Abstract
Although regulation of histone methylation is believed to contribute to embryonic stem cell (ESC) self-renewal, the mechanisms remain obscure. We show here that the histone H3 trimethyl lysine 4 (H3K4me3) demethylase, KDM5B, is a downstream Nanog target and critical for ESC self-renewal. Although KDM5B is believed to function as a promoter-bound repressor, we find that it paradoxically functions as an activator of a gene network associated with self-renewal. ChIP-Seq reveals that KDM5B is predominantly targeted to intragenic regions and that it is recruited to H3K36me3 via an interaction with the chromodomain protein MRG15. Depletion of KDM5B or MRG15 increases intragenic H3K4me3, increases cryptic intragenic transcription, and inhibits transcriptional elongation of KDM5B target genes. We propose that KDM5B activates self-renewal-associated gene expression by repressing cryptic initiation and maintaining an H3K4me3 gradient important for productive transcriptional elongation.
Keywords: chromatin, epigenetics, histone demethylase, self-renewal, transcriptional elongation
Introduction
Embryonic stem cells (ESCs) derived from the inner cell mass (ICM) of pre-implantation embryos can differentiate into all somatic lineages and have an unlimited capacity for self-renewal (Niwa, 2007a). ESCs serve as a model for the study of development specification and as an important resource for cell replacement therapy. Insight into the transcriptional and epigenetic mechanisms that regulate ESC self-renewal is essential for their use in medicine.
The transcription factors Oct4 and Nanog have a central role in the initiation and maintenance of ESC pluripotency and self-renewal (Chambers et al, 2003; Mitsui et al, 2003; Do and Scholer, 2009). Nanog is also essential for the acquisition of pluripotency in the embryo and during reprogramming (Silva et al, 2009). ESC pluripotency is believed to depend on an epigenetic state that facilitates unlimited self-renewal and prevents developmental gene silencing. ESCs are characterized by a relaxed chromatin conformation (Niwa, 2007b) and unique bivalent domains (Bernstein et al, 2007). During reprogramming of somatic cells, extensive epigenetic remodelling occurs, including loss of repressive histone and DNA methylation and re-establishment of bivalent histone modifications (Maherali et al, 2007). Although Nanog has been reported to interact with chromatin complexes (Liang et al, 2008), the downstream mechanisms by which Nanog regulates initiation and maintenance of pluripotency remain unclear. We utilized a ChIP-Seq screen to identify the histone demethylase KDM5B as a major downstream target of Nanog.
KDM5B is an H3K4me2/3 demethylase that is up-regulated in a wide range of human cancers and enhances cancer self-renewal (Lu et al, 1999; Yamane et al, 2007; Hayami et al, 2010; Roesch et al, 2010). Forced KDM5B expression also increases histone H3 Ser10 phosphorylation in ESCs and reduces neural progenitor cell differentiation (Dey et al, 2008). Nevertheless, the molecular mechanisms by which KDM5B regulates self-renewal have not been well characterized. Although KDM5B is believed to function as a transcriptional repressor by removing promoter-associated H3K4me3, we show here that KDM5B also functions as an activator of self-renewal-associated gene expression. ChIP-Seq analysis shows that KDM5B occupancy is highly correlated with H3K36me3, a chromatin mark associated with active transcriptional elongation. KDM5B is recruited to intragenic H3K36me3 at least, in part, via its interaction with the chromodomain protein MRG15. Moreover, we demonstrate that KDM5B safeguards transcriptional elongation by repressing spurious intragenic transcription. Our study reveals that compartmentalization of a histone demethylase to a distinct chromatin domain results in an unexpected role in gene regulation.
Results
KDM5B is a transcriptional target of Nanog and contributes to ESC self-renewal
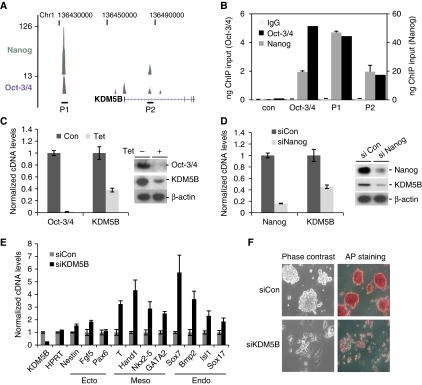
Nanog and Oct4 are core transcriptional regulators of ESC self-renewal. Analysis of ChIP-Seq data showed that Nanog and Oct4 co-occupy the KDM5B genomic locus in mouse ESCs (Figure 1A). ChIP-PCR confirmed Nanog and Oct4 occupancy at these regions in both mouse and human ESCs (Figure 1B; Supplementary Figure S1A). Tetracycline-mediated depletion of Oct4 in ZHBTc4 ESCs (Niwa et al, 2000) markedly reduced KDM5B protein and mRNA levels (Figure 1C). Likewise, depletion of endogenous Nanog or Oct4 in ESCs attenuated KDM5B expression (Figure 1D; data not shown). Transient knockdown of KDM5B increased expression of lineage-associated genes (Figure 1E). These genes are likely indirectly regulated because the majority of them are not associated with KDM5B occupancy as assessed by ChIP-Seq (Supplementary Table S1). Prolonged knockdown of KDM5B triggered morphological differentiation, loss of alkaline phosphatase activity, and a reduction in a subset of pluripotency-associated genes (Figure 1F; Supplementary Figure S1B). Consistent with its proposed role in cancer stem cell self-renewal (Roesch et al, 2010), KDM5B depletion markedly decreased ESC proliferation (Supplementary Figure S2A). In particular, S phase was reduced and the percentage of cells in G1 increased (Supplementary Figure S2B). These data suggest that KDM5B contributes to ESC self-renewal.
Figure 1.
KDM5B is a Nanog and Oct4 target and critical for ESC self-renewal. (A) UCSC genome browser track depicting Oct4 and Nanog ChIP-Seq occupancy in the vicinity of the murine KDM5B genomic locus. (B) Confirmation of Oct4 and Nanog occupancy at KDM5B (P1 and P2) by ChIP-PCR. The Oct4 enhancer region is a positive control. (C, D) ZHBTc4 ESCs were treated with tetracycline (Tet) for 48 h to deplete Oct4 (C). J1 ESCs were transfected with siNanog or non-targeting control (siCon) (D). KDM5B cDNA and protein levels were measured by RT–PCR (left) and immunoblot (right). (E) RT–PCR analysis of lineage-associated marker gene expression following siKDM5B in ESCs. (F) ESCs were transfected with control (siCon) or KDM5B siRNA (siKDM5B); 72 h post-transfection cells were stained for alkaline phosphatase activity (AP).
KDM5B is an activator of self-renewal-associated gene expression
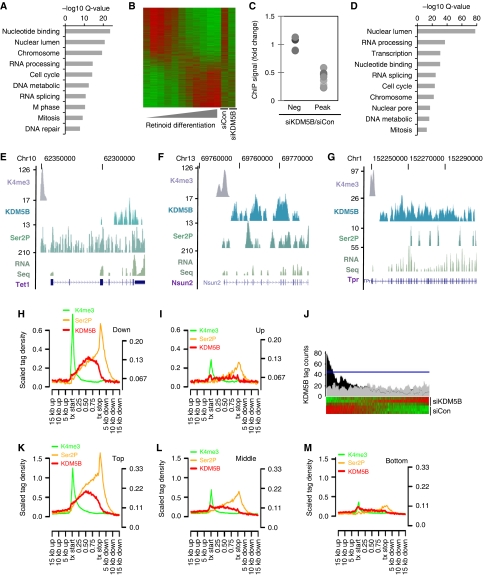
Previous studies suggest that KDM5-family demethylases repress genes involved in developmental processes (Christensen et al, 2007; Dey et al, 2008; Lopez-Bigas et al, 2008; Pasini et al, 2008). We explored the mechanisms by which KDM5B regulates ESC self-renewal by profiling gene expression following siRNA-mediated KDM5B knockdown. Surprisingly, the majority of significantly regulated genes showed decreased expression (Supplementary Table S2). Moreover, gene ontology (GO) analyses revealed that the most significantly regulated categories were exclusively associated with genes down-regulated by KDM5B knockdown (Figure 2A). Significant GO categories were not associated with developmental processes, but were linked to mitosis, chromatin, and nucleotide metabolism. Interestingly, genes down-regulated by KDM5B depletion were significantly (KS test P<1 × 106) correlated with genes whose expression decreases during ESC differentiation (Figure 2B). Real-time PCR confirmed the down-regulation of a subset of these genes following KDM5B knockdown (Supplementary Figure S3). These data strongly suggest that KDM5B functions as an activator of genes associated with self-renewal.
Figure 2.
An unexpected role for KDM5B in transcriptional activation. (A) Gene ontology (GO) analysis of microarray data showing categories of genes significantly down-regulated following siKDM5B. No up-regulated gene categories were detected at Q<1 × 10−6. (B) Heatmap comparison of microarray data from ESC retinoic acid (RA) differentiation and KDM5B knockdown experiments. (C) ChIP analysis of KDM5B ChIP-Seq peaks and negative control regions (Neg) following KDM5B knockdown (siKDM5B). Data are expressed as the ratio of siKDM5B ChIP signal over siCon. Each dot represents a distinct ChIP-Seq locus (pairwise comparisons to Peak data P<0.01). (D) GO analysis of RefSeq genes within 5 kb of a KDM5B ChIP-Seq peak 5′ end. (E–G) UCSC genome browser tracks depicting H3K4me3, KDM5B, and Ser 2 phosphorylated PolII (Ser2P) ChIP-Seq peaks at representative gene loci. The RNA-Seq track depicts 3′-directed RNA-Seq data from ESCs. The numbers on the left axes indicate peak amplitude. (H, I) ChIP-Seq tag density relative to significantly down- (H) and up-regulated (I) genes following KDM5B knockdown. All Ref-Seq gene lengths were scaled to 1. The left y axis corresponds to H3K4me3 ChIP-Seq data, while the right y axis represents other data. (J) Histogram of KDM5B ChIP-Seq peak tag density relative to ESC RefSeq microarray data rank ordered by fold-change following KDM5B knockdown. The black profile above the heat map depicts average number of significant ChIP-Seq KDM5B peak tags found per gene. The grey profile represents a randomized control and the blue line delineates an FDR of P<0.01. (K–M) ChIP-Seq tag density relative to the top (K), middle (L), and bottom third (M) of RefSeq genes expressed in ESCs. All Ref-Seq gene lengths were scaled to 1. The left y axis corresponds to H3K4me3 ChIP-Seq data, while the right y axis represents other data.
KDM5B occupies transcribed regions of self-renewal-associated genes
The ability of KDM5B to function as an activator could result from indirect regulation. To address this question, we utilized ChIP-Seq to profile the genome-wide occupancy of KDM5B in an unbiased manner. The 8 425 313 ChIP-Seq tags were sequenced of which 3 864 894 mapped to a single genomic locus. A sliding-window algorithm identified 11 142 KDM5B ChIP-Seq peaks significantly enriched at an FDR of 5% (Supplementary Table S1). The KDM5B antibody used for ChIP-Seq identified only one protein by western blot (Supplementary Figure S4A) and ChIP-PCR confirmed occupancy of a randomly selected subset of peaks with antibodies to distinct epitopes (Supplementary Figure S4B and C). Moreover, siRNA-mediated depletion of KDM5B attenuated KDM5B ChIP signal (Figure 2C). GO analysis of genes adjacent to KDM5B ChIP-Seq peaks generated categories that were highly similar to those associated with genes down-regulated following KDM5B knockdown (Figure 2A and D). Surprisingly, unbiased ChIP-Seq revealed that 84% of KDM5B ChIP-Seq peaks were present in intragenic regions (Wilcoxon rank-sum P<1 × 10−6; Figure 2E–G), while only 36% of random loci and 40% of Oct4 ChIP-Seq peaks were located inside genes.
The accumulation of KDM5B in transcribed regions suggested an association with transcriptional activity. We utilized ChIP-Seq to profile occupancy of a phosphorylated form of PolII associated with elongation (Ser2P) and the promoter-associated epigenetic mark, H3K4me3. We found that ∼59% of KDM5B peaks were within 2 kb of a Ser2P peak (Wilcoxon rank-sum P<1 × 10−6; Figure 2E–G), while only 6% of random loci and 13% of Oct4 ChIP-seq peaks were adjacent to a Ser2P peak. KDM5B was not significantly associated with H3K4me3 (Wilcoxon rank-sum P=0.23). These data strongly suggest that KDM5B functions as a transcriptional activator. Consistent with this idea, KDM5B ChIP-Seq tags accumulated in intragenic regions of genes positively regulated by KDM5B, but not in intragenic regions of genes repressed by KDM5B (Figure 2H and I). Comparison of ChIP-Seq tag accumulation relative to ranked microarray data reveals that KDM5B occupancy is significantly correlated with genes decreased by KDM5B knockdown (Figure 2J; Supplementary Figure S5). Consistent with this finding, KDM5B occupies intragenic regions of highly expressed RefSeq genes, but was not significantly detected in genes expressed at lower levels (Figure 2K–M). The lack of ChIP-Seq peak correlation with genes increased following KDM5B knockdown shows that KDM5B functions predominantly as an activator of gene expression. Both microarray and ChIP-Seq studies identify an overlapping set of genes that are transcriptionally activated by KDM5B. Taken together, these data indicate that KDM5B regulates self-renewal by functioning as a direct activator of a self-renewal-associated gene network. Analyses of microarray and ChIP-Seq data identified a large set of KDM5B targets associated with cell-cycle progression and DNA biosynthesis, including Ccnb1, Ccna2, Cdc25a, Pola1, Mcm3/4/5/6/7, Cdc45, and Orcl1/2/3/5/6 (Supplementary Figure S6A; data not shown). We confirmed that these genes were directly occupied by KDM5B (Supplementary Figure S6B). These data indicate that KDM5B regulates a novel gene network that directly regulates ESC self-renewal.
Demethylation of intragenic H3K4me3 domains by KDM5B
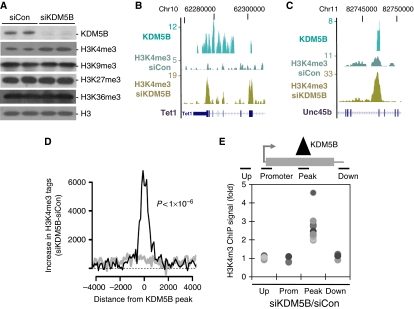
Although the majority of H3K4me3 is localized near promoters (Mikkelsen et al, 2007), our data indicate that KDM5B is predominantly localized to intragenic regions. This led us to examine whether KDM5B functions as a demethylase at intragenic loci. Knockdown of KDM5B modestly increased global H3K4me3, but not H3K9me3, H3K27me3, or H3K36me3 in ESCs (Figure 3A). To test whether KDM5B specifically regulates intragenic H3K4me3 at its targets, we utilized ChIP-Seq to examine global changes in H3K4me3 following knockdown of KDM5B. Interestingly, we observed highly localized increases in H3K4me3 near KDM5B peaks following knockdown of KDM5B (Figure 3B and C). Importantly, the increase in H3K4me3 at KDM5B ChIP-Seq peaks was highly significant (Wilcoxon rank-sum P<1 × 10−6) and occurred on a genome-wide basis (Figure 3D). We utilized ChIP-PCR to confirm this observation by showing that KDM5B knockdown increased H3K4me3 at ChIP-Seq peaks, but not at upstream or downstream regions (Figure 3E). These data show that KDM5B functions to erase intragenic H3K4me3 specifically at its target sites.
Figure 3.
KDM5B removes local domains of intragenic H3K4me3. (A) Immunoblot analysis of bulk histone modifications from ESCs transfected with control/KDM5B siRNAs. (B, C) Genome browser tracks depicting KDM5B ChIP-Seq peaks and H3K4me3 ChIP-Seq peaks from control (siCon) or KDM5B knockdown cells (siKDM5B). The numbers on the left axes indicate peak amplitude. (D) Histogram depicts the difference of H3K4me3 ChIP-Seq tag counts following knockdown of KDM5B (siKDM5B) versus control (siCon). ChIP-Seq tag counts are plotted relative to the centre of mass of KDM5B ChIP-Seq peaks (black line) or randomized controls (grey line). The ChIP-Seq difference was normalized by total tag counts and tags within 1.5 kb of RefSeq gene 5′ ends were not included. The difference of H3K4me3 ChIP-Seq tag counts following knockdown of KDM5B was highly significant (Wilcoxon rank-sum P<1 × 10−6). (E) Scatter plot depicts H3K4me3 ChIP data following KDM5B knockdown (siKDM5B) at KDM5B ChIP-Seq peaks (Peak), upstream regions (Up), associated promoters (Prom), and downstream regions (Down). Data are expressed as the ratio of H3K4me3 ChIP signal from siKDM5B over siCon. Each dot represents a distinct ChIP-Seq peak locus and all comparison to Peak data P<0.01.
Recruitment of KDM5B to chromatin depends on H3K36me3
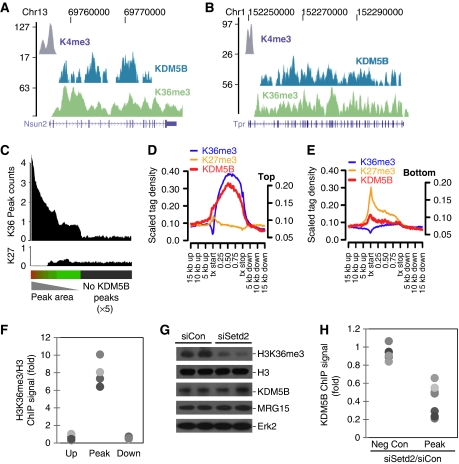
The enrichment of KDM5B in transcriptionally active regions is reminiscent of the distribution of H3K36me3, an epigenetic mark deposited by elongating PolII (Li et al, 2007). We compared the localization of KDM5B to H3K4me3 ChIP-Seq data generated in this study and published H3K36me3 and H3K27me3 ChIP-Seq data (Mikkelsen et al, 2007) (Figure 4A and B). A total of 83% of KDM5B regions were within 5 kb of H3K36me3 peak (Wilcoxon rank-sum P<1 × 10−6), while only ∼18% of KDM5B peaks were near H3K4me3 peaks (Wilcoxon rank-sum P=0.23). As the majority of KDM5B ChIP-Seq peaks (84%) are localized to RefSeq intragenic regions, we examined the density of H3K36me3 relative to RefSeq genes ordered by KDM5B ChIP-Seq peak area. H3K36me3 density was highly correlated with genes containing the most KDM5B occupancy (Figure 4C). Importantly, the spatial profile of KDM5B occupancy in highly expressed genes mirrored that of H3K36me3 (Figure 4D and E). ChIP-PCR confirmed that H3K36me3 was specifically enriched at KDM5B ChIP-Seq peaks, but not at down- or upstream regions (Figure 4F). The colocalization of KDM5B and H3K36me3 raised the possibility that KDM5B is directly recruited by this epigenetic mark. We tested this by examining KDM5B occupancy following knockdown of the Setd2 H3K36me3 methyltransferase (Figure 4G). Setd2 depletion markedly decreased KDM5B occupancy (Figure 4H) and induced a localized increase in H3K4me3 (Supplementary Figure S7A). These data show that H3K36me3 deposition facilitates recruitment of KDM5B. As H3K36me3 is deposited by elongating PolII, KDM5B recruitment should also be dependent on transcriptional elongation. Inhibition of PolII elongation markedly attenuated occupancy of KDM5B and elongating PolII, while sparing promoter-bound PolII (Supplementary Figure S7B and C). These data strongly suggest that KDM5B is recruited by elongation-associated H3K36 methylation.
Figure 4.
H3K36me3 recruits KDM5B to transcriptionally active intragenic regions. (A, B) UCSC genome browser tracks depicting H3K4me3, KDM5B, and H3K36me3 ChIP-Seq peaks at representative gene loci. The numbers on the left axes indicate peak amplitude. (C) Histogram depicting H3K36me3, and H3K27me3 (K27) ChIP-Seq peak density in RefSeq genes rank ordered by KDM5B peak area. The grey bar indicates genes without KDM5B peaks (× 5 scaling). (D, E) ChIP-Seq tag density relative to 5000 highly expressed RefSeq genes (D) and 5000 genes expressed at low levels (E). All Ref-Seq gene lengths were scaled to 1. The left y axis corresponds to H3K4me3 ChIP-Seq data, while the right y axis represents other data. (F) Scatter plot depicts ChIP-PCR for H3K36me3 at KDM5B ChIP-Seq peaks (Peak), and control regions (Up and Down). Each dot represents a distinct ChIP-Seq locus and pairwise comparisons to Peak data P<0.01. (G) Immunoblot of ESC extracts with the indicated antibodies following knocking down H3K36 methyltransferase Setd2. (H) KDM5B ChIP-PCR signal at negative control regions (Up and Down) and KDM5B peaks following knockdown of Setd2 (siSetd2). Data are expressed as the ratio of KDM5B ChIP signal of siSetd2 over siCon. Each dot represents a distinct locus and pairwise comparisons to Peak data P<0.01.
An Rpd3S-like complex recruits KDM5B to H3K36me3
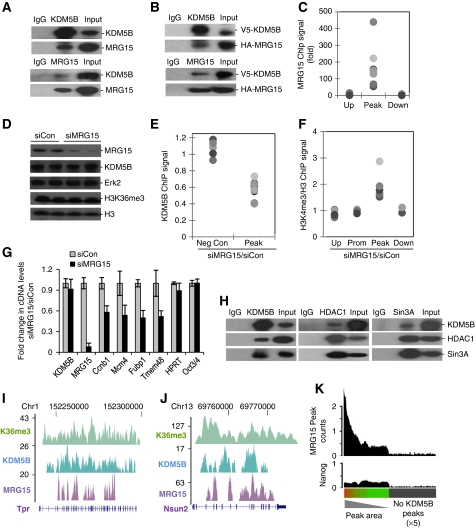
The KDM5B orthologue, little imaginal discs (Lid), interacts with the chromodomain H3K36me3-binding protein MRG15 in Drosophila (Lee et al, 2009b). Interestingly, mammalian KDM5B also interacts with MRG15 in ESCs (Figure 5A). Similar results were seen with epitope-tagged KDM5B and MRG15 in a heterologous expression system (Figure 5B). Moreover, ChIP-PCR analysis showed that MRG15 was selectively enriched at KDM5B peaks, but not at distal regions (Figure 5C). Knockdown of MRG15 modestly decreased KDM5B occupancy, increased intragenic H3K4me3, and reduced expression of KDM5B target genes (Figure 5D–G). These data indicate that MRG15 contributes to recruitment of KDM5B to H3K36me3. The yeast MRG15 orthologue Eaf3 functions to recruit the Rpd3S histone deacetylase and Sin3 to transcriptionally active chromatin. Thus, we asked whether KDM5B interacts with a mammalian Rpd3S-like complex. Pull down experiments show that KDM5B interacts with mammalian Rpd3S components, HDAC1 and Sin3A (Figure 5H). KDM5B was not detected in HDAC1-containing Mi-2/NURD immunoprecipitates (data not shown). Importantly, KDM5B knockdown did not affect H4 acetylation or H3K36me3 (Supplementary Figure S8). We next utilized ChIP-Seq to test whether MRG15 colocalized with KDM5B on a genome-wide scale. MRG15 ChIP-Seq signal was highly correlated with KDM5B occupancy (Figure 5I and J) and 49% of KDM5B ChIP-Seq peaks were within 5 kb of an MRG15 peak (Wilcoxon rank-sum P<1 × 10−6). Like KDM5B, MRG15 was predominantly associated with intragenic transcription (71% of MRG15 peaks are inside RefSeq genes; Wilcoxon rank-sum P<1 × 10−6) and the spatial profile of MRG15 occupancy overlapped with that of KDM5B and H3K36me3 (Supplementary Figure S9). Moreover, MRG15 occupancy was highly correlated with KDM5B, but not Nanog intragenic occupancy (Figure 5K). Although the Nanog negative control showed no correlation with KDM5B occupancy, a general enrichment for Nanog (over regions without KDM5B) is expected due to the tendency of KDM5B to demarcate transcriptionally active chromatin. These data strongly suggest that KDM5B can be recruited to H3K36me3 via its interaction with MRG15. As not all KDM5B peaks are associated with MRG15 occupancy, additional recruitment mechanisms likely also exist.
Figure 5.
MRG15 mediates recruitment of KDM5B. (A, B) Endogenous (A) or epitope-tagged (B) KDM5B and MRG15 immunoprecipitates from ESCs were immunoblotted with the indicated antibodies. (C) ChIP-PCR analysis of MRG15 occupancy at KDM5B peaks (Peak) and control regions (Up and Down). (D) Immunoblot analysis of KDM5B and MRG15 from ESC whole cell extracts following MRG15 knockdown (siMRG15). (E, F) KDM5B or H3K4me3 ChIP-PCR analysis at negative control regions (Up and Down) and KDM5B peaks following MRG15 knockdown (siMRG15). Data are expressed as the ratio of ChIP signal from siMRG15 over siCon. Each dot represents a distinct ChIP-Seq locus and pairwise comparisons to Peak data P<0.01. (G) RT–PCR analysis of randomly picked KDM5B target genes (Ccnb1, Mcm4, Fubp1, Tmem48) and control (HPRT, Oct4) following siRNA-mediated MRG15 knockdown. (H) KDM5B, HDAC1, and SIN3A immunoprecipitates from ESCs were immunoblotted with the indicated antibodies. (I, J) UCSC genome browser tracks depict H3K36me3, KDM5B, and MRG15 ChIP-Seq peaks at representative gene loci. The numbers on the left axes indicate peak amplitude. (K) Histogram depicting MRG15 and Nanog (control) ChIP-Seq peak density in RefSeq genes rank ordered by KDM5B peak area. The grey bar indicates genes without KDM5B peaks (× 5 scaling).
KDM5B represses cryptic transcription by removing intragenic H3K4me3
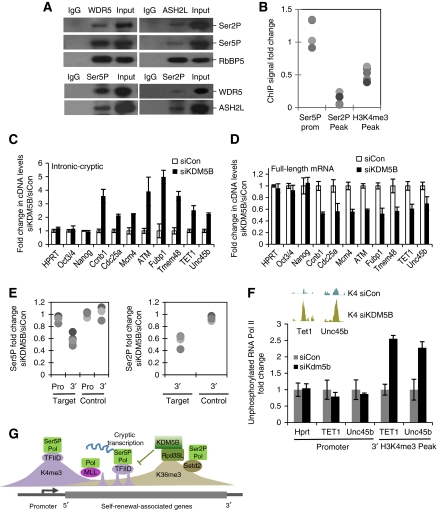
Rpd3, the catalytic core of the Rpd3S complex, is believed to repress cryptic transcription by erasing histone acetylation deposited by elongating polymerase (Carrozza et al, 2005; Joshi and Struhl, 2005; Keogh et al, 2005). Our observation that KDM5B interacts with mammalian orthologues of Rpd3S complex suggests that KDM5B functions in a manner analogous to Rpd3. In support of this hypothesis, we found that mammalian H3K4me3 methyltransferase subunits form a complex with elongating RNA PolII (Figure 6A). Consequently, we examined whether intragenic H3K4me3 deposition was dependent on transcriptional activity. Treatment of ESCs with the inhibitor of PolII elongation, DRB, significantly decreased intragenic H3K4me3 at both 1 h (Figure 6B) and 6 h time points (data not shown). Unphosphorylated PolII is tethered to sites of initiation, at least in part, via an interaction with TFIID/TBP (Usheva et al, 1992; Nikolov et al, 1995). Interestingly, KDM5B knockdown markedly stimulated recruitment of unphosphorylated PolII to intragenic H3K4me3 peaks strongly suggesting that these sites represent sites of cryptic initiation (Figure 6F). As H3K4me3 is highly correlated with transcriptional start sites and recruits the PolII preinitiation complex via interactions with TFIID (Vermeulen et al, 2007), we hypothesized that KDM5B repressed cryptic transcription by preventing intragenic initiation.
Figure 6.
KDM5B safeguards transcriptional elongation by repressing cryptic transcription. (A) Immunoblot analysis of immunoprecipitated WDR5, Ash2L, Ser5P, and Ser2P with indicated antibodies. (B) ChIP-PCR analysis of Ser2P, Ser5P, and H3K4me3 at KDM5B ChIP-Seq peaks and associated promoters following 1 h DRB treatment. Data are expressed as fold change versus untreated. Each dot represents a distinct ChIP-Seq locus. (C, D) RT–PCR measurements of cryptic transcription (left panel) and full-length mRNAs (right panel) following knocking down of KDM5B (siKDM5B) at negative control genes (HPRT, Oct4, Nanog) and KDM5B target genes (Ccnb1, Cdc25a, ATM, Mcm4, Fubp1, Tmem48, TET1, Unc45b). (E) ChIP-PCR analysis of Ser5P and Ser2P occupancy at promoter region (Pro) and 3′ end (3′) of KDM5B target genes (Target) near KDM5B ChIP-Seq peaks. The promoters and 3′end of non-KDM5B targets genes (house-keeping genes and Oct4, Nanog) were used as control (Control). Data are expressed as the ratio of PolII ChIP signal from siKDM5B over siCon. Each dot represents a distinct ChIP-Seq locus. (F) ChIP analyses of un-phosphorylated PolII occupancy (8WG16 mAb, Covance) at control promoter regions and intragenic H3K4me3 peaks following KDM5B knockdown (siKDM5B) in J1 ESCs. (G) Deposition of intragenic H3K4me3 by PolII results in initiation of cryptic transcription. The KDM5B demethylase is recruited to H3K36me3 via interaction with an Rpd3S-like (Rpd3SL) complex. KDM5B removes intragenic H3K4me3, represses cryptic initiation, and safeguards productive mRNA elongation.
Knockdown of KDM5B markedly increased expression of cryptic unspliced transcripts (Figure 6C). Moreover, ChIP and global ChIP-Seq analyses show that the increase in cryptic transcription was associated with a localized increase of H3K4me3 (Figure 3B–E), but not H4Ace or H3K36me3 (Supplementary Figure S8). These non-coding transcripts did not originate from the canonical promoter because we detected no change in Ser5P PolII recruitment at promoter regions after KDM5B knockdown (Figure 6E). The position of intragenic H3K4me3 at the 3′ end of the gene led us to examine whether some of this cryptic transcription was antisense to the coding strand. We found that both Tet1 and Unc45b contained cryptic transcripts antisense to the coding strand (Supplementary Figure S10). These cryptic transcripts did not originate from other transcripts because KDM5B-regulated transcription was not detected outside the transcribed locus (data not shown). Moreover, full-length (intron-spanning) transcription of KDM5B targets was always down-regulated following KDM5B depletion (Figure 6D). Recruitment of Ser5P to promoter regions was unaffected by KDM5B depletion, but Ser5P and Ser2P recruitment was selectively decreased from the 3′ ends of KDM5B target genes, suggesting a defect in later phases of transcriptional elongation (Figure 6E). We observed similar effect for MRG15 knockdown (data not shown). These data strongly suggest that KDM5B functions to prevent cryptic transcription by removing intragenic H3K4me3 (Figure 6F) and that KDM5B safeguards expression of a gene network associated with self-renewal by maintaining an H3K4me3 gradient that favours productive elongation (Figure 6G; data not shown).
Discussion
KDM5-family demethylases are believed to repress transcription by removing promoter-associated H3K4me3 (Christensen et al, 2007; Klose et al, 2007; Yamane et al, 2007). Surprisingly, we show that KDM5B paradoxically functions as a transcriptional activator of genes linked to ESC self-renewal. ChIP-Seq analyses reveal that KDM5B predominantly occupies actively transcribed chromatin and is recruited to domains enriched for elongating PolII. KDM5A has been shown to function as an activator or repressor depending on gene context (Chan and Hong, 2001; Benevolenskaya et al, 2005) and apparently occupies promoters and intragenic regions (Christensen et al, 2007; Hayakawa et al, 2007). Interestingly, the Drosophila KDM5B orthologue Lid is associated with transcriptionally active chromatin, positively regulates gene expression, and antagonizes heterochromatin formation (Lee et al, 2007; Secombe et al, 2007; Lloret-Llinares et al, 2008). Thus, KDM5B's role in gene activation may be evolutionarily conserved.
Nanog is required for the initiation and maintenance of pluripotency (Chambers et al, 2003; Mitsui et al, 2003; Silva et al, 2009). We utilized ChIP-Seq data to identify KDM5B as a Nanog target gene. KDM5B depletion attenuated ESC self-renewal and decreased expression of genes associated with mitosis and nuclear metabolism. In particular, KDM5B regulated core DNA biosynthesis machinery, including Pola1, Cdc45, Mcm3–7, Orc1/2/3/5/6, Cdc25a, and Ccnb1. Interestingly, Mcm4 and Mcm6 contribute to rapid early embryonic proliferation (Coue et al, 1996) and both Mcm4 and Cdc25a null embryos fail to expand ICM (Shima et al, 2007; Lee et al, 2009a). Interestingly, many KDM5B-regulated genes are not targets of core pluripotency-associated transcription factors. Thus, KDM5B regulates ESC self-renewal by activating a novel gene network that regulates ESC proliferation and DNA synthesis.
We show here that KDM5B recruitment depends on H3K36me3, a histone modification associated with transcriptional elongation (Li et al, 2007). The KDM5B orthologue Lid interacts with MRG15, a chromodomain protein that recognizes H3K36me2/3 (Zhang et al, 2006; Lee and Shilatifard, 2007). We show that KDM5B interacts with MRG15 and that knockdown of MRG15 reduced KDM5B recruitment. Genome-wide MRG15 ChIP-seq analysis shows a high degree of colocalization with KDM5B. MRG15 is an orthologue of Eaf3, a yeast Rpd3S histone deacetylase complex component (Carrozza et al, 2005; Keogh et al, 2005) and has also been linked to transcriptionally active genes in Drosophila (Filion et al, 2010). Interestingly, KDM5B also interacts with Rpd3S complex orthologues, HDAC1 and Sin3A. Similarly, Drosohpila Lid and mammalian KDM5A also interact with an Eaf3/MRG15, Sin3, and Rpd3/HDAC1 Rpd3S-like complex (Hayakawa et al, 2007; Moshkin et al, 2009). Our data indicate that KDM5B can be tethered to chromatin by H3K36me2/3 via association with an Rpd3S-like complex. As ∼17% of KDM5B ChIP-Seq peaks are not adjacent to H3K36me3, other mechanisms for recruitment may also exist. Interestingly, Setd2 knockdown and inhibition of PolII elongation induced a more robust decrease in KDM5B occupancy than MRG15 knockdown, suggesting recruitment to H3K36me3 may involve additional mechanisms. Recent work has identified a family of PWWP domain proteins that also interact with H3K36me3 in mammalian cells (Vermeulen et al, 2010; Vezzoli et al, 2010).
Recruitment of Rpd3S by H3K36me3 is believed to prevent cryptic transcription by removal of transcription-associated histone acetylation (Carrozza et al, 2005; Keogh et al, 2005). Mutation of Set2 or Rpd3S subunits increases cryptic intragenic transcription in Saccharomyces cervisiae and antisense transcription in Schizosaccharomyces pombe (Carrozza et al, 2005; Keogh et al, 2005; Nicolas et al, 2007). Interestingly, elongating PolII also interacts with the Set1 H3K4 methyltransferase in yeast (Krogan et al, 2003; Ng et al, 2003; Dehe et al, 2006) and cryptic intragenic deposition of H3K4me3 represses gene expression in yeast (Pinskaya et al, 2009). We show that elongating PolII interacts with core subunits of the H3K4 methyltransferase and that inhibition of polymerase elongation reduces intragenic H3K4me3 deposition. As H3K4me3 has been proposed to serve as a nucleation site for PolII (Vermeulen et al, 2007), we propose that KDM5B recruitment functions to prevent cryptic transcription. Consistent with this idea, KDM5B knockdown triggered a marked increase in H3K4me3 specifically at intragenic target sites and ChIP-Seq analyses confirmed that KDM5B knockdown is correlated with localized increase in H3K4me3. Importantly, knockdown of KDM5B increased spurious transcription in KDM5B target genes. Cryptic transcription induced by KDM5B knockdown is associated with a marked decrease in the transcription of functional full-length mRNAs. Decreased production of functional transcripts was associated with repression of elongation because PolII recruitment was specifically reduced in intragenic regions, but not at sites of initiation. We also show that some cryptic transcripts specifically originate from the antisense strand. Cryptic transcription could have a role in repressing productive transcription via inhibition of PolII processivity, altering nucleosomal structure, or post-transcriptional mechanisms, such as RNAi. The recent observation that MRG15 regulates PTB-dependent alternative splicing also suggests a possible role for KDM5B's regulation of intragenic H3K4me3 in splicing (Luco et al, 2010).
Accumulation of H3K4me3 is believed to generate an epigenetic landscape conducive to high levels of transcriptional activity (Kouzarides, 2007; Li et al, 2007) via its ability to recruit initiating PolII (Vermeulen et al, 2007). Our data suggest that active demethylation of intragenic H3K4me3 by KDM5B is another mechanism by which this H3K4me3 gradient is established. Interestingly, the human H3K4me2/me1 demethylase, LSD2, also accumulates in intragenic regions presumably via its interaction with elongating PolII (Fang et al, 2010). Nevertheless, several lines of evidence suggest that KDM5B and LSD2 have distinct functions. Unlike H3K4me3, LSD2's substrate (H3K4me2/me1) is not associated with recruitment of initiating polymerase. Moreover, KDM5B but not LSD2 interacts with the Rpd3S component HDAC1, while LSD2 but not KDM5B directly interacts with elongating PolII (Fang et al, 2010). These experiments suggest that LSD2 and KDM5B exist in different complexes and are recruited to active genes via different mechanisms. A recent study also found that the H3K36 demethylase, KDM2A, is recruited to non-methylated CpG islands and maintains low levels of H3K36me at these regions (Blackledge et al, 2010). As CpG islands are associated with promoter regions, a phenomenon similar to the action of KDM5B may serve to repress accumulation of H3K36me at promoters.
Recent studies suggest that initiation of elongation is a rate-limiting step for ESC transcriptional activity. In all, 30% of human ESC genes undergo transcriptional initiation without evidence of productive elongation (Guenther et al, 2007) and PolII is paused at the promoters of many ESC genes (Rahl et al, 2010). Our data show that KDM5B safeguards productive elongation complements data linking c-Myc to initiation of transcriptional elongation (Rahl et al, 2010) and suggest that regulation of elongation may contribute to ESC self-renewal. Regulators of transcriptional elongation and KDM5-family demethylases have also been implicated in cancer. Nup98 fusions to the NSD1 H3K36 methyltransferase and KDM5A induce leukaemia (Wang et al, 2007, 2009). Moreover, KDM5B is up-regulated in human cancers (Xiang et al, 2007; Yamane et al, 2007; Hayami et al, 2010) and contributes to self-renewal of a subpopulation of melanoma cells required for tumour growth (Roesch et al, 2010). It would be interesting to determine whether epigenetic regulation of transcriptional elongation by KDM5B contributes to cancer cell renewal.
Materials and methods
Plasmids
Mouse KDM5B was amplified using PCR from murine cDNA and cloned into pUB6-V5 (Invitrogen). HA-tagged MRG15 vector is a gift from Dr Kaoru Tominaga and Dr Olivia M Pereira-Smith. All vectors were verified by sequencing.
Cell culture and reagents
J1 and ZHBTc4 mouse ESCs were cultured at 37°C with 5% CO2. All cells were maintained on gelatin-coated dishes in Dulbecco's modified Eagle medium (Invitrogen), supplemented with 10% ESC-tested foetal bovine serum (PAA), 0.1 mM MEM NEAA (Invitrogen), 2 mM L-glutamine (Invitrogen), 100 U/ml/100 μg/ml Pen Strep (Invitrogen), 0.055 mM β-mercaptoethanol (Invitrogen), and 1000 U/ml LIF (Chemicon). Human ESCs (H1) were cultured on matrigel-coated plates in mTeSR1 medium (StemCell Technologies) and passaged using collagenase IV according to the manufacturer's protocol (StemCell Technologies). The 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB, 50 μM; Sigma) was directly applied to ESC medium for indicated periods of time. AP activity was monitored using the commercial Alkaline Phosphatase Detection kit (Millipore) according to the manufacturer's instructions.
Antibodies
Antibodies used for immunoblotting include KDM5B (Abcam 50958 or Santa Cruz sc-67035), Oct4 (Santa Cruz sc-5279), Nanog (Chemicon AB5731), Histone 3 (Abcam ab1791), H3K4me3 (Active Motif 39159), H3K9me3 (Abcam ab8898-100), H3K27me3 (Abcam ab6002-100), H3K36me3 (Abcam ab9050-100), MRG15 (gift from Dr Kaoru Tominaga and Dr Olivia M Pereira-Smith), RbBP5 (Bethyl A300-109A), WDR5 (Abcam, ab22512-100), and Ash2L (Bethyl A300-112A). Antibodies utilized for ChIP or IP: KDM5B (Abcam ab50958 and Santa Cruz sc-67035), Nanog (Chemicon, AB5731), Oct4 (Santa Cruz, sc-5279), Histone 3 (Abcam, ab1791), H3K4me3 (Active Motif, 39159), H3K36me3 (Abcam, ab9050-100), MRG15 (AVIVA Systems Biology, ARP32832-T100), non-phosphorylated RNAP (Covance 8GW16), Ser2P RNAP (Abcam ab5095-100 and Covance H5), Ser5P RNAP (Covance H14), WDR5 (Abcam, ab22512-100), and Ash2L (Bethyl A300-112A).
RNA interference
RNA interference of J1 ESCs was conducted as previously described (Loh et al, 2006). The following 19 mer sequences were cloned into the pSUPER.puro backbone according to the manufacturer's instructions (Oligoengine). We used at least two shRNA for each knockdown experiment to minimize off-target effectsKDM5B-1: 5′-TCTTTGCCCTCGGTGTGAC-3′; KDM5B-2: 5′-GAGATGCACTCCGATACAT-3′; MRG15-1: 5′-GAGTACCATCGGAAAGCCG-3′; MRG15-2: 5′-ACAATATGCAGAGGGCAAG-3′; Setd2-1: 5′-GTCTTAAGCTCATTCAGAA-3′; Setd2-2: 5′-GGGAGTGTCTGATGTTGAA-3′. The 19-mer sequences for GFP, Oct4, and Nanog were previously described (Loh et al, 2006). Puromycin (Sigma) selection was introduced 1 day after transfection at 1 μg/ml, and maintained for 2–3 days prior to harvesting.
RNA isolation, reverse transcription, and quantitative real-time PCR
Total or nuclear RNA was isolated using the Trizol (Invitrogen) method following manufacture's protocol and reverse transcribed using MMLV (Invitrogen). Real-time PCR was conducted as described previously (Impey et al, 2004). All quantitation utilized standard curve real-time PCR. For detection of cryptic transcripts, we performed gene-specific and strand-specific RT–PCR. Briefly, primers were designed toward introns of validated KDM5B target genes where we observed significant increase of H3K4me3 following KDM5B knockdown. Additionally, we designed primers spanning a terminal 3′ intron of corresponding KDM5B target genes to detect functional full-length transcripts. Primers will be provided upon request.
Proliferation assay and cell-cycle analysis
Equal numbers of J1 ESCs (500 000) were plated in six-well plates and transfected with the indicated plasmids. And three wells were counted with a hemocytometer at the indicated times. For FACS analyses, ESCs were trypsinized, washed with PBS, fixed with 70% ice-cold ethanol, and stained with propidium iodide (10 μg/ml, Sigma). DNA cell-cycle analysis was conducted using an FACS Calibur instrument (Becton Dickinson) in the Oregon Stem Cell Center Flow Cytometry Core. Cell-cycle compartments were deconvoluted from single-parameter DNA histograms of 20 000 cells and cell-cycle data were analysed using ModFit (Verity Software House).
Chromatin immunoprecipitation assay
ChIP was conducted as described previously (Impey et al, 2004). Briefly, ESCs were fixed with 1% formaldehyde for 10 min at room temperature. Chromatin was sonicated and immunoprecipitated using indicated antibodies overnight. All ChIP-PCR analysis was conducted using real-time PCR. Primers will be provided upon request.
Microarray analysis
Control and KDM5B siRNA (designed from Darmacon) treated samples were run on MOE430 2.0 Affymetrix chips using standard procedures. ESC retinoic acid time course microarray data were obtained from GEO (GSE2972). CEL file data were processed using RMA and significant genes were isolated using the limma (heatmap analysis) or SAM (gene lists) R packages.
ChIP-Seq and RNA-Seq analysis
For RNA-Seq, ∼10 ng of Poly(A) RNA was converted to double-stranded cDNA as described previously (Nagalakshmi et al, 2008). ChIP and cDNA samples were sequenced using standard Solexa protocols. In total, 25 bp reads were mapped to the mouse genome (UCSC mm9) and unique tags were selected for analysis. For ChIP-Seq, areas of enrichment at an FDR of 0.05 were determined using a custom sliding-window approach based on previous work (Fejes et al, 2008), but implemented in C++. For RNA-Seq analyses, counts of mapped tags in RefSeq exons were determined and significantly regulated genes were selected using an FDR-adjusted χ2 statistic. Statistical analyses were carried out in the R programming environment. H3K36me3 and H3K36me27 ChIP-Seq data used in this study were generated from Solexa/Illumina output from a previous study (Mikkelsen et al, 2007).
Bioinformatic and statistical analyses
Microarray heatmaps were generated by selecting significantly regulated genes at an FDR of 0.1 using the Bioconductor limma package and sorting based on fold-change (Gentleman et al, 2004). The RNA-Seq heatmap was generated in R by selecting genes with an FDR<0.001 and a fold-change >2 and sorting based on the FDR-adjusted χ2 P-value. All heatmap data were log scaled and row scaled. Pie charts were generated using custom annotation scripts and statistical significance was assessed with the Wilcoxon rank-sum test. Profile plots of ChIP-Seq data relative to RefSeq genes were generated in R by setting all gene lengths equal to 1 and plotting tag density. Profile plots of histone methylation relative to Ref-Seq genes sorted by KDM5B Peak area were generated by selecting equal sets of histone methylation ChIP-Seq peaks and plotting peaks counts that either overlap with a Ref-Seq gene or are within 1 kb of a gene end. ChIP-Seq profile plots relative to heatmaps were generated by displaying averaged ChIP-Seq peak counts that overlap with at least 1 bp of a Ref-Seq genes present in the heatmap (black profile). A ‘background’ for ChIP-Seq profile plots was generated by randomizing the position of ChIP-Seq peaks and the blue significance line was generated using a permutation test. The UCSC genome browser was used to visualize RNA-Seq and ChIP-Seq data. Gene ontology analysis was conducted using DAVID (Dennis et al, 2003). Comparison of low-throughput biological data utilized the Student's t-test or ANOVA followed by a post-test where appropriate. The Storey Q-value was used for all FDR adjustments (Storey and Tibshirani, 2003). ChIP-Seq, RNA-Seq, and microarray data have been deposited in GEO (accession number will be added).
Supplementary Material
Acknowledgments
We thank Ian Chambers and Austin Smith for providing ZHBTc4 ES cells. We also thank Kaoru Tominaga and Olivia M Pereira-Smith for providing MRG15 anti-sera and HA-tagged MRG15 expression plasmid. We thank the Oregon Stem Cell Center Flow Cytometry Core for assistance with flow cytometry data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG Jr (2005) Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 18: 623–635 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ (2010) CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Chan SW, Hong W (2001) Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J Biol Chem 276: 28402–28412 [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K (2007) RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076 [DOI] [PubMed] [Google Scholar]
- Coue M, Kearsey SE, Mechali M (1996) Chromotin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J 15: 1085–1097 [PMC free article] [PubMed] [Google Scholar]
- Dehe PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, Rosaleny LE, Tordera V, Chavez S, Stewart AF, Geli V (2006) Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 281: 35404–35412 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C (2008) The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol 28: 5312–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do JT, Scholer HR (2009) Regulatory circuits underlying pluripotency and reprogramming. Trends Pharmacol Sci 30: 296–302 [DOI] [PubMed] [Google Scholar]
- Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, Shi YG (2010) Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell 39: 222–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ (2008) FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24: 1729–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J (2007) RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cell 12: 811–826 [DOI] [PubMed] [Google Scholar]
- Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R (2010) Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH (2004) Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG Jr (2007) The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128: 889–900 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Lee G, White LS, Hurov KE, Stappenbeck TS, Piwnica-Worms H (2009a) Response of small intestinal epithelial cells to acute disruption of cell division through CDC25 deletion. Proc Natl Acad Sci USA 106: 4701–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shilatifard A (2007) A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res 618: 130–134 [DOI] [PubMed] [Google Scholar]
- Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2009b) The H3K4 demethylase lid associates with and inhibits histone deacetylase Rpd3. Mol Cell Biol 29: 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2007) The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol 14: 341–343 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10: 731–739 [DOI] [PubMed] [Google Scholar]
- Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F (2008) Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucl Acids Res 36: 2852–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV (2008) Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell 31: 520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, Taylor-Papadimitriou J (1999) A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem 274: 15633–15645 [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T (2010) Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K (2007) Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP (2009) Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell 35: 782–793 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI (2007) Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 14: 372–380 [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK (1995) Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377: 119–128 [DOI] [PubMed] [Google Scholar]
- Niwa H (2007a) How is pluripotency determined and maintained? Development 134: 635–646 [DOI] [PubMed] [Google Scholar]
- Niwa H (2007b) Open conformation chromatin and pluripotency. Genes Dev 21: 2671–2676 [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev 22: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A (2009) H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J 28: 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA (2010) c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M (2010) A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN (2007) The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev 21: 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC (2007) A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet 39: 93–98 [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A (2009) Nanog is the gateway to the pluripotent ground state. Cell 138: 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y (1992) Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell 69: 871–881 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M (2010) Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol 17: 617–619 [DOI] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP (2007) NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9: 804–812 [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD (2009) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD (2007) JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA 104: 19226–19231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25: 801–812 [DOI] [PubMed] [Google Scholar]
- Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J (2006) Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucl Acids Res 34: 6621–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.