OCRL1539–901 with an excess of non-fluorescent Rab8a:GppNHp (Supplementary Figure S1). In agreement with previous reports, no complex formation was observed with Rab7 protein (Fukuda et al, 2008).
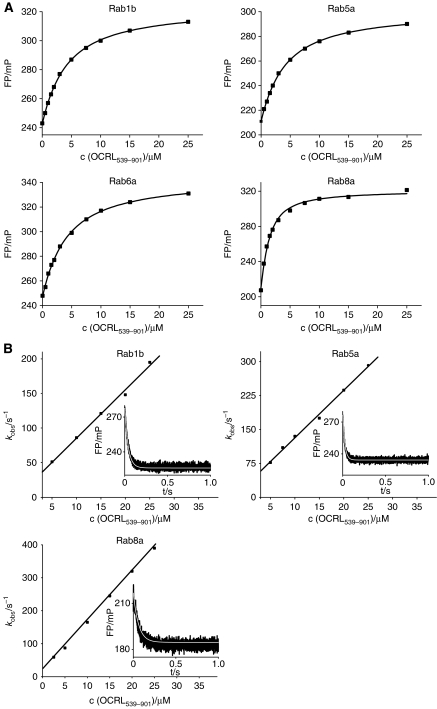
Since the binding specificity of OCRL1 for Rab proteins is rather broad, we were interested whether a certain Rab is preferably bound in vitro. Hence, we determined the affinity of various Rab:OCRL1539–901 complexes (Rab1b, Rab5a, Rab6a, Rab8a) using equilibrium titrations combined with fluorescence polarization as an indicator of complex formation (Figure 2A). Among the tested Rab proteins, Rab8a:mantGppNHp revealed the highest affinity towards OCRL1539–901, with a dissociation equilibrium constant (KD) of 0.9 μM. Rab1b, Rab5a and Rab6a were bound less strongly with KD values of about 4 μM (Figure 2A). Thus, Rab8a appears to be the preferred in vitro binding partner of OCRL1 among the Rabs tested. Strong interaction of OCRL1 with Rab8a is also supported by significant co-localization of both proteins in Hela cells (Supplementary Figure S2). Previously, the relative affinities to OCRL1 had been reported for Rab8a, Rab6a, Rab5a and Rab1a with Rab8a showing the lowest relative affinity to OCRL1 and Rab6a having the highest (Hyvola et al, 2006). Our findings differ from this report, possibly due to the different techniques applied. Thus, we measured binding affinities in solution, while the previous report used surface-immobilized OCRL1 for its binding study. In addition, we used wild-type Rab:mantGppNHp proteins for our interaction studies, whereas the other report applied GST-fusion proteins of constitutively active Rab mutants. Also, an indirect influence of the mant-group at the ribose moiety of mantGppNHp cannot be excluded and might have a minor effect on the binding of the derivatized Rab protein to OCRL1. The KD values obtained in the described equilibrium titrations in the present study were confirmed by the kinetic investigations described in the next section.
Figure 2.
OCRL1 binds different exocytic and endocytic Rab proteins with moderate affinities. (A) Fluorescence polarization based equilibrium titration of 1 μM Rab1b:mantGppNHp, Rab5a:mantGppNHp, Rab6a:mantGppNHp or Rab8a:mantGppNHp with OCRL1539–901, respectively. (B) Association and dissociation kinetics of Rab1b:mantGppNHP, Rab5a:mantGppNHp and Rab8a:mantGppNHp with OCRL1539–901, respectively, monitored by the change in fluorescence polarization using stopped-flow apparatus (c(RabX:mantGppNHp)=1 μM). The individual observed association rate constants are linearly dependent on the OCRL1539–901 concentration and were fitted to a straight line, yielding the association rate constant. (Insets) The dissociation of fluorescent RabX:mantGppNHp (2 μM) from its complex with OCRL1539–901 was monitored by displacement with a 10-fold molar excess of non-fluorescent Rab using the stopped-flow apparatus. The time-dependent decrease in fluorescence polarization, indicative of complex dissociation, was fitted to a single exponential function (white line) to determine the dissociation rate constants. FP, fluorescence polarization; mP, milli-polarization units.
Using these Rab:mantGppNHp complexes, we also investigated the dynamics of Rab:OCRL1539–901 complexes by determining their association and dissociation rate constants (kon and koff, respectively). The association process was monitored by the time-dependent change in fluorescence polarization in a stopped-flow apparatus. Single exponential fitting of individual traces yielded the observed pseudo first-order rate constant of the association reaction at a specific OCRL1539–901 concentration. Plotting the observed rate constants (kobs) against the OCRL1539–901 concentration gave a straight line, with the slope equal to the association rate constant (kon) for each Rab protein (Rab1b: kon=7.0 × 106 M−1 s−1; Rab5a: kon=1.05 × 107 M−1 s−1; Rab8a: kon=1.5 × 107 M−1 s−1) (Figure 2B). The dissociation rate constants (koff) were determined by displacing the fluorescent Rabs from their complex with OCRL1539–901 with a 10-fold molar excess of non-fluorescent Rab:GppNHp (Rab1b: koff=25 s−1 Rab5a: koff=40.1 s−1 ; Rab8a: koff=16.2 s−1) (Figure 2B). From the association and dissociation rate constants, KD values for the affinity of the Rab:mantGppNHp–OCRL1539–901 interaction (Rab1b: KD=3.7 μM; Rab5a: KD=3.8 μM; Rab8a: KD=1.1 μM) could be calculated. As shown in Table I and Figure 2, Rab8a:mantGppNHp displays the highest affinity toward OCRL1539–901, while the other evaluated Rabs are bound about four-fold more weakly.
Table 1. Summary of equilibrium constants (KD) of Rab proteinsa.
| KD (equilibrium titration)/μM | KD (from kinetics)/μM | |
|---|---|---|
| Rab1b | 3.7 | 3.7 |
| Rab5a | 3.6 | 3.8 |
| Rab6a | 3.7 | N/A |
| Rab8a | 0.9 | 1.1 |
| N/A, not assessed. | ||
| aOCRL1539–901 was used to determine the kinetics. | ||
Previous reports suggested that Rab proteins bind to OCRL1 within the linker region between the 5-phosphatase and RhoGAP-like domains (Figure 1). To further localize the Rab-binding site, we produced various N-terminal and C-terminal truncations of OCRL1 and analysed the binding to Rab8a:GppNHp by analytical gel filtration (Supplementary Figure S3). The constructs OCRL1540–678 and OCRL1555–678 were able to bind Rab8a:GppNHp, but the OCRL1560–678 fragment failed to interact with active Rab8a. The construct OCRL1555–668 could not bind to Rab8a:GppNHp since the protein was prone to aggregation, indicating an unfolded ASH domain. Hence, OCRL1555–678 represents the minimal Rab-binding construct.
Crystal structure of the Rab8a6–176:GppNHp: OCRL1540–678 complex
In order to understand the molecular basis of the interaction between Rab proteins and OCRL1, we determined the crystal structure of a complex between active, GppNHp-loaded Rab8a (amino acids 6–176, Rab8a6–176) and OCRL1 (amino acids 540–678, OCRL1540–678) at 2.0 Å resolution. The crystal structure contains four individual complexes per asymmetric unit (for data collection and refinement statistics, see Table II). Some residues in the N-terminal regions of two OCRL1540–678 molecules (residues 540–548 of chain B and D) were not visible in electron density, and thus were omitted from the final model. Since the mode of interaction among the four complexes in the asymmetric unit is highly similar (excluding small differences due to crystal packing) (Supplementary Figure S4), we discuss only one representative complex structure here.
Table 2. Data collection and refinement statistics for Rab8a6–176:OCRL1540–678.
| Data collection a | |
| Space group | C2 |
| Cell dimensions | |
| A, B, C (Å) | 157.15, 55.34, 173.84 |
| α, β, γ (deg) | 90.00, 91.93, 90.00 |
| Resolutions (Å) | 20–2.0 (2.1–2.0) |
| Rmerge (%)b | 5.5 (33.4) |
| I/σ(I) | 15.4 (4.2) |
| Completeness (%) | 98.3 (97.0) |
| Redundancy | 3.2 (3.0) |
| Refinement | |
| No. of reflections | 320 388 (39 313) |
| Rwork/Rfree | 20.9/24.1 |
| No. of atoms | |
| Protein | 9954 |
| Ligand/ion | 142 |
| Water | 324 |
| B-factors | |
| Protein | 38 |
| Ligand/ion | 28 |
| Water | 39 |
| R.m.s. deviation | |
| Bond lengths (Å) | 0.014 |
| Bond angles (deg) | 1.412 |
| Values in parentheses refer to the highest resolution shell. | |
| aThe data set was collected from one single crystal on beamline X10SA of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). | |
| bCalculated as defined by Diederichs and Karplus. | |
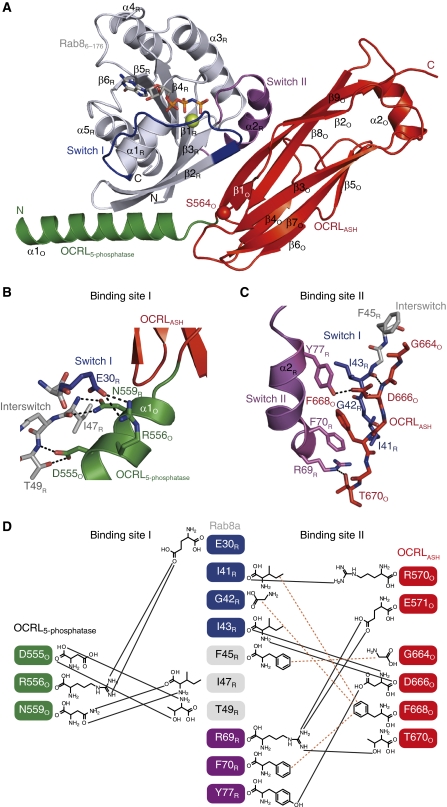
A cartoon representation of the Rab8a6–176:OCRL1540–678 complex is shown in Figure 3A. In the complex, Rab8a6–176 adopts the common GTPase fold, which consists of six central β-strands (β1R–β6R) surrounded by five α-helices (α1R–α5R) (subscripts ‘R’ and ‘O’ denote elements of Rab8a and OCRL1, respectively). OCRL1540–678 consists of the ASH domain and an elongated helix (α1O in green) belonging to the 5-phosphatase domain. The ASH domain of OCRL1540–678 forms two layers of β-sheets fold and is closely related to the members of the family of immunoglobulin-like β-sandwiches (Erdmann et al, 2007).
Figure 3.
Crystal structure of the Rab8a6–176:OCRL1540–678 complex (A) Cartoon representation of the Rab8a6–176:OCRL1540–678 complex. Rab8a6–176 is shown in grey, with Switch I in blue and Switch II in magenta. OCRL1540–670 is in green (helix α1O of the 5-phosphatase domain) and red (ASH domain). The position of S564O (S564PO causes Lowe syndrome) is indicated with a red sphere. (N, N-termini; C, C-termini; sticks, non-hydrolyzable GTP-analogue GppNHp; green sphere, Mg2+, colouring is kept consistent throughout all panels). (B, C) Detailed view of interactions between Rab8a6–176 with α1O-helix (binding site I, (B)) and the ASH domain (binding site II, (C)) of OCRL1540–678. The Switch I region of Rab8a and the β9O-strand of OCRL1 are in stick representation to show the main-chain interactions (black dashes: polar interactions). (D) Schematic view of Rab8a–OCRL1 interactions (black lines: hydrogen bonds, brown dashes: hydrophobic contacts).
The Rab8a:OCRL1 complex interface
Complex formation leads to the burying of 1733 Å2 of solvent-accessible surface area. The majority of contacts in the complex are mediated by the α1O-helix and the β-strand 9 (β9O) of OCRL1, and Switch I, Switch II and the Interswitch region of Rab8a (Figure 3A).
The complex crystal structure reveals a Rab-binding site in OCRL1540–678 that consists of two separate elements: The first site of protein–protein contact (binding site I) in OCRL1 is located in the linker region between the ASH domain and 5-phosphatase domain. The interactions between Rab8a and OCRL1 in binding site I are mostly of polar nature (Figure 3B). The amino acids D555O to N559O in the N-terminal region of OCRL1 are essential for binding Rab proteins (Supplementary Figure S3). D555O and R556O form side chain interactions with T49R in the Interswitch region and E30R in Switch I, respectively. In addition, the adjacent residue N559O binds strongly with the main chain of I47R in the Interswitch region via a hydrogen bond. Hence, the significance of the amino acids 555–559 of OCRL1 for Rab binding (Supplementary Figure S3) is apparent from the structure of the complex interface.
All known structurally characterized Rab-effector proteins bind to their respective Rab proteins mainly via 1–2 α-helices (Zhu et al, 2004; Eathiraj et al, 2005, 2006; Kawasaki et al, 2005; Wu et al, 2005; Jagoe et al, 2006; Chavas et al, 2008; Kukimoto-Niino et al, 2008). In contrast to these established binding modes, OCRL1 Rab8a interacts with the Rab-binding motif of OCRL1 via a β-strand (β9O), which forms the Rab-binding site II. The interaction between Rab8a and OCRL1 in binding site II is mediated by both hydrophobic and polar contacts. The side chain of F668O in OCRL1 penetrates into a hydrophobic core of Rab8a, which is formed by I41R and G42R in the Switch I region and F70R in the Switch II region. Moreover, G664O of OCRL1 Interacts with F45R in the Rab8a Interswitch region. Besides these hydrophobic interaction, D666O forms hydrogen bonds with the main chain of I43R of Switch I and the hydroxyl oxygen of Y77R of Switch II. An additional polar interaction is seen between T670O and R69R of the Rab8a Switch II region. Furthermore, R570O and E571O from the N-terminal region of the ASH domain interact with the main chain of I42R and side chain of R69R, respectively (Figure 3C and D).
The molecular interactions seen in the Rab8a:OCRL1 complex also provide a structural understanding of previous Rab-binding studies (D555E, S568G, S564P, G664D, N606K; Supplementary Figure S5) (Hyvola et al, 2006). According to the complex structure, D555 and G664 are responsible for the interaction with the Interswitch region by forming polar and hydrophobic interactions, respectively. Consequently, the D555E and G664D substitutions generated by Hyvola et al (2006) reduced the Rab-binding activity of OCRL1 in vitro and impaired OCRL1 membrane localization in vivo, hence supporting their implication in Rab8a–OCRL1 complex formation. The amino acids S568 and N606 are not involved in establishing the complex interface and their substitutions (S568G and N606K) do not influence Rab binding or OCRL1 membrane recruitment (Hyvola et al, 2006). Variant S564P had the strongest effect on Rab binding, abolishing it completely (Hyvola et al, 2006). Based on our complex structure, the substitution S564P is expected to destabilize the conformation of binding site II, since this residue appears to be an integral part of the beginning of the ASH domain, leading to complete loss of Rab-binding activity. Additionally, the S564P mutation could impair the correct relative arrangement of the α1-helix and the ASH domain, since proline has reduced conformational freedom compared with serine. Since S564 is at the C-terminal end of the α1-ASH linker region, its mutation to proline possibly affects Rab binding by impairing the orientation of helix α1 (Rab-binding site I) and the ASH domain (Rab-binding site II). Thus, the in vivo and in vitro effects of OCRL1 point mutations reported previously perfectly correlate with their relevance for Rab8a:OCRL1 complex formation as suggested by the complex crystal structure.
The Rab8a:OCRL1 interaction mode in comparison to other Rab effectors
In the active state, Rab proteins recruit diverse effectors to specific subcellular compartments. A number of Rab-effector crystal structures have been reported. These are (1) Rab3:Rabphilin, Rab27a:Slp2a and Rab27b:Slac2-a, which are associated with secretory vesicles (Ostermeier and Brunger, 1999; Chavas et al, 2008; Kukimoto-Niino et al, 2008); (2) Rab7:RILP (Rab7-interacting lysosomal protein), which is involved in regulating fusion between lysosomes and endosomes (Wu et al, 2005); (3) Rab4:Rabenosyn-5, Rab22:Rabenosyn-5 and Rab5:EEA1 (early endosomal autoantigen 1), which coordinate endosomal traffic (Eathiraj et al, 2005; Mishra et al, 2010); (4) Rab5:Rabaptin5, which is implicated in early endosome fusion (Zhu et al, 2004); (5) Rab11:FIP2 (Rab11-family interacting protein 2) and Rab11:FIP3, which are involved in regulating endocytic recycling (Eathiraj et al, 2006; Jagoe et al, 2006); (6) Rab6:Rab6IP1 (Rab6-interacting protein 1); and (7) Rab6:GCC185, which regulate endosome to Golgi transport (Burguete et al, 2008; Recacha et al, 2009). In all of the previously reported structures of Rab-effector complexes, the RBD is exclusively formed by an α-helical region in the effector (Lee et al, 2009). In contrast to these helix-dominated RBDs, OCRL1 interacts with Rab8a not only via an α-helix (α1O) but also via β-strand β9O from the all-β ASH domain in binding site II. The general binding interfaces of Rabs include the Switch and Interswitch regions and are either centred on or overlap with an invariant hydrophobic triad (F45R and W62R in the Interswitch and Y77R in Switch II according to the sequence of Rab8a) (Merithew et al, 2001; Lee et al, 2009). However, in the Rab8a6–176:OCRL1540–678 complex, not all of the three aromatic residues of the hydrophobic core participate in the binding interface. W62R in the Interswitch region is not involved in OCRL1 binding, while Y77R in the Switch II region associates with D666O only via a hydrogen bond interaction, but not a hydrophobic interaction as in other Rab-effector complexes. Only F45R in the Interswitch region interacts with G664O in OCRL1.
When comparing the OCRL1/Rab-binding mode with that of other Rab effectors, it seems that the invariant hydrophobic triad may not be such a critical requirement for the Rab binding of OCRL1 as for other Rab-effector interactions (reviewed in Lee et al, 2009; Itzen and Goody, 2011). Although there are numerous effectors that can bind to several different Rab proteins, an effector binding as broad as observed for OCRL1, which includes Rab proteins from completely different subfamilies, is unusual. Based on previous interaction studies, different Rab proteins bind to the same site in OCRL1, although with subtle differences (Hyvola et al, 2006). It is therefore appropriate to consider why OCRL1 binds to such a broad variety of different Rab proteins. It has been suggested that the hydrophobic triad of Rab proteins represents a main determinant for Rab subfamily specific effector binding (Merithew et al, 2001). The side chain conformations of this conserved hydrophobic triad (F45R, W62R and Y77R) in the active state are similar for members of a Rab subfamily but different among subfamilies, and this structural plasticity is suggested to be an important determinant for Rab-effector specificity (Merithew et al, 2001; Grosshans et al, 2006). In the case of the Rab–OCRL1 interaction, the hydrophobic triad appears to have only a minor role in OCRL1 binding, since the central W62R residue in Rab8a is not involved in any interactions with residues of OCRL1. Thus, the minor involvement of the hydrophobic triad of Rabs in OCRL1 binding might serve as an explanation for the low Rab-binding specificity of OCRL1. It will be interesting to see whether effectors with a wide Rab-binding range comparable to OCRL1 (Fukuda et al, 2008) employ a similar structural mechanism, that is by omitting the hydrophobic triad of Rabs as a main binding determinant to achieve interaction with multiple Rabs.
The broad Rab-binding specificity of OCRL1 contributes to its diverse subcellular localization. siRNA experiments have demonstrated that knockdown of Rab1 together with Rab6 leads to a significantly reduced binding of OCRL1 to the Golgi apparatus. It was also demonstrated that constitutively expressed Rab5 can translocate OCRL1 from the cytosol to enlarged endosomes. Point mutations abolishing binding to Rab proteins lead to complete delocalization of OCRL1 to the cytosol (Hyvola et al, 2006). However, other targeting mechanisms might exist, for example transient localization of OCRL1 to late clathrin-coated pits has been demonstrated to rely on its two clathrin-binding motifs (Mao et al, 2009).
APPL1/Ses1 peptides do not influence the Rab8a:OCRL1 interaction
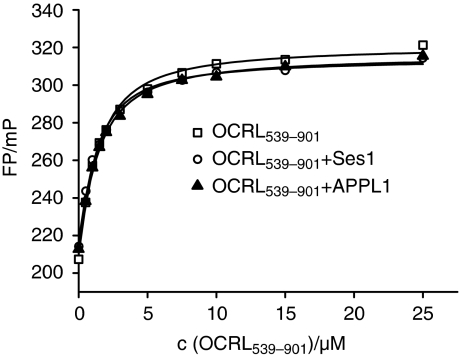
The ASH domain is not only required for binding to Rab proteins but also for binding to the Rab5-effector protein APPL1 (Erdmann et al, 2007), as well as to the two novel endocytic proteins Ses1/2 (Swan et al, 2010). APPL1/Ses interactions with OCRL1 are both mediated by a conserved short amino-acid motif (Phe and His [F&H] motif) in APPL1/Ses1. We asked whether binding of APPL1 and Ses1 modulates the affinity of OCRL1 for Rab proteins. To do this, we determined the affinity of Rab8a:mantGppNHp to OCRL1539–901 by equilibrium titration as described above in the presence of the APPL1 and Ses1 OCRL1 binding peptides (SFQQRHESLRP and PFARLHECYGQEI, respectively) (Swan et al, 2010) (Figure 4). The concentrations used are orders of magnitude higher than the known KD values for the peptides for OCRL1, ensuring binding under these conditions. The binding activity of the synthesized peptides to OCRL1539–901 has been independently confirmed by isothermal titration calorimetry, yielding results that are comparable to the ones reported by Swan et al (2010). The KD of the complex between Rab8a:mantGppNHp and OCRL1539–901 was unchanged by the presence of APPL1 or Ses1 peptides, suggesting they have no modulatory effect on the stability of the Rab8a:OCRL1 complex (Figure 4). Apparently, the interaction of Rab with OCRL1 is independent of the simultaneous binding of the APPL1 or Ses1 peptides to OCRL1, indicating no communication of the APPL1/Ses1-binding site with the Rab-binding site.
Figure 4.
The binding of APPL1 and Ses1 peptides does not alter the dissociation equilibrium constant of the Rab8:mantGppNHp–OCRL1539–901 interaction. The experimental setup described in Figure 2A was repeated in the presence of 200 μM APPL1 peptide or 100 μM Ses1 peptide with 1 μM Rab8a:mantGppNHp and varying concentrations of OCRL1539–901 (FP, fluorescence polarization; mP, milli-polarization units).
A structural basis for Lowe syndrome
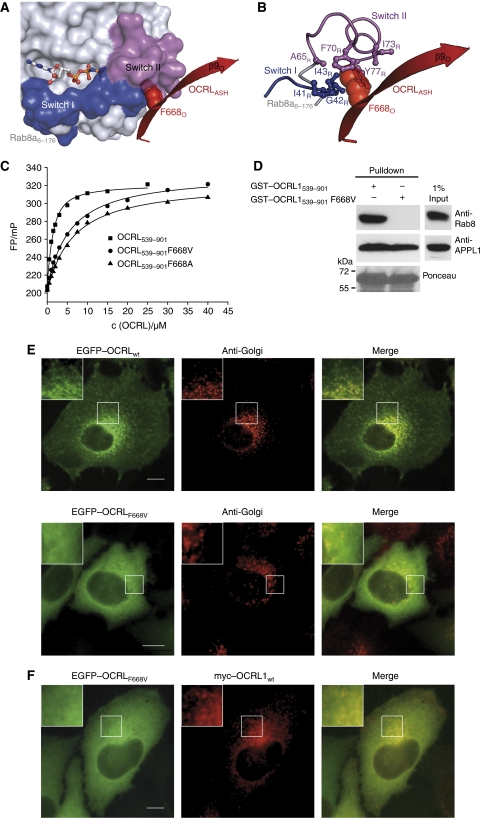
Two missense mutations causing Lowe syndrome are responsible for amino-acid substitutions in the minimal Rab-binding region of OCRL1 (L634P and F668V). Substitution L634P is likely to destabilize the ASH domain fold, which is in line with complete loss of binding to APPL1 and Ses proteins. However, F668V retained the binding to APPL1 and Ses1/2, making an APPL1/Ses1 interaction deficiency unlikely to account for disease development. Thus, the molecular basis for this disease-causing mutation remained unclear (Swan et al, 2010). In the Rab8a6–176:OCRL1540–678 structure, F668 participates in the complex interface by establishing a multitude of contacts within a hydrophobic groove of Rab8a (Figure 5A and B). We speculated that the F668V substitution impairs the interaction with Rab8a due to the lower hydrophobicity of valine compared with phenylalanine. Furthermore, this effect should be even more pronounced with an F668A substitution in OCRL1. This hypothesis was confirmed by determining the dissociation constants between active Rab8a:mantGppNHp and the OCRL1539–901 variants (F668V or F668A) by equilibrium titration (Figure 5C). The affinity of OCRL1539–901 F668V was decreased 5.8-fold compared with the wild-type protein (KD,Rab8:OCRL1=0.9 μM, KD,Rab8:OCRL1 F668V=5.2 μM). The affinity of the F668A variant was decreased by a factor of 6.7 compared with the wild-type protein (KD,Rab8:OCRL1 F668A=6.0 μM). Thus, the variants F668V and F668A of OCRL1 weaken the binding towards active Rab8a. In addition, the amino-acid substitutions F668V and F668A also significantly impair the binding to Rab1b, Rab5a and Rab6a (Supplementary Figure S6). To confirm weakened binding of OCRL1 (F668V) to Rab8a by an independent assay, we performed pull-down experiments using GST-fusion proteins of OCRL1539–901 and OCRL1539–901 F668V. Using this assay, binding of Rab8a to OCRL1539–901 was detected; however, no binding could be detected between Rab8a and OCRL1539–901 F668V. In contrast, both OCRL1 constructs bound APPL1 efficiently, indicating selective impairment of binding to Rab8a for OCRL1539–901 F668V (Figure 5D). It has been previously shown that interaction with Rab proteins is crucial for OCRL1 membrane recruitment. Thus, we tested whether the Lowe syndrome mutation F668V changes the subcellular localization of OCRL1. Transfection of GFP–OCRL1 F668V into Hela cells revealed that the mutation leads to a large increase in cytosolic localization of OCRL1 and to a significant reduction at the Golgi apparatus and vesicular structures (Figure 5E). Co-transfection experiments of Myc-tagged wild-type OCRL1 together with GFP–OCRL1 F668V also demonstrated significant localization differences between mutant and wild-type OCRL1 for example reduced localization at subcellular membranes and a higher cytosolic pool for mutant OCRL1 (Figure 5F). The observed differences in binding affinity and subcellular localization are completely in line with our predictions based on the Rab8a:OCRL1 complex structure.
Figure 5.
Structural and thermodynamic basis for the Lowe syndrome causing mutation F668V. (A) The residue F668O (red spheres) of the β9O-strand of OCRL1 (red cartoon) is binding within a hydrophobic pocket of Rab8a (surface representation) located between Switch I and Switch II (the nucleotide is shown as ball and stick representation). (B) F668O is penetrating into a hydrophobic pocket of seven hydrophobic residues (I41R, G42R, I43R, A65R, F70R, I73R and Y77R). (C) Fluorescence polarization based equilibrium titrations of Rab8a:mantGppNHp with different OCRL1539–901 variants (wild type, F668VO and F668AO), demonstrating the significance of the Lowe disease related mutant F668O for Rab binding. The mutations F668VO and F668AO of OCRL1 decrease the affinity to Rab8a (KD,Rab8:OCRL1 wt=0.9 μM, KD,Rab8:OCRL1 F668V=5.2 μM, KD,Rab8:OCRL1 F668A=6.0 μM). (D) Pull-down experiment of Rab8a with GST–OCRL1 fusion proteins. Indicated GST-fusion proteins were incubated with Hela cell lysate, bound proteins were separated by SDS–polyacrylamide gel electrophoresis transferred to nitrocellulose and detected using anti-Rab8a or anti-APPL1 antibody, equal loading of GST-fusion proteins is indicated by ponceau staining. (E) Subcellular localization of OCRL1 F668V. Hela cells expressing GFP-tagged full-length OCRL1 or OCRL1 point mutant F668V were analysed by immunofluorescence microscopy with antibodies to the Golgi 58K protein. Insets show an enlargement of the Golgi area. (F) Comparison of subcellular localizations of OCRL1 wt with OCRL1 F668V. Hela cells were co-transfected with constructs for EGFP–OCRL1 F668V and Myc-tagged wild-type OCRL1 and analysed using immunofluorescence microscopy with antibodies against the Myc epitope (Bar, 10 μm).
In summary, the point mutation F668V is so far the only Lowe syndrome causing variant, which selectively affects the Rab-binding properties of OCRL1, giving direct in vivo evidence for the importance of Rab interaction for OCRL1 function(s).
Structural model of the interaction of OCRL1 and Rab8a at the membrane interface
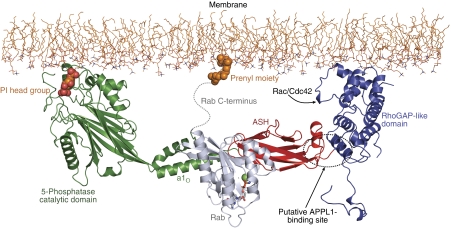
The Rab8a6–176:OCRL1540–678 complex structure allows us to expand the previous structural model of OCRL1 at the membrane interface (Erdmann et al, 2007) (Figure 6): Superimposition of the C-terminal α-helix of the inositol polyphosphate 5-phosphatase domain of the yeast homolog of synaptojanin (Tsujishita et al, 2001) with the helix α1O of OCRL1 from our complex structure together with a superimposition of the ASH domain of the ASH-RhoGAP (Erdmann et al, 2007) structure yields an approximate model of OCRL1. The possible relative orientation of this virtual construct towards a membrane is obtained from the position of inositol-1,4-bisphosphate bound to the inositol polyphosphate 5-phosphatase domain in the X-ray structure of the synaptojanin yeast homolog (Tsujishita et al, 2001). In this model, Rab8a is centrally located between the inositol polyphosphate 5-phosphatase and the RhoGAP domain. The nucleotide-binding pocket of Rab8a is pointing away from the membrane and faces the cytosol, whereas the structurally flexible C-terminus of the GTPase is oriented towards the lipid bilayer, thereby leading to undisturbed interaction of the single C-terminal geranylgeranyl moiety of Rab8a (Joberty et al, 1993) with the membrane. Since Switch I and Switch II are oriented towards the cytosol in our model, an interaction with additional Rab regulators and/or effectors is in principle possible, which could further modulate the Rab–OCRL1 interaction.
Figure 6.
Structural model of the interaction of OCRL1 and Rab8a at the membrane interface. The inositol-5-phosphatase domain (Tsujishita et al, 2001) together with the ASH and RhoGAP-like domains (Erdmann et al, 2007) are docked at the membrane interface as reported previously. The N-terminal region of OCRL1 is not included. The Rab protein binds centrally between the 5-phosphatase and RhoGAP domains, with the nucleotide-binding pocket (sticks, GppNHp; green sphere, Mg2+) facing towards the cytosol. In this model, binding of Rac/Cdc42 to the RhoGAP domain and APPL1 to OCRL1 (dashed circle) simultaneously with Rab8a is conceivable.
In this study, we have presented the first structure of the RBD of OCRL1 in complex with a Rab protein (Rab8a). Based on our results we have rationalized the structural consequences of known Lowe syndrome causing mutations leading to substitutions in the RBD. However, the identification of Lowe syndrome mutations is still ongoing, and recently 51 novel mutations were reported (Hichri et al, 2010). Our results will help to understand the functional consequences of newly identified mutations in the future. Finally, our data extend the current structural understanding of OCRL1 and support the idea that impairment of Rab binding contributes to the complex disease mechanism in Lowe syndrome.
Materials and methods
Protein expression and purification
Human Rab8a and OCRL1 genes were synthesized as codon-optimized DNA for expression in Escherichia coli (MR GENE, Regensburg, Germany). The truncated Rab8a6–176 was subcloned into a modified pET19 vector that contained an N-terminal hexa-histidine (His6) tag and a Tobacco Etch Virus (TEV) protease cleavage sequence. The expression and purification of Rab8a6–176 were performed as described for full-length Rab8a (Bleimling et al, 2009). OCRL1539–901 was cloned into a modified pMAL vector containing an N-terminal His6-maltose binding protein (MBP)-tag and TEV protease cleavage sequence. OCRL1540–678, OCRL1555–678, OCRL1560–678 and OCRL1555–668 were cloned by the Dortmund Protein Facility (http://www.mpi-dortmund.mpg.de/misc/dpf/) into a pOPINM-vector (N-terminal His6-MBP tag followed by a PreScission protease cleavage sequence) by the in-fusion cloning method (Berrow et al, 2007). OCRL1 constructs were expressed in Escherichia coli BL21 (DE3) overnight at 20°C after induction with 0.2 mM isopropyl-β-dithiogalactopyranoside (IPTG). Proteins were purified from bacterial lysate using nickel-nitrilotriacetic acid (Ni-NTA) column pre-equilibrated with buffer A (50 mM HEPES, pH 8.0, 500 mM NaCl, 2 mM β-mercaptoethanol) and eluted with a linear imidazole gradient (0–500 mM imidazole). Fractions containing the desired proteins were pooled, and the His6-MBP-tag of OCRL1 was cleaved off overnight at 4°C with TEV protease or PreScission protease, respectively. Uncleaved protein, His6-MBP and protease were removed via a Ni-NTA column, and the sample further purified by gel filtration over Superdex 75 column (GE Healthcare) pre-equilibrated with buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl and 2 mM dithioerythritol (DTE). Fractions containing OCRL1 fragments were pooled and concentrated. The Rab8a6–176:GppNHp:OCRL1540–678 complex was formed with 1.5-fold molar excess of Rab8a6–176, purified via a Superdex 75 16/60 column (GE Healthcare), and concentrated Rab8a6–176:GppNHp:OCRL1540–678 to 17 mg/ml.
Crystallization and structure determination
The crystallization screening of the Rab8a6–176:GppNHp:OCRL1540–678 complex was carried out with the JCSG Core I-IV (Qiagen), using the sitting-drop vapour diffusion method at 20°C. Diffraction-quality crystals of the complex were grown against a reservoir solution containing 20% (w/v) polyethylene glycol 4000, 20% (v/v) glycerol, 0.16 M ammonium sulphate and 0.1 M sodium acetate at pH 4.6 by the hanging-drop vapour diffusion method. Diffraction data were collected at beamline X10SA of the Swiss Light Source and processed with XDS for indexing and data reduction (Kabsch, 1993). The structure of the complex was solved by molecular replacement in Phaser (McCoy et al, 2007) with poly-alanine derived from the coordinates of the Rab5:GppNHp (PDB entry code 1R2Q (Terzyan et al, 2004)) and the partial structure of OCRL1 (PDB entry code 2QV2 (Erdmann et al, 2007)) as search models. The initial crystallographic model was improved with iterative cycles of manual building in Coot (Emsley et al, 2010) and refinement in Refmac5 (Murshudov et al, 1997). Data collection and refinement statistics are shown in Table II. Structural figures were prepared with the program PyMol (Delano, 2002).
Preparative nucleotide exchange
Rab GTPases were incubated with a 20-fold molar excess of guanine nucleotides (mantGppNHp or GppNHp) and five-fold molar excess of EDTA over MgCl2 for 2 h at 4°C. Unbound nucleotides were removed by NAP5 column (GE Healthcare) with the elution buffer consisting of 20 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 2 mM DTE, 10 μM nucleotide. The efficiency of nucleotide exchange was determined by the reversed phase HPLC using a C-18 column under isocratic conditions (running buffer 50 mM potassium phosphate (pH 6.6), 10 mM tributylammonium bromide, and 8 or 25% (V/V) acetonitrile for GppNHp or mantGppNHp, respectively).
Fluorescence measurements
All fluorescence measurements were carried out at 25°C in buffer containing 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM MgCl2 and 5 mM DTE. Fluorescence equilibrium titrations were performed with a Fluoromax-3 fluorescence spectrometer (Horiba Jobin Yvon). The binding of OCRL1 to mantGppNHp-loaded Rabs was monitored by the change in fluorescence polarization (excitation 365 nm, emission 440 nm). The association and dissociation kinetics of Rab loaded with mantGppNHp were measured with a stopped-flow apparatus (Applied Photophysics) using the fluorescence polarization change (excitation at 365 nm, emission with a 420-nm cutoff filter). To monitor the influence of the APPL1/Ses1 peptides on Rab8a:mantGppNHp:OCRL1 affinity, the KD of Rab8a:mantGppNHp with OCRL1 was determined as described above in the presence of 200 μM APPL1 peptide (sequence: SFQQRHESLRP) or 100 μM Ses1 peptide (sequence: PFARLHECYGQEI), respectively.
Analysis of fluorescence titrations
The concentration-dependent change in fluorescence polarization upon interaction of OCRL1 with Rab:mantGppNHp was fitted to a quadratic equation describing the binding isotherm of a reaction R+O=RO (R: Rab, O: OCRL1, RO: Rab:OCRL1 complex). The titration can be described by the following equation
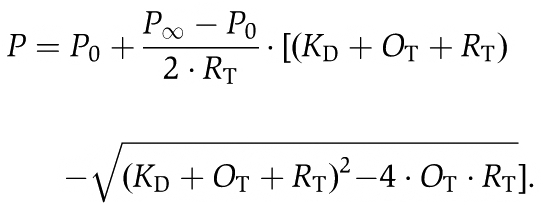 |
KD is the equilibrium dissociation constant, OT is the total (free and bound) concentration of OCRL1 at each titration step, RT is the total (free and bound) concentration of Rab:mantGppNHp in the cuvette, P is the fluorescence polarization. P0 and P∞ are the fluorescence polarization values at OT=0 and OT=∞, respectively.
Immunofluorescence microscopy
Hela cells were plated on glass coverslips and transfected with the indicated expression constructs using Fugene 6 transfection reagent. At 48 h after transfection, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 for 15 min at RT and blocked with 2% BSA for 30 min at RT. Cells were incubated with the following antibodies for 1 h at RT: mouse anti-Golgi 58K protein (Sigma-Aldrich, 1:100) or rabbit anti-c-myc (Santa Cruz, 1:40). After three washes with PBS, secondary Alexa Fluor 555-conjugated antibody (Molecular Probes, 1:400) was used for 1 h at RT. Cells were mounted on glass slides with Vectashield Mounting Medium (Axxora) and fluorescence was visualized with an Olympus inverted microscope.
Pull-down experiments and immunoblotting
Hela cells were lysed in lysis buffer (1 × PBS, 0.5% Triton X-100, protease inhibitors) and the cleared lysate was incubated with equal amounts of GST-fusion proteins bound to Glutathione Sepharose 4B (GE Healthcare) for 3 h at 4°C. After three washing steps with lysis buffer, bound proteins were eluted in 2 × Laemmli buffer for 10 min at 95°C and separated by SDS–polyacrylamide gel electrophoresis (SDS–PAGE). Proteins were transferred to nitrocellulose and incubated with mouse anti-Rab8a antibody (BD Biosciences, 1:1000) or goat anti-APPL1 antibody (Abcam, 1:2000) overnight at 4°C, followed by incubation with appropriate secondary HRP-conjugated antibodies. The blot was developed using the chemiluminescent Super Signal West Pico Chemilumiscent Substrate (Pierce).
Supplementary Material
Acknowledgments
We thank N Bleimling for invaluable technical assistance, the staff of Beamline X10SA at the Paul Scherrer Institute (Villingen, Switzerland), and the X-ray communities at the Max-Planck-Institute (MPI) Dortmund and the MPI Heidelberg (Germany). Sascha Gentz and Florian Seebeck are acknowledged for the APPL1 and Ses1 peptide synthesis. The Dortmund Protein Facility is acknowledged for assistance in cloning and protein expression/purification. This work was supported by a Grant from the Deutsche Forschungsgemeinschaft SFB642, projects A4 and A17 to RSG and KSE, respectively.
Author contributions: XH determined the Rab8:OCRL1 complex crystal structure, performed the kinetics and wrote the paper. NH performed immunofluorescence, GST pull downs and immune blot analysis. SS analysed OCRL1 truncations. WB supervised the crystallographic analysis. RSG wrote the manuscript. KSE and AI designed the experiments and wrote the manuscript. Structural coordinates have been deposited in the PDB with the accession code 3QBT.
Footnotes
The authors declare that they have no conflict of interest.
References
- Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL, Lewis RA, McInnes RR, Nussbaum RL (1992) The Lowe Oculocerebrorenal Syndrome Gene Encodes a Protein Highly Homologous to Inositol Polyphosphate-5-Phosphatase. Nature 358: 239–242 [DOI] [PubMed] [Google Scholar]
- Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res 35: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleimling N, Alexandrov K, Goody R, Itzen A (2009) Chaperone-assisted production of active human Rab8A GTPase in Escherichia coli. Protein Expr Purif 65: 190–195 [DOI] [PubMed] [Google Scholar]
- Burguete AS, Fenn TD, Brunger AT, Pfeffer SR (2008) Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell 132: 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavas LM, Ihara K, Kawasaki M, Torii S, Uejima T, Kato R, Izumi T, Wakatsuki S (2008) Elucidation of Rab27 recruitment by its effectors: structure of Rab27a bound to Exophilin4/Slp2-a. Structure 16: 1468–1477 [DOI] [PubMed] [Google Scholar]
- Choudhury R, Diao A, Zhang F, Eisenberg E, Saint-Pol A, Williams C, Konstantakopoulos A, Lucocq J, Johannes L, Rabouille C, Greene LE, Lowe M (2005) Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell 16: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M (2009) Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem 284: 9965–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon BG, Mukherjee D, Hanna CB, Riese DJ, Lowe M, Aguilar RC (2009) Lowe syndrome patient fibroblasts display Ocrl1-specific cell migration defects that cannot be rescued by the homologous Inpp5b phosphatase. Hum Mol Genet 18: 4478–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL (2002) The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC [Google Scholar]
- Delleman JW, Bleeker-Wagemakers EM, van Veelen AW (1977) Opacities of the lens indicating carrier status in the oculo-cerebro-renal (Lowe) syndrome. J Pediatr Ophthalmol 14: 205–212 [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Eathiraj S, Mishra A, Prekeris R, Lambright DG (2006) Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J Mol Biol 364: 121–135 [DOI] [PubMed] [Google Scholar]
- Eathiraj S, Pan X, Ritacco C, Lambright DG (2005) Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 436: 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P (2007) A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell 13: 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucherre A, Desbois P, Satre V, Lunardi J, Dorseuil O, Gacon G (2003) Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum Mol Genet 12: 2449–2456 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T (2008) Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics 7: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P (2006) Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA 103: 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri H, Rendu J, Monnier N, Coutton C, Dorseuil O, Poussou RV, Baujat G, Blanchard A, Nobili F, Ranchin B, Remesy M, Salomon R, Satre V, Lunardi J (2010) From Lowe syndrome to Dent disease: correlations between mutations of the OCRL1 gene and clinical and biochemical phenotypes. Hum Mutat (doi:10.1002/humu.21391) [DOI] [PubMed] [Google Scholar]
- Hoopes RR Jr, Shrimpton AE, Knohl SJ, Hueber P, Hoppe B, Matyus J, Simckes A, Tasic V, Toenshoff B, Suchy SF, Nussbaum RL, Scheinman SJ (2005) Dent disease with mutations in OCRL1. Am J Hum Genet 76: 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M (2006) Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J 25: 3750–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen A, Goody RS (2011) GTPases involved in vesicular trafficking: structures and mechanisms. Semin Cell Dev Biol 22: 48–56 [DOI] [PubMed] [Google Scholar]
- Jagoe WN, Lindsay AJ, Read RJ, Mccoy AJ, McCaffrey MW, Khan AR (2006) Crystal structure of Rab11 in complex with Rab11 family interacting protein 2. Structure 14: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Janne PA, Suchy SF, Bernard D, MacDonald M, Crawley J, Grinberg A, Wynshaw-Boris A, Westphal H, Nussbaum RL (1998) Functional overlap between murine Inpp5b and Ocrl1 may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J Clin Invest 101: 2042–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson AB, Majerus PW (1995) Properties of type-Ii inositol polyphosphate 5-phosphatase. J Biol Chem 270: 9370–9377 [DOI] [PubMed] [Google Scholar]
- Joberty G, Tavitian A, Zahraoui A (1993) Isoprenylation of Rab proteins possessing a C-terminal Caax Motif. FEBS Lett 330: 323–328 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kawasaki M, Nakayama K, Wakatsuki S (2005) Membrane recruitment of effector proteins by Arf and Rab GTPases. Curr Opin Struct Biol 15: 681–689 [DOI] [PubMed] [Google Scholar]
- Kenworthy L, Charnas L (1995) Evidence for a discrete behavioral phenotype in the oculocerebrorenal syndrome of Lowe. Am J Med Genet 59: 283–290 [DOI] [PubMed] [Google Scholar]
- Kleta R (2008) Fanconi or not Fanconi? Lowe syndrome revisited. Clin J Am Soc Nephrol 3: 1244–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S (2008) Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure 16: 1478–1490 [DOI] [PubMed] [Google Scholar]
- Lee MTG, Mishra A, Lambright DG (2009) Structural mechanisms for regulation of membrane traffic by Rab GTPases. Traffic 10: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M (2005) Structure and function of the Lowe syndrome protein OCRL1. Traffic 6: 711–719 [DOI] [PubMed] [Google Scholar]
- Mao YX, Balkin DM, Zoncu R, Erdmann KS, Tomasini L, Hu FH, Jin MM, Hodsdon ME, De Camilli P (2009) A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J 28: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Nelson WJ (2008) Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol 9: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merithew E, Hatherly S, Dumas JJ, Lawe DC, Heller-Harrison R, Lambright DG (2001) Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition. J Biol Chem 276: 13982–13988 [DOI] [PubMed] [Google Scholar]
- Mishra A, Eathiraj S, Corvera S, Lambright DG (2010) Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of early endosomal autoantigen 1 (EEA1). Proc Natl Acad Sci USA 107: 10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead AM, Jung JY, Smirnov A, Kaufer S, Scidmore MA (2010) Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect Immun 78: 1990–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Ooms LM, Horan KA, Rahman P, Seaton G, Gurung R, Kethesparan DS, Mitchell CA (2009) The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem J 419: 29–49 [DOI] [PubMed] [Google Scholar]
- Ostermeier C, Brunger AT (1999) Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell 96: 363–374 [DOI] [PubMed] [Google Scholar]
- Recacha R, Boulet A, Jollivet F, Monier S, Houdusse A, Goud B, Khan AR (2009) Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure 17: 21–30 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gabin AG, Ortiz E, Demoliner K, Si Q, Almazan G, Larocca JN (2010) Interaction of Rab31 and OCRL-1 in oligodendrocytes: its role in transport of Mannose 6-phosphate receptors. J Neurosci Res 88: 589–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N (2001) Ypt/Rab GTPases: regulators of protein trafficking. Sci STKE 2001: re11. [DOI] [PubMed] [Google Scholar]
- Shrimpton AE, Hoopes RR Jr, Knohl SJ, Hueber P, Reed AA, Christie PT, Igarashi T, Lee P, Lehman A, White C, Milford DV, Sanchez MR, Unwin R, Wrong OM, Thakker RV, Scheinman SJ (2009) OCRL1 mutations in Dent 2 patients suggest a mechanism for phenotypic variability. Nephron Physiol 112: p27–p36 [DOI] [PubMed] [Google Scholar]
- Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Swan LE, Tomasini L, Pirruccello M, Lunardi J, De Camilli P (2010) Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1. Proc Natl Acad Sci USA 107: 3511–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzyan S, Zhu GY, Li GP, Zhang XJC (2004) Refinement of the structure of human Rab5a GTPase domain at 1.05 angstrom resolution. Acta Crystallogr D Biol Crystallogr 60: 54–60 [DOI] [PubMed] [Google Scholar]
- Tsujishita Y, Guo SL, Stolz LE, York JD, Hurley JH (2001) Specificity determinants in phosphoinositide dephosphorylation: Crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell 105: 379–389 [DOI] [PubMed] [Google Scholar]
- Ungewickell A, Ward ME, Ungewickell E, Majerus PW (2004) The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc Natl Acad Sci USA 101: 13501–13506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsch B, Bokenkamp A, Benz MR, Besbas N, Dotsch J, Franke I, Frund S, Gok F, Hoppe B, Karle S, Kuwertz-Bröking E, Laube G, Neb M, Nuutinen M, Ozaltin F, Rascher W, Ring T, Tasic V, van Wijk JA, Ludwig M (2006) Novel OCRL1 mutations in patients with the phenotype of Dent disease. Am J Kidney Dis 48: 942.e1–942.e14 [DOI] [PubMed] [Google Scholar]
- Wu M, Wang T, Loh E, Hong W, Song H (2005) Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J 24: 1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GY, Zhai P, Liu J, Terzyan S, Li GP, Zhang XJC (2004) Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol 11: 975–983 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.