Abstract
This Nano Focus article highlights recent advances in RNA nanotechnology as presented at the First International Conference of RNA Nanotechnology and Therapeutics, which took place in Cleveland, OH, USA (October 23–25, 2010) (http://www.eng.uc.edu/nanomedicine/RNA2010/), chaired by Peixuan Guo and co-chaired by David Rueda and Scott Tenenbaum. The conference was the first of its kind to bring together more than 30 invited speakers in the frontier of RNA nanotechnology from France, Sweden, South Korea, China, and throughout the United States to discuss RNA nanotechnology and its applications. It provided a platform for researchers from academia, government, and the pharmaceutical industry to share existing knowledge, vision, technology, and challenges in the field and promoted collaborations among researchers interested in advancing this emerging scientific discipline. The meeting covered a range of topics, including biophysical and single-molecule approaches for characterization of RNA nanostructures; structure studies on RNA nanoparticles by chemical or biochemical approaches, computation, prediction, and modeling of RNA nanoparticle structures; methods for the assembly of RNA nanoparticles; chemistry for RNA synthesis, conjugation, and labeling; and application of RNA nanoparticles in therapeutics. A special invited talk on the well-established principles of DNA nanotechnology was arranged to provide models for RNA nanotechnology. An Administrator from National Institutes of Health (NIH) National Cancer Institute (NCI) Alliance for Nanotechnology in Cancer discussed the current nanocancer research directions and future funding opportunities at NCI. As indicated by the feedback received from the invited speakers and the meeting participants, this meeting was extremely successful, exciting, and informative, covering many groundbreaking findings, pioneering ideas, and novel discoveries.
The meeting was launched with an introductory keynote address by Peixuan Guo (University of Cincinnati), the chair of the organizing committee. Dr. Guo introduced the topic of RNA nanotechnology, its history, approaches, current status, and future prospects, emphasizing that living organisms possess a wide variety of natural nanomachines, elegantly patterned arrays, and highly ordered structures performing diverse biological functions. There are numerous intriguing configurations that have inspired biomimetic stategies. He noted that macromolecules of DNA, RNA, and proteins have intrinsically defined features at the nanometer scale and can serve as powerful building blocks for bottom-up fabrication of nanostructures. The rapid advances in DNA nanotechnology have created unexpected bridges between material engineering and synthetic structural biology.1−3 Dr. Guo emphasized that the field of RNA nanotechnology is new and rapidly emerging. Over the last five years, there has been a burst of publications on RNA nanostructures, indicating increasing interest in RNA nanotechnologies in diverse fields such as microbiology, biochemistry, biophysics, chemistry, structural biology, nanomedicine, and cell biology. RNA-based nanoscaffolds are therefore expected to have great impact in the near future especially with regard to diagnostics and therapeutics.(4)
Guo noted that RNA, as a cousin of DNA, has recently emerged as an important nanotechnology platform due to its extraordinary diversity in structure and function. RNA nanoparticles can be fabricated with a level of simplicity characteristic of DNA, plus they possess versatile tertiary structure and catalytic functions that can mimic some forms of proteins.5−8 RNA is unique in comparison to DNA by virtue of its high thermodynamic stability,9,10 the formation of both canonical and noncanonical base pairings,11−15 the capability of base stacking,9,10 and distinctive in vivo attributes.16−23 The remarkable modularity of RNA tertiary motifs can be encoded at the level of an RNA sequence to specify complex three-dimensional (3D) architectures exhibiting helices, loops, bulges, stems, hairpins, and pseudoknots. Further, a large variety of single-stranded loops are suitable for inter- and intramolecular interactions, serving as a mounting dovetail in self-assembly. Taking advantage of these unique characteristics, Dr. Guo presented highlights from his pioneering work in 1998, which demonstrated that RNA dimer, trimer, and hexamer nanopaticles can be fabricated by re-engineering RNA molecules using the model of motor pRNA (packaging RNA), a component that gears the DNA packaging motor of bacteriophage phi29.(22) He showed that the pRNA can be used as a building block or scaffold for constructing a variety of RNA nanostructures with functional entities as delivery vehicles or imaging tools. He further described the application of RNA nanomotors in various aspects of cellular and molecular biology as a tool for potential therapeutics. He pointed out that the sensitivity of RNA to RNase degradation has previously made many scientists shy away from RNA nanotechnology. The demonstration that RNA nanoparticles can be produced to drive viral DNA packaging motors using chemically modified RNA suggests that it is possible to produce RNase-resistant, biologically active, and stable RNA for applications in nanotechnology.(24) Dr. Guo also discussed the significant contributions of Dr. Eric Westhoff, Dr. Neocles Leontis, and Dr. Luc Jaeger for their fundamental structural studies of RNA motifs and important contributions to RNA technology.13,25−28
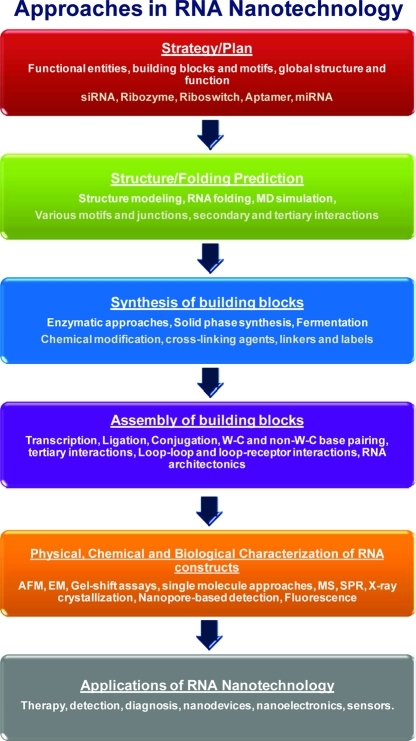
Lastly, Dr. Guo summarized the key approaches in RNA nanoparticles construction(4) starting with the conception step, whereby the desired properties of the nanoparticles are defined and the global structure of the particle and application are considered. A computational approach is then applied to predict the folding of the building blocks and the consequences of inter-RNA interactions in the assembly of the final RNA quaternary structure. The building blocks are then generated (in vitro transcription or chemical synthesis), and the individual subunits are assembled (templated or nontemplated) into quaternary architectures. The resulting RNA nanoparticles are characterized by the atomic force microscope (AFM), electron microscope (EM), gel electrophoresis, chromatography, or fluorescence experiments to ensure proper folding that is consistent with the desired structural and functional capabilities. After thorough assessment, RNA nanoparticles are used for a variety of applications including the treatment and diagnosis of diseases and the regulation of cellular functions. We have arranged the following sections of this article to match the six steps of RNA nanoparticle construction, which served as the overarching themes of the meeting (1).
Figure 1.
Approaches in RNA nanotechnology. Figure modified with permission from ref (4). Copyright 2010 Nature Publishing Group.
RNA Structures for Nanoparticle Construction
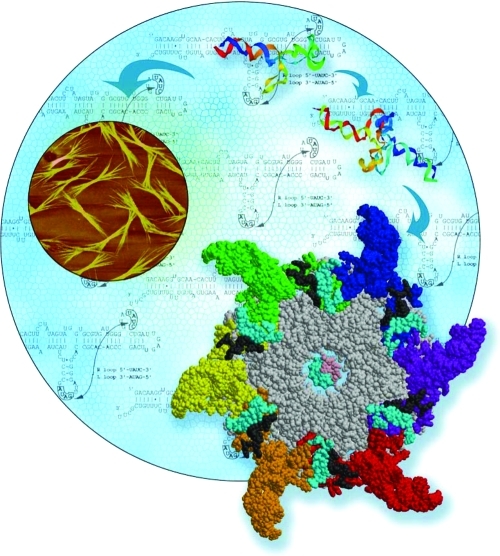
RNA typically contains various single-stranded loops that interact to form 3D biologically important RNA structures via inter- and intramolecular interactions. Several naturally occurring diverse RNA loops and motifs with defined 3D architectures exist that can serve as structural scaffolds in RNA nanotechnology. One such example is based on the structural features of the pRNA of the bacteriophage phi29 DNA packaging motor,22,29 which uses a hexameric RNA ring to gear the machine.30,31 Dr. Guo’s group has extensively re-engineered pRNA to form dimers, trimers, tetramers, hexamers (with proteins), and arrays via hand-in-hand or foot-to-foot interactions between two interlocking loops.32,33 The dimer and trimer nanoparticles have been used successfully as polyvalent vehicles to deliver a variety of therapeutic molecules32,33 as well as for constructing RNA arrays (2).(32)
Figure 2.
Role of phi29 DNA packaging RNA (pRNA) in RNA nanotechnology. Six pRNA assemble into a hexameric ring to gear the DNA translocating machine. pRNA monomers can be re-engineered to fold into well-defined structures, such as dimers, hexamers, and arrays via hand-in-hand or foot-to-foot interactions between two interlocking loops. pRNA-based nanoparticles have been successfully constructed with functional modules and used as polyvalent vehicles to deliver a variety of therapeutic/detection molecules to cancer- and viral-infected cells. Image courtesy of YinYin Guo. Figure adapted with permission from ref (136). Copyright 2011 Elsevier.
In his keynote address, Luc Jaeger (University of California, Santa Barbara) described two complementary approaches for the design and engineering of versatile, programmable, 3D RNA-based nanoscaffolds with precise control over their geometry, size, and composition and possibility of further functionalization. The first approach takes advantage of known RNA structural elements. This approach, called “RNA architectonics”, achieves a remarkable degree of structural control by using prefolded RNA structural motifs as building blocks for bottom-up assemblies.34−37 This strategy is based on the rational design of artificial 3D RNA architectures that are formed by inverse folding processes. The second approach, based only on canonical Watson–Crick interactions, utilizes relatively short (26–48 nt) single-stranded RNAs. This approach employs computer-aided techniques to engineer 3D RNA, RNA/DNA, and DNA nanoscaffolds.(38) His group, in collaboration with Bruce Shapiro (National Cancer Institute), developed a range of strategies for obtaining self-assembling RNA architectures with unprecedented control and precise positioning of functional groups in space, such as 3D RNA nanocubic scaffolds,(38) tRNA polyhedrons,(39) and nanorings.(34)
Another approach in RNA nanoparticle construction is to utilize existing RNA structures with known functions as building blocks. Within the cell, there exist small RNAs, such as riboswitches with regulatory functions.40−43 Herve Isambert’s group (Institut Curie–Center de Recherche, France) developed pioneering experimental and computational approaches for the development of small regulatory circuits based mainly on RNA interactions.(44) In addition, he presented a specific example of a small bacterial regulatory RNA, DsrA, which self-assembles into long filamentous-like structures, as well as more extended 2D/3D nanostructures with unique switching properties.(45) This important finding suggests novel self-assembly principles to design RNA nanoparticles with structural switching capabilities. An online RNA folding server (http://kinefold.curie.fr) has more than 40 000 RNA folding simulation studies available.
In related work, Li Niu (State University of New York, Albany) presented an exciting structural folding property of an RNA aptamer sequence (identified by SELEX), which can assume two different structures with different functions. The two structures assemble during transcription and act collaboratively to inhibit the expression of GluR2, one of the glutamate ion channel receptor subunits.(46) The results highlight the adaptive nature of native RNA molecules to assume alternative stable structures with different functions.
Nuclear magnetic resonance (NMR) studies can elucidate the 3D structure of RNA motifs based on sequence. Recently, the NMR structure of a piece of the pRNA component (E-loop) was reported.(47) Thomas Leeper’s group (University of Akron) also presented solution NMR analysis on the Ku motif derived from the telomerase RNA component, which is a highly flexible scaffold that recruits multiple protein components.(48) Such architectural motifs can potentially be used for designing multifunctional RNA nanoparticles.
Importing from DNA Nanoarchitectures
Since DNA and RNA share some common structural and chemical features, a special seminar on the well-established principles of DNA nanotechnology (for recent reviews, see refs (1−3)) was arranged to provide viable models for the developing RNA nanotechnology community. DNA self-assembly is programmable and complex nanostructures and patterns can be created with full addressability. DNA-directed self-assembly is an efficient and precise way to organize nanomaterials on a molecular level, as demonstrated elegantly by branched DNA tiles, tensegrity triangles (rigid structures in perioidic array form),(49) algorithmic self-assembled Sierpinski triangles (aperiodic arrays of fractal patterns),(50) nanotubes, helix bundles,(51) polycatenated DNA ladders,(52) and 3D cubes, polyhedrons, prisms, and buckyballs.3,53
A striking demonstration of the addressable and programmable properties of DNA is Paul Rothemund’s DNA origami.(54) More recently, Yan Liu’s group (Arizona State University) in collaboration with Hao Yan’s group (Arizona State University) was able to use DNA to build a reconfigurable Möbius strip (topological surface with only one side and only one boundary) based on DNA fold-and-cut methodology (DNA kirigami).(55) Catenane (interlocking ring-like structures) was assembled efficiently without enzymatic ligation. In other work, the two groups in collaboration with William Shih’s group (Harvard University) were able to design and construct multilayered, 3D DNA nanostructures (DNA helices packed on square-lattice geometries) using the scaffolded-DNA-origami strategy process.(56) Their goal is to utilize programmable, 3D DNA nanoassemblies to direct the assembly of materials, such as proteins arrays, as a tool in proteomics, crystallography, and bioagent screening processes.
Although the folding properties of RNA and DNA differ to some degree, the fundamental principles in DNA nanotechnology are applicable to RNA nanotechnology. The use of three-way junctions (3WJ) and four-way junctions (4WJ)13,37 to build novel and diverse RNA architectures is very similar to the branching approaches in DNA.1,3 Both RNA and DNA polymers can be developed into bundles32,45,57 and jigsaw puzzles.36,58
RNA Computation and Modeling
Unlike DNA, RNA molecules display diverse structures mediated by both canonical and noncanonical base pairing and further stabilized by tertiary interactions, pseudoknots, kissing loops, stem stacking, etc. The use of computational methodologies can significantly lessen the time and expense required to bring the design of RNA-based nanoconstructs to experimental fruition. However, prediction of RNA structure or folding for particle assembly remains a great challenge, due to the unusual folding properties involving noncanonical interactions. Single base modifications can result in folding alterations and functional loss. Currently, using the RNA two-dimensional (2D) prediction program by Zuker, typically only 70% of the 2D folding prediction is accurate based on experimental data.59,60 Clearly, predicting the RNA 3D and 4D structures is even more elusive. Over the past decade, several groups have developed computational algorithms to predict the secondary and tertiary structures of RNA.
Christoph Flamm (Universitaet Wien, Austria) presented theoretical background based on the graph-coloring problem for the design of RNA switches and an approach that enables analysis of the kinetic behavior of the RNA folding energy landscapes. The main focus of the presentation was a computational approach for the design of RNA sequences that can fold into two or more metastable secondary structures that can further be induced to fold into one of the designed conformations based on an external trigger (e.g., temperature).61,62
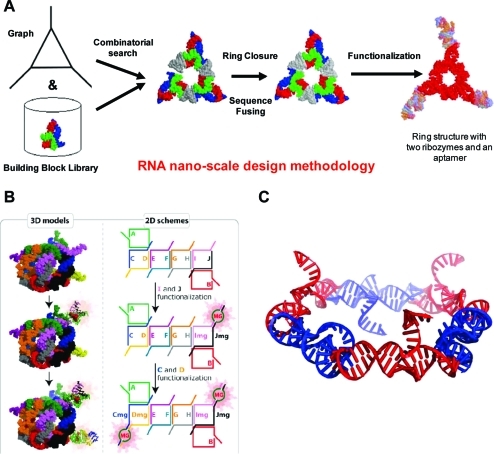
Computational strategies that permit the design of RNA nanoarchitectures were detailed by Bruce Shapiro (National Cancer Institute). He discussed novel strategies that predict nucleotide sequences that can self-assemble into 3D RNA nanocomplexes (also see http://www-ccrnp.ncifcrf.gov/∼bshapiro/).34,38 Dr. Shapiro presented elements of a computational RNA nanodesign pipeline,63,64 discussing the RNAJunction Database,(19) NanoTiler,(65) RNA2D3D algorithms,(66) and RNA Dynamics,67,68 which are used to build RNA nanoconstructs that incorporate individual RNA motifs adhering to defined user specifications (3).(69) Several of the designed constructs, such as nanocubes and hexagonal nanorings (3), have been shown to self-assemble in vitro and have therapeutic potentials.34,38
Figure 3.
(A) Representation of part of the computational RNA nanodesign pipeline associated with NanoTiler.63−65 What is depicted is one result, of many, which produces a closed-ring structure generated with the NanoTiler software that was derived in a totally automated fashion from a combinatorial search of motifs found in the RNAJunction database.(19) Sequences that were predicted for the shown structure were ultimately shown to be able to experimentally self-assemble into the depicted form. Panel A adapted with permission from ref (65). Copyright 2008 Elsevier. (B) Depiction of 10 stranded RNA cubes that were computer-designed with NanoTiler and later shown to be able to self-assemble.38,65 The layouts of the interacting strands are shown on the right. It should also be noted that the 10 stranded cubes, as shown here, were assembled in three forms: without a malachite green aptamer, with one malachite green aptamer, and with two malachite green aptamers. A properly formed aptamer fluoresces in the presence of a triarylmethane dye. The formation of the aptamers was used to confirm the cubes’ self-assembly and designed functionalization. Panel B adapted with permission from ref (38). Copyright 2011 Nature Publishing Group. (C) Depiction of the computer-designed, and later shown to be able to self-assemble, RNA hexagonal ring.34,68,69 Each corner of the ring contains the motif derived from the RNAIi/RNAIIi kissing loop interaction that forms an angle of about 120°, which is conducive to hexagonal ring formation. Sides and dangling ends can be further functionalized. Panel C adapted from ref (69). Copyright 2011 American Chemical Society.
A computational coarse-grained approach for predicting RNA 3D structure was presented by Nikolay Dokholyan (University of North Carolina). His group was able to predict the native folding of large RNA molecules by combining experimental and computational approaches70,71 and also predicted the folding of small RNAs using discrete coarse-grained molecular dynamics (DMD).72,73
Irina Novokova from Neocles Leontis’s group (Bowling Green State University) described a combined experimental and modeling approach for the assembly of tecto-RNAs using pairs of GNRA loop/loop-receptor interaction motifs. Closed programmable assembled complexes were shown to form dimers, trimers, and tetramers as well as open forms. Experimental evidence supports the formation of these complexes with high resistance to nucleases.(74) Additional functionalities can be incorporated in the helical regions, thus rendering these nanocomplexes as potential therapeutic delivery agents.
Rhiju Das (Stanford University) made a case for a novel algorithm for high-resolution 3D structure modeling of RNA. He presented a full-atom refinement framework for predicting and designing noncanonical RNA motifs that stabilize the overall conformation via tertiary interactions.(75) He further presented an alternative RNA modeling method, StepWise Ansatz (SWA), that can identify noncanonical RNA motifs and has the potential to characterize conformation landscapes of this complex biomolecule.
A novel computational method was developed by Svetlana Shabalina’s group (National Institutes of Health, Bethesda, USA) addressing a significant challenge in the RNAi field, namely, the prediction of efficient oligonucleotides for RNA interference. The algorithm can design and validate siRNA and shRNA associated with high silencing efficiency, and targeted specificity can be achieved by optimizing parameters such as terminal duplex asymmetry and duplex stability.(76) This novel approach considers the predicted local secondary structure of the target and antisense RNAs, and it is powerful enough for genome-wide si/shRNA screening. The program and numerous RNAi experiments with various siRNA and shRNA constructs are publicly available at ftp://ftp.ncbi.nlm.nih.gov/pub/shabalin/siRNA/si_shRNA_selector/.
Finally, protein–RNA interactions are common in protein synthesis, binding transfer RNA (tRNA), and ribosomal RNA (rRNA), as well as binding messenger RNA (mRNA) and small nuclear RNA (snRNA), which are involved in RNA modification. Jarek Meller (University of Cincinnati) discussed the potential of sequence-based solvent accessibility prediction for mapping RNA interaction sites in proteins.
Biophysical and Single-Molecule Approaches in RNA Nanotechnology
RNA as an intermediary molecule in the complex cascade of gene expression manifests diverse arrays of structures and participates in numerous tertiary interactions. To synthesize functionally relevant and deliverable RNA nanoparticles, an understanding of how RNA structures are formed and stabilized via higher order secondary structure and tertiary interactions is of paramount importance.
Single-molecule imaging of RNA molecules is a powerful method that can significantly contribute to better understanding of RNA structures, their folding properties, and associated interactions within the quaternary complexes. Peixuan Guo (University of Cincinnati) demonstrated the utility of single-molecule fluorescence resonance energy transfer (smFRET) in understanding pRNA-based nanostructures. His group was able to predict conformational changes of pRNA upon binding to the phage procapsid.(77) Furthermore, the application of single-molecule high-resolution imaging with photobleaching (SHRImp)(78) in distance measurements of two pRNAs bound to procapsid was also discussed.
Elvin Aleman from David Rueda’s lab (Wayne State University) discussed the use of 2-aminopurine (2AP), a fluorescent nucleotide, to study local conformational changes using the single-molecule approach. They developed a click-chemistry-based surface immobilization approach(79) to study 2AP fluorescence dynamics when it is incorporated in a ssDNA and a dsDNA. This method can also be applied to RNA.
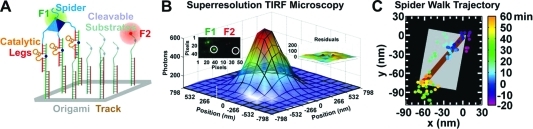
Advances in single-particle tracking will serve as a promising tool in DNA and RNA nanotechnology. Nils Walter’s group (University of Michigan), in collaboration with Eric Winfree’s (Caltech), Hao Yan’s (Arizona State University), and Milan Stojanovic’s (Columbia University) laboratories, developed experimental approaches to study the molecular motion of nucleic-acid-based nanoscale molecular assemblies, called “spiders”, using real-time single-particle fluorescence microscopy.80,81 They were able to observe directional motion of the molecular robot along a 2D DNA origami substrate (4).
Figure 4.
Watching the movement of single-molecular spiders at super-resolution (Nils Walter group). A multi-legged molecular nanoassembly walks along a prescribed substrate track on a DNA origami (A), where its fluorophore label (F1) is tracked at super-resolution relative to a label on the end of the track (F2) by total internal reflection fluorescence (TIRF) microscopy (B).80,81 The resulting trajectory compares well with a scale-drawn schematic of the origami (C). Figure modified with permission from ref (81). Copyright 2010 Elsevier.
Mass spectrometry (MS)-based approaches are an elegant way to characterize oligonucleotides, RNA-mediated interactions, and effects of common ligands in these interactions. Patrick Limbach’s group (University of Cincinnati) is applying high-resolution, high-mass-accuracy MS to characterize oligonucleotides and RNA-based nanoparticle building blocks as a means for diagnostic applications and as a drug development platform.82,83 Daniele Fabris’s group (State University of New York, Albany) is also applying MS-based approaches to decipher RNA structures and investigate tertiary/quaternary interactions and modulatory effects of small ligands as potential therapeutic targets.(84)
Scott Tenenbaum’s group (State University of New York, Albany) is pioneering an RNA-based nanoswitch technology, called “structurally interacting RNAs” (sxRNA) for real-time detection of specific microRNAs (miRNA) in cells; this technology has tremendous potential to be developed as a method in diagnostics and therapeutics.
Meni Wanunu in Marija Drndic’s group (University of Pennsylvania), in collaboration with New England Biolabs, presented an electronic-detection platform using synthetic nanopores for detecting and quantifying miRNA enriched from biological tissue.(85) This single-molecule approach provides an attractive low-cost alternative and, combined with ultralow sample volumes (nL), has the potential to exceed the detection limits of conventional methods of microarray-based miRNA profiling, fluorescence, and radioactive gel electrophoresis assays.
Nicolas Spinelli (Université Joseph Fourier, France) presented an approach to direct the assembly of oligonucleotides into G-quadruplex architectures (four-stranded structure of stacked guanine tetrads) using a peptide scaffold. Surface plasma resonance (SPR) was then used to evaluate the affinity, selectivity, and binding mode of various ligands with the immobilized G-quadruplex complexes of various topologies,(86) as a means for screening viable drug targets.(87)
Blaine Moore’s group (University of Oklahoma Health Sciences Center) is working toward determining the crystal structure of a fragment of guide RNAs (gRNA) that are involved in post-transcriptional editing of mRNA (uridine insertion/deletion), a requirement for the subsequent expression of several mitochondrial proteins. The results provide valuable insight on the conformation and dynamics of the U-helix, which is common among all gRNA/mRNA duplexes and is a valuable drug target.(88)
RNA Nanoparticle Assembly
Self-assembly of RNA building blocks in a predefined manner to form larger multidimensional structures is a prominent bottom-up approach and represents an important means by which biological techniques and biomacromolecules can be successfully integrated into nanotechnology.32,89,90 Within the realm of self-assembly, there are two main subcategories: templated and nontemplated assembly. Templated assembly involves the interaction of RNAs with one another under the influence of a specific external force, structure, or spatial constraint. RNA transcription, hybridization, replication, molding, and phi29 pRNA hexameric ring formation are within this category. Nontemplated assembly involves the formation of a larger structure by individual components without any external influence. Examples include ligation, chemical conjugation, covalent linkages, loop/loop interactions of RNA such as the HIV kissing loop, and phi29 pRNA dimer or trimer formation.25,32,33,89,90
Natural RNA molecules form unique and intriguing multimers with special functionalities. Examples include retroviral kissing loops that facilitate dimerization,91,92 pRNA of the bacteriophage phi29 DNA packaging motor that assembles into dimers and hexamers via hand-in-hand interactions between right- and left-interlocking loops,(21) and bicoid mRNA of Drosophila embryos that form dimers via hand-in-arm interactions.(93) In addition, numerous RNA molecules contain motifs with fixed structure and tightly folded domains. These natural properties have been utilized to assemble varieties of RNA nanoparticles with diverse biological properties and wide-ranging functions.
Peixuan Guo’s group (University of Cincinnati) reported the construction of chemically modified stable pRNA-based nanoparticles that were RNase-resistant, while retaining their biological activity in procapsid binding and gearing the phi29 DNA packaging motor.(24) He further demonstrated the scale-up synthesis of stable RNA nanoparticles greater than 100 nt,(94) which overcomes the limitation of currently available commercial synthesis of RNA oligonulceotides (up to 80 nt with low yield). His group generated bipartite chimeric pRNA constructs that can assemble into the full-length functional pRNA, while retaining their biological activity and proficiency for incorporating therapeutic and diagnostic modules.
RNA aptamers are a class of oligonucleotides that can recognize specific ligands through the formation of binding pockets.95−97 SELEX is typically used to screen for the aptamers from randomized RNA pools.95,96,98 Chaoping Chen (Colorado State University) has been developing innovative approaches to tackle Flaviviral infections including Dengue, West Nile, and Yellow Fever, for which there are no effective therapeutics currently available. Using SELEX, her group was able to identify RNA aptamers that were found to bind with RNA-capping enzymes of Flaviviruses. Using in vitro and in vivo experiments, she demonstrated the therapeutic potential of novel tRNA–aptamer fusions in treatment of viral infections. In related work, Hua Shi (State University of New York, Albany) discussed bottom-up approaches that utilize RNA aptamers to construct protein-like molecules with predetermined functions.99,100
Porphyrin macrocycles have been studied for their roles in oxygen transport in blood and photosynthesis. By virtue of porphines’ interaction with light, this class of molecules has been exploited in medicine. Magnus Bergkvist (State University of New York, Albany) presented work demonstrating the oligonucleotide-mediated packaging of photoactive porphyrin into bacteriophage MS2 capsid. His group has been working on virion-derived nanocontainers that are capable of delivering nonviral RNA/DNA for therapeutics.
RNA Nanoparticles in Therapeutics
Over the past decade, RNA-based therapeutics have been investigated extensively for treatment and detection of cancer, viral ailments, and genetic diseases. Several natural and synthetic RNA molecules are being actively pursued, including, but not limited to (1) design and construction of antisense and siRNA for silencing genes with high efficiency and specificity; (2) assembly of multivalent RNA nanoparticles with receptor-binding aptamers for targeted delivery; (3) incorporation of ribozymes for intercepting and cleaving target RNA substrates; and (4) conjugation of drugs and chemical ligands for therapy and detection.
Ningsheng Shao’s group (Beijing Institute of Basic Medical Sciences, China) is developing a wide range of SELEX techniques for identifying tumor markers as a means for early diagnosis and therapy. Subtractive cell SELEX procedure, developed in his lab in 2003, can select aptamers that can distinguish differentiated cells (e.g., carcinoma cells) from undifferentiated epithelial cells.(101) A similar strategy, using whole-bacterium-based SELEX, can select ssDNA aptamers for detection of pathogens, such as the Staphylococcus aureus.(102) The group has also developed an in situ tissue-slide-based SELEX method that can select for aptamers specific to all tissue fractions (such as extracellular matrix, membrane components, and intracellular targets) and can distinguish cancer cells from normal cells in clinical specimens.(103)
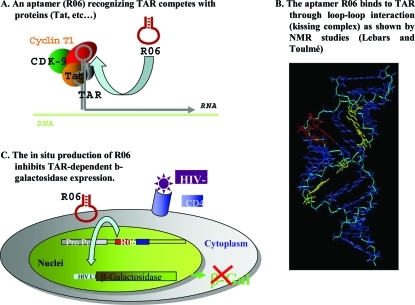
Jean-Jacques Toulmé (Université de Bordeaux, France) presented work on a novel aptamer identified via SELEX that folds into a hairpin structure and can target viral RNA (HIV and HCV) structural elements by forming stable kissing complexes.(104) Such a kissing aptamer targeted to the trans-activating responsive (TAR) element of HIV-1 displayed both high affinity and exquisite specificity (5). RNA aptamers produced in situ in HeLaP4 cells from an expression vector inhibit TAR-dependent expression of a reporter gene. Chemically modified aptamers also reduce TAR-dependent expression upon transfection of recipient cells.
Figure 5.
Complex between the trans-activating responsive RNA element TAR, viral (Tat), and cell proteins (cyclin T1, CDK-9) is required for efficient transcription of the HIV-1 genome. Disruption of this complex leads to abortive transcription of the retroviral DNA. A high-affinity RNA hairpin aptamer (R06) specifically recognizes TAR. The transcription of a R06 construct in recipient cells reduces the production of β-galactosidase whose expression is driven by the HIV-1 promoter containing TAR, in response to retroviral infection. Therefore, targeting RNA hairpins offers an alternative to targeting proteins for the design of artificial modulators of gene expression.
RNA interference (RNAi)-based therapeutics using siRNA have the potential to silence disease-causing genes. However, significant challenges remain, especially with regard to in vivo delivery of siRNA. Muthiah Manoharan (Alnylam Pharmaceutical, USA) delineated the prospects of novel lipid-like nanoparticles (LNP) mediated in vivo delivery of RNAi therapeutics.(105) He also discussed the potential of chemically modified RNA and various siRNA-based therapeutics currently in clinical trials or under development. Likewise, Weikang Tao (Merck Research Laboratories, USA) presented current work on in vivo siRNA therapeutics, their potential toxicities, and therapeutic efficacies. He provided guidance on how to optimize the development of LNP-formulated siRNA for therapeutics and ways to minimize immuno-stimulatory activities for improving their safety profiles.(106)
Thomas Hermann’s (University of California, San Diego) group has characterized a new RNA drug target in the hepatitis C virus’s internal ribosome entry site (IRES) and established a conformational mechanism of action for a small molecule inhibitor that binds to this RNA.(107) He was able to obtain the crystal structure at 2.2 Å resolution of a square-shaped, double-stranded RNA construct derived from HCV IRES domain. He also presented a novel supramolecular approach for cellular delivery of siRNAs using a specially designed β-cyclodextrin peptide conjugate, which acts as a ligand for complex formation with the RNA (RGO Biosciences LLC).
Use of multimerized siRNA conjugate complexes for cell delivery and gene silencing was presented by Tae Gwan Park (Korea Advanced Institute of Science and Technology, South Korea). He reviewed the use of charged cationic polymers, lipids, and peptides for intracellular delivery of siRNAs. He also presented work from his laboratory focused on the development and delivery of self-cross-linked and multimerized siRNAs via cleavable chemical linkages. Efficacy of these multimerized siRNAs in gene silencing without invoking immune induction was presented.(108)
Dong-Ki Lee’s group (Sungkyunkwan University, South Korea) presented promising new results on the development of a branched RNA duplex structure that has the potential to target and to silence multiple genes implicated in cancer cell proliferation and survival.(109)
Henry Li (Kylin Therapeutics, USA) presented compelling evidence on the therapeutic prospects of pRNA-based RNA nanoparticles. pRNA nanoparticles have a defined structure (modular design that enables multiple functionalities), are thermodynamically and metabolically stable, have an optimum diameter of 10–50 nm, and display superior in vivo pharmacological profiles.(110) Malak Kotb’s group (University of Cincinnati) is using pRNA as a scaffold for constructing chimeric pRNA-siRNA-CD4 aptamer nanoparticles to block the growth of lymphocytic leukemia and lymphoma cells without affecting the survival of normal lymphocytes.
John Rossi (Beckman Research Institute, USA) presented an elegant study in which three endogenous targets were silenced by siRNA cocktails in both in vitro cell cultures and in vivo animal models. This approach utilized a combination of siRNAs targeting expression of host and viral genes. Dr. Rossi’s research demonstrates that siRNAs are able to inhibit HIV replication in a humanized HSC-CD34 mouse model and prevent HIV-mediated T-cell depletion characteristic of AIDS.111,112 Mei Zhang (Case Western Reserve University, USA) also presented the efficacy of siRNA-mediated inhibition of mammary tumor growth and metastasis in a 4T1 mouse model.
Numerous proteins are associated with innate immunity (initial immune response to pathogens), including RNA-activated protein kinase (PKR). These classes of proteins have the ability to discriminate self-RNA from foreign, pathogenic RNA via pathogen-associated molecular patterns.(113) Philip Bevilacqua’s group (Pennsylvania State University) is developing mRNA-based therapeutics by designing nucleoside modifications for regulating the activity of PKR.(114)
RNA Chemistry for Nanoparticle Synthesis, Conjugation, and Labeling
For the field of RNA nanotechnology and therapeutics, it is vital to develop efficient methods for (1) preparing large amounts of pure RNA oligonucleotide sequences and (2) incorporating a large diversity of conjugates and labels for functionalizing and probing the RNA construct at specific positions. Fundamentally, RNA oligonucleotides can be prepared by either enzymatic transcription reactions or automated solid-phase synthesis. Enzymatic approaches can provide access to relatively long transcripts in significant quantities; however, their outcome is frequently sequence-dependent. The heterogeneity of the 3′-end has been an issue,(115) which can be addressed by extending the transcribed sequence beyond the intended end and then cleaving the RNA at the desired site using ribozymes, DNAzymes, or RNase H.115−117 Transcription reactions are also limited in their ability to furnish labeled or sequence specifically modified RNA transcripts. To meet this challenge, GMP or AMP derivatives, which can only be used for transcription initiation but not for chain elongation, have been used. Fluorescent RNA can also be easily synthesized in vitro with T7 RNA polymerase using a new agent tCTP.(118) To generate longer RNA, two short, synthetic RNA fragments can be further ligated using RNase ligase II or T4 DNA ligases.(116)
Standard solid-phase phosphoramidite chemistry for the synthesis of DNA and RNA oligonucleotides has advanced significantly in recent years, particularly when it comes to the synthesis of relatively long RNA oligonucleotides.119,120 In addition to “classical” approaches relying on the tert-butyldimethylsiloxy (TBDMS) protecting group for the 2′-hydroxyl, which were somewhat limited in their ability to provide rather long sequences, two “modern” approaches have been introduced for RNA synthesis: (1) the 5′-O-DMT-2′-O-[(trisisopropylsilyl)oxy]methyl (2′-O-TOM) protecting scheme developed by Pitsch,(121) and (2) the 5′-O-silyl-2′-O-orthoester (2′-ACE) protecting group combination, introduced by Scaringe and Caruthers.(122) These protection schemes facilitate the synthesis of labeled and modified RNA oligonucleotides for diverse applications. However, challenges remain when it comes to the preparation and incorporation of novel nucleosides, linkers, and labels, as their compatibility with current synthetic and protection schemes needs to be empirically determined.
Natural RNA is sensitive to RNase and is especially unstable in serum. The relative instability of RNA has long hindered its application as a construction material. Improvements in RNA stability have progressed rapidly, including (1) chemical modification of the base (e.g., 5-Br-Ura and 5-I-Ura); (2) modification of the phosphate linkages (e.g., phosphothioate, boranophosphate); (3) C2′ (e.g., 2′-fluorine, 2′-O-methyl or 2′-amine);(123) (4) peptide nucleic acids (PNA), locked nucleic acids (LNA) and derivatives;124,125 (5) 3′-end-capping;(126) and use of cross-linking agents (psoralen, nitrogen mustard derivatives, and transition metal compounds).(127) Adam Mazur (Girindus America Inc., USA) discussed the commercial availability of a wide variety of these chemically modified RNAs and their advantages in RNA-based therapeutics. He also discussed the challenges in manufacturing processes, from optimizing small-scale sequence-directed synthesis to purification schemes and large-scale production of the oligonucleotides.
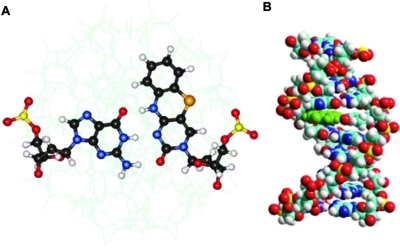
In probing and monitoring the structure and structural changes of RNA nanoconstructs and therapeutics, the use of external and/or internal fluorescence reporter molecules (6) is frequently central. Fluorescent labels are also important for detecting and determining the strength of the interaction between RNA and, for example, other macromolecules and ligands. There is continuous development of this group of reporter molecules and a rapidly growing subgroup is the fluorescent nucleic acid base analogues. Marcus Wilhelmsson (Chalmers University of Technology, Sweden) discussed the advantage of fluorescent analogues of the tricyclic cytosine family128,129 in calculating accurate distances and orientation changes in nucleic acid systems using FRET.(130) Since nucleic acids are practically nonfluorescent in nature, artificial fluorescent base analogues are important for investigating DNA or RNA systems. Fluorescent base analogues are morphologically similar to natural nucleobases and have the ability to form hydrogen bonds with a natural nucleobase in the complementary strand. Furthermore, these artificial bases retain the structure of DNA (or RNA) and can be incorporated internally and therefore have an advantage over covalently attached end-labeled dyes (fluorescein, rhodamines, Cy or Alexa dyes).
Figure 6.
(A) Internal nucleic acid reporter group, represented here by the fluorescent base analogue tC in pair with guanine looking down the long axis of a DNA duplex. (B) Same internal reporter group looking along the short axis of a DNA duplex.
Yitzhak Tor’s group (University of California, San Diego) is working on RNA folding and RNA drug binding. He discussed the design, synthesis, and incorporation of novel fluorescent nucleobase analogues and described the advantages in developing fluorescence-based discovery and detection systems.131−133 For different classes of fluorophores, their characteristic profiles, and applications, see recent reviews.134,135
Funding Opportunities in Nanotechnology for Therapeutics
Sara Hook (National Cancer Institute, USA) presented the tremendous resources and infrastructure available through the Alliance for Nanotechnology in Cancer established by the National Cancer Institute (NCI) to support cancer-relevant nanotechnology endeavors. She described how the alliance supports interdisciplinary research efforts between biologists, clinicians, and physical scientists with the goal of advancing prevention, diagnosis, and treatment efforts. The NCI alliance will continue to support collaborations by providing infrastructure as well as research and training grants for establishing a diverse portfolio of new technologies supporting a range of applications. In addition, several centers of cancer nanotechnology across the United States will be established. Detailed information regarding the NCI supported Alliance for Nanotechnology in Cancer and other funding opportunities is available at the NCI Web site (http://nano.cancer.gov). A fully equipped Nanotechnology Characterization Laboratory (http://ncl.cancer.gov) is also available for characterizing potential therapeutic nanoparticles, such as their physical attributes (e.g., size, shape, surface chemistry, and solubility), in vitro biological properties (e.g., pharmacology and cytotoxicity), and in vivo compatibility assays (e.g., efficacy and safety). At present, the challenging areas include molecular imaging and early detection (high spatial and temporal resolution, as well as sensitivity), in vivo nanotechnology imaging systems (minimal or noninvasive methods), and multifunctional therapeutics, especially with regard to cancers with low survival rates such as brain, lung, pancreas, and ovarian cancers.
Future Outlook
Significant challenges remain with regard to delivery of RNA-based nanoparticles. From a commercial perspective, the manufacturing process can be complex and inconsistent. The production/synthesis of oligonucleotides with functional moieties has to be a scalable process (chemically). The RNA nanoscaffold has to retain its folding properties and display a stable nature. The RNA construct needs to have a modular design, such that the complex can be self-assembled from the basic building blocks. Moreover, the cost of manufacturing and associated analytical characterizations of nanoparticles needs to be reasonable.
From a structural point of view, there are concerns over the stoichiometry and particle size, robustness of functional modules, and stability (thermodynamic, chemical, and metabolic) of the RNA nanoparticles. Effective computational approaches accounting for noncanonical base pairing, long-range tertiary interactions, and equilibrium folding of the final complex are in great demand for the design and construction of RNA nanoparticles (using interacting, architectural, and ligand-binding motifs) with functional modalities (therapeutic, targeting, and diagnostic). The flexibility of RNA molecules and unpredictability of RNA folding presents a challenge for simulation and modeling.
From an in vivo delivery point of view, the nanoparticles need to display favorable pharmacological profiles, biodistribution, pharmokinetic profiles, minimum immunogenicity, relatively high efficiency of disease tissue/cell targeting, and remain intact at extremely low concentrations. Furthermore, the RNAi-based therapeutics need to be internalized by the target cell and escape from the endosomes if they enter via receptor-mediated endocytosis. Endosomal escape for release of the therapeutic nanoparticles remains a challenge in the field.
As outlined in this Nano Focus article, several groups have made significant strides with regard to aptamer- and dendrimer-based delivery approaches to therapeutic RNAs. Combinatorial therapy using aptamers and siRNA has proved useful for achieving maximal efficiency and efficacy in targeting. In addition, structural and functional aspects of RNA nanoparticles have made rapid progress with respect to their design and delivery. Finally, chemical labeling of RNA using fluorescent analogues has progressed remarkably. The meeting highlighted current research and future prospects of RNA nanotechnology. Applications and the impact of RNA nanotechnology in the assembly and delivery of siRNAs, pRNA, therapeutic ribozymes, RNA aptamers, and riboswitches are becoming a reality. RNA nanotechnology is already making a significant impact in drug delivery and therapeutics as many siRNA-based products are in the pipeline for potential treatment of respiratory synchitial viral infections, hepatitis C virus, liver cancer, and Huntington’s Disease. Collaborative endeavors between academia, government, and industry are likely necessary to advance RNA nanotechnology into a practical tool for drug discovery and delivery in the near future.
Acknowledgments
We thank the participants and sponsors of the meeting. We also wish to thank Thomas Hermann, Mark Laskovics, Christoph Flamm, Chad Schwartz, Randall Reif, and Daniel Binzel for their insightful comments. We apologize to participants whose work could not be reviewed in this article due to space limitations. The meeting was supported and sponsored by funding or donations in part from Kylin Therapeutics, Inc., Girindus, Trilink BioTechnologies, Agilent Technologies, NIH Nanomedicine Development Center of Phi29 DNA Packaging Motor for Nanomedicine (PN2 EY 018230), NIH grants EB003730, GM059944, CA151648, and funding from the laboratories of the invited speakers and the co-authors. P.G. is a co-founder and contractor of Kylin. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Funding Statement
National Institutes of Health, United States
References
- Lin C.; Liu Y.; Yan H. Designer DNA Nanoarchitectures. Biochemistry 2009, 48, 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaye F. A.; Palmer A. L.; Sleiman H. F. Assembling Materials with DNA as the Guide. Science 2008, 321, 1795–1799. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. Nanomaterials Based on DNA. Annu. Rev. Biochem. 2010, 79, 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. On Finding All Suboptimal Foldings of an RNA Molecule. Science 1989, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- Pleij C. W.; Bosch L. RNA Pseudoknots: Structure, Detection, and Prediction. Methods Enzymol. 1989, 180, 289–303. [DOI] [PubMed] [Google Scholar]
- Guo P. RNA Nanotechnology: Engineering, Assembly and Applications in Detection, Gene Delivery and Therapy. J. Nanosci. Nanotechnol. 2005, 5, 1964–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isambert H. The Jerky and Knotty Dynamics of RNA. Methods 2009, 49, 189–196. [DOI] [PubMed] [Google Scholar]
- Sugimoto N.; Nakano S.; Katoh M.; Matsumura A.; Nakamuta H.; Ohmichi T.; Yoneyama M.; Sasaki M. Thermodynamic Parameters To Predict Stability of RNA/DNA Hybrid Duplexes. Biochemistry 1995, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- Searle M. S.; Williams D. H. On the Stability of Nucleic Acid Structures in Solution: Enthalpy–Entropy Compensations, Internal Rotations and Reversibility. Nucleic Acids Res. 1993, 21, 2051–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa Y.; Tsuda K.; Matsumura S.; Inoue T. De Novo Synthesis and Development of an RNA Enzyme. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 13750–13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S.; Ohmori R.; Saito H.; Ikawa Y.; Inoue T. Coordinated Control of a Designed Trans-Acting Ligase Ribozyme by a Loop-Receptor Interaction. FEBS Lett. 2009, 583, 2819–2826. [DOI] [PubMed] [Google Scholar]
- Leontis N. B.; Lescoute A.; Westhof E. The Building Blocks and Motifs of RNA Architecture. Curr. Opin. Struct. Biol. 2006, 16, 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder K. T.; McPhee S. A.; Ouellet J.; Lilley D. M. A Structural Database for k-Turn Motifs in RNA. RNA 2010, 16, 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Horiya S.; Harada K. An Efficient Thermally Induced RNA Conformational Switch as a Framework for the Functionalization of RNA Nanostructures. J. Am. Chem. Soc. 2006, 128, 4035–4040. [DOI] [PubMed] [Google Scholar]
- Laurenti E.; Barde I.; Verp S.; Offner S.; Wilson A.; Quenneville S.; Wiznerowicz M.; MacDonald H. R.; Trono D.; Trumpp A. Inducible Gene and shRNA Expression in Resident Hematopoietic Stem Cells In Vivo. Stem Cells 2010, 28, 1390–1398. [DOI] [PubMed] [Google Scholar]
- Hoeprich S.; Zhou Q.; Guo S.; Qi G.; Wang Y.; Guo P. Bacterial Virus Phi29 pRNA as a Hammerhead Ribozyme Escort To Destroy Hepatitis B Virus. Gene Ther. 2003, 10, 1258–1267. [DOI] [PubMed] [Google Scholar]
- Chang K. Y.; Tinoco I. Jr. Characterization of a “Kissing” Hairpin Complex Derived from the Human Immunodeficiency Virus Genome. Proc. Natl. Acad. Sci. U.S.A. 1994, 91, 8705–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindewald E.; Hayes R.; Yingling Y. G.; Kasprzak W.; Shapiro B. A. RNAJunction: A Database of RNA Junctions and Kissing Loops for Three-Dimensional Structural Analysis and Nanodesign. Nucleic Acids Res. 2008, 36, D392–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C.; Ehresmann C.; Ehresmann B.; Brunel C. Mechanism of Dimerization of Bicoid mRNA: Initiation and Stabilization. J. Biol. Chem. 2004, 279, 4560–4569. [DOI] [PubMed] [Google Scholar]
- Chen C.; Sheng S.; Shao Z.; Guo P. A Dimer as a Building Block in Assembling RNA: A Hexamer That Gears Bacterial Virus Phi29 DNA-Translocating Machinery. J. Biol. Chem. 2000, 275, 17510–17516. [DOI] [PubMed] [Google Scholar]
- Guo P.; Zhang C.; Chen C.; Trottier M.; Garver K. Inter-RNA Interaction of Phage Phi29 pRNA To Form a Hexameric Complex for Viral DNA Transportation. Mol. Cell 1998, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Lemieux S.; Wu X.; St.-Arnaud S.; McMurray C. T.; Major F.; Anderson D. Function of Hexameric RNA in Packaging of Bacteriophage Phi29 DNA In Vitro. Mol. Cell 1998, 2, 141–147. [DOI] [PubMed] [Google Scholar]
- Liu J.; Guo S.; Cinier M.; Shlyakhtenko L.; Shu Y.; Chen C.; Shen G.; Guo P. Fabrication of Stable and RNase-Resistant RNA Nanoparticles Active in Gearing the Nanomotors for Viral DNA Packaging. ACS Nano 2011, 5, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L.; Leontis N. B. Tecto-RNA: One Dimensional Self-Assembly through Tertiary Interactions. Angew. Chem., Int. Ed. 2000, 39, 2521–2524. [DOI] [PubMed] [Google Scholar]
- Jaeger L.; Westhof E.; Leontis N. B. TectoRNA: Modular Assembly Units for the Construction of RNA Nano-Objects. Nucleic Acids Res. 2001, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B.; Westhof E. Analysis of RNA Motifs. Curr. Opin. Struct. Biol. 2003, 13, 300–308. [DOI] [PubMed] [Google Scholar]
- Jaeger L; Chworos A. The Architectonics of Programmable RNA and DNA Nanostructures. Curr. Opin. Struct. Biol. 2006, 16, 531–543. [DOI] [PubMed] [Google Scholar]
- Guo P.; Erickson S.; Anderson D. A Small Viral RNA Is Required for In Vitro Packaging of Bacteriophage Phi29 DNA. Science 1987, 236, 690–694. [DOI] [PubMed] [Google Scholar]
- Xiao F.; Demeler B.; Guo P. Assembly Mechanism of the Sixty-Subunit Nanoparticles via Interaction of RNA with the Reengineered Protein Connector of Phi29 DNA-Packaging Motor. ACS Nano 2010, 4, 3293–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Zhang H.; Jin J.; Guo P. Counting of Six pRNAs of Phi29 DNA-Packaging Motor with Customized Single Molecule Dual-View System. EMBO J. 2007, 26, 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Moll W. D.; Deng Z.; Mao C.; Guo P. Bottom-Up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology. Nano Lett. 2004, 4, 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Huang L.; Hoeprich S.; Guo P. Construction of Phi29 DNA-Packaging RNA (pRNA) Monomers, Dimers and Trimers with Variable Sizes and Shapes as Potential Parts for Nano-Devices. J. Nanosci. Nanotechnol. 2003, 3, 295–302. [DOI] [PubMed] [Google Scholar]
- Grabow W. W.; Zakrevsky P.; Afonin K. A.; Chworos A.; Shapiro B. A.; Jaeger L. Self-Assembling RNA Nanorings Based on RNAI/II Inverse Kissing Complexes. Nano Lett. 2011, 11, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L. Defining the Syntax for Self-Assembling RNA Tertiary Architectures. Nucleic Acids Symp. Ser. 2009, 53, 83–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chworos A.; Severcan I.; Koyfman A. Y.; Weinkam P.; Oroudjev E.; Hansma H. G.; Jaeger L. Building Programmable Jigsaw Puzzles with RNA. Science 2004, 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- Severcan I; Geary C; Verzemnieks E.; Arkadiusz A.; Jaeger L. Square-Shaped RNA Particles from Different RNA Folds. Nano Lett. 2009, 9, 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin K. A.; Bindewald E.; Yaghoubian A. J.; Voss N.; Jacovetty E.; Shapiro B. A.; Jaeger L. In Vitro Assembly of Cubic RNA-Based Scaffolds Designed in Silico. Nat. Nanotechnol. 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severcan I.; Geary C.; Chworos A.; Voss N.; Jacovetty E.; Jaeger L. A Polyhedron Made of tRNAs. Nat. Chem. 2010, 2, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R. R. Complex Riboswitches. Science 2008, 319, 1795–1797. [DOI] [PubMed] [Google Scholar]
- Fabian M. R.; Sonenberg N.; Filipowicz W. Regulation of mRNA Translation and Stability by MicroRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [DOI] [PubMed] [Google Scholar]
- Zhang C. Novel Functions for Small RNA Molecules. Curr. Opin. Mol. Ther. 2009, 11, 641–651. [PMC free article] [PubMed] [Google Scholar]
- Marvin M. C.; Engelke D. R. Broadening the Mission of an RNA Enzyme. J. Cell Biochem. 2009, 108, 1244–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid A.; Cayrol B.; Isambert H. RNA Synthetic Biology Inspired from Bacteria: Construction of Transcription Attenuators under Antisense Regulation. Phys. Biol. 2009, 6, 025007. [DOI] [PubMed] [Google Scholar]
- Cayrol B.; Nogues C.; Dawid A.; Sagi I.; Silberzan P.; Isambert H. A Nanostructure Made of a Bacterial Noncoding RNA. J. Am. Chem. Soc. 2009, 131, 17270–17276. [DOI] [PubMed] [Google Scholar]
- Huang Z.; Pei W.; Han Y.; Jayaseelan S.; Shekhtman A.; Shi H.; Niu L. One RNA Aptamer Sequence, Two Structures: A Collaborating Pair That Inhibits AMPA Receptors. Nucleic Acids Res. 2009, 37, 4022–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.; Schroeder S. J. Nuclear Magnetic Resonance Structure of the Prohead RNA E-Loop Hairpin. Biochemistry 2010, 49, 5989–5997. [DOI] [PubMed] [Google Scholar]
- Zappulla D. C.; Cech T. R. Yeast Telomerase RNA: A Flexible Scaffold for Protein Subunits. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 10024–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.; Wang M.; Deng Z.; Walulu R.; Mao C. Tensegrity: Construction of Rigid DNA Triangles with Flexible Four-Arm DNA Junctions. J. Am. Chem. Soc. 2004, 126, 2324–2325. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W.; Papadakis N.; Winfree E. Algorithmic Self-Assembly of DNA Sierpinski Triangles. PLoS Biol. 2004, 2, e424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. H.; Barish R.; Li H.; Reif J. H.; Finkelstein G.; Yan H.; LaBean T. H. Three-Helix Bundle DNA Tiles Self-Assemble into 2D Lattice or 1D Templates for Silver Nanowires. Nano Lett. 2005, 5, 693–696. [DOI] [PubMed] [Google Scholar]
- Weizmann Y.; Braunschweig A. B.; Wilner O. I.; Cheglakov Z.; Willner I. A Polycatenated DNA Scaffold for the One-Step Assembly of Hierarchical Nanostructures. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldaye F. A.; Sleiman H. F. Modular Access to Structurally Switchable 3D Discrete DNA Assemblies. J. Am. Chem. Soc. 2007, 129, 13376–13377. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA To Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. [DOI] [PubMed] [Google Scholar]
- Han D.; Pal S.; Liu Y.; Yan H. Folding and Cutting DNA into Reconfigurable Topological Nanostructures. Nat. Nanotechnol. 2010, 5, 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y.; Douglas S. M.; Liu M.; Sharma J.; Cheng A.; Leung A.; Liu Y.; Shih W. M.; Yan H. Multilayer DNA Origami Packed on a Square Lattice. J. Am. Chem. Soc. 2009, 131, 15903–15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H.; Douglas S. M.; Shih W. M. Folding DNA into Twisted and Curved Nanoscale Shapes. Science 2009, 325, 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M.; Sugita T.; Katsuda Y.; Hidaka K.; Sugiyama H. Programmed-Assembly System Using DNA Jigsaw Pieces. Chemistry 2010, 16, 5362–5368. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham N. R.; Zuker M. UNAFold: Software for Nucleic Acid Folding and Hybridization. Methods Mol. Biol. 2008, 453, 3–31. [DOI] [PubMed] [Google Scholar]
- Hofacker I. L.; Flamm C.; Heine C.; Wolfinger M. T.; Scheuermann G.; Stadler P. F. BarMap: RNA Folding on Dynamic Energy Landscapes. RNA 2010, 16, 1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm C.; Hofacker I. L.; Maurer-Stroh S.; Stadler P. F.; Zehl M. Design of Multistable RNA Molecules. RNA 2001, 7, 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro A.; Ohtsu T.; Nakamura Y. An Aptamer-Based Biosensor for Mammalian Initiation Factor Eukaryotic Initiation Factor 4A. Anal. Biochem. 2009, 388, 102–107. [DOI] [PubMed] [Google Scholar]
- Shapiro B. A.; Bindewald E.; Kasprzak W.; Yingling Y. Protocols for the In Silico Design of RNA Nanostructures. Methods Mol. Biol. 2008, 474, 93–115. [DOI] [PubMed] [Google Scholar]
- Bindewald E.; Grunewald C.; Boyle B.; O’Connor M.; Shapiro B. A. Computational Strategies for the Automated Design of RNA Nanoscale Structures from Building Blocks Using NanoTiler. J. Mol. Graphics Modell. 2008, 27, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez H. M.; Maizel J. V.; Shapiro B. A. RNA2D3D: A Program for Generating, Viewing, and Comparing 3-Dimensional Models of RNA. J. Biomol. Struct. Dyn. 2008, 25, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzak W.; Bindewald E.; Kim T. J.; Jaeger L.; Shapiro B. A.. Use of RNA Structure Flexibility Data in Nanostructure Modeling. Methods 2010, DOI: 10.1016/j.ymeth.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliy M.; Melnik R.; Shapiro B. A. Molecular Dynamics Study of the RNA Ring Nanostructure: A Phenomenon of Self-Stabilization. Phys. Biol. 2009, 6, 046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling Y. G.; Shapiro B. A. Computational Design of an RNA Hexagonal Nanoring and an RNA Nanotube. Nano Lett. 2007, 7, 2328–2334. [DOI] [PubMed] [Google Scholar]
- Lavender C. A.; Ding F.; Dokholyan N. V.; Weeks K. M. Robust and Generic RNA Modeling Using Inferred Constraints: A Structure for the Hepatitis C Virus IRES Pseudoknot Domain. Biochemistry 2010, 49, 4931–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe C. M.; Leonard C. W.; Ding F.; Dokholyan N. V.; Weeks K. M. Native-Like RNA Tertiary Structures Using a Sequence-Encoded Cleavage Agent and Refinement by Discrete Molecular Dynamics. J. Am. Chem. Soc. 2009, 131, 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F.; Sharma S.; Chalasani P.; Demidov V. V.; Broude N. E.; Dokholyan N. V. Ab Initio RNA Folding by Discrete Molecular Dynamics: From Structure Prediction to Folding Mechanisms. RNA 2008, 14, 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.; Ding F.; Dokholyan N. V. iFoldRNA: Three-Dimensional RNA Structure Prediction and Folding. Bioinformatics 2008, 24, 1951–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova I. V.; Hassan B. H.; Mirzoyan M. G.; Leontis N. B. Engineering Cooperative Tecto-RNA Complexes Having Programmable Stoichiometries. Nucleic Acids Res. 2011, 39, 2903–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R.; Karanicolas J.; Baker D. Atomic Accuracy in Predicting and Designing Noncanonical RNA Structure. Nat. Methods 2010, 7, 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva O. V.; Kang Y.; Spiridonov A. N.; Saetrom P.; Nemtsov V. A.; Ogurtsov A. Y.; Nechipurenko Y. D.; Shabalina S. A. Optimization of Duplex Stability and Terminal Asymmetry for shRNA Design. PLoS One 2010, 5, e10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Zhang H.; Petrenko R.; Meller J.; Guo P. Dual-Channel Single-Molecule Fluorescence Resonance Energy Transfer To Establish Distance Parameters for RNA Nanoparticles. ACS Nano 2010, 4, 6843–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. P.; Ha T.; Selvin P. R. Single-Molecule High-Resolution Imaging with Photobleaching. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 6462–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman E. A.; Pedini H. S.; Rueda D. Covalent-Bond-Based Immobilization Approaches for Single-Molecule Fluorescence. ChemBioChem 2009, 10, 2862–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund K.; Manzo A. J.; Dabby N.; Michelotti N.; Johnson-Buck A.; Nangreave J.; Taylor S.; Pei R.; Stojanovic M. N.; Walter N. G.; et al. Molecular Robots Guided by Prescriptive Landscapes. Nature 2010, 465, 206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti N.; de Silva S. C.; Johnson-Buck A. E.; Manzo A. J.; Walter N. G. A Bird’s Eye View Tracking Slow Nanometer-Scale Movements of Single Molecular Nano-Assemblies. Methods Enzymol. 2010, 475, 121–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castleberry C. M.; Limbach P. A. Relative Quantitation of Transfer RNAs Using Liquid Chromatography Mass Spectrometry and Signature Digestion Products. Nucleic Acids Res. 2010, 38, e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castleberry C. M.; Lilleness K.; Baldauff R.; Limbach P. A. Minimizing 18O/16O Back-Exchange in the Relative Quantification of Ribonucleic Acids. J. Mass Spectrom. 2009, 44, 1195–1202. [DOI] [PubMed] [Google Scholar]
- Fabris D. A Role for the MS Analysis of Nucleic Acids in the Post-Genomics Age. J. Am. Soc. Mass Spectrom. 2010, 21, 1–13. [DOI] [PubMed] [Google Scholar]
- Wanunu M.; Dadosh T.; Ray V.; Jin J.; McReynolds L.; Drndic M. Rapid Electronic Detection of Probe-Specific MicroRNAs using Thin Nanopore Sensors. Nat. Nanotechnol. 2010, 5, 807–814. [DOI] [PubMed] [Google Scholar]
- Murat P.; Bonnet R.; Van der H. A.; Spinelli N.; Labbe P.; Monchaud D.; Teulade-Fichou M. P.; Dumy P.; Defrancq E. Template-Assembled Synthetic G-Quadruplex (TASQ): A Useful System for Investigating the Interactions of Ligands with Constrained Quadruplex Topologies. Chemistry 2010, 16, 6106–6114. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S.; Neidle S. G-Quadruplex Nucleic Acids as Therapeutic Targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Nosrati M.; Kashani-Sabet M. Knockdown of Telomerase RNA Using Hammerhead Ribozymes and RNA Interference. Methods Mol. Biol. 2007, 405, 113–131. [DOI] [PubMed] [Google Scholar]
- Guo S.; Tschammer N.; Mohammed S.; Guo P. Specific Delivery of Therapeutic RNAs to Cancer Cells via the Dimerization Mechanism of Phi29 Motor pRNA. Hum. Gene Ther. 2005, 16, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled A.; Guo S.; Li F.; Guo P. Controllable Self-Assembly of Nanoparticles for Specific Delivery of Multiple Therapeutic Molecules to Cancer Cells Using RNA Nanotechnology. Nano Lett. 2005, 5, 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J. L.; Wong M. L.; Parslow T. G. Requirements for Kissing-Loop-Mediated Dimerization of Human Immunodeficency Virus RNA. J. Virol. 1996, 70, 5902–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujeeb A.; Clever J. L.; Billeci T. M.; James T. L.; Parslow T. G. Structure of the Dimer Initiation Complex of HIV-1 Genomic RNA. Nat. Struct. Biol. 1998, 5, 432–436. [DOI] [PubMed] [Google Scholar]
- Wagner C.; Palacios I.; Jaeger L.; St. Johnston D.; Ehresmann B.; Ehresmann C.; Brunel C. Dimerization of the 3′UTR of Bicoid mRNA Involves a Two-Step Mechanism. J. Mol. Biol. 2001, 313, 511–524. [DOI] [PubMed] [Google Scholar]
- Shu Y.; Cinier M.; Fox S. R.; Ben-Johnathan N.; Guo P.. Assembly of Therapeutic pRNA-siRNA Nanoparticles Using Bipartite Approach. Mol. Ther. 2011, DOI: 10.1016/j.ymeth.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- Mi J.; Liu Y.; Rabbani Z. N.; Yang Z.; Urban J. H.; Sullenger B. A.; Clary B. M. In Vivo Selection of Tumor-Targeting RNA Motifs. Nat. Chem. Biol. 2010, 6, 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D. Back to the Future of Nucleic Acid Self-Amplification. Nat. Chem. Biol. 2009, 5, 200–201. [DOI] [PubMed] [Google Scholar]
- Mallik P. K.; Nishikawa K.; Millis A. J.; Shi H. Commandeering a Biological Pathway Using Aptamer-Derived Molecular Adaptors. Nucleic Acids Res. 2010, 38, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Shepard J. R.; Shi H. An RNA-Based Transcription Activator Derived from an Inhibitory Aptamer. Nucleic Acids Res. 2010, 38, 2378–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Zhang M.; Yang G.; Zhang D.; Ding H.; Wang H.; Fan M.; Shen B.; Shao N. Single-Stranded DNA Aptamers That Bind Differentiated but Not Parental Cells: Subtractive Systematic Evolution of Ligands by Exponential Enrichment. J. Biotechnol. 2003, 102, 15–22. [DOI] [PubMed] [Google Scholar]
- Cao X.; Li S.; Chen L.; Ding H.; Xu H.; Huang Y.; Li J.; Liu N.; Cao W.; Zhu Y.; et al. Combining Use of a Panel of ssDNA Aptamers in the Detection of Staphylococcus aureus. Nucleic Acids Res. 2009, 37, 4621–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Xu H.; Ding H.; Huang Y.; Cao X.; Yang G.; Li J.; Xie Z.; Meng Y.; Li X.; et al. Identification of an Aptamer Targeting hnRNP A1 by Tissue Slide-Based SELEX. J. Pathol. 2009, 218, 327–336. [DOI] [PubMed] [Google Scholar]
- Watrin M.; Von P. F.; Dausse E.; Schroeder R.; Toulme J. J. In Vitro Selection of RNA Aptamers Derived from a Genomic Human Library Against the TAR RNA Element of HIV-1. Biochemistry 2009, 48, 6278–6284. [DOI] [PubMed] [Google Scholar]
- Akinc A.; Querbes W.; De S.; Qin J.; Frank-Kamenetsky M.; Jayaprakash K. N.; Jayaraman M.; Rajeev K. G.; Cantley W. L.; Dorkin J. R.; et al. Targeted Delivery of RNAi Therapeutics with Endogenous and Exogenous Ligand-Based Mechanisms. Mol. Ther. 2010, 18, 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W.; Mao X.; Davide J. P.; Ng B.; Cai M.; Burke P. A.; Sachs A. B.; Sepp-Lorenzino L. Mechanistically Probing Lipid-siRNA Nanoparticle-Associated Toxicities Identifies Jak Inhibitors Effective in Mitigating Multifaceted Toxic Responses. Mol. Ther. 2011, 19, 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.; Castaldi M. P.; Dutta S.; Dibrov S. M.; Wyles D. L.; Hermann T. Conformational Inhibition of the Hepatitis C Virus Internal Ribosome Entry Site RNA. Nat. Chem. Biol. 2009, 5, 823–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H.; Mok H.; Jo S.; Hong C. A.; Park T. G. Dual Gene Targeted Multimeric siRNA for Combinatorial Gene Silencing. Biomaterials 2011, 32, 2359–2368. [DOI] [PubMed] [Google Scholar]
- Chang C. I.; Kang H. S.; Ban C.; Kim S.; Lee D. K. Dual-Target Gene Silencing by Using Long, Synthetic siRNA Duplexes without Triggering Antiviral Responses. Mol. Cells 2009, 27, 689–695. [DOI] [PubMed] [Google Scholar]
- Abdelmawla S.; Guo S.; Zhang L.; Pulukuri S.; Patankar P.; Conley P.; Trebley J.; Guo P.; Li Q. X.. Pharmacological Characterization of Chemically Synthesized Monomeric pRNA Nanoparticles for Systemic Delivery. Mol. Ther. 2011, DOI: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.; Li M. J.; Palmer B.; Remling L.; Li S.; Yam P.; Yee J. K.; Rossi J.; Zaia J.; Akkina R. Safety and Efficacy of a Lentiviral Vector Containing Three Anti-HIV Genes—CCR5 Ribozyme, tat-rev siRNA, and TAR Decoy—in SCID-hu Mouse-Derived T Cells. Mol. Ther. 2007, 15, 1182–1188. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Swiderski P.; Li H.; Zhang J.; Neff C. P.; Akkina R.; Rossi J. J. Selection, Characterization and Application of New RNA HIV gp 120 Aptamers for Facile Delivery of Dicer Substrate siRNAs into HIV Infected Cells. Nucleic Acids Res. 2009, 37, 3094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M. Innate Sensing of Self and Non-Self RNAs by Toll-Like Receptors. Trends Mol. Med. 2006, 12, 167–176. [DOI] [PubMed] [Google Scholar]
- Nallagatla S. R.; Toroney R.; Bevilacqua P. C. Regulation of Innate Immunity through RNA Structure and the Protein Kinase PKR. Curr. Opin. Struct. Biol. 2011, 21, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne T.; Bertrand J. R.; Vasseur J. J.; Debart F. A Base-Labile Group for 2′-OH Protection of Ribonucleosides: A Major Challenge for RNA Synthesis. Chemistry 2008, 14, 9135–9138. [DOI] [PubMed] [Google Scholar]
- Solomatin S.; Herschlag D. Methods of Site-Specific Labeling of RNA with Fluorescent Dyes. Methods Enzymol. 2009, 469, 47–68. [DOI] [PubMed] [Google Scholar]
- Hoeprich S.; Guo P. Computer Modeling of Three-Dimensional Structure of DNA-Packaging RNA(pRNA) Monomer, Dimer, and Hexamer of Phi29 DNA Packaging Motor. J. Biol. Chem. 2002, 277, 20794–20803. [DOI] [PubMed] [Google Scholar]
- Stengel G.; Urban M.; Purse B. W.; Kuchta R. D. Incorporation of the Fluorescent Ribonucleotide Analogue tCTP by T7 RNA Polymerase. Anal. Chem. 2010, 82, 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. S.; Kaiser R. J. Recent Advances in the High-Speed Solid Phase Synthesis of RNA. Curr. Opin. Chem. Biol. 2004, 8, 222–229. [DOI] [PubMed] [Google Scholar]
- Reese C. B. The Chemical Synthesis of Oligo- and Poly-Nucleotides: A Personal Commentary. Tetrahedron 2002, 58, 8893–8920. [Google Scholar]
- Pitsch S.; Weiss P. A.; Jenny L.; Stutz A.; Wu X. Reliable Chemical Synthesis of Oligoribonucleotides (RNA) with 2′-O-[(Trisisopropylsilyl)oxy]methyl (2′-O-tom)-Protected Phosphoramidites. Helv. Chim. Acta 2001, 84, 3773–3795. [Google Scholar]
- Scaringe S. A.; Wincott F. E.; Caruthers M. H. Novel RNA Synthesis Method Using 5′-O-Silyl-2′-O-Orthoester Protecting Groups. J. Am. Chem. Soc. 1998, 120, 11820–11821. [Google Scholar]
- Watts J. K.; Deleavey G. F.; Damha M. J. Chemically Modified siRNA: Tools and Applications. Drug Discovery Today 2008, 13, 842–855. [DOI] [PubMed] [Google Scholar]
- Madhuri V.; Kumar V. A. Design, Synthesis and DNA/RNA Binding Studies of Nucleic Acids Comprising Stereoregular and Acyclic Polycarbamate Backbone: Polycarbamate Nucleic Acids (PCNA). Org. Biomol. Chem. 2010, 8, 3734–3741. [DOI] [PubMed] [Google Scholar]
- Mathe C.; Perigaud C. Recent Approaches in the Synthesis of Conformationally Restricted Nucleoside Analogues. Eur. J. Org. Chem. 2008, 1489–1505. [Google Scholar]
- Patra A.; Richert C. High Fidelity Base Pairing at the 3′-Terminus. J. Am. Chem. Soc. 2009, 131, 12671–12681. [DOI] [PubMed] [Google Scholar]
- Efimov V. A.; Fediunin S. V.; Chakhmakhcheva O. G. Cross-Linked Nucleic Acids: Formation, Structure, and Biological Function. Bioorg. Khim. 2010, 36, 56–80. [DOI] [PubMed] [Google Scholar]
- Sandin P.; Wilhelmsson L. M.; Lincoln P.; Powers V. E.; Brown T.; Albinsson B. Fluorescent Properties of DNA Base Analogue tC upon Incorporation into DNA—Negligible Influence of Neighbouring Bases on Fluorescence Quantum Yield. Nucleic Acids Res. 2005, 33, 5019–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin P.; Borjesson K.; Li H.; Martensson J.; Brown T.; Wilhelmsson L. M.; Albinsson B. Characterization and Use of an Unprecedentedly Bright and Structurally Non-Perturbing Fluorescent DNA Base Analogue. Nucleic Acids Res. 2008, 36, 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borjesson K.; Preus S.; El-Sagheer A. H.; Brown T.; Albinsson B.; Wilhelmsson L. M. A Nucleic Acid Base Analog FRET-Pair Facilitating Detailed Structural Measurements in Nucleic Acid Containing Systems. J. Am. Chem. Soc. 2009, 131, 4288–4293. [DOI] [PubMed] [Google Scholar]
- Sinkeldam R. W.; Wheat A. J.; Boyaci H.; Tor Y. Emissive Nucleosides as Molecular Rotors. ChemPhysChem 2011, 12, 567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Dix A. V.; Tor Y. Antibiotic Selectivity for Prokaryotic vs. Eukaryotic Decoding Sites. Chem. Commun. 2010, 46, 5542–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Dix A. V.; Tor Y. FRET Enabled Real Time Detection of RNA–Small Molecule Binding. J. Am. Chem. Soc. 2009, 131, 17605–17614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson L. M. Fluorescent Nucleic Acid Base Analogues. Q. Rev. Biophys. 2010, 43, 159–183. [DOI] [PubMed] [Google Scholar]
- Sinkeldam R. W.; Greco N. J.; Tor Y. Fluorescent Analogs of Biomolecular Building Blocks: Design, Properties, and Applications. Chem. Rev. 2010, 110, 2579–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.Special Issue: RNA Nanotechnology. Methods 2011, 54, Cover illustration. [DOI] [PubMed] [Google Scholar]