This article synthesizes discussions held during an internationalmeeting, “Surveillance forDecisionMaking: The Example of 2009 Pandemic Influenza A/H1N1,” held at Harvard School of Public Health in June 2010. It defines the needs for surveillance in terms of the key decisions thatmust be made in response to a pandemic: how large a response to mount and which control measures to implement, for whom, and when. The article describes the sources of surveillance and other population-based data that can form the basis for such evidence, and the interpretive tools needed to process raw surveillance data. It concludes with key lessons of the 2009 pandemic for designing and planning surveillance in the future.
Abstract
This article synthesizes and extends discussions held during an international meeting on “Surveillance for Decision Making: The Example of 2009 Pandemic Influenza A/H1N1,” held at the Center for Communicable Disease Dynamics (CCDD), Harvard School of Public Health, on June 14 and 15, 2010. The meeting involved local, national, and global health authorities and academics representing 7 countries on 4 continents. We define the needs for surveillance in terms of the key decisions that must be made in response to a pandemic: how large a response to mount and which control measures to implement, for whom, and when. In doing so, we specify the quantitative evidence required to make informed decisions. We then describe the sources of surveillance and other population-based data that can presently—or in the future—form the basis for such evidence, and the interpretive tools needed to process raw surveillance data. We describe other inputs to decision making besides epidemiologic and surveillance data, and we conclude with key lessons of the 2009 pandemic for designing and planning surveillance in the future.
The first year of the 2009-10 influenza A/H1N1 pandemic was the first test of the local, national, and global pandemic response plans developed since the reemergence of human cases of avian influenza H5N1 in 2003. The plans specified that response decisions be based on estimates of the transmissibility and, in some cases, the severity of the novel infection. Although public health surveillance provided a critical evidence base for many decisions made during this phase, its implementation also revealed the practical challenges of gathering representative data during an emerging pandemic1 and showed that such decisions, with their significant public health and economic consequences, must often be made before many key data are available.2
In this article, we reflect on the nature and timing of decisions made during the course of the first year of the pandemic and on the corresponding urgent need to gather surveillance data and process it into useful evidence. Our goal is to suggest how surveillance systems can be improved to provide better, more timely data to estimate key parameters of a future pandemic, with the goal of improving management of that pandemic. We also attempt to identify the human and technical capabilities needed to process and interpret these data to maximize their value for decision makers. While we cite examples from many countries, we focus to some extent in this article on the United States experience. More generally, no attempt has been made to be exhaustive in citing the literature, which includes more than 2,000 references with just the keywords “H1N1 AND surveillance” between the start of the pandemic and the end of 2010.
We use the term surveillance here to encompass ongoing monitoring of disease activity and gathering of clinical and epidemiologic data that define the disease, such as comparative severity in different risk groups. In section 1 we describe the key decisions made during the 2009 pandemic, emphasizing early decisions about matters of broad strategy and tactics,3 and their evidentiary inputs. Section 2 identifies sources of surveillance data. In section 3 we discuss methods of processing surveillance data into usable evidence, and in section 4 we examine inputs, other than surveillance or epidemiologic inputs, that can inform, or indeed disrupt, public health decisions. Section 5 concludes with lessons learned for future pandemics.
1. Pandemic Response Decisions and Their Evidentiary Inputs
1.1 Decision to Respond
As a novel influenza virus emerges, the first set of decisions required of global, national, and local authorities involves whether to progress beyond routine monitoring and devote resources to a large-scale response. The World Health Organization (WHO) and ProMED-mail together report more than 3,000 alerts of human infectious diseases annually.4 Consequently, devoting extraordinary public health resources to tracking and preparing a response must depend on the estimated risk that the outbreak will reach a given jurisdiction and cause widespread, serious illness. Unfortunately, the extent of transmission—and therefore the severity of the disease—may be unclear during the early stages of a pandemic. For example, infection in Mexico was already widespread by late April 2009 when the link was made between the unusual cases of pneumonia reported in March and April and a novel strain of influenza.5,6 Specialized laboratory tests and epidemiologic follow-up of individual cases ultimately provided the critical information that confirmed the novelty of the H1N1 virus and its presence in Mexico and U.S. border states.
1.2 Overall Scale of Response
Once a novel strain of influenza establishes widespread human-to-human transmission, global spread will be rapid, warranting an escalated response. This may include deploying or reassigning public health workers to pandemic-related activities, acquiring and deploying supplies (such as antivirals, antibiotics, vaccines, ventilators, and personal protective equipment), and commencing nonpharmaceutical interventions (eg, school closures, border screenings). For each of these measures, decision makers must balance the costs against the likely benefits, both of which depend on the epidemiologic characteristics of the novel infection and on the expected scale of damage arising from an unmitigated pandemic—estimated as the number of individuals infected multiplied by the probability that each infection will lead to severe illness, hospitalization, or death. Predicting the timing of such events could also inform decision making. Control measures may be cost-effective, for example, if they avert a sharp, acute increase in demand on healthcare services in a population, but less so if an unmitigated pandemic were less peaked and hence less disruptive.
Rapidly generated transmissibility and severity estimates are essential for predicting the scale and time course of a pandemic.7 With influenza, most important are measures of severity per infected individual—that is, the probability of death, hospitalization, or other severe outcome. Historically, influenza pandemics have led to symptomatic infection in between 15% and 40% of the population.8 This variability is minor compared with variation in severity per symptomatic case as measured, for example, by the case-fatality ratio, which varies by orders of magnitude among pandemics. Thus, a key task for surveillance early in a pandemic is to estimate the per-infection severity of the new pandemic strain precisely enough to define the appropriate scale of response. This proved challenging in 2009 up until late summer, because early estimates indicated an uncertainty ranging from the mildest possible pandemic envisaged in preparedness planning to a level of severity requiring highly stringent interventions.5,9–12 As we discuss further in section 4.1, another form of uncertainty was whether the severity, drug-sensitivity, or other characteristics of the infection might change as the pandemic progressed; unlike the first form of uncertainty, this question could not be resolved using better data.
As a pandemic unfolds, surveillance data and epidemiologic studies may provide early indications of the impact of public health interventions. Ideally, data on the economic costs (including indirect costs for socially disruptive measures such as school dismissals) and the public health and economic benefits of interventions would be formally weighed within a cost-benefit or cost-effectiveness framework to inform policy decisions. Yet, we know of no such evaluations having been undertaken during a pandemic. Instead, more informal comparisons have typically informed decision making, including in 2009.
1.3 Measures to Protect Individuals
Another set of decisions involves interventions to protect individuals against infection and against severe morbidity or mortality following infection. Guidelines or policies are needed to help clinicians and health systems determine whom to test for infection, whom to treat and under what circumstances, whom to advise on early treatment when a high risk of complications exists, and whether to make antiviral drugs more readily available for such individuals.13 Decisions about how much vaccine to purchase are based in part on the severity of infection in population subgroups and on ensuring adequate supplies for at least those in greatest need.
Comparative severity of infection is estimated from risk factor studies, in which the frequency of particular demographic characteristics or particular comorbidities in the general population is compared with the frequency among people with severe outcomes (death, ICU admission, hospitalization).14–16 Ideally, one would also have estimates of the effectiveness of prevention and treatment in the high-risk groups, since prioritizing groups for intervention presupposes that the interventions will help them. In practice, data on intervention effectiveness in high-risk groups may be difficult to obtain in time, but may be inferred from experience with seasonal influenza,14,17 despite epidemiologic differences between pandemic and seasonal disease.
1.4 Measures to Slow Transmission
Decisions also must be made about interventions to prevent transmission. In the early phases of a pandemic, these may include screening travelers as they enter or leave a jurisdiction and isolating those who are symptomatic. Mathematical models indicate that such measures are unlikely to delay global spread of influenza by more than about 2 weeks.18–20 In 2009, some jurisdictions judged this delay adequate to justify stringent border controls, and evidence shows that the controls measurably delayed the start of local transmission of the H1N1 virus in several regions.21 Once community transmission is established, however, the measures are less useful, so a key role for surveillance is to inform decision makers about the extent of infection within their jurisdictions.
Other nonpharmaceutical interventions to slow transmission in a community include school dismissals and cancellation of public gatherings; these measures are often undertaken reactively based on transmission in an individual school or district. These decisions specifically rely on fine-grained local data (see sidebar: Local Surveillance Data).
Local Surveillance Data: When Is It Necessary?
In an ideal situation, every public health authority could access its own high-quality, local data to synthesize into evidence relevant for its disease control decisions. Gathering, processing, and interpreting data, however, cost money, time, and expertise, and few jurisdictions worldwide can undertake these activities independently. Decision makers must instead rely on a heterogeneous mix of local, regional, and global data sources.
Local authorities, particularly elected officials, may want data on the progress of the local epidemic for a variety of reasons. In addition, from a public health and evidence-based decision-making perspective, 3 basic arguments favor geographically distributed surveillance.
First, infection may remain geographically focal for weeks to months. In 2009, transmission remained focal within Mexico for at least 1 month, probably longer. Once the virus spread to other countries, transmission was again initially confined to certain areas. When early transmission is geographically circumscribed, the first estimates of key parameters that serve to inform decision makers worldwide must rely on data from areas of ongoing transmission. In the UK, even by the end of the summer 2009 wave of transmission, seroprevalence was higher in London and the West Midlands than elsewhere.65
Second, even when infection spreads virtually everywhere, its characteristics may differ across regions, resulting in different local priorities for control. In 2009, many risk factors for severe outcome—race/ethnicity,45,135 income,35 comorbidities,14,135 healthcare access,14,135 and exposure to bacterial co-infections76—varied geographically on scales ranging from neighborhood to continent. Global awareness of these and other risk factors depends on having some form of surveillance available in populations where risk factors are concentrated. Awareness of local disparities in infection rates or severe disease35,71 within a jurisdiction can also improve resource allocation.
Third, some decisions, such as school dismissals (and reopenings) in response to within-school transmission, require very detailed, real-time local data. During the 2009 pandemic, these decisions had to be made based on limited data with known biases. One example is school absences, which can be due either to influenza infection or to fear of acquiring influenza at school. More generally, decisions about specific tactics to control transmission in particular settings—such as schools, hospitals, or other institutions, or aircraft or other vehicles—necessarily rely on local data. Surveillance to trigger and guide such interventions has been called “control-focused” surveillance, in contrast to the “strategy-focused” surveillance3 that is discussed throughout most of this article.
These factors favoring local data must be balanced against resource limitations and competing public health priorities. For many questions—specifically, those for estimating overall severity—high-quality data from within a country or a group of countries with broadly similar health systems are likely adequate and often more reliable than local data from an individual jurisdiction. As noted in section 3.1, sharing of data across hemispheres can be particularly valuable, given the seasonality of transmission.86,87
To avoid making misleading comparisons, consumers of local surveillance data should also know the factors that differ among jurisdictions. In the spring-summer of 2009, the city of Milwaukee, Wisconsin, confirmed about 3 times as many cases as did New York City, even though New York has about 8 times the population. This was due not to a greater incidence in Milwaukee, but to different decisions regarding whom to test. Milwaukee tested many mildly ill individuals, while New York City focused on hospitalized cases.44 Clearly, without knowledge of surveillance differences, it is easy to misinterpret differences in case numbers.
The above considerations suggest that to prepare for future pandemics:
decision makers should be educated about the limitations of local data and the cost-benefit trade-off of gathering high-quality data at the local level;
national public health agencies should maintain the epidemiologic and laboratory capacity to study focal outbreaks, including those in areas that lack high-quality routine surveillance or local capacity for such investigations; and
high-quality routine surveillance systems should be geographically distributed within and among countries to improve the likelihood that some systems will be in place in populations that experience early waves of infection. This approach should improve the timeliness of estimates of key parameters for national or international use.
Vaccination can also be used to slow transmission and protect nonvaccinated people. Appropriately targeting vaccines for this purpose requires identifying the groups most likely to become infected and then infect others in the population.22,23 Although school-age children play a key role in this process, their importance often declines as a pandemic wave progresses. Changes in the age distribution of cases can help track this trend.22
1.5 Investment Allocation
Decision makers must allocate limited resources between measures to protect individuals and measures to reduce transmission. In addition to money, resource limitations can involve public health personnel, public attention, and supplies. For example, the supply of antiviral drugs is limited, as is the virus's susceptibility to them, which can also be considered a limited “resource.”24 Consequently, using antivirals to slow transmission may hinder their future use for treatment.19,25
Limited vaccine supplies create a trade-off between vaccinating those at highest risk and those most likely to transmit infection (see sidebar: Prioritizing Vaccination). The key questions for prioritization are: Who is at highest risk, and how readily can they be identified? On the other hand, who are the transmitters? When vaccine becomes available in substantial quantities, will transmission be ongoing at a high level? Will the group driving transmission early in the pandemic (eg, schoolchildren) still be the key driver, or will susceptible members of that group have been largely exhausted, making their vaccination less effective?22
Prioritizing Vaccination.
Vaccines have 2 effects: direct protection of the vaccinated individual against infection and its consequences and indirect protection of the population, in which certain individuals are vaccinated to reduce their risk of becoming infected and subsequently passing on the infection. The ability of immune individuals to protect others against infection is often called herd immunity.
When vaccine availability is limited, the choice of whom to vaccinate first is partly a strategic decision to focus on either direct protection or on herd immunity. Table 1 describes key differences in these strategies and in the information required to implement them.
Predictions of the likely timing and magnitude of the peak (or peaks) of disease incidence would also facilitate response planning and resource allocation by anticipating likely periods of intense stress on healthcare providers. Optimally targeting vaccination depends on the number of doses available prior to the peak of transmission (see sidebar: Prioritizing Vaccination). Raw surveillance data by definition provides information about the past, not predictions of the future course of the pandemic. We argue in section 3.4, however, that carefully designed surveillance programs, combined with mathematical and statistical modeling to infer the number of infected individuals, could provide considerable insight and help decision makers prioritize particular scenarios.
Decisions about prioritizing interventions will need to balance projected benefits of prioritizing particular groups against important considerations of logistics and public acceptance. As in all matters of public health prioritization, the calculation of projected benefits depends on one's assumptions about the relative value of preventing morbidity versus mortality, and about the relative value of preventing mortality in various groups. This question has been heavily debated,26 and, in the case of pandemic influenza, it is clear that unless vaccines are so plentiful that transmission can be completely or nearly halted,27 policies to minimize total mortality may differ from those to minimize years of life lost or disability-adjusted years of life lost.28–30 Moreover, efforts to target particular groups may result in underuse of available supplies, be difficult to implement, or provoke negative public response if some individuals disagree with the choice of whom to prioritize. In the U.S., a public engagement process in 2006 documented public preferences for groups that should be prioritized in the event of a pandemic.31
1.6 Timing of Responses
Finally, decision makers must determine when to set policies in motion, when to change existing policies, and which decisions to delay. A key lesson of the 1976 swine flu outbreak was that certain decisions can and should be delayed until evidence accumulates.32 Decisions that cannot wait can be revised in light of new evidence on severity and intervention effectiveness. During the 2009 pandemic, for example, early decisions to close schools in the U.S. in April were quickly revised as evidence grew to suggest an illness severity in the U.S. lower than that first reported in Mexico. (Later, it became clear that the severity in Mexico was also lower than it initially appeared.33)
Table 1.
Considerations in the Use of Vaccines for Direct Protection of Vulnerable People vs. Herd Immunity
| Strategy | ||
|---|---|---|
| Direct Protection | Herd Immunity | |
| Goal | To protect the vaccinated directly against infection, illness, hospitalization, or death | To protect the population (including the nonvaccinated) against infection (and its consequences) by reducing transmission |
| Criteria for who should receive priority for vaccination | Individuals who will benefit most from the vaccine's effects: groups at high per capita risk of severe outcomes (infection risk × severity) | Individuals at high risk of becoming infected and transmitting infection to others (initially, schoolchildren; also, certain healthcare workers) |
| Data used to identify priority groups | Predictors of high risk of severe outcome (eg, risk factors for death or hospitalization, compared with the general population) Evidence that the vaccine is effective in the high-risk groups (difficult to obtain in the pandemic setting, but possible to extrapolate from seasonal vaccines) |
Incidence rate and/or force of infection by age group22 Estimates of potentially infectious contacts per day in different groups136,137 |
| Factors favoring the strategy | Convincing data on who is at highest risk (Note: Predictors of high risk need not be causal, only reliable markers of high risk.) Good immunogenicity of the vaccine in the high-risk groups Limited quantities of vaccine Late availability of vaccine |
High-risk groups are unknown or vaccine has limited effectiveness in them (eg, the elderly during seasonal influenza17) Large supplies of vaccine available in time to significantly reduce transmission7,22,23 Evidence available on the key transmitters22 (Note: This may change over time, as most affected groups become increasingly immune and contribute less to transmission as the epidemic progresses.) |
2. Sources of Surveillance and Epidemiologic Data
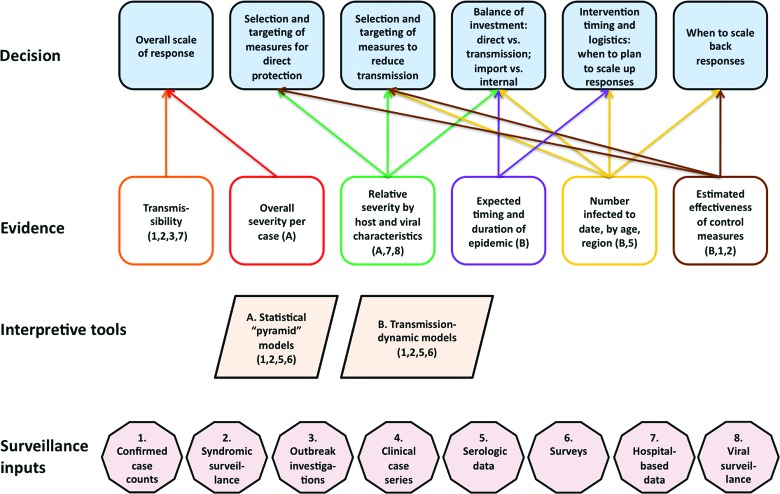
This section describes data sources—currently or potentially available—that provide the evidentiary basis for the decisions outlined in section 1 and shown in Figure 1.
Figure 1.
A schematic view of the public health decisions required in a pandemic response, the evidence needed to make these decisions in an informed fashion, and the sources of data and interpretive tools necessary to generate this evidence. The numbers under “surveillance inputs” follow the order of section 2. Color images available online at www.liebertonline.com/bsp
2.1. Confirmed Cases
Awareness of the novel influenza strain first arose from its detection in young patients in California who presented with influenzalike illness (ILI) and were tested for influenza as part of routine surveillance. Soon after, laboratory data on cases of severe atypical pneumonia in young adults in Mexico confirmed the presence of the pandemic strain of H1N1 (pH1N1). These early cases confirmed that pH1N1 could cause severe lower respiratory tract infection in young adults, a characteristic of previous pandemics.
The earliest quantitative surveillance data specific to 2009 H1N1 were daily reports of laboratory-confirmed or probable cases of the infection. As public awareness spread and more laboratories acquired the capacity to test for the new virus, testing in many jurisdictions, including the U.S., became increasingly common, including for mild cases. By one estimate, the fraction of cases detected in the U.S. was increasing 10% per day during late April to early May.34
Case reports to public health officials in some countries included age, gender, comorbidities, outcome (ie, hospitalization, ICU admission, recovery, or death), date of symptom onset, geographic location, and the like. By mid-May, however, the proportion of cases tested had declined in the U.S. because of testing fatigue and a lack of resources to consistently test a growing case burden.1 Similar changes occurred worldwide, prompting WHO to recommend the cessation of routine testing of all suspect cases. Counting confirmed cases of hospitalized, ICU-admitted, or fatal H1N1 infection became more feasible and thus the focus in some systems, including the U.S. Emerging Infections Program (hospitalizations), New York City,35,36 and the later phases of surveillance in Hong Kong.35–37
As case counts grew, aggregate reporting replaced individual case reports in most jurisdictions, so details of individual patients were often no longer available. Furthermore, most symptomatic cases were not tested, confirmed, or reported,38 and the proportion tested varied geographically and over time.
Early in the 2009 pandemic, population-wide case count data were useful for estimating transmissibility5,34,39–43 and served as a key input for WHO's declaration of Pandemic Phase 5 and the decision by many countries to undertake a large-scale response. These data were also used to make initial severity estimates,11,44,45 although biases were recognized, leading to considerable uncertainty in the estimates. Combining case counts among travelers to Mexico returning to their home countries with estimates of travel volume and assumptions about their exposure led to early estimates that the number of confirmed cases in Mexico was several orders of magnitude lower than the total number of infections.5,6 This showed that severity was considerably lower than a simple ratio of deaths to confirmed cases would have suggested. Overall, however, the varied rates of testing and reporting reduced the usefulness of case count data alone to estimate key parameters.
2.2. Syndromic Surveillance
Perhaps the most widely used sources of data for monitoring the course of the 2009 pandemic were syndromic surveillance systems, which track visits to primary care providers or emergency departments for a defined syndrome, such as influenzalike illness or acute respiratory illness.
Some systems, including ILINet in the U.S. and similar systems elsewhere, routinely track seasonal and pandemic influenza. Others, such as emergency department surveillance systems, were originally designed to detect natural disease outbreaks and acts of bioterrorism. To our knowledge, the earliest appearance of the pandemic did not trigger a quantitative alert in any of these systems, although 4 of the earliest cases in the U.S. presented at providers who were members of ILINet and so were tested and flagged for attention. At a later stage of pandemic spread, both the purpose-built influenza syndromic surveillance systems and the more general, detection-oriented systems proved very valuable for tracking the relative level of ILI over time, across age groups, and across geographic areas—data that were important for many of the decisions described in section 1. With several assumptions, ILI surveillance can be transformed into symptomatic influenza case estimates (see sidebar: From Syndromic Surveillance to Estimates of Symptomatic Influenza).
From Syndromic Surveillance to Estimates of Symptomatic Influenza.
Can syndromic data be used to make reliable estimates of influenza-attributable symptomatic disease? The answer depends on one's ability to accurately estimate several key quantities that define the relationship between syndromic surveillance outputs and the underlying number of pandemic influenza-attributable symptomatic infections. These quantities may change over time, so a system that provides reliable estimates in one situation may or may not remain reliable from month to month.
Here we make these relationships explicit and identify the key quantities that must be estimated to convert syndromic data into estimates of pandemic influenza-attributable symptomatic illness. In this box, the term symptomatic is used to mean “meeting the definition of influenzalike illness (ILI): fever, and either cough or sore throat.”
We define the following quantities for week w:
Cw = the number of ILI consultations per 100,000 population per week
Fw = the number of people with ILI whose symptoms are caused by pandemic influenza per 100,000 persons per week
Pw = the probability that an individual with ILI caused by pandemic influenza seeks medical attention and is diagnosed with ILI
Nw = the number of individuals with ILI seeking care per 100,000 population per week, whose symptoms are not caused by pandemic influenza
The relationship among these quantities is:
Cw = Fw Pw + Nw
where Fw is the number we would like to estimate, since this represents the true incidence of ILI due to pandemic influenza.
Cw can be measured using data from a variety of sources, such as the sentinel systems in France,138 the United Kingdom,139 and New Zealand,45 and other general-purpose systems, such as those based in health maintenance organizations in the U.S. Weekly data on the proportion of ILI among primary care consultations can be obtained from systems like the ILINet in the U.S.; unlike the population-based systems mentioned above, this is not an incidence rate.
In settings where an incidence rate Cw of ILI consultations is available, the challenge in determining Fw is to estimate Pw and Nw.
Nw may be negligible, especially in adults, if the pandemic occurs outside the normal winter respiratory infection season, as happened in the northern hemisphere in 2009. However, this need not be the case. In Mexico, the start of the pandemic overlapped with the end of seasonal influenza, and in the southern hemisphere, pandemic H1N1 transmission coincided with the normal winter season.
Pw measures the probability that a symptomatic individual in week w who is truly infected with pandemic influenza seeks care and is diagnosed with ILI. Pw appears to vary, at least geographically and possibly over time, perhaps in response to levels of public concern. In the U.S., Pw has been consistently estimated at about 40% to 60%;38 during the 2009 pandemic in New Zealand, by contrast, it was estimated at 5.5%.45
However, at times of greater public concern about influenza, both Pw and Nw—the incidence of consultations for ILI caused by something other than actual pandemic influenza—will rise. Thus, for example, New York City saw spikes in consultations by the worried ill, and sometimes by the worried well, following news reports about outbreaks or deaths. The use of specific case definitions—for example, the requirement for measured fever to define ILI—should reduce variability due to the worried well but does not prevent changes in syndromic counts due to increases in the number of worried ill—that is, increases in Pw and Nw. Even more specific case definitions, such as emergency department consultation resulting in hospital admission for ILI, should also reduce the impact of the worried ill; this approach was used in New York City in 2009.
One approach to estimate Fw is to make assumptions (based on telephone or web surveys and knowledge of other causes of ILI) about Pw and Nw.
Another approach is to estimate weekly the proportion of all medically attended ILI caused by pandemic influenza. In our notation, this proportion is Fw/C w. This proportion can be estimated by testing a representative subset of symptomatic individuals meeting the syndromic case definition (here, medically attended ILI) for pandemic influenza infection.1 Such an estimate can give a consistent relative measure of symptomatic pandemic influenza infection, but not a rate per population, since it does not directly estimate Pw. To limit the laboratory burden, the total number of cases tested must be limited, even as case numbers grow (perhaps by testing a fixed number of random samples weekly). Even so, testing will be limited to populations where sufficient laboratory capacity is available.
Syndromic measures of ILI consultations are easy to understand, relatively inexpensive and scalable. In the U.S., coverage of emergency department surveillance increased significantly during the pandemic because multiple jurisdictions contributed age-stratified data to the Distribute Network,46 which was updated in nearly real-time on the Web. Syndromic surveillance also was used to compare trends in medically attended ILI cases in 2 neighboring jurisdictions, one of which implemented school dismissal while the other did not, to make a nearly real-time assessment of whether school dismissal affected transmission.47
The 2009 experience did provide 2 clear examples where syndromic data were misleading. The first was a brief surge in ILI encounters observed in many syndromic systems in the U.S. during weeks 17 and 18, coinciding with intense media coverage of early cases and outbreaks. Syndromic data are sensitive to changes in healthcare-seeking behavior, and the tendency of mildly ill patients—the so-called worried ill or worried well—to seek care during periods of heightened concern can trigger false signals.48 In addition to being subject to misinterpretation, these false alarms perturb natural baseline patterns in the data, making it more difficult to detect subsequent increases representing real illness. In some jurisdictions during spring 2009, the initial surge in worried well visits was still subsiding just as ILI activity began accelerating, making these systems less useful for detecting the onset of community-wide illness.
A second example was the temporary slowing in the growth of ILI activity in the U.S. around week 38 (mid-September).49 Data gathered from most U.S. regions for that time showed the trend in consultations for ILI becoming almost flat over a 2- to 3-week period before accelerating to reach a true peak about 4 weeks later. This false peak remains unexplained, but it was credible at the time because of its replication throughout the country.
2.3. Outbreak Investigations
Investigations of outbreaks in defined populations are a classic tool of public health practice, usually designed to discover the cause of an outbreak, identify risk factors, and assess intervention effectiveness. In an influenza pandemic, however, outbreak investigations offer key advantages over routine, population-based surveillance in defining characteristics of the new infection. They can identify focal pockets of infectious transmission weeks or months before the eventual global spread of the infection, since initial seeding into particular geographic areas occurs with the arrival of 1 or a few infected people, and transmission may be concentrated in particular groups, such as schools or universities. In the 2009 pandemic, for example, early outbreaks were observed in contained settings such as schools,50–53 military camps,54 and universities.55
Outbreak investigations in sufficiently large but localized populations can also provide relatively unbiased estimates of case severity, because severe outcomes (hospitalizations, deaths: the numerators for severity estimates) can be determined with high reliability, while symptomatic attack rates (the denominator) can be estimated using surveys. One of the earliest compelling indications of the relatively low symptomatic case-fatality and case-hospitalization ratios in young adults in 2009 came from the University of Delaware outbreak.55 Such studies provide rapid, reliable data, although estimates may be limited to certain demographic groups.
Another advantage of studying outbreaks, particularly in nonresidential settings, is that once a case is identified, household contacts can be tested for infection (through virus detection and serology), monitored for symptoms and outcomes, and later tested again serologically. Viral shedding or serologic data, or both, can be combined with symptom data to estimate the proportion of infectious or infected individuals with particular symptom profiles, including asymptomatic infection, and thus aid in evaluating case definitions. These prospective studies can also estimate the distribution of shedding times.56,57 As noted earlier, confirmed cases identified as a result of people having sought medical care often represent a biased sample skewed toward severe cases. Identifying possible cases through exposure, such as by household contact tracing, yields a less biased sample. In 2009, a study of household contacts of cases from a Pennsylvania school outbreak provided estimates of household transmission rates and further evidence on severity.58
Outbreak investigations could be improved through advanced planning of study designs and by having epidemiologically trained personnel conduct the studies. Since not every outbreak can be fully investigated, planners should concentrate resources on investigations likely to generate the most useful and generalizable data. Investigation plans should also be adaptable to the location judged most likely to be informative, which cannot be predicted until the pandemic is under way. Collaborations between public health authorities and experts in statistical analysis of outbreak data58 can help quantify epidemiologic parameters of the pandemic and define how they are used to model the future spread of the infection.
2.4. Clinical Case Series
Descriptive data on mild, hospitalized, and fatal cases are valuable for many of the evidentiary needs described in section 1. These data comprise line lists, with each line describing at least an individual's demographics (age, sex, place of residence), preexisting medical conditions, and outcome, and ideally including data on the course of the illness and its treatment. Data for severe cases are collected by state and local health departments and often come from hospital records,14,35 while data for milder cases could be obtained from primary care providers, albeit with more difficulty in a setting facing intense healthcare demands.
Assessing risk factors requires data on the frequency of the same preexisting conditions and demographic traits in the general population. Remarkably, comorbidity frequencies in the general population are often hard to obtain, even in resource-rich countries, particularly for rare diseases (such as neurologic disorders) or for co-occurrence of more than one condition.59 Nonetheless, combining comorbidity data from a few hundred cases with estimates of population frequencies can suggest the factors associated with large excess risks. In addition, these hospitalized case series provide one element of the severity “pyramid” (section 3.1) by defining the proportion of hospitalized cases that require ICU admission or mechanical ventilation, and the proportion that are fatal.
2.5. Serologic Data
As described in section 1.3, measures of severity per infected individual are extremely valuable for informing decisions about the scale and targeting of response to an emerging pandemic. To estimate severity per infected individual, it is of course necessary to estimate the number of infected people. The gold standard for detecting infection is testing paired serum samples (preexposure and convalescent on the same person) for virus neutralization or, more often, hemagglutination inhibition.60 The rapid initiation of prospective studies required to collect paired samples is challenging in a pandemic. Therefore, cross-sectional studies of convalescent sera provide a viable alternative. Although other kinds of data—estimates of the number of symptomatic, medically attended, or virologically confirmed infections in a population—are useful for estimating the number of infections, these measures are difficult to interpret because the “multiplier” relating any of them to true infections likely varies by time, population, and characteristics (eg, age) within a population. All the sources of uncertainty noted in the sidebar on syndromic surveillance contribute to uncertainty in the ratio between serological infections and symptomatic or medically attended infections. If serologic data are not available, these sources of uncertainty combine with uncertainty about the proportion of symptomatic infections to result in wide confidence bounds on the ratio of severe outcomes to infection.61
Another potentially highly valuable use of serologic data is to estimate parameters for transmission-dynamic models to project the course of an epidemic.62,63 Transmissibility can be estimated from the early growth rate of case numbers, which does not depend on the proportion of cases reported (as long as that proportion stays constant or changes are accounted for34,37). To determine the timing and magnitude of the peak in incidence, the estimated transmissibility must be combined with an estimate of the absolute number of individuals in age groups who have been infected at a particular time. If sufficient resources are available, the number infected can be measured continuously in almost real time by an ongoing serologic study.64 More economically, one could estimate, at one point for a defined population, the proportion of true infections (serologically determined) that result in medically attended illness, hospitalization, or other more convenient surveillance measure. Syndromic or hospitalization surveillance can thereafter be a proxy for ongoing serological surveillance.
The 2009 pandemic illustrated the challenges of using serology to detect infection. Development of specific serologic markers of pandemic H1N1 infection was hampered by cross-reactions with antibodies resulting from prior seasonal influenza infection. In addition, the experts needed to develop and optimize such assays were in demand for other tasks, including developing assays for vaccine candidate evaluation. Nevertheless, serologic data were obtained on statistically meaningful samples of the population in the United Kingdom65 and Hong Kong.56,64 Workers in Hong Kong gathered symptomatic and serologic data within the same defined population, in the context of a household study56 that yielded a multiplier between symptomatic and serologic cases. More detailed quasi-population-based serologic studies have been described from Hong Kong in recent meetings.64 The value of improving the capacity to conduct large-scale serosurveillance in many populations for future pandemics engendered lively debate at the Symposium, as it has in other recent influenza meetings (see sidebar: Debating the Value of Large-Scale Serosurveillance). For early estimates of seroprevalence, serosurveys may be needed in populations where laboratory capacity for processing samples is absent or inadequate. Collaborative arrangements may be needed (and ideally should be set up in advance) to ensure timely processing of samples and dissemination of results.
Debating the Value of Large-Scale Serosurveillance.
Perhaps the most spirited debate at the CCDD Symposium and, indeed, among the authors of this report, concerned the priority and feasibility of conducting serosurveillance during a pandemic. Both sides agree on the importance of estimating the cumulative incidence of infection over time as the pandemic unfolds, for reasons described in sections 1 and 2. The argument in favor of serosurveillance61 emphasizes that serologic testing is the gold standard for such estimates and that all other approaches suffer from considerable uncertainty. The argument against serosurveillance emphasizes the expense and logistical difficulty of wide-scale serologic testing, the unavailability and limitations of early serologic tests (which add uncertainty to estimates), and the possibility of obtaining estimates of infection from nonserologic sources. In short, a large-scale prepandemic investment would be needed to improve current influenza serologic assay technology sufficiently so that valid serologic tests could be developed quickly at the start of the next pandemic.
As noted, challenges in serologic testing include: the need to obtain ethical approval for serologic testing in some locations; poor sensitivity and specificity of tests for some novel viruses (including many early tests for 2009 H1N1); the variable time to seroconversion, which adds variance to estimates of the proportion positive at any one time; and the labor involved in extensive testing, especially repeat testing. Proponents of serosurveillance note that statistical models can be used to adjust for the sensitivity, specificity, and variation in time to seroconversion; in fact, work is under way on such models. They also stress that tests with low sensitivity and specificity, which might be poor tools for clinical diagnosis of an individual, can still provide valuable information about the proportion of the population infected, given the appropriate statistical adjustment.
With regard to using nonserologic approaches to estimating the fraction of the population already infected (eg, data on patient visits), supporters of serosurveillance note that in 2009 such efforts resulted in broad uncertainty spanning several orders of magnitude and lasting until just before the peak of transmission, at which point predicting the peak was no longer very useful.62,63 However, those skeptical of committing resources to serosurveillance argue that these calculations could have had narrower ranges of uncertainty. They note that in pandemic and seasonal flu, the proportion of infected people who are asymptomatic or whose symptoms fall below the standard definition of ILI has been repeatedly estimated at between 25% and 75%,140–147 thereby defining a limited range for the conversion factor between estimates of population-based, influenza-attributable ILI incidence (cases per capita) and estimates of infection incidence. In the U.S., estimates of the incidence of symptomatic pandemic influenza had a range of uncertainty of about 2.5-fold.81 Combining this with an uncertainty of about 3-fold in the multiplier between symptomatic cases and infections, one obtains about a 4-fold uncertainty in the number of infections.
Further work is needed, perhaps based on data from the 2009 pandemic, to assess how well such proxies can approximate retrospectively collected serologic data, as well as how nonserologic data sources could be improved to optimize the measurement of absolute incidence. In addition to the immediate benefits of serologic surveillance, the benefits to future epidemiologic studies of being able to retrospectively track the spread of infection in a population should be considered. The costs and benefits of enhanced nonserologic surveillance providing such proxies could then be compared against those for serologic surveillance.
2.6. Telephone and Web-Based Surveys
Telephone surveys, including the U.S. Centers for Disease Control and Prevention's (CDC's) nationwide Behavioral Risk Factors Surveillance System (BRFSS),66 and a more limited telephone survey focused on ILI in New York City,67 provided estimates of several quantities of interest to decision makers, including symptomatic infection incidence over a month-long recall window, the probability that a symptomatic individual would seek medical attention, and vaccine coverage. Such a telephone survey led to one of the earliest robust estimates of the symptomatic case-fatality ratio.67
Telephone surveys can be performed rapidly and at reasonable cost proportionate to the number of individuals sampled, and standard methods exist to adjust such surveys to reflect the population as a whole.66 However, with questions that cover long recall periods (eg, “Have you had this symptom in the past month?”), the concern is how well individuals can recount their illness history. While the estimate of 12% of New Yorkers reporting ILI during peak spring transmission is plausible,67 a similar study performed outside the influenza season indicated 18% to 20% of New Yorkers with self-reported ILI.68 Clearly, statistical adjustment for recall bias is required, as is more work to ascertain how well these surveys reflect true symptomatic incidence. If these issues were addressed, surveys of illness in populations with well-ascertained severe outcomes could become a very valuable tool for rapid severity assessment that should be incorporated into pandemic plans.
Web-based surveys are a novel way to obtain estimates of symptomatic incidence. Influenzanet,69 a Web cohort survey that tracks influenza, is used in 5 countries.70 Individuals are invited to join—and invite friends to join—a cohort surveyed weekly for influenza symptoms. The Web cohort design has several advantages: The low marginal cost of including more subjects and more frequent queries can yield a large sample. Better recall results are likely, since individuals describe symptoms from a shorter time period, such as the prior week. Furthermore, repeatedly surveying the cohort is a cost-effective way to collect fairly detailed demographic and other data for comparing risk profiles of symptomatic and asymptomatic individuals.
Since raw data from a Web cohort are unrepresentative of the overall regional population, it will be important to assess how well incidence estimates from Web cohorts track true population-based estimates. As with many other surveys, it should be possible to develop techniques that weight raw data from Web surveys to create nationally representative estimates, but these corrections will be population-specific and may be difficult in jurisdictions with less extensive demographic data.
2.7. Hospital- and ICU-Based Data
Hospitalized patients can be characterized in greater detail than most patients with mild influenza, offering the opportunity to identify risk factors and perform detailed clinical studies. Some jurisdictions, such as New York City, focused much of their surveillance effort on hospitalized patients—having judged that the smaller volume of case reports and relatively consistent case definition over time would allow careful characterization of a defined subset of more severe cases. High ascertainment of hospitalized cases enabled the calculation of population-based hospitalization rates, which could be compared across populations to reveal important epidemiologic features of the pandemic, including the disproportionate impact of the pandemic on high-poverty neighborhoods in New York City35 and on racial/ethnic minorities in Wisconsin.71
In 2009, several jurisdictions decided that hospitalizations were the most reliable basis for estimating both cumulative case numbers and weekly trends.37,38 This decision reflects the judgment that the rate of hospitalizations is less affected by changing levels of concern in the population than other measures (such as physician consultation for ILI). As with ILI data (see sidebar: Syndromic Surveillance), hospitalization data can be converted into estimates of total cases only by making assumptions about the fraction of cases hospitalized.
Combining Syndromic Surveillance with Viral Testing.
The value of syndromic surveillance increases when combined with viral testing of a representative subset of individuals with a particular case definition,1 as this can generate a data-based estimate of the proportion of consultations caused by H1N1 influenza. Although not necessary for every case, this testing should be performed according to an algorithm that minimizes clinician discretion to ensure that cases tested represent a defined syndromic population.1
To limit the laboratory burden of testing, the total number of cases tested must be limited even as case numbers grow (perhaps by testing a fixed number or fixed proportion of random samples per week). Even so, testing will be limited to certain populations where sufficient laboratory capacity is available.
This approach was applied in the outpatient setting as a special study, in which patients with ILI in 8 states and approximately 40 practices were tested with influenza rapid antigen tests and PCR to determine the proportion of outpatient ILI visits caused by influenza.148 To determine the incidence of influenza-associated hospitalizations, patients with ILI admitted to the 200 hospitals in the U.S. Emerging Infections Program were also tested with RT-PCR.49
Even without assumptions about how many infections each hospitalization represents, the number and rate of hospitalizations in various age groups can measure the relative burden of severe influenza disease during a pandemic. Some jurisdictions maintain hospitalization surveillance during seasonal influenza. In the U.S. during 2009, age-specific influenza hospitalization rates reported by the Emerging Infections Program (EIP) Influenza Network were an important indicator of pandemic severity72,73 because they could be compared against estimates from prior nonpandemic seasons.74,75
The use of hospital-based data can present several challenges, the most important being the variation across hospitals in the automation and timeliness of computerized medical records. Also, in many settings, considerable effort is required to extract and analyze data on hospitalized patients, and protocols for testing for influenza infection can vary and depend on clinician discretion. The sensitivity and specificity of even “gold standard” PCR-based tests may be suboptimal.76 Many clinicians initially lacked access to these tests and were limited to using less sensitive, rapid tests,77 leading to underestimation of influenza-positive cases. Once the rapid tests were known to be low-sensitivity, their utility in clinical decisions was reduced, lowering the incentives for clinicians to test at all and further hampering ascertainment.38,44 Despite these caveats, hospital-based surveillance proved useful for many purposes in the 2009 pandemic.
2.8. Virologic Surveillance
In the U.S., virologic surveillance takes 2 forms: submission of influenza viral specimens and submission of data on respiratory samples tested for respiratory viruses, along with the number and percent positive for influenza. Worldwide, specimens are collected and tested by WHO Collaborating Centers.
In a pandemic setting, enhanced diagnostic sample collection makes available many more viruses for testing within such systems. Other sources of virus strains include hospital-based surveillance systems, such as the U.S. Emerging Infections Program (EIP) Influenza Project and ILINet in the U.S., whereby virus samples taken for clinical care are further characterized for surveillance purposes.
Since the method of choosing isolates for testing is not standardized, the representativeness of the tested strains is uncertain.1 Nonetheless, these samples can generally illustrate the proportion of cases that, at a given level of severity, are positive for pandemic influenza infection (or for other strains of influenza), while further testing of selected strains can characterize genetic and phenotypic changes that perhaps involve drug susceptibility, antigenicity, and virulence.
Each week in the U.S., the WHO and the National Respiratory and Enteric Virus Surveillance System (NREVSS) laboratories submit data from sample testing of respiratory viruses to CDC, with each laboratory reporting the number of samples submitted for respiratory virus testing and the number testing positive for influenza. During the influenza season and the 2009 pandemic, the percentage of samples positive for influenza provided information about the location and intensity of influenza circulation. Data from more than 900,000 tests were reported during the pandemic—almost 4 times as many test results as CDC receives during an average influenza season.
Global virologic surveillance and national systems outside the U.S. have similar objectives and in many cases use similar systems. WHO's FluNet reports weekly data from National Influenza Centers on the number of specimens positive for influenza A (by subtype and sometimes lineage within subtype) and for influenza B.78
2.9. Surveillance in Resource-Limited Settings
The challenges of conducting surveillance during a pandemic are magnified in settings with limited (and uneven) access to health care and limited surveillance infrastructure. As this summary was being written, WHO was attempting to estimate the total burden of the pandemic worldwide. Unfortunately, for many parts of the world, representative data are simply unavailable, even after the fact.
Several approaches should be considered for future planning. In middle-income countries with significant public health infrastructure, it should be feasible to expand population-based surveillance for respiratory illness that provides a baseline for comparison with an emerging pandemic.79,80 It should also be possible to plan rapid outbreak investigations and hospital-based surveillance to characterize a pandemic's severity, clinical course, and risk groups. Whether such preparations would be an efficient use of public health resources in middle-income areas remains to be determined. In some cases, it may be preferable to invest in different priorities and depend on other jurisdictions and WHO for guidance.
In the developing world, developing comprehensive national surveillance is difficult and may be a poor use of limited public health funds. However, most countries have access to influenza laboratory capacity, either in country in the form of a national influenza center or in a nearby country. Many low-income countries also currently perform hospital- and clinic-based surveillance for mild and severe respiratory disease in large urban centers and can provide valuable data on the relative severity. It would be valuable to set up a network that combines data from areas in the developing world with unusually good surveillance resources that could include demographic surveillance system (DSS) sites, as well as medical study sites funded by institutions like the U.S. CDC, the UK Medical Research Council, the Wellcome Trust, and the Pasteur Institute's RESPARI network. Such a network could also provide timely evidence on the characteristics of a novel influenza strain in developing country settings, including those with a greater burden of other infections, including HIV, TB, and pneumococcal disease.
3. Interpreting Surveillance Data for Decision Making
Raw surveillance data on a novel influenza strain, especially from established systems with background data on nonpandemic influenza, can broadly illustrate the trajectory of symptomatic infections in time and space. To inform many decisions, however, surveillance data must be processed to estimate particular quantities—for example, transmissibility and severity measures or the cumulative proportion of the population infected to date—and to define the pandemic's possible course through formal prediction or plausible planning scenarios.
3.1. Estimating Severity and Disease Burden
Severity estimation and disease burden estimation are different approaches to answering interrelated questions: How many cases? How many deaths (burden)? What is the ratio of deaths to cases (severity)?
As described in section 1, case-fatality or other case-severity ratios are probably the most important quantitative inputs for early decision making. However, estimating both the numerator and denominator of these ratios is challenging. For discussion purposes here, we focus on symptomatic case-fatality ratios, where the numerator is fatalities and the denominator is symptomatic cases.
Symptomatic case number estimates can come from surveys (with some correction for the proportion of symptomatic cases truly due to pandemic virus infection67,81,82) or from data at other levels of the “severity and reporting pyramid,”38 such as confirmed cases38 or hospitalizations81 combined with an estimate of the proportion of symptomatic cases hospitalized.81 If one made assumptions about the proportion of asymptomatic cases, these estimates could be converted into estimates per infection (see sidebar: Large-Scale Serosurveillance).
Case ascertainment will always be less than 100% and will vary over space and time in the pandemic. If unaccounted for, ascertainment can bias severity estimates. In the earliest phases of a pandemic, symptomatic case numbers can be biased by the preferential detection of the most severe cases, leading to substantial overestimates of severity, since these cases are more likely than typical cases to be fatal.
For example, as of May 5, 2009, Mexico had reported almost 1,100 confirmed H1N1 cases and 42 deaths from H1N1,10 a crude case-fatality proportion of about 4%. In hindsight, this high apparent severity was largely, or entirely, attributable to the underascertainment of mild cases, of which there were probably several orders of magnitude more than the number confirmed.5,6 As public health efforts scaled up and an increasing number of milder cases were detected, this bias declined,34 but it did not disappear, showing how even with the most intense efforts, only a minority of symptomatic cases may be virologically confirmed.38
The numerator—fatalities—can be directly estimated in jurisdictions with routine viral testing of fatal cases. Experience in 2009 showed that some fatal cases are diagnosed only on autopsy,36,76 posing a risk for underestimating the numerator. A second source of variability is differences between jurisdictions in the definition of influenza deaths, which may include all fatalities in individuals in whom the virus was detected or only those in whom the virus was judged to have caused the death. Another potential source of error in estimating the number of deaths is the delay from symptom onset to death from pandemic influenza, which can be a week or longer.11,82,83 For this reason, deaths counted at time t may not correspond to all the cases up to time t but to the cases that had occurred up to a week or more before t. In the exponentially growing phase of the pandemic, there may be many recently infected individuals who will die but have not yet died; they are counted in the denominator but not the numerator. If unaccounted for, this “censoring bias” can lead to an underestimate of severity as much as about 3-fold to 6-fold during the growing phase of a flu pandemic.11 Two basic approaches can address this bias: One is to correct for it based on the growth rate in disease incidence and the lag time between case reporting and death reporting.11 Another is to perform analyses after transmission has subsided in a population, by which time most deaths will have been registered in the data set.44
Notwithstanding these sources of bias, it is particularly challenging to precisely estimate the case-fatality proportion when the true proportion is low. In any population with a statistically robust number of deaths (eg, more than 10 cases) and a symptomatic case-fatality proportion of 1 in 10,000 (0.01%), it would require 100,000 documented symptomatic cases (or a correspondingly large number of confirmed cases) to directly estimate the ratio—an impractical approach.1 The 2009 pandemic highlighted the need for other approaches.
One alternative is to conduct surveys within defined outbreak populations to estimate the number who are ill and relate this number to the directly measured number of severe outcomes. For example, in an early H1N1 outbreak at the University of Delaware, 10% of student respondents and 5% of faculty and staff on a campus of 29,000 reported ILI that resulted in 4 hospitalizations but no deaths.55 While this could not yield a precise estimate of the symptomatic case-fatality or case-hospitalization proportions, it provided useful upper bounds. A telephone survey yielded similar estimates in New York City.67
Another approach to estimating case-fatality proportion is to decompose the severity “pyramid,” instead relying on some types of surveillance to estimate the ratio of deaths to hospitalizations and on other types to estimate the ratio of hospitalizations to symptomatic cases.44 Bayesian evidence synthesis methods84 are a natural framework for combining the uncertainty in the inputs to such estimates into a single estimate of uncertainty in severity measures.44
Overall, the presence of countervailing biases (the underascertainment of both numerator and denominator) made initial severity assessment challenging in the 2009 pandemic. Although both biases were recognized, it was difficult at the time to identify the more severe bias. In retrospect, censoring bias was minor compared to the underascertainment of mild cases, making early estimates of severity higher than current estimates based on more complete data. There was also important uncertainty about whether estimates differed between populations (eg, U.S. versus Mexico) because the severity was truly different or because ascertainment patterns differed. These conclusions are outbreak-specific; in SARS, for example, ascertainment was relatively complete, but censoring bias—perhaps more acute than in 2009 because of the longer delay from symptom onset to death—led to substantial underestimates of severity until the bias was corrected for.85
All of these considerations, described in the context of attempting to estimate overall risk of mortality, are relevant as well to more complex measures of severity, such as years of life lost.29
Because influenza is seasonal, experiences in one hemisphere can—and did—inform planners and decision makers in the other. In 2009, the southern hemisphere was the first to have a full, uninterrupted winter season with the novel H1N1 virus. Rapid reviews of the experience in the southern hemisphere's winter season86,87 provided evidence for northern hemisphere planners that the capacity of intensive care units would likely be adequate overall, though local shortages might occur.
3.2. Interpreting Clinical Data
Data on the characteristics of severe clinical cases are directly relevant to decisions about prioritizing prevention (eg, vaccination) and using scarce resources to treat those most likely to benefit. Choosing priority groups for such preventive measures as vaccination should depend in part on the per capita relative risk of having various groups suffer severe outcomes without vaccination.30 For a particular group—pregnant women, for example—this risk can be estimated by dividing the proportion of pregnant women among individuals with severe outcomes by the proportion of pregnant women in the general population.
This same measure of comparative severity applies to prioritizing other measures that are distributed to uninfected people. In 2009, it was proposed that certain groups might benefit from predispensed (or easier access to) antiviral drugs to aid in early treatment. The potential benefits of such a policy depend mainly on the per capita risk of severe outcomes in the priority groups compared to the general population.88
To prioritize treatment of symptomatic individuals, the relevant measure of comparative risk is severity per case, not per capita, since the decision involves a person with presumed or known infection, not a randomly chosen group member. The distinction between these 2 measures is that per capita severity is equal to per case severity times the risk of becoming a case. For example, in most countries, people over age 50 showed considerably higher severity per case, but only modestly higher per capita severity, because they were less likely than younger people to be infected.
The goal for all the purposes outlined above is to identify predictors of severe outcome rather than understand why the predictors are associated with the severe outcome.88 In the 2009 pandemic, morbid obesity was identified in some case series as a predictor of hospitalization and death,89 prompting much discussion about whether morbid obesity itself caused the outcomes or whether it was a marker of other conditions that did so. This question of etiology is unimportant when allocating resources; if high-risk individuals can be identified, they can receive priority for prevention or treatment and the benefit will be the same, regardless of whether the identifying factor is causal or only a marker.
3.3. Estimating Transmissibility
A standard summary measure of transmission intensity is its reproductive number—the mean number of secondary cases caused per primary case. When this exceeds 1, incidence grows; below 1, incidence declines. Absent mitigation measures, an estimate of the reproductive number of a strain at baseline can inform how intensely transmission must be reduced to slow or stop the growth in the number of cases.90 To halt growth, the critical proportion of transmission events that must be blocked is given by 1 minus the reciprocal of the reproduction number. Estimates of changes in the reproduction number over time91,92 can indicate the impact of control measures37 or of intrinsic changes in transmissibility due to depletion of susceptible individuals, seasonality, or other changes.93
A common approach to estimate transmissibility of a newly emerging infection relies on estimates of 2 quantities: the exponential growth rate of the number of cases and the distribution of the serial interval or generation time—that is, the time from infection to transmission.94 The minimal data required to derive the reproductive number from these 2 estimates are a time series of the number of new cases (ideally, a daily time series) and an estimate of the serial interval distribution,92 which can come from early outbreak investigations.5,95 The assumption is that the distribution of intervals between symptom onsets approximates that of the intervals between times of infections.96 Remarkably, with certain assumptions, one can infer both the serial interval distribution and the reproductive number using only the time series of new cases.34,97
Key challenges in estimating reproduction numbers from epidemic curve time series include changes over time in the fraction of cases ascertained—which can affect apparent growth rates and therefore bias reproductive number estimates—and reporting delays, whereby, even in a growing epidemic, recent case incidence will appear to drop off due to recent, unreported cases.7 There is growing methodological and applied literature on addressing these challenges34,37,94,96,98 and obtaining corrected estimates or bounds for the reproductive number. Analyzing viral sequence data, discussed below, can provide a partially independent estimate of transmissibility and validate conclusions made from purely case-based estimates.
3.4. Real-Time Predictive Modeling
Transmission-dynamic models can be used to predict the possible future course of an epidemic (eg, the number of infections per day in various groups) given certain assumptions. These assumptions, or model inputs, include such quantities as the reproductive number of the infection, the relative susceptibility and infectiousness of different groups in the population, the natural history of infectiousness, and the nature and timing of possible interventions. Transmission-dynamic models have been widely used as planning tools to assess the likely effectiveness of interventions for pandemic influenza19,25,99–101 and many other infections.102,103 In these cases, the models are applied to hypothetical epidemics, and the input assumptions are taken from past epidemics of similar viruses. In this section, we consider a distinct though related application of transmission-dynamic models: predicting the dynamics of a pandemic as it unfolds by using real-time data on the incidence and prevalence of infection to date in various population subgroups.
Since reliable predictions of a pandemic's time course are tremendously helpful for response planning and decision making (sections 1.5–1.6), it would be valuable if transmission-dynamic models were employed in real time to make and update predictions of the course of transmission. This would require 3 ingredients: (a) a sufficiently accurate mathematical model of the key processes that influence transmission; (b) data on the current and past incidence of infection with the pandemic strain, population immunity, and other parameters needed to set initial model conditions; and (c) an assumption that the biological properties of the influenza virus would not change within the time scale of prediction.
Transmission-dynamic models are now computationally capable of including virtually unlimited amounts of detail in the transmission process,104 but knowledge of some of their inputs is limited. For example, while much is known about the factors that affect influenza transmission, areas of uncertainty remain, including the exact contributions of household, school, and community transmission;104 the contribution of school terms, climatic factors, and other drivers to transmission seasonality;93,105–107 and the role of long- and short-term immune responses to infection with other strains in susceptibility to pandemic infection.108 Our understanding of behavioral responses to pandemics is also at an early stage.109
During a pandemic, incidence data are imperfect and subject to substantial uncertainty in the “multiplier” between observed measures of incidence and true infection (see sidebars: Syndromic Surveillance and Large-Scale Serosurveillance). Since infection and resulting immunity drive the growth, peaking, and decline of epidemics, this conversion factor is crucial to setting model parameters. Finally, changes in the antigenicity, virulence, or drug resistance of a circulating strain could invalidate otherwise reliable model predictions.
Efforts at real-time modeling in the 2009 pandemic showed that uncertainty in the number of individuals infected in various age groups at any given time hampered efforts to forecast the pandemic using transmission-dynamic models. Despite this limitation, the 2009 experience illustrated the potential of predictive models to provide policy guidance by generating plausible scenarios and, as important, by showing that certain scenarios are less plausible and thus of lower priority for planning.
In one published case study, Ong and colleagues set up an ad hoc monitoring network among general practitioners in Singapore for influenzalike illness and used the reported numbers of daily visits for ILI to estimate, in real time, the parameters of a simple, homogeneously mixed transmission-dynamic model, which they then used to predict the course of the outbreak.62 Early predictions of this model were extremely uncertain and included the possibility of an epidemic much larger than that which occurred. This uncertainty reflected the limitation of the input data (here, physician consultations). Without a known multiplier, it was impossible to scale the number of infections anticipated by the model to the number of consultations. By late July, the growth in the incidence of new cases had slowed, providing the needed information to scale the observed data to the dynamics of infection, allowing for more accurate and more precise predictions.62
Data on healthcare-seeking behavior were used in a similar effort in the United Kingdom, but with a more detailed, age-stratified transmission-dynamic model. Here, too, the timing and magnitude of the peak were difficult to predict because of uncertainty in the conversion factor between observed consultations and true infections—although in this case the authors, by their own description, had made a guess that was roughly accurate63 when tested against serologic data.65
A third effort was made in late 2009 to assess the likelihood of a winter wave in U.S. regions. Based on estimates of the rate of decline of influenza cases detected by CDC surveillance in November to December, combined with estimates of the possible boost in transmissibility that might occur due to declining absolute humidity,107 it was anticipated that any winter wave would be modest and likely geographically limited. Further analysis after the fact showed that the southeastern U.S. was the region most likely to experience further transmission due to a seasonal boost in transmissibility, a finding consistent with observations.93
These experiences indicate that real-time predictive modeling is possible but will also include considerable uncertainty if undertaken responsibly. For transmission-dynamic modeling, the most important source of uncertainty lies in the multiplier between cases observed in surveillance systems and infections. Modelers must therefore seek out the best data to estimate parameters for models by paying careful attention to publicly available data and by building relationships with those who conduct surveillance prior to pandemics, so that information transfer is facilitated in the midst of an event. As noted by the authors of the studies described above, and in reviews by several consortia of transmission modelers,7,61 the factor that could most improve the reliability of real-time models is having nearly real-time estimates of cumulative incidence—whether through serosurveillance or other means.
The real-time, predictive modeling described above is one of the developing frontiers within the broader scope of transmission-dynamic modeling. These approaches differ from scenario-based modeling, which may be performed before or during a pandemic to provide robust estimates of the possible effects of interventions under particular assumptions rather than predict the short-term dynamics of the infection. Scenario-based modeling18,19,23,25,90,104,110 has significantly improved our understanding of epidemic dynamics and the likely responses to interventions, but it is most helpful when predictions are robust to variations in assumptions that may be difficult to pin down during an epidemic.
3.5. Interpreting Virologic Data
Simple virologic confirmation of pandemic H1N1 infection was an integral part of case-based surveillance, especially early on and for severe cases. The proportion of viral samples that are positive, alone or in combination with ILI data, provides a measure of incidence (with the caveats described in previous sections). A more novel approach is to use viral sequence data to time the origin, rate of growth, and other characteristics of a pandemic,5 applying methods that rely on the coalescent theory developed in recent decades in population genetics, in which the quantitative history of a population can be inferred from the pattern of branching in a phylogenetic tree.111
Work is still under way to assess the quality of inferences made from methods for viral sequence data, although very early transmissibility estimates were broadly consistent with those from case-count data.5 However, one early finding is that the strength of inferences can be improved by consistently associating epidemiologic data (in particular, data on the geographic and temporal origin of a strain) with sequence data. Unfortunately, while easy to gather, these “metadata” often do not appear together with sequences in online databases.
3.6. Detecting Changes in the Virus
On the minds of many analysts and decision makers in 2009 was the 1918 experience, in which a wave of clinically mild infection with a pandemic virus occurred in the spring, followed by a wave of much more virulent influenza in the fall.112–114 The appearance of a more virulent virus strain is one of several hypotheses for how such a change occurred. Speculation aside, such an event reinforces the public health importance of detecting changes in drug resistance or antigenicity during a pandemic. Ongoing sampling of viral isolates from diverse sources, along with surveillance to detect unusual clusters of severe illness, are valuable in maintaining awareness of any variation in a virus that could be biologically and epidemiologically significant.
The 2009 pandemic showed it is possible to detect mutations that, on biological grounds, may affect virulence and transmissibility, but it also illustrated the challenge of interpreting such genetic changes. The E627K mutation in the PB2 gene, detected in several isolates of pH1N1, was expected to dramatically increase virulence; however, animal studies showed the mutation had little effect in the genetic background of the 2009 strain, and it has not appeared to spread widely in the viral population.115 In contrast, the hemagglutinin D222G substitution, which alters receptor binding,116 was associated in several populations with more severe disease.117–119 The exact mechanistic consequences of this mutation remain uncertain, and it has not replaced the wild-type sequence in the viral population. It would be valuable to develop more specific strategies for obtaining and characterizing novel or unusual variants of pandemic viruses that may be associated with important phenotypic changes and to implement and test these principles during interpandemic periods. Targeted approaches, including systematic sampling from a defined mix of mild, severe, treated, and untreated infections, could be one component of such a strategy.1
4. Other Considerations in Decision Making
The progression from data to evidence to evidence-based decisions, described up to now and portrayed in Figure 1, is an idealization. In reality, the decision-making process is not so simple. Evidence is neither always perfect nor available when required. Even if it were, sources other than surveillance and epidemiologic data should and do influence decision making.
4.1 Historical Experience
When confident estimates of the absolute and relative severity, transmissibility, and other key parameters of a novel influenza strain are unavailable, historical experience of pandemics can provide valuable evidence for planning.120 All 3 influenza pandemics that occurred in the twentieth century—in 1918, 1957, and 1968—shared a set of features: greater burden of severe disease in younger people than seen during seasonal flu; persistence of this pattern into subsequent years;121 and transmission outside of normal flu season in temperate regions. These common characteristics informed public health planning during the 2009 pandemic, especially during the early stages.120
Also valuable was knowledge of the differences among the pandemics: major variation in overall severity, variation in the impact on the elderly, and increased severity in the fall wave versus the spring wave (which occurred only in 1918). Awareness of these differences expanded the range of possibilities for which to plan. Even as evidence accumulated on overall severity of the 2009 pandemic and as estimates of the case-fatality ratio and other measures of severity declined, historical considerations justified planning for the contingency that severity could drastically increase.
Decisions to react aggressively to the pandemic were made when data could not reliably estimate per-case severity, so the possibilities consistent with the early data ranged from an outcome considerably milder than a typical flu season to one comparable to a severe flu season, or worse. In the face of this uncertainty, developed countries had to decide whether to invest billions of dollars in vaccine procurement. Such investments are cost-effective in average influenza seasons. Thus, the expectation they would be so even in a mild pandemic was justified—all the more so if severity were higher than a typical flu season or if the virulence of the virus changed.
Even if decision makers had known the pandemic strain's severity in 2009 was lower than envisioned in pandemic planning scenarios, we suspect vaccines would have been procured and that this would have been a sound decision. Suppose, for example, data had been available in April 2009 to definitively show a case-fatality ratio of well below 1 in 10,000 in all groups—a ratio below the lower end of current estimates for the 2009 pandemic virus in developed countries38,44,45,67—but that the virus was transmissible from person to person and was spreading widely. Would it have been responsible for public health authorities to defer vaccine procurement, given the historical precedent for a mild infection to turn virulent in a matter of months?112–114 Arguably, historical experience would trump contemporary evidence from surveillance and other sources and call for an investment in prevention, regardless of the estimated severity of the disease at the time of the decision.
4.2 Public Opinion
Public opinion affects policy decisions about pandemic response in at least 2 ways. First, since policymakers are ultimately responsible to the population, they must take into account a number of factors besides the (uncertain) projected public health benefits of a decision. A policy may receive little public support, even if it most efficiently uses resources to solve a public health problem, while another decision with little immediate benefit may be judged desirable.
Second, public opinion may constrain the range of policy options available to decision makers because these policies rely on voluntary decisions made by individuals. The use of an adjuvant-containing vaccine, for example, might be a prudent public health decision for maximizing the number of available doses, but public opposition might have made such a decision impractical in the U.S. even if regulatory concerns had been met. Political consequences aside, public opposition could also reduce uptake of the vaccine, potentially leading to counterproductive outcomes.
4.3 Logistics
As evidence accumulates, it may be desirable to change policies; however, some factors may restrict such changes. First, implementing decisions and disseminating recommendations takes time—particularly when policies require hiring and training staff for “surge” operations—and such delays can render a policy change ineffective. Second, even if a policy could be changed, its benefit must be weighed against any potential undesirable effects, such as confusing the public, clinicians, or other recipients with a revised public health message. In extreme cases, officials can even lose their credibility if guidance is perceived as being inconsistent.
4.4 Cognitive Limitations
Limitations in how analysts and decision makers process, evaluate, and prioritize information also hinder the incorporation of surveillance evidence into decision making. Although the 2009 experience was not as severe as other scenarios considered in pandemic planning exercises, the response was nevertheless an extreme escalation in public health agency activity, with decisions made under conditions of stress, fatigue, and time pressure, as well as with limited information.
Such conditions make cognitive errors more likely.122,123 At the Symposium, decision makers discussed the difficulties of authenticating and balancing conflicting information and in prioritizing the many decisions required. They also reported that certain forms of data were a distraction. For example, following the very early stages of the 2009 pandemic, case counts were an unreliable indicator of infectious spread because of inconsistencies in testing and reporting.1
Spatial variation in case confirmation further challenged data interpretation. During the spring-summer wave, Milwaukee and the state of Wisconsin devoted more effort to case testing and confirmation than most other jurisdictions. However, the differences in their efforts were not known to all data recipients, creating confusion about how much of the geographic variation in reported pandemic flu activity was real and how much of it was due to differences in ascertainment.
All users of the data confronted these challenges, but they were particularly acute for decision makers, such as elected officials, who lacked direct access to the primary data. Generally, as new data are gathered, it is difficult to determine what biases exist and how much they distort the evidence. For example, in the first week of May 2009, an approximate 40-fold difference existed in the ratio of deaths to cases in data from the U.S. (about 0.1%) and Mexico (about 4%). While both figures were biased, it was unclear which (if either) accurately reflected case severity.
We have discussed the importance of mathematical and statistical modeling in surveillance and epidemiology data processing. Some of these techniques are unfamiliar to many public health officials, and the outputs of these models depend strongly on the quality of their inputs, which are often uncertain in a pandemic. Consequently, the greater the sophistication or complexity of a method, the more difficult it may be for decision makers to understand the strengths and limitations of the evidence provided.
Given the time constraints and other pressures just described, the usual scientific checks and balances of peer review, replication, and debate can be compressed into a very short time period for findings that are presented rapidly after their generation. Thus, in such a setting, there is an added responsibility for those who present decision makers with results of complex analyses to highlight the limitations and assumptions of their models, as well as to identify particularly robust predictions. As important, modelers need data to calibrate models and estimate their parameters, but they may lack an understanding of the biases and limitations of data collected in an emerging epidemic and about the possible changes over time in the ascertainment of cases. An additional responsibility in the 3-way interaction among public health agencies that gather data, decision makers, and modelers (some individuals may have more than one of these roles) is to ensure that these limitations are understood and accounted for in the process of modeling or other “processing” of the data.
It should also be noted that seasonal influenza epidemics occurring outside of pandemics offer opportunities to develop, disseminate, and explain new methods to the planners who will rely on them during an emergency. A complementary strategy, used in several places in 2009, is to “embed” mathematical modelers and statisticians with skills in the analysis of epidemic data within public health agencies during a pandemic, to facilitate rapid exchange of data, questions, and analyses.
5. Lessons for the Future
The response to the 2009 pandemic was successful in many ways, thanks to the extraordinary public health resources mobilized to confront the novel H1N1 virus. This response relied on existing infrastructure built for surveillance and for epidemiologic and virologic studies of seasonal influenza, and expanded in many cases to meet the needs of the pandemic. Pandemic planning, a major focus of public health agencies over the previous half decade, improved the ability of public health agencies to rapidly scale up a response.
Nevertheless, the 2009 experience also highlighted the limitations of our ability to respond to a pandemic virus. The most important of them—the inability to manufacture a vaccine fast enough to immunize the population before peak influenza activity in either hemisphere—is a technological problem outside the scope of this article. However, the successes and challenges of influenza surveillance in 2009 yielded clear lessons, some of which are discussed in this section along with recommendations for improving preparedness and response in future pandemics.
5.1. Overall Response
Despite its extensive spread throughout Mexico, the pH1N1 influenza virus was discovered and characterized before most other countries experienced significant transmission. Estimates of transmissibility and the likely broad extent of infection within Mexico were published within 1 month of the initial public health response and have generally proven consistent with later estimates based on more complete data.124 Existing surveillance systems in developed countries provided estimates of the geographic and temporal trajectory of disease incidence, although only in rare cases could these estimates be defined as true incidence rates (numbers of clinical cases per person per unit of time). The mobilization of public health efforts at all levels led to the swift creation of systems that tracked the trajectory of infection at various geographic levels. Some systems, such as the Distribute project network of emergency department surveillance,46 grew dramatically during the pandemic and should continue as low-cost, useful resources for monitoring seasonal influenza and future pandemics.
5.2. Severity Estimation
As discussed earlier, per case severity is perhaps the most important quantitative input to decision making. By midsummer, published estimates of the case-fatality ratio (with different denominators) ranged from several deaths per thousand cases, down to 1 death in 250,000 cases.11,12,55,67 While each estimate was reasonably inferred from the data used, it was difficult at the time to quantify the biases in each data set. How ambiguities could have been resolved much earlier remains unclear.
For future reference, it would be valuable to have a set of principles for evaluating severity—with strengths and weaknesses of each method defined—and a formal process to compare estimates. Such a framework would require analysts to precisely define the numerators and denominators of each data set. These definitions could facilitate data interpretation and thus aid in the understanding of seemingly conflicting information. As the 2009 experience showed, disparate severity estimates may often reflect different denominators (confirmed cases, severe cases, symptomatic cases) rather than truly different severity. A precise framework would also allow the natural incorporation of disparate data types into an overall estimate of severity via Bayesian evidence synthesis.44,62,84 Several public health authorities, including the U.S. and European CDCs and WHO, are currently developing such frameworks.
5.3. Timeliness of Information
The dissemination of information in the 2009 pandemic has been judged successful overall.9,125 We share this assessment, but we note that several factors slowed the gathering and dissemination of important information on the virus's epidemiology. For example, although there was sharing of clinical experience via networks of clinicians set up by WHO and other organizations, the first large-scale (hundreds of cases) quantitative analysis of risk factors for hospitalization for 2009 was published online on October 8, almost 4 months after the data were gathered.14 Addressing sources of delay in gathering, computerizing, aggregating, analyzing, and reporting emerging data should lead to a more efficient and productive public health response in future pandemics.
For example, many records, especially at hospitals and local public health departments, were created on paper, requiring significant data entry prior to analysis.126 Improved computerization of medical and public health data, along with computer system integration and databases that can combine surveillance, epidemiologic, and laboratory data, would have been valuable for this aspect of the response.
Once in computerized form, raw data must be processed into evidence on risk factors, treatment effectiveness, and other information, with analyses conducted by individuals with statistical and epidemiologic analysis skills, as well as knowledge of the subject matter and an intuitive sense for identifying potential problems in the data. Because qualified analysts were in high demand for many other response activities, the timeliness of data analysis lagged in certain jurisdictions.
Such personnel shortages are almost certain to recur in future pandemics. Advance planning to define priorities for surveillance and other public health activities, as well as the personnel requirements to accomplish them, would help expose areas where particular skills are needed. Ensuring surge capacity, perhaps through collaboration with academic centers, would also be helpful.
Once analyses are available, dissemination should be rapid. Many jurisdictions published data summaries on a daily or weekly basis on their websites. For many aspects of the data obtained in pandemics, including severity estimates, public health officials face conflicting pressures in communicating current knowledge in a timely fashion. Transparency demands immediate dissemination of evidence—and clear statements of uncertainties surrounding it. However, releasing every potentially conflicting piece of information risks confusing the public and decision makers outside public health. It may also undermine the credibility of those providing the evidence. Properly balancing these demands requires sensitivity and good judgment.
Another delay in disseminating information in 2009 was the internal clearance processes at some (but not all) public health agencies, and the peer-review process, which at some journals took months even for information with urgent clinical implications. The internal clearance process should be streamlined in jurisdictions where it created significant delay. The peer-review bottleneck was recognized during the 2009 pandemic (and before), and several promising approaches were tried. The journal Eurosurveillance provided extremely rapid peer review and publication,127 thereby acquiring a reputation as a home for reports deemed important enough for rapid dissemination. The Public Library of Science (PLoS) teamed with Google to create PLoS Currents Influenza, an online-only “journal,” to which scientific papers, opinion pieces, or other analyses could be submitted for moderation “to determine as rapidly as possible if … [the submission] is a legitimate work of science and does not contain any obvious methodological, ethical or legal violations.” PLoS Currents Influenza provided a useful forum for quickly sharing results. The U.S. CDC's Morbidity and Mortality Weekly Report, which is not formally peer reviewed, also provided a forum for rapid dissemination of both data and recommendations.
Crucial to the success of these forums was that articles could be referenced on PubMed, making them easy to find in literature searches, and they were available free of charge. In the early phases of the pandemic, PLoS Currents Influenza also allowed articles published online to be submitted to the peer-reviewed journals of PLoS, meaning that researchers did not have to choose between rapid dissemination and peer review, thus keeping open the possibility of publication in a well-regarded journal. The leading subject-specific journal, Influenza and Other Respiratory Viruses, adopted the same policy.
PLoS has since changed this policy: Today, PLoS Currents Influenza publication is considered final, with no option to submit for further publication. Eurosurveillance remains peer reviewed and rapidly evaluates articles (though not as quickly as during the pandemic). For future pandemics, other leading journals could consider modifying embargo rules and enhancing peer-review systems to encourage swift dissemination of key data.128
Finally, many population-wide data sources, such as hospitalization databases129 and cause of death records,130 are made available with a delay of several years. At present, all-cause mortality reporting in Europe131 and all-cause and pneumonia and influenza mortality reporting in the U.S. from 122 cities132 provide timely signals of large-scale trends, but it should be possible to increase the timeliness of more detailed data collected electronically on age- and cause-specific hospitalizations and deaths at various geographic levels.
5.4 Integration of Information
Even if available in a timely fashion, information must be presented in a way that allows decision makers to evaluate it quickly and effectively. Thus, it is critical that information be clear, consistent, and in a concise format that summarizes key knowledge and includes both uncertainty about the major questions and any report updates. No less important is providing decision makers and those responsible for implementation with a common source of information that fosters an equal understanding of the problem among all involved. One approach could be a computerized “dashboard” that supplies summary estimates and additional details on specific topics. A complementary solution could be the dissemination of narrative, yet quantitative, planning scenarios that translate surveillance data into a small number of possible outcomes to facilitate decision making. Such coordination would best be accomplished at the national level, but shared with local jurisdictions. Data inputs to such a summary would come from local and (in the U.S.) state health authorities, from local and national investigations and surveillance systems, and from other sources. Analytic capacity to process these inputs might come from a combination of government and academic experts. The severity frameworks described in section 5.2 could be a prototype for quantitative or narrative synthesis with a broader scope.
5.5. Information Sharing
During the pandemic, communication improved among decision makers, data analysts (including transmission-dynamic modelers and epidemiologists), and data gatherers at the local, national, and global levels. At their best, these linkages enhanced the exchange of information and ideas, often through informal channels. Collaborations across these levels led to rapid and sometimes novel data analyses (often by academics collaborating with public health authorities).7
However, the novelty of many connections and competing demands on the time and attention of participants meant that information exchange was often incomplete. Improving relationships—especially those between government decision makers and technical experts within their own public health agencies, and those between public health agencies and academics—can pay dividends in future pandemics. Joint appointments between medical or public health schools and public health agencies can be particularly helpful if appointees are influential within the agencies and maintain robust research groups that can assist with the issues described in this article. In whatever administrative manner these connections are established, a clear understanding by participants of each other's capabilities and requirements for data or evidence inputs is necessary to improve the efficiency of the future response.
5.6. Serologic Studies
During the 2009 pandemic, population-based serologic studies were rare. The first large seroprevalence study was completed and published only after the spring and autumn waves occurred in the northern hemisphere. As with epidemiologic data analysis, the development of serologic tests suffered from a competition for skilled personnel. Many who could have been optimizing seroepidemiologic assays were instead occupied with other urgent tasks, including developing assays to test vaccine lots for immunogenicity.
Given the views of some61 that large-scale serosurveys are essential to surveillance in a pandemic, but in light of the substantial barriers to conducting these with current technology and resources (sidebar: Serosurveillance), formal efforts are needed to assess the value that such studies would add to situational awareness, transmission-dynamic model parameterization, improved decision making, and better understanding of the pandemic after the fact. If an assessment concludes that serologic data provide significant benefits over other forms of data, then investments should be made for improving serologic assays, improving statistical methods to interpret these assays despite imperfect sensitivity and specificity, and ensuring surge capacity for personnel to undertake them. The utility of serologic investigation of close contacts of early confirmed cases is more clear-cut. Even relatively small cohorts of close contacts could provide valuable information for the infection severity pyramid—for example, upper bounds on the proportion of infections requiring hospitalization.
5.7. Novel Surveillance Tools
Web-based cohorts,69 mining of managed-care organization and hospital databases, and perhaps even surveys conducted using mobile phones or other devices133 may substantially contribute to situational awareness and decision making in future pandemics. The common feature of many novel surveillance approaches is that once they are in place, the additional cost to increase their coverage is relatively small.
There are several requirements for these approaches to be useful. Baseline data collected during seasonal influenza and outside of influenza season are needed to validate and calibrate these systems. Concerns about the representativeness of survey respondents must be addressed by targeting underrepresented groups or by statistical adjustment, or both. For approaches with high fixed costs, such as those involving privately held hospital or other healthcare databases, it should be determined whether multiple applications of the data can justify the investment or whether lower-cost arrangements can be devised. For reasons described in the sidebar Combining Syndromic Surveillance with Viral Testing, these novel tools will contribute additional value if geographically widespread.
Novel approaches to processing data may also improve the reliability of estimates of disease burden. Capture-recapture methods, for example, were used in New Zealand to estimate the coverage of different reporting systems.134 Further work on the statistical methods for data analysis during emerging epidemics may contribute to more robust estimates in future pandemics.
5.8. Surveillance in Developing Countries
Surveillance in the developing world was limited in 2009; thus, many countries made decisions without the support of national or even regional data on the extent of infection in their populations. Current efforts to estimate the overall global impact of the pandemic are hampered by these large geographic pockets of limited data. Some sentinel sites—hospitals, demographic surveillance system sites, or entire cities—exist in many parts of the developing world, but in 2009, their data were not integrated and communicated in a way that could provide disease burden estimates in representative parts of the developing world. Assessment of the ability of existing sites to provide a global picture of pandemic spread, expansion of or development of additional sites, and development of plans to share data during a pandemic would improve the response to the next pandemic, in addition to their ongoing benefits in nonpandemic times. As in the developed world, these sentinel data would be much more useful if accompanied by virological and serologic testing of carefully designed subsamples.1,61
Pandemic responses will always require decision making with limited data. Good judgment and political considerations will compel decision makers with sufficient resources to err on the side of caution until the severity and extent of transmission become clear. Moreover, the changeability of influenza argues for a precautionary approach even once severity is established, as it could change in an unpredictable fashion. Advance efforts to tailor surveillance systems and analytic capacities to decisions that must be made will reduce uncertainty and help decision makers respond effectively despite any remaining uncertainty.
Contributor Information
Collaborators: for the 2009 H1N1 Surveillance Group
Acknowledgments
We thank Mary Kalamaras for assistance with editing of the manuscript and Alissa Scharf for assistance with the manuscript and expert organization of the Symposium on which it was based. The Symposium, as well as preparation of this manuscript, were supported by Award Number U54GM088558 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Conflict of Interest
Dr. Lipsitch reports that he has received consulting income or honoraria from Novartis Vaccines and Diagnostics, Pfizer/Wyeth Vaccines, and the Avian/Pandemic Flu Registry (Outcome Sciences), which is funded by Roche.
2009 H1N1 Surveillance Group.
Michael G. Baker
Department of Public Health
University of Otago
Wellington, New Zealand
Paul A. Biedrzycki
City of Milwaukee Health Department
Milwaukee, Wisconsin
Benjamin J. Cowling
School of Public Health
University of Hong Kong
Pokfulam
Hong Kong Special Administrative Region China
Daniela De Angelis
Health Protection Agency
Health Protection Services, London, UK;
Medical Research Council Biostatistics Unit
Cambridge, United Kingdom
Nathan Eagle
MIT Media Laboratory
Annie D. Fine
New York City Department of Health and
Mental Hygiene
Christophe Fraser
MRC Centre for Outbreak Analysis and Modelling
Department of Infectious Disease Epidemiology
Imperial College London, United Kingdom
Richard J. Hatchett
Biomedical Advanced Research and Development Authority
Katrin S. Kohl
Division of Global Migration and Quarantine
National Center for Emerging and Zoonotic Infectious Diseases
Centers for Disease Control and Prevention
George Korch
Assistant Secretary for Preparedness and Response
Department of Health & Human Services
Lawrence C. Madoff
Massachusetts Department of Public Health
Division of Infectious Diseases and Immunology
University of Massachusetts Medical School
Donald R. Olson
International Society for Disease Surveillance
New York, New York
Steven Riley
MRC Centre for Outbreak Analysis and Modelling
School of Public Health
Imperial College London, United Kingdom
Lone Simonsen
Department of Global Health
George Washington University
Maria D. Van Kerkhove
MRC Centre for Outbreak Analysis and Modelling
Department of Infectious Disease Epidemiology
Imperial College London, United Kingdom
Sander van Noort
Instituto Gulbenkian de Ciência
Portugal
Cécile Viboud
Fogarty International Center
National Institutes of Health
Jacco Wallinga
Centre for Infectious Disease Control
National Institute for Public Health and the Environment, Bilthoven, Netherlands;
Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht
Utrecht, Netherlands
Laura F. White
Department of Biostatistics
Boston University School of Public Health
Marc-Alain Widdowson
Influenza Division
Centers for Disease Control and Prevention
References
- 1.Lipsitch M. Hayden FG. Cowling BJ. Leung GM. How to maintain surveillance for novel influenza A H1N1 when there are too many cases to count. Lancet. 2009 Aug 11;374:1209–1211. doi: 10.1016/S0140-6736(09)61377-5. [DOI] [PubMed] [Google Scholar]
- 2.Lipsitch M. Riley S. Cauchemez S. Ghani AC. Ferguson NM. Managing and reducing uncertainty in an emerging influenza pandemic. N Engl J Med. 2009 Jul 9;361(2):112–115. doi: 10.1056/NEJMp0904380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker MG. Easther S. Wilson N. A surveillance sector review applied to infectious diseases at a country level. BMC Public Health. 2010;10:332. doi: 10.1186/1471-2458-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Health Map. http://healthmap.org/en/ [Apr 21;2011 ]. http://healthmap.org/en/
- 5.Fraser C. Donnelly CA. Cauchemez S, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009 Jun 19;324(5934):1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipsitch M. Lajous M. O'Hagan JJ, et al. Use of cumulative incidence of novel influenza A/H1N1 in foreign travelers to estimate lower bounds on cumulative incidence in Mexico. PLoS One. 2009;4(9):e6895. doi: 10.1371/journal.pone.0006895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathematical modelling of the pandemic H1N1 2009. Wkly Epidemiol Rec. 2009 Aug 21;84(34):341–348. [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control. Lessons from Previous Pandemics. May, 2009. http://ecdc.europa.eu/en/healthtopics/Documents/0905_Influenza_AH1N1_Lessons_from_Previous_Pandemics.pdf. [Apr 25;2011 ]. http://ecdc.europa.eu/en/healthtopics/Documents/0905_Influenza_AH1N1_Lessons_from_Previous_Pandemics.pdf
- 9.Washington, DC: Executive Office of the President; Aug 7, 2009. [Apr 21;2011 ]. [Google Scholar]
- 10.Defunciones: Descripciones Preliminar. http://portal.salud.gob.mx/descargas/pdf/influenza/graficas_defunciones060509.pdf. [Apr 21;2011 ]. http://portal.salud.gob.mx/descargas/pdf/influenza/graficas_defunciones060509.pdf
- 11.Garske T. Legrand J. Donnelly CA, et al. Assessing the severity of the novel influenza A/H1N1 pandemic. BMJ. 2009;339:b2840. doi: 10.1136/bmj.b2840. [DOI] [PubMed] [Google Scholar]
- 12.Wilson N. Baker MG. The emerging influenza pandemic: estimating the case fatality ratio. Euro Surveill. 2009;14(26) pii = 19255. [PubMed] [Google Scholar]
- 13.Goldstein E. Miller JC. O'Hagan J. Lipsitch M. Predispensing of antivirals to high-risk individuals in an influenza pandemic. PLoS Curr Influenza. 2009:RRN1007. doi: 10.1371/currents.RRN1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain S. Kamimoto L. Bramley AM, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N Engl J Med. 2009 Nov 12;361(20):1935–1944. doi: 10.1056/NEJMoa0906695. [DOI] [PubMed] [Google Scholar]
- 15.Louie JK. Acosta M. Winter K, et al. Factors associated with death or hospitalization due to pandemic 2009 influenza A(H1N1) infection in California. JAMA. 2009 Nov 4;302(17):1896–1902. doi: 10.1001/jama.2009.1583. [DOI] [PubMed] [Google Scholar]
- 16.Van Kerkhove MD. Vandemaele KAH. Shinde V, et al. Risk factors for severe outcomes following 2009 influenza A (H1N1) infection: a global pooled analysis. PLoS Med in review. doi: 10.1371/journal.pmed.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonsen L. Taylor RJ. Viboud C. Miller MA. Jackson LA. Mortality benefits of influenza vaccination in elderly people: an ongoing controversy. Lancet Infect Dis. 2007 Oct;7(10):658–666. doi: 10.1016/S1473-3099(07)70236-0. [DOI] [PubMed] [Google Scholar]
- 18.Cooper BS. Pitman RJ. Edmunds WJ. Gay NJ. Delaying the international spread of pandemic influenza. PLoS Med. 2006 Jun;3(6):e212. doi: 10.1371/journal.pmed.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson NM. Cummings DA. Fraser C. Cajka JC. Cooley PC. Burke DS. Strategies for mitigating an influenza pandemic. Nature. 2006 Jul 27;442(7101):448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scalia Tomba G. Wallinga J. A simple explanation for the low impact of border control as a countermeasure to the spread of an infectious disease. Math Biosci. 2008 Jul-Aug;214(1–2):70–72. doi: 10.1016/j.mbs.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Cowling BJ. Lau LL. Wu P, et al. Entry screening to delay local transmission of 2009 pandemic influenza A (H1N1) BMC Infect Dis. 2010;10:82. doi: 10.1186/1471-2334-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallinga J. van Boven M. Lipsitch M. Optimizing infectious disease interventions during an emerging epidemic. Proc Natl Acad Sci U S A. 2010 Jan 12;107(2):923–928. doi: 10.1073/pnas.0908491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dushoff J. Plotkin JB. Viboud C, et al. Vaccinating to protect a vulnerable subpopulation. PLoS Med. 2007 May;4(5):e174. doi: 10.1371/journal.pmed.0040174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Althouse BM. Bergstrom TC. Bergstrom CT. Evolution in health and medicine. Sackler colloquium: a public choice framework for controlling transmissible and evolving diseases. Proc Natl Acad Sci U S A. 2010 Jan 26;107(Suppl 1):1696–1701. doi: 10.1073/pnas.0906078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipsitch M. Cohen T. Murray M. Levin BR. Antiviral resistance and the control of pandemic influenza. PLoS Med. 2007 Jan;4(1):e15. doi: 10.1371/journal.pmed.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persad G. Wertheimer A. Emanuel EJ. Principles for allocation of scarce medical interventions. Lancet. 2009 Jan 31;373(9661):423–431. doi: 10.1016/S0140-6736(09)60137-9. [DOI] [PubMed] [Google Scholar]
- 27.Medlock J. Galvani AP. Optimizing influenza vaccine distribution. Science. 2009 Sep 25;325(5948):1705–1708. doi: 10.1126/science.1175570. [DOI] [PubMed] [Google Scholar]
- 28.Miller MA. Viboud C. Olson DR. Grais RF. Rabaa MA. Simonsen L. Prioritization of influenza pandemic vaccination to minimize years of life lost. J Infect Dis. 2008 Aug 1;198(3):305–311. doi: 10.1086/589716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viboud C. Miller M. Olson D. Osterholm M. Simonsen L. Preliminary estimates of mortality and years of life lost associated with the 2009 A/H1N1 pandemic in the US and comparison with past influenza seasons. PLoS Curr. 2010:RRN1153. doi: 10.1371/currents.RRN1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein E. Lipsitch M. H1N1 vaccination and adults with underlying health conditions in the US. PLoS Curr Influenza. 2009:RRN1132. doi: 10.1371/currents.RRN1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.U.S. Department of Health and Human Services, U.S. Department of Homeland Security. Guidance on Allocating and Targeting Pandemic Influenza Vaccine. No date. www.flu.gov/individualfamily/vaccination/allocationguidance.pdf. [Apr 21;2011 ]. www.flu.gov/individualfamily/vaccination/allocationguidance.pdf
- 32.Neustadt RE. Fineberg H. The Epidemic that Never Was: Policy-making and the Swine Flu Affair. New York: Random House; 1982. [Google Scholar]
- 33.Echevarria-Zuno S. Mejia-Arangure JM. Mar-Obeso AJ, et al. Infection and death from influenza A H1N1 virus in Mexico: a retrospective analysis. Lancet. 2009 Dec 19;374(9707):2072–2079. doi: 10.1016/S0140-6736(09)61638-X. [DOI] [PubMed] [Google Scholar]
- 34.White LF. Wallinga J. Finelli L, et al. Estimation of the reproductive number and the serial interval in early phase of the 2009 influenza A/H1N1 pandemic in the USA. Influenza Other Respi Viruses. 2009 Nov;3(6):267–276. doi: 10.1111/j.1750-2659.2009.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balter S. Gupta LS. Lim S. Fu J. Perlman SE. Pandemic (H1N1) 2009 surveillance for severe illness and response, New York, New York, USA, April–July 2009. Emerg Infect Dis. 2010 Aug;16(8):1259–1264. doi: 10.3201/eid1608.091847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee EH. Wu C. Lee EU, et al. Fatalities associated with the 2009 H1N1 influenza A virus in New York City. Clin Infect Dis. 2010 Jun 1;50(11):1498–1504. doi: 10.1086/652446. [DOI] [PubMed] [Google Scholar]
- 37.Wu JT. Cowling BJ. Lau EH, et al. School closure and mitigation of pandemic (H1N1) 2009, Hong Kong. Emerg Infect Dis. 2010 Mar;16(3):538–541. doi: 10.3201/eid1603.091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed C. Angulo FJ. Swerdlow DL, et al. Estimates of the prevalence of pandemic (H1N1) 2009, United States, April-July 2009. Emerg Infect Dis. 2009 Dec;15(12):2004–2007. doi: 10.3201/eid1512.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowling BJ. Lau MS. Ho LM, et al. The effective reproduction number of pandemic influenza: prospective estimation. Epidemiology. 2010 Nov;21(6):842–846. doi: 10.1097/EDE.0b013e3181f20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiura H. Roberts MG. Estimation of the reproduction number for 2009 pandemic influenza A(H1N1) in the presence of imported cases. Euro Surveill. 2010;15(29) doi: 10.2807/ese.15.29.19622-en. [DOI] [PubMed] [Google Scholar]
- 41.Nishiura H. Chowell G. Safan M. Castillo-Chavez C. Pros and cons of estimating the reproduction number from early epidemic growth rate of influenza A (H1N1) 2009. Theor Biol Med Model. 2010;7:1. doi: 10.1186/1742-4682-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyers SM. Gile JD. Mallard reproductive testing in a pond environment: a preliminary study. Arch Environ Contam Toxicol. 1986 Nov;15(6):757–761. doi: 10.1007/BF01054923. [DOI] [PubMed] [Google Scholar]
- 43.Pourbohloul B. Ahued A. Davoudi B, et al. Initial human transmission dynamics of the pandemic (H1N1) 2009 virus in North America. Influenza Other Respi Viruses. 2009 Sep;3(5):215–222. doi: 10.1111/j.1750-2659.2009.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Presanis AM. De Angelis D. Hagy A, et al. The severity of pandemic H1N1 influenza in the United States, from April to July 2009: a Bayesian analysis. PLoS Med. 2009 Dec;6(12):e1000207. doi: 10.1371/journal.pmed.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker MG. Wilson N. Huang QS, et al. Pandemic influenza A(H1N1)v in New Zealand: the experience from April to August 2009. Euro Surveill. 2009;14(34) doi: 10.2807/ese.14.34.19319-en. pii = 19319. [DOI] [PubMed] [Google Scholar]
- 46.International Society for Disease Surveillance. Proportion of Emergency Department Visits for Influenza-like Illness (ILI) per Week. http://isdsdistribute.org/ [Apr 22;2011 ]. http://isdsdistribute.org/
- 47.Copeland D. Basurto-Davila R. Chung W, et al. Impact of a school district closure for pandemic influenza A (H1N1) on respiratory infections in the community. Options for the Control of Influenza VII. Hong Kong. 2010 O-877. [Google Scholar]
- 48.Reis BY. Kohane IS. Mandl KD. An epidemiological network model for disease outbreak detection. PLoS Med. 2007 Jun;4(6):e210. doi: 10.1371/journal.pmed.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.U.S. Centers for Disease Control and Prevention. FluView. 2009-2010 Influenza Season Week 20 Ending May 22, 2010. http://www.cdc.gov/flu/weekly/weeklyarchives2009-2010/weekly20.htm. [Apr 22;2011 ]. http://www.cdc.gov/flu/weekly/weeklyarchives2009-2010/weekly20.htm
- 50.Lessler J. Reich NG. Cummings DA. Nair HP. Jordan HT. Thompson N. Outbreak of 2009 pandemic influenza A (H1N1) at a New York City school. N Engl J Med. 2009 Dec 31;361(27):2628–2636. doi: 10.1056/NEJMoa0906089. [DOI] [PubMed] [Google Scholar]
- 51.Smith A. Coles S. Johnson S. Saldana L. Ihekweazu C. O'Moore E. An outbreak of influenza A(H1N1)v in a boarding school in South East England, May–June 2009. Euro Surveill. 2009 Jul 9;14(27) doi: 10.2807/ese.14.27.19263-en. [DOI] [PubMed] [Google Scholar]
- 52.Calatayud L. Kurkela S. Neave PE, et al. Pandemic (H1N1) 2009 virus outbreak in a school in London, April-May 2009: an observational study. Epidemiol Infect. 2009 Feb;138(2):183–191. doi: 10.1017/S0950268809991191. [DOI] [PubMed] [Google Scholar]
- 53.Shimada T. Gu Y. Kamiya H, et al. Epidemiology of influenza A(H1N1)v virus infection in Japan, May–June 2009. Euro Surveill. 2009 Jun 18;14(24) doi: 10.2807/ese.14.24.19244-en. [DOI] [PubMed] [Google Scholar]
- 54.Lee VJ. Yap J. Cook AR, et al. Oseltamivir ring prophylaxis for containment of 2009 H1N1 influenza outbreaks. N Engl J Med. 2010 Jun 10;362(23):2166–2174. doi: 10.1056/NEJMoa0908482. [DOI] [PubMed] [Google Scholar]
- 55.Iuliano AD. Reed C. Guh A, et al. Notes from the field: outbreak of 2009 pandemic influenza A (H1N1) virus at a large public university in Delaware, April–May 2009. Clin Infect Dis. 2009 Dec 15;49(12):1811–1820. doi: 10.1086/649555. [DOI] [PubMed] [Google Scholar]
- 56.Cowling BJ. Chan KH. Fang VJ, et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med. 2010 Jun 10;362(23):2175–2184. doi: 10.1056/NEJMoa0911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lau LL. Cowling BJ. Fang VJ, et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis. 2010 May 15;201(10):1509–1516. doi: 10.1086/652241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cauchemez S. Donnelly CA. Reed C, et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med. 2009 Dec 31;361(27):2619–2627. doi: 10.1056/NEJMoa0905498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmerman RK. Lauderdale DS. Tan SM. Wagener DK. Prevalence of high-risk indications for influenza vaccine varies by age, race, and income. Vaccine. 2010 Sep 7;28(39):6470–6477. doi: 10.1016/j.vaccine.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lennette EH. Lennette DA. Lennette ET. Diagnostic Procedures for Viral, Rickettsial, and Chlamydial Infections. 7th. Washington, DC: American Public Health Association; 1995. [Google Scholar]
- 61.Van Kerkhove MD. Asikainen T. Becker NG, et al. Studies needed to address public health challenges of the 2009 H1N1 influenza pandemic: insights from modeling. PLoS Med. 2010;7(6):e1000275. doi: 10.1371/journal.pmed.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ong JB. Chen MI. Cook AR, et al. Real-time epidemic monitoring and forecasting of H1N1-2009 using influenza-like illness from general practice and family doctor clinics in Singapore. PLoS One. 2010;5(4):e10036. doi: 10.1371/journal.pone.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baguelin M. Hoek AJ. Jit M. Flasche S. White PJ. Edmunds WJ. Vaccination against pandemic influenza A/H1N1v in England: a real-time economic evaluation. Vaccine. 2010 Mar 11;28(12):2370–2384. doi: 10.1016/j.vaccine.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Wu JT. Cowling BJ. Riley S, et al. A serial cross-sectional serologic survey of 2009 pandemic (H1N1) in Hong Kong: implications for future influenza surveillance. Options for the Control of Influenza VII. Hong Kong. 2010 P-351. [PubMed] [Google Scholar]
- 65.Miller E. Hoschler K. Hardelid P. Stanford E. Andrews N. Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet. 2010 Mar 27;375(9720):1100–1108. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 66.Reporting of influenza-like illness during the 2009 H1N1 influenza pandemic through the Behavioral Risk Factor Surveillance System—United States, September 2009–March 2010. MMWR Morb Mortal Wkly Rep. 2010 in press. [Google Scholar]
- 67.Hadler JL. Konty K. McVeigh KH, et al. Case fatality rates based on population estimates of influenza-like illness due to novel H1N1 influenza: New York City, May–June 2009. PLoS One. 2010;5(7):e11677. doi: 10.1371/journal.pone.0011677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Metzger KB. Hajat A. Crawford M. Mostashari F. How many illnesses does one emergency department visit represent? Using a population-based telephone survey to estimate the syndromic multiplier. MMWR Morb Mortal Wkly Rep. 2004 Sep 24;53(Suppl):106–111. [PubMed] [Google Scholar]
- 69.van Noort SP. Muehlen M. Rebelo de Andrade H. Koppeschaar C. Lima Lourenco JM. Gomes MG. Gripenet: an internet-based system to monitor influenza-like illness uniformly across Europe. Euro Surveill. 2007 Jul;12(7):E5–6. doi: 10.2807/esm.12.07.00722-en. [DOI] [PubMed] [Google Scholar]
- 70.Influenzanet. http://www.influenzanet.com/influenzanet/ [Apr 22;2011 ]. http://www.influenzanet.com/influenzanet/
- 71.Truelove SA. Chitnis AS. Heffernan RT. Karon AE. Haupt TE. Davis JP. Comparison of patients hospitalized with pandemic 2009 influenza A(H1N1) virus infection during the first two pandemic waves in Wisconsin. J Infect Dis. 2011;203(6):828–837. doi: 10.1093/infdis/jiq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shrestha SS. Swerdlow D. Borse RH, et al. Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009–April 2010) Clin Infect Dis. 2011;52(suppl 1):S75–S82. doi: 10.1093/cid/ciq012. [DOI] [PubMed] [Google Scholar]
- 73.Armstrong GL. Brammer L. Finelli L. Timely assessment of the severity of the 2009 H1N1 influenza pandemic. Clin Infect Dis. 2011;52(suppl 1):S83–S89. doi: 10.1093/cid/ciq013. [DOI] [PubMed] [Google Scholar]
- 74.Dao CN. Kamimoto L. Nowell M, et al. Adult hospitalizations for laboratory-positive influenza during the 2005-2006 through 2007-2008 seasons in the United States. J Infect Dis. 2010 Sep 15;202(6):881–888. doi: 10.1086/655904. [DOI] [PubMed] [Google Scholar]
- 75.Dawood FS. Fiore A. Kamimoto L, et al. Influenza-associated pneumonia in children hospitalized with laboratory-confirmed influenza, 2003-2008. Pediatr Infect Dis J. 2010 Jul;29(7):585–590. doi: 10.1097/inf.0b013e3181d411c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shieh WJ. Blau DM. Denison AM, et al. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol. 2010 Jul;177(1):166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uyeki TM. Prasad R. Vukotich C, et al. Low sensitivity of rapid diagnostic test for influenza. Clin Infect Dis. 2009 May 1;48(9):e89–92. doi: 10.1086/597828. [DOI] [PubMed] [Google Scholar]
- 78.World Health Organization. Global Alert and Response (GAR) http://www.who.int/csr/disease/influenza/influenzanetwork/flunet/en/index.html. [Apr 22;2011 ]. http://www.who.int/csr/disease/influenza/influenzanetwork/flunet/en/index.html
- 79.Lindblade KA. Arvelo W. Gray J, et al. A comparison of the epidemiology and clinical presentation of seasonal influenza A and 2009 pandemic influenza A (H1N1) in Guatemala. PLoS One. 2010;5(12):e15826. doi: 10.1371/journal.pone.0015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olsen SJ. Thamthitiwat S. Chantra S, et al. Incidence of respiratory pathogens in persons hospitalized with pneumonia in two provinces in Thailand. Epidemiol Infect. 2010 Dec;138(12):1811–1822. doi: 10.1017/S0950268810000646. [DOI] [PubMed] [Google Scholar]
- 81.U.S. Centers for Disease Control and Prevention. 2009 H1N1 Flu: Situation Update. http://www.cdc.gov/h1n1flu/update.htm. [Apr 22;2011 ]. http://www.cdc.gov/h1n1flu/update.htm
- 82.Klugman KP. Astley CM. Lipsitch M. Time from illness onset to death, 1918 influenza and pneumococcal pneumonia. Emerg Infect Dis. 2009 Feb;15(2):346–347. doi: 10.3201/eid1502.081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mills CE. Robins JM. Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004 Dec 16;432(7019):904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goubar A. Ades AE. De Angelis D, et al. Estimates of human immunodeficiency virus prevalence and proportion diagnosed based on Bayesian multiparameter synthesis of surveillance data. Journal of the Royal Statistical Society, Series A. 2008;171(3):1–27. [Google Scholar]
- 85.Donnelly CA. Ghani AC. Leung GM, et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003 May 24;361(9371):1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Webb SA. Pettila V. Seppelt I, et al. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med. 2009 Nov 12;361(20):1925–1934. doi: 10.1056/NEJMoa0908481. [DOI] [PubMed] [Google Scholar]
- 87.U.S. Department of Health and Human Services. Assessment of the 2009 Influenza A (H1N1) Pandemic on Selected Countries in the Southern Hemisphere: Argentina, Australia, Chile, New Zealand and Uruguay. 2009. http://www.flu.gov/professional/global/final.pdf. [Apr 22;2011 ]. http://www.flu.gov/professional/global/final.pdf [DOI] [PMC free article] [PubMed]
- 88.Goldstein E. Miller JC. O'Hagan JJ. Lipsitch M. Pre-dispensing of antivirals to high-risk individuals in an influenza pandemic. Influenza Other Respi Viruses. 2010 Mar;4(2):101–112. doi: 10.1111/j.1750-2659.2009.00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morgan OW. Bramley A. Fowlkes A, et al. Morbid obesity as a risk factor for hospitalization and death due to 2009 pandemic influenza A(H1N1) disease. PLoS One. 2010;5(3):e9694. doi: 10.1371/journal.pone.0009694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fraser C. Riley S. Anderson RM. Ferguson NM. Factors that make an infectious disease outbreak controllable. Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):6146–6151. doi: 10.1073/pnas.0307506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cauchemez S. Boelle PY. Thomas G. Valleron AJ. Estimating in real time the efficacy of measures to control emerging communicable diseases. Am J Epidemiol. 2006 Sep 15;164(6):591–597. doi: 10.1093/aje/kwj274. [DOI] [PubMed] [Google Scholar]
- 92.Wallinga J. Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol. 2004 Sep 15;160(6):509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shaman J. Goldstein E. Lipsitch M. Absolute humdity and pandemic versus epidemic influenza. Am J Epidemiol. 2011;173(2):127–135. doi: 10.1093/aje/kwq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wallinga J. Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007 Feb 22;274(1609):599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donnelly C. Finelli L. Cauchemez S, et al. Serial intervals and the temporal distribution of secondary infections within households of 2009 pandemic influenza A (H1N1)—Implications for influenza control recommendations. Clin Infect Dis. 2011;52(Suppl 1):S123–S130. doi: 10.1093/cid/ciq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goldstein E. Dushoff J. Ma J. Plotkin JB. Earn DJ. Lipsitch M. Reconstructing influenza incidence by deconvolution of daily mortality time series. Proc Natl Acad Sci U S A. 2009 Dec 22;106(51):21825–21829. doi: 10.1073/pnas.0902958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.White LF. Pagano M. A likelihood-based method for real-time estimation of the serial interval and reproductive number of an epidemic. Stat Med. 2008 Jul 20;27(16):2999–3016. doi: 10.1002/sim.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.White LF. Pagano M. Reporting errors in infectious disease outbreaks, with an application to pandemic influenza A/H1N1. Epidemiol Perspect Innov. 2010;7:12. doi: 10.1186/1742-5573-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ferguson NM. Cummings DA. Cauchemez S, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005 Sep 8;437(7056):209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 100.Germann TC. Kadau K. Longini IM., Jr Macken CA. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A. 2006 Apr 11;103(15):5935–5940. doi: 10.1073/pnas.0601266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu JT. Leung GM. Lipsitch M. Cooper BS. Riley S. Hedging against antiviral resistance during the next influenza pandemic using small stockpiles of an alternative chemotherapy. PLoS Med. 2009 May 19;6(5):e1000085. doi: 10.1371/journal.pmed.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anderson RM. May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford University Press; 1991. [Google Scholar]
- 103.Keeling MJ. Rohani P. Modeling Infectious Diseases in Humans and Animals. Princeton: Princeton University Press; 2008. [Google Scholar]
- 104.Halloran ME. Ferguson NM. Eubank S, et al. Modeling targeted layered containment of an influenza pandemic in the United States. Proc Natl Acad Sci U S A. 2008 Mar 25;105(12):4639–4644. doi: 10.1073/pnas.0706849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cannell JJ. Vieth R. Umhau JC, et al. Epidemic influenza and vitamin D. Epidemiol Infect. 2006 Dec;134(6):1129–1140. doi: 10.1017/S0950268806007175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cauchemez S. Ferguson NM. Wachtel C, et al. Closure of schools during an influenza pandemic. Lancet Infect Dis. 2009 Aug;9(8):473–481. doi: 10.1016/S1473-3099(09)70176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shaman J. Pitzer VE. Viboud C. Grenfell BT. Lipsitch M. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol. 2010;8(2):e1000316. doi: 10.1371/journal.pbio.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hancock K. Veguilla V. Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009 Nov 12;361(20):1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 109.Epstein JM. Parker J. Cummings D. Hammond RA. Coupled contagion dynamics of fear and disease: mathematical and computational explorations. PLoS One. 2008;3(12):e3955. doi: 10.1371/journal.pone.0003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu JT. Lee CK. Cowling BJ. Yuen KY. Logistical feasibility and potential benefits of a population-wide passive-immunotherapy program during an influenza pandemic. Proc Natl Acad Sci U S A Feb. 16;107(7):3269–3274. doi: 10.1073/pnas.0911596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wakeley J. Coalescent Theory: An Introduction. Greenwood Village: Roberts & Company Publishers; 2008. [Google Scholar]
- 112.Barry JM. Viboud C. Simonsen L. Cross-protection between successive waves of the 1918-1919 influenza pandemic: epidemiological evidence from US Army camps and from Britain. J Infect Dis. 2008 Nov 15;198(10):1427–1434. doi: 10.1086/592454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andreasen V. Viboud C. Simonsen L. Epidemiologic characterization of the 1918 influenza pandemic summer wave in Copenhagen: implications for pandemic control strategies. J Infect Dis. 2008 Jan 15;197(2):270–278. doi: 10.1086/524065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Olson DR. Simonsen L. Edelson PJ. Morse SS. Epidemiological evidence of an early wave of the 1918 influenza pandemic in New York City. Proc Natl Acad Sci U S A. 2005 Aug 2;102(31):11059–11063. doi: 10.1073/pnas.0408290102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Herfst S. Chutinimitkul S. Ye J, et al. Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. J Virol. 2010 Apr;84(8):3752–3758. doi: 10.1128/JVI.02634-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chutinimitkul S. Herfst S. Steel J, et al. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol. 2010 Nov;84(22):11802–11813. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mak GC. Au KW. Tai LS, et al. Association of D222G substitution in haemagglutinin of 2009 pandemic influenza A (H1N1) with severe disease. Euro Surveill. 2010;15(14):19534. [PubMed] [Google Scholar]
- 118.Kilander A. Rykkvin R. Dudman SG. Hungnes O. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009-2010. Euro Surveill. 2010 Mar 4;15(9):19498. doi: 10.2807/ese.15.09.19498-en. [DOI] [PubMed] [Google Scholar]
- 119.Preliminary review of D222G amino acid substitution in the haemagglutinin of pandemic influenza A (H1N1) 2009 viruses. Wkly Epidemiol Rec. 2010 Jan 22;85(4):21–22. [PubMed] [Google Scholar]
- 120.Miller MA. Viboud C. Balinska M. Simonsen L. The signature features of influenza pandemics—implications for policy. N Engl J Med. 2009 Jun 18;360(25):2595–2598. doi: 10.1056/NEJMp0903906. [DOI] [PubMed] [Google Scholar]
- 121.Simonsen L. Clarke MJ. Schonberger LB. Arden NH. Cox NJ. Fukuda K. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J Infect Dis. 1998 Jul;178(1):53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- 122.Perrow C. Normal Accidence: Living with High Risk Technologies. Princeton, NJ: Princeton University Press; 1999. [Google Scholar]
- 123.Weick KE. Making Sense of the Organization. Malden, MA: Blackwell; 2001. The vulnerable system: an analysis of the Tenerife air disaster; pp. 125–147. [Google Scholar]
- 124.Transmission dynamics and impact of pandemic influenza A (H1N1) 2009 virus. Wkly Epidemiol Rec. 2009 Nov 13;84(46):481–484. [PubMed] [Google Scholar]
- 125.Leung GM. Nicoll A. Reflections on pandemic (H1N1) 2009 and the international response. PLoS Med. 2010;7(10) doi: 10.1371/journal.pmed.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goldstein E. Cowling BJ. O'Hagan JJ, et al. Oseltamivir for treatment and prevention of pandemic influenza A/H1N1 virus infection in households, Milwaukee, 2009. BMC Infect Dis. 2010 Jul 20;10(1):211. doi: 10.1186/1471-2334-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eurosurveillance website. http://www.eurosurveillance.org/Public/ForAuthors/ForAuthors.aspx. [Apr 22;2011 ]. http://www.eurosurveillance.org/Public/ForAuthors/ForAuthors.aspx
- 128.Xing W. Hejblum G. Leung GM. Valleron AJ. Anatomy of the epidemiological literature on the 2003 SARS outbreaks in Hong Kong and Toronto: a time-stratified review. PLoS Med. 2010;7(5):e1000272. doi: 10.1371/journal.pmed.1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.U.S. Department of Health and Human Services. Healthcare Cost and Utilization Project (HCUP) http://www.ahrq.gov/data/hcup/ [Apr 22;2011 ]. http://www.ahrq.gov/data/hcup/
- 130.U.S. Centers for Disease Control and Prevention. National Vital Statistics System. Mortality Data. http://www.cdc.gov/nchs/deaths.htm. [Apr 22;2011 ]. http://www.cdc.gov/nchs/deaths.htm
- 131.Mazick A. Gergonne B. Wuillaume F, et al. Higher all-cause mortality in children during autumn 2009 compared with the three previous years: pooled results from eight European countries. Euro Surveill. 2010 Feb 4;15(5) [PubMed] [Google Scholar]
- 132.U.S. Centers for Disease Control and Prevention. Seasonal Influenza. Flu Activity and Surveillance. http://www.cdc.gov/flu/weekly/fluactivitysurv.htm. [Apr 22;2011 ]. http://www.cdc.gov/flu/weekly/fluactivitysurv.htm
- 133.Lajous M. Danon L. Lopez-Ridaura R, et al. Mobile messaging as surveillance tool during pandemic (H1N1) 2009, Mexico. Emerg Infect Dis. 2010 Sep;16(9):1488–1489. doi: 10.3201/eid1609.100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jackson G. Thornley S. Burden of novel influenza A virus (H1N1) in Auckland and Counties Manukau District Heath Boards (July 2009): a capture-recapture analysis. N Z Med J. 2009 Aug 21;122(1301):66–69. [PubMed] [Google Scholar]
- 135.Zarychanski R. Stuart TL. Kumar A, et al. Correlates of severe disease in patients with 2009 pandemic influenza (H1N1) virus infection. CMAJ. 2010 Feb 23;182(3):257–264. doi: 10.1503/cmaj.091884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Goldstein E. Apolloni A. Lewis B, et al. Distribution of vaccine/antivirals and the ‘least spread line’ in a stratified population. J R Soc Interface. 2010 May 6;7(46):755–764. doi: 10.1098/rsif.2009.0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mossong J. Hens N. Jit M, et al. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 2008 Mar 25;5(3):e74. doi: 10.1371/journal.pmed.0050074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boussard E. Flahault A. Vibert JF. Valleron AJ. Sentiweb: French communicable disease surveillance on the World Wide Web. BMJ. 1996 Nov 30;313(7069):1381–1382. doi: 10.1136/bmj.313.7069.1381. discussion 2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Health Protection Agency. HPA National Influenza Weekly Reports Archive. http://www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/SeasonalInfluenza/EpidemiologicalData/05influsWeeklyinfluenzareportsarchive/ [Apr 22;2011 ]. http://www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/SeasonalInfluenza/EpidemiologicalData/05influsWeeklyinfluenzareportsarchive/
- 140.Carrat F. Vergu E. Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008 Apr 1;167(7):775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 141.Hayden FG. Atmar RL. Schilling M, et al. Use of the selective oral neuraminidase inhibitor oseltamivir to prevent influenza. N Engl J Med. 1999 Oct 28;341(18):1336–1343. doi: 10.1056/NEJM199910283411802. [DOI] [PubMed] [Google Scholar]
- 142.Jordan WS., Jr Denny FW., Jr Badger GF, et al. A study of illness in a group of Cleveland families. XVII. The occurrence of Asian influenza. Am J Hyg. 1958 Sep;68(2):190–212. doi: 10.1093/oxfordjournals.aje.a119962. [DOI] [PubMed] [Google Scholar]
- 143.Monto AS. Koopman JS. Longini IM., Jr Tecumseh study of illness. XIII. Influenza infection and disease, 1976-1981. Am J Epidemiol. 1985 Jun;121(6):811–822. doi: 10.1093/oxfordjournals.aje.a114052. [DOI] [PubMed] [Google Scholar]
- 144.Monto AS. Robinson DP. Herlocher ML. Hinson JM., Jr Elliott MJ. Crisp A. Zanamivir in the prevention of influenza among healthy adults: a randomized controlled trial. JAMA. 1999 Jul 7;282(1):31–35. doi: 10.1001/jama.282.1.31. [DOI] [PubMed] [Google Scholar]
- 145.Noble GR. Epidemiological and clinical aspects of influenza. In: Beare AS, editor. Basic and Applied Influenza Research. Washington, DC: Public Health Service; 1982. [Google Scholar]
- 146.Foy HM. Cooney MK. Allan I. Longitudinal studies of types A and B influenza among Seattle schoolchildren and families, 1968–74. J Infect Dis. 1976 Oct;134(4):362–369. doi: 10.1093/infdis/134.4.362. [DOI] [PubMed] [Google Scholar]
- 147.Welliver R. Monto AS. Carewicz O, et al. Effectiveness of oseltamivir in preventing influenza in household contacts: a randomized controlled trial. JAMA. 2001 Feb 14;285(6):748–754. doi: 10.1001/jama.285.6.748. [DOI] [PubMed] [Google Scholar]
- 148.Fowlkes A. Dasgupta S. Chao E, et al. Influenza Incidence Project: estimating the incidence of influenza-like illness and influenza in the US [abstract 4487]. Program and abstracts of the 2010 Annual Conference Portland, OR. Atlanta, GA: Council of State and Territorial Epidemiologists; 2010. [Google Scholar]