Abstract
The structural basis of the tRNA selection process is investigated by cryo-electron microscopy of ribosomes programmed with UGA codons and incubated with ternary complex (TC) containing the near-cognate Trp-tRNATrp in the presence of kirromycin. Going through more than 350 000 images and employing image classification procedures, we find ∼8% in which the TC is bound to the ribosome. The reconstructed 3D map provides a means to characterize the arrangement of the near-cognate aa-tRNA with respect to elongation factor Tu (EF-Tu) and the ribosome, as well as the domain movements of the ribosome. One of the interesting findings is that near-cognate tRNA's acceptor stem region is flexible and CCA end becomes disordered. The data bring direct structural insights into the induced-fit mechanism of decoding by the ribosome, as the analysis of the interactions between small and large ribosomal subunit, aa-tRNA and EF-Tu and comparison with the cognate case (UGG codon) offers clues on how the conformational signals conveyed to the GTPase differ in the two cases.
Keywords: cryo-EM, ribosome, ternary complex, translation, tRNA incorporation
Introduction
The translation of the genetic code into proteins is a task carried out universally, and with high fidelity, by the ribosome. The decoding process relies on the complementary Watson-Crick base matching between the codon of the messenger RNA (mRNA) and the anticodon of the incoming aminoacyl-tRNA (aa-tRNA), delivered to the ribosome in a ternary complex (TC) with GTP and the GTPase elongation factor Tu (EF-Tu). For the required accuracy to be achieved, the ribosome must recognize the cognate aa-tRNA species and reject those near- and non-cognate species that form non-Watson-Crick pairing interactions with the mRNA (one and at least two mismatches, respectively). The ability of the protein synthesis machinery to differentiate between these species is fundamental to protein synthesis in all life forms. While the difference in free energy of binding between cognate and non-cognate aa-tRNAs is large enough, this difference is not sufficiently large to discriminate cognate versus near-cognate species. Moreover, we know that the events leading to discrimination must take place on a relatively fast time scale that is commensurate with the known rates of protein synthesis (Zaher and Green, 2009). This conundrum was resolved when kinetic studies showed that the ribosomal machinery uses a kinetic discrimination process to exclusively accelerate the two major steps of tRNA selection, GTPase activation and accommodation, for cognate aa-tRNA species (Pape et al, 1998, 1999; Blanchard et al, 2004; Gromadski and Rodnina, 2004; Cochella and Green, 2005; Gromadski et al, 2006), thus promoting rapid and high-fidelity tRNA selection.
The differences in forward rates are attributed to an induced-fit mechanism (Pape et al, 1999), wherein instrumental conformational changes are induced only by binding of the cognate species. Some of these rearrangements have been characterized by structural approaches. First, a number of cryo-electron microscopy (cryo-EM) studies established (Stark et al, 2002; Valle et al, 2002, 2003; Li et al, 2008; Schuette et al, 2009; Villa et al, 2009) that the incoming cognate aa-tRNA is deformed, relative to the X-ray structure of free tRNA, during the early steps of tRNA selection before EF-Tu:GDP is released (in a state referred to as A/T). The details of this particular distortion of the tRNA have recently been confirmed by the first crystal structure of ribosome-bound TC (Schmeing et al, 2009). The deformation is characterized by a partial bending of the tRNA structure that permits interaction between the anticodon of the tRNA and the mRNA in the decoding centre while the aa-tRNA is still bound to EF-Tu. Second, X-ray crystallography of 30S subunits soaked with anticodon stem loops (ASLs) has demonstrated that the cognate codon–anticodon match triggers localized changes in the universally conserved G530, A1492 and A1493 of the 16S rRNA (Ogle et al, 2001). These nucleotides strictly monitor the Watson-Crick complementarity between the codon and the anticodon at the first two positions, but not the third (i.e. the wobble position). In addition to these local rearrangements, global concerted changes in the head and shoulder regions of the 30S subunit also appear to be triggered upon cognate codon–anticodon pairing. These conformational changes result in an overall more closed conformation of the 30S subunit (Ogle et al, 2002).
Despite these many insights, a complete understanding of the structural basis for the induced-fit mechanism remains elusive, as fully functional 70S ribosomes in complex with near-cognate TCs have not been visualized to date. To address this issue, we have determined the structure of Escherichia coli ribosomes programmed with a near-cognate codon by means of cryo-EM single-particle reconstruction. Comparison with a ribosome complex programmed with the cognate species provides structural evidence that the near-cognate aa-tRNA is differently positioned. We further describe how the structural differences affect EF-Tu positioning and the structural context of the ribosome, ultimately bringing about high-fidelity discrimination.
Results
Overview of the structures
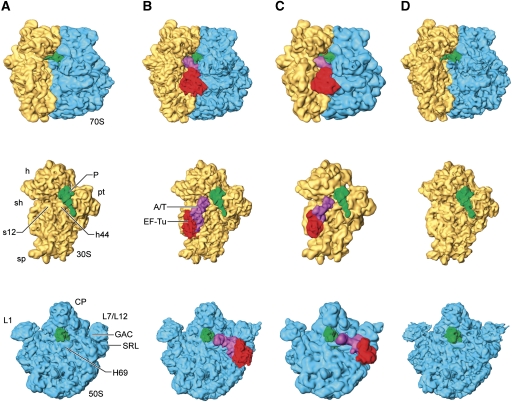
Cryo-EM maps were generated from ribosomes programmed with an AUG codon in the P site and either a cognate (UGG) or near-cognate (UGA/stop) codon in the A site. These ribosome complexes were subsequently loaded with fMet-tRNAfMet and kirromycin-stalled TCs, Trp-tRNATrp•EF-Tu•GTP (see Materials and methods section). Kirromycin arrests the TC on the ribosome after the hydrolysis of GTP, but before the conformational change that takes place in EF-Tu (Parmeggiani and Swart, 1985). The study of this kirromycin-arrested conformer is of particular importance because the antibiotic is thought to lock EF-Tu in a state that resembles the earlier, pre-hydrolysis GTPase-activated state (Rodnina et al, 1994). This state is regarded as a state that comes after, but is still very close to, the GTP hydrolysis transition state. All the complexes were prepared at low Mg2+ concentration (3.5 mM) as it is well documented that excess amounts of Mg2+ increase the errors of decoding, thus altering the overall selectivity of the process (Gromadski et al, 2006). Due to the expected lower stability and higher dissociation rates of the tRNA in the near-cognate codon situation (Rodnina et al, 1996), which leads to decreased levels of A/T-site occupancy, we applied supervised classification methodology to sort the original data set into two major subsets (see Materials and methods section). To minimize any risk of reference bias in the final structures, the selected particles were then re-aligned using the map of an ‘empty’ ribosome (i.e. without TC and P-site tRNA) as a starting reference and refined iteratively during multiple rounds. This procedure allows structures to be obtained that diverge from the initial references due to the intrinsic structural information contained in the corresponding subsets. The first group (shown in Figure 1C) was composed of merely ∼8% of the initial particle set, and corresponded to ribosomes with A/T-bound near-cognate TCs (Figure 1C), similar to those shown in the cognate case (Figure 1B). The second group, composed of the remaining 92%, corresponded to ribosomes bearing a single tRNA in the P site (i.e. without A/T-bound TC in the A site) (Figure 1D). The conformation of this latter reconstructed ribosome was found to be virtually identical to that of the initiation 70S ribosome with the initiator P-site fMet-tRNAfMet generated as a control (Figure 1A). The final resolution of the maps was 8.25 Å (TC on cognate codon), 13.2 Å (TC on near-cognate codon), 8.05 Å (vacant near-cognate A site) and 8.65 Å (initiation-like 70S ribosome). Resolution was determined by 0.5 cutoff in the Fourier shell correlation (Supplementary Figure S1).
Figure 1.
Three-dimensional cryo-EM structures of 70S ribosomes. Cryo-EM densities are shown for the small (yellow) and large (blue) subunits, A/T- and P-site tRNAs (magenta and green, respectively) and EF-Tu (red). (A) Initiation 70S ribosome. Ribosomes bearing ternary complexes (B) with cognate codon and (C) with near-cognate codon. (D) Initiation-like ribosomes showing a vacant A site. Labels and landmarks: sp (spur), h (head), sh (shoulder), pt (platform), CP (central protuberance), L1 (L1 stalk), L7/L12 (L7/L12 stalk), h44 (small subunit helix 44), SRL (large subunit sarcin-ricin loop), GAC (GTPase-associated centre), H69 (large subunit helix 69), s12 (small subunit protein 12).
In the reconstructions displayed in Figure 1, the densities of the TCs both for cognate (Figure 1B) and near-cognate (Figure 1C) species show EF-Tu bound to the base of the L7/L12 stalk of the large subunit, in proximity to the universally conserved α-sarcin-ricin loop (SRL; formed by helix H95 of the 23S rRNA), while the aa-tRNA binds to the GTPase-associated centre (GAC; formed by L11 and 58 nucleotides from H43 and H44) and H69 through its T loop (elbow region) and D arm, respectively. In the 30S subunit, the TCs interact with the shoulder region and helices h4, h5 and h15 of the 16S rRNA (note convention designating helices of 23S rRNA by ‘H’ and those of 16S rRNA by ‘h’). Ribosomal protein S12 is consistently visualized interacting with cognate and near-cognate tRNA species in the vicinity of the D stem. In both cognate and near-cognate cases, the tRNA contacts the tip of h44 (i.e. the decoding centre) via its anticodon region, but the interaction appears distinct in the near-cognate case. While the resolution of the reconstruction precluded any detailed characterization of the decoding site interactions based on the density alone, the flexibly fitted atomic models of the complexes show differences in the decoding centre (data to be introduced below). It is noteworthy that, in our 3D reconstruction, the acceptor stem of the near-cognate tRNA is not as well defined as the ASL, indicating some degree of flexibility (see Figure 2B and C). In addition, the structure is apparently completely disordered at the very 3′ CCA end, as the ultimate string of nucleotides forming the tRNA structure is completely blurred in the experimental density map. These apparent differences summarized here do persist when the two structures are displayed at the same resolution, ∼13.5 Å (Supplementary Figure S2).
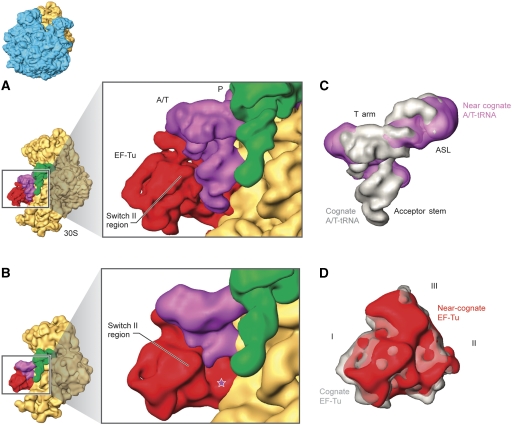
Figure 2.
Interaction of ternary complexes with the ribosome. Cryo-EM densities are shown for the small (yellow), A/T- and P-site tRNAs (magenta and green, respectively) and EF-Tu (red). Small subunits bearing ternary complexes on (A) cognate and (B) near-cognate codons. Superimposition of the experimental cryo-EM densities corresponding to (C) A/T-tRNA and (D) EF-Tu on cognate and near-cognate codons. Cognate tRNA and EF-Tu are shown in grey, whereas near-cognate tRNA and EF-Tu are displayed in magenta and red, respectively (see coloured labels). The star in (B) indicates the missing CCA region of the near-cognate tRNA. Labels and landmarks: acceptor stem, T arm and ASL of tRNA, domains I, II and III of EF-Tu. All other labels are introduced in Figure 1. The orientation of the subunit is shown as a thumbnail on the left.
The structures reveal distinct conformations of A/T-tRNA and EF-Tu
To adopt the A/T state of binding, the tRNA must be distorted at its ASL. The superimposition of cryo-EM densities (Figure 2C) reveals that the A/T-tRNA in near-cognate complexes differs from that seen in the cognate case. First, the hinge region at the junction of the ASL and D stems exhibits a different arrangement. Second, there are differences in the T- and D-loop regions. These changes bring the resolved part of the near-cognate tRNA's acceptor stem into a different geometry at the binding interface with EF-Tu.
To further analyse the results, we built atomic models from the experimental cryo-EM maps using molecular dynamics-based flexible fitting (MDFF) (Trabuco et al, 2008) (described in the Materials and methods section). This allowed us to interpret our density maps in molecular terms to the extent allowed by the resolution. For the comparison of tRNA conformations, structures resulting from MDFF refinements in water were used, since the secondary structure restraints necessary for MDFF performed in vacuo preclude local rearrangements. This approach permitted movements within the tRNA bodies to be more accurately characterized. In Figure 3A, we show the atomic models inferred for both TCs. We see for the near-cognate complex that the tRNA density is partially blurred, indicating the tRNA is more dynamic. In particular, the positions of the terminal portion of the acceptor stem and the 3′ CCA sequence cannot be determined unambiguously. In contrast, the density corresponding to EF-Tu in the near-cognate cryo-EM map is well defined, delineating the location of domains and secondary structure and allowing a meaningful three-dimensional model to be built (Figure 3B and C).
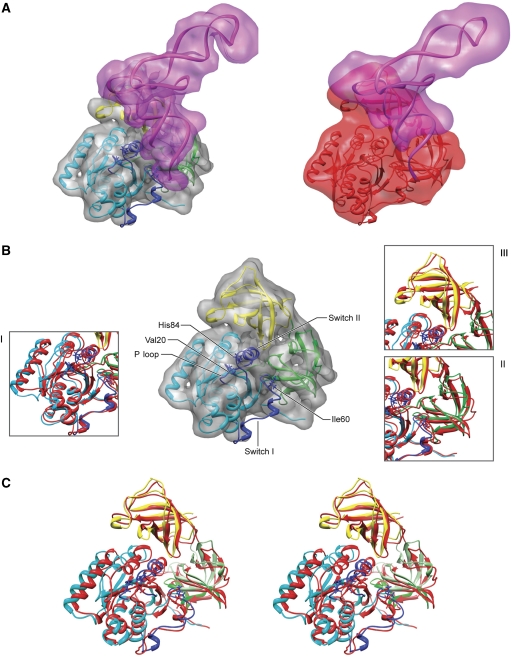
Figure 3.
Fitting of atomic structures into the cryo-EM densities for the stalled ternary complexes. (A) Cryo-EM densities are shown for cognate and near-cognate A/T and EF-Tu in transparent magenta (tRNA) and grey/red (for cognate and near-cognate EF-Tu, respectively). The fitted structures are shown in ribbons. (B) Close-up view of superimposed fitted structures for cognate (coloured by domains) and near-cognate EF-Tu (in red). (C) Stereo-view representation of EF-Tu structures. Cognate EF-Tu fragments 1–202 (domain I), 203–300 (domain II) and 301–393 (domain III) are shown in cyan, green and yellow, respectively. Switch I, II and P loop of domain I are represented in darker blue. Fragments 219–226 and 256–273 of domain II are represented in darker green. The position of residues Val20, Ile60 and His84 is highlighted for comparison in both fitted structures. The EF-Tu complexes are aligned with respect to the 70S ribosomes. Supplementary Figure S3 depicts the stereo-view representation of the same EF-Tu complexes but aligned with respect to each other.
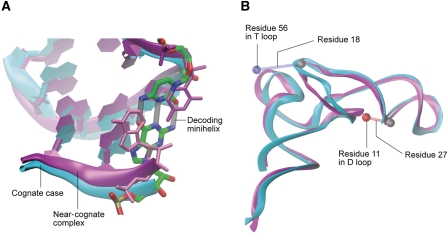
The superimposition of the ASL part of the A/T-tRNA shows that the overall ASL structure is similar for the cognate and near-cognate cases, but there are clear differences in the details. An interesting observation is that codon–anticodon interactions remain stable throughout the entire fitting process for the cognate maps, but break apart at the third basepair position already at the very beginning of the fitting for the near-cognate map (Figure 4A). Moreover, a reorganization of the T and D loops can be seen (Figure 4B). In particular, the D stem is shifted closer to the ASL (distances between phosphorus atoms of residues 11 and 27 are 9 and 13 Å for cognate and near-cognate complexes, respectively) and the T loop shifts away from the D loop in the near-cognate complex (distances between phosphorus atoms of residues 18 and 56 are 11 and 18 Å for cognate and near-cognate complexes, respectively). Apart from the shift of the T loop, the internal structure of the acceptor stem in the cognate and near-cognate complexes is similar. The largest differences are observed in the 3′ terminal part, though since this region is not resolved in the near-cognate complex, no rigorous comparison is possible.
Figure 4.
Comparison of the modelled A/T-tRNA in cognate and near-cognate complexes. (A) The superimposition of the ASL shows the basepair at the third position of the decoding minihelix (grey cylinders) in the cognate case (shown in cyan), which is disrupted in the near-cognate complex (shown in magenta). The alignment of the two structures was performed using the backbone atoms of tRNA residues 31–39. (B) Superimposition of the full tRNAs shows the differences in the elbow region between cognate and near-cognate case. Atoms used for calculation of internal distances are shown as spheres. The blue sphere corresponds to residue 56 in the T loop, while the red sphere shows residue 11 in the D loop. The calculated distance is indicated by a cylinder at the reference point, which is residue 18 and 27 for the T and D loop, respectively. The alignment of the structures was performed using the backbone of the resolved part of the tRNA, that is residues 1–70.
The relative orientation between the various EF-Tu domains in the present structures is similar to that seen in the previously characterized GTP-bound form (Abel et al, 1996; Polekhina et al, 1996). However, due to the high flexibility of the acceptor stem in the near-cognate complex and subsequent modification of the interface between the aa-tRNA and EF-Tu, several changes are noticeable (Figures 2D and 3C; Supplementary Figure S3). In domain I, there are two main features to be noticed. First, the reconstructions show that the density corresponding to switch I (the effector loop, residues 40–62) is not completely resolved, suggesting its dynamic nature. It is worth noting that this region is better represented in the near-cognate than in the cognate complex when the maps are displayed at low threshold (data not shown). In addition, the atomic models suggest, supported by experimental density between residues 55 and 62 in both cases, that the switch I is positioned closer to the rest of domain I in the near-cognate case (Figure 3B). Considering that the P loop containing residue 20 is kept in place by the SRL in an identical manner in both complexes, this shift renders the ‘hydrophobic gate’ (formed by residues 20 and 60 and thought to control the access of the conserved catalytic His84 to GTP) (see Daviter et al (2003) and references therein) more closed in the near-cognate than in the cognate case (distance between residues forming the gate is 14 and 19 Å, respectively). Nevertheless, the gate is still more open in the near-cognate case than in the isolated TC (8 Å, PDB 1OB2), which may explain the reduced but still measurable rate of GTP hydrolysis of near-cognate species. We note that these results are qualitative due to the limited resolution of the EM data, and it is likely that the ribosome populations on which the reconstructions are based are to some extent heterogeneous with regards to the switch I position. Our results are nevertheless worth bringing into this forum as a contribution to the discussion.
The switch II region (containing the critical His84 and extending from residues 80–100) is also distinctly structured in cognate versus near-cognate species. The differences in the orientation and positioning of the region of switch II are particularly well seen for the junction between domains I and III (see Figure 2D), and they probably reflect the higher flexibility/freedom of the switch II region in the near-cognate complex in the absence of a correctly structured acceptor stem. Indeed, cryo-EM and X-ray data have shown that upon binding to the ribosome the switch II loop is shifted toward the tRNA's acceptor stem (Schmeing et al, 2009; Villa et al, 2009). Our fitted structures show that residue Asp86 in particular moves by about 5 Å, inserting itself between the SRL and the acceptor stem of the tRNA in the near-cognate complex. This observation would be consistent with the more dynamic properties of the acceptor stem in the near-cognate case. However, we cannot exclude the possibility that this observed shift in the inferred model is an artifact of MDFF fitting in areas of the density affected by conformational averaging: the tRNA density is poorly resolved in this region for the near-cognate complex, so it may be that the residues around Asp86 were drawn into the density between the SRL and the acceptor stem.
Finally, while domain III in the near-cognate case is rotated counter-clockwise when compared with its cognate counterpart, domain II, the region interacting with the 16S rRNA, displays no major rearrangements when aligned (see Supplementary Figure S3). Atomic data suggest that local changes in domain II may be implicated in GTPase activation (Schmeing et al, 2009), as residues 256–273 and 219–226, composing a β loop and presenting the highly conserved Gly222 (Vorstenbosch et al, 1996), shift toward the small subunit when compared with their position in solution. The conformation of domain II in our fitted structures is very similar as the same residues are found to interact with the 16S rRNA region formed by helices h4, h5 and h15.
Exploring the domain closure of the small subunit
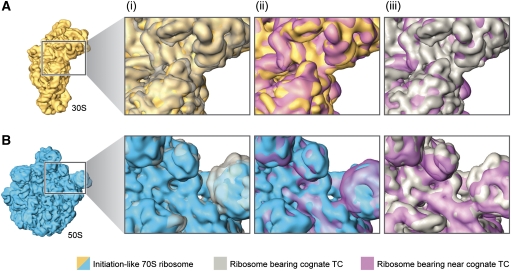
As previously mentioned, initial X-ray crystallography of the 30S subunit identified local changes in the decoding centre that result in more global conformational changes. These large-scale movements of the head and shoulder, referred to as domain closure, involve the rotation of the head and shoulder toward the subunit interface where decoding takes place (Ogle et al, 2002). During this rearrangement, new contacts between S12 and 16S rRNA (h44 and h27) are formed. To investigate whether the binding of near-cognate TCs changes the global conformation of the small subunit, we compared the structures of cognate and near-cognate decoding 30S subunits by superimposing the relevant cryo-EM densities. The results presented in Figure 5A indicate that changes in the relative position of the head and shoulder regions of the small subunit occur only in the cognate situation.
Figure 5.
Domain closure and GAC movement during aa-tRNA incorporation. Cryo-EM densities are shown for the small (A) and large subunits (B). From left to right, close-up views of the superimposed cryo-EM densities from (i) initiation ribosome (30S and 50S subunits shown in yellow and blue, respectively) and ribosome bearing cognate (grey) ternary complex, (ii) initiation ribosome and ribosome bearing near-cognate (purple) ternary complex (see coloured labels) and (iii) ribosomes bearing cognate (grey) and near-cognate (purple) ternary complexes.
To further analyse and quantify the details of the domain closure motion, we also compared the extent of this rearrangement for several existing X-ray structures with the atomic models inferred from our cryo-EM reconstructions (Table I). In particular, we considered the structures of the isolated 30S subunit, either in its apo form (Ogle et al, 2001) or in complex with cognate, near-cognate and non-cognate tRNA ASLs (Ogle et al, 2001, 2002). In addition, we analysed the structures of 70S ribosomes with an empty A site (Korostelev et al, 2006) or in complex with either A/T- (Schmeing et al, 2009) or A/A-tRNAs (Selmer et al, 2006). Tensors of inertia were computed for individual domains of the small subunit (see Materials and methods section and Supplementary Figure S4), which enabled us to characterize the movements of the body with respect to the platform (BPlopcl), tilt of the head domain towards the subunit interface (Htilt), opening/closing of the head relative to the shoulder (Hopcl) and swiveling of the head domain (Hswiv). We note that the variations in the values of the measured domain motions are large, even within the set of X-ray structures. However, the trends of motion, that is, which of the complexes is more closed or more open, is a robust descriptor.
Table 1. Measurements of 30S subunit domain displacements.
| PDB | BPlopcl | Htilt | Hopcl | Hswiv | Note |
|---|---|---|---|---|---|
| 2AVY | −0.53 | −1.87 | 0.44 | −2.97 | eco, 70S, apo |
| 2AW7 | −0.85 | −6.40 | −4.27 | −7.09 | eco, 70S, apo |
| 3I1O | 0.37 | −3.31 | −0.02 | −2.26 | eco, 70S, apo |
| 3I1S | 0.23 | −0.25 | 0.78 | −0.01 | eco, 70S, P-site fMet-ASL |
| 2I2U | 0.00 | 0.00 | 0.00 | 0.00 | eco, 70S, P-site Phe-ASL |
| 2I2P | 0.01 | 0.01 | 0.00 | −0.02 | eco, 70S, P-site Phe-ASL |
| 3I21 | 1.21 | 0.59 | 1.08 | 0.71 | eco, 70S, P-site Phe-ASL, A-site Phe-ASL |
| 2OW8 | 1.88 | 1.54 | 2.13 | 1.70 | tth, 70S, P-site Phe, E-site mix |
| 2J00 | 3.22 | 2.06 | 1.59 | 2.53 | tth, 70S, P-site fMet, A-site Phe-ASL, E-site noncog+PAR |
| 2J02 | 3.80 | 2.23 | 1.73 | 2.82 | tth, 70S, P-site fMet, A-site Phe-ASL, E-site noncog+PAR |
| 2WRN | 3.66 | 1.64 | 1.47 | 2.19 | tth, 70S, P-site Phe, A/T Thr/tern, E-site Phe noncog+PAR |
| 2WRQ | 3.72 | 1.57 | 1.66 | 2.32 | tth, 70S, P-site Phe, A/T Thr/tern, E-site Phe noncog+PAR |
| EM5036 | 4.30 | 2.03 | 1.50 | 2.23 | Refitted to the same density as 3FIH |
| 3FIH | 4.31 | 1.90 | 1.34 | 2.42 | eco, 70S, P-site Phe, A/T Phe/tern, E-site Phe noncog |
| cog | 3.93 | 1.96 | 1.41 | 2.44 | This work |
| nearcog | 3.42 | 1.62 | 1.19 | 2.13 | This work |
| 1IBM | 2.88 | −0.81 | −0.71 | 1.80 | tth, 30S, cog ASL in A site |
| 1N34 | 1.01 | −2.52 | −0.26 | 0.78 | tth, 30S, nearcog ASL in A site |
| 1N36 | 1.08 | −3.06 | −0.13 | 0.26 | tth, 30S, noncog ASL in A site |
| 1IBK | 2.08 | −0.47 | −0.92 | 1.97 | tth, 30S, apo+PAR |
| 1IBL | 4.03 | 0.14 | 0.42 | 2.35 | tth, 30S, cog ASL in A site+PAR |
| 1N32 | 3.96 | 0.01 | 0.43 | 2.29 | tth, 30S, nearcog ASL in A site+PAR |
| 1N33 | 3.51 | −0.53 | 0.11 | 1.91 | tth, 30S, noncog ASL in A site+PAR |
| 1J5E | 1.78 | −0.96 | −1.12 | 1.79 | tth, 30S, apo, spur in P site |
| BPlopcl, body/platform opening, larger angle indicates a relative closing of the two domains; Htilt, tilt of the head domain towards the subunit interface, larger angle corresponds to stronger tilt; Hopcl, opening/closing of the head relative to the shoulder, larger angle corresponds to a more closed structure; Hswiv, swiveling of the head domain, larger angle corresponds to a structure in which the beak rotates away from the inter-subunit side; PAR, paramomycin. | |||||
| The values reported are average values obtained from the last ns of MDFF simulation trajectories. In this phase of the calculation, the total RMSD does not change by >0.1 Å/ns and the fit is regarded as converged. The absolute values represent the angle formed between one of the vectors of the inertia tensor of one domain with a vector of the inertia tensor of another domain. The difference between the measured angle and the reported angle is a constant offset, which corresponds to the measured value in the reference structure. | |||||
A look at the table indicates that in the presence of paromomycin, the small subunit closes in response to the mere presence of a tRNA ASL. Interestingly, no difference is seen in the extent of closing between the cognate and near-cognate complexes, consistent with the observation that paromomycin generally increases miscoding (Pape et al, 2000). In contrast, in the absence of paromomycin, the cognate and near-cognate complexes can be clearly distinguished, the latter being similar to the non-cognate case. In 70S complexes, a similar trend is observed: ribosome complexes with either cognate A/T- or cognate A/A-tRNAs exhibit a larger extent of small subunit closure than a ribosome with an empty A site. This is true for both E. coli and Thermus thermophilus complexes.
A more systematic analysis of the existing small subunit shows that, apart from the relative motion of the body and platform, the head domain changes its relative orientation with respect to the rest of the small subunit as a function of the type of tRNA in the A site (cognate versus near-cognate). In agreement with the earlier findings (Ogle et al, 2002), the results in Table I show that for all of the analysed complexes, the tilt angle—that is, the inclination of the head toward the subunit interface—is consistently larger in cognate complexes as compared with apo, non-cognate or near-cognate cases. Consistent with this observation, the A-minor interactions between the P-site tRNA and residues 1338 and 1339 of the head domain are lost in the near-cognate complex in the present reconstruction. Interestingly, in the presence of paromomycin it is not possible to distinguish cognate from near-cognate complexes based on the head tilt. Another type of head movement, the swiveling, shows the same trend. In contrast, a less clear picture is seen in the opening and closing of the head domain relative to the body. According to the observed trends in this motion, the structures can be divided into two classes: (1) X-ray 70S E. coli structures, T. thermophilus 30S structures with paromomycin, and the present cryo-EM reconstructions and (2) the X-ray 70S structures with paromomycin and 30S subunit structures from T. thermophilus. The functional implications of this observation, if any, are not clear.
The position of the GAC
Binding of the TCs to the ribosome generates conformational changes in the L7/L12 stalk. Two distinct regions in the vicinity of this stalk, the SRL and GAC, form the main interaction sites for TCs on the 50S subunit. While no conformational changes are observed in the lobe formed by ribosomal protein L6 and H95 (i.e. the SRL, the region implicated in the stimulation of GTPase activity of EF-Tu), the altered disposition of the body of the tRNA affects the position of the other region, the GAC, in the near-cognate complex when compared with its cognate counterpart. When the maps are superimposed so as to produce maximal correlation, and the position of the L7/L12 stalk base (where the GAC is located) is compared (Figure 5B), we observe that only in the case of the cognate species is the stalk base displaced toward protein L16 of the large subunit, as previously described in a cognate situation (Valle et al, 2003). In contrast, the position of the GAC region with respect to L16 is similar in both near-cognate and initiation complexes. Thus, the change observed in the GAC region, previously described as the ‘closed’ state of GAC (Valle et al, 2003), is a primary characteristic that distinguishes cognate from near-cognate tRNA binding after initial codon recognition and GTPase activation. It is noteworthy that, despite the conformational changes in the L7/L12 region, nucleotide A1067, located in the GAC and implicated in TC binding and GTP hydrolysis activity (Saarma et al, 1997; Li et al, 2006), is seen within interaction distance of the elbow region of the tRNA, at position 56, in both fitted cognate and near-cognate atomic models.
Discussion
The kinetic model for aa-tRNA incorporation states that selection is based on the higher stability of the cognate 70S-TC complex in comparison with the near-cognate complex and, more importantly, on the acceleration of GTPase and accommodation rates in the cognate case through conformational rearrangements (Rodnina and Wintermeyer, 2001). It is important to note that in the near-cognate situation, GTP is also hydrolyzed, but at much lower rates (i.e. the ‘correctness’ of the codon–anticodon pair determines the GTPase activation and accommodation rates). These rate differentials are used by the ribosome to discriminate between cognate and near-cognate species.
In this study, we present cryo-EM reconstructions of the complete E. coli 70S-TC complex containing a near-cognate aa-tRNA, as well as its cognate counterpart, with an initiation complex used as a control. These new data provide a structural framework for understanding the mechanism of induced fit during aa-tRNA incorporation, as a comparison of the structures reveals distinct configurations for the different TCs. These differences are likely linked to the way the ribosome and TC work together to facilitate kinetic discrimination during aa-tRNA selection. From our results, we propose that changes in the positions of the GAC of the 50S subunit and the head and shoulder of the 30S subunit, coupled with the distinct conformation shown by the tRNA acceptor stem, result in a suboptimal configuration and positioning of EF-Tu in the near-cognate situation. These structural differences are then ultimately manifested in kinetic differences in GTPase activation shown by cognate versus near-cognate Trp-tRNATrp•EF-Tu TCs (Cochella and Green, 2005). We note that the structural analysis of near-cognate TCs with different aa-tRNAs will provide definitive evidence on how general these structural differences are, as well as on the binding features that might be unique to any particular species. Along these lines, an open question is if the disordering of the near-cognate tRNA CCA end, which prevails despite of its interactions with EF-Tu, is related to the nature of the tRNA species analysed or, alternatively, to the use of kirromycin, which might also participate in the stabilization of the near-cognate EF-Tu structure. On the other hand, the fact that (1) interactions between the activated amino acid of the aa-tRNA and certain residues of EF-Tu protect the ester bond between the amino acid and the 3′ CCA string and (2) the CCA end is totally smeared out in our reconstruction due to its mobility, poses the interesting question if the labile ester bond is still preserved in the observed near-cognate environment. Unfortunately, an answer to this type of question cannot be obtained from the current data due to the limited resolution of the maps.
The antibiotic kirromycin employed in this work stalls the ribosome just after GTP is hydrolyzed, but presumably before any major change reverting/altering the GTPase-activated state has occurred. Therefore, the changes we observe in our structures can be readily linked to aa-tRNA selection and GTPase activation, but not necessarily to the accommodation process. However, as we have clear indications that the acceptor stem, and mainly the CCA end, are not stabilized in our map, we can speculate that correct alignment of the cognate tRNA after the hydrolysis of GTP might result in optimal transit through the accommodation corridor toward the A site of the large subunit. In contrast, the dynamic nature of the CCA end in the near-cognate species would allow unproductive motion to occur in the acceptor stem region that precludes direct and rapid accommodation (Sanbonmatsu, 2006). Indeed, recent simulations point out the critical role of the configurational entropy of the CCA region in the accommodation process (Whitford et al, 2010).
To determine whether or not additional changes affecting the 30S subunit occur during accommodation, ribosomes with near-cognate A/T- and fully accommodated A/A-tRNAs must be compared. Along these lines, X-ray structures of 70S ribosomes soaked with cognate and near-cognate tRNAs have been obtained as this work was completed (Jenner et al, 2010). These structures show conformational variability of the 70S in response to the binding state of the A site; however, while the authors' comparison of empty and cognate A/A states has also brought out conformational changes related to the head closure of the 30S subunit, their near-cognate structure challenges the idea of head closure as a conformational response unique to cognate binding, as near-cognate binding appears to induce a rearrangement that is very similar to that observed with the cognate tRNA species. This goes against the notion that head closure might also facilitate the accommodation of the tRNA. Interestingly, the near-cognate A/A-tRNA in this structure is also partially blurred. However, as their 70S ribosome complexes carry an initiator tRNAMet in the A site of the near-cognate complex, instead of an elongator tRNA, a comparison of these results with ours is not straightforward. Moreover, the analysis of existing structures (see Table I) shows that the position of the head with respect to the rest of the small subunit reflects the presence of cognate, near-cognate or non-cognate tRNA in the decoding centre. Our analysis reveals a picture which, in essence, is consistent with the observations made from the X-ray structures of 30S subunits (Ogle and Ramakrishnan, 2005): a stronger closing of the body and platform domains in the cognate than in the near-cognate case (see Supplementary Movies 1 and 2). As these conformational changes lead to a more closed state of the small subunit, the cognate TC is poised to interact with both small and large subunits as part of the interaction network that defines the productive configurations for aa-tRNA and EF-Tu domains.
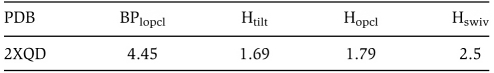
Note: After this work was completed, we became aware of the paper by Voorhees et al (2010), reporting a new X-ray structure of 70S-TC complexes in the presence of a non-hydrolyzable GTP analogue. This new structure, which represents a pre-hydrolysis state, shows a completely structured switch I loop and the activated conformation of His84. The overall conformation of the A/T-tRNA and the rest of EF-Tu are very similar to that shown by the GDP-kirromycin structures. Our analysis and quantification of the domain closure motion in the structure reported by Voorhees and coworkers gives the following values, using the nomenclature of Table I:
These values are very similar to the values obtained for the cognate GDP-kirromycin structures, except that the body is closed to a larger extent, which may indicate a slight relaxation upon hydrolysis. A functional role of this movement cannot be discarded; however, a note of caution should be introduced here. First, only one structure is reported, and as the comparison of the values reported in Table I shows, the variation of structures within the same crystal can be significant. Thus, to provide a robust analysis, we have compared trends only and not the particular values. In addition, the structure reported by Voorhees and coworkers contains an A/T-G24A-Trp-tRNATrp (i.e. the miscoding Hirsh mutant, a tRNATrp variant that decodes both UGG and UGA stop codons). It is well established that while this mutant tRNA, which shows a substitution in the D arm, increases the GTP hydrolysis rate on the near-cognate codon, both wild-type and mutant species activate hydrolysis on the cognate codon to the same extent (Cochella and Green, 2005). However, it is possible that the D-arm substitution might have an effect on the deformability of the tRNA even in cognate environments, and therefore have an influence in the further closing of the body as measured in structure 2XQD.
Finally, we note that new biochemical data (Mittelstaet et al, 2011) were published while this manuscript was under revision. In this work, the timing and the extent of the tRNA distortion upon reading cognate or near-cognate codons is analysed by means of fluorescence quenching. Overall, the results presented strongly support our structural interpretation.
Materials and methods
Sample preparation
The ribosomal complexes were prepared using an in vitro translation system in HiFi buffer (50 mM Tris–HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 0.5 mM spermidine, 8 mM putrescine, 2 mM DTT) as previously described (Cochella and Green, 2005). Kirromycin was added (final concentration: 0.25 mM) to stall the TC on the ribosome.
EM and image processing
Carbon-coated holey grids were prepared using standard cryo-procedures (Grassucci et al, 2007). The AutoEMation program (Lei and Frank, 2005) was used to acquire CCD images on an FEI Tecnai Polara at 300 kV at an effective magnification of × 100 000 and a final pixel size of 1.5 Å/pixel. The 3D reconstructions followed standard SPIDER protocols for reference-based reconstruction (Shaikh et al, 2008). A lower-resolution reconstruction of the ribosomes bearing the wt TC on cognate codon was previously introduced in Li et al (2008). The total number of particles used was 294 671 (ribosomes programmed on cognate codon), 359 223 (ribosomes programmed on near-cognate codon) and 186 732 (initiation-like 70S ribosomes). Supervised classification methodology was applied to sort the data set programmed with near-cognate ribosome in two subsets. The two 3D references employed differ only in the presence versus absence of a TC. To this end, a soft Gaussian mask was applied to remove the wt TC from the ribosome map shown in Figure 1B. Finally, the data were assigned to the two classes based on the reference that gave the highest correlation coefficient (CC) value. The first group was composed of ∼8% (26 873 particles) that showed higher CC against the reference presenting the TC. The second group was composed of the remaining 92% of the initial particles and corresponded to ribosomes that had rejected the incoming tRNA (332 410 particles).
Fitting of atomic structures into EM maps
Atomic models were obtained by fitting crystallographic structures into the cryo-EM densities employing MDFF (Trabuco et al, 2008). All modelling was performed with VMD (Humphrey et al, 1996) and NAMD 2.7 (Phillips et al, 2005). Complete atomic models of E. coli ribosome and tRNAPhe containing all modified ribonucleosides, together with the necessary force field parameters, were introduced as previously described (Trabuco et al, 2010). The initial EF-Tu structure was taken from PDB 3FIH (Villa et al, 2009), and a homology model of E. coli tRNATrp was built by mutating a Bos taurus structure (PDB 2DR2; Shen et al, 2006) and subsequently optimizing through molecular dynamics simulations. In contrast to the original MDFF protocol (Trabuco et al, 2008), secondary structure restraints were specified for individual RNA helices and proteins, that is, codon–anticodon interactions and tertiary interactions between different RNA structural elements were not restrained, which permitted domain movements within the rRNAs to be more accurately characterized. As an initial step, the 70S structure together with tRNAPhe were fitted using MDFF in vacuo with a coupling constant of 0.3 (Trabuco et al, 2008). The fittings were considered converged when the RMSD change was <0.1 Å/ns and the change of the RMSD running average (1 ns window) was <0.01 Å/ns, which was achieved after 6–8 ns of MDFF simulations. The resulting structures were energy minimized for 2000 steps using MDFF with a coupling constant of 1.0.
In order to obtain a more realistic picture of the interactions at the interfaces of the different components of the system, refinement MDFF simulations were performed in explicit solvent (Trabuco et al, 2009). To refine the local interactions of the TCs with the ribosome, the system size was reduced, and the tRNAPhe was replaced by a pre-fitted tRNATrp, the type of tRNA employed in the experiment (see Sample preparation section). Each final system contained the TC and all the residues of the ribosome within 22 Å. After placement of Mg2+ ions and completion of their solvation shell, the systems were solvated and potassium chloride was added yielding a 100-mM solution. A multi-step protocol controlled the refinement simulation, in which first water and ions (0.5 ns), then side chains (0.5 ns), and finally all residues but those at the boundaries of the system were free to move (18 ns). All free residues were coupled to the cryo-EM map with a coupling constant of 0.5.
The angles describing the rotational displacements of the individual domains of the 30S ribosomal small subunit were measured based on the corresponding tensors of inertia (see Supplementary Figure S4). The frame of reference was defined as the inertia tensor of the 16S ribosomal RNA. In the following it is implied that the axes of inertia are ordered according to magnitude of their corresponding moments, that is, the first axis, x, corresponds to the largest moment of inertia. In the case of the 16S rRNA, the x axis is roughly normal to the interface of the subunits, the y axis connects shoulder and platform and the z axis is roughly parallel to h44 and the neck.
The determination of displacement angles for a given part of the ribosome relative to a reference structure is illustrated here for the case of the swiveling of the head domain of the small subunit. The inertia tensor of the reference structure defines the internal frame of reference (x, y and z axes). After an initial alignment of the 16S rRNA to the reference structure, the inertia tensor is calculated for the head domain, defining axes x′, y′ and z′. The axis roughly parallel to the beak, z′, is then projected onto the plane spanned by x and y. The swiveling angle corresponds to the angle between this projection and the y axis of the frame of reference. To prevent contaminations of the measurements by internal structural changes, the inertia tensor for the head is calculated from a reference structure of the head domain aligned to the actual structure. In the present case, we used the 16S rRNA from PDB 2I2U (Berk et al, 2006) for all references.
The programs Amira (http://amira.zib.de), Chimera (Pettersen et al, 2004) and VMD (Humphrey et al, 1996) assisted in the visualization of cryo-EM densities and atomic models.
Accession numbers
The EM maps have been deposited in the 3D-EM database, EMBL-European Bioinformatics Institute, Cambridge under the following accession numbers: EMD-1849 (70S•fMet-tRNAfMet•EF-Tu•Trp-tRNATrp on cognate codon), EMD-1850 (70S•fMet-tRNAfMet•EF-Tu•Trp-tRNATrp on near-cognate codon). The atomic coordinates generated by MDFF for the large/small subunits have been deposited in the Protein Data Bank with PDB ID codes 3IZU/3IZW and 3IZT/3IZV for cognate and near-cognate complexes, respectively.
Supplementary Material
Acknowledgments
We thank Robert A Grassucci for assistance with the image collection and Melissa Thomas for assistance with the preparation of the illustrations. We also thank T Martin Schmeing, Rebecca M Voorhees, and Venki Ramakrishnan for providing coordinates of the Thermus thermophilus cognate ternary complex prior to deposition. XA is a recipient of a ‘Ramon y Cajal’ fellowship from the Spanish Government (RYC-2009-04885), ES is recipient of a Feodor Lynen fellowship from the Alexander von Humboldt Foundation, and LGT is a recipient of an European Molecular Biology Organization long-term fellowship. This study was supported by HHMI and NIH grants R01 GM55440 and R37 GM29169 (to JF), NIH grant R01 GM059425 (to RG), and NIH P41-RR005969 and NSF PHY0822613 (to KS). Computer time for MDFF simulations was provided by NSF through the National Resources Allocation Committee (MCA93S028).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abel K, Yoder MD, Hilgenfeld R, Jurnak F (1996) An alpha to beta conformational switch in EF-Tu. Structure 4: 1153–1159 [DOI] [PubMed] [Google Scholar]
- Berk V, Zhang W, Pai RD, Cate JH (2006) Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA 103: 15830–15834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD (2004) tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R (2005) An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV (2003) Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol 332: 689–699 [DOI] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Frank J (2007) Preparation of macromolecular complexes for cryo-electron microscopy. Nat Protoc 2: 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV (2006) A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell 21: 369–377 [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV (2004) Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- Jenner L, Demeshkina N, Yusupova G, Yusupov M (2010) Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol 17: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF (2006) Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Lei J, Frank J (2005) Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J Struct Biol 150: 69–80 [DOI] [PubMed] [Google Scholar]
- Li W, Agirrezabala X, Lei J, Bouakaz L, Brunelle JL, Ortiz-Meoz RF, Green R, Sanyal S, Ehrenberg M, Frank J (2008) Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. EMBO J 27: 3322–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sengupta J, Rath BK, Frank J (2006) Functional conformations of the L11-ribosomal RNA complex revealed by correlative analysis of cryo-EM and molecular dynamics simulations. RNA 12: 1240–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstaet J, Konevega AL, Rodnina MV (2011) Distortion of tRNA upon near-cognate codon recognition on the ribosome. J Biol Chem (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V (2005) Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M (1999) Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV (1998) Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J 17: 7490–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV (2000) Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol 7: 104–107 [DOI] [PubMed] [Google Scholar]
- Parmeggiani A, Swart GW (1985) Mechanism of action of kirromycin-like antibiotics. Annu Rev Microbiol 39: 557–577 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polekhina G, Thirup S, Kjeldgaard M, Nissen P, Lippmann C, Nyborg J (1996) Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure 4: 1141–1151 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Wintermeyer W (1994) Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 33: 12267–12275 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Pape T, Fricke R, Kuhn L, Wintermeyer W (1996) Initial binding of the elongation factor Tu.GTP.aminoacyl-tRNA complex preceding codon recognition on the ribosome. J Biol Chem 271: 646–652 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Saarma U, Remme J, Ehrenberg M, Bilgin N (1997) An A to U transversion at position 1067 of 23 S rRNA from Escherichia coli impairs EF-Tu and EF-G function. J Mol Biol 272: 327–335 [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu KY (2006) Alignment/misalignment hypothesis for tRNA selection by the ribosome. Biochimie 88: 1075–1089 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V (2009) The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM (2009) GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J (2008) SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3: 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Guo L, Yang B, Jin Y, Ding J (2006) Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res 34: 3246–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M (2002) Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat Struct Biol 9: 849–854 [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K (2010) The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J Mol Biol 402: 741–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K (2008) Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K (2009) Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J (2003) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J (2009) Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA 106: 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V (2010) The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstenbosch E, Pape T, Rodnina MV, Kraal B, Wintermeyer W (1996) The G222D mutation in elongation factor Tu inhibits the codon-induced conformational changes leading to GTPase activation on the ribosome. EMBO J 15: 6766–6774 [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY (2010) Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA 16: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R (2009) Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.