Abstract
RB is a key substrate of Cdks and an important regulator of the mammalian cell cycle. RB either represses E2Fs that promote cell proliferation or enhances the activity of cell-specific factors that promote differentiation, although the mechanism that facilitates this dual interaction is unclear. Here, we demonstrate that RB associates with and stabilizes pancreatic duodenal homeobox-1 (Pdx-1) that is essential for embryonic pancreas development and adult β-cell function. Interestingly, Pdx-1 utilizes a conserved RB-interaction motif (RIM) that is also present in E2Fs. Point mutations within the RIM reduce RB–Pdx-1 complex formation, destabilize Pdx-1 and promote its proteasomal degradation. Glucose regulates RB and Pdx-1 levels, RB/Pdx-1 complex formation and Pdx-1 degradation. RB occupies the promoters of β-cell-specific genes, and knockdown of RB results in reduced expression of Pdx-1 and its target genes. Further, RB-deficiency in vivo results in reduced pancreas size due to decreased proliferation of Pdx-1+ pancreatic progenitors, increased apoptosis and aberrant expression of regulators of pancreatic development. These results demonstrate an unanticipated regulatory mechanism for pancreatic development and β-cell function, which involves RB-mediated stabilization of the pancreas-specific transcription factor Pdx-1.
Keywords: Cdk, pancreas, Pdx-1, RB, stability
Introduction
In addition to its well-characterized role in regulating cell-cycle progression by restraining the E2F family of transcription factors, the Retinoblastoma protein (RB) performs an important albeit less studied function via regulating the differentiation program of diverse cell types (Khidr and Chen, 2006; van den Heuvel and Dyson, 2008). Cdk-mediated phosphorylation modifies association of RB with its myriad binding partners that include E2Fs, chromatin modifiers and cell-type-specific transcription factors (Morris and Dyson, 2001). Repression of E2F transcriptional activity is regarded as a signature role performed by RB and one that involves recruitment of chromatin modifying enzymes to the E2F target promoters. In addition to repressing E2F, RB appears to perform an equally complex, yet poorly understood, role in promoting cell differentiation by acting on cell and tissue-specific transcription factors. Intriguingly, in contrast to its repressive actions on E2F, RB appears to activate differentiation-specific transcription factors, such as Id2, the myogenic transcription factors MyoD and MEF2, the adipocyte differentiation transcription factor C/EBP, the osteogenic transcription factor Cbfa1/Runx2, the melanocyte transcription factor Mitf, the macrophage differentiation regulator PU.1, and the androgen and glucocorticoid nuclear receptors. Restraining deregulated E2F activity is regarded as an important tumour-suppressor function attributed to RB. In addition, by virtue of its role in activating cell-specific differentiation factors, it is equally plausible that the differentiation-promoting functions of RB are integral to its tumour-suppressor role (Burkhart and Sage, 2008). However, the mechanism by which RB activates differentiation-promoting factors in a cell-type-specific manner is poorly understood. Moreover, it is not entirely clear how RB can dually function as a transcription repressor of genes (such as E2Fs) and as an activator of differentiation-specific genes and we address this in the manuscript.
Previously, we and others reported that inactivating the Cdk4 locus in mice results in severe β-cell hypoplasia and insulin-deficient diabetes (Rane et al, 1999; Tsutsui et al, 1999). In contrast, mice that express an activating Cdk4R24C kinase (Cdk4R/R mice), that is refractory to inhibition by the CKI p16Ink4a, develop β-cell hyperplasia (Rane et al, 1999, 2002). While these studies emphasized the crucial role played by Cdk4 in regulating pancreatic β-cell mass, the underlying mechanisms remained unknown. Considering RB's role in promoting differentiation, we postulated that RB might regulate pancreas-specific factors and that Cdk4-mediated phosphorylation could modulate such interactions. Here, we report that RB associates with and stabilizes a key pancreas-specific transcription factor, pancreatic duodenal homeobox-1 (Pdx-1). Interestingly, we show that Pdx-1 and E2Fs share a common RB-binding motif and indeed they compete for RB interaction. Moreover, we demonstrate that RB occupies the promoters of pancreas-specific genes and ablation of RB in vivo leads to defects in early pancreatic development revealing an important regulatory role of RB in pancreas biology.
Results
Pdx-1 associates with RB via a conserved binding motif and competes with E2F for RB binding
The E2F transcription factors are the most extensively characterized RB-binding proteins (van den Heuvel and Dyson, 2008). The RB/E2F complex is cell cycle regulated wherein E2F associates strongly with hypophosphorylated forms of RB and demonstrates little affinity with hyperphosphorylated RB. Many RB-binding proteins feature a conserved LXCXE motif although not all proteins with LXCXE motifs bind RB (Morris and Dyson, 2001). E2F proteins lack an LXCXE motif; however, a conserved RB-interaction motif (RIM), with an amino-acid sequence YX7EX3DLF, is embedded in the transactivation domain of all E2F proteins (Helin et al, 1992; Shan et al, 1996). Sequence analysis of known pancreas-specific transcription factors (Kim and MacDonald, 2002; Jorgensen et al, 2007) revealed an amino-acid sequence YTRAQLLELEKEFLF in the Pdx-1 transcription factor that is essential for embryonic pancreas development and adult β-cell function (McKinnon and Docherty, 2001). Interestingly, the YTRAQLLELEKEFLF sequence in Pdx-1 is similar to the RIM sequence (YX7EX3DLF) present in E2F proteins (Helin et al, 1992; Shan et al, 1996) with high degree of sequence conservation in the core residues that confer RB binding (Figure 1A). These findings suggested a strong possibility of a RB/Pdx-1 interaction and we designed experiments to test this hypothesis. Co-immunoprecipitation analysis of HA-tagged RB and myc-tagged Pdx-1 showed evidence of RB/Pdx-1 interaction in Cos7 cells (Figure 1B). Western blot analyses showed that RB and Pdx-1 are abundantly expressed in β-cell lines, MIN6 and β-HC9 (Supplementary Figure S1) and co-immunoprecipitation experiments revealed endogenous RB/Pdx-1 complex formation in MIN6 (Figure 1C and D) and β-HC9 cells (Supplementary Figure S1).
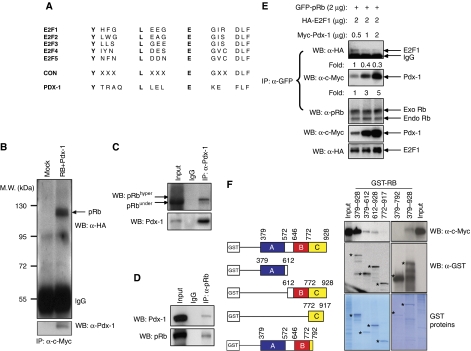
Figure 1.
Pdx-1 associates with RB. (A) Amino-acid sequence of the conserved RIM in E2F and Pdx-1 proteins. CON, consensus sequence. (B) Association of RB and Pdx-1 in Cos7 cells. Protein extracts from Cos7 cells either untransfected (Mock) or transfected with HA-RB and c-myc-Pdx-1 were immunoprecipitated with anti-c-myc antibodies followed by immunoblot with HA-antibody and Pdx-1 antibodies. (C, D) Protein extracts from MIN6 cells were immunoprecipitated with anti-Pdx-1 (C) or anti-RB (D) antibodies followed by immunoblot with anti-Pdx-1 or anti-RB antibodies to detect endogenous RB/Pdx-1 complexes. (E) Pdx-1 competes with E2F1 for RB binding. Cos7 cells were transfected with GFP-RB, HA-E2F1 and c-myc-Pdx-1. Cells were immunoprecipitated with GFP antibody followed by immunoblotting with antibodies against HA (E2F1), c-myc (Pdx-1) or RB (to detect exogenous GFP-RB and endogenous RB). Fold change in level of RB-associated protein is provided below respective lanes. (F) Pdx-1 associates with ‘large pocket’ (A+B+C domains) of RB. GST-tagged mutants of RB were expressed in Cos7 cells transfected with myc-tagged Pdx-1 followed by GST-pull down and immunoblot with anti-myc or anti-GST antibody. Coomasie blue-stained gel shows the expression levels of GST-RB proteins. Specific proteins are indicated with asterisks. Western blots were repeated multiple times and different exposures were taken to ensure linearity of the signal and fold changes were determined upon analyses of a representative experiment.
Considering the similar RIMs, we postulated that Pdx-1 and E2F may compete for RB binding and we examined this possibility utilizing equal concentration of RB and E2F1 and varying the levels of Pdx-1 in a binding assay. These analyses revealed that Pdx-1 and E2F1 indeed compete for RB binding as increasing levels of Pdx-1 displaced E2F1 from RB association (Figure 1E). RB contains several functional domains (Morris and Dyson, 2001). Domains A and B interact with each other along an extended inter-domain interface to form the central ‘small pocket’ of RB. The C-domain harbours Cdk-phosphorylation sites and along with domains A and B forms the ‘large pocket’ of RB. The ‘large pocket’ regulates E2F1 association, transcriptional repression, cell-cycle inhibition and RB's subcellular localization (Jiao et al, 2006). The ‘large pocket’ is critical to the tumour-suppressor function of RB and is disrupted by most naturally occurring germ-line mutations in hereditary retinoblastoma patients and by most tumour-derived mutations. Binding proteins require the A-domain (amino acids 379–572) and B-domain (amino acids 646–772) of RB for interaction (Morris and Dyson, 2001), and we performed assays in Cos7 cells using myc-tagged Pdx-1 and GST-tagged domain mutants of RB to determine the regions on RB that specify Pdx-1 binding. Co-immunoprecipitation analyses revealed a strong interaction between GST-RB (379–928; comprising the A+B+C domain large pocket) and Pdx-1 (Figure 1F). In comparison, relatively weak Pdx-1 binding was observed with the A-domain GST-RB (379–612) and the B+C domain GST-RB (612–928). Further, Pdx-1 failed to associate with the C-domain GST-RB (772–917) or the A+B domain GST-RB (379–792) of RB (Figure 1F). Taken together, these results suggest that the entire ‘large pocket’ of RB, comprising the A+B+C domains, is important for Pdx-1 interaction.
Pdx-1 interacts preferentially with phosphorylated forms of RB
Sixteen potential Cdk-phosphorylatable serine (Ser)/threonine (Thr) residues span the RB protein and we next examined the contribution of Cdk-mediated phosphorylation in RB/Pdx-1 interaction. Incubation of MIN6 cells with a small molecule Cdk4 inhibitor resulted in reduced phosphorylation of RB on Ser780, Ser807 and Ser811 (Figure 2A) and co-immunoprecipitation analyses revealed a reduction in RB/Pdx-1 complexes in MIN6 cells treated with the Cdk4 inhibitor (Figure 2B). Increased phosphorylation of RB on Ser780, Ser807 and Ser811 was observed in Cdk4R/R islets, compared with Cdk4WT islets (Figure 2C). Co-immunoprecipitation analyses revealed increased RB/Pdx-1 interaction in Cdk4R/R islets, compared with that observed in Cdk4WT islets (Figure 2D). Interestingly, Pdx-1 was unable to associate with underphosphorylated forms of RB when assayed using an antibody that specifically recognizes the underphosphorylated RB (Supplementary Figure S2). Co-immunoprecipitation assays in MIN6 cells showed that Pdx-1 efficiently associated with RB phosphorylated on Serine 780, but not with RB phosphorylated on Serine 807/811 (Figure 2E). To further examine the relative importance of the phospho-residues in RB/Pdx-1 complex formation, we engineered serine to alanine mutations on Ser780, Ser807 and Ser811 by site-directed mutagenesis. Co-immunoprecipitation assays using these mutants revealed a weak interaction of Pdx-1 with the RB-S780A (mouse S773A) phospho-mutant, but normal association with the RB-S807/811A (mouse S800/804A) mutant (Figure 2F), suggesting that the RB/Pdx-1 interaction is regulated by the Cdk4-phosphorylatable Ser780 residue of RB. We observe that the overall levels of RB are moderately reduced following Cdk4 inhibition (Figure 2A) or increased in the Cdk4R/R islets (Figure 2C), suggesting that, in addition to the effects due to phosphorylation, the RB–Pdx-1 interaction may be regulated by RB protein levels. Together, these results suggest that Pdx-1 preferentially interacts with phosphorylated forms of RB, particularly via the Cdk4-dependent phosphorylation of the serine 780 residue of RB.
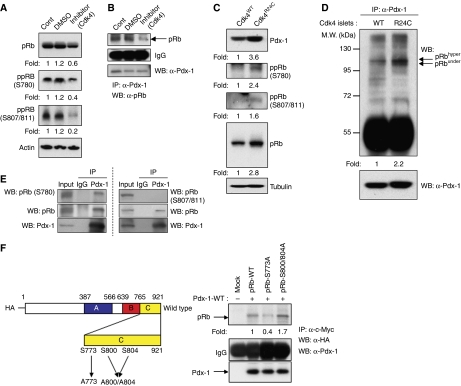
Figure 2.
Pdx-1 associates with phosphorylated RB. (A) MIN6 cells were left untreated or incubated with vehicle (DMSO) or a CDK4 inhibitor for 24 h followed by immunoblot analysis to detect total and phosphorylated forms of RB. Actin levels serve as protein loading control. (B) Protein lysate from MIN6 cells treated with the CDK4 inhibitor were immunoprecipitated with an anti-Pdx-1 antibody followed by western blots with RB and Pdx-1 antibodies. (C) Expression of Pdx-1, RB, RB-Ser780, RB-Ser807/811 and tubulin in Cdk4 wild-type (WT) and Cdk4R24C/R24C (R24C) islets. (D) Increased RB/Pdx-1 complexes in R24C islets, compared with WT islets, upon immunoprecipitation with anti-Pdx-1 antibodies followed by immunoblot with anti-RB antibodies. Position of under and hyperphosphorylated forms of RB are indicated with arrows. Levels of immunoprecipitated Pdx-1 protein are provided in the lower panel. (E) Protein lysate from MIN6 cells were immunoprecipitated with control IgG or an anti-Pdx-1 antibody followed by western blots with Pdx-1, total RB, RB-Ser780, RB-Ser807/811 antibodies. Input lanes serve as positive control. (F) HA-tagged site-directed RB mutants of the phospho-Ser773 and phospho-Ser800/804 residues were expressed in Cos7 cells along with c-myc-Pdx-1. Immunoprecipitation with anti-c-myc antibody was followed by immunoblot assays with anti-HA and anti-Pdx-1 antibodies. Western blots were repeated multiple times and different exposures were taken to ensure linearity of the signal and fold changes were determined upon analyses of a representative experiment.
RIM domain Pdx-1 mutants are unstable and degraded by the ubiquitin-proteasome pathway
We next conducted experiments to examine the importance of the RIM domain in the RB/Pdx-1 interaction. This motif is conserved in E2F proteins and facilitates RB/E2F complex formation (Helin et al, 1992) and E2F proteins with mutations in five amino-acid residues within the RIM show impaired RB/E2F complex formation (Shan et al, 1996). We therefore engineered similar mutations in the RIM domain of Pdx-1 by site-directed mutagenesis to generate site-specific Pdx-1 mutants (Y154H, L159P, E163K, L167P/F168L) and Pdx-1-M (comprising mutations at all five sites). Co-immunoprecipitation assays revealed weak, albeit variable, association of RB with the Pdx-1 mutants compared with the wild-type RB/Pdx-1 interaction (Figure 3A and B). Interestingly, western blot analyses revealed that the expression levels of Pdx-1 mutants were substantially reduced compared with the levels of wild-type Pdx-1 (Figure 3C). Although we made efforts to use equal amounts of Pdx-1 wild-type and mutant proteins in the co-immunoprecipitation assays (Figure 3A and B), it is plausible that the reduced affinity between RB and Pdx-1 mutants is, at least in part, attributable to the reduced expression of the mutant Pdx-1 proteins. Treatment with ubiquitin-proteasome pathway inhibitors, lactacystin or MG-132, restored Pdx-1 mutant protein levels, suggesting a role for proteasome-mediated degradation (Figure 3D and E). Further, mutant Pdx-1-M protein exhibited increased levels of ubiquitination that was further enhanced upon treatment with lactacystin (Figure 3F). Interestingly, the wild-type Pdx-1 protein also showed evidence of substantial ubiquitination after lactacystin treatment suggestive of ongoing ubiquitination of wild-type Pdx-1 protein.
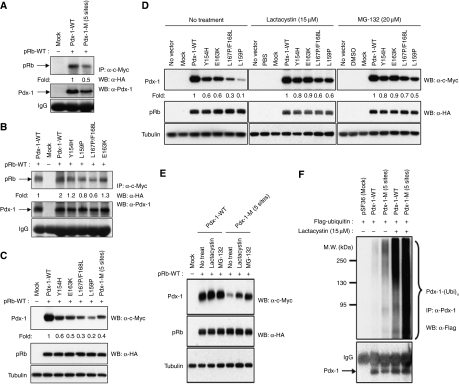
Figure 3.
RIM domain mutants of Pdx-1 are degraded via the ubiquitin-proteasome pathway. (A, B) c-Myc-tagged wild-type and RIM mutants of Pdx-1, (A) Pdx-1-M (mutations in five sites), and (B) Y154H, L159P, L167P/F168L and E163K, were transfected in Cos7 cells along with HA-RB followed by immunoprecipitation with anti-c-myc antibodies and immunoblot with anti-HA antibodies. Mock-transfected Cos7 cells are shown as controls and fold change of RB/Pdx-1 complex formation is indicated along with control IgG levels. Levels of Pdx-1 protein are provided. (C) Levels of expression of c-myc-tagged wild-type and RIM mutants of Pdx-1, Pdx-1-M, Y154H, L159P, L167P/F168L and E163K in Cos7 cells. (D, E) c-Myc-tagged wild-type and Pdx-1 RIM individual site-mutants Y154H, L159P, L167P/F168L and E163K (D) and the RIM five site-mutant Pdx-1-M (E), were transfected along with HA-RB in Cos7 cells that were left untreated or incubated with proteasome inhibitors, lactacystin or MG-132 for 6 h followed by immunoblot assays with anti-c-myc and anti-HA antibodies to detect expression of Pdx-1 and RB, respectively. (F) Cos7 cells transfected with empty vector (Mock) and wild-type Pdx-1 or the RIM mutant Pdx-1-M and Flag-tagged ubiquitin were either left untreated or incubated with lactacystin for 6 h followed by immunoprecipitation with anti-Pdx-1 antibodies and immunoblot with anti-Flag (to detect ubiquitinated Pdx-1) and anti-Pdx-1 antibodies (lower panel; arrow points to Pdx-1 under the IgG band). Note the increased ubiquitinated Pdx-1 in the last three lanes. Western blots were repeated multiple times and different exposures were taken to ensure linearity of the signal and fold changes were determined upon analyses of a representative experiment.
RB stabilizes Pdx-1 protein and regulates its half-life
We next examined the effect of RB on Pdx-1 protein half-life and stability. Treatment of MIN6 cells with siRNA that knockdown RB resulted in suppression of endogenous Pdx-1 protein level (Figure 4A). Further, infection of MIN6 cells and normal wild-type mouse islets with lentiviruses carrying RB shRNA (Supplementary Figure S3), resulted in reduced expression of Pdx-1 protein (Figure 4B). We then conducted experiments to measure the half-file of Pdx-1 protein. First, normal wild-type islets were infected with control lentiviruses or lentiviruses carrying RB shRNA in the presence or absence of the protein translation inhibitor, cycloheximide (CHX). We observed reduced Pdx-1 protein levels in islets infected with RB shRNA and Pdx-1 levels were further reduced upon CHX treatment (Figure 4C). Next, we examined the effect of the Cdk4 inhibitor on Pdx-1 protein half-life, which again showed a dramatically reduced Pdx-1 protein level in islets exposed to the Cdk4 inhibitor (Figure 4D). Finally, we examined the relative Pdx-1 half-life in islets isolated from Cdk4WT mice and Cdk4R/R mice. Reduced Pdx-1 protein levels were observed within 7 h of CHX treatment in Cdk4WT islets, whereas, Pdx-1 levels were comparatively maintained in response to CHX treatment in the Cdk4R/R islets (Figure 4E). Taken together, these results suggest that reduction of RB levels or Cdk4 inhibition reduces Pdx-1 protein half-life, and, in contrast, increased Cdk4-mediated phosphorylation of RB stabilizes Pdx-1 and increases its half-life.
Figure 4.
RB regulates half-life and stability of Pdx-1 protein. (A) Treatment of MIN6 cells with RB siRNA (+), but not control siRNA (−), results in reduced level of RB and Pdx-1 protein. Tubulin levels serve as control. (B) Infection of MIN6 or wild-type mouse islets with RB shRNA lentiviruses, but not control scrambled lentiviruses, results in reduced RB and Pdx-1 protein levels. (C) Wild-type islets were infected with control (−shRB) or RB shRNA (+shRB) for 72 h followed by no treatment (Cont), or incubation for 7 h with DMSO or cycloheximide (CHX). Islet protein lysates were immunoblotted with Pdx-1 and tubulin antibodies. (D) Wild-type islets were incubated with DMSO (−) or Cdk4 inhibitor (+). Islet protein lysates were immunoblotted with Pdx-1 and tubulin antibodies. (E) Cdk4WT or Cdk4R24C islets were incubated for the indicated hours with cycloheximide (CHX). Islet protein lysates were immunoblotted with Pdx-1 and tubulin antibodies. (F) Empty vector (Mock), wild-type Pdx-1 or the RIM mutant Pdx-1-M was transfected in Rb-deficient Saos-2 cells along with vector alone pcDNA3 or HA-RB followed by immunoblot with anti-c-myc, anti-HA and tubulin antibodies. (G) Rb-deficient Saos-2 cells were transfected with empty vector (Mock), wild-type Pdx-1 or the RIM mutant Pdx-1-M and Flag-tagged ubiquitin without (lanes 2–4) or with (lanes 5–7) HA-RB. Ubiquitinated Pdx-1 was assessed by immunoprecipitation with anti-Pdx-1 antibody followed by immunoblot with anti-Flag (to detect ubiquitinated Pdx-1) and anti-Pdx-1 antibodies (lower panel; arrow points to Pdx-1 under the IgG band) to detect levels of Pdx-1.
To directly probe RB's role in the ubiquitination process, wild-type and Pdx-1-M were transfected along with HA-tagged RB in the RB-null Saos-2 cell line. In the absence of RB, Pdx-1-M was undetectable in Saos-2 cells, whereas, RB transfection moderately restored Pdx-1-M expression (Figure 4F). Interestingly, transfection of HA-RB also increased levels of wild-type Pdx-1, which suggests that RB stabilizes both the wild-type and mutant Pdx-1 proteins. Moreover, in the absence of RB we observed substantial ubiquitinated Pdx-1-M even though Pdx-1 protein was low or undetectable indicative of a rapid turnover of Pdx-1-M protein (Figure 4G). Restoration of RB expression resulted in higher levels of wild-type and mutant Pdx-1 proteins (Figure 4G). Interestingly, wild-type and mutant Pdx-1 proteins remained ubiquitinated upon RB addition, which suggests that although RB may allow ubiquitination it precludes ubiquitinated Pdx-1 proteins from degradation. Further, it is interesting that although it interacts weakly with RB (Figure 3A), the Pdx-1-M mutant gets stabilized by RB which is suggestive of alternate points of contact between RB and Pdx-1 (see below).
RB–Pdx-1 interaction is regulated by glucose levels
Glucose regulates β-cell function and we next examined the effects of glucose levels on RB/Pdx-1 interaction. First, we examined relative expression levels of RB and Pdx-1 proteins in MIN6 cells during conditions of glucose stimulation and glucose withdrawal (Figure 5A). For the glucose-stimulation experiment, MIN6 cells were cultured for 24 h without glucose and then stimulated by medium containing 25 mM glucose. For the glucose-withdrawal condition, MIN6 cells were cultured in glucose-containing medium for 24 h, followed by culture in glucose-free medium for 24 h. Levels of RB and Pdx-1 proteins were assayed by western blots at various time points. We observed that RB protein levels increased under glucose-removal conditions, whereas RB protein levels reduced when MIN6 cells were stimulated by glucose (Figure 5B). Interestingly, we observed a similar expression pattern for Pdx-1 proteins upon glucose withdrawal or addition (Figure 5B) Consistent with this, we detected increased levels of ubiquitinated Pdx-1 in the presence of high glucose (Figure 5C and D). These results suggested that high-glucose levels destabilize RB/Pdx-1 complexes, and we next examined this possibility. We cultured MIN6 cells in glucose-free media for 16 h followed by incubation in low (3 mM) or high (25 mM) glucose-containing medium for 2 or 24 h (Figure 5E). For the 2-h glucose-stimulation condition, co-immunoprecipitation analyses revealed RB/Pdx-1 interaction in cells deprived of glucose or those cultured in 3 mM glucose, whereas a weak RB/Pdx-1 interaction was observed in cells stimulated with 25 mM glucose (Figure 5F). Interestingly, culture for 24 h in either 3 or 25 mM glucose resulted in reduced RB/Pdx-1 interaction (Figure 5G). Of note, in both the 2- and 24-h experimental conditions, Pdx-1 was observed to associate with the slower migrating phosphorylated form of RB and not with underphosphorylated RB (Figure 5F and G). Taken together, these observations indicate that glucose regulates the levels of RB and Pdx-1 proteins and consequently the degree of RB/Pdx-1 interaction.
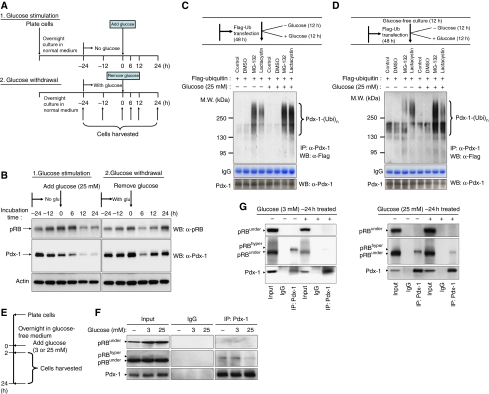
Figure 5.
Glucose regulates RB/Pdx-1 interaction. (A) Schematic of glucose-stimulation/withdrawal experiment (see Materials and methods section). (B) Protein lysate from cells harvested in the glucose-addition/withdrawal experiment (A) was subjected to immunoblot analyses using antibodies against RB, Pdx-1 and actin (control). (C) MIN6 cells were transfected with Flag-tagged ubiquitin for 48 h in normal glucose-containing media and subsequently cultured for an additional 12 h in glucose-free or glucose-containing media. (D) MIN6 cells were transfected with Flag-tagged ubiquitin for 48 h in normal glucose-containing media followed by culture in glucose-free media for 12 h and subsequently incubated for an additional 12 h in glucose-free or glucose-containing media. For both (C, D), prior to harvesting, cells were incubated with proteasome inhibitors MG-132 or lactacystin for 6 h. Endogenous Pdx-1 was immunoprecipitated using anti-Pdx-1 antibodies followed by immunoblot with anti-Flag antibodies (to detect ubiquitinated Pdx-1) and Pdx-1 antibodies. (E–G) MIN6 cells were deprived of glucose for 16 h followed by stimulation with 3 or 25 mM glucose for 2 h (F) and 24 h (G) followed by immunoprecipitation with control IgG or anti-Pdx-1 antibodies and immunoblot with anti-total RB, anti-underphosphorylated RB or anti-Pdx-1 antibodies.
RB occupies β-cell-specific gene promoters and regulates expression of Pdx-1-target genes
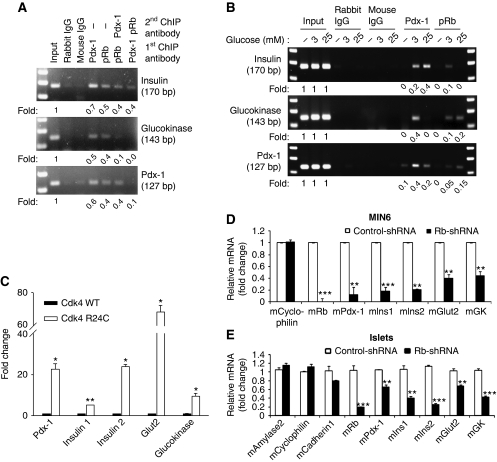
β-cell-specific inactivation of Pdx-1 results in age-dependent loss of β-cell function and diabetes (Ahlgren et al, 1996; Brissova et al, 2005). Pdx-1 regulates adult β-cell function by binding and transactivating the promoters of key β-cell-specific genes, specifically, Insulin, Glucokinase and Pdx-1 itself (McKinnon and Docherty, 2001). Chromatin immunoprecipitation (ChIP) assays revealed that RB and Pdx-1 occupy the Insulin, Glucokinase and Pdx-1 promoters in MIN6 cells (Figure 6A). Whereas these results showed that RB, like Pdx-1, could occupy promoters of Pdx-1-target genes, they do not confirm the existence of promoter-bound RB/Pdx-1 complexes. To address this possibility, we performed sequential ChIP experiments in which immunoprecipitated RB–chromatin or Pdx-1–chromatin complexes were eluted and then subjected to a second round of immunoprecipitation with either Pdx-1 or RB antibodies, respectively. Analyses of eluates from the sequential ChIP provided evidence of RB/Pdx-1 complexes on the Insulin, Glucokinase and Pdx-1 promoters (Figure 6A). Together, these results unequivocally demonstrated that RB can occupy key β-cell-specific gene promoters, either individually or in complex with Pdx-1. For assaying the functional consequence of RB binding, we conducted ChIP assays using cells that were cultured in glucose-free media and subsequently stimulated with low-glucose (3 mM) or high-glucose (25 mM) concentration. Minimal RB and Pdx-1 binding was seen in glucose-deprived cells, whereas glucose stimulation promoted variable and promoter-dependent DNA binding of RB and Pdx-1 on the insulin, glucokinase and Pdx-1 promoters (Figure 6B).
Figure 6.
RB regulates expression of Pdx-1 and its target genes. (A) ChIP assays in MIN6 cells show evidence of Pdx-1 and RB binding to the promoters of Pdx-1-target genes, insulin, glucokinase and Pdx-1 itself. Sequential ChIP assays demonstrate binding of RB/Pdx-1 complexes to the three promoters, albeit at varying levels. (B) ChIP assays done in glucose-deprived (−) or 3 or 25 mM glucose-treated MIN6 cells show binding of RB and Pdx-1. Fold change in promoter binding compared with input is shown and immunoprecipitation with rabbit IgG and mouse IgG serves as negative control for the ChIP assay. (C) mRNA expression of Pdx-1-target genes is elevated in Cdk4R24C/R24C (R24C) islets, compared with Cdk4 wild-type (WT) islets. (D, E) RB-shRNA (closed bars), but not control shRNA (open bars), infection in MIN6 cells (D) and wild-type mouse islets (E) represses Rb mRNA and β-cell-specific Pdx-1-target genes, whereas, non-specific genes such as Cyclophilin, amylase2, Cadherin1 are not affected. Western blots were repeated multiple times and different exposures were taken to ensure linearity of the signal and fold changes were determined upon analyses of a representative experiment. Graphs represent real-time PCR assays that were repeated multiple times. *P<0.05; **P<0.001; and ***P<0.0001.
Expression of Pdx-1 and its target genes, insulin 1, insulin 2, glut2 and glucokinase was significantly upregulated in Cdk4R/R islets compared with Cdk4WT islets (Figure 6C). In agreement, RB induced activation of a luciferase-linked insulin reporter, whereas a phospho-RB mutant was unable to activate the insulin reporter (Supplementary Figure S4). Further, infection of MIN6 cells and normal wild-type mouse islets with lentiviruses carrying RB shRNA resulted in reduced expression of Pdx-1-target genes, pdx-1, insulin1, insulin2, glut2 and glucokinase (Figure 6D and E). Thus, we find that RB occupies the promoters of Pdx-1-target genes and regulates their expression.
Abnormal pancreas development in Rb−/− embryos
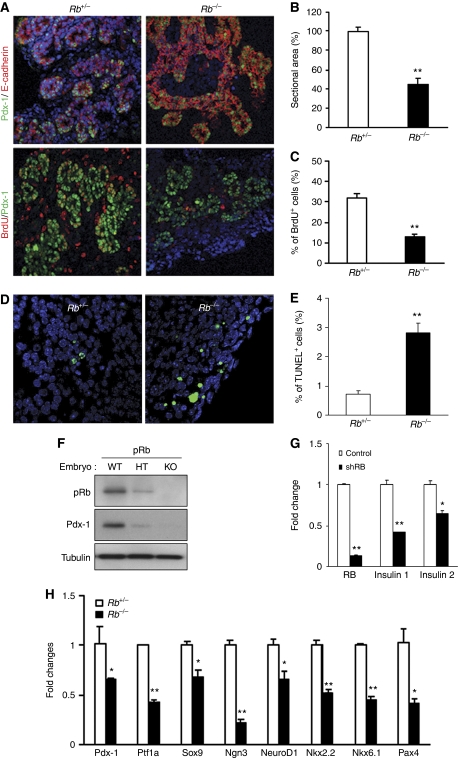
Pdx-1 has a critical role during early pancreatic development and is essential for pancreatic outgrowth (Jonsson et al, 1994; Ahlgren et al, 1996; Offield et al, 1996; Holland et al, 2002). As our results suggested an important role for RB in regulating Pdx-1 stability, we next inquired whether RB is essential during pancreas development. RB deficiency results in embryonic lethality such that Rb−/− embryos die by embryonic day E16 due to defects in neurogenesis and hematopoiesis (Clarke et al, 1992; Jacks et al, 1992; Lee et al, 1992). Mouse pancreas begins to form by E8.5, co-incident with the appearance of Pdx-1+ cells, followed at E12.5 by a secondary transition where the pancreatic epithelium undergoes differentiation to the endocrine, exocrine or ductal lineages (Kim and MacDonald, 2002). Analysis of the E13.5 pancreas revealed significantly reduced pancreas tissue in Rb−/− embryos, compared with Rb+/− embryos (Figure 7A and B), and significantly reduced numbers of proliferating Pdx-1+ pancreatic progenitor cells (Figure 7C). RB is a critical regulator of the cell cycle and regulates cell apoptosis by virtue of its interactions with E2F transcription factors (Harbour and Dean, 2000; Dick and Dyson, 2003). Examination of embryonic pancreas tissue by TUNEL immunofluorescence assays revealed elevated numbers of TUNEL+ cells in the Rb−/− pancreas (Figure 7D and E), suggestive of increased cell death as a result of RB deficiency.
Figure 7.
Rb−/− embryos exhibit defects in pancreas development. (A) Immunofluorescence analyses of pancreas tissue sections from E13.5 Rb+/− and Rb−/− mice. Sections shown in top panel were stained with antibodies against Pdx-1 (green) and the epithelial cell marker, E-cadherin (red). In the bottom panel, sections from BrdU incorporating pancreas with stained with antibodies against Pdx-1 (green) and BrdU (red) to detect proliferating Pdx-1+:BrdU+ double-positive cells. DAPI staining identifies nuclei. (B) Pancreatic sectional area identified by Pdx-1+ cells as shown in (A), was quantified in E13.5 Rb+/− and Rb−/− mice. (C) BrdU-labelled Pdx-1+ proliferating cells were quantified in E13.5 Rb+/− and Rb−/− mice. (D, E) TUNEL staining was performed in Rb+/− and Rb−/− embryonic pancreas at E13.5. The loss of RB in early pancreas induces apoptosis compared with control. (E) The percentages of TUNEL+ cells (green) are to total number of Pdx-1+ cells. In all sections, DAPI identifies nuclei. **P<0.001. (F) Relative Pdx-1 protein expression of E12.5 Rb+/+, Rb+/− and Rb−/− embryonic pancreas. Results are shown relative to tubulin and RB protein expression. (G) Relative levels of gene expression of Rb, Insulin1 and Insulin2 were examined in control (open bars) and shRb (closed bars) lentiviruses infected normal wild-type embryonic pancreas rudiment cells. Results are normalized with 18S expression and are shown as relative fold change compared with control cells. *P<0.05 and **P<0.001. (H) Relative levels of gene expression of pancreatic developmental markers were examined in E11.5 Rb+/− and Rb−/− pancreas. Results are shown relative to 18S expression. Graphs represent experiments (real-time PCR, BrdU and TUNEL assays) that were repeated multiple times. *P<0.05 and **P<0.001.
Consistent with the data in this manuscript, suggestive of RB-mediated stabilization of Pdx-1 protein, we observed dramatically reduced Pdx-1 protein levels in Rb−/− embryonic pancreas, when compared with age-matched Rb+/− pancreas or Rb+/+ pancreas (Figure 7F). It is plausible that the observed defects in pancreas development could be due to the placental deficits caused by RB deletion in the extra-embryonic trophoblast stem cells (Wu et al, 2003; Wenzel et al, 2007). Placental rescue of RB mutant mice resulted in a lack of obvious defects in neurogenesis and erythropoiesis—two main reasons for embryonic lethality in Rb−/− fetuses (Clarke et al, 1992; Jacks et al, 1992; Lee et al, 1992). However, the placental-rescued Rb mutant mice did exhibit marked defects in other tissues such as the lens and skeletal muscle (Wu et al, 2003). It thus appears that RB has a cell-autonomous and non-cell-autonomous role depending on cell and tissue type. Effect of RB loss on the pancreas, with or without normal placental compensation, was not addressed in either the original (Clarke et al, 1992; Jacks et al, 1992; Lee et al, 1992) or the subsequent placental rescue studies (Wu et al, 2003; Wenzel et al, 2007). To examine the effects of RB loss on pancreatic development without the placental contribution, we established a pancreatic rudiment ex vivo culture system. To this end, we infected pancreatic rudiments established from wild-type E14.5 embryonic pancreas with GFP-tagged RB shRNA carrying lentiviruses or with control GFP lentiviruses (Supplementary Figure S5). The rudiments were analysed after 3 days when we observed a significant downregulation of the Pdx-1-target genes, insulin1 and insulin2 (Figure 7G). The data support the notion that RB loss leads to suppression of an important Pdx-1-target gene, insulin, which is critical for normal β-cell function. Further, as shown in Figure 7H, E11.5 Rb−/− pancreas, compared with E11.5 Rb+/− pancreas, displayed significantly reduced expression of genes associated with early pancreas development (Kim and MacDonald, 2002; Jorgensen et al, 2007) such as Pdx-1, Ptf1a, Sox9, Ngn3, NeuroD1, Nkx2.2, Nkx6.1 and Pax4. Taken together, these studies suggest that RB is required during pancreas development, specifically, for optimal proliferation of Pdx-1+ pancreatic progenitors, suppressing apoptosis and subsequently for mature β-cell function.
Discussion
In addition to its well-characterized role in controlling cell-cycle progression via its repressive effects on E2F transcription factors, RB has an important role in regulating cell differentiation by controlling cell-type-specific transcription factors. However, the precise mechanism that facilitates RB's dual interaction with cell proliferation factors such as E2Fs and the cell-type-specific transcription factors that promote differentiation has been obscure. We show here that RB associates with Pdx-1, a transcription factor that is essential for pancreas development and islet β-cell function, via a conserved interacting motif that is present in E2F proteins (Figure 8). The fact that this mechanism involves direct association with a cell-type-specific transcription factor via a motif that is integral to RB's association with E2Fs is of interest to understanding RB-dependent proliferation and differentiation. Importantly, interaction with RB appears to be crucial to Pdx-1's stability as loss of RB association results in reduced half-life of Pdx-1 protein and its degradation via the ubiquitin-proteasome pathway. Moreover, RB-deficiency in vivo results in pancreas developmental defects due to reduced proliferation of Pdx-1+ pancreatic progenitors, combined with increased apoptosis and reduced expression of key pancreatic developmental genes.
Figure 8.
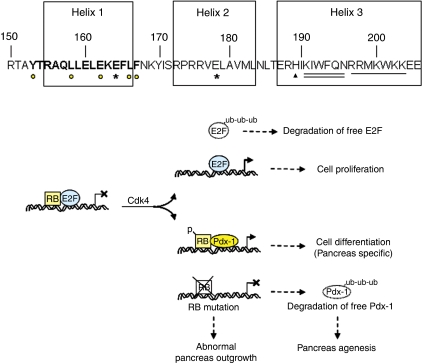
Proposed model for RB/Pdx-1 interaction. Schematic of the Pdx-1 homeodomain with the triple helices. Helix 1 houses the putative RIM (bold letters) with the five critical residues identified (yellow circles under the letters). The histidine residue (triangle symbol; aa 189) and the KIWFQN motif (double underline) required for DNA binding and the nuclear localization signal (underline) is in Helix 3. Mutation of two amino acids in Helices 1 and 2 (E164 and E178; asterisks) that lead to an unstable Pdx-1 protein are involved in human pancreas agenesis. Model for RB-mediated stabilization of Pdx-1. Underphosphorylated RB associates with E2F and precludes activation of E2F-target genes. Cdk4-mediated phosphorylation of RB triggers release of E2F and free E2F gets ubiquitinated (ub) and degraded by the ubiquitin-proteasome pathway. Phosphorylated RB associates with Pdx-1 and participates in activation of pancreas-specific genes to promote pancreas differentiation. Mutation of RB results in impaired pancreas outgrowth. Also, loss of RB binding destabilizes Pdx-1 that gets degraded by the ubiquitin-proteasome pathway. Pdx-1, with mutations in the RIM, is unstable and associated with human pancreas agenesis.
Previously, we described the important role of Cdk4 in the regulation of β-cell mass and β-cell regeneration (Rane et al, 1999, 2002; Rane and Reddy, 2000; Mettus and Rane, 2003; Lee et al, 2010). The results described here provide a molecular mechanism underlying the unique role played by Cdk4 and RB in pancreas biology. We demonstrate that Cdk4-mediated phosphorylation of RB regulates its association with Pdx-1. It appears that Pdx-1 associates with hyperphosphorylated RB with the Cdk4-specific RB-Ser780 residue playing an important role in facilitating the RB/Pdx-1 interaction. In contrast, E2F preferentially associates with underphosphorylated RB and phosphorylation of RB-Ser780 leads to dissociation of the RB/E2F1 complex (van den Heuvel and Dyson, 2008). Pdx-1 and E2Fs lack the LXCXE motif present in many RB-binding proteins. Instead, RB/Pdx-1 complex formation, similar to the RB/E2F association, depends on the interaction between a conserved RIM domain on the E2F and Pdx-1 transcription factors and RB. Further, Pdx-1 and E2F1 compete for RB binding and Pdx-1 can displace RB-bound E2F1. The large pocket of RB (A+B+C domains comprising amino acids 379–928) is sufficient for stable interaction with E2F (Qin et al, 1992). In addition, it was more recently shown that RB has an alternate binding region for E2F1 in its C-domain (Dick and Dyson, 2003). Thus, RB interacts with E2Fs via at least two distinct domains: one spanning the entire ‘large pocket’ and the other one in the C-domain. Crystal structural studies confirmed that the RIM domain of E2F contacts directly to amino acids in both the A and B domains of the ‘small pocket’ of RB (Lee et al, 2002; Xiao et al, 2003). Interestingly, the C-domain site of RB contacts regions on E2F distinct from the RIM domain (Dick and Dyson, 2003). It is plausible that the RB/Pdx-1 interaction, similar to the RB/E2F-1 association, also utilizes multiple binding regions, both on Pdx-1 and on RB. Indeed, we find that the ‘small pocket’ (A+B) region and C-domain are both necessary, but not sufficient for Pdx-1 interaction; instead, the entire ‘large pocket’, comprising the A+B+C domains, is required for Pdx-1 association.
Mutations in the RIM significantly reduces although does not completely abolish RB/Pdx-1 interaction and it is possible that Pdx-1 interacts with RB in at least one other location. Interestingly, residual RB/E2F association is also maintained despite mutations in the RIM on E2F perhaps due to RB's association with the ‘marked-box’ region on E2F protein (O’Connor and Hearing, 1994). Similar to the RB–E2F interaction, we believe that the RB–Pdx-1 interaction is also regulated by multiple interactions with the RB pocket and phosphorylation sites on RB. Phosphorylation of RB regulates its binding to DNA-bound E2F and consequently the E2F-mediated promoter repression (Brown et al, 1999). It is also known that binding to free E2F is regulated by dual mechanisms involving phosphorylation either at a number of C-terminal sites or at sites 8 and 9 in the insert domain of RB protein (Knudsen and Wang, 1997). Further, phosphorylation of RB by distinct cyclin–Cdk complexes initiates sequential intramolecular interactions between the C-terminal region and the pocket domain of RB that results in a progressive change in RB's association with its interacting proteins and consequently a change in RB function (Lundberg and Weinberg, 1998; Harbour et al, 1999; Xiao et al, 2003). It is plausible that disruption of these intramolecular interactions opens up binding sites for other proteins (like Pdx-1), with affinity for phosphorylated forms of RB. Importantly, interaction of the core binding fragment of E2F with RB is inhibited by cyclin–Cdk phosphorylation (Xiao et al, 2003). We infer that Cdk4-mediated phosphorylation of RB-S780A (mouse S773A) modulates RB/Pdx-1 association. However, the RB-S780A (mouse S773A) mutation does not completely eliminate RB/Pdx-1 interaction supporting the reliance on the A+B domain for binding. Taken together, these studies further confirm that the A+B domain (small pocket) and the C-domain (that houses the critical phospho-serine 780 residue) of RB are required for complete Pdx-1 binding.
We show that glucose levels regulate the ability of RB to occupy the promoter of Pdx-1 and of its target genes. In addition, we show that glucose regulates the RB/Pdx-1 interaction, with high glucose able to dissociate the RB/Pdx-1 complex. Further, we find that high-glucose levels promote Pdx-1 ubiquitination and also reduce levels of Pdx-1 and RB proteins. It is worth noting that, the effect of glucose on Pdx-1 in our study is at odds with published literature in the same MIN6 cell line in which Pdx-1 half-life is acutely stabilized under high-glucose conditions and destabilized under low-glucose conditions (Humphrey et al, 2010). The different findings could be a consequence of differences in the experimental design. We examined the half-life of endogenous Pdx-1 and its glucose-dependent interaction with endogenous RB using MIN6 cells. The study by Humphrey et al used an overexpression system to monitor the effects of GSK and AKT kinases on exogenous Pdx-1 levels in MIN6 cells transfected with cytomegalovirus promoter-driven expression vectors encoding HA-tagged wild-type and mutant Pdx-1. Further, our glucose-stimulation condition also differs from the study by Humphrey et al. We starved MIN6 cells in glucose-free media for 24 h, primarily to ensure that MIN6 cells are in a synchronized cell-cycle state in consideration of the role of RB and Cdk4 in cell-cycle progression. In addition, the purpose of preincubating the cells under glucose-starvation conditions was to optimize the opportunity to observe the effects of glucose addition on the Pdx-1 levels, and RB/Pdx-1 interactions.
The overlapping nature of critical RB association sequences with putative Pdx-1 ubiquitination targeting domains and/or proteasome-mediated degradation regions is consistent with a simple occlusion model for inhibiting Pdx-1 degradation, whereby RB blocks the ubiquitination apparatus and/or the proteasome machinery from either recognizing Pdx-1 or precluding its degradation (Figure 8). We believe that the absence of RB allows ubiquitination and subsequent degradation of the ubiquitinated Pdx-1 protein. However, when RB is present, Pdx-1 although ubiquitinated, does not get degraded in the proteasomal system. This is similar to adding a proteasomal inhibitor to the assay, when ubiquitinated forms are observed. We infer that RB interaction protects Pdx-1 from getting degraded, although it may not necessarily protect it from getting ubiquitinated. Recently, the Stoffers’ group elucidated that the Pcif1/SPOP protein interacts with Pdx-1 and that the Pcif1/SPOP–Cul3 complex targets Pdx-1 protein for ubiquitination and proteasomal degradation (Claiborn et al, 2010). It remains to be seen whether the RB/Pdx-1 interaction and the effects on Pdx-1 stability, that we report here, modify the ability of Pcif1/SPOP–Cul3 complexes to target Pdx-1.
Interestingly, RIM-mediated interaction with RB protects E2F1 from getting degraded by the ubiquitin-proteasome pathway (Hofmann et al, 1996; Campanero and Flemington, 1997), suggesting that proteins that interact with RB using the RIM domain may do so to maintain protein stability. Whether other cell-type-specific factors that interact with RB (i) have the RIM sequence, (ii) compete with E2Fs for RB binding and (iii) utilize the RB association to protect from degradation is not known. Pdx-1 has a characteristic triple-helical homeodomain that is involved in DNA-binding, protein–protein interaction and nuclear localization (McKinnon and Docherty, 2001). Interestingly, the RIM that controls Pdx-1 stability is embedded within Helix 1 of the homeodomain (Figure 8). Thus, the RB-binding site and the sequences that participate in homeodomain functions and self-degradation of Pdx-1 are vicinal to each other, suggesting steric shielding by RB of Pdx-1 stability and function. A highly conserved histidine at position 189 that is unique to the Pdx-1 homeodomain and a KIWFQN motif within Helix 3 appear to be important in DNA binding (Lu et al, 1996). In addition, the nuclear localization signal RRMKWKK is also housed in Helix 3. These observations are consistent with a particularly important role for Helix 3 in DNA binding (Figure 8). The RIM domain in Helix 1 is well separated from the DNA-binding functions of Helix 3 that could potentially enable Pdx-1 to bind DNA and simultaneously interact with RB.
PDX-1 mutations are associated with early-onset (MODY4; pro63fsdelC) and late-onset forms of type 2 diabetes (Stoffers et al, 1997a, 1997b; Hani et al, 1999; Macfarlane et al, 1999). Interestingly, two mutations within the PDX-1 homeodomain are associated with human pancreas agenesis (Schwitzgebel et al, 2003). Importantly, the E164D mutation is embedded within the RIM that controls Pdx-1 stability (Figure 8), and, moreover, both the E164D and E178K mutations significantly decrease the mutant Pdx-1 protein's half-life (Schwitzgebel et al, 2003). In addition, the E178K mutation is also recently implicated in neonatal diabetes (Nicolino et al, 2010). Whether, these mutations interfere with the ability of Pdx-1 to bind RB remains to be determined. Further, optimal Pdx-1 levels are important for proper pancreas formation (Fujitani et al, 2006), suggesting that mechanisms that control Pdx-1 expression and its stability are vital during pancreas development. We illustrate that complete RB deficiency leads to defects in pancreas outgrowth and differentiation perhaps due to reduced proliferation of Pdx-1+ progenitors and suppression of genes associated with pancreas development.
In addition, we observe that RB deficiency results in increased apoptosis in the RB knockout embryonic pancreas. These observations, taken together with the reduced proliferation of Pdx-1+ pancreas progenitors, allows us to conclude that the reduced embryonic pancreas size in the RB knockout embryonic pancreas is due to a combination of limited proliferation of pancreas progenitors and increased apoptosis. However, it is unclear whether the two processes, defective proliferation and increased apoptosis, are both exclusively due to the unstable Pdx-1, or, at least in part, a result of a more general cell cycle role played by RB. Interestingly, Pdx-1 deficiency enhances β-cell susceptibility to ER stress-associated apoptosis (Sachdeva et al, 2009), and it will be important to inquire whether this phenomenon is dependent on RB. Interestingly, β-cell-specific deletion of RB, using a rat-insulin promoter Cre-mediated excision, results in minimal defects in β-cell mass and function most likely due to the inactivation of RB later during pancreas development (Vasavada et al, 2007), although RB and its related family member p130 regulate the G1/S transition in pancreatic β-cells (Harb et al, 2009). These results suggest that the timing of RB expression (before and/or during the secondary transition) or its expression in pancreatic cell types other than β-cells is important for optimal pancreas development. It remains to be determined whether mutations in RB predispose to pancreas agenesis or diabetes in humans. In conclusion, these results strongly suggest that RB has an important role in pancreas development and in β-cell function by physically interacting with and stabilizing Pdx-1 thereby precluding its degradation via the ubiquitin-proteasome machinery. Moreover, this study elucidates a unique yet simple mechanism utilized by RB to dually control factors that promote cell proliferation and cell differentiation.
Materials and methods
Cell culture, mouse islet preparation and reagents
MIN6, β-HC9, Cos7 cells and 293ET cells were cultured in DMEM/10% FBS. RB-deficient Saos-2 cells were maintained in RPMI 1640/10% FBS. MG-132 and lactacystin were from Peptide international, Inc. Cdk4 inhibitor II was from Calbiochem and final concentration used was 0.4 μM. Mouse islets were prepared as described previously (Lin et al, 2009). Following ductal injection of collagenase, the pancreas was digested. Islets were purified by Ficoll gradient and cultured in RPMI with 10% FBS, 1% Pen/Strep and 5.5 mM Glucose overnight before experiments.
Viral infection and creating stable cell lines
The GFP-tagged pSICOR and pSICOR-shRb lentiviral vectors containing two different shRNA constructs (shRb#3; shRb#5) targeting mouse Rb were used. For lentiviral packaging, 293ET cells were cultured 16 h before transfection. In all, 0.5 ml/plate of viral supernatants together with 4 μg/ml (final) of polybrene (Sigma) was added to the cultures. Cells were transferred to fresh medium after 48 h of infection and allowed to grow for an additional 24 h before analyses.
Pancreatic rudiments culture
Embryonic pancreases were harvested from normal wild-type embryos at E14.5 and digested with 0.2 mg/ml of Liberase RI (Roche) for 8 min at 37°C. The digested pancreatic rudiments are referred as fetal pancreatic (FP) cells. FP cells were grown in DMEM with 10% FBS. Cells were plated in 24-well tissue culture plates 24 h before infection with GFP-tagged control or shRb lentiviruses. Imaging of these cells to detect GFP fluorescence was done 72 h after infection using an inverted microscope Axio Observer.Z1 (Zeiss).
Plasmids and transient transfection
HA-E2F1, c-myc Pdx-1 (a gift of Dr S Ozcan, University of Kentucky), HA-RB, GFP-RB and Flag-Ubiquitin (gift of Dr Yihong Ye, NIDDK) were transfected in MIN6, Cos7 or Saos-2 cells using FuGENE 6 (Roche, IN). After 48 h incubation, cell lysates were prepared. For effect of MG-132 or lactacystin, cells were treated for 6 h before harvesting of the cells.
Site-directed mutagenesis
Site-directed mutagenesis in HA-RB and c-myc Pdx-1 were generated using Quick-Change Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). All mutant plasmids were verified by DNA sequencing by Agencourt Bioscience Co. (Beverly, MA).
Purification of GST-RB recombinant and GST-pull down assay
Expression and purification of recombinant GST proteins were as described elsewhere (Chittenden et al, 1991; Kaelin et al, 1991; Chew et al, 1998; Gagrica et al, 2004). Escherichia coli BL21 cells with various GST-fusion expression plasmids were cultured at 37°C and expression of recombinant proteins was induced by 0.1 mM final concentration of IPTG for 3 h. GST-tagged mutants were expressed in Cos7 cells transfected with myc-tagged Pdx-1 followed by GST-pull down and immunoblot with anti-myc or anti-GST antibody.
CHX experiments
Islets were cultured with final concentration of 10 μM CHX for the indicated times. Lentivirus infection was 48 h before 10 μM CHX treatment for 7 h. Islets were pretreated for 24 h with 0.4 μM Cdk4 inhibitor prior to 10 μM CHX treatment for 7 h.
Glucose experiments
In the glucose-stimulation condition, MIN6 cells were cultured overnight in normal medium and were then cultured for 24 h in glucose-free media when two time points (−24 and −12 h) were taken. After the 24-h glucose-deprivation period (time 0), the cells were moved to glucose-containing medium and cells were harvested at 6, 12 and 24 h after glucose stimulation. In the glucose-withdrawal condition, MIN6 cells cultured overnight in normal medium and were then cultured for 24 h in glucose-containing media when two time points (−24 and −12 h) were taken. After the 24-h glucose-addition period (time 0), the cells were moved to glucose-free medium and cells were harvested at 6, 12 and 24 h after glucose withdrawal. MIN6 cells were transfected with Flag-tagged ubiquitin for 48 h followed by culture in glucose-containing or glucose-free media for 12 h and subsequently cultured for an additional 12 h in glucose-free or glucose-containing media. Prior to harvesting, cells were incubated with proteasome inhibitors MG-132 or lactacystin for 6 h.
Western blotting and co-immunoprecipitation
Western blot analysis used protocols as described previously (Lin et al, 2009). Antibodies used are anti-RB monoclonal antibodies (BD Biosciences, San Jose, CA), anti-Pdx-1 polyclonal antibodies (Millipore), mouse monoclonal c-Myc antibodies (Santa Cruz Biotechnology, CA), mouse monoclonal HA epitope antibodies (Roche, MA), phospho-RB (Ser780) and phospho-RB (Ser807/Ser811) were from Cell Signaling (Beverly, MA). The same membranes were reblotted with monoclonal anti-α-tubulin and β-actin antibodies (Sigma, Saint Louis, MO) to estimate the amount of protein loaded.
ChIP assay
The ChIP assays were performed according to the manufacturer's recommendation (Active Motif, CA), using protocols described previously (Lin et al, 2009), with antibodies to RB (BD Biosciences, CA), Pdx-1 antibodies (Millipore, IN) and normal rabbit/mouse IgG (Vector Lab, CA) and promoter-specific primers (information is available upon request) to amplify the Pdx-1-binding region. Sequential ChIP protocols were kindly provided by Dr M Szyf (McGill University).
Animals
Rb+/+ and Rb+/− mice and Cdk4 mice are described elsewhere (Jacks et al, 1992; Rane et al, 1999). Rb+/+ and Rb+/− mice were maintained on a C57BL/6 background. Timed matings were conducted, with E0.5 as midday of the day of discovery of a vaginal plug. For BrdU labelling, pregnant females were injected i.p. with 50 μg of BrdU per gram of body weight, and embryos were harvested 1 h after injection.
Immunofluorescence analysis
Harvesting of embryonic tissues was followed by immunostaining overnight at 4°C in 10 mM PBS with the following primary antibodies: 1:800 rabbit anti-Pdx-1 (Millipore, CA), 1:1000 mouse anti-E cadherin (BD Biosciences, CA) and 1:50 mouse anti-BrdU (Dako, IN). All secondary antibodies were obtained from Invitrogen and were used at 1:1000 (Alexa Fluor 488) or 1:1000 (Alexa Fluor 568) dilutions, and coverslips were mounted with Vectashield (Vector Laboratories, CA). Slides were imaged on a confocal microscope LSM510 (Zeiss, NY).
TUNEL assay
To evaluate the apoptosis in embryonic pancreases at E13.5, we performed the TUNEL labelling using the DeadEnd Fluorometric TUNEL System (Promega) following the manufacturer's instructions. To determine the percentage of apoptotic cells, we counted the percentage of TUNEL+ cells to total Pdx-1+ cells. At least 10–12 sections were analysed per genotype.
Real-time PCR
Total RNA was prepared from cells, islets or embryonic pancreases from Rb+/− and Rb−/− mice at E11.5 by using the RNAqueous-Micro kit (Ambion, TX) and subjected to real-time RT–PCR as described elsewhere (Lin et al, 2009). Reactions were performed in triplicate, and relative amounts of cDNA were normalized to 18 s rRNA.
Supplementary Material
Acknowledgments
We thank Drs Yihong Ye, Kai Ge, Neha Rane and David Harlan for advice and critical reading of the manuscript. We thank Drs Stephan Gaubatz, Sabire Ozcan, William Kaelin, M Szyf, Tyler Jacks, Julien Sage and Sibylle Mittnacht for generous gifts of reagents and technical advice. William Neidermyer was supported in part by the Colgate University-NIH internship program. This work was supported by NIDDK, NIH intramural funds awarded to SGR.
Author contributions: YK performed the majority of the biochemistry and molecular biology studies. SK performed experiments with the RB mutant embryos. JMG performed islet isolations. HY performed real-time PCR experiments. WN created the site-directed mutant constructs. AK performed the lentivirus experiments. SR conceived the hypothesis, advised on experiments, supervised the project and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahlgren U, Jonsson J, Edlund H (1996) The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development (Cambridge, England) 122: 1409–1416 [DOI] [PubMed] [Google Scholar]
- Brissova M, Blaha M, Spear C, Nicholson W, Radhika A, Shiota M, Charron MJ, Wright CV, Powers AC (2005) Reduced PDX-1 expression impairs islet response to insulin resistance and worsens glucose homeostasis. Am J Physiol Endocrinol Metab 288: E707–E714 [DOI] [PubMed] [Google Scholar]
- Brown VD, Phillips RA, Gallie BL (1999) Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol 19: 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J (2008) Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanero MR, Flemington EK (1997) Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA 94: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew YP, Ellis M, Wilkie S, Mittnacht S (1998) pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene 17: 2177–2186 [DOI] [PubMed] [Google Scholar]
- Chittenden T, Livingston DM, Kaelin WG Jr (1991) The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell 65: 1073–1082 [DOI] [PubMed] [Google Scholar]
- Claiborn KC, Sachdeva MM, Cannon CE, Groff DN, Singer JD, Stoffers DA (2010) Pcif1 modulates Pdx1 protein stability and pancreatic beta cell function and survival in mice. J Clin Invest 120: 3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H (1992) Requirement for a functional Rb-1 gene in murine development. Nature 359: 328–330 [DOI] [PubMed] [Google Scholar]
- Dick FA, Dyson N (2003) pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell 12: 639–649 [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV (2006) Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev 20: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagrica S, Hauser S, Kolfschoten I, Osterloh L, Agami R, Gaubatz S (2004) Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J 23: 4627–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hani EH, Stoffers DA, Chevre JC, Durand E, Stanojevic V, Dina C, Habener JF, Froguel P (1999) Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest 104: R41–R48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb G, Vasavada RC, Cobrinik D, Stewart AF (2009) The retinoblastoma protein and its homolog p130 regulate the G1/S transition in pancreatic beta-cells. Diabetes 58: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Dean DC (2000) Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2: E65–E67 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC (1999) Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98: 859–869 [DOI] [PubMed] [Google Scholar]
- Helin K, Lees JA, Vidal M, Dyson N, Harlow E, Fattaey A (1992) A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell 70: 337–350 [DOI] [PubMed] [Google Scholar]
- Hofmann F, Martelli F, Livingston DM, Wang Z (1996) The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev 10: 2949–2959 [DOI] [PubMed] [Google Scholar]
- Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ (2002) Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci USA 99: 12236–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey RK, Yu SM, Flores LE, Jhala US (2010) Glucose regulates steady-state levels of PDX1 via the reciprocal actions of GSK3 and AKT kinases. J Biol Chem 285: 3406–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA (1992) Effects of an Rb mutation in the mouse. Nature 359: 295–300 [DOI] [PubMed] [Google Scholar]
- Jiao W, Datta J, Lin HM, Dundr M, Rane SG (2006) Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via Cdk phosphorylation-dependent nuclear export. J Biol Chem 281: 38098–38108 [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H (1994) Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371: 606–609 [DOI] [PubMed] [Google Scholar]
- Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J (2007) An illustrated review of early pancreas development in the mouse. Endocr Rev 28: 685–705 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr, Pallas DC, DeCaprio JA, Kaye FJ, Livingston DM (1991) Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell 64: 521–532 [DOI] [PubMed] [Google Scholar]
- Khidr L, Chen PL (2006) RB, the conductor that orchestrates life, death and differentiation. Oncogene 25: 5210–5219 [DOI] [PubMed] [Google Scholar]
- Kim SK, MacDonald RJ (2002) Signaling and transcriptional control of pancreatic organogenesis. Curr Opin Genet Dev 12: 540–547 [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY (1997) Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol 17: 5771–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Chang JH, Lee HS, Cho Y (2002) Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev 16: 3199–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A (1992) Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359: 288–294 [DOI] [PubMed] [Google Scholar]
- Lee JH, Jo J, Hardikar AA, Periwal V, Rane SG (2010) Cdk4 regulates recruitment of quiescent beta-cells and ductal epithelial progenitors to reconstitute beta-cell mass. PloS One 5: e8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HM, Lee JH, Yadav H, Kamaraju AK, Liu E, Zhigang D, Vieira A, Kim SJ, Collins H, Matschinsky F, Harlan DM, Roberts AB, Rane SG (2009) Transforming growth factor-beta/Smad3 signaling regulates insulin gene transcription and pancreatic islet beta-cell function. J Biol Chem 284: 12246–12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Miller C, Habener JF (1996) Functional regions of the homeodomain protein IDX-1 required for transactivation of the rat somatostatin gene. Endocrinology 137: 2959–2967 [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Weinberg RA (1998) Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol 18: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane WM, Frayling TM, Ellard S, Evans JC, Allen LI, Bulman MP, Ayres S, Shepherd M, Clark P, Millward A, Demaine A, Wilkin T, Docherty K, Hattersley AT (1999) Missense mutations in the insulin promoter factor-1 gene predispose to type 2 diabetes. J Clin Invest 104: R33–R39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon CM, Docherty K (2001) Pancreatic duodenal homeobox-1, PDX-1, a major regulator of beta cell identity and function. Diabetologia 44: 1203–1214 [DOI] [PubMed] [Google Scholar]
- Mettus RV, Rane SG (2003) Characterization of the abnormal pancreatic development, reduced growth and infertility in Cdk4 mutant mice. Oncogene 22: 8413–8421 [DOI] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ (2001) Retinoblastoma protein partners. Adv Cancer Res 82: 1–54 [DOI] [PubMed] [Google Scholar]
- Nicolino M, Claiborn KC, Senee V, Boland A, Stoffers DA, Julier C (2010) A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes 59: 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RJ, Hearing P (1994) Mutually exclusive interaction of the adenovirus E4-6/7 protein and the retinoblastoma gene product with internal domains of E2F-1 and DP-1. J Virol 68: 6848–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV (1996) PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development (Cambridge, England) 122: 983–995 [DOI] [PubMed] [Google Scholar]
- Qin XQ, Chittenden T, Livingston DM, Kaelin WG Jr (1992) Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev 6: 953–964 [DOI] [PubMed] [Google Scholar]
- Rane SG, Cosenza SC, Mettus RV, Reddy EP (2002) Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol Cell Biol 22: 644–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, Reddy EP, Barbacid M (1999) Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat Genet 22: 44–52 [DOI] [PubMed] [Google Scholar]
- Rane SG, Reddy EP (2000) Cell cycle control of pancreatic beta cell proliferation. Front Biosci 5: D1–D19 [DOI] [PubMed] [Google Scholar]
- Sachdeva MM, Claiborn KC, Khoo C, Yang J, Groff DN, Mirmira RG, Stoffers DA (2009) Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proc Natl Acad Sci USA 106: 19090–19095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzgebel VM, Mamin A, Brun T, Ritz-Laser B, Zaiko M, Maret A, Jornayvaz FR, Theintz GE, Michielin O, Melloul D, Philippe J (2003) Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J Clin Endocrinol Metab 88: 4398–4406 [DOI] [PubMed] [Google Scholar]
- Shan B, Durfee T, Lee WH (1996) Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc Natl Acad Sci USA 93: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers DA, Ferrer J, Clarke WL, Habener JF (1997a) Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet 17: 138–139 [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF (1997b) Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet 15: 106–110 [DOI] [PubMed] [Google Scholar]
- Tsutsui T, Hesabi B, Moons DS, Pandolfi PP, Hansel KS, Koff A, Kiyokawa H (1999) Targeted disruption of CDK4 delays cell cycle entry with enhanced p27(Kip1) activity. Mol Cell Biol 19: 7011–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Vasavada RC, Cozar-Castellano I, Sipula D, Stewart AF (2007) Tissue-specific deletion of the retinoblastoma protein in the pancreatic beta-cell has limited effects on beta-cell replication, mass, and function. Diabetes 56: 57–64 [DOI] [PubMed] [Google Scholar]
- Wenzel PL, Wu L, de Bruin A, Chong JL, Chen WY, Dureska G, Sites E, Pan T, Sharma A, Huang K, Ridgway R, Mosaliganti K, Sharp R, Machiraju R, Saltz J, Yamamoto H, Cross JC, Robinson ML, Leone G (2007) Rb is critical in a mammalian tissue stem cell population. Genes Dev 21: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett LA, Weinstein M, Cross JC, Robinson ML, Leone G (2003) Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421: 942–947 [DOI] [PubMed] [Google Scholar]
- Xiao B, Spencer J, Clements A, Ali-Khan N, Mittnacht S, Broceno C, Burghammer M, Perrakis A, Marmorstein R, Gamblin SJ (2003) Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proc Natl Acad Sci USA 100: 2363–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.