Abstract
The SR proteins constitute a family of nuclear phosphoproteins, which are required for constitutive splicing and also influence alternative splicing regulation. Initially, it was suggested that SR proteins were functionally redundant in constitutive splicing. However, differences have been observed in alternative splicing regulation, suggesting unique functions for individual SR proteins. Homology searches of the Caenorhabditis elegans genome identified seven genes encoding putative orthologues of the human factors SF2/ASF, SRp20, SC35, SRp40, SRp75 and p54, and also several SR-related genes. To address the issue of functional redundancy, we used dsRNA interference (RNAi) to inhibit specific SR protein function during C.elegans development. RNAi with CeSF2/ASF caused late embryonic lethality, suggesting that this gene has an essential function during C.elegans development. RNAi with other SR genes resulted in no obvious phenotype, which is indicative of gene redundancy. Simultaneous interference of two or more SR proteins in certain combinations caused lethality or other developmental defects. RNAi with CeSRPK, an SR protein kinase, resulted in early embryonic lethality, suggesting an essential role for SR protein phosphorylation during development.
Keywords: Caenorhabditis elegans/pre-mRNA splicing/RNA interference/SR proteins
Introduction
The SR proteins are a family of structurally and functionally related proteins that have a dual role in pre-mRNA splicing. They are essential splicing factors involved in several steps of the splicing reaction, and they also regulate alternative splicing in a concentration-dependent manner (for reviews, see Fu, 1995; Manley and Tacke, 1996; Válcarcel and Green, 1996; Cáceres and Krainer, 1997). The SR proteins are highly conserved proteins found throughout metazoans, and in plants (Zahler et al., 1992; Lazar et al., 1995; Lopato et al., 1996a,b, 1999). Individual members of this family of proteins show higher homology across species than to other family members within the same species. In addition to the SR protein family of splicing regulators, a class of related RS domain-containing proteins, termed SR protein-related polypeptides (SRrps) or SR-like proteins, are also involved in splicing regulation (for a review, see Fu, 1995).
The SR proteins have a modular structure that consists of one or two RNA recognition motifs (RRMs) and a C–terminal domain rich in arginine and serine residues, known as the RS domain. The individual domains in SR proteins are functional modules (Cáceres et al., 1997; Chandler et al., 1997; Mayeda et al., 1999); whereas the coordinated action of the RNA recognition motifs determines their RNA binding specificity, the RS domains function as activators of splicing and are functionally interchangeable (J.Wang et al., 1998). The RS domains do not contribute to splicing specificity and are able to activate splicing even when fused to a heterologous RRM (Graveley and Maniatis, 1998).
The SR proteins are antagonized in alternative splicing regulation by members of the hnRNP A/B family of proteins (Mayeda and Krainer, 1992; Cáceres et al., 1994; Mayeda et al., 1994; Yang et al., 1994), and in certain cases by other SR proteins (Gallego et al., 1997; Jumaa and Nielsen, 1997). There are tissue-specific variations in the total and relative amounts of SR proteins (reviewed in Cáceres and Krainer, 1997) and, in particular, the molar ratio of SF2/ASF to its antagonist, hnRNPA1, varies considerably in different rat tissues (Hanamura et al., 1998). Thus, the relative abundance of members of these two families of antagonistic splicing factors may be crucial in regulating the patterns of alternative splicing in a tissue-specific, or developmentally regulated manner.
An emerging question is whether SR proteins have unique or redundant functions. Each individual SR protein can complement an inactive S100 cytosolic extract that contains all the components required for splicing activity except for SR proteins, suggesting redundant functions in constitutive splicing (Cáceres and Krainer, 1997; Tacke and Manley, 1999). However, several differences have been observed in the ability of these proteins to regulate alternative splicing, both in vitro and in vivo (Zahler et al., 1993; Screaton et al., 1995; Wang and Manley, 1995). In addition, individual SR proteins are able to commit different pre-mRNAs to the splicing pathway, suggesting unique functions in splicing regulation (Fu, 1993).
The notion that SR proteins have unique functions is supported further by the finding that B52, the Drosophila melanogaster homologue of the SRp55 gene, and SF2/ASF are both essential genes. A B52 null allele results in lethality during development, indicating that B52 provides at least one non-redundant function necessary for proper development (Ring and Lis, 1994). No general splicing defects were found in the null background, either for constitutive or alternatively spliced genes (Ring and Lis, 1994); however, recent observations suggest that the lethality of the B52 deletion strain is a consequence of splicing defects in tissues in which B52 is normally the major SR protein (Hoffman and Lis, 2000). In addition, genetic evidence has established a critical role for B52 in pre–mRNA splicing in vivo (Peng and Mount, 1995). SF2/ASF was inactivated by homologous recombination in a chicken cell line, DT40, and was shown to be essential for cell survival (Wang et al., 1996). More importantly, the lack of SF2/ASF could not be rescued by overexpression of other SR proteins, confirming SF2/ASF as an essential protein with non-redundant functions (Wang et al., 1996). Recently, it was shown that SRp20 is required for mouse development, since mouse embryos lacking SRp20 died at the morula stage, with a block in blastocyst formation (Jumaa et al., 1999).
We decided to use the nematode Caenorhabditis elegans as a model organism to characterize the SR family of proteins functionally, and to address the important issue of functional redundancy. An advantage of the C.elegans system is the availability of the complete genome sequence, and also the possibility of inhibiting gene expression by RNA interference (RNAi) (for reviews, see Fire, 1999; Hunter, 1999; Sharp, 1999). RNAi has now been established as a rapid and convenient method for interfering selectively with gene expression, not only in C.elegans, but also in plants (Voinnet et al., 1998; Waterhouse et al., 1998), in Trypanosoma brucei (Ngo et al., 1998) and in Drosophila (Kennerdell and Carthew, 1998). It has been shown recently that RNAi is also an effective tool in mammals, as shown by specific interference with gene function obtained with double-stranded RNA (dsRNA) in early mouse development (Wianny and Zernicka-Goetz, 2000). Introduction of dsRNA corresponding to a particular gene causes interference with that gene's functional expression. This process is highly sequence specific and, for a number of genes, has been shown to phenocopy strong loss-of-function or null alleles of the gene (Fire et al., 1998; Wianny and Zernicka-Goetz, 2000). In C.elegans, injection of dsRNA into the worm generally leads to the disappearance of the corresponding gene mRNA both from the somatic cells of the injected adult animal and from its F1 progeny. However, the F2 progeny from RNAi-treated animals generally revert to wild-type phenotype. RNAi is a highly efficient process since only a few molecules per cell are sufficient to produce gene-specific suppression, suggesting the existence of catalytic or amplification components in the interference process (Fire et al., 1998). It has been proposed that RNAi causes gene-specific degradation of mRNA from the targeted gene in both nucleus and cytoplasm (Montgomery et al., 1998).
Two different types of splicing, cis- and trans-splicing, occur in C.elegans (for a review, see Blumenthal and Thomas, 1998). Approximately one-quarter of all C.elegans genes are organized in operons containing 2–8 genes, which are transcribed polycistronically (Zorio et al., 1994; for a review, see Blumenthal, 1998). Pre-mRNAs are processed by trans-splicing, resulting in the addition of the SL1 leader near the 5′ end, and the SL2 leader at internal trans-splice sites of polycistronic pre-mRNAs (Spieth et al., 1993; Blumenthal, 1998). The SR proteins are required for trans-splicing in vitro (Bruzik, 1996; Sanford and Bruzik, 1999a), which was suggested originally by the ability of SR proteins to promote the formation of splicing products even when the 5′ and 3′ splice sites were located on separate transcripts (Bruzik and Maniatis, 1995; Chiara and Reed, 1995). Interestingly, a novel SR protein, TSR1, has been identified in T.brucei, an organism that exclusively trans-splices (Ismaili et al., 1999).
Here we report the use of dsRNA interference to inhibit SR protein gene expression specifically in C.elegans. A search of the C.elegans genome databases identified seven homologues of human SR genes, and also several SR-related genes including the homologues of Tra-2β, SRm160 and SRPK, an SR protein-specific kinase. RNAi with CeSRPK and CeSF2/ASF led to early and late embryonic lethality, respectively, suggesting that these genes have essential functions during C.elegans development. Surprisingly, RNAi with other individual SR genes resulted in no obvious phenotype, which strongly suggests the existence of gene functional redundancy for the SR family of proteins in C.elegans. However, simultaneous suppression of two or more SR proteins in different combinations caused lethality or other severe phenotypes. Finally, some SR-related proteins displayed less severe phenotypes affecting primarily gut and gonad functions, suggesting more specific roles for these genes during development.
Results
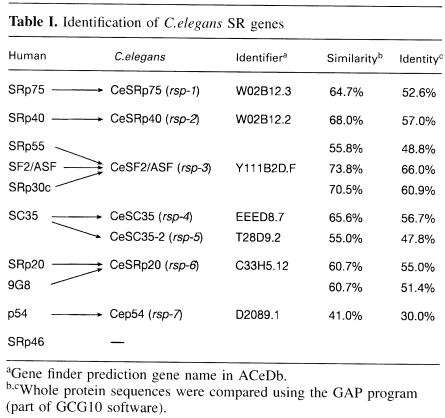
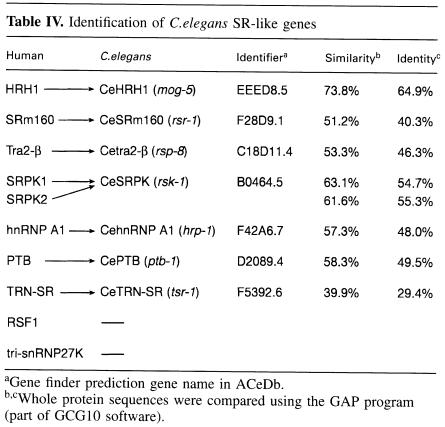
A BLAST search of the C.elegans genome identified seven homologues of human SR genes. We describe the predicted encoded C.elegans SR proteins according to their putative human SR protein orthologue and give the formal C.elegans gene name in parentheses. They are CeSRp75 (rsp-1), CeSRp40 (rsp-2), CeSF2/ASF (rsp-3), CeSC35 (rsp-4), CeSC35-2 (rsp-5), CeSRp20 (rsp-6) and p54 (rsp-7) (Table I). In the case of SC35, two C.elegans genes displaying 56.7 and 47.8% identity were identified, which we termed CeSC35 (rsp-4) and CeSC35-2 (rsp-5), respectively. The predicted CeSRp20 displays the highest homology to two human SR proteins, SRp20 (55% identity) and 9G8 (51.4% identity); however, it lacks the zinc knuckle motif present in human 9G8 (Cavaloc et al., 1994), which suggests that it is structurally more related to human SRp20. The nematode factor corresponding to human p54 (Chaudhary et al., 1991) displays the weakest sequence homology observed for a nematode SR protein (Table I). No predicted orthologues for the human SRp55 or SRp30c (Screaton et al., 1995) were identified in the C.elegans genome. In addition, we did not find an orthologue for SRp46, a human SR protein encoded by an SC35 retropseudogene, which has no homologue in the mouse (Soret et al., 1998). We have also identified several SR-related genes in C.elegans, including probable orthologues of Tra-2β (rsp-8), and SRPK (rsk-1), an SR protein kinase, and also homologues of hnRNP A1 (hrp–1) and PTB (ptb-1) (Table IV, see below). Both U2AF genes are also present in C.elegans, and it was demonstrated recently by RNAi that uaf-1 (which encodes U2AF65) and uaf-2 (which encodes U2AF35) are required for viability (Zorio and Blumenthal, 1999).
Table I. Identification of C.elegans SR genes.
aGene finder prediction gene name in ACeDb.
b,cWhole protein sequences were compared using the GAP program (part of GCG10 software).
Table IV. Identification of C.elegans SR-like genes.
aGene finder prediction gene name in ACeDb.
b,cWhole protein sequences were compared using the GAP program (part of GCG10 software).
We sought to study the effects of loss of function for individual SR genes and we used RNAi in order to interfere selectively with SR protein gene expression. The most efficient way of causing RNA interference in C.elegans is to microinject dsRNA into adult worms. It has also been shown that feeding worms with dsRNA or simply soaking the worms in dsRNA can also induce specific interference. However, both soaking and feeding are reported to have a lower efficiency (Tabara et al., 1998; Timmons and Fire, 1998); therefore, we decided to use microinjection of dsRNA. Although the site of injection is not critical, we carried out the injections into the gonads or the gut of young adult hermaphrodites (Bristol strain N2). Injected animals were left to recover and to lay any eggs present in utero prior to injection, for 16 h, and were then transferred onto individual plates and allowed to egg lay. This allowed us to observe the effect of RNAi only in F1 progeny that were produced after the RNAi treatment.
CeSF2/ASF and other C.elegans SR proteins
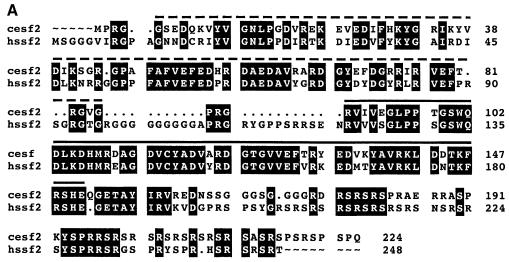
Human SF2/ASF is the prototype of the SR family of proteins and was identified simultaneously by two groups, looking either for an essential splicing factor able to complement an inactive S100 cytosolic extract (Krainer et al., 1990, 1991) or for a splicing activity able to modulate splice site selection (Ge and Manley, 1990; Ge et al., 1991). It was shown later that inactivation of SF2/ASF in a chicken cell line caused cell lethality, demonstrating that this splicing factor is essential (Wang et al., 1996). Caenorhabditis elegans SF2/ASF protein is highly conserved, displaying 73.8% similarity and 66% identity to the human protein (Table I). This high degree of conservation is evident throughout the different motifs of the protein, including the RS domain. In particular, both RNP-2 and RNP-1 submotifs within RRM1 are highly conserved, and the SWQDLKD motif, an invariant signature of those human SR proteins with a second RRM, is also present (Figure 1A) (Birney et al., 1993).
Fig. 1. (A) Sequence comparison between human and C.elegans SF2/ASF proteins. Sequences were compared using a gap program (GCG10 software) and the output was produced using prettybox (GCG10 software). Identical residues are highlighted in black and dsRNA fragments corresponding to two different dsRNA probes are shown. dsRNA fragment 1 is represented by a solid line and dsRNA fragment 2 by a dashed line. (B) RNA interference with the CeSF2/ASF gene. RNAi with fragment 1 (a and b) and fragment 2 (c) resulted in the same late embryonic lethal phenotype. Embryos are partially elongated and organs are formed, but morphogenesis failed. Wild-type embryo developmental stages (d, 1.5-fold; e, 2–fold; f, 3-fold) are shown for comparison. Each embryo is ∼50 μm in length.
When dsRNA corresponding to CeSF2/ASF was injected, a late embryonic lethal phenotype was observed in F1 progeny. Although organogenesis had occurred as indicated by the presence of major tissue types, the embryos were morphogenically defective (Figure 1B), clearly demonstrating that CeSF2/ASF is an essential protein in C.elegans, and that it is required for at least one non-redundant function. The RNAi effect was specific and extensive; whereas CeSF2/ASF mRNA was greatly depleted as shown by RT–PCR analysis in the affected embryos, CeSRp20 and CeSRp40 mRNAs were present at levels comparable to wild-type embryos (Figure 2A; data not shown). Moreover, identical phenotypes were obtained when two different dsRNA fragments corresponding to the second RRM (Figure 1A and 1B, panels a and b) or to the first RRM of CeSF2/ASF (Figure 1A and B, panel c) were used for RNAi.
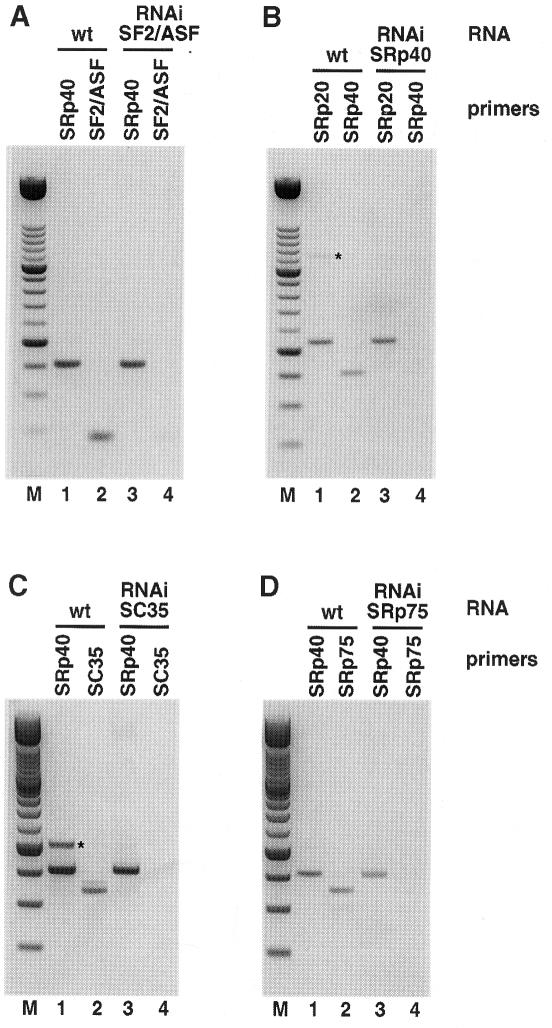
Fig. 2. Efficiency of RNAi treatment. RT–PCR analysis of total RNA isolated from wild-type and RNAi-treated embryos was performed as described in Materials and methods. In each panel, lanes 1 and 3 show the level of the control SR gene RNA. Lanes 2 and 4 show the RNA levels of RNAi-treated SR genes. Specific RNA is depleted in RNAi-treated embryos (lane 4) as compared with RNA from the same gene in wild-type embryos (lane 2). RT–PCR analysis of (A) CeSF2/ASF RNAi, (B) CeSRp40 RNAi, (C) CeSC35 RNAi and (D) CeSRp75 RNAi. This figure shows a negative of an ethidium bromide-stained agarose gel. M: 100 bp ladder DNA size marker. Bands marked with an asterisk are due to genomic DNA contamination, and only appear in certain wild-type RNA preparations.
SF2/ASF has been shown to be essential for cell viability in a chicken B cell line (Wang and Manley, 1996). CeSF2/ASF RNAi caused late embryonic lethality, with the affected embryos going through organogenesis, but failing morphogenesis. This would suggest that CeSF2/ASF is not cell essential, but is required for development. We cannot determine whether the differences observed are species specific, or whether any potential contribution of maternal SF2/ASF protein could be responsible for a delayed phenotype. However, the way in which the RNAi experiments are performed make this unlikely (see Discussion).
When dsRNA fragments corresponding to the other C.elegans SR proteins, CeSRp75, CeSRp40, CeSC35, CeSC35-2 or CeSRp20, were injected individually, no discernible phenotypes were observed (Table II). These results suggested that SR proteins have redundant functions and that the absence of a particular CeSR protein, other than CeSF2/ASF, can be rescued by the presence of other SR proteins. We analysed the effectiveness of the RNAi treatment by looking at the level of the specific transcripts following dsRNA injections. Total RNA from F1 embryos of worms injected with dsRNA fragments corresponding to individual SR proteins was extracted. RT–PCR analysis showed that CeSF2/ASF, CeSRp40, CeSC35 and CeSRp75 RNAs were greatly depleted in the affected embryos (Figure 2), whereas RNAs for control SR proteins were present at levels comparable to wild-type embryos (Figure 2, compare lanes 2 and 4 in each panel). Similar results were obtained for CeSRp20 and PTB (data not shown). This experiment strongly suggests that the RNAi treatment is very effective. Moreover, these experiments argue against the possibility that the lack of phenotype observed when interfering with individual SR proteins (other than SF2/ASF) is due to inefficient RNA removal, and supports the notion of functional redundancy. In addition, when two different dsRNA fragments were used for RNAi of CeSC35 and CeSC35-2, no discernible phenotypes were observed either (not shown). It is not clear why SF2/ASF produces a different phenotype from the rest of the C.elegans SR proteins, but we believe that we have ruled out RNAi artefacts (see above), strongly suggesting that CeSF2/ASF has unique properties and is the only nematode SR protein that is essential for development.
Table II. RNAi phenotypes for C.elegans SR genes.
| Genes | RNAi phenotype |
|---|---|
| SF2/ASF | late embryonic lethal, organs formed, embryos partially elongated, failed morphogenesis (100%) |
| SRp20 | no phenotype |
| SC35 | no phenotype |
| SC35-2 | no phenotype |
| SRp40 | no phenotype |
| SRp75 | no phenotype |
| p54 | nd |
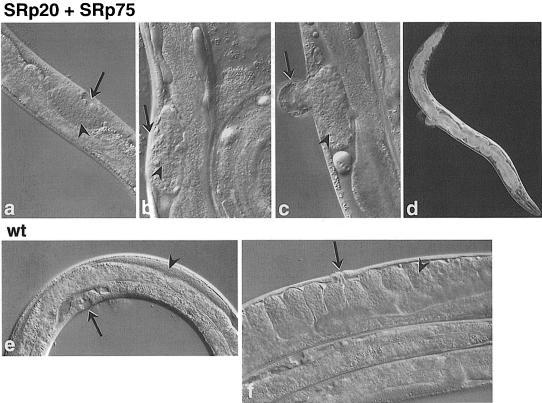
RNAi allows the simultaneous introduction of fragments corresponding to different genes. In order to test further the functional redundancy of C.elegans SR proteins, we injected dsRNA fragments corresponding to all SR genes, except CeSF2/ASF. A late embryonic lethal phenotype was observed, in which organs formed, but again, morphogenesis was defective (Figure 3). This phenotype is more severe than the one observed with CeSF2/ASF alone and strongly suggests that in addition to CeSF2/ASF, other SR proteins are also required for proper development. We then decided to inject dsRNAs corresponding to individual SR genes in different combinations, and the results are summarized in Table III. Whereas individual interference of CeSRp20 or CeSRp75 showed no obvious phenotype, the simultaneous interference with these two C.elegans SR genes resulted in a distinct phenotype. The F1 animals developed to adulthood and showed a combination of vulval defects, such as aberrant or missing vulva, and were sterile with severely underdeveloped gonads. They were also notably slow growing, and frequently had partially blocked guts (Figure 4). The cells most affected by this RNAi include those most active in post-embryonic cell divisions in C.elegans. This opens up the possibility that CeSRp20 and CeSRp75 may have specific functions necessary either for post-embryonic cell divisions in C.elegans or for specific developmental processes in the affected cell types. Simultaneous suppression of CeSRp75 and CeSC35-2 genes resulted in reduced motility in the F1 progeny. However, the worms responded to touch normally, and no other abnormalities were observed (Table III). Interestingly, RNAi with other combinations of C.elegans SR proteins, such as CeSC35 + CeSRp40, CeSC35 + CeSC35-2 and CeSRp20 + CeSC35-2 gave no discernible phenotype (Table III). However, when a dsRNA fragment corresponding to CeSRp20 was added to the CeSC35 + CeSRp40 mix, the simultaneous interference of CeSRp20, CeSC35 and CeSRp40 genes resulted in slow growth, and also a low frequency of ruptured vulva and dumpy phenotype was observed (Table III).
Fig. 3. Multiple RNA interference for CeSR proteins. RNAi with all CeSR proteins apart from CeSF2/ASF resulted in late embryonic lethal phenotype (a and b). Embryos are partially elongated and some organs are formed, but morphogenesis has failed. This phenotype is more severe than that observed for the CeSF2/ASF gene alone. Wild-type embryo developmental stages (c, 1.5–fold; d, 2–fold; e, 3–fold) are shown for comparison. Each embryo is ∼50 μm in length.
Table III. RNAi phenotypes for multiple suppression of C.elegans SR genes.
| Genes | RNAi phenotype |
|---|---|
| SRp20 + SRp75 | vulval defects or vulvaless, sterile, blocked gut, slow growth |
| SC35-2 + SRp75 | reduced motility |
| SRp40 + SC35 | no phenotype |
| SC35 + SC35-2 | no phenotype |
| SRp20 + SC35-2 | no phenotype |
| SRp20 + SRp40 + SC35 | slow growth, variable phenotype: ruptured vulva, dumpy (low frequency) |
| SRp20 + SC35 + SC35-2 + SRp40 + SRp75 | late embryonic lethal, organs formed, failed morphogenesis (100%) |
Fig. 4. Simultaneous RNAi with CeSRp20 and CeSRp75 genes. The phenotype included underdeveloped gonad and vulva in L4 stage larvae (a). In adults, RNAi effects included underdeveloped gonads with only a small number of germline nuclei and missing vulva (b), aberrant vulva (c) and blocked gut (d). Wild-type animals are shown for comparison: L4 (e) and adult (f). Arrow = vulva, arrowhead = gonad.
Phosphorylation of SR proteins
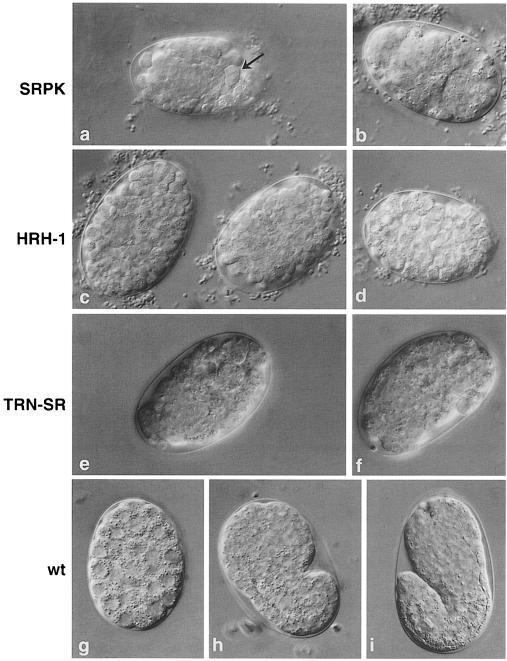
RNA interference of CeSRPK (rsk-1), which is the only SR protein-specific kinase predicted in C.elegans, resulted in an early embryonic lethal phenotype (Figure 5a and b). Development of the embryos arrested probably around gastrulation, although some muscle and neuronal development occurred as indicated by movement of the dying embryos. The presence of some unusually large cells also suggests a cell cycle defect. These findings demonstrate that phosphorylation of SR proteins is essential for proper development in C.elegans. It has recently been shown in a different nematode, Ascaris lumbricoides, that the state of phosphorylation of SR proteins is critical for their activity in vitro. Drastic changes in SR protein phosphorylation occur during early development of this nematode and these differences in phosphorylation correlate with changes in SR protein activity in both cis- and trans-splicing (Sanford and Bruzik, 1999b).
Fig. 5. RNA interference with SR-related genes. RNAi of CeSRPK (a and b) resulted in an early embryonic lethal phenotype. Embryos are arrested around gastrulation, judged by comparison with wild-type embryos (g). The presence of some unusually large cells (a, arrow) suggests a possible cell cycle defect. RNAi of CeHRH1 resulted in an early embryonic lethal phenotype (c and d). RNAi of CeTRN–SR also resulted in early embryonic lethal phenotype (e and f). Wild-type embryos are shown for comparison (g, gastrulated; h, comma; i, 1.5–fold). Each embryo is ∼50 μm in length.
SR-related and hnRNP proteins
We next searched for C.elegans homologues of SR-related and hnRNP proteins, which have important roles in splicing regulation. Homologies were found for the splicing activator SRm160 (Blencowe et al., 1998), for human Tra2–β (Beil et al., 1997), for the human helicase HRH1 (Ono et al., 1994), for hnRNP A1 (Biamonti et al., 1989) and for PTB (Gil et al., 1991; Patton et al., 1991). A BLAST homology search with human full-length transportin SR (TRN–SR), a nuclear import receptor for SR proteins (Kataoka et al., 1999), identified a C.elegans protein with 39.9% similarity and 29.4% identity (Table IV). However, no homologue was found for RSF1, a splicing repressor identified in Drosophila that antagonizes SR protein function (Labourier et al., 1999). No identifiable homologue for the tri-snRNP27 K protein, an SR protein that is a component of the U4/U6⋅U5 tri-snRNP, was found (Fetzer et al., 1997).
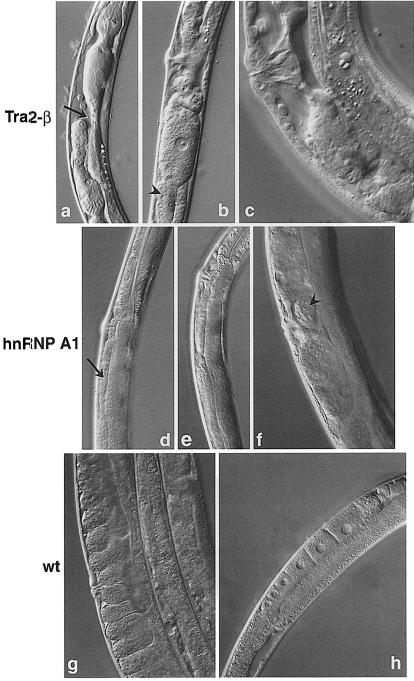
HRH1, a human RNA helicase-like protein homologous to the yeast splicing factor Prp22, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome (Ono et al., 1994; Ohno and Shimura, 1996). RNAi with CeHRH1 (mog-5) or CeTRN–SR (tsr-1) gave early embryonic lethal phenotypes, demonstrating that these are essential proteins (Figure 5c–f). RNA interference with Tra2-β (rsp-8) resulted in some larval lethality, but most F1 worms reached adulthood. These animals were slow growing and stunted. The complex phenotype included a dysfunctional gut with severe constrictions, and full of undigested bacteria. F1 animals had underdeveloped gonads and were almost sterile (Figure 6a–c). In Drosophila, Tra2 is a non-essential protein since flies with a null mutation are viable; however, sexual differentiation is affected and female flies are transformed into sterile phenotypical males (Baker and Ridge, 1980).
Fig. 6. RNA interference with SR-related genes. RNAi of Cetra2-β resulted in a complex phenotype including dysfunctional gut with severe constrictions (a, arrow), underdeveloped gonad (b, arrowhead) and sterility. Detail of an adult vulva and gonad with no developed embryos (c) is shown. RNAi of CehnRNP A1 resulted in defect in oogenesis (d, arrow, and e) and much reduced fertility. Detail of an adult with only one aberrant embryo (f, arrowhead) is shown. Wild-type animals are shown for comparison (g, adult with normal vulva and normal gonad full of embryos; h, young adult with normal progression of oogenesis).
hnRNP A/B proteins antagonize SR proteins in splice site selection, and they also promote splicing inhibition by binding to exonic splicing silencers (Caputi et al., 1999; Del Gatto-Konczak et al., 1999). They are proposed to have a role in many other aspects of RNA metabolism, such as mammalian telomere biogenesis (LaBranche et al., 1998) and mRNA export (Izaurralde et al., 1997). We found that worms that were interfered for hnRNP A1 (hrp-1) expression were viable, although they displayed a distinct phenotype. RNAi resulted in a defect in oogenesis with aberrant oocytes, much reduced fertility, and the F1 animals were longer and thinner when compared with wild-type animals (Figure 6d–f). hnRNP A1 has been shown previously to be a non-esential protein in an erythroleukaemia cell line (Ben-David et al., 1992), and a null mutant of an hnRNP A1-like protein in Drosophila, hrp36, is also viable (Zu et al., 1996). Thus, despite being non-essential for cell survival, inactivation of hnRNP A1 causes important developmental defects when analysed in a complete organism. The abundant hnRNP protein, PTB, has been shown to be involved in the regulation of several splicing events, mostly as a negative regulator, such as in the case of neural genes (Zhang et al., 1999), α–tropomyosin (Gooding et al., 1998), β–tropomyosin (Mulligan et al., 1992) and c–src (Chan and Black, 1997) (for a review, see Válcarcel and Gebauer, 1997). By RNA interference, we demonstrate that CePTB (ptb-1) is not an essential protein, as there was no discernible phenotype despite the disappearance of PTB RNA following RNAi (Table V and data not shown). Finally, RNA interference for the splicing factor for SRm160 (srr-1) (Blencowe et al., 1998) showed no obvious phenotype, suggesting that this protein has redundant functions (Table V).
Table V. RNAi phenotypes for C. elegans SR-like genes.
| Gene | RNAi phenotype |
|---|---|
| SRPK | early embryonic lethal, some tissues developed |
| (76.4 ± 5.0%) | |
| TRN-SR | early embryonic lethal (100%) |
| HRH-1 | early embryonic lethal (100%) |
| hnRNP A1 | defect in oogenesis, aberrant oocytes, much reduced fertility, worms long and thin |
| Tra2-β | some larval lethality, dysfunctional gut, underdeveloped gonad, almost sterile, slow growth |
| PTB | no phenotype |
| SRm160 | no phenotype |
Splicing defects
Since SR proteins are involved in the regulation of both constitutive and alternative splicing, we decided to investigate whether the RNAi phenotypes observed were due to general defects in splicing or changes in alternative splicing. We analysed the splicing patterns of three constitutively spliced genes: hlh-1 (Krause et al., 1990), cpr–5 (Larminie and Johnstone, 1996) and ama-1 (Bird and Riddle, 1989); and of three genes that undergo alternative splicing: that for DNA topoisomerase-1 (Lee et al., 1998), unc-52 (Mullen et al., 1999) and uaf-1 (Zorio et al., 1997). Total RNA was extracted from SRp20, SC35, SC35-2, SRp20 + SRp75, SC35 + SRp40, SRp20 + SC35 + SRp40 and PTB-injected worms and analysed by RT–PCR with specific primers. No changes in constitutive or alternative splicing were detected, which suggests that the absence of tested SR and SR-related genes does not have a global effect on splicing (data not shown). This, of course, does not rule out a role for SR proteins in splicing regulation but rather reflects the fact that SR proteins are involved in the regulation of subsets of genes in certain tissues.
Discussion
The completion of the C.elegans genome sequencing project (Wilson, 1999) allows the easy identification of all members of a gene family, such as the SR family of splicing regulators, and issues of genetic redundance can be studied. Here we have identified probable C.elegans orthologues for all human SR proteins known to date. However, it is speculated that there are many more human SR proteins yet to be identified. Therefore, in the future, it would be necessary to confirm the total number of C.elegans SR genes by a comprehensive motif search of C.elegans databases.
We have used C.elegans as a model system to characterize functionally SR, SR-related and hnRNP proteins, which are involved in splicing regulation. A major aim of this study was to address the issue of functional redundancy, and for this purpose we have used RNAi to interfere selectively with their gene expression. Although initially SR proteins were considered to have redundant functions in constitutive splicing, growing evidence has shown that they could have unique functions in enhancer-dependent splicing, and also in the regulation of alternative splicing. Moreover, the finding that gene knock-outs of SF2/ASF in a chicken cell line, B52 (SRp55) in Drosophila and SRp20 in mice are required for viability or development suggests unique functions for individual SR proteins. This is supported further by the fact that SR proteins have distinct RNA binding specificities determined by their unique RNA recognition motifs, and this will most likely contribute to their unique functions.
Injection of dsRNA into C.elegans leads to interference with expression of the gene to which the RNA is homologous, producing phenotypes that resemble a null mutation in that gene (Fire et al., 1998). RNA interference has been widely adopted recently as a powerful tool to interfere selectively with gene expression. By RNAi, we have demonstrated that CeSF2/ASF is an essential protein and is required early in development. In contrast, when the expression of every other nematode SR protein was suppressed individually by RNAi, no phenotypes were observed. This strongly suggests functional redundancy among the different members of this family of proteins. However, certain considerations should be kept in mind when interpreting RNAi results; for instance, when a phenotype is observed following injection of dsRNA, a major concern is that the dsRNA fragment used could have caused cross-interference, suppressing the expression of additional related genes. In the case of CeSF2/ASF, identical phenotypes were observed after injection of two different dsRNA fragments corresponding to distinct regions of the protein (Figure 1). In addition, exclusive depletion of CeSF2/ASF mRNA, but not of RNAs coding for other CeSR proteins, as shown by RT–PCR analysis of the affected embryos, argues against cross-interference (Figure 2 and data not shown). Although we cannot be certain that all of the RNA corresponding to the targeted gene has been eliminated, we can assume that the phenotype is due to at least a vast reduction in the levels of endogenous mRNA. In the cases where no phenotypes were observed, such as with CeSR proteins other than SF2/ASF, one major concern is that the dsRNA fragment used is not causing adequate interference with the targeted gene. To rule out this possibility, we analysed the effectiveness of RNAi by looking at the level of residual RNA following RNAi by RT–PCR. We observed a drastic reduction in the level of the specific SR protein RNAs in every case (Figure 2 and data not shown), even though RNAi of individual SR proteins gave no observable phenotypes. This experiment demonstrates that the lack of phenotype when interfering with individual SR proteins is not due to inefficient RNA removal, and strongly suggests the existence of partial redundancy of individual SR proteins in a physiological context. In addition, we used dsRNA fragments corresponding to different regions of the targeted gene. For example, in the case of CeSC35 and CeSC35-2, RNAi produced with two different dsRNA fragments resulted in no phenotype in either case, arguing against inefficient RNAi with certain dsRNAs. A further indication that RNAi is working and that the absence of phenotype is not due to inefficient RNA removal is best illustrated by the results obtained with multiple interferences. For example, individual interference with CeSRp20 or CeSRp75 produced no discernible phenotype, but when interfered simultaneously, a very strong, highly penetrant and specific phenotype was observed (Figure 4).
The fact that RNAi with different combinations of SR proteins results in different phenotypes can be explained in several ways. One possibility is that the levels of SR protein gene expression vary drastically among individual members of this family. Thus, removing certain combinations of SR proteins that are relatively more abundant will reduce the overall level of SR proteins below an acceptable threshold, giving rise to a phenotype. We favour the alternative explanation, that the simultaneous suppression of certain SR proteins, such as CeSRp20 and CeSRp75, results in a distinct phenotype because of a common function for those proteins. Thus, when SR proteins that cooperate in the regulation of an essential splicing event are depleted together, a phenotype becomes evident. In the case of SR proteins that display no phenotype when interfered with simultaneously, such as CeSC35 and CeSRp40, the implication is that they do not share common essential functions and their removal by RNAi can be rescued by the presence of the other remaining SR proteins.
It is unlikely that residual maternal proteins are affecting the results we observed. RNAi interferes with maternal mRNA, but it will not eliminate maternal protein present in the mother at the time of RNAi. It is therefore possible that some maternal protein will be present in RNAi-treated embryos. To circumvent this problem, worms were injected with specific dsRNA and left overnight to recover and to allow egg laying to proceed. The injected worms were then transferred to fresh plates where they continued egg laying. Any residual maternal protein of a gene being targeted by RNAi would be present in the first eggs layed, but would reduce progressively in later embryos. This is indeed what we observed. For genes that give a lethal RNAi phenotype, the first embryos laid by the mother were frequently viable. The phenotypes we described are those obtained from embryos laid at least 16 h after the injection, and are consistent between a very large number of later laid embryos.
SR protein phosphorylation
It has been shown that spliceosome formation requires protein phosphorylation, and that a dephosphorylation event is also required to complete the splicing reaction (Mermoud et al., 1994). In agreement with this, a cycle of phosphorylation and dephosphorylation of SF2/ASF has been shown to be required for pre-mRNA splicing in vitro (Cao et al., 1997). Several protein kinases that are able to phosphorylate SR proteins have been identified: SRPK1 (Gui et al., 1994a,b), SRPK2 (Kuroyanagi et al., 1998; H.Y.Wang et al., 1998), Clk/Sty (Colwill et al., 1996) and DNA topoisomerase I (Rossi et al., 1996), but their functions in vivo have only recently begun to emerge. In Drosophila, SR protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination (Du et al., 1998).
A precise level of phosphorylation is a critical factor in splicing regulation, since both hyper- and hypophosphorylation of SR proteins by the Clk/Sty kinase inhibit splicing activity (Prasad et al., 1999). SR protein phosphorylation mediates protein–protein interactions within the spliceosome (Xiao and Manley, 1997, 1998; for a review, see Misteli, 1999), and is also required for their recruitment from the nuclear speckles to sites of transcription in vivo (Misteli et al., 1998).
By RNA interference, we have shown that CeSRPK is essential for viability. We have observed a very early embryonic lethal phenotype, demonstrating the critical role of phosphorylation of SR and SR-related proteins for proper development. In the nematode, A.lumbricoides, the state of phosphorylation of SR proteins is also critical and determines the splicing activity at discrete stages of the developmental programme (Sanford and Bruzik, 1999b).
In summary, we have used RNA interference to interfere selectively with SR protein gene expression in the nematode C.elegans. We have shown that CeSF2/ASF has unique properties and is essential for viability, and that functional redundancy exists for other nematode SR proteins. Future experiments combining RNAi with transgenesis will be used to determine the requirement for individual domains and to investigate further the role of individual SR proteins in splicing regulation.
Materials and methods
dsRNA preparation and microinjection
Templates for RNA synthesis were generated by PCR from C.elegans genomic DNA using gene-specific primers with T3 and T7 promoter sequences added on to forward (F) and reverse (R) primers, respectively. Where possible, the region amplified corresponds to a large exon or an exon-rich part of the gene. For CeSRp20, the whole coding region was amplified. Oligonucleotide primers were purchased from Genosys (Cambridge). PCR conditions using Vent DNA polymerase (New England Biolabs) were as follows: (i) 98°C 5 min, once; (ii) 98°C 30 s, 58°C 50 s, 72°C 1 min, 30 times; and (iii) 72°C 10 min, once.
The primers used were: T3 sequence: attaaccctcactaaagggaag; T7 sequence: taatacgactcactatagg; CeSRp75F: T3 + ggcagctcgaatttacatc; CeSRp75R: T7 + agagattctcgacgacgac; CeSRp40F: T3 + gtttgccaaatagagcatc; CeSRP40R: T7 + tgccaactgtaacgagtag; CeSF2/ASFF: T3 + cgagtgatcgttgaaggtc; CeSF2/ASFR: T7 + ctcatgtgatctgaacttgg; CeSF2/ASFF2: T3 + cggctcagaggaccaaaaag; CeSF2/ASFR2: T7 + actccacgagtgaactcgac; CeSC35F: T3 + caatggtctaacttcgctg; CeSC35R: T7 + tatcttggagatctggagc; CeSC35F2: T3 + atttcagatcccgctcacc; CeSC35R2: T7 + ttggaacggctacgacttg; CeSC35-2F: T3 + caattgtcacgaacgcgac; CeSC35-2R: T7 + cttctcggacgactttcac; CeSC35-2F2: T3 + acttcgcgcaaacatatcg; CeSC35-2R2: T7 + tttggagatcctggagacg; CeSRp20F: T3 + tggacgccaaggtgtacgtc; CeSRp20R: T7 + agtgcggagaagcagaacgg; CeHRH1F: T3 + aacgaaaccgttcaagaag; CeHRH1R: T7 + aatcaggcatatctgtggc; CeSRm160F: T3 + tctctgagcctcaacaaag; CeSRm160R: T7 + agctctgcctcttcatgac; CeTraF: T3 + atcgtgaaaatccacagcc; CeTraR: T7 + accgctatttccagatccg; CeSRPKF: T3 + atagaaccacgctgactcc; CeSRPKR: T7 + caaatggtaacaagaacgg; CeSRPKF2: T3 + gatggctcaatggcttcag; CeSRPKR2: T7 + gacacgacgccaaatcttc; CehnRNPA1F: T3 + tcaaacaccaccgatgacc; CehnRNPA1R: T7 + tttgaagaacgcattgatcc; CePTBF: cacccagcaacaaccaaac; CePTBR: T7 + ttggaaaccaggatgaccg; CeTRNSRF: T3 + atcaactcaaaatgcgctg; and CeTRN–SRR: T7 + atccaaagcataggccgtg.
PCR products were gel purified and used as templates for in vitro RNA synthesis with T3 and T7 RNA polymerase (Boehringer Mannheim) following instructions from the manufacturers. RNA was dissolved in sterile water with 0.4 U/μl RNase Inhibitor (Boehringer Mannheim) to reach a final concentration of 0.5 μg/μl. Double-stranded RNA was assembled by mixing equal amounts of sense and antisense RNA followed by incubation at 68°C for 10 min and then 37°C for 30 min. For each gene, 10–15 young adult hermaphrodites (Bristol strain N2) were injected with dsRNA into the gut or gonad and allowed to recover for 16 h. Then, animals were transferred onto individual plates, and the phenotype was observed in the F1 progeny generated after transfer. F1 progeny were scored for embryonic lethality, slow progression through larval stages, size of adults, abnormal organ development in adults, abnormalities in feeding and movements, and sterility. The affected progeny were examined using DIC microscopy.
RT–PCR
Total RNA from embryos was prepared as follows. Approximately 20 gravid hermaphrodites, either wild type or previously injected with dsRNA, were dissolved in a 1:10 solution of bleach in 1 M NaOH. Embryos were collected and washed twice in 1 ml of phosphate-buffered saline (PBS), pelleted and resuspended in 200 μl of 0.5% SDS, 5% β–mercaptoethanol, 10 mM EDTA, 10 mM Tris–HCl pH 7.5 and 0.5 mg/ml proteinase K. Samples were incubated at 55°C for 1 h, and processed further using Total RNA Isolation Reagent (Advanced Biotechnologies Ltd) following the manufacturer's instructions. RT–PCR was performed using SuperScript™ One-Step™ RT–PCR System (Gibco-BRL) following the manufacturer's instructions. To test the efficiency of RNAi treatment by RT–PCR, 100 μl reactions were prepared for wild-type and RNAi-treated embryos. These reactions were split into two identical fractions and RT–PCR analysis was performed to compare RNA levels corresponding to either a control SR gene or the gene targeted in the RNAi experiment. After 30 cycles of amplification, RT–PCR products were loaded on an ethidium bromide-stained agarose gel. Primers used for RT–PCR analysis were the same as those used for preparation of dsRNA fragments without the T3 and T7 sequences.
Splicing defects were analysed by RT–PCR as described above, using the following primers: hlh-1F: ggcaacaatgcgtgagagac; hlh-1R: ggttggaggagttgttcgtc; cpr-5F: agctctccgctattcttctc; cpr-5R: agtatggcttgcatccgaac; ama-1F: tgtcagtggctcatgtcgag; ama-1R: gggtcatctggcattccttc; topo-1F: atgagccaatggcatcagac; topo-1R: cgatagctcgaaaatcgcac; unc-52F: caagtgttcagctcacgttc; unc-52R: gctgtgcaagtgtagtctcc; uaf-1F: cttgtcaatcacgtcgtctc; and uaf-1R, catcagctttcagctcatcc.
Acknowledgments
Acknowledgements
We thank Ian Hope (University of Leeds) and Martin Taylor for discussions, and Kathryn Newton for technical help. We would like to acknowledge Tim Schedl (Washington University) for useful comments on the RNAi phenotypes displaying gonad and vulval defects. We are grateful to Nick Hastie, Wendy Bickmore and Gavin Screaton for critical reading of the manuscript. We acknowledge support from the Medical Research Council (J.F.C. and D.L.); I.L.J. was supported by an MRC Senior Fellowship.
References
- Baker B.S. and Ridge, K.A. (1980) Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics, 94, 383–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beil B., Screaton, G. and Stamm, S. (1997) Molecular cloning of htra2-β-1 and htra2-β-2, two human homologs of tra-2 generated by alternative splicing. DNA Cell Biol., 16, 679–690. [DOI] [PubMed] [Google Scholar]
- Ben-David Y., Bani, M.R., Chabot, B., De Koven, A. and Bernstein, A. (1992) Retroviral insertions downstream of the heterogeneous nuclear ribonucleoprotein A1 gene in erythroleukemia cells: evidence that A1 is not essential for cell growth. Mol. Cell. Biol., 12, 4449–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G., Buvoli, M., Bassi, M.T., Morandi, C., Cobianchi, F. and Riva, S. (1989) Isolation of an active gene encoding human hnRNP protein A1. Evidence for alternative splicing. J. Mol. Biol., 207, 491–503. [DOI] [PubMed] [Google Scholar]
- Bird D.M. and Riddle, D.L. (1989) Molecular cloning and sequencing of ama-1, the gene encoding the largest subunit of Caenorhabditis elegans RNA polymerase II. Mol. Cell. Biol., 9, 4119–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Kumar, S. and Krainer, A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Issner, R., Nickerson, J.A. and Sharp, P.A. (1998) A coactivator of pre-mRNA splicing. Genes Dev., 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T. (1998) Gene clusters and polycistronic transcription in eukaryotes. BioEssays, 20, 480–487. [DOI] [PubMed] [Google Scholar]
- Blumenthal T. and Thomas,J. (1998) Cis and trans mRNA splicing in C.elegans. Trends Genet., 4, 305–308. [DOI] [PubMed] [Google Scholar]
- Bruzik J.P. (1996) Splicing glue: a role for SR proteins in trans splicing? Microb. Pathogen., 21, 149–155. [DOI] [PubMed] [Google Scholar]
- Bruzik J.P. and Maniatis, T. (1995) Enhancer-dependent interaction between 5′ and 3′ splice sites in trans. Proc. Natl Acad. Sci. USA, 92, 7056–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres J.F. and Krainer,A.R. (1997) Mammalian pre-mRNA splicing factors. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. IRL Press at Oxford University Press, Oxford, UK, pp. 174–212. [Google Scholar]
- Cáceres J.F., Stamm, S., Helfman, D.M. and Krainer, A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- Cáceres J.F., Misteli, T., Screaton, G.R., Spector, D.L. and Krainer, A.R. (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol., 138, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Jamison, S.F., Garcia-Blanco, M.A. (1997) Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA, 3, 1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Mayeda, A., Krainer, A.R. and Zahler, A.M. (1999) hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J., 18, 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y., Popielarz, M., Fuchs, J.P., Gattoni, R. and Stevenin, J. (1994) Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J., 13, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.C. and Black, D.L. (1997) The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol., 17, 4667–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler S.D., Mayeda, A., Yeakley, J.M., Krainer, A.R. and Fu, X.D. (1997) RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc. Natl Acad. Sci. USA, 94, 3596–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary N., McMahon, C. and Blobel, G. (1991) Primary structure of a human arginine-rich nuclear protein that colocalizes with spliceosome components. Proc. Natl Acad. Sci. USA, 88, 8189–8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara M.D. and Reed, R. (1995) A two-step mechanism for 5′ and 3′ splice-site pairing. Nature, 375, 510–513. [DOI] [PubMed] [Google Scholar]
- Colwill K., Pawson, T., Andrews, B., Prasad, J., Manley, J.L., Bell, J.C. and Duncan, P.I. (1996) The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J., 15, 265–275. [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Olive, M., Gesnel, M.C. and Breathnach, R. (1999) hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol., 19, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., McGuffin, M.E., Dauwalder, B., Rabinow, L. and Mattox, W. (1998) Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol. Cell, 2, 741–750. [DOI] [PubMed] [Google Scholar]
- Fetzer S., Lauber, J., Will, C.L. and Luhrmann, R. (1997) The [U4/U6⋅U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA, 3, 344–355. [PMC free article] [PubMed] [Google Scholar]
- Fire A. (1999) RNA-triggered gene silencing. Trends Genet., 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fu X.D. (1993) Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature, 365, 82–85. [DOI] [PubMed] [Google Scholar]
- Fu X.D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- Gallego M.E., Gattoni, R., Stevenin, J., Marie, J., Expert-Bezancon, A. (1997) The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the β-tropomyosin alternative exon 6A. EMBO J., 16, 1772–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H. and Manley, J.L. (1990) A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell, 62, 25–34. [DOI] [PubMed] [Google Scholar]
- Ge H., Zuo, P. and Manley, J.L. (1991) Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell, 66, 373–382. [DOI] [PubMed] [Google Scholar]
- Gil A., Sharp, P.A., Jamison, S.F., Garcia-Blanco, M.A. (1991) Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev., 5, 1224–1236. [DOI] [PubMed] [Google Scholar]
- Gooding C., Roberts, G.C. and Smith, C.W. (1998) Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA, 4, 85–100. [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R. and Maniatis, T. (1998) Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell, 1, 765–771. [DOI] [PubMed] [Google Scholar]
- Gui J.F., Lane, W.S. and Fu, X.D. (1994a) A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature, 369, 678–682. [DOI] [PubMed] [Google Scholar]
- Gui J.F., Tronchere, H., Chandler, S.D. and Fu, X.D. (1994b) Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl Acad. Sci. USA, 91, 10824–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A., Cáceres, J.F., Mayeda, A., Franza, B.R., Jr and Krainer, A.R. (1998) Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA, 4, 430–444. [PMC free article] [PubMed] [Google Scholar]
- Hoffman B.E. and Lis, J.T. (2000) Pre-mRNA splicing by the essential Drosophila protein B52: tissue and target specificity. Mol. Cell. Biol., 20, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.P. (1999) Genetics: a touch of elegance with RNAi. Curr. Biol., 9, R440–R442. [DOI] [PubMed] [Google Scholar]
- Ismaili N., Perez-Morga, D., Walsh, P., Mayeda, A., Pays, A., Tebabi, P., Krainer, A.R. and Pays, E. (1999) Characterization of a SR protein from Trypanosoma brucei with homology to RNA-binding cis-splicing proteins. Mol. Biochem. Parasitol., 102, 103–115. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Jarmolowski, A., Beisel, C., Mattaj, I.W., Dreyfuss, G. and Fischer, U. (1997) A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J. Cell Biol., 137, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H. and Nielsen, P.J. (1997) The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J., 16, 5077–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H., Wei, G. and Nielsen, P.J. (1999) Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol., 9, 899–902. [DOI] [PubMed] [Google Scholar]
- Kataoka N., Bachorik, J.L. and Dreyfuss, G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol., 145, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J.R. and Carthew, R.W. (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- Krainer A.R., Conway, G.C. and Kozak, D. (1990) Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev., 4, 1158–1171. [DOI] [PubMed] [Google Scholar]
- Krainer A.R., Mayeda, A., Kozak, D. and Binns, G. (1991) Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell, 66, 383–394. [DOI] [PubMed] [Google Scholar]
- Krause M., Fire, A., White Harrison, S., Priess, J. and Weintraub, H. (1990) CeMyoD accumulation defines the body wall muscle cell fate during C.elegans embryogenesis. Cell, 63, 907–919. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi N., Onogi, H., Wakabayashi, T. and Hagiwara, M. (1998) Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem. Biophys. Res. Commun., 242, 357–364. [DOI] [PubMed] [Google Scholar]
- Labourier E., Bourbon, H.M., Gallouzi, I.E., Fostier, M., Allemand, E. and Tazi, J. (1999) Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev., 13, 740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche H., Dupuis, S., Ben-David, Y., Bani, M.R., Wellinger, R.J. and Chabot, B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nature Genet., 19, 199–202. [DOI] [PubMed] [Google Scholar]
- Larminie C.G. and Johnstone, I.L. (1996) Isolation and characterization of four developmentally regulated cathepsin β-like cysteine protease genes from the nematode Caenorhabditis elegans. DNA Cell Biol., 15, 75–82. [DOI] [PubMed] [Google Scholar]
- Lazar G., Schaal, T., Maniatis, T. and Goodman, H.M. (1995) Identification of a plant serine–arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc. Natl Acad. Sci. USA, 92, 7672–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Jang, Y.J., Koo, H.-S. (1998) Alternative splicing in the Caenorhabditis elegans DNA topoisomerase I gene. Biochim. Biophys. Acta, 1396, 207–214. [DOI] [PubMed] [Google Scholar]
- Lopato S., Waigmann, E. and Barta, A. (1996a) Characterization of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell, 8, 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S., Mayeda, A., Krainer, A.R. and Barta, A. (1996b) Pre-mRNA splicing in plants: characterization of Ser/Arg splicing factors. Proc. Natl Acad. Sci. USA, 93, 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A.R. and Barta, A. (1999) atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev., 13, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J.L. and Tacke, R. (1996) SR proteins and splicing control. Genes Dev., 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Krainer, A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Munroe, S.H., Cáceres, J.F. and Krainer, A.R. (1994) Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J., 13, 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A., Screaton, G.R., Chandler, S.D., Fu, X.D. and Krainer, A.R. (1999) Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol., 19, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud J.E., Cohen, P.T. and Lamond, A.I. (1994) Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J., 13, 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. (1999) RNA splicing: what has phosphorylation got to do with it?Curr. Biol., 9, R198–R200. [DOI] [PubMed] [Google Scholar]
- Misteli T., Cáceres, J.F., Clement, J.Q., Krainer, A.R., Wilkinson, M.F. and Spector, D.L. (1998) Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol., 143, 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery M.K., Xu, S. and Fire, A. (1998) RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 95, 15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen G.P., Rogalski, T.M., Bush, J.A., Gorji, P.R. and Moerman, D.G. (1999) Complex patterns mediate the spatial and temporal distribution of Perlecan/UNC–52 in Caenorhabditis elegans. Mol. Biol. Cell, 10, 3205–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan G.J., Guo, W., Wormsley, S. and Helfman, D.M. (1992) Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J. Biol. Chem., 267, 25480–25487. [PubMed] [Google Scholar]
- Ngo H., Tschudi, C., Gull, K. and Ullu, E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M. and Shimura, Y. (1996) A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev., 10, 997–1007. [DOI] [PubMed] [Google Scholar]
- Ono Y., Ohno, M. and Shimura, Y. (1994) Identification of a putative RNA helicase (HRH1), a human homolog of yeast Prp22. Mol. Cell. Biol., 14, 7611–7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J.G., Mayer, S.A., Tempst, P., Nadal-Ginard, B. (1991) Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev., 5, 1237–1251. [DOI] [PubMed] [Google Scholar]
- Peng X. and Mount, S.M. (1995) Genetic enhancement of RNA-processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol. Cell. Biol., 15, 6273–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad J., Colwill, K., Pawson, T. and Manley, J.L. (1999) The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol., 19, 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring H.Z. and Lis, J.T. (1994) The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol., 14, 7499–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F. et al. (1996)Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature, 381, 80–82. [DOI] [PubMed] [Google Scholar]
- Sanford J.R. and Bruzik, J.P. (1999a) SR proteins are required for nematode trans-splicing in vitro. RNA, 5, 918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R. and Bruzik, J.P. (1999b) Developmental regulation of SR protein phosphorylation and activity. Genes Dev., 13, 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton G.R., Cáceres, J.F., Mayeda, A., Bell, M.V., Plebanski, M., Jackson, D.G., Bell, J.I. and Krainer, A.R. (1995) Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J., 14, 4336–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.A. (1999) RNAi and double-strand RNA. Genes Dev., 13, 139–141. [PubMed] [Google Scholar]
- Soret J. et al. (1998)Characterization of SRp46, a novel human SR splicing factor encoded by a PR264/SC35 retropseudogene. Mol. Cell. Biol., 18, 4924–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J., Brooke, G., Kuersten, S., Lea, K. and Blumenthal, T. (1993) Operons in C.elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell, 73, 521–532. [DOI] [PubMed] [Google Scholar]
- Tabara H., Grishok, A. and Mello, C.C. (1998) RNAi in C.elegans: soaking in the genome sequence. Science, 282, 430–431. [DOI] [PubMed] [Google Scholar]
- Tacke R. and Manley, J.L. (1999) Determinants of SR protein specificity. Curr. Opin. Cell Biol., 11, 358–362. [DOI] [PubMed] [Google Scholar]
- Timmons L. and Fire, A. (1998) Specific interference by ingested dsRNA. Nature, 395, 854. [DOI] [PubMed] [Google Scholar]
- Valcárcel J. and Gebauer, F. (1997) Post-transcriptional regulation: the dawn of PTB. Curr. Biol., 7, R705–R708. [DOI] [PubMed] [Google Scholar]
- Valcárcel J. and Green, M.R. (1996) The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci., 21, 296–301. [PubMed] [Google Scholar]
- Voinnet O., Vain, P., Angell, S. and Baulcombe, D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Lin, W., Dyck, J.A., Yeakley, J.M., Songyang, Z., Cantley, L.C. and Fu, X.D. (1998) SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol., 140, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. and Manley, J.L. (1995) Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA, 1, 335–346. [PMC free article] [PubMed] [Google Scholar]
- Wang J., Takagaki, Y. and Manley, J.L. (1996) Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev., 10, 2588–2599. [DOI] [PubMed] [Google Scholar]
- Wang J., Xiao, S.H. and Manley, J.L. (1998) Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev., 12, 2222–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse P.M., Graham, M.W. and Wang, M.B. (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl Acad. Sci. USA, 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F., Zernicka-Goetz, M. (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nature Cell Biol., 2, 70–75. [DOI] [PubMed] [Google Scholar]
- Wilson R.K. (1999) How the worm was won. The C.elegans genome sequencing project. Trends Genet., 15, 51–58. [DOI] [PubMed] [Google Scholar]
- Xiao S.H. and Manley, J.L. (1997) Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev., 11, 334–344. [DOI] [PubMed] [Google Scholar]
- Xiao S.H. and Manley, J.L. (1998) Phosphorylation–dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J., 17, 6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Bani, M.R., Lu, S.-J., Rowan, S., Ben-David, Y. and Chabot, B. (1994) The A1 and A1B proteins of heterogenous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl Acad. Sci. USA, 91, 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A.M., Lane, W.S., Stolk, J.A. and Roth, M.B. (1992) SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev., 6, 837–847. [DOI] [PubMed] [Google Scholar]
- Zahler A.M., Neugebauer, K.M., Lane, W.S. and Roth, M.B. (1993) Distinct functions of SR proteins in alternative pre-mRNA splicing. Science, 260, 219–222. [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu, W. and Grabowski, P.J. (1999) Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA, 5, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D.A. and Blumenthal, T. (1999) U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA, 5, 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio D.A., Cheng, N.N., Blumenthal, T. and Spieth, J. (1994) Operons as a common form of chromosomal organization in C.elegans. Nature, 372, 270–272. [DOI] [PubMed] [Google Scholar]
- Zorio D.A., Lea, K. and Blumenthal, T. (1997) Cloning of Caenorhabditis U2AF65: an alternatively spliced RNA containing a novel exon. Mol. Cell. Biol., 17, 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu K., Sikes, M.L., Haynes, S.R. and Beyer, A.L. (1996) Altered levels of the Drosophila HRB87F/hp36 hnRNP protein have limited effects on alternative splicing in vivo. Mol. Biol. Cell, 7, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]