Abstract
Efficient splicing of higher plant pre-mRNAs depends on AU- or U-rich sequences in introns. Moreover, AU-rich sequences present in 3′–untranslated regions (3′–UTRs) may play a role in 3′ end processing of plant mRNAs. Here, we describe the cloning and characterization of a Nicotiana plumbaginifolia nuclear protein that can be cross-linked to U-rich intron and 3′–UTR sequences in vitro, and associates with nuclear poly(A)+ RNA in vivo. The protein, UBP1, strongly enhances the splicing of otherwise inefficiently processed introns when overexpressed in protoplasts. It also increases the accumulation of reporter mRNAs that contain suboptimal introns or are intronless. The enhanced accumulation is apparently due to UBP1 interacting with the 3′–UTR and protecting mRNA from exonucleolytic degradation. The effect on mRNA accumulation but not on mRNA splicing was found to be promoter specific. The fact that these effects of UBP1 can be separated suggests that they represent two independent activities. The properties of UBP1 indicate that it is an hnRNP protein that functions at multiple steps to facilitate the nuclear maturation of plant pre-mRNAs.
Keywords: hnRNP proteins/mRNA splicing/plant RNA processing/RNA-binding protein/3′-UTR
Introduction
Messenger RNA undergoes multiple processing steps in the course of its maturation from a nascent transcript to a capped, spliced and polyadenylated RNA that is exported from the nucleus. The mechanism by which these events occur and the nature of the complexes that execute them are well documented (Krämer, 1996; Burge et al., 1999; Wahle and Ruegsegger, 1999). Transcription, splicing and polyadenylation can be separated as distinct processes in vitro, but their interdependence and interactions in vivo are increasingly apparent (Bentley, 1999; Wahle and Ruegsegger, 1999). For example, many proteins that function in different processing reactions associate with the C–terminal domain (CTD) of the RNA polymerase II (pol II) complex at transcription initiation, to facilitate coordinated maturation of the RNA transcripts (Bentley, 1999).
The recognition substrate for the various processing machineries consists of the pre-mRNA bound by RNA-binding proteins in a ribonucleoprotein complex. These proteins, referred to as hnRNP proteins (Dreyfuss et al., 1993; Krecic and Swanson, 1999), associate with the nascent transcript and often remain complexed with it throughout its nuclear maturation (Visa et al., 1996). Some of them shuttle between the nucleus and the cytoplasm, playing a role in the nuclear export of mRNA (Nakielny and Dreyfuss, 1997). hnRNP proteins are not merely passive chaperones of pre-mRNA, but instead function dynamically and specifically as regulators of diverse processing events including mRNA splicing, transport and stability (Krecic and Swanson, 1999). The same protein can effect regulated RNA processing at more than one level (Krecic and Swanson, 1999). The best-studied example is the hnRNP protein A1, which functions in alternative splicing and export of mRNA in metazoa (Mayeda and Krainer, 1992; Michael et al., 1995; Visa et al., 1996).
The basic processing of pre-mRNAs in plants resembles the mechanisms employed by other eukaryotes. For instance, multiple functional homologues of spliceosomal components are conserved in plants (Simpson and Filipowicz, 1996; Brown and Simpson, 1998; for recent examples, see Domon et al., 1998; Lopato et al., 1999), and features of the mechanism by which introns are excised in complex multi-intron transcripts also appear to be conserved (Brown and Simpson, 1998). However, there are important differences, best exemplified by the fact that mammalian pre-mRNAs are processed neither accurately nor efficiently in plant cells (reviewed by Simpson and Filipowicz, 1996). This distinction is not due to differences in either the 5′ or 3′ splice sites (5′ss and 3′ss), or the branch point (BP) consensus sequence, since these elements are similar between plant and mammalian introns. Instead, the distinguishing feature of plant pre-mRNA splicing is a compositional bias for AU- or U-rich sequences that typically are distributed throughout the entire length of plant introns. Such sequences are required for efficient intron processing (Goodall and Filipowicz, 1989, 1991; Luehrsen and Walbot, 1994a,b; Gniadkowski et al., 1996; Ko et al., 1998) and can also influence splice site selection (Lou et al., 1993; McCullough et al., 1993). Notably, 3′–untranslated regions (3′–UTRs) in plant mRNAs are also generally AU-rich, and it has been found that enrichment in A+U, or specific U-rich sequences, may enhance the 3′ end processing of pre-mRNAs (Luehrsen and Walbot, 1994b; Rothnie et al., 1994).
We have shown previously that efficient and authentic processing of synthetic introns can be recapitulated in vivo by the incorporation of consensus 5′ss and 3′ss, a consensus BP and random intron sequence that exhibits an A+U bias. The processing of related introns that lack an elevated AU composition is accurate, but inefficient (Goodall and Filipowicz, 1989; Gniadkowski et al., 1996). The inefficient processing of GC–rich introns can be rescued by the addition of short segments of U-rich, but not A-rich, sequence at positions close to either the 5′ss, the 3′ss or the centre of the intron (Gniadkowski et al., 1996). This indicates that it is the U-rich sequence that is recognized as the promotive signal, a conclusion supported by other experiments (Baynton et al., 1996; Ko et al., 1998). The ability of U-rich elements inserted at alternative sites in an intron to promote processing in plant cells indicates that they act in a manner distinct from that of the BP- and 3′ss-proximal polypyrimidine tract that characterizes metazoan introns (Krämer, 1996). Since the processing of GC–rich introns can occur accurately, but inefficiently (Gniadkowski et al., 1996), it suggests that U-rich sequences function as enhancers of processing. It seems likely that hnRNP proteins recognize the U-rich sequences and facilitate spliceosomal association; however, no plant hnRNP proteins have been characterized to date.
In this report, we describe the characterization of a nuclear Nicotiana plumbaginifolia protein, named UBP1, that can be UV cross-linked to plant introns and 3′–UTRs in vitro and which associates with nuclear poly(A)+ RNA in vivo. Transient overexpression of UBP1 in plant cells results in a significant enhancement of processing of inefficiently spliced pre-mRNAs and in an increase of abundance of reporter RNAs that is both intron and promoter dependent. These results identify UBP1 as a novel and important component of the plant pre-mRNA processing machinery.
Results
Cloning of UBP1, a nuclear protein that can be cross-linked to plant pre-mRNA introns in vitro
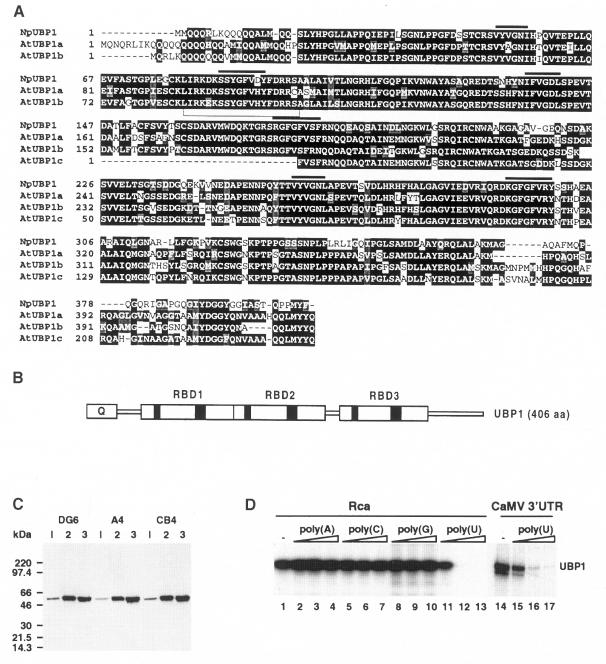
We have previously isolated cDNAs encoding fragments of RBD-type RNA-binding proteins from Nicotiana tabacum (Mieszczak et al., 1992). Complete cDNAs were then obtained by screenings of an N.plumbaginifolia library. The protein, named UBP1, encoded by one of the clones, contains three RBD-type RNA-binding domains (Dreyfuss et al., 1993) and a glutamine-rich N–terminus (Figure 1A and B). UBP1 has a calculated molecular mass of 44 kDa, but Western analysis with specific α-UBP1 monoclonal antibodies (mAbs) indicates that it migrates with an apparent mass of ∼50 kDa (Figure 1C). Northern analysis indicated that the 1.8 kb UBP1 mRNA is expressed at similar levels in the leaves, roots and stems of N.plumbaginifolia (data not shown). Database searches identified expressed sequence tags (ESTs) encoding three variants of UBP1 in both Arabidopsis thaliana and rice. The Arabidopsis cDNAs were sequenced completely and the deduced proteins (AtUBP1s) were found to exhibit 72–75% identity with UBP1 (Figure 1A). Comparison of UBP1 with proteins from non-plant organisms revealed sequence similarity (40–52% identity in RBD1 and RBD2 domains, and 33–35% identity in the RBD3 domain) with the metazoan TIA proteins, implicated in apoptosis, germ cell development and regulation of translation (Gueydan et al., 1999, and references therein), and the yeast Pub1p, a protein associated with poly(A)+ RNA in the nucleus and the cytoplasm (Krecic and Swanson, 1999, and references therein).
Fig. 1. Properties of UBP1. (A) Alignment of N.plumbaginifolia UBP1 with A.thaliana homologues. Amino acids identical or similar in >50% of analysed sequences have a black and grey background, respectively. RNP2 (six amino acids) and RNP1 (eight amino acids) motifs of RBD domains are overlined and shown in black in (B). Genomic sequences encoding AtUBP1a (AC006577) and b (AC007843) have recently been deposited in DDBJ/EMBL/GenBank. (B) Schematic structure of UBP1. (C) Specificity of α-UBP1 mAbs. Cell extracts were prepared from N.plumbaginifolia mesophyll (lanes 1), cell suspension (lanes 2) or N.tabacum BY-2 (lanes 3) protoplasts. DG6, A4 and CB4 mAbs recognize different epitopes in UBP1. (D) Competition of ribopolymers with UV cross-linking of the 32P-labelled Rca RNA (lanes 1–13) or CaMV 3′–UTR (lanes 14–17) to recombinant UBP1. The polymers indicated were added at 5-, 25- and 100–fold excess (calculated in moles of nucleotides) over labelled RNA. Lane –, cross-linking with no competitor added.
A preliminary investigation of the RNA-binding specificity of UBP1 was performed with purified recombinant protein. RNAs corresponding to intron 3 of the Arabidopsis Rubisco activase gene (Rca, Figure 1D) and a synthetic AU-rich intron (Syn7) that is processed efficiently in plant cells (Goodall and Filipowicz, 1989; data not shown) were employed in UV cross-linking experiments where binding was competed by the addition of ribohomopolymers. These experiments revealed that UBP1 is an oligouridylate-binding protein, hence the name. UBP1 also cross-linked efficiently to the cauliflower mosaic virus (CaMV) 3′–UTR, which is likewise AU-rich (Figure 1D).
The nucleotide-binding specificity of UBP1 and its electrophoretic mobility closely resemble those of the 50 kDa poly(U)-binding protein identified previously in nuclear extracts of N.plumbaginifolia as a protein that can be cross-linked efficiently by UV light to plant intron sequences in vitro (Gniadkowski et al., 1996). Immunoprecipitation experiments have demonstrated that UBP1 and the 50 kDa nuclear extract protein are indeed identical or highly related (Figure A, Supplementary material; the supplementary data are available in The EMBO Journal Online).
UBP1 is a nuclear protein
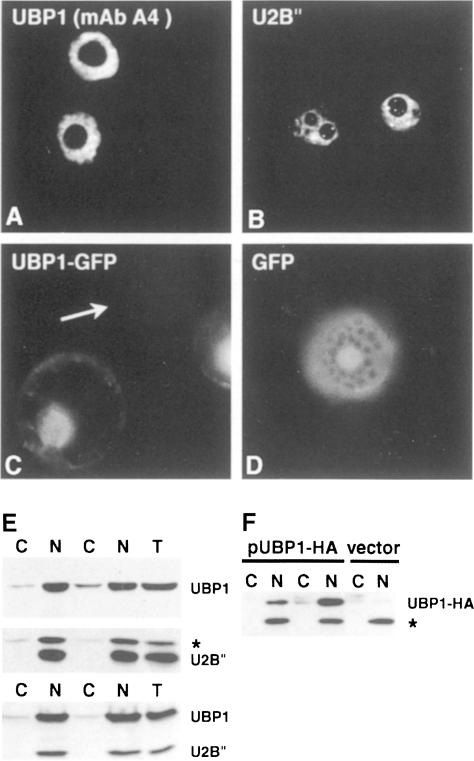
Endogenous UBP1 was localized in tobacco BY2 cells by indirect immunofluoresence, using specific primary mAbs A4 (Figure 2A) and DG6 (not shown), and a Cy3-labelled secondary antibody. UBP1 is nuclear localized, with no significant staining of the cytoplasm (Figure 2A). Its distribution within the nucleus is diffuse, with no staining of defined structures such as coiled bodies [compare with Figure 2B: localization of the spliceosomal protein U2B″ (Beven et al., 1995)] or nucleoli. A similar localization pattern was observed when UBP1 tagged with either the influenza haemagglutinin (HA) epitope (not shown, but see below) or green fluorescent protein (GFP) (Figure 2C) was expressed in protoplasts of N.plumbaginifolia.
Fig. 2. Localization of proteins by indirect immunofluorescence in BY–2 cells (A and B) by GFP fluorescence in living N.plumbaginifolia protoplasts (C and D) and by cell fractionation (E and F). (A) Localization of endogenous UBP1, studied with mAb A4. (B) U2B″ protein visualized with mAb 4G3. (C) Expression of the UBP1–GFP fusion; the arrow indicates a non-transfected protoplast. (D) Expression of GFP alone. (E) Fractionation of N.plumbaginifolia mesophyll protoplasts (upper panel) or N.tabacum BY-2 protoplasts (lower panel). Western blots were analysed using mAbs DG6 (UBP1) and 4G3 (U2B″). A protein marked with an asterisk cross-reacts unspecifically with mouse α–U2B″ mAb. (F) Fractionation of N.plumbaginifolia protoplasts transfected with pUBP1-HA or the empty vector pGGS5. Blots were analysed with mouse α–HA mAb 12CA5. The 40 kDa nuclear protein marked with an asterisk, and cytoplasmic proteins of 50–55 kDa cross-react unspecifically with α–HA mAb. T, total cell extract; C, cytoplasmic fraction; N, nuclear fraction. Duplicate fractionations are shown.
The intracellular distribution of UBP1 was also studied by separating lysates of N.plumbaginifolia and tobacco BY2 protoplasts into nuclear and cytoplasmic fractions. Western analysis of the nuclear U2B″ protein (Figure 2E) and the cytosolic isoform of cysteine synthase (not shown) indicated that there is no appreciable contamination of the cytoplasmic fraction with nuclear proteins or vice versa. Western analysis with α-UBP1 mAbs indicated that only a minor portion of the reactive material is recovered in the cytoplasm (Figure 2E). Even less UBP1 was detected in the cytoplasmic fraction when distribution of the transiently expressed UBP1–HA fusion was analysed with α–HA mAbs (Figure 2F).
UBP1 associates with nuclear poly(A)+ RNA in vivo
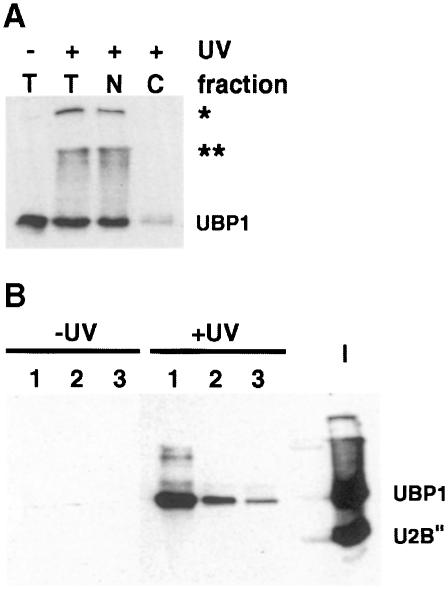
We investigated whether UBP1 associates with pre-mRNA in vivo. Protoplasts prepared from cells grown in suspension were irradiated with UV and extracts from whole cells or nuclear preparations were subjected to oligo(dT)–cellulose chromatography in the presence of SDS. Western analysis of proteins co-eluting with poly(A)+ RNA, isolated from either preparation (Figure 3; data not shown), indicated that UBP1 is associated with poly(A)+ RNA in vivo. The validity of this approach is supported by the absence of the spliceosomal protein U2B″ from the poly(A)+ fractions (Figure 3B) (U2B″ binds specifically to U2 snRNA and not pre-mRNA).
Fig. 3. UV-induced cross-linking of UBP1 to poly(A)+ RNA in vivo. (A) Western blot of extracts prepared from UV-treated (+) and control (–) protoplasts. T, total protoplast extract; N, nuclear extract; C, cytoplasmic fraction. The tops of the stacking and separating gels are marked with asterisks. (B) Detection of UBP1 but not U2B″ in oligo(dT)–cellulose eluates obtained from nuclear extracts originating from UV-irradiated (+UV) but not control (–UV) protoplasts. The blot was probed with a mixture of DG6 and 4G3 mAbs. Numbers indicate consecutive fractions eluted from oligo(dT). Lane I, input nuclear extract from control protoplasts prior to oligo(dT) fractionation.
Overexpression of UBP1 enhances the splicing efficiency of suboptimal introns
We sought to determine whether UBP1 functions in pre-mRNA maturation as a factor contributing to efficient intron recognition. To this end, plasmids designed to express different reporter pre-mRNAs were co-transfected into protoplasts with increasing amounts of plasmid encoding UBP1. The total amount of plasmid DNA was kept constant by the addition of the empty expression vector, pDEDH/Nco. The efficiency of processing was assessed by RNase A/T1 protection, using gene-specific probes complementary to the unspliced forms of the RNA. Endogenous U2 snRNA was monitored to facilitate quantification.
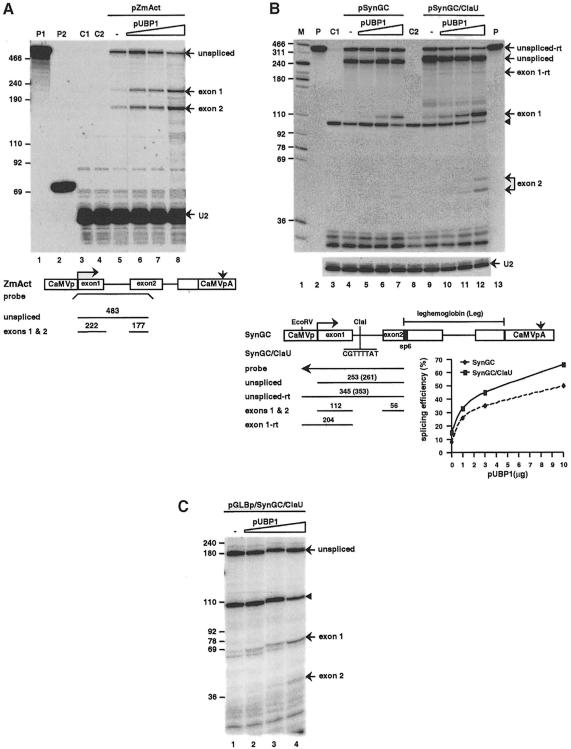
We first studied the processing of introns known to be spliced inefficiently in N.plumbaginifolia proto– plasts: intron 1 of the maize actin gene [ZmAct; this intron is moderately (62%) AU-rich (Goodall and Filipowicz, 1991)] and synthetic GC–rich introns that either lack U–rich sequence altogether or possess one short U-rich island [SynGC and SynGC/ClaU, respectively (Gniadkowski et al., 1996)]. The addition of a plasmid encoding UBP1 stimulated, in a concentration-dependent manner, the splicing efficiency of ZmAct from 44 to 80% (Figure 4A; for quantitation, see legend), of SynGC from 8 to 50% and of SynGC/ClaU from 15 to 65% [Figure 4B; for explanation of the identity of the protected bands ‘unspliced-rt’ and ‘exon1-rt’, diagnostic of RNAs that read through the CaMV polyadenylation (pA) site, see legend]. In addition to the test introns, the reporter transcripts contained a second intron [originating from the leghaemoglobin gene (Leg) positioned downstream; Figure 4; Gniadkowski et al., 1996]. The processing efficiency of the Leg intron present in the SynGC transcript also increased from 47 to 70% in the presence of pUBP1 (data not shown). However, the enhancement of splicing of the upstream SynGC intron was not dependent on the presence of the downstream intron: the processing of SynGC expressed in a pre-mRNA lacking the Leg intron (SynGCΔLeg) increased from 8 to 33% (Figure B, Supplementary material).
Fig. 4. Effect of overexpression of UBP1 on splicing of reporter RNAs expressed in N.plumbaginifolia protoplasts. Schemes of transfected genes and sizes of probes and fragments protected by unspliced and spliced RNAs are indicated in the lower portions of the panels. Protoplasts were always co-transfected with 5 μg of the reporter plasmid and with either empty vector alone (lanes –) or with increasing amounts of pUBP1 (1, 3 and 10 μg). Lanes M and P, size markers and aliquots of undigested probes. (A) Analysis of splicing of the ZmAct intron 1. RNA from protoplasts was analysed with a mixture of ZmAct probe and U2 RNA probe, which detects a 50 nucleotide 5′–terminal fragment of endogenous U2 RNA. Lanes C1 and C2, control mappings of RNA from protoplasts transfected with 15 μg of the empty vector pDEDH/Nco, and with 5 μg of pDEDH/Nco and 10 μg of pUBP1, respectively. Calculated splicing efficiencies are 44, 57, 64 and 80% for transfections with 0, 1, 3 and 10 μg of pUBP1, respectively. (B) Splicing of SynGC and SynGC/ClaU RNAs. Lanes C1 and C2, RNA from protoplasts transfected with pDEDH/Nco (5 μg) and pUBP1 (10 μg) analysed with SynGC and SynGC/ClaU probes, respectively. Mappings with the U2 probe were performed in separate reactions. Protected fragments diagnostic of spliced and unspliced RNAs, and of respective readthrough (rt) RNAs are indicated. [As verified experimentally, some transcripts initiated at the CaMV promoter are not cleaved at the CaMV pA site, and their elongation continues around the plasmid, yielding rtRNAs that encompass the promoter and the SynGC region. Since SynGC–specific probes extend into the promoter region, they detect rtRNAs and yield longer protected fragments (see scheme).] Rt transcripts from pDEDH/Nco and pUBP1 protect a 103 nucleotide fragment, marked with an arrowhead [note that rt transcription is more pronounced with pDEDH/Nco than with pUBP1 (see also Figure 6)]. Scheme: transcription initiation and pA sites, SP6 promoter, Leg regions and insertion of eight nucleotides in the ClaI site of SynGC/ClaU are indicated. Sizes of fragments diagnostic of unspliced and unspliced-rt SynGC/ClaU RNAs are in parentheses. Calculated splicing efficiencies of SynGC and SynGC/ClaU RNAs are shown. The accuracy of splicing of SynGC constructs was confirmed additionally by RT–PCR and sequencing (Gniadkowski et al., 1996). (C) Splicing of SynGC/ClaU RNA initiated from the GLB promoter. Splicing efficiencies are 9, 28, 39 and 52% for transfections with 0, 1, 3 and 10 μg pUBP1, respectively. Yields of RNA are 1.0, 1.0 and 1.2 for transfections with 1, 3 and 10 μg pUBP1.
The processing of single-intron RNAs, containing either SynGC or SynGC/ClaU introns, but expressed from constructs in which the CaMV 35S (referred to as CaMV) promoter was replaced by the tobacco β–glucanase gene (GLB) promoter, was also stimulated by UBP1. The splicing efficiency of SynGC increased from 5 to 20% and that of SynGC/ClaU from 9 to 52% (Figure 4C; data not shown). The addition of pUBP1 also increased the efficiency of processing of the natural ZmAct intron in RNA expressed from the GLB promoter construct from 20 to 60% (data not shown).
We have also examined whether UBP1 affected the processing of otherwise efficiently processed introns: the synthetic AU-rich intron Syn7, present either in a di-intronic transcript (Goodall and Filipowicz, 1989) or in a single intron transcript (Syn3/ivs), and the tobacco chitinase gene present in its natural context (all expressed from the CaMV promoter constructs). These introns were processed at up to 90% efficiency, but co-transfection with UBP1 had no measurable effect on their processing (data not shown). Note that transiently expressed plant pre-mRNAs are never spliced with efficiencies higher than 85–90% (Goodall and Filipowicz, 1989, 1991).
We conclude that the transient overexpression of UBP1 stimulates excision of otherwise inefficiently processed introns in transcripts expressed from either the CaMV or the GLB promoter.
Overexpression of UBP1 increases the abundance of mRNAs in a promoter- and intron-dependent manner
The quantitation of total transcript levels, represented by the sum of unspliced and spliced RNAs analysed in Figure 4, revealed that the phenotype of transient overexpression of UBP1 is more complex. In addition to enhancing splicing, co-transfection of UBP1 also increased steady-state levels of reporter RNAs containing suboptimal introns by 2- to 3–fold. Remarkably, this increase was only apparent when expression of pre-mRNAs was driven by the CaMV (Table I) but not the GLB promoter (Figure 4C and its legend, and see below).
Table I. Effect of UBP1 on abundance of different reporter RNAs initiated at the CaMV promoter.
| Reporter RNA | Average stimulationa | Range (n)b |
|---|---|---|
| Intronless | ||
| Syn3 | 3.5 | 2.5–6 (10) |
| β-globin | 2.1 | 1.4–2.5 (6) |
| GUS | 2.5 | 1.7–3.8 (6) |
| CAT | 2.0 | 1.5–2.2 (3) |
| Chitinase cDNA | 1.0 | 0.7–1.4 (3) |
| Strong introns | ||
| Chitinase gene | 0.7 | 0.6–0.7 (3) |
| Syn7 | 1.0 | 0.8–2.0 (8) |
| Syn3/ivs | 0.6 | 0.5–0.7 (3) |
| Weak introns | ||
| ZmAct | 2.5 | 1.7–2.7 (4) |
| SynGC | 2.3 | 1.4–2.6 (4) |
| SynGC/ClaU | 1.8 | 1.3–2.7 (4) |
aFold increase of RNA level in the presence of 10 μg of co-transfected pUBP1. All quantifications are based on PhosphorImager data corrected for the endogenous U2 snRNA recovery.
bn, number of independent experiments.
We further investigated the effect of UBP1 overexpression on mRNA levels by analysing whether it would affect RNAs that did not contain introns. We found that, with the exception of the chitinase cDNA, the accumulation of all reporter RNAs expressed from the CaMV promoter increased 2- to 3–fold upon co-transfection with UBP1 (Figure 5A and B, lanes 1–4; Table I). Where examined, we found a corresponding 2.5–fold increase in the amount of reporter protein expressed, indicating that UBP1 neither inhibited nor enhanced the translation of the reporter mRNAs (data not shown).
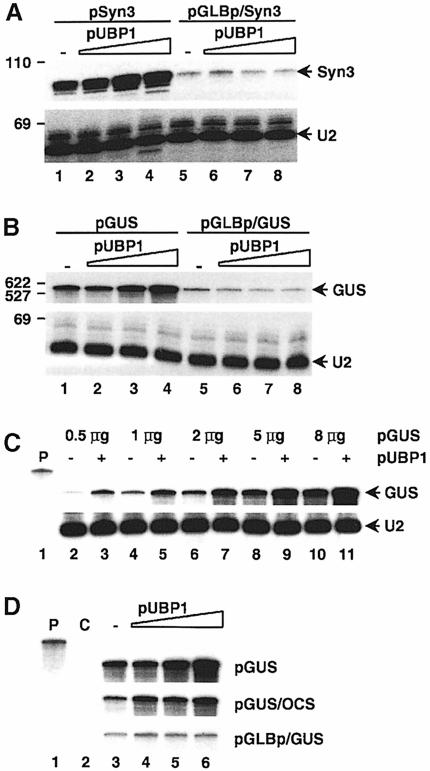
Fig. 5. Effect of UBP1 on accumulation of intronless RNAs is promoter specific. (A and B) Effect of UBP1 on accumulation of Syn3 RNA (A) and GUS mRNA (B) initiated at the CaMV (lanes 1–4) and GLB (lanes 5–8) promoters. Yields of RNA expressed from pGLBp/Syn3 are 1.2, 0.7 and 0.8 for transfections with 1, 3 and 10 μg pUBP1. Respective values for pGLBp/GUS are 0.6, 0.5 and 0.5. (C) Effect of UBP1 on the accumulation of GUS mRNA expressed from pGUS is independent of the initial level of RNA expressed in protoplasts. Different amounts of pGUS were co-transfected with 10 μg of either empty expression vector pDEDH/Nco (lanes –) or pUBP1 (lanes +). (D) Effect of UBP1 on GUS mRNA accumulation is independent of the nature of the pA site. Protoplasts were transfected with pGUS (upper row), pGUS/OCS (middle row) or pGLBp/GUS (lower row) and increasing concentrations of pUBP1. Lane C, mapping of RNA from protoplasts transfected by the empty vector. U2-protected bands are not shown. Yields of RNA for transfections with 1, 3 and 10 μg pUBP1 are, respectively, 1.4, 2.2 and 2.6 for pGUS, 2.5, 2.2 and 4.5 for pGUS/OCS, and 1, 0.9 and 1.1 for pGLBp/GUS. All panels in each figure represent portions of the same gel.
To investigate the promoter specificity of the UBP1-mediated enhancement of RNA accumulation, we have compared the effect of UBP1 on the accumulation of Syn3 and GUS transcripts initiated from the GLB and CaMV promoters. The respective pairs of genes containing different promoters were constructed such that they yield transcripts of identical sequences. As shown in Figure 5A and B, overexpression of UBP1 did not increase levels of RNAs expressed from the GLB promoter (lanes 5–8), in marked contrast to RNAs driven by the CaMV promoter (lanes 1–4; for quantification, see legend). Likewise, accumulation of the poly(A)+ form of U2 RNA was increased when its synthesis was initiated from the CaMV (see below) but not the GLB promoter (data not shown).
We have verified that the promoter-specific effect of UBP1 is not related to the different amounts of RNAs produced from CaMV and GLB promoters. Higher levels of RNA expressed from the CaMV constructs could, for example, lead to post-transcriptional gene silencing, a phenomenon that can be triggered by overexpression of transgenes (Vaucheret et al., 1998), and this process potentially could be reversed by UBP1. As shown in Figure 5C, the effect of UBP1 on the accumulation of RNA transcribed from pGUS was independent of the starting level of GUS mRNA expressed in protoplasts. This is supported further by the experiment with the construct pGUS/OCS, in which the CaMV pA site is replaced by the site originating from the octopine synthase gene. Expression of GUS mRNA from pGUS/OCS and GLBp/GUS was found to be comparable, but response to UBP1 was only observed for the gene containing the CaMV promoter (Figure 5D). This experiment also indicates that the effect of UBP1 on RNA accumulation is independent of the nature of the pA site.
As summarized in Table I, the effect of UBP1 on mRNA accumulation is intron dependent. Although an increase in mRNA levels was observed with mRNAs lacking introns or possessing inefficiently processed introns, we have never observed an increase in RNA amount for transcripts bearing efficiently processed introns, either natural or synthetic. In fact, overexpression of UBP1 decreased the yield of some of these RNAs.
In conclusion, we find that the overexpression of UBP1 can increase the levels of transcripts in a manner dependent on the promoter driving their expression and on the quality of the intron present in the pre-mRNA.
UBP1 does not stimulate transcription or the 3′ end cleavage/polyadenylation reaction
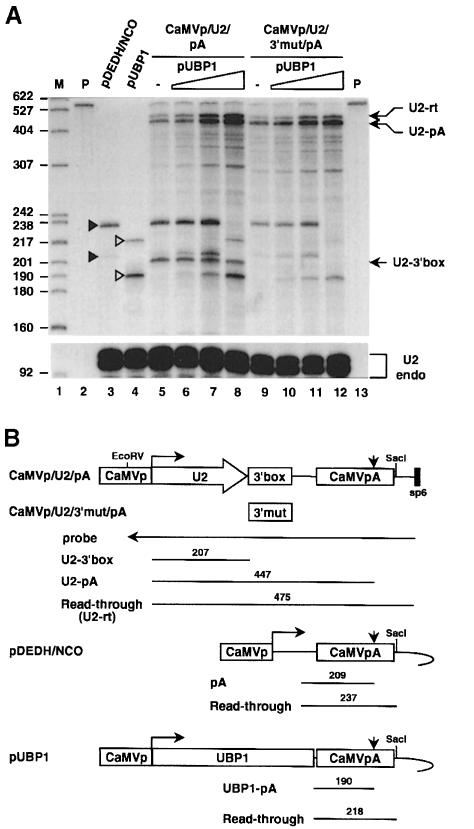
We investigated the stage of pre-mRNA maturation at which UBP1 exerts its effect on mRNA levels. It seemed unlikely that this was simply the result of transcriptional stimulation since mRNA accumulation was seen for only some RNAs driven by the CaMV promoter (Table I). To address this question more rigorously, we used a modified U2 snRNA gene as a reporter. In higher plants, snRNAs are transcribed from specific pol II promoters and are terminated at a sequence downstream of the coding region (3′ box). However, they can also be expressed from conventional mRNA pol II promoters. In such a case, pol II partially reads through the 3′ box and transcripts can be cleaved and polyadenylated at an mRNA pA signal placed downstream (Connelly and Filipowicz, 1993). We examined the effect of UBP1 on expression of U2 driven by the CaMV promoter and bearing a functional mRNA pA signal positioned downstream of either the wild-type or a debilitated 3′ box (constructs CaMVp/U2/pA and CaMVp/U2/3′mut/pA, respectively; Figure 6B). If UBP1 affects transcription initiation, accumulation of both kinds of RNAs, one terminated at the 3′ box and one extending to the CaMV pA site, would be stimulated. However, if UBP1 instead exerts its activity via the 3′ end of an mRNA, then only the poly(A)+ U2 RNA should accumulate in the presence of UBP1. As shown in Figure 6, the results are consistent with the latter prediction and argue against UBP1 acting through transcriptional stimulation.
Fig. 6. UBP1 increases accumulation of U2 RNA processed at an mRNA pA site but not that terminated at the 3′ box. (A) RNase mapping. Lanes 3 and 4, RNA from protoplasts transfected with 10 μg of the empty vector pDEDH/Nco or pUBP1. The identity of protected fragments diagnostic of RNAs expressed from these plasmids (marked with filled and open triangles, respectively) and of U2 RNA-specific fragments is explained in (B). Note that expression of UBP1 has no effect on the ratio of correctly processed to readthrough RNAs expressed from pDEDH/Nco and pUBP1. (B) Schemes of transfected plasmids showing the sizes of RNA fragments protected by the probe used. Calculated ratios of U2-pA to U2-rt for lanes 5–8 are 2.2, 1.7, 1.5 and 1.6, respectively, and for lanes 9–12 they are 3.6, 3.5, 2.3 and 2.8, respectively.
Because of the apparent specificity for an mRNA 3′ end, we next asked whether UBP1 enhances the 3′ end cleavage/polyadenylation reaction. We used constructs containing mutations in cis-acting elements of the CaMV pA site to investigate the effect of UBP1 further. Test pA sites were inserted downstream of the CAT reporter gene and were followed by another pA site from the nopaline synthase gene, which acts as a trap for transcripts not processed at the mutant CaMV site (Rothnie et al., 1994). Although co-transfection of UBP1 increased the steady-state levels of CAT mRNA ∼2–fold, the efficiency of cleavage/polyadenylation at the CaMV site in transcripts containing different mutations in either the AAUAAA hexamer or upstream UUUGUA enhancer elements was not increased (Supplementary Figure C; and data not shown). Consistent with the lack of effect on CaMV pA site utilization, co-transfection of UBP1 did not affect the ratio of correctly processed to readthrough RNAs in any of the other co-transfection experiments we have performed (see, for example, Figure 6 and its legend).
In summary, the results presented above indicate that UBP1 stimulates neither transcription nor the 3′ end cleavage/polyadenylation reaction. They suggest, however, that UBP1 requires an mRNA 3′ end to exert its effect on RNA accumulation.
UBP1 may act by protecting mRNAs against exonucleolytic degradation
We attempted to examine the influence of UBP1 on the stability of reporter RNAs. However, the half-life of Syn3 RNA (selected because of its strongest response to UBP1) was found to be very long (>10 h) in both the presence and absence of UBP1 (data not shown), making the protoplast transient assay system unsuitable to assess the effect of UBP1 on RNA stability.
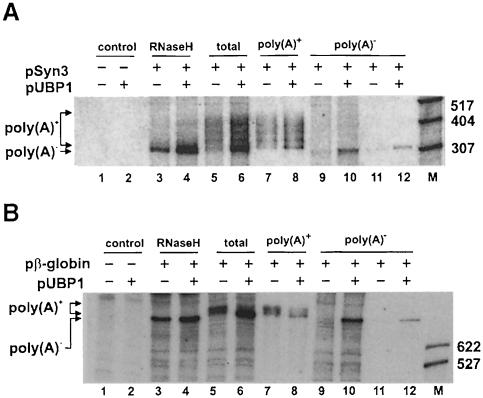
We have investigated whether UBP1 has an effect on the status of the 3′ end of reporter RNAs expressed in protoplasts. Northern analysis of Syn3 transcripts separated on polyacrylamide gels revealed that UBP1 not only increased the yield of the Syn3 poly(A)+ RNA, but also led to the appearance of an additional shorter RNA. Co-migration of this additional RNA species with the appropriate size marker obtained by the treatment of Syn3 RNA with oligo(dT) and RNase H, and its presence in poly(A)– but not poly(A)+ RNA fractions, indicated that the RNA corresponds to the deadenylated ‘body’ of Syn3 (Figure 7A). Similarly, when the β–globin mRNA was expressed in protoplasts, its deadenylated form accumulated in the presence of UBP1 (Figure 7B).
Fig. 7. Effect of UBP1 on the status of the 3′ end of Syn3 (A) and β–globin (B) mRNAs. RNA from protoplasts transfected with the plasmids indicated was fractionated on a 6% polyacrylamide–8 M urea gel and analysed by Northern blots. Lanes 1, 2, 5 and 6, total RNA; lanes 3 and 4, RNA treated with RNase H; lanes 7 and 8, poly(A)+ RNA; lanes 9–12, poly(A)– RNA either not retained on (9 and 10) or washed off from (11 and 12) oligo(dT)–beads.
The most straightforward interpretation of these results is that UBP1 binds to the 3′–UTR and protects the mRNA against exonucleolytic degradation.
Discussion
Many aspects of nuclear pre-mRNA maturation are highly conserved between higher plants and other organisms. However, a fundamental distinguishing feature remains the requirement for U-rich intron sequences for efficient splicing of plant pre-mRNAs (see Introduction). With the aim of elucidating the molecular basis for this requirement, we have cloned a cDNA encoding UBP1, the protein previously identified in plant nuclear extracts as the major product that UV cross-links to plant introns in vitro (Gniadkowski et al., 1996). The properties of UBP1 are consistent with its expected general role in pre-mRNA maturation. (i) It is a nuclear protein having a strong preference for U-rich RNAs. (ii) It associates with poly(A)+ RNA in the nucleus of plant cells. (iii) It is constitutively expressed and conserved in other plant genomes. We have demonstrated that UBP1 strongly enhances splicing of otherwise inefficiently processed introns when transiently overexpressed in protoplasts. However, we have found that UBP1 also increases accumulation of reporter mRNAs in a manner that is both intron and promoter dependent. The enhanced accumulation appears to be due to UBP1 interacting with the 3′–UTR and protecting mRNA from degradation. The observation that stimulatory activities on RNA splicing and accumulation can be uncoupled from each other suggests that they represent two independent effects of UBP1. The data are consistent with UBP1 being an hnRNP protein that functions to facilitate, at multiple steps, the maturation of pre-mRNAs in nuclei of plant cells.
Transient overexpression of UBP1 in transfected protoplasts resulted in strong (up to 6–fold) improvement in the efficiency of intron excision from pre-mRNAs which are otherwise spliced inefficiently. The enhancement of splicing was independent of the promoter used to drive transcription, and the accuracy of splicing was preserved. Importantly, the processing of U-rich and otherwise efficiently processed introns was unaffected by UBP1. The effect on pre-mRNA splicing is specific to UBP1; we have cloned several other higher plant nuclear RNA-binding proteins having a binding preference for U-rich RNAs, but none exhibits this effect when examined in the same way (Z.Lorkovic and W.Filipowicz, unpublished data).
In addition to an effect on splicing, co-transfection with UBP1 led to an increased accumulation of many reporter mRNAs, which were either intronless or possessed inefficiently processed introns. mRNA abundance was not increased when UBP1 was co-transfected with plasmids expressing transcripts bearing efficiently spliced introns. In contrast to the promoter-independent effect on splicing, UBP1 increased the levels of mRNA derived from the viral CaMV 35S promoter but not the cellular GLB promoter. Again, we have verified that the observed effect on RNA accumulation does not result from the non-specific overexpression of an RNA-binding protein: with the exception of a protein that interacts with UBP1 (see below), none of the other RNA-binding proteins we have examined exhibits this phenotype. In addition, there does not appear to be a widespread, non-specific interference of gene expression associated with UBP1 overexpression; we found no evidence for an effect of UBP1 on the transcription, polyadenylation or translation of reporter RNAs.
The fact that co-transfection of UBP1 can promote both efficient pre-mRNA splicing and mRNA accumulation raises the possibility that the effect on splicing is indirect. However, the data demonstrating that the two phenomena can be separated strongly argue against this. First, the promotion of efficient pre-mRNA splicing was detectable even when the GLB promoter was used to drive expression of the test transcripts, and yet no effect of UBP1 on mRNA accumulation was ever observed when this promoter was used. Secondly, we have identified a novel RNA-binding protein that interacts with UBP1 and, when overexpressed in protoplasts, has a stimulatory effect on the accumulation of reporter mRNAs that resembles that of UBP1, but has no effect on splicing (M.Lambermon and W.Filipowicz, unpublished data). These findings reveal that an effect on mRNA accumulation need not necessarily result in an apparent effect on pre-mRNA splicing.
The mechanism by which UBP1 effects the enhancement of splicing efficiency is not clear. The effect was observed with natural introns that are moderately AU-rich (maize actin and soybean leghaemoglobin gene introns) and with synthetic introns that are GC–rich. Although, based on the in vitro binding specificity, it seems unlikely that the effect of UBP1 is mediated through a direct association with GC–rich RNA, its sequence preference could be different in the cell. More probably, overexpression of UBP1 might increase the likelihood of making contacts with other proteins that subsequently facilitate stable recognition of pre-mRNA by the spliceosome. In a similar way, the addition of excess SR proteins can dispense with the need for U1 snRNA in 5′ss recognition (Crispino et al., 1994; Tarn and Steitz, 1994) and excess SC35 can eliminate the need for U2AF binding to the polypyrimidine tract (MacMillan et al., 1997).
We have investigated the mechanism responsible for the stimulatory effect of UBP1 on mRNA abundance. We found no evidence that UBP1 acts through increasing the efficiency of transcription or the 3′ end cleavage/polyadenylation reaction. However, analysis of Syn3 and β–globin transcripts isolated from protoplasts co-expressing UBP1 identified abundant RNA forms corresponding to the poly(A)– body of these RNAs. It is probable that these and other heterologous or intronless reporter RNAs used in this study lack optimal mRNA maturation signals. As such, these RNAs may be processed inefficiently in plant nuclei and be liable to degradation by intracellular nucleases. Like higher plant introns, the 3′–UTRs of plant pre-mRNAs are generally AU-rich (Luehrsen and Walbot, 1994b; Rothnie, 1996), and we have found that UBP1 can be efficiently UV cross-linked to the CaMV 3′–UTR in vitro. We propose that it is the binding of UBP1 to the 3′–UTR of suboptimal pre-mRNAs that results in their increased accumulation. This association with UBP1 might prevent 3′→5′ exonuclease-mediated degradation from continuing beyond the poly(A) tail and may make the RNAs available for readenylation and export from the nucleus to the cytoplasm. Our finding that the poly(A)– RNA accumulating upon expression of UBP1 is localized in the nucleus (unpublished results) is consistent with this interpretation. Importantly, the poly(A)+ Syn3 RNA isolated from protoplasts appears to have a periodic distribution of poly(A) tails, with prominent species differing in length by ∼30 nucleotides (Figure 7). This ladder probably represents decay intermediates resulting from the association of the poly(A)-binding protein with the poly(A) tail (Körner and Wahle, 1997). This supports the suggestion that the poly(A)– mRNA forms that accumulate upon co-transfection with UBP1 represent deadenylated RNAs rather than RNAs that underwent cleavage but no polyadenylation of the 3′ end. It should be noted that cleavage and polyadenylation are believed to be tightly coupled in vivo (Wahle and Ruegsegger, 1999).
The available evidence is consistent with UBP1 functioning in the nucleus. This conclusion is supported by the nuclear localization of the protein and its association with the nuclear poly(A)+ RNA in vivo, and also by its stimulatory effect on splicing and the intron dependence of the effect on mRNA accumulation. A comparison of the influence of UBP1 on the expression of Syn3 and Syn3/ivs is particularly important in this regard. Syn3/ivs differs from Syn3 only by the presence of the optimal AU-rich intron interrupting the Syn3 RNA coding region. The spliced Syn3/ivs transcript and Syn3 RNA are therefore identical. Since UBP1 increases the abundance of Syn3 RNA, but not the identical RNA formed as a result of splicing, it is unlikely that UBP1 acts as a factor affecting RNA turnover in the cytoplasm [but see Matsumoto et al. (1998) for a discussion of the possible effects of the nuclear history of pre-mRNA on its activity in the cytoplasm]. In addition, we found that inhibition of transcription does not alter the nuclear–cytoplasmic distribution of UBP1 (unpublished results), indicating that UBP1 does not shuttle between the two compartments in a transcription-dependent manner (Nakielny and Dreyfuss, 1997).
The stimulatory effect of UBP1 on RNA accumulation was found to be promoter specific; it occurred when transcription was driven by the CaMV but not the GLB promoter. Many protein factors participating in different RNA processing reactions in yeast and mammals recently have been reported to associate with the CTD domain of the large subunit of pol II at transcription initiation (Bentley, 1999). For some mammalian SR proteins, it was shown that their recruitment to pre-mRNA is modulated by the promoter structure (Cramer et al., 1999). One likely explanation of the promoter specificity described in this work is that association of UBP1 with RNA transcripts similarly requires its prior interaction with the transcription complex. We have investigated whether UBP1 can interact with the CTD domain. However, we found no significant interaction in vitro with either the unphosphorylated or phosphorylated form of the yeast CTD nor any evidence for an interaction with the Arabidopsis CTD in the yeast two-hybrid system (unpublished data). Since the UBP1 effect on accumulation is promoter dependent, it is possible that its interaction with the transcription machinery involves other, more specific, transcription components, as has been described recently for the ASF/SF2 splicing factor in mammalian cells (Ge et al., 1998). Alternatively, UBP1 might interact with the transcription machinery only as part of a complex with other proteins. The scenario that UBP1 has to associate with specific proteins in order to exert its different functions would help to explain why only its effect on RNA accumulation and not that on splicing is promoter specific.
It is well established that the presence of introns can dramatically affect the expression levels of mRNAs in both mammals (Liu and Mertz, 1995, and references therein) and plants (Callis et al., 1987; Simpson and Filipowicz, 1996). In mammalian cells, many natural intronless mRNAs contain sequence elements that enable efficient intron-independent processing and nuclear export (Liu and Mertz, 1995; Huang et al., 1999). The processing of herpes simplex virus thymidine kinase mRNA, for example, is facilitated by the specific association of the hnRNP L protein (Liu and Mertz, 1995). The output of the Arabidopsis genome project indicates that ∼20% of genes lack introns. Our observation that overexpression of UBP1 affects the accumulation of only intronless or otherwise suboptimal pre-mRNAs in a promoter-dependent manner suggests that UBP1 may play a role in regulating the expression of certain classes of pre-mRNAs.
We recognize that it can be difficult to interpret the significance of overexpression data in terms of the normal function of the protein under investigation. The continued absence of competent plant cell-derived in vitro systems to investigate RNA processing reactions has made it impossible for us to study the significance of the association of UBP1 with plant pre-mRNAs under defined conditions in vitro. In addition, attempts to generate stable transgenic tobacco lines expressing UBP1, which might have facilitated an analysis of its effect on mRNA metabolism, failed, presumably due to ultimate lethality of the effects of its overexpression. This phenotype is consistent with UBP1 exerting a profound effect on the normal balance of gene expression in planta and would suggest that UBP1 levels normally are tightly controlled. Indeed, the importance of maintaining appropriate UBP1 levels in the cell is implied by our findings that the protein can autoregulate its expression by interacting with the unusually U-rich 5′–UTR of its own message (unpublished data).
In conclusion, the expression, conservation and RNA-binding properties of UBP1, taken together with its effects on pre-mRNA splicing and accumulation, are consistent with UBP1 being an hnRNP protein that plays an important role, at more than one level, in plant nuclear pre-mRNA maturation. The promoter dependence and pre-mRNA specificity of some UBP1 effects suggest that, in addition to its more general role in splicing, the protein also functions as a regulator of expression of certain classes of mRNAs. Such a combination of activities makes UBP1 a mediator of gene expression of widespread importance.
Materials and methods
Cloning of UBP1 cDNA
Screening of the λ-ZAPII cDNA library of N.plumbaginifolia and the RBD12 cDNA probe were as described by Mieszczak et al. (1992). The longest of four isolated clones is referred to as pUBP1-SK. The cDNA sequence has been deposited in the EMBL database (accession No. AJ272011). Arabidopsis thaliana EST clones (T21032, T88403 and R30154, encoding AtUBP1a, b and c, respectively) were obtained from the Arabidopsis Biological Resource Center, Ohio State University.
Plasmid constructions
To construct pET11D-UBP1-His6, pET11d (Novagene) was modified by inserting the UBP1-coding sequence followed by a His6 tag. pUBP1-HA contains the UBP1-coding sequence fused at the C–terminus with an in-frame HA epitope tag (MYPYDVPDYA) in the plant expression vector pGGS5 (Genschik et al., 1997). In pUBP1-GFP, the HA epitope tag is replaced by the GFP coding sequence. pDEDH/Nco is the plant expression vector pDEDH (Goodall and Filipowicz, 1989) containing an NcoI site at an ATG in an optimal translation initiation context. pUBP1 was generated by cloning the UBP1 cDNA fragment into pDEDH/Nco. The plasmid pZmAct corresponds to pDEmac described by Goodall and Filipowicz (1991). The constructs pSynGC and pSynGC/ClaU were described by Gniadkowski et al. (1996) and the constructs CaMV/U2/pA and CaMV/U2/3′mut/pA by Connelly and Filipowicz (1993). For pSyn7, see Goodall and Filipowicz (1989). pSyn3 and pSyn3/ivs correspond to CaMVp/cDNA and CaMVp/syn, respectively, described by Connelly and Filipowicz (1993). To prepare pGUS, the GUS coding sequence was cloned into pDEDH/Nco. pGUS/OCS was constructed by replacing the CaMV pA site fragment from pGUS with the OCS pA fragment from Bin-Hyg-Top (DDBJ/EMBL/GenBank accession No. Z37515). To create pGLBp/Syn3, the GLB region of pGLB/U2/pA (Connelly and Filipowicz, 1993) was PCR amplified and cloned into pSyn3 in place of the CaMV promoter. pGLBp/SynGC, pGLBp/SynGC/ClaU, pGLB/ZmAct and pGLB/GUS were created by removing the Syn3 sequence from pGLB/Syn3 and replacing it with SynGC, SynGC/ClaU, maize actin and GUS reporter genes, respectively. The chitinase cDNA and gene (a kind gift of Dr F.Meins of this Institute) were cloned into pDEDH/Nco. Constructs used for testing the effect of UBP1 in the 3′ cleavage/polyadenylation reaction correspond to R-CAT* (used also in RNA accumulation studies), ΔAATAAA, GTATTC, 1T, Δ45-32 and (TTTGTA)2 described by Rothnie et al. (1994). To create pβ-globin, human β-globin cDNA was inserted into the BamHI site of pDEDH. Details of cloning are available on request.
Transfection of protoplasts and RNA analysis
Protoplasts of N.plumbaginifolia were transfected by the PEG method (Goodall et al., 1990). Unless indicated otherwise, 5 μg of the reporter plasmid and 1–10 μg of pUBP1 were used. The total amount of DNA was kept constant by the addition of the empty expression vector, pDEDH/Nco. RNA was extracted from the protoplasts using the RNeasy Plant Mini Kit (Qiagen) and treated with DNase I. Probes for RNase A/T1 mapping were prepared by in vitro transcription using appropriately linearized plasmids containing cDNA or gene inserts as templates. The template for the synthesis of the U2 RNA probe contains a conserved 5′–terminal 50 bp region of the U2 gene. All probes were labelled with [α–32P]UTP. RNase A/T1 mappings and quantitation were performed as described earlier (Goodall and Filipowicz, 1991; Gniadkowski et al., 1996). Unless indicated otherwise, all mappings were repeated using RNAs originating from at least three different transfection experiments. RNA yields are given as ratios of transcript levels seen in the presence and absence of pUBP1.
For Northern analysis, RNA was separated into poly(A)+ and poly(A)– fractions using oligo(dT)25 Dynabeads (Dynal) according to the manufacturer's protocol. RNase H cleavage, 6% PAGE and electroblotting were as described in Brown and Sachs (1998). Northern hybridizations were as described by Genschik et al. (1997).
Western blotting
Proteins, separated by 10–12% SDS–PAGE, were electroblotted to a PVDF membrane. Antibodies were diluted as follows: mAbs DG6, A4, CB2, 4G3 (α-U2B″, Cappel) and 12CA5 (mouse α-HA, Boehringer), 1:100; horseradish peroxidase-conjugated goat α-mouse (Jackson ImmunoResearch Laboratories) 1:5000.
Immuno- and GFP fluorescence
Treatment of cells and confocal microscopy were as described by Genschik et al. (1997). Nicotiana plumbaginifolia protoplasts transfected with UBP1–GFP were examined 6–24 h after transfection. Photographs were taken using the Zeiss Axiophot microscope.
Overexpression of UBP1, preparation of antibodies, plant cells, protoplasts, cell fractionation, and in vivo and in vitro UV cross-linking
These are provided as supplementary material and are also available on request.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank J.Petruska for excellent technical assistance, H.Rothnie and S.Buratowski for constructs, A.Beven and P.Shaw for help in introducing us to confocal microscopy, Z.Lorkovic for discussions, and H.Rothnie, F.Meins, M.Konarska and Z.Lorkovic for critical comments on the manuscript. G.G.S. was supported by the Royal Society (London) and EMBO.
References
- Baynton C.E., Potthoff, S.J., McCullough, A.J. and Schuler, M.A. (1996) U-rich tracts enhance 3′ splice site recognition in plant nuclei. Plant J., 10, 703–711. [DOI] [PubMed] [Google Scholar]
- Bentley D. (1999) Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol., 11, 347–351. [DOI] [PubMed] [Google Scholar]
- Beven A.F., Simpson, G.G., Brown, J.W. and Shaw, P.J. (1995) The organization of spliceosomal components in the nuclei of higher plants. J. Cell Sci., 108, 509–518. [DOI] [PubMed] [Google Scholar]
- Brown C.E. and Sachs, A.B. (1998) Poly(A) tail length control in S.cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol., 18, 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W.S. and Simpson, C.G. (1998) Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiol. Plant Mol. Biol., 49, 77–95. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560. [Google Scholar]
- Callis J., Fromm, M. and Walbot, V. (1987) Introns increase gene expression in cultured maize cells. Genes Dev., 1, 1183–1200. [DOI] [PubMed] [Google Scholar]
- Connelly S. and Filipowicz, W. (1993) Activity of chimeric U small nuclear RNA (snRNA)/mRNA genes in transfected protoplasts of N.plumbaginifolia: U snRNA 3′–end formation and transcription initiation can occur independently in plants. Mol. Cell. Biol., 13, 6403–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Caceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E. and Kornblihtt, A.R. (1999) Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell, 4, 251–258. [DOI] [PubMed] [Google Scholar]
- Crispino J.D., Blencowe, B.J. and Sharp, P.A. (1994) Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science, 265, 1866–1869. [DOI] [PubMed] [Google Scholar]
- Domon C., Lorkovic, Z.J., Valcarcel, J. and Filipowicz, W. (1998) Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J. Biol. Chem., 273, 34603–34610. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis, M.J., Pinol-Roma, S. and Burd, C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Ge H., Si, Y. and Wolffe, A.P. (1998) A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell, 2, 751–759. [DOI] [PubMed] [Google Scholar]
- Genschik P., Hall, J. and Filipowicz, W. (1997) Cloning and characterization of the Arabidopsis cyclic phosphodiesterase which hydrolyzes ADP-ribose 1′′,2′′-cyclic phosphate and nucleoside 2′,3′–cyclic phosphates. J. Biol. Chem., 272, 13211–13219. [DOI] [PubMed] [Google Scholar]
- Gniadkowski M., Hemmings-Mieszczak, M., Klahre, U., Liu, H.X. and Filipowicz, W. (1996) Characterization of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in N.plumbaginifolia.Nucleic Acids Res., 24, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G.J. and Filipowicz, W. (1989) The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell, 58, 473–483. [DOI] [PubMed] [Google Scholar]
- Goodall G.J. and Filipowicz, W. (1991) Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J., 10, 2635–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C., Droogmans, L., Chalon, P., Huez, G., Caput, D. and Kruys, V. (1999) Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem., 274, 2322–2326. [DOI] [PubMed] [Google Scholar]
- Huang Y., Wimler, K.M. and Carmichael, G.G. (1999) Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J., 18, 1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C.H., Brendel, V., Taylor, R.D. and Walbot, V. (1998) U-richness is a defining feature of plant introns and may function as an intron recognition signal in maize. Plant Mol. Biol., 36, 573–583. [DOI] [PubMed] [Google Scholar]
- Körner C.G. and Wahle, E. (1997) Poly(A) tail shortening by a mammalian poly(A)-specific 3′–exoribonuclease. J. Biol. Chem., 272, 10448–10456. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Krecic A. and Swanson, M.S. (1999) HnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Liu X. and Mertz, J.E. (1995) HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev., 9, 1766–1780. [DOI] [PubMed] [Google Scholar]
- Lopato S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A.R. and Barta, A. (1999) atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev., 13, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H., McCullough, A.J. and Schuler, M.A. (1993) 3′ Splice site selection in dicot plant nuclei is position dependent. Mol. Cell. Biol., 13, 4485–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K.R. and Walbot, V. (1994a) Addition of A- and U-rich sequence increases the splicing efficiency of a deleted form of a maize intron. Plant Mol. Biol., 24, 449–463. [DOI] [PubMed] [Google Scholar]
- Luehrsen K.R. and Walbot, V. (1994b) Intron creation and polyadenylation in maize are directed by AU-rich RNA. Genes Dev., 8, 1117–1130. [DOI] [PubMed] [Google Scholar]
- MacMillan A.M., McCaw, P.S., Crispino, J.D. and Sharp, P.A. (1997) SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl Acad. Sci. USA, 94, 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Wassarman, K.M. and Wolffe, A.P. (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J., 17, 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A. and Krainer, A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- McCullough A.J., Lou, H. and Schuler, M.A. (1993) Factors affecting authentic 5′ splice site selection in plant nuclei. Mol. Cell. Biol., 13, 1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W.M., Choi, M. and Dreyfuss, G. (1995) A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell, 83, 415–422. [DOI] [PubMed] [Google Scholar]
- Mieszczak M., Klahre, U., Levy, J.H., Goodall, G.J. and Filipowicz, W. (1992) Multiple plant RNA binding proteins identified by PCR: expression of cDNAs encoding RNA binding proteins targeted to chloroplasts in N.plumbaginifolia.Mol. Gen. Genet., 234, 390–400. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss, G. (1997) Nuclear export of proteins and RNAs. Curr. Opin. Cell Biol., 9, 420–429. [DOI] [PubMed] [Google Scholar]
- Rothnie H.M. (1996) Plant mRNA 3′–end formation. Plant Mol. Biol., 32, 43–61. [DOI] [PubMed] [Google Scholar]
- Rothnie H.M., Reid, J. and Hohn, T. (1994) The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′–end formation in plants. EMBO J., 13, 2200–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G. and Filipowicz, W. (1996) Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol., 32, 1–41. [DOI] [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz, J.A. (1994) SR proteins can compensate for the loss of U1 snRNP functions in vitro.Genes Dev., 8, 2704–2717. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Mourrain, P., Palauqui, J.C. and Vernhettes, S. (1998) Transgene-induced gene silencing in plants. Plant J., 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Visa N., Alzhanova-Ericsson, A.T., Sun, X., Kiseleva, E., Bjorkroth, B., Wurtz, T. and Daneholt, B. (1996) A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell, 84, 253–264. [DOI] [PubMed] [Google Scholar]
- Wahle E. and Ruegsegger, U. (1999) 3′–End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]