Abstract
Histones are structural and functional components of the eukaryotic chromosome, and their function is essential for normal cell cycle progression. In this work, we describe the characterization of two Saccharomyces cerevisiae cold-sensitive histone H2A mutants. Both mutants contain single amino acid replacements of residues predicted to be on the surface of the nucleosome and in close contact with DNA. We show that these H2A mutations cause an increase-in-ploidy phenotype, an increased rate of chromosome loss, and a defect in traversing the G2–M phase of the cell cycle. Moreover, these H2A mutations show genetic interactions with mutations in genes encoding kinetochore components. Finally, chromatin analysis of these H2A mutants has revealed an altered centromeric chromatin structure. Taken together, these results strongly suggest that histone H2A is required for proper centromere–kinetochore function during chromosome segregation.
Keywords: centromere/chromatin/histones/kinetochore/ploidy
Introduction
Each time a eukaryotic cell divides, a precise sequence of events takes place in order to segregate its replicated chromosomes. These events require that a complex set of structures, including the mitotic chromosome, centromere–kinetochore complex (hereafter centromere) and mitotic spindle must assemble and work in concert to ensure the accurate transmission of chromosomes to daughter cells. Chromosomes themselves undergo major structural changes and movement during mitosis: chromatin is decondensed for DNA replication, the newly replicated DNA is detangled and condensed into sister chromatids, the chromatids attach to the mitotic spindle via the centromere, the chromatids disjoin, and the chromosomes segregate into daughter cells. Chromosome segregation must occur with extreme fidelity, since mistakes in segregation often lead to aneuploidy, polyploidy and cell death.
Several studies in Saccharomyces cerevisiae have suggested that chromatin structure plays a critical role in chromosome segregation and centromere function (Schulman and Bloom, 1991). Chromatin analysis has revealed a distinct chromatin structure over centromeres not observed at other chromosomal sites (Bloom and Carbon, 1982). Genetic studies have shown that either increased histone levels (Meeks-Wagner and Hartwell, 1986) or depletion of histone H2B or H4 (Han et al., 1987; Kim et al., 1988) impairs mitotic chromosome segregation. Moreover, histone depletion causes increased nuclease sensitivity in centromeric DNA, as well as an altered nuclease sensitivity in chromatin surrounding the centromere, suggesting a direct role for histones in centromere function (Saunders et al., 1990). Finally, analysis of other classes of histone mutants also suggests that chromatin structure plays a critical role in segregation. Deletion of the conserved N-terminal domains of H3 and H4 delays the cell cycle at the G2–M phase (Morgan et al., 1991), a phenotype often observed for segregation mutants (Hartwell and Smith, 1985). Interestingly, mutations in the conserved lysine residues on the H4 N-terminus caused a marked delay during the G2–M phase of the cell cycle, apparently as a response to DNA damage sensed by the RAD9-dependent checkpoint (Megee et al., 1995). Another H4 mutant also causes arrest at G2–M and has an increased rate of chromosome loss (Smith et al., 1996). Together, these studies suggest a prominent role for histones in chromosome segregation. The studies of H4 mutants indicate that different H4 mutations may alter different aspects of chromosome segregation, suggesting that this may be true for the other core histones as well. However, little is known about the roles of other histones in centromere function.
In this work we describe the characterization of two S.cerevisiae cold-sensitive histone H2A mutants. We show that these two mutations confer both an increase-in-ploidy phenotype as well as an increased rate of chromosome loss. In addition, these mutations cause defects in traversing the G2–M phase of the cell cycle and show genetic interactions with mutations in genes encoding kinetochore components. Finally, chromatin analysis has revealed an altered chromatin structure flanking the centromere. Taken together, these results suggest that histone H2A is required for proper centromere function during chromosome segregation.
Results
Mutations in a histone H2A gene cause an increase in ploidy
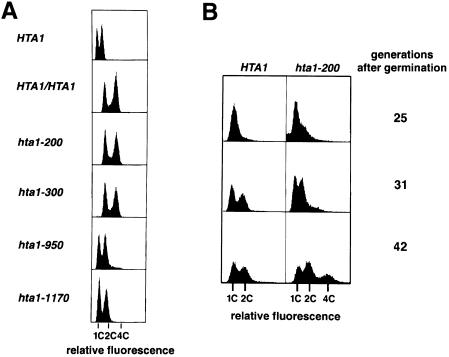
A set of fourteen mutations in HTA1, which encodes histone H2A, were isolated previously and identified based on transcriptional defects (Hirschhorn et al., 1995). Characterization of these hta1 mutants has been done in the absence of the second H2A gene, HTA2. Further analysis has demonstrated that two of these mutants are cold sensitive (Cs–) for growth at 13°C. These mutants, hta1–200 and hta1–300, contain single amino acid replacements at evolutionarily invariant positions, serine 20→phenylalanine (S20F) and glycine 30→aspartic acid (G30D), respectively. Based on the crystallographic data for both the histone octamer and the nucleosome, the location of these residues is predicted to be on the surface of the nucleosome and in close contact with DNA (Figure 1; E.Moudrianakis, personal communication; Luger et al., 1997). Genetic analysis suggested that the hta1–200 and hta1–300 mutants are unable to maintain the haploid state. Although these mutant strains were initially constructed by a gene replacement method in a wild-type haploid strain, extremely poor spore germination in crosses of these mutants with wild-type haploid strains and the good germination in crosses with diploid strains suggested that the strains had become diploids (Campbell et al., 1981; I.Pinto, J.Hirschhorn and F.Winston, unpublished). To examine the ploidy and DNA content of these mutants more directly, we performed flow cytometric analysis. The wild-type haploid (HTA1) shows two peaks representing the G1 (1C) and G2–M (2C) DNA content. Correspondingly, the diploid (HTA1/HTA1) shows peaks representing G1 (2C) and G2–M (4C) DNA content. Both hta1 mutants have 2C and 4C peaks, a DNA content consistent with their being diploids (Figure 2A).
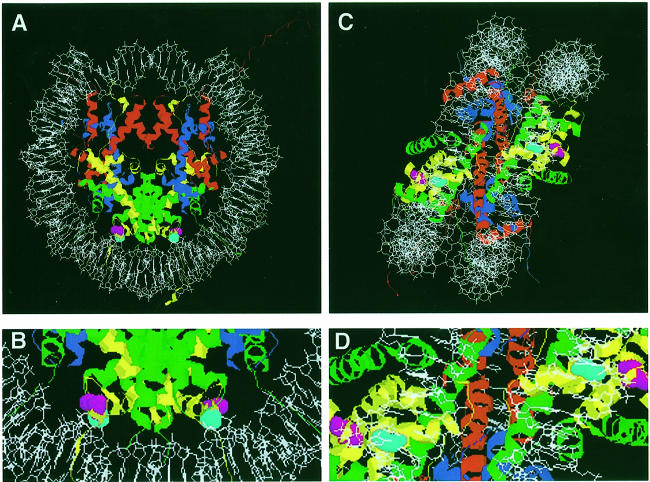
Fig. 1. Nucleosome model showing the position of the altered amino acids in the hta1 mutants. The positions of the single amino acid replacements in the hta1–200 and hta1–300 mutants, S20 and G30, respectively, are indicated in the model of the nucleosome based on the crystal structure generated by Luger et al. (1997). The atomic coordinates file 1aoi was obtained from Brookhaven Protein Databank (PDB) and visualized with RasMac v2.6. The DNA phosphodiester backbone is shown in white. The eight histone protein chains forming the histone octamer are shown as ribbon traces in different colors: H4, blue; H3, red; H2B, green; and H2A, yellow. Amino acids S20 (magenta) and G30 (turquoise) are evolutionarily invariant and correspond to positions S18 and G28 in the amphibian protein used for the X-ray crystallography. (A) Top view, down the DNA superhelix axis. (B) Detail from (A) showing the amino acids S20 and G30 in close contact with DNA. (C) Side view, perpendicular to the DNA superhelix axis. (D) Detail from (C), showing amino acids S20 and G30.
Fig. 2. Flow cytometric analysis of DNA content in the hta1 mutants. (A) Comparison of exponentially growing hta1–200 (FY987), hta1–300 (FY988), hta1–950 (FY990) and hta1–1170 (FY991) mutant strains with wild-type HTA1 haploid (FY604) and HTA1/HTA1 diploid (FY604 × FY605) strains. (B) Appearance of diploids in a growing YPD culture of spores germinated from an HTA1/hta1–200 heterozygous diploid (a cross of FY605 × FY1819 followed by loss of plasmid pSAB6). The first 25 generations are present in the colony formed from the germinated spore, where most cells are arrested in G1 (1C). Flow cytometric analysis was performed on aliquots of the cultures at the indicated number of cell generations after germination.
To test the possible allele specificity of this phenotype among our collection of hta1 mutants, we also analyzed two other hta1 alleles isolated in the original screen. The other two mutants, hta1–950 and hta1–1170, maintain the haploid state (Figure 2A). Thus, the increase-in-ploidy phenotype is allele specific, strongly suggesting that it is distinct from the transcriptional defect by which these mutants were originally isolated.
To study the kinetics of the ploidy increase in the hta1 mutants, we monitored the DNA content of newly germinated, haploid hta1–200 mutants by flow cytometric analysis (Materials and methods). In the experiment shown (Figure 2B), we monitored the DNA content of the four progeny from one tetrad of an hta1–200 × HTA1 cross. The two HTA1 progeny behaved as expected for stable haploids, with 1C and 2C peaks, indicating the DNA content of cells before and after DNA replication. In contrast, for the two hta1–200 progeny a 4C peak comprising ∼30% of the population became evident by 42 generations (Figure 2B). In repeat experiments, the actual percentage of diploid cells in an hta1 mutant culture after 42 generations varied, ranging from very few to 100% diploids (data not shown), which is likely to reflect fluctuation with respect to when diploidization occurred after germination. These results demonstrate that the hta1 spores germinate as haploids and remain haploid for several generations before diploids form. Thus, the hta1 mutations cause an increase in ploidy from haploid to diploid.
To determine whether the mutant strains had become stable diploids, we tested their ability to give rise to viable meiotic progeny. Diploids are expected to yield a high percentage of viable meiotic progeny, while aneuploids are expected to yield a much lower frequency, at best 50% for a 2n–1 aneuploid. To examine meiotic progeny, we first switched the mating type of the putative MATα/MATα diploid to MATa/MATα (Materials and methods) and then sporulated these strains. For both hta1–200 and hta1–300 mutants, 20 tetrads were dissected and scored for mating type. As observed previously for other hta1 and htb1 mutants, germination was poor (Norris and Osley, 1987); for both hta1 mutants, eight out of 20 tetrads yielded four viable spores, while the remainder yielded three viable spores. For each mutant, seven of the eight complete tetrads displayed 2 MATa:2 MATα segregation. The viability pattern and the segregation of MAT, in conjunction with the flow cytometric analysis described above, strongly suggest that the hta1–200 and hta1–300 strains form stable diploids at 30°C.
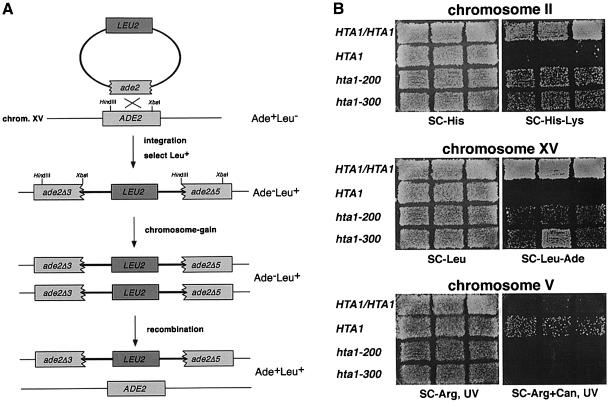
To measure the ploidy increase by an independent approach, we monitored the copy number of three chromosomes by genetic tests. First, we monitored the copy number of chromosomes II and XV using the method of Chan and Botstein (1993). In this assay, one nutritional marker is used to disrupt a second nutritional marker (Figure 3A), such that the disrupted gene can only regain function by homologous recombination, resulting in loss of the inserted gene. Thus, only those cells that have gained an extra copy of the chromosome can become prototrophic for both markers. Using this assay, both the hta1–200 and hta1–300 mutants had a significantly increased number of cells with extra copies of chromosomes II and XV (Figure 3B; Table I). We used a second assay to monitor the copy number of chromosome V (Schild et al., 1981). This assay takes advantage of the fact that mutations in CAN1 cause resistance to canavanine (CanR). Because the CanR phenotype is recessive, haploid cells become CanR at a much greater frequency than diploid cells. In this assay, the hta1–200 and hta1–300 mutants gave rise to very few CanR papillae, behaving similarly to the diploid control (Figure 3B). These genetic tests demonstrate that the hta1 mutants have an extra copy of at least three chromosomes, consistent with the flow cytometric analysis and tetrad analysis that suggested they had become diploid.
Fig. 3. Analysis of ploidy increase in the hta1 mutants. (A) Genetic events that result from chromosome gain in the strains marked at ade2 on chromosome XV, leading to Ade+ Leu+ papillae. Equivalent events occur on chromosome II marked at lys2. Details of the assay are explained in Materials and methods and Chan and Botstein (1993). (B) Papillation assay showing chromosome gain in the indicated strains marked at chromosomes II and XV. Chromosome V was monitored by the appearance on CanR papillae induced by UV irradiation. Strains used were: HTA1/HTA1 (FY1820), HTA1 (FY1821), hta1–200 (FY1823) and hta1–300 (FY1824).
Table I. Chromosome gain and recombination in hta1 mutants.
| Strain | Frequency of chromosome gaina (×10–6) |
Frequency of recombinationb (×10–4) |
Frequency of cells with an extra chromosomec |
||||||
|---|---|---|---|---|---|---|---|---|---|
| II | XV | II + XV | II | XV | II + XV | II | XV | II + XV | |
| Lys+ His+ | Ade+ Leu+ | Lys+ His+ Ade+ Leu+ | Lys+ | Ade+ | Lys+ Ade+ | ||||
| HTA1/HTA1 | 100 | 830 | 22.0 | 1.1 | 9.9 | 0.2 | 0.9 | 0.8 | 1.1 |
| HTA1 (a) | <0.2 | 0.2 | <0.2 | 0.3 | 15.3 | 0.1 | <0.01 | <0.01 | <0.02 |
| HTA1 (α) | <0.2 | 0.2 | <0.2 | 0.3 | 14.5 | 0.1 | <0.01 | <0.01 | <0.02 |
| hta1–200 | 35.0 | 160 | 3.2 | 2.2 | 5.3 | 0.1 | 0.2 | 0.3 | 0.3 |
| hta1–300 | 64.0 | 409 | 6.8 | 2.8 | 11.1 | 0.1 | 0.2 | 0.4 | 0.7 |
The strains used were FY1820 (HTA1/HTA1), FY1821 (HTA1 MATa), FY1822 (HTA1 MATα), FY1823 (hta1–200) and FY1824 (hta1–300). Colonies were scored after 3 (wild type) or 4 days (mutants) at 30°C. Frequencies of recombination and chromosome gain were obtained by fluctuation analysis (method of the median; Lea and Coulson, 1949) from six independent cultures of each strain.
aThe frequency of chromosome gain was determined for each of the marked chromosomes (II and XV) individually, or together (II + XV column), by scoring colony formation on the media indicated on top of each column. Growth on this media requires two copies of the tested chromosome to allow for recombination events that result in restoration of the disrupted marker (LYS or ADE) while maintaining prototrophy for the integrated marker (HIS or LEU).
bThe frequency of recombination was determined for each of the marked chromosomes (II and XV) individually, or together (II + XV column), by scoring colony formation on the media indicated on top of each column. Growth on this media requires homologous recombination between the two tandem copies of the disrupted marker (LYS or ADE), regardless of the copy number of the marked chromosome.
cThe frequency of cells with an extra copy of the marked chromosome can be inferred by dividing the frequencies of chromosome gain by the frequencies of recombination for each or both marked chromosomes. As a reference parameter, the expected value for the wild-type diploid (HTA1/HTA1) is 1 (two copies of each chromosome).
We used the first assay to determine the frequencies of chromosome gain for chromosomes II and XV (Table I). The frequency of chromosome gain is the product of the recombination frequency at the marked locus and the frequency of cells with an extra copy of the marked chromosome. In the hta1 mutants, chromosome gain frequencies were significantly higher than in the HTA1 haploids, consistent with the papillation assay. When the observed values of recombination and chromosome gain were used to determine the frequency of cells with an extra copy of the marked chromosome, the values for the wild-type HTA1/HTA1 diploid were close to the expected 100%. For the hta1 mutants, the frequency of cells with an extra copy of chromosome II, chromosome XV and both chromosomes was similar, suggesting that the hta1 mutants gained the extra copies of chromosomes II and XV simultaneously. To confirm this prediction, the colonies selected for gain of an extra copy of chromosome II were tested for an extra copy of chromosome XV, and vice versa. For >100 colonies tested, in all cases the other chromosome was also present in extra copy (data not shown), consistent with the concurrent gain of chromosomes II and XV. Thus, the transition from haploid to diploid appears to occur in one step, rather than as a gradual gain of chromosomes.
The hta1 mutations cause decreased viability and a G2–M cell cycle delay
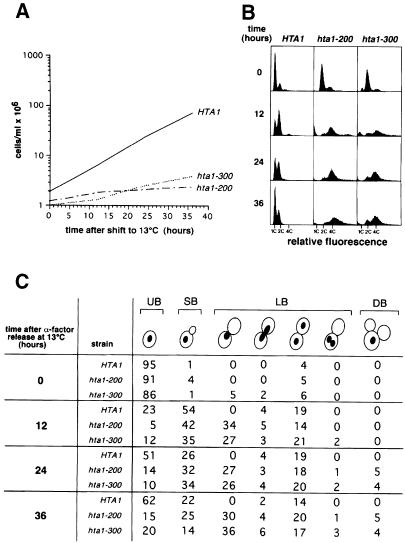
To learn more about the defects caused by these hta1 mutations, we studied their viability and cell cycle phenotypes during incubation at the non-permissive temperature. Asynchronously growing cultures of wild-type and hta1 mutant strains were shifted from the permissive (30°C) to the non-permissive temperature (13°C) and cells were scored for viability and morphology at increasing times after the shift. At 13°C, the generation time for the wild-type strain is 6 h. After 24 h at 13°C, both hta1 mutant cultures had significantly greater numbers of large-budded cells, 60%, compared with wild type (30%) (data not shown). A large-budded morphology is characteristic of cells arrested or delayed in traversing the G2–M phase of the cell cycle. In addition, the hta1 mutant cells were larger than HTA1 cells, a characteristic of cell division cycle (cdc) mutants that are blocked in cell division but continue protein synthesis (Hartwell, 1967; Johnston et al., 1977). The appearance of large-budded cells correlates with decreased viability for both hta1 mutants (data not shown), suggesting that the hta1 mutations affect an essential function required for progression through mitosis.
To examine the cell cycle defect of the hta1 mutants more accurately, we grew cells at the permissive temperature (30°C), arrested them in G1 with α–factor, and released them from the block at the non-permissive temperature (13°C). Cell growth, DNA content and cell morphology were then examined for the wild-type and mutant cultures. In such an experiment, the hta1 mutant cells are diploid at the time of the shift, whereas the wild-type cells are haploid. After the release, growth was severely impaired for the hta1 mutant, as we had observed with asynchronous cultures (Figure 4A). Flow cytometric analysis of DNA content per cell revealed that the mutant culture accumulated diploid cells with a G2 DNA content (4C) by 12 h at 13°C (Figure 4B). Most of the cells also had a G2 DNA content at 24 and 36 h. However, at 12, 24 and 36 h, there was also a low percentage of cells with both lower and higher DNA content, ranging as high as 8C. The small peak of G1 cells (2C) is likely to represent those that escaped the G2–M arrest; alternatively this peak might represent a small percentage of haploids. Because flow cytometric analysis does not distinguish between nuclear and mitochondrial DNA, it was important to determine whether mitochondrial DNA replication uncoupled from cell cycle control would contribute to the apparent nuclear G2 DNA content. Therefore, we repeated the experiment with hta1 mutants lacking mitochondrial DNA (rho° mutants) and obtained the same results (data not shown), indicating that the accumulation of cells with G2 DNA content is cell cycle dependent. The appearance of wider and less discrete G1 and G2 peaks in the hta1 mutants is likely to be due to the presence of aneuploid cells generated at the restrictive temperature and is consistent with the loss of viability.
Fig. 4. hta1 mutants arrest at G2–M. Exponentially growing cells were synchronized in G1 with α–factor and released at 13°C. Samples were taken at the times indicated and analyzed. Strains used were: HTA1 (FY605), hta1–200 (FY1817) and hta1–300 (FY1818). (A) Cell growth in YPD. (B) DNA content by flow cytometry. (C) Quantitation of the nuclear and bud morphology. UB, unbudded; SB, small budded; LB, large budded; DB, double budded. The same cells were used for flow cytometric analysis in (B).
Analysis of cell morphology and nuclear division also indicated an accumulation of G2 cells in the mutant cultures after 12 h at 13°C, 34% of hta1–200 and 27% of hta1–300 cells were large budded with a single nucleus, while none were seen in the wild-type control (Figure 4C). Interestingly, the large-budded cells with an undivided nucleus remained ∼30% after 36 h for both alleles, while the unbudded cells initially decreased (after 12 h), as expected for cells going into the cycle, and then increased up to 15% (hta1–200) and 20% (hta1–300) after 36 h. This increase is likely to be the result of cells that escaped the G2–M block and completed cytokinesis. Both hta1 mutant cultures contained a small percentage of cells with divided nuclei in one bud, or double-budded cells with either one or two nuclei. If it is assumed that 30–50% of the small-budded cells have completed DNA replication, the morphology data correlate well with the flow cytometry data, indicating that the majority of the hta1 cells arrest in G2–M with replicated DNA.
Microtubules and spindle pole bodies are normal in the hta1 mutants
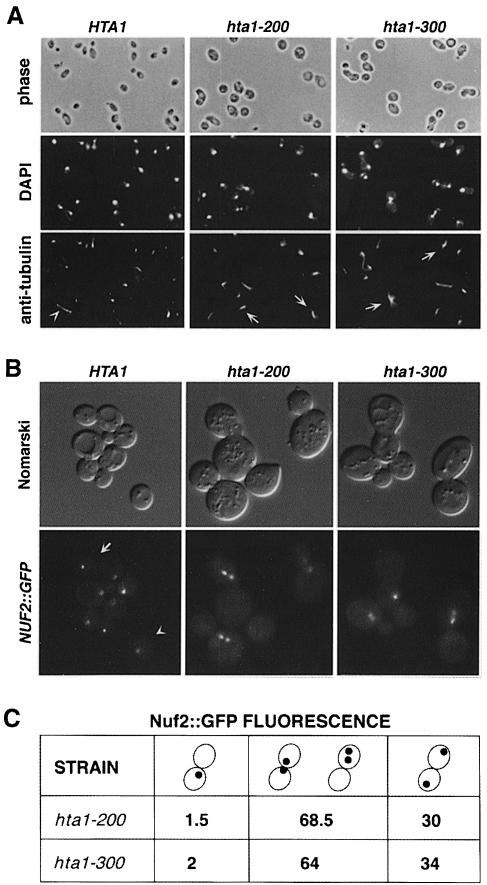
Cells defective in microtubule assembly or spindle formation show phenotypes similar to those of the hta1 mutants, including increase in ploidy, chromosome missegregation and G2–M delay (Schild et al., 1981; Thomas and Botstein, 1986; Rose and Fink, 1987; Huffaker et al., 1988; Schatz et al., 1988; Snyder and Davis, 1988; Winey et al., 1991). Therefore, we analyzed the spindle morphology in the hta1 mutants by staining microtubules with anti-tubulin antibodies. The results indicate that these mutants appear to polymerize microtubules similarly to the wild-type cells (Figure 5A). Most large-budded cells with an undivided nucleus arrested with a short spindle, characteristic of cells blocked at G2–M, and those that contained a divided nucleus appeared with a normal elongated spindle. Because cold-sensitive tubulin mutants also have phenotypes similar to these hta1 mutants (Huffaker et al., 1988; Schatz et al., 1988), we tested whether the mRNA levels of any of the tubulin genes (TUB1, TUB2 and TUB3) were altered in the hta1 mutants. No differences were found by Northern analysis, and none of the TUB genes were able to suppress the cold sensitivity of the hta1 mutants when present on high-copy-number plasmids (data not shown). Thus, we conclude that the hta1 mutant phenotypes are not caused by an indirect effect on microtubule assembly or function.
Fig. 5. Microtubule and spindle pole body analysis in synchronized cells shifted to 13°C. (A) Microtubule staining by indirect immuno- fluorescence with anti-tubulin antibodies of cells taken 24 h after the shift to 13°C. Nuclear DNA was stained with DAPI. The hta1 mutant shows large-budded cells with an undivided nucleus and a short spindle. The arrowhead indicates a wild-type cell in anaphase with an elongated spindle. Arrows indicate hta1 large-budded cells with a short spindle. Strains used were: HTA1 (FY605), hta1–200 (FY1817) and hta1–300 (FY1818). (B) Spindle pole body staining by green fluorescence on live cells marked with NUF2::GFP. Cells were synchronized with α–factor and released at 13°C; a sample was taken 24 h after the shift to 13°C and analyzed by fluorescence microscopy. In the wild-type cells a single dot corresponding to an undivided spindle pole body is present in an unbudded cell (arrowhead), or two dots at the opposite poles of a large-budded cell (arrow) are present in a cell at anaphase. The two dots shown in mutants correspond to the spindle pole body that has duplicated and separated a short distance within a large-budded cell, consistent with a large-budded cell with an undivided nucleus. In some cases only one dot appears in the focal plane of the picture. (C) Quantitation of the GFP signal in the hta1–200 and hta1–300 strains. Numbers represent percentage values. Strains used were: HTA1 (FY1825), hta1–200 (FY1826) and hta1–300 (FY1827).
We also monitored the behavior of the spindle pole body, since defects in genes that affect its structure or function also cause similar phenotypes, including an increase in ploidy (Schild et al., 1981; Thomas and Botstein, 1986; Rose and Fink, 1987; Snyder and Davis, 1988; Winey et al., 1991). The spindle pole body was examined in growing cells using a functional NUF2::GFP fusion integrated at the NUF2 locus (Kahana et al., 1998). Nuf2 has been shown previously to localize to the spindle pole body (Osborne et al., 1994). The spindle pole body appears to duplicate normally in the hta1 mutants, although in many large-budded cells the spindle pole bodies remain in one cell, indicating an undivided nucleus (Figure 5B). Quantitation of the GFP signals present in the large-budded cells of the hta1 strains revealed that 1–2% of the cells had a single GFP signal, which could represent an undivided spindle pole body. One-third of the cells contained normally separated spindle pole bodies, one in each cell. The majority of the cells contained separated spindle pole bodies within one cell or near the neck of the bud (Figure 5B and C), a good correlation with the number of large-budded cells containing an undivided nucleus (Figure 4C). Therefore, the defects observed in the hta1 mutants are not caused indirectly by either a defect in microtubule assembly or in spindle pole body duplication.
The hta1 mutations impair chromosome segregation
The combination of mutant phenotypes of the hta1 mutants strongly suggested that they may have a chromosome segregation defect. Therefore, to monitor chromosome segregation defects, the frequency of chromosome loss in the hta1 mutants was measured as described in Materials and methods. This assay also allowed us to determine the frequency of mitotic recombination, which can be used as a monitor of DNA damage or defects in DNA replication (Hartwell and Smith, 1985). Briefly, diploids were constructed that are heterozygous at the HIS4 locus by integration of URA3 into HIS4 on one homolog, creating HIS4/his4Δ::URA3 diploids. Either loss of the chromosome that contains the his4Δ::URA3 allele or mitotic recombination between CEN3 and HIS4 would result in 5′–fluoro-orotic acid (5–FOA)-resistant cells. Chromosome loss and mitotic recombination can be distinguished by scoring the mating type (see Materials and methods). In this assay, both hta1–200 and hta1–300 homozygous diploids exhibited a significantly increased frequency of chromosome loss at 30°C, and an even greater increase after incubation at 13°C, ranging from a 10- to a 13–fold effect (Table II). The mutants display a 3- to 6–fold increase in recombination only at 13°C. Thus, the hta1 mutations confer chromosome segregation defects, consistent with a role in mitosis.
Table II. Chromosome III loss and recombination in hta1 mutants.
| Genotype | Recombination frequencya (×10–6) |
Chromosome loss frequencyb (×10–6) |
||
|---|---|---|---|---|
| 30°C | 13°C | 30°C | 13°C | |
| HTA1/HTA1 | 2.4 | 4.2 | 3.6 | 6.8 |
| hta1–200/hta1–200 | 4.5 | 13 | 24 | 71 |
| hta1–300/hta1–300 | 6.6 | 23 | 48 | 90 |
Frequencies of mitotic recombination and loss of chromosome III in diploids were determined as described in Materials and methods. Frequencies were derived by fluctuation analysis from 10 individual cultures of each strain. The strains used were: FY1828 (HTA1/HTA1), FY1829 (hta1–200/hta1–200) and FY1830 (hta1–300/hta1–300).
aThe frequency of mitotic recombination was determined by scoring for the loss of the URA3 marker (growth on 5–FOA medium) at the his4 locus, located on the right arm of chromosome III, and the maintenance of both MATa/MATα loci (non-mater) on the left arm on chromosome III.
bThe frequency of chromosome loss was determined by scoring for the loss of the URA3 marker (growth on 5–FOA medium) at the his4 locus, located on the right arm of chromosome III, in conjunction with the loss of the respective MAT locus (mater) on the left arm of chromosome III.
Altered chromosome segregation leads to the ploidy increase in the hta1 mutants
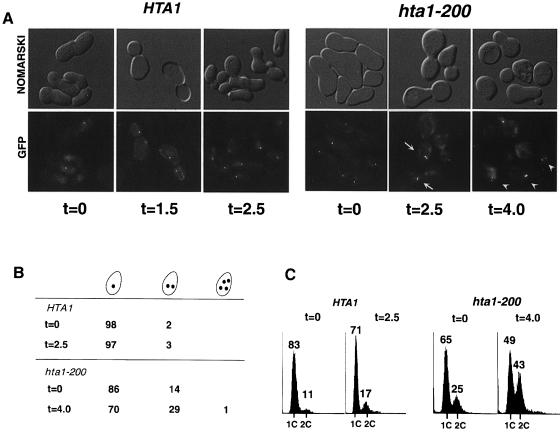
A ploidy increase is usually associated with defects in sister chromatid separation or in chromosome segregation. To investigate whether the hta1 mutants are defective in one of these steps, we constructed wild-type and hta1 mutant strains marked at CEN3 with GFP, as described in Materials and methods (Straight et al., 1996). We monitored sister chromatid separation by microscopy and DNA content by flow cytometry. Soon after germination (∼25 generations), when most of the hta1 mutants are still haploid, the cells were arrested in G1 with α–factor and released at permissive temperature (30°C). To prevent cells from entering the next cell cycle, we added α–factor back to the culture at two-thirds of their generation time after the initial release from α–factor. At the beginning of the cycle (t = 0) most wild-type cells show a single green dot corresponding to the unreplicated chromosome (Figure 6A and B). At the end of anaphase the replicated chromatids have separated to opposite poles between mother and daughter cells, showing one green dot on each cell (t = 1.5). After the cells have completed cytokinesis they arrest in G1 (t = 2.5) showing one green dot per cell. The hta1–200 mutant shows a quite different behavior. At the beginning of the cycle, 14% of the unbudded cells contain two green dots, which is likely to be a reflection of the cells that have already diploidized in the colony. At the end of anaphase (t = 2.5), there is a significant increase in the number of cells with two dots that remain in one cell, which is consistent with the final outcome (t = 4.0) of 29% of the single cells with two green dots, indicating that the sister chromatids were able to separate but did not segregate properly to opposite poles, giving rise to cells carrying two copies of the chromosome. These numbers correlate very well with the DNA content of the cells (Figure 6C), indicating that the segregation defect takes place at once in all the chromosomes, giving rise to diploid cells. Thus, we conclude that the ploidy increase is the result of a defect in chromosome segregation rather than in sister chromatid separation. Therefore, the same defect may be responsible for the increased rate of chromosome loss and the aneuploidy seen at the restrictive temperature (13°C).
Fig. 6. Sister chromatid separation in the hta1–200 mutant. Newly germinated wild-type HTA1 and hta1–200 cells were scored for sister chromatid separation and DNA content during a single cell cycle. (A) Phenotypes of HTA1 and hta1–200 cells released from α–factor arrest at 30°C (t = 0) and observed after one generation time (1.5 and 2.5 h, respectively) and 1.7 generation times (2.5 and 4.0 h, respectively). These strains carry lactose operators integrated near the centromere of chromosome III. CEN3 was visualized by fluorescence microscopy after induction of GFP–lacI. At the beginning of the cycle (t = 0) the unbudded cells contain one GFP dot representing the unreplicated chromosomes. By the end of the cycle HTA1 cells have separated their sister chromatids to opposite poles (t = 1.5), while hta1–200 cells contain cells where both sisters separate at one pole (t = 2.5, arrows). After completion of cytokinesis HTA1 cells contain single dots (t = 2.5), while hta1–200 cells contain both sisters (t = 4.0, arrowheads). (B) Single cells containing one, two or four GFP dots were scored at the beginning and end of the cycle. At least 200 cells were scored for each time point. Cells with no GFP signal (aploids) were not scored. Numbers represent percentage values of total cells with GFP signal. (C) DNA content of the samples used in (A) and (B) was analyzed by flow cytometry.
Genetic interactions with kinetochore components
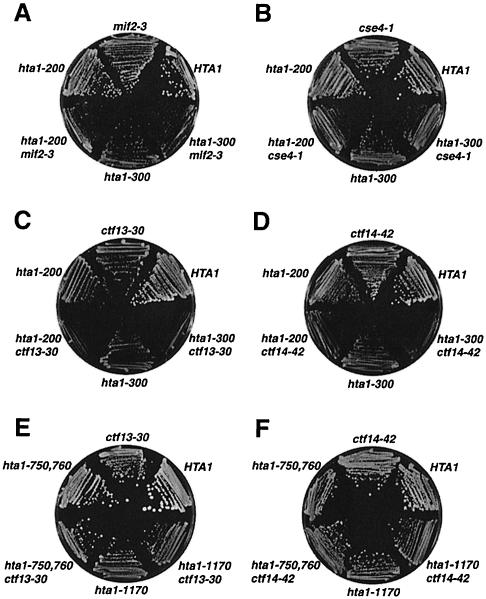
The results presented so far strongly suggest that the hta1 mutants are defective in a function required for chromosome segregation. However, our results have ruled out defects in either microtubule function or spindle pole body duplication. Therefore, to test whether the hta1 mutants might be defective in centromere function, we assayed for genetic interactions between the hta1 mutations and mutations in genes encoding kinetochore components. We constructed double mutants between the hta1 mutations and temperature-sensitive mutations in CTF13, NDC10/CTF14/CBF2/CEP2, MIF2 and CSE4. CTF13 encodes the p58 subunit of CBF3, the protein complex that specifically binds the CDEIII region of centromere DNA. NDC10 encodes the p110 subunit of CBF3 (Hyman and Sorger, 1995; Hoyt and Geiser, 1996). Both Cse4 and Mif2 have been genetically and biochemically implicated in centromere function (Meeks-Wagner et al., 1986; Brown et al., 1993; Stoler et al., 1995; Meluh and Koshland, 1997; Meluh et al., 1998). In each case, the double mutant grew more slowly than the single mutants, ranging from a mild effect for hta1 mif2 double mutants to a severe growth defect for hta1 ndc10/ctf14 double mutants (Figure 7A–D). To test whether the interaction between hta1–200, hta1–300 and the ctf13 and ndc10/ctf14 alleles was specific for these two hta1 alleles, we also constructed and tested double mutants between hta1–1170 and the ctf mutants. None of these double mutants have growth defects (Figure 7E and F). This allele specificity indicates that the growth defects are specific for the two cold-sensitive hta1 mutants that are also defective in chromosome segregation. Therefore, these data suggest that histone H2A interacts with components of the kinetochore and that the hta1 mutants affect centromere function.
Fig. 7. Double-mutant phenotypes caused by hta1 mutants in combination with the kinetochore mutants indicated. The genotypes correspond to the following strains: HTA1 (FY605), hta1–200 (FY1817), hta1–300 (FY1818), hta1–1170 (L992), mif2-3 (L986), hta1–200 mif2-3 (L987), hta1–300 mif2-3 (L988), cse4-1 (L989), hta1–200 cse4-1 (L990), hta1–300 cse4-1 (L991), ctf13-30 (L980), hta1–200 ctf13-30 (L981), hta1–300 ctf13-30 (L982), ctf14-42 (L983), hta1–200 ctf14-42 (L984), hta1–300 ctf14-42 (L985), hta1–1170 ctf13-30 (L993) and hta1–1170 ctf14-42 (L994). Strains were grown on YPD for 2 days at 30°C (A, B and C), for 4 days at 23°C (D) or for 4 days at 26°C (E and F).
Centromeric chromatin is altered in the hta1 mutants
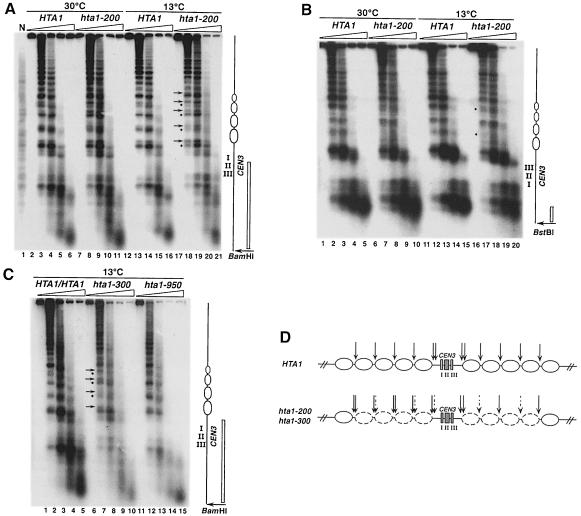
One direct way that the hta1 mutants could impair centromere function would be by altering centromeric chromatin structure. To test this possibility, we analyzed the chromatin structure around CEN3 by indirect end-labeling analysis of micrococcal nuclease (MNase)-digested chromatin. We examined a region of ∼3 kb around the centromere of a wild-type haploid, a wild-type diploid and three hta1 mutants (Figure 8). Both the wild-type and hta1 mutant strains show the previously described nuclease-resistant centromeric core, flanked by arrays of positioned nucleosomes for at least 1.5 kb (Bloom and Carbon, 1982; Figure 8). We find no differences in the digestion pattern of the wild-type haploid and diploid strains. However, in both the hta1–200 and hta1–300 mutants the chromatin surrounding the nuclease-resistant core has an altered pattern of cleavage by MNase. This change is most evident in cells grown at 13°C. In contrast, an hta1–950 mutant, which maintains the haploid state (Figure 2A), shows the same pattern of MNase digestion as the wild-type strains (Figure 8C, lanes 12–15). For the hta1–200 and hta1–300 mutants grown at 13°C, the MNase digestion sites between nucleosomes are enhanced at some positions and diminished at others. This difference is most easily observed for the region to the left of CEN3 (5′ to CDEI) (Figure 8A, compare lanes 13–15 with 18–20; and Figure 8C, compare lanes 2–3 with 7–8). For each of the two internucleosomal cuts observed on each side of the protected nucleosome, the intensity of the lower band is always diminished, and the upper band is always enhanced in the hta1–200 and hta1–300 mutants. There are also differences in the MNase digestion pattern between the wild type and the hta1–200 mutant to the right of CEN3 (3′ to CDEIII) (Figure 8B). In this case, MNase analysis usually detects only one site between each nucleosome. Digestion at these sites appears diminished at a minimum of two internucleosomal sites near the centromere (Figure 8B, compare lanes 12–14 with 17–19). Similar results were seen for the hta1–300 mutant (data not shown). We do not understand why the cleavage pattern to the left of CEN3 appears different from that to the right. However, the periodicity observed on both patterns suggests that the nucleosomal structure is different and perhaps a higher-order structure may be altered in the hta1–200 and hta1–300 mutants. In summary, there are significant differences in the chromatin structure of the CEN3 region in the hta1–200 and hta1–300 mutants compared with the wild type, and these differences are specific for the hta1–200 and hta1–300 mutants. An altered MNase pattern extends over at least 600 bp of DNA on each side of CEN3, encompassing a total of eight positioned nucleosomes (Figure 8D). These results suggest that changes in the chromatin structure flanking the centromere affect the function of the centromere–kinetochore complex.
Fig. 8. The hta1–200 and hta1–300 mutants have altered chromatin structure over the CEN3 region. Nuclei were isolated from wild-type haploid, diploid and hta1 mutant strains after growth at permissive (30°C) or restrictive (13°C for 24 h) conditions, digested with increasing concentrations of MNase and subjected to indirect end-labeling analysis as described in Materials and methods. (A and C) BamHI-digested DNA hybridized with a radiolabeled 616 bp DNA fragment adjacent to the restriction site. (B) BstBI-digested DNA hybridized with a radiolabeled 173 bp DNA fragment adjacent to the restriction site. Positions of the CEN3 nuclease-resistant core and the flanking nucleosomes altered in the mutant are indicated in the diagram to the right. Positions of the probes are indicated as open bars. Arrows and bullets represent enhanced and diminished MNase digestion in the hta1–200 and hta1–300 mutants, respectively, compared with wild type. Strains are as follows: N, naked DNA from FY604, HTA1 (FY604), HTA1/HTA1 (FY604/FY605), hta1–200 (FY987), hta1–300 (FY988) and hta1–950 (FY990). Strains FY987 and FY988 are homogeneous diploid populations. (D) Summary of the MNase digestion pattern shown by the hta1–200 and hta1–300 mutants at the CEN3 locus. Dashed arrows represent diminished digestion and thicker arrows represent enhanced digestion. The eight nucleosomes altered in the hta1 mutant are depicted by dashed ovals.
Dosage suppressors define different functions for histone H2A
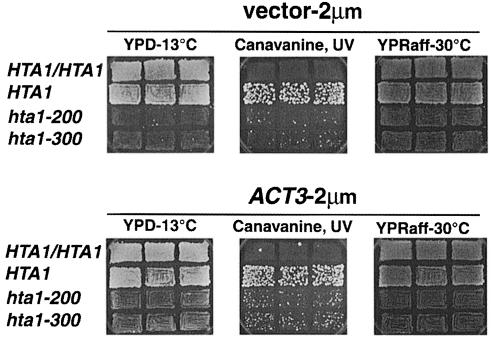
The allele specificity associated with the increase-in-ploidy, cold-sensitive and transcriptional defects suggests that these phenotypes represent different H2A functions. Alternatively, they could all be an indirect consequence of a transcriptional defect (Hirschhorn et al., 1995). To learn more about the relationship among these phenotypes we performed two high-copy-number suppressor screens, one for suppressors of the cold sensitivity and another for suppressors of the SUC2 transcriptional defect. Different genes were found to suppress the two different phenotypes. MSN1 (Estruch and Carlson, 1990) in high copy suppressed the Raf– phenotype caused by the SUC2 transcriptional defect, but did not suppress the cold sensitivity. Conversely, ACT3 (Harata et al., 1994) in high copy partially suppressed the cold sensitivity but not the Raf– phenotype. Although we do not know how these proteins function to suppress the H2A defects, the specificity of suppression suggests that the Cs– and Raf– phenotypes represent two different H2A functions. ACT3 in high copy was also able to suppress partially the increase-in-ploidy phenotype (Figure 9), suggesting that the cold sensitive and increase-in-ploidy phenotypes are related to a specific H2A function. Act3 has recently been shown to interact with histones (Harata et al., 1999). The distinct suppression patterns strongly suggest that these hta1 mutations impair distinct functions: transcription and centromere activity.
Fig. 9. High-copy-number suppression of the cold sensitivity and increase in ploidy of the hta1 mutants. Strains containing the 2 μm vector pRS425 or ACT3 carried on the same vector were grown as patches on 5–FOA-Leu to allow the hta1 mutant strains to lose the episomal wild-type HTA1 gene carried on a URA3-based plasmid, and then replica printed on the media indicated. Cold sensitivity was monitored by growth on YPD plates for 2 weeks at 13°C, increase in ploidy was monitored by the appearance on CanR papillae induced by UV irradiation after 4 days at 30°C, SUC2 expression was monitored by growth on raffinose plates grown at 30°C for 2 days. Strains used were: HTA1/HTA1 (FY604 × FY605), HTA1 (FY604), hta1–200 (FY1819) and hta1–300 (FY1897).
Discussion
This study provides strong evidence that histone H2A is required for normal chromosome segregation by playing a role in centromere function. This conclusion is based on six sets of results analyzing a specific class of hta1 mutants. First, the hta1 mutants are unable to maintain the haploid state, forming diploids soon after germination. Secondly, the hta1 mutants have a delayed cell cycle at G2–M. Thirdly, the hta1 mutants have an increased rate of chromosome loss. Fourthly, the hta1 mutants increase ploidy as a result of unipolar segregation of sister chromatids. Fifthly, the hta1 mutations show genetic interactions with mutations in genes encoding kinetochore proteins. Finally, the hta1 mutants have an altered chromatin structure over centromeric DNA. This structural alteration, taken together with the genetic results, strongly suggests a critical role for histone H2A in centromere function.
Two functions for histone H2A
The hta1–200 and hta1–300 alleles were originally isolated in a screen for hta1 mutants defective in transcription of the SUC2 gene (Hirschhorn et al., 1995). Their cold-sensitive and ploidy instability phenotypes set them apart from the rest of the mutants, suggesting that perhaps another histone function was also altered in these mutants, resulting in the additional phenotypes. Our finding of high-copy-number suppressors that are specific for the SUC2 transcriptional defect or for the cold-sensitive and increase-in-ploidy defects strongly suggests that the latter phenotypes are related to a histone H2A function that is distinct from the defect at SUC2. Moreover, neither MSN1 nor ACT3 have decreased mRNA levels in the hta1–200 and hta1–300 mutants (data not shown), indicating that the dosage suppression is not a compensation of decreased levels of transcription of the suppressor genes. Thus, these results suggest that the hta1–200 and hta1–300 mutants are defective in a previously unstudied function of histone H2A.
Histone H2A is necessary for maintaining a haploid state
Analysis of the hta1 mutants revealed that they are unable to maintain the haploid state and become full, stable diploids at permissive temperature. This transition from haploid to diploid has been observed for hta1 meiotic progeny from HTA1/hta1 heterozygotes and for hta1 haploids following loss of a wild-type copy of HTA1 on a plasmid. The frequency of increasing from one to two copies of two different chromosomes suggests that the transition from haploid to diploid occurs in one step, rather than a gradual gain of chromosomes. This apparent jump in ploidy, as well as the stability of the diploid state, may reflect a combination of a chromosome segregation defect and a strong selective advantage for diploids versus aneuploids.
Several genes have been implicated in the control of ploidy. Mutations in one class fail to regulate DNA replication properly, allowing re-replication within one cell cycle (Heichman and Roberts, 1996; Singer et al., 1996; Liang and Stillman, 1997). Mutations in a second class impair functions required for chromosome segregation, such as spindle pole body duplication, kinetochore attachment and microtubule formation (Schild et al., 1981; Thomas and Botstein, 1986; Rose and Fink, 1987; Baum et al., 1988; Snyder and Davis, 1988; Winey et al., 1991; McGrew et al., 1992; Vallen et al., 1992; Chan and Botstein, 1993). In these cases, ploidy increases are due to non-disjunction events in which replicated sister chromatids segregate to the same pole at anaphase. Interestingly, similarly to the hta1 mutants that we have studied, most of these mutants also have increased rates of chromosome loss, indicating that a single defect can produce both polyploid and aneuploid cells. Moreover, mutations in centromeric DNA that affect centromere function can also cause both ploidy increases and chromosome loss (Saunders et al., 1988; McGrew et al., 1989; Densmore et al., 1991). Taken together, our data suggest that the hta1 mutants analyzed here increase their ploidy as a result of defective centromere function impairing chromosome segregation. These mutants are unusual in that the ploidy increase occurs under conditions permissive for growth, a behavior that, to our knowledge, has been described for only one other mutant, mob1 (Luca and Winey, 1998).
Recent studies of another mutant with both increase-in-ploidy and chromosome segregation defects has shed some light on the regulation of centromere function (Biggins et al., 1999). Temperature-sensitive ipl1 mutants increase in ploidy (Chan and Botstein, 1993) and have defects in chromosome segregation, resulting in unipolar segregation of sister chromatids (Biggins et al., 1999), similarly to the hta1–200 and hta1–300 mutants. It has been proposed that the Ipl1 kinase regulates kinetochore function through the phosphorylation of Ndc10, the p110 subunit of the CBF3 complex. Therefore, segregation of sister chromatids to one pole of the spindle can be explained by models of kinetochore malfunction. The hta1 mutants could be defective in centromere function due to an altered chromatin structure (see section below) that would result in the formation of a defective kinetochore. Such a defect could lead, for example, to the preferential attachment of sister kinetochores to microtubules from the same pole, resulting in segregation of the sister chromatids to the same cell.
Genetic evidence that histone H2A has a role in centromere function
Genetic properties of the hta1–200 and hta1–300 mutants suggest that they impair centromere function. First, the growth defects observed for double mutants have established genetic interactions between histone H2A and kinetochore components. The severity of the growth defect in the hta1 ctf14 double mutants was particularly striking. Strong genetic interactions among mutations in genes encoding centromere-binding proteins have been observed previously and are likely to reflect an accumulated defect in the kinetochore structure (Meluh and Koshland, 1995; Baker et al., 1998). In addition, the frequencies of chromosome loss and the leakiness of the cell cycle arrest phenotype in the hta1 mutants are similar to the defects observed for kinetochore mutants, especially ctf13, ctf14/ndc10 (Doheny et al., 1993), cse4 (Stoler et al., 1995) and mif2 (Brown et al., 1993). In particular, the ndc10-1 mutant displays a similar DNA content profile at the restrictive temperature, with cells of higher and lower DNA content, interpreted as the results of non-disjunction events (Goh and Kilmartin, 1993). Taken together, these results strongly suggest that the hta1 mutants are defective in some aspect of centromere function.
Centromere chromatin is altered in an hta1 mutant
Analysis of the chromatin structure of the CEN3 region by MNase digestion has demonstrated a clear difference between a wild-type strain and the hta1–200 and hta1–300 mutants. In all of the strains the nucleosomes appear to be positioned, but the internucleosomal cuts differ between wild type and mutants. These changes appear on both sides of the centromere, extending over a total distance of at least eight nucleosomes. Previous analysis of the chromatin structure at the SUC2 promoter revealed some differences between wild type and the hta1–200 and hta1–300 mutants (Hirschhorn et al., 1995). This result, combined with our finding at CEN3, suggests that some loci may be more sensitive than others to a common change in chromatin structure caused by the altered nucleosomes present throughout the genome, perhaps due to interactions with locus-specific factors. Alternatively, the chromatin changes at centromeres may be particular to this region and represent a specialized chromatin structure, perhaps some form of centromeric heterochromatin. It has been proposed that the centromere–kinetochore complex is assembled on a specialized centromeric nucleosome, which contains Cse4, the S.cerevisiae counterpart of the mammalian centromere protein CENP-A (Meluh et al., 1998). While the presence of any core histone at the centromere has not been demonstrated, genetic data suggest interactions between H4 and Cse4 (Smith et al., 1996). Our results suggest that histone H2A in the nucleosomes flanking the centromere affects centromere function, even though the centromere core itself may not be affected. In a recently proposed model for S.cerevisiae centromere structure, the centromeric DNA wraps around a Cse4-containing nucleosome (Meluh et al., 1998). By this model, the flanking nucleosomes would be in close contact with kinetochore proteins, and therefore would be likely to play a prominent role with respect to centromere function. We propose that the interaction between histone H2A present in the nucleosomes flanking the centromere and kinetochore proteins is essential for normal centromere function.
The only other histone mutant known to have similar defects in mitotic chromosome segregation is a temperature-sensitive histone H4 mutant (Smith et al., 1996). Like the H2A mutants we have studied, the H4 mutants arrest at G2–M and have an increased rate of chromosome loss at the restrictive temperature. In contrast to the H2A mutants, chromosome segregation is not affected at the temperature permissive for growth, and the H4 mutant was not reported to diploidize. Interestingly, the temperature sensitivity of this H4 mutant is suppressed by increased expression of CSE4, establishing a connection with centromere function. (Our hta1 mutations are not suppressed by increased expression of CSE4; data not shown.) If a specialized chromatin structure exists at the centromere, the H4 mutant could affect centromere function via a faulty interaction with Cse4, as part of a modified nucleosome (Smith et al., 1996). Alternatively, the H4 mutant could affect centromere function by altering the interaction of kinetochore proteins with flanking nucleosomes, similarly to our model for the H2A mutants. These studies clearly reveal a role for individual histones in mitotic chromosome segregation, and the phenotypic differences of these mutants may reflect different roles in these processes.
The X–ray crystallographic data for both the histone octamer and the nucleosome predict that the amino acids S20 and G30 of H2A are located on the surface of the nucleosome in close contact with DNA (E.Moudrianakis, personal communication; Luger et al., 1997). Both of these amino acids are evolutionarily conserved and S20 is a potential site for phosphorylation (van Holde, 1988). The amino acid changes encoded by hta1–200 and hta1–300, S20F and G30D, respectively, are likely to result in steric hindrance on the surface of the nucleosome and in changes of the local charge, which could disrupt the DNA–histone interaction. Thus, these changes might affect the path of the DNA over the octamer, revealed by the altered pattern of MNase digestion that we have observed. Moreover, these changes in the nucleosome surface may affect specific interactions between H2A and kinetochore proteins that result in a defective centromere function. In support of this idea, we find normal CEN3 chromatin and normal haploid maintenance in the hta1–950 mutant, which encodes a serine replacement at position L95. This amino acid is predicted to be located toward the center of the octamer, without making direct contact with DNA.
In conclusion, we have presented genetic and molecular data strongly suggesting that histone H2A is involved directly in centromere function. While we cannot rule out an indirect effect, the change in chromatin structure around a centromere combined with the several mutant phenotypes that have established a centromere defect, the allele specificity of these phenotypes and the phenotype specificity of high-copy suppressors make a direct effect the most likely explanation. The normal role of H2A and nucleosomes is likely to be in the proper assembly or function of the centromere. In the hta1 mutants, a defective centromere might result in defective microtubule attachment or tension. Such a defect could lead to the increase-in-ploidy and chromosome instability phenotypes that we have observed. Further genetic and biochemical studies will be necessary to elucidate the precise functions of histone H2A in chromosome segregation. Because of the evolutionary conservation of histones, understanding their role in chromosome transmission in yeast will contribute to the understanding of chromosome segregation in more complex eukaryotes.
Materials and methods
Yeast strains, genetic methods, growth and media
The S.cerevisiae strains used in this study are listed in Table III. Strains designated FY are isogenic to S288C and are GAL2+ (Winston et al., 1995). Strains designated L are derived from crosses between FY strains and other genetic backgrounds. Lower case letters indicate a recessive mutant allele, and upper case indicates the wild-type allele. Strain construction and other genetic manipulations were carried out by standard methods (Guthrie and Fink, 1991). All yeast media, including yeast extract/peptone/dextrose (YPD), synthetic minimal (SD), synthetic complete (SC), omission media (SC–) and media containing 5–FOA were made as described previously (Rose et al., 1990). Cell viability was determined by plating cells on YPD and monitoring colony formation, as described in Brown et al. (1993). Mating-type switching was performed in the hta1–200 (FY987) and hta1–300 (FY988) mutants by transient expression of the HO gene using pGAL-HO, as described (Guthrie and Fink, 1991). Cultures were grown overnight on 0.5% glucose followed by induction on 2% galactose (SC-Ura) for 6 h. Colonies growing on YPD were screened for mating. Non-mater MATa/MATα candidates were sporulated, dissected and scored for the segregation of the MAT locus.
Table III. Saccharomyces cerevisiae strains.
| Strain | Genotype |
|---|---|
| FY604 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 |
| FY605 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 |
| FY987 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–200 |
| FY988 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–300 |
| FY990 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–950 |
| FY991 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–1170 |
| FY1817 | MATa his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–200 |
| FY1818 | MATa his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–300 |
| FY1819 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–200 <pSAB6> |
| FY1820 | MATα/MATa his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 ura3-52/ura3-52trp1Δ63 /trp1Δ63 (hta2-htb2)Δ::TRP1/(hta2-htb2)Δ::TRP1 ade2Δ101::LEU2::ade2Δ102/ade2Δ101::LEU2::ade2Δ102 lys2Δ101::HIS3::lys2Δ102/lys2Δ101::HIS3::lys2Δ102 <pSAB6> |
| FY1821 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 ade2Δ101::LEU2::ade2Δ102 lys2Δ101::HIS3::lys2Δ102 <pSAB6> |
| FY1822 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 ade2Δ101::LEU2::ade2Δ102 lys2Δ101::HIS3::lys2Δ102 <pSAB6> |
| FY1823 | MATα his3Δ200 leu2Δ1ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–200 ade2Δ101::LEU2::ade2Δ102 lys2Δ101::HIS3::lys2Δ102 <pSAB6> |
| FY1824 | MATα his3Δ200 leu2Δ1ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–300 ade2Δ101::LEU2::ade2Δ102 lys2Δ101::HIS3::lys2Δ102 <pSAB6> |
| FY1825 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 NUF2::GFP |
| FY1826 | MATa his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–200 NUF2::GFP |
| FY1827 | MATa his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–300 NUF2::GFP |
| FY1828 | MATα/MATa his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 ura3-52/ura3-52 trp1Δ63/trp1Δ63 (hta2-htb2)Δ::TRP1/(hta2-htb2)Δ::TRP1 HIS4/his4Δ10::URA3 |
| FY1829 | MATα/MATa his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2-128δ/lys2-128δ ura3-52/ura3-52 trp1Δ63/trp1Δ63 (hta2-htb2)Δ::TRP1/(hta2-htb2)Δ::TRP1 HIS4/his4Δ10::URA3 hta1–200/hta1–200 |
| FY1830 | MATα/MATa his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 ura3-52/ura3-52trp1Δ63/trp1Δ63 (hta2-htb2)Δ::TRP1/(hta2-htb2)Δ::TRP1 HIS4/his4Δ10::URA3 hta1–300/hta1–300 |
| FY1897 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–300 <pSAB6> |
| FY1898 | MATa ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 his3-205::GFP-lacI-HIS3 leu2Δ1:: lacO-LEU2 |
| FY1899 | MATα lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 his3-205::GFP-lacI-HIS3 leu2Δ1:: lacO-LEU2 hta1–200 <pSAB6> |
| L980 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 or trp1Δ1(hta2-htb2)Δ::TRP1 ctf13-30 |
| L981 | MATa his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 or trp1Δ1(hta2-htb2)Δ::TRP1 ctf13-30 hta1–200 |
| L982 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 or trp1Δ1(hta2-htb2)Δ::TRP1 ctf13-30 hta1–300 |
| L983 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 ctf14-42 |
| L984 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 ctf14-42 hta1–200 |
| L985 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 ctf14-42 hta1–300 |
| L986 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 mif2-3 |
| L987 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 mif2-3 hta1–200 |
| L988 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 mif2-3 hta1–300 |
| L989 | MATa his3Δ200 or his3-11,15 leu2Δ1 ura3-52 trp1Δ63 or trp1Δ901 (hta2-htb2)Δ::TRP1 cse4-1 |
| L990 | MATa his3Δ200 or his3-11,15 leu2Δ1 ura3-52 trp1Δ63 or trp1Δ901 (hta2-htb2)Δ::TRP1 cse4-1 hta1–200 |
| L991 | MATα his3Δ200 or his3-11,15 leu2Δ1 ura3-52 trp1Δ63 or trp1Δ901 (hta2-htb2)Δ::TRP1 cse4-1 hta1–300 |
| L992 | MATα his3Δ200 leu2Δ1 lys2-128δ ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 hta1–300 hta1–1170 |
| L993 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 or trp1Δ1(hta2-htb2)Δ::TRP1 ctf13-30 hta1–1170 |
| L994 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 (hta2-htb2)Δ::TRP1 ctf14-42 hta1–1170 |
Bacterial strains and plasmids
Plasmids were amplified and isolated from Escherichia coli strains HB101 or DH5α, according to standard procedures (Ausubel et al., 1988). pIP65 was constructed by ligating the 548 bp HindIII–XbaI fragment of ADE2, obtained from pRS402 (Brachmann et al., 1998), into the HindIII–XbaI sites of pRS305 (Sikorski and Hieter, 1989). pIP65 contains a unique StuI site within ADE2, which was used to linearize the plasmid for homologous integration into ADE2.
Assays to measure gain in chromosome copy number
Saccharomyces cerevisiae cells that have gained genetically marked chromosomes were detected by two different papillation assays. In both assays, the hta1 strains are initially maintained as haploids by carrying pSAB6, an episomal copy of HTA1 on a URA3-ARS-CEN plasmid (Hirschhorn et al., 1995). The mutants are then assayed following growth of the strains on media containing 5–FOA, to allow the testing of mutants lacking the plasmid.
In the first assay, the copy number of chromosomes II and XV was monitored as described previously (Chan and Botstein, 1993; Figure 3). Chromosome II was marked by integrating the HIS3 gene into the LYS2 locus using the linearized plasmid pRB1210 (Chan and Botstein, 1993). Chromosome XV was marked by integrating LEU2 into ADE2 using plasmid pIP65 linearized with StuI. The integrations disrupt and result in a partial duplication of LYS2 and ADE2, respectively, rendering the strains phenotypically Lys– His+ and Ade– Leu+. Lys+ or Ade+ colonies can be generated by homologous recombination between the duplicated sequences of the disrupted gene, in this case the HIS3 or LEU2 marker is lost. If a marked chromosome duplicates, then Lys+His+ and Ade+ Leu+ derivatives can be obtained via a recombination event on one of the two homologs for each marked chromosome. Such events are detected as papillae after replica plating to selective media. Patched cells grown on 5–FOA-Leu-His plates were replica plated onto SC-Lys-His and SC-Ade-Leu media to test for chromosome-gain.
The frequencies of recombination and chromosome loss were determined as follows. Wild-type and mutant strains carrying the episomal HTA1 gene on pSAB6 were grown on SC-His-Leu to allow the cells to lose the plasmid and then grown on 5–FOA-His-Leu at 30°C. Single colonies were excised from the plates using a sterile scalpel and resuspended in water. Aliquots were appropriately diluted, briefly sonicated, and plated onto SC medium for cell count, onto SC-Lys, SC-Ade and SC-Lys-Ade plates to determine recombination rates, and onto SC-Lys-His, SC-Ade-Leu and SC-Lys-His-Ade-Leu to determine chromosome-gain rates. Colonies were scored after 3 days (for wild-type strains) or 4 days (for hta1 mutants) at 30°C. The frequencies were determined by fluctuation analysis (method of the median; Lea and Coulson, 1949) from six independent cultures of each strain.
In the second assay, chromosome V ploidy was assayed by monitoring the CAN1 gene as described (Schild et al., 1981). Since canavanine resistance is conferred by recessive mutations in the CAN1 gene, the frequency of CanR mutants is much greater among haploids than among diploids or among strains with two copies of chromosome V. In this assay, strains were replicated onto SC-Arg plates either with or without canavanine, the cells were mutagenized by UV irradiation (300 ergs/mm2), and plates were incubated at 30°C for 3 days.
Flow cytometry
The DNA content of yeast cells was determined on cells fixed with ethanol, treated with RNase and stained with propidium iodide as described previously (Lew et al., 1992). Yeast cells from aliquots taken at various time points were pelleted and fixed with 70% ethanol until all the samples were taken. For each sample, the DNA content of at least 15 000 individual cells was measured using a fluorescence-activated cell sorter (Ortho Diagnostics) at 488 nm excitation. To analyze the kinetics of the ploidy increase, an HTA1/hta1–200 diploid was sporulated and dissected. The spore colonies were excised from the YPD plate with a sterile scalpel and resuspended in 1 ml of YPD. Cells were counted with a hemocytometer and an aliquot was taken and prepared for DNA content analysis by flow cytometry. The remainder of the cells were inoculated into 1 l of YPD and grown at 30°C overnight. Cells were counted and another aliquot was taken for DNA content analysis. The procedure was repeated until the cells had divided for 42 generations. DNA content of cells shifted to 13°C was determined using the same procedure.
Chromosome loss and recombination assays
Diploids homozygous for HTA1, hta1–200 and hta1–300 were marked at the HIS4 locus on the right arm of chromosome III by the integration of URA3 into HIS4. The resulting strains, FY1828, FY1829 and FY1830, are HIS4/his4Δ10::URA3. Each strain was grown overnight on SC-Ura, streaked for single colonies on a YPD plate and incubated at 30°C for 2 (for HTA1 strains) or 5 (for hta1 strains) days. Ten colonies were excised from each plate using a sterile scalpel and resuspended in 1 ml of YPD. For the analysis at restrictive temperature, the cultures were transferred to a 13°C shaker for 24 h. After brief sonication, the cells were counted and dilutions were plated on 5–FOA at 30°C. 5–FOAR colonies can result from either chromosome III loss or a mitotic recombination event between CEN3 and his4Δ10::URA3, loosing the URA3 gene. To distinguish between the two events, we assayed the MAT locus, located on the left arm of chromosome III, by testing the 5–FOAR colonies for their mating phenotype. 5–FOAR colonies resulting from chromosome loss will be either MATa or MATα and therefore should mate, whereas mitotic recombinants will remain non-mating MATa/MATα diploid strains. To confirm the structure of the MAT locus, 12 colonies of each class were analyzed by PCR using MATa- and MATα-specific primers as described (Huxley et al., 1990). In all cases the PCR result was consistent with the result of the mating assay. In these chromosome loss tests, two isolates from each strain were assayed. The rates of chromosome III loss and recombination were calculated by the method of the median (Lea and Coulson, 1949).
Fluorescence microscopy
Exponentially growing cells were synchronized with 0.9 μM α–factor for 2.5 h, washed twice with fresh YPD and shifted to 13°C. Aliquots were taken and prepared for indirect immunofluorescence as described (Rose et al., 1990). The rat anti-yeast α–tubulin monoclonal antibody YOL1/34 (MAS 078b; Accurate, Westbury, NY) was diluted 1:200. Cy3-conjugated goat anti-rat antibody (Jackson ImmunoResearch, West Grove, PA) was used at a 1:500 dilution. Nuclei were visualized with 4′,6–diamidino-2–phenylindole (DAPI; Sigma, St Louis, MO) added to the last wash before mounting the slides. Cells were viewed on a Zeiss Axiophot microscope equipped with epifluorescence and photographed with Kodak TMAX-400 film. GFP fluorescence was visualized in live cells using a fluorescein filter and a Nikon microscope equipped with a digital camera (Kahana et al., 1998). Quantitation of the GFP signal was done on at least 200 cells from each strain. Photographs were prepared using Photoshop software (Adobe).
Preparation of yeast nuclei and indirect end-labeling analysis
Yeast nuclei preparations, MNase digestions and indirect end-labeling analysis of CEN3 chromatin were performed as described previously (Hirschhorn et al., 1992). Following MNase digestion and DNA purification, the DNA was digested to completion with BamHI as described previously (Bloom and Carbon, 1982). CEN3 DNA was detected using a 616 bp CEN3 probe generated by PCR with the following oligonucleotides: 5′–CAGCGCCAAACAATATGGA–3′ (forward) and 5′–CCCGGGTGGGAAACTGAAG–3′ (reverse). This probe allows detection of the centromere core and nucleosomes to the left (5′ to CDEI) of CEN3. The analysis of CEN3 chromatin from the opposite direction (to the right of CEN3, 3′ to CDEIII) was performed after digestion of the MNase-treated samples with BstBI and hybridization with a 173 bp probe adjacent to the BstBI site, ∼200 bp left of CDEI. The oligonucleotides used as primers to generate this probe were: 5′–CTAACACTTGTCAAACAGAATATAAGG–3′ (forward) and 5′–GTATAGGTACTGTACTATAAGCGGAAGG–3′ (reverse). Radiolabeled probes were generated by PCR using standard procedures (Ausubel et al., 1988).
Isolation of hta1 dosage suppressors
Strain FY987 (hta1–200) was transformed with a high-copy Yep13-based library (2μm-LEU2). Cells were plated on SC-Leu medium and incubated at 15°C. Approximately 27 000 transformants gave rise to 87 Cs+ colonies. From these, two clones were able to re-transform and were neither HTA1–HTB1 nor HTA2–HTB2. One of these clones was subcloned and sequenced. The suppressor was identified as ACT3/ARP4, an essential gene encoding an actin-related protein (Harata et al., 1994). In a similar experiment we isolated high-copy suppressors of the defect in SUC2 expression, by replica printing the Leu+ transformants on YPRaf medium and isolating colonies that grew on raffinose. One of these clones was identified as MSN1, a gene previously isolated as suppressor of temperature-sensitive alleles of SNF1 and SNF4, two transcriptional activators of SUC2 (Estruch and Carlson, 1990).
Analysis of sister chromatids by GFP tagging of chromosome III
Strains were tagged with GFP at the centromere of chromosome III essentially as described by Straight et al. (1996). Wild-type HTA1 and mutant hta1–200 strains were transformed with plasmid pAFS59 linearized with EcoRV to integrate the Lac operator repeats at LEU2, marking CEN3, and pAFS135 linearized with NheI to integrate the GFP–LacI-I12 fusion under the control of the HIS3 promoter at HIS3. The resulting strains, FY1898 and FY1899, were mated to obtain a heterozygous HTA1/hta1–200 diploid that was sporulated and dissected. Spores were germinated and grown for ∼25 generations at 30°C. Wild-type and hta1–200 MATa colonies were picked and resuspended in liquid YPD, arrested with α–factor for 2 h, switched to SC-His containing α–factor and 10 mM 3–aminotriazole to induce the HIS3 promoter (Straight et al., 1996), and the incubation was continued for 1 h. Cells were washed and released from the arrest into YPD lacking α–factor. Samples were taken at 0, 1 and 1.7 generation times for each strain, fixed for 30 min in 3.7% formaldehyde and saved at 4°C for fluorescence microscopy. To prevent cells from entering the next cell cycle, we added α–factor back to the culture at two-thirds of their generation time after the initial release from α–factor. At the same time points, aliquots were taken and processed for analysis of DNA content by flow cytometry.
Acknowledgments
Acknowledgements
We are grateful to Clarence Chan, Molly Fitzgerald-Hayes, Leland Hartwell, Phil Hieter, Jason Kahana and Pamela Silver for strains and plasmids. We thank Evangelos Moudrianakis and Connie Holm for valuable discussions. We thank Aimée Dudley and Mary Bryk for critical reading of the manuscript, and Juanita Campos-Torres for expert technical assistance with the flow cytometry. We are grateful to Paul Ferrigno, Jason Kahana and Pamela Silver for help with the GFP microscopy, and Aaron Straight for plasmids and advice with the GFP tagging experiment. I.P. was supported by a grant from the American Cancer Society (PF-4112) and a Charles King Trust Fellowship from The Medical Foundation. This work was supported by NIH grant GM32967 to F.W. I.P. dedicates this work to the memory of Víctor Manuel Pinto.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1988) Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley-Interscience, New York, NY. [Google Scholar]
- Baker R.E., Harris, K. and Zhang, K. (1998) Mutations synthetically lethal with cep1 target S.cerevisiae kinetochore components. Genetics, 149, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Yip, C., Goetsch, L. and Byers, B. (1988) A yeast gene essential for regulation of spindle pole duplication. Mol. Cell. Biol., 8, 5386–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin, F.F., Bhalla, N., Sassoon, I., Hyman, A.A. and Murray, A.W. (1999) The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev., 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K.S. and Carbon, J. (1982) Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell, 29, 305–317. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P. and Boeke, J.D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae 288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Brown M.T., Goetsch, L. and Hartwell, L. (1993) MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol., 123, 387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D., Doctor, J.S., Feursanger, J.H. and Doolittle, M.M. (1981) Differential mitotic stability of yeast disomes derived from triploid meiosis. Genetics, 98, 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S.M. and Botstein, D. (1993) Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics, 135, 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densomore L., Payne, W.E., Fitzgerald-Hayes, M. (1991) In vivo genomic footprint of a yeast centromere. Mol. Cell. Biol., 11, 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny K.F., Sorger, P.K., Hyman, A.A., Tugendreich, S., Spencer, F. and Hieter, P. (1993) Identification of essential components of the S.cerevisiae kinetochore. Cell, 73, 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F. and Carlson, M. (1990) Increased dosage of the MSN1 gene restores invertase expression in yeast mutants defective in the SNF1 protein kinase. Nucleic Acids Res., 18, 6959–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P.-Y. and Kilmartin, J.V. (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol., 121, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (1991) Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA. [Google Scholar]
- Han M., Chang, M., Kim, U.-J. and Grunstein, M. (1987) Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication and transcription. Cell, 48, 589–597. [DOI] [PubMed] [Google Scholar]
- Harata M., Karwan, A. and Wintersberger, U. (1994) An essential gene of Saccharomyces cerevisiae coding for an actin-related protein. Proc. Natl Acad. Sci. USA, 91, 8258–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harata M., Oma,Y., Mizuno,S., JiangY.W., Stillman,D.J. and Wintersberger,U. (1999) The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol. Biol. Cell, 10, 2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. (1967) Macromolecular synthesis in temperature-sensitive mutants of yeast. J. Bacteriol., 93, 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. and Smith, D. (1985) Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S.cerevisiae. Genetics, 110, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichman K.A. and Roberts, J.M. (1996) The yeast CDC16 and CDC27 genes restrict DNA replication to once per cell cycle. Cell, 85, 39–48. [DOI] [PubMed] [Google Scholar]
- Hirschhorn J.N., Brown, S.A., Clark, C.D. and Winston, F. (1992) Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev., 6, 2288–2298. [DOI] [PubMed] [Google Scholar]
- Hirschhorn J.N., Bortvin, A.L., Ricupero-Hovasse, S.L. and Winston, F. (1995) A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol., 15, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A. and Geiser, J.R. (1996) Genetic analysis of the mitotic spindle. Annu. Rev. Genet., 30, 7–33. [DOI] [PubMed] [Google Scholar]
- Huffaker T.C., Thomas, J.H. and Botstein, D. (1988) Diverse effects of β-tubulin mutations on microtubule formation and function. J. Cell Biol., 106, 1997–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C., Green, E.D. and Dunham, I. (1990) Rapid assessment of S.cerevisiae mating type by PCR. Trends Genet., 6, 236. [DOI] [PubMed] [Google Scholar]
- Hyman A.A. and Sorger, P.K. (1995) Structure and function of kinetochores in budding yeast. Annu. Rev. Cell Dev. Biol., 11, 471–495. [DOI] [PubMed] [Google Scholar]
- Johnston G.C., Pringle, J.R. and Hartwell, L.H. (1977) Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res., 105, 79–98. [DOI] [PubMed] [Google Scholar]
- Kahana J.A., Schlenstedt, G., Evanchuk, D.M., Geiser, J.R., Hoyt, M.A. and Silver, P.A. (1998) The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell, 9, 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U.-J., Han, M., Kayne, P. and Grunstein, M. (1988) Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J., 7, 2211–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D.E. and Coulson, C.A. (1949) The distribution of number of mutants in bacterial populations. J. Genet., 49, 264–284. [DOI] [PubMed] [Google Scholar]
- Lew D.J., Marini, N.J. and Reed, S.I. (1992) Different G1 cyclins control the timing of cell cycle commitment in mother and daughter cells of the budding yeast S.cerevisiae. Cell, 69, 317–327. [DOI] [PubMed] [Google Scholar]
- Liang C. and Stillman, B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F.C. and Winey, M. (1998) MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell, 9, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder, A., Richmond, R., Sargent, D. and Richmond, T. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- McGrew J.T., Xiao, Z., Fitzgerald-Hayes, M. (1989) Saccharomyces cerevisiae mutants defective in chromosome segregation. Yeast, 5, 271–284. [DOI] [PubMed] [Google Scholar]
- McGrew J.T., Goetsch, L., Byers, B. and Baum, P. (1992) Requirement for ESP1 in the nuclear division of Saccharomyces cerevisiae. Mol. Biol. Cell, 3, 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D. and Hartwell, L.H. (1986) Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission in S.cerevisiae. Cell, 44, 43–52. [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner D., Wood, J.S., Garvik, B. and Hartwell, L.H. (1986) Isolation of two genes that affect mitotic chromosome transmission in S.cerevisiae. Cell, 44, 53–63. [DOI] [PubMed] [Google Scholar]
- Megee P.C., Morgan, B.A. and Smith, M.M. (1995) Histone H4 and the maintenance of genome integrity. Genes Dev., 9, 1716–1727. [DOI] [PubMed] [Google Scholar]
- Meluh P.B. and Koshland, D. (1995) Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Cell. Biol., 6, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B. and Koshland, D. (1997) Budding yeast centromere composition and assembly as revealed by in vivo crosslinking. Genes Dev., 11, 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang, P., Glowczewski, L., Koshland, D. and Smith, M.M. (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell, 94, 607–613. [DOI] [PubMed] [Google Scholar]
- Morgan B., Mittman, B. and Smith, M. (1991) The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol., 11, 4111–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris D. and Osley, M.A. (1987) The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell. Biol., 7, 3473–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne M.A., Schlenstedt, G., Jinks, T. and Silver, P.A. (1994) Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol., 125, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D. and Fink, G.R. (1987) KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell, 48, 1047–1060. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Saunders M., Fitzgerald-Hayes, M. and Bloom, K. (1988) Chromatin structure of altered yeast centromeres. Proc. Natl Acad. Sci. USA, 85, 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M.J., Yeh, E., Grunstein, M. and Bloom, K. (1990) Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol. Cell. Biol., 10, 5721–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P.J., Solomon, F. and Botstein, D. (1988) Isolation and characterization of conditional-lethal mutations in the TUB1 α-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics, 120, 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Ananthaswamy, H.N. and Mortimer, R.K. (1981) An endomitotic effect of a cell cycle mutation of Saccharomyces cerevisiae. Genetics, 97, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman I. and Bloom,K.S. (1991) Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu. Rev. Cell Biol., 311–336. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J.D., Manning, B.M. and Formosa, T. (1996) Coordinating DNA replication to produce one copy of the genome requires genes that act in ubiquitin metabolism. Mol. Cell. Biol., 16, 1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.M., Yang, P., Santisteban, M.S., Boone, P.W., Goldstein, A.T. and Megee, P.C. (1996) A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol., 16, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. and Davis, R.W. (1988) SPA1: a gene important for chromosome segregation and other mitotic functions in S.cerevisiae. Cell, 54, 743–754. [DOI] [PubMed] [Google Scholar]
- Stoler S., Keith, K.C., Curnick, K.E., Fitzgerald-Hayes, M. (1995) A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev., 9, 573–586. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Belmont, A.S., Robinett, C.C. and Murray, A.W. (1996) GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol., 6, 1599–1608. [DOI] [PubMed] [Google Scholar]
- Thomas J.H. and Botstein, D. (1986) A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell, 44, 65–76. [DOI] [PubMed] [Google Scholar]
- Vallen E.A., Scherson, T.Y., Roberts, T., van Zee, K. and Rose, M.D. (1992) Asymmetric mitotic segregation of the yeast spindle pole body. Cell, 69, 505–515. [DOI] [PubMed] [Google Scholar]
- van Holde K. (1988) Chromatin. Springer-Verlag, New York, NY. [Google Scholar]
- Winey M., Goetsch, L., Baum, P. and Byers, B. (1991) MPS1 and MPS2: novel yeast genes defining distinct steps spindle pole body duplication. J. Cell Biol., 114, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard, C., Ricupero-Hovasse, S.L. (1995) Construction of a set of convenient S.cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]