Abstract
In all eukaryotic nuclei, the La autoantigen binds nascent RNA polymerase III transcripts, stabilizing these RNAs against exonucleases. Here we report that the La protein also functions in the assembly of certain RNA polymerase II-transcribed RNAs into RNPs. A mutation in a core protein of the spliceosomal snRNPs, Smd1p, causes yeast cells to require the La protein Lhp1p for growth at low temperatures. Precursors to U1, U2, U4 and U5 RNAs are bound by Lhp1p in both wild-type and mutant cells. At the permissive temperature, smd1–1 cells contain higher levels of stable U1 and U5 snRNPs when Lhp1p is present. At low temperatures, Lhp1p becomes essential for the accumulation of U4/U6 snRNPs and for cell viability. When U4 RNA is added to extracts, the pre-U4 RNA, but not the mature RNA, is bound by Smd1p. These results suggest that, by stabilizing a 3′-extended form of U4 RNA, Lhp1p facilitates efficient Sm protein binding, thus assisting formation of the U4/U6 snRNP.
Keywords: La autoantigen/Saccharomyces cerevisiae/Sm proteins/snRNP assembly/spliceosomal snRNPs
Introduction
All eukaryotic cells contain numerous small RNA molecules that play crucial roles in cell metabolism. These include the nuclear U snRNAs, which function in mRNA processing events, such as pre-mRNA splicing (U1, U2, U4/U6 and U5 snRNAs) and 3′ end processing of histone mRNAs (U7 snRNA) (reviewed by Yu et al., 1998). The best characterized of the cytoplasmic small RNAs, tRNAs and 5S rRNA, function in protein synthesis. While much is known about the functions of many small RNAs, far less is known about the very early events in their biogenesis, such as how they are processed from larger precursors, folded and assembled into functional RNA–protein complexes.
One protein that binds many nascent small RNAs is a nuclear phosphoprotein known as the La protein. First described as an autoantigen in patients with rheumatic disease, the La protein is the first protein that binds all newly synthesized RNA polymerase III transcripts. These RNAs include precursors to tRNAs, 5S rRNA and U6 snRNA (Rinke and Steitz, 1982, 1985). Part of the binding site for the La protein on these RNAs is the sequence UUUOH, which is the 3′ end of all nascent RNA polymerase III transcripts (Stefano, 1984).
Genetic analyses in the yeast Saccharomyces cerevisiae have revealed that binding by the La protein Lhp1p (La homologous protein 1) to U6 RNA and pre-tRNAs facilitates the correct fate of these RNAs. Lhp1p stabilizes newly synthesized U6 RNA against degradation (Pannone et al., 1998). Binding by Lhp1p to pre-tRNAs is required for the normal pathway of tRNA maturation (Yoo and Wolin, 1997). While Lhp1p is not essential in wild-type cells, it becomes required when cells contain a mutation that disrupts the anticodon stem of an essential tRNASer. As restoration of base pairing in the stem eliminates the requirement for LHP1, Lhp1p may stabilize the mutant pre-tRNA in the correct structure (Yoo and Wolin, 1997). Lhp1p also stabilizes certain pre-tRNAs in yeast strains that fail to carry out the 1-methyladenosine modification (Calvo et al., 1999).
To learn more about the function of Lhp1p, we carried out genetic screens to identify mutations that cause yeast cells to require LHP1. Cells containing a mutation in SMD1, which encodes an Sm core protein of the spliceosomal U1, U2, U4 and U5 snRNPs (Rymond, 1993), require Lhp1p for growth at low temperature. We demonstrate that Lhp1p functions in the assembly of the RNA polymerase II-transcribed U RNAs into snRNPs. In vertebrate cells, newly transcribed U RNAs are exported to the cytoplasm, where they assemble with Sm proteins and undergo hypermethylation of the 5′ cap (Mattaj, 1988). Two proteins, SMN and SIP1, that bind Sm proteins in the cytoplasm are required for U snRNP assembly (Pellizzoni et al., 1998). In contrast, less is known about the snRNP biogenesis pathway in budding yeast. It is unclear whether yeast U snRNAs transit to the cytoplasm, or whether assembly takes place entirely within the nucleus. While SIP1 is distantly related to yeast Brr1, which functions in U snRNP biogenesis (Noble and Guthrie, 1996; Liu et al., 1997), SMN has not been identified in S.cerevisiae. We show that Lhp1p binds precursors to the U1, U2, U4 and U5 snRNAs in both wild-type and smd1–1 cells. Although smd1–1 cells that lack LHP1 are viable at 30°C, Lhp1p is essential at lower temperatures (16°C) for the formation of stable U4/U6 snRNPs and for cell viability. Interestingly, when in vitro synthesized U4 RNA is incubated in extracts, only the pre-U4 RNA, not the mature RNA, is bound by Smd1p. Our results suggest a model in which stabilization of pre-U4 RNA by Lhp1p facilitates binding by Sm proteins.
Results
Yeast cells containing a mutation in a core Sm protein require Lhp1p for growth
To identify additional roles of Lhp1p, we searched for mutants that required LHP1 for growth. Briefly, an ade2 strain lacking LHP1 (lhp1::LEU2) was transformed with a centromeric plasmid containing the LHP1, TRP1 and ADE2 genes. Cells that retain the plasmid are white, while cells that lose the plasmid are red due to a pigment that accumulates in ade2 strains. As LHP1 is not essential, the starting strain forms colonies containing red sectors. Following mutagenesis with ethylmethane sulfonate, 165 000 colonies were screened for the inability to lose the plasmid at 25°C. Five strains were identified that required LHP1 for wild-type growth at 25°C.
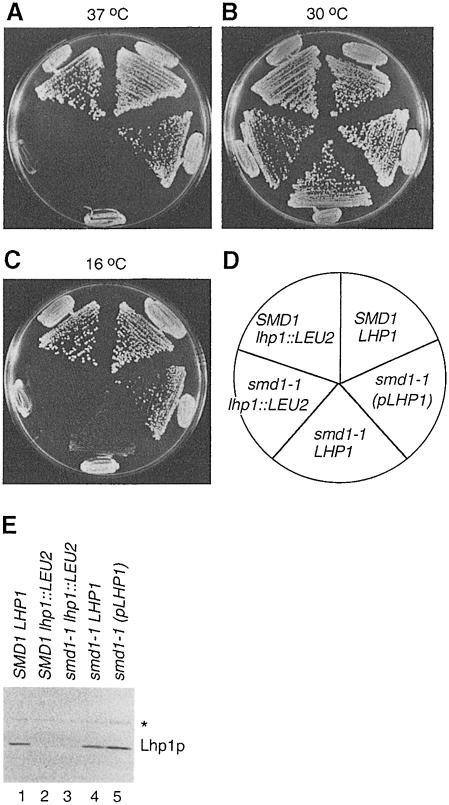
One strain, which we refer to as smd1–1, was unusual in that the requirement for LHP1 depended upon the temperature at which the cells were grown. At both 25°C (not shown) and 30°C (Figure 1B), smd1–1 lhp1::LEU2 cells had only a slight growth defect compared with wild-type cells. However, these cells were inviable at 37°C (Figure 1A) and 16°C (Figure 1C). In the presence of chromosomal LHP1, smd1–1 cells were able to grow at 16°C, although far more slowly than wild-type cells (Figure 1C). When the sole copy of LHP1 in the smd1–1 cells was present on the centromeric plasmid, the cells grew nearly as well as wild-type cells at both high (Figure 1A) and low (Figure 1C) temperatures. Thus, while the genomic copy of LHP1 is required for viability of smd1–1 cells at 16°C, LHP1 is also a low copy suppressor of the temperature-sensitive and cold-sensitive phenotypes. Western blotting of cell extracts revealed that the levels of Lhp1p when LHP1 was supplied on the plasmid were at most 2-fold higher than when the gene was present in the chromosome (Figure 1E, lanes 4 and 5). Thus, a small excess of Lhp1p resulted in dramatic increases in the growth of the smd1–1 strain at both temperatures.
Fig. 1. LHP1 is required for growth of smd1–1 cells at low temperatures. (A–D) Wild-type cells (SMD1 LHP1), cells lacking LHP1 (SMD1 lhp1::LEU2) and smd1–1 cells carrying either no LHP1 (smd1–1 lhp1::LEU2), chromosomal LHP1 (smd1–1 LHP1) or LHP1 on a centromeric plasmid [smd1–1 (pLHP1)] were streaked to single colonies on YPD agar and grown at 37°C (A), 30°C (B) and 16°C (C). The position of each strain is shown in (D). (E) Extracts were prepared from the strains and subjected to Western blotting. The asterisk denotes an unrelated protein detected by the anti-Lhp1p serum.
The mutant gene was cloned based on complementation of the LHP1 requirement and cold sensitivity. The SMD1 gene, which encodes the Sm D1 core protein of the spliceosomal U1, U2, U4 and U5 snRNPs (Rymond, 1993), complemented both phenotypes. To confirm that the mutation resided in SMD1, a HIS3 gene was integrated adjacent to SMD1 in an lhp1::LEU2 strain. Crossing of the HIS3-marked strain to the smd1–1 lhp1::LEU2 strain revealed that HIS3 segregated with the ability to lose the LHP1-containing plasmid. Sequencing of the smd1 gene from the mutant strain revealed a G to A mutation that converts the conserved glycine at position 89 to serine.
The sizes and levels of pre-U RNAs depend upon the amount of Lhp1p present
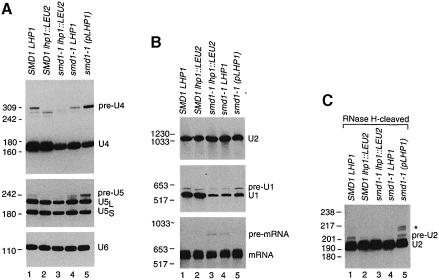
The finding that smd1–1 cells required LHP1 was unexpected, as the U RNAs bound by Smd1p are transcribed by RNA polymerase II (Dahlberg and Lund, 1988), and Lhp1p had only been demonstrated to bind polymerase III transcripts (Yoo and Wolin, 1994). To determine which snRNAs were affected by the mutation, we isolated RNA from wild-type and smd1–1 strains grown at 30°C and performed Northern blots. In smd1–1 cells, PhosphorImager quantitation revealed that the levels of U4 RNA (Figure 2A, top panel) and U1 RNA (Figure 2B, middle panel) were reduced to ∼30 and 25% of wild-type levels, respectively (compare lanes 3–5 in each panel with lane 1). Also, the levels of U5 RNA were reduced to ∼45% of wild-type levels in smd1–1 lhp1::LEU2 cells (Figure 2A, middle panel, lanes 1 and 3). However, the levels of U2 RNA were unaffected (Figure 2B, top panel). As a control, the blots were probed for U6 RNA, which is not bound by Smd1p. The levels of U6 RNA were similar in all strains (Figure 2A, lower panel).
Fig. 2. The levels of 3′-extended U RNAs depend upon the amount of Lhp1p present. (A) RNA extracted from wild-type (lane 1), SMD1 lhp1::LEU2 (lane 2) and smd1–1 cells containing either no LHP1 (lane 3), chromosomal LHP1 (lane 4) or plasmid LHP1 (lane 5) was fractionated in denaturing gels and subjected to Northern analysis using oligonucleotides complementary to U4 (top), U5 (middle) or U6 (bottom panel). To visualize pre-U5 RNA, the autoradiograph was overexposed, making the difference in mature U5 levels between wild-type and smd1–1 lhp1::LEU2 cells less apparent. (B) RNA was fractionated as in (A) except that the gel was run to maximize resolution of larger RNAs. The blot was probed to detect U2 RNA (top panel), U1 RNA (middle panel) and CRY1 mRNA (bottom panel). (C) The RNA was subjected to oligonucleotide-directed RNase H cleavage to generate a 3′ fragment of U2 RNA of ∼195 nucleotides. Samples were analyzed by Northern blotting using an oligonucleotide complementary to the 3′ end of U2 RNA.
We also detected longer forms of the U1, U4 and U5 RNAs in the Northern blots. Notably, the sizes and levels of some of the longer RNAs varied with the amount of Lhp1p present. The band labeled ‘pre-U4’ in Figure 2A (top panel) was shorter in SMD1 lhp1::LEU2 cells (lane 2) than in wild-type cells (lane 1), and was undetectable in smd1–1 lhp1::LEU2 cells (lane 3). The band labeled ‘pre–U5’ was undetectable in lhp1::LEU2 strains (Figure 2A, lanes 2 and 3), but present in strains with LHP1 (lanes 1 and 4). In contrast, the level of pre-U1 RNA was unchanged in SMD1 lhp1::LEU2 cells (Figure 2B, lane 2), but was slightly decreased in smd1–1 cells (lanes 3 and 4). Expression of LHP1 on the plasmid increased the levels of all the longer RNAs (Figure 2A and B, lane 5), and also slightly raised the levels of mature U1 RNA (Figure 2B, middle panel, lane 5).
Reprobing the blots with oligonucleotides complementary to sequences 3′ of the U1, U4 and U5 RNA coding regions revealed that the longer RNAs were extended at the 3′ end (data not shown). To determine the sizes of the extensions, we performed site-directed cleavage using RNase H and 2′-O-methyl RNA–DNA chimeric oligonucleotides (Inoue et al., 1987), followed by Northern analysis of the cleavage products (not shown). The pre-U1 RNA contained a 76 nucleotide extension, and thus corresponds to a U1 RNA precursor (U1β) described by Seipelt et al. (1999). The U4 RNA contains an ∼130 nucleotide extension, and the U5 RNA is 16 nucleotides longer than mature U5L RNA. The longer U4 and U5 RNAs are both similar to precursors recently described by Allmang et al. (1999). Each of the longer RNAs ends in a run of uridylates that lies 4–10 nucleotides downstream of the RNase III cleavage sites for these RNAs (Chanfreau et al., 1997; Allmang et al., 1999; Seipelt et al., 1999).
Because U2 RNA in yeast is 1175 nucleotides, a precursor could be difficult to resolve from the mature RNA. We performed oligonucleotide-directed RNase H cleavage to generate a 3′ fragment that could be resolved from the mature RNA. This analysis revealed a 3′-extended U2 RNA (Figure 2C, lane 1) that was reduced in lhp1::LEU2 cells (lane 2) and in cells containing the smd1–1 mutation (lanes 3 and 4). In the presence of plasmid LHP1, the levels of this RNA were restored and several larger RNAs became apparent (asterisk, lane 5). On longer exposures, the larger RNAs were also detected in wild-type cells (see Figure 3C). Mapping of the 3′ ends revealed that the major band contained an ∼10 nucleotide extension, and that the longer RNAs contained ∼20 add– itional nucleotides (data not shown). Thus, the major band probably corresponds to the pre-U2 RNA described by Noble and Guthrie (1996). Similarly to the longer U1, U4 and U5 RNAs, the pre-U2 RNA ends in uridylates.
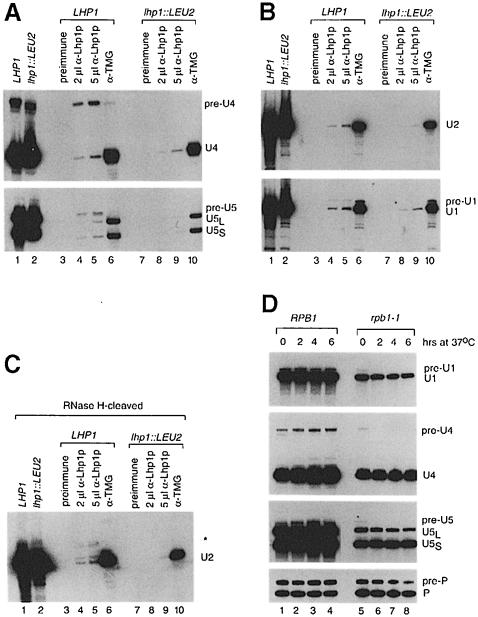
Fig. 3. The pre-U RNAs are bound by Lhp1p and transcribed by RNA polymerase II. (A) Wild-type (lanes 3–6) and lhp1::LEU2 extracts (lanes 7–10) were phenol extracted (lanes 1 and 2) or incubated with pre-immune serum (lanes 3 and 7), anti-Lhp1p serum (lanes 4, 5, 8 and 9) or antibodies against the TMG cap of U RNAs (lanes 6 and 10). RNAs in the immunoprecipitates (lanes 3–10) and an equivalent amount of extract (lanes 1 and 2) were subjected to Northern analysis to detect U4 RNA (top panel) and U5 RNA (bottom panel). (B) The RNAs in (A) were subjected to Northern blotting to detect U2 RNA (top panel) and U1 RNA (bottom panel). (C) The RNAs in (A) were subjected to oligonucleotide-directed RNase H cleavage to generate a 3′ fragment of mature U2 RNA of ∼195 nucleotides. Samples were analyzed by Northern blotting using an oligonucleotide complementary to the 3′ end of U2 RNA. Cleavage of mature U2 RNA generates two closely spaced bands. The asterisk designates the 3′-extended RNA. (D) Wild-type (lanes 1–4) and rpb1–1 cells (lanes 5–8) were grown at 30°C. After shifting to 37°C, cells were removed at the indicated times and subjected to Northern blotting to detect U1, U4, U5 and RNase P RNAs.
To examine the effects of the smd1–1 mutation on pre-mRNA splicing, the blot in Figure 2B was reprobed to detect CRY1 mRNA, which encodes ribosomal protein S14A. Both smd1–1 lhp1::LEU2 cells and smd1–1 cells carrying chromosomal LHP1 accumulated low levels of unspliced mRNA, although the splicing defect was slightly more severe in smd1–1 lhp1::LEU2 cells (Figure 2B, bottom panel, lanes 3 and 4). In the presence of plasmid LHP1, the unspliced mRNA was undetectable (lane 5), consistent with the idea that LHP1 is also a low copy suppressor of the smd1–1 mutation.
Lhp1p binds precursors to spliceosomal snRNAs
To determine whether Lhp1p bound the longer U RNAs, we performed immunoprecipitations. Northern analysis revealed that ∼40% of the pre-U4 RNA and ∼20% of the pre-U5 RNA were contained within anti-Lhp1p immunoprecipitates at the highest level of serum (Figure 3A, lane 5). Higher amounts of serum did not increase the levels of these RNAs (not shown). Although only a small fraction of the pre-U1 RNA was detected in anti-Lhp1p immunoprecipitates from wild-type cells (Figure 3B, lanes 4 and 5), none was detected in immunoprecipitates from lhp1::LEU2 cells (lanes 8 and 9). To detect pre-U2, we subjected the immunoprecipitated RNA to oligonucleotide-directed RNase H cleavage. The major pre-U2 RNAs in the immunoprecipitates corresponded to RNAs that were more apparent in the presence of plasmid LHP1 (compare Figure 3C, lanes 4 and 5 with Figure 2C). Small amounts of mature U RNAs were also detected in the anti-Lhp1p immunoprecipitates (Figure 3A–C, lanes 4 and 5). As these RNAs were also present at lower levels in immunoprecipitates from lhp1::LEU2 cells (lanes 8 and 9), it is unclear whether their presence is significant.
Since Lhp1p had only been demonstrated to bind RNA polymerase III transcripts (Yoo and Wolin, 1994), we confirmed that the longer RNAs were synthesized by RNA polymerase II, rather than representing anomalous transcripts made by polymerase III. We examined their synthesis in a strain containing a temperature-sensitive mutation in the large subunit of RNA polymerase II (rpb1–1; Nonet et al., 1987). Even at the permissive temperature, both the mature U RNAs and the 3′-extended RNAs were decreased in rpb1–1 cells, consistent with transcription by polymerase II (Figure 3D, lane 5). Within 2 h at 37°C, the pre-U4, pre-U5 and pre-U1 RNAs were undetectable (lane 6). RNase H cleavage revealed that pre-U2 RNA also did not accumulate in the rpb1–1 strain at 37°C (data not shown). However, the levels of the mature RNAs were unchanged (lanes 5–8). This was noted for U1 RNA in a similar experiment and suggested to be due to the high stability of small RNAs (Chapon et al., 1997). Reprobing to detect RNase P RNA, which is synthesized by polymerase III (Lee et al., 1991), revealed that the precursor and mature RNA remained stable during this period (lane 6). Thus, the 3′-extended U RNAs are transcribed by RNA polymerase II. Also, the rapid turnover of these RNAs, compared with the mature RNAs, is consistent with the idea that they are precursors. These experiments, coupled with findings that the pre-U2 RNA is processed to mature U2 RNA in vivo (Noble and Guthrie, 1996), and that the pre-U1 RNA is processed to mature U1 RNA in vitro (Seipelt et al., 1999), suggest that the four 3′-extended RNAs represent authentic precursors.
smd1-1 cell extracts contain higher levels of stable U1 and U5 snRNPs when Lhp1p is present
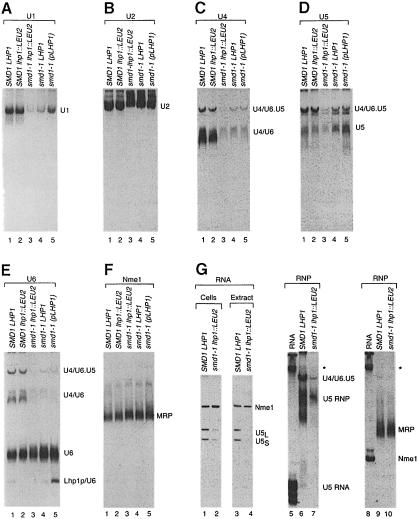
To understand why smd1–1 cells require LHP1, we examined the various U snRNP-containing particles. We grew the wild-type and smd1–1 strains at 30°C and prepared whole-cell extracts. Following electrophoresis in native gels, the U1-, U2-, U4- and U5-containing particles were detected by Northern blotting. Both U4 and U5 RNAs are present in multiple particles: U4 RNA base-pairs with U6 RNA to form the U4/U6 snRNP and associates with the U5 snRNP to form the U4/U6⋅U5 tri-snRNP (Staley and Guthrie, 1998). Our analysis revealed that smd1–1 cells contained lower amounts of the U1-, U4- and U5-containing particles than SMD1 cells (Figure 4A, C and D). This was expected for the U1 and U4 snRNPs, as these RNAs were decreased in smd1–1 cells (Figure 2). However, the levels of U5 RNA were only reduced to 45% of wild-type levels in smd1–1 lhp1::LEU2 cells when RNA was extracted by lysing cells in hot phenol and SDS (Figure 2A). PhosphorImager quantitation revealed that the U5 snRNP particles were decreased to 22% of wild-type levels when smd1–1 lhp1::LEU2 cells were lysed in aqueous buffers and the extract supernatants fractionated in native gels (Figure 4D, lane 3). Similarly, while U1 RNA levels were reduced to 25% of wild-type levels when cells were lysed in phenol–SDS (Figure 2), the relative level of U1 snRNP particles in the native gels was 14% (Figure 4A, lane 3).
Fig. 4. smd1–1 cell extracts contain reduced levels of stable U1 and U5 snRNPs when Lhp1p is absent. (A–F) Extracts from wild-type (lane 1), SMD1 lhp1::LEU2 (lane 2) and smd1–1 cells containing either no LHP1 (lane 3), chromosomal LHP1 (lane 4) or plasmid LHP1 (lane 5) were fractionated in 4% polyacrylamide native gels and subjected to Northern analysis. Blots were probed to detect U1 snRNPs (A), U2 snRNPs (B), U4–containing snRNPs (C), U5-containing snRNPs (D), U6 snRNPs (E) and the Nme1 RNA-containing MRP (F). (G) Wild-type and smd1–1 lhp1::LEU2 cells were lysed in aqueous buffer as described in Materials and methods. The relative levels of U5 and Nme1 RNAs in the extract supernatants (lanes 3 and 4) are compared with the relative levels of these RNAs when the cells are lysed in hot phenol (lanes 1 and 2). Fractionation of the extract supernatants in native gels is shown in lanes 6, 7, 9 and 10. Lanes 5–7 are probed to detect U5 snRNPs and lanes 8–10 are probed to detect RNase MRP. To confirm that naked RNA remained on the gel, lanes 5 and 8 contain total RNA from wild-type cells. The band denoted by the asterisk may represent aggregated RNA, as the RNA was not heated prior to loading.
To investigate this phenomenon, we examined the levels of U1 and U5 RNA in the extracts. In extract supernatants, the levels of U5 RNA in smd1–1 lhp1::LEU2 cells were reduced to 24% of wild-type levels (Figure 4G, lanes 3 and 4) and were comparable with the levels of U5 RNPs detected when the same extracts were fractionated in native gels (lanes 6 and 7). Reprobing the blots to detect U1 RNA also revealed decreased levels of U1 in the extract supernatants (data not shown). However, the relative levels of a control RNA, Nme1 (lanes 3 and 4), and U4 RNA were largely unaffected (data not shown). As misfolded proteins can form aggregates that are insoluble in non-ionic detergent (e.g. Cheng et al., 1989), incorrectly assembled U1 and U5 RNA–protein complexes could form similar aggregates. However, solubilization of the pellets in hot phenol–SDS failed to recover the missing species (data not shown). Other extract preparation protocols, such as those used to prepare splicing extracts (Ansari and Schwer, 1995), gave similar results (not shown). Thus, a fraction of the U1 and U5 RNAs in smd1–1 lhp1::LEU2 cells may be unstable when cells are lysed in aqueous buffers.
As controls, we probed the native gels for U6 RNA and Nme1 RNA, the RNA component of the rRNA processing endonuclease MRP (Schmitt and Clayton, 1992). In addition to the free U6 snRNP, the U4/U6 snRNP and the U4/U6⋅U5 tri-snRNP, newly synthesized U6 RNA is bound by Lhp1p (Pannone et al., 1998). In smd1–1 lhp1::LEU2 cells, less U6 RNA assembled into the U4/U6 and U4/U6⋅U5 complexes than in wild-type cells, consistent with the decreased U4 and U5 snRNPs in these cells. Concomitant with these changes, the levels of the free U6 snRNP increased (Figure 4E, lane 3). When LHP1 was present in smd1–1 cells, the levels of the tri-snRNP increased (Figure 4E, lanes 3–5; also Figure 4C, lanes 3–5), consistent with the elevated levels of U5 snRNPs in the extracts. While expression of LHP1 on the plasmid did not raise the levels of the tri-snRNP further, the levels of the Lhp1p–U6 complex increased (Figure 4E, lane 5). Reprobing to detect Nme1 revealed that the levels of MRP were similar in all strains (Figure 4F).
Lhp1p is required for the accumulation of U4/U6 snRNPs in smd1-1 cells at low temperatures
To understand why smd1–1 cells required Lhp1p at low temperatures, we examined the fate of the U RNAs when wild-type and smd1–1 cells were shifted to 16°C. As observed on agar plates, cells containing the smd1–1 mutation grew more slowly in liquid than SMD1 cells (Figure 5A). Also, the rate at which smd1–1 cells grew at 16°C was dependent upon the amount of Lhp1p. Specifically, smd1–1 lhp1::LEU2 cells grew more slowly than smd1–1 LHP1 cells, and smd1–1 cells containing plasmid LHP1 grew better than cells containing chromosomal LHP1. As even at the starting temperature of 30°C smd1–1 lhp1::LEU2 cells grow more slowly than wild-type cells, it was difficult to pinpoint the exact time at which the cells slowed further in growth. However, between 12 and 24 h at 16°C, there was a substantial slowdown (Figure 5A).
Fig. 5. Analysis of U RNA levels during growth of smd1–1 cells at 16°C. (A) The growth of wild-type cells, SMD1 lhp1::LEU2 cells and smd1–1 cells containing either no LHP1 (smd1–1 lhp1::LEU2), chromosomal LHP1 (smd1–1 LHP1) or plasmid LHP1 [smd1–1 (pLHP1)] is compared. Cells were grown to OD = 0.3 at 30°C and switched to 16°C at time 0. (B and C) At intervals after the switch to 16°C, RNA was extracted from the strains and subjected to Northern analysis. The blot was probed to detect U1 and U2 RNAs (B) or U4, U5 and U6 RNAs (C).
To determine which U RNAs were affected by growth at 16°C, RNA was extracted from the cells at intervals and analyzed by Northern blotting. The levels of U2 RNA were unchanged during the experiment (Figure 5B). Similarly, while U1 RNA is present at lower levels in smd1–1 strains, the levels of this RNA did not change during growth in the cold (Figure 5B). However, by 24 h, the levels of U4 RNA decreased ∼2-fold in the smd1–1 lhp1::LEU2 strain (Figure 5C, lanes 6 and 7). A small decrease in U4 RNA levels was also evident in smd1–1 cells carrying chromosomal LHP1 by 39 h at 16°C (lane 12). Interestingly, in smd1–1 cells carrying plasmid LHP1, the levels of the pre-U4 RNA, the pre-U5 RNA and the longer form of U5, U5L, increased during growth in the cold (Figure 5C, lanes 13–17). One explanation is that processing of these RNAs slows at 16°C, and that binding by Lhp1p stabilizes the pre-U RNAs.
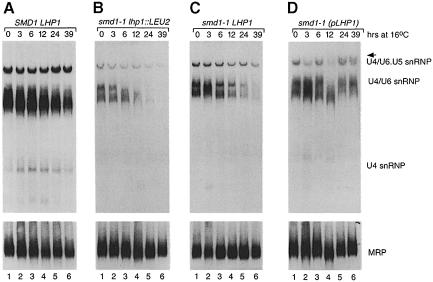
To examine whether Lhp1p was required for the stable accumulation of particular U snRNP particles, we prepared extracts at intervals during the cold shift. Native gel analyses revealed that the levels of the U1, U2 and U5 RNA-containing particles were not significantly affected by growth at 16°C (data not shown). However, in smd1–1 lhp1::LEU2 cells, the levels of both the U4/U6 snRNP and the U4/U6⋅U5 tri-snRNP declined (Figure 6B, lanes 1–6). The U4/U6 snRNP was most affected, as this complex was altered in electrophoretic mobility within 6 h (lane 3) and was undetectable by 39 h (lane 6). Reprobing to detect U6 snRNPs revealed that while the U4/U6 and tri-snRNP levels declined, the free U6 snRNP remained stable (not shown). The decrease in U4/U6 levels was observed to a lesser degree in smd1–1 cells carrying chromosomal LHP1 (Figure 6C). However, in smd1–1 cells carrying plasmid LHP1, the U4-containing particles were unaffected (Figure 6D). In these cells, the tri-snRNP migrated as a doublet by 24 h of growth at 16°C (Figure 6D, lanes 5 and 6, arrow). Reprobing with an oligonucleotide complementary to the 3′ extension of U4 RNA revealed that the pre-U4 RNA was contained within the top band of the doublet (data not shown). Thus, the pre-U4 RNA that accumulates in these cells at 16°C assembles into the tri-snRNP. These data suggest that, at low temperatures, binding by Lhp1p to pre-U4 RNA is required for the accumulation of U4/U6 snRNPs.
Fig. 6. Lhp1p is required for the accumulation of U4/U6 snRNPs in smd1–1 cells at 16°C. (A–D) Aliquots of wild-type cells (A) or smd1–1 cells carrying either no LHP1 (B), chromosomal LHP1 (C) or plasmid LHP1 (D) were removed at intervals after the switch to 16°C. Extracts were fractionated in 4% polyacrylamide gels and probed to detect U4 RNA-containing RNPs. In (D), the tri-snRNP appears as a doublet. Reprobing using an oligonucleotide specific for the pre-U4 RNA revealed that this RNA was contained primarily within the upper band (arrow). As a control, blots were reprobed to detect MRP (A–D, bottom).
Only pre-U4 RNA, not mature U4 RNA, is bound by Smd1p in vitro
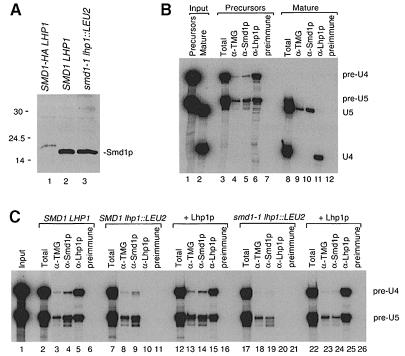
To dissect the role played by Lhp1p in U4/U6 snRNP formation, we examined the binding of Smd1p to U RNAs in extracts. Western blotting using an anti-Smd1p serum revealed that the levels of Smd1p were similar in wild-type and smd1–1 strains (Figure 7A, lanes 2 and 3). Using T7 RNA polymerase, we synthesized RNAs corresponding to U4 and U5L RNAs and their 3′-extended forms. Upon incubation in a wild-type extract, pre-U4 RNA, but not mature U4, assembled with Smd1p to form immunoprecipitable RNPs (Figure 7B, lanes 5 and 10). Also, only pre-U4 RNA underwent trimethylation of the cap, as judged by immunoprecipitation with anti-2,2,7-trimethylguanosine (TMG) antibodies (lanes 4 and 9). In contrast, both mature U5L and the pre-U5 RNA bound Smd1p and underwent cap modification. Thus, if pre-U4 RNA is the preferred substrate for Smd1p binding in vivo, Lhp1p could facilitate assembly of U4/U6 snRNPs by stabilizing the most efficient substrate for Sm protein binding.
Fig. 7. Binding of Smd1p to pre-U RNAs in extracts. (A) Extracts from wild-type (lane 2) and smd1–1 lhp1::LEU2 cells (lane 3) were subjected to Western blotting to detect Smd1p. Lane 1 contains extract from a strain in which an influenza HA epitope tag was added to Smd1p (Seto et al., 1999). (B) Mixtures of 32P-labeled pre-U4 and pre-U5 RNAs (lanes 1 and 3–7) or mature U4 and U5L RNAs (lanes 2 and 8–12) were incubated in wild-type extracts. Extracts were aliquoted and phenol extracted (lanes 3 and 8) or subjected to immunoprecipitation with the indicated sera. The pre-immune serum is from the anti-Lhp1p rabbit. Lanes 1 and 2 show the input RNAs. As mature U4 RNA terminates in UUUOH, it is bound by Lhp1p in extracts (lane 11). (C) 32P-Labeled pre-U4 and pre-U5 RNAs were incubated in wild-type (lanes 2–6), SMD1 lhp1::LEU2 (lanes 7–16) or smd1–1 lhp1::LEU2 extracts (lanes 17–26). Extracts were aliquoted and subjected to phenol extraction (lanes 2, 7, 12, 17 and 22) or immuno- precipitation as in (B). In lanes 12–16 and 22–26, 100 ng of Lhp1p (an amount equivalent to that in the wild-type extract) were included in the reactions. Lane 1 shows the input RNA. Both full-length pre-RNAs and their shorter degradation products were included in the quantitation of the data.
To examine whether Lhp1p plays a direct role in snRNP assembly, we compared the binding of Smd1p to pre-U4 and pre-U5 RNAs in extracts lacking Lhp1p. In SMD1 lhp1::LEU2 extracts, the level of Smd1p-bound pre-U4 RNA was 69% of wild-type levels (Figure 7C, compare lanes 4 and 9), as measured with a PhosphorImager. Addition of purified Lhp1p increased the binding to 93% of wild-type levels (lane 14). Thus, Lhp1p has a small effect on the binding of Smd1p to pre-U4 RNA in wild-type extracts. The levels of pre-U5 RNA in the immunoprecipitates were unaffected by the presence of Lhp1p (lanes 4, 9 and 14). In smd1–1 lhp1::LEU2 cells, the amounts of pre-U4 and pre-U5 RNAs bound by Smd1p were reduced to 5 and 34% of wild-type levels, respectively (lane 19). Addition of Lhp1p had a small but reproducible effect on the binding of the mutant Smd1p to pre-U4, raising the immunoprecipitable RNA to 21% of wild-type levels (compare lanes 19 and 24). Again, the levels of pre-U5 in the anti-Smd1p immunoprecipitate were unaffected by Lhp1p. Thus, while the major role of Lhp1p is likely to be the stabilization of pre-U4 RNA, it plays a small but detectable role in directly facilitating Smd1p binding.
Overexpression of U1 and U4 RNA in smd1-1 cells eliminates the requirement for LHP1
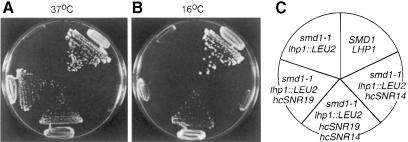
To confirm that the requirement for LHP1 in smd1–1 cells was due to a role for Lhp1p in U snRNP biogenesis, we asked whether raising the number of U RNA genes in the smd1–1 lhp1::LEU2 strain could eliminate the requirement. We introduced high copy plasmids containing the U1, U2, U4 and U5 genes into the smd1–1 lhp1::LEU2 strain and examined the growth of the transformants. Overexpression of U1 RNA, but not of U2, U4 or U5 RNA, restored the ability of the smd1–1 lhp1::LEU2 cells to grow at 37°C, although not to wild-type levels (Figure 8A; data not shown). Expression of all combinations of these genes in the smd1–1 lhp1::LEU2 cells did not increase the level of growth (not shown). Thus, part of the failure of smd1–1 lhp1::LEU2 cells to grow at 37°C may be due to defects in U1 snRNP biogenesis.
Fig. 8. smd1–1 lhp1::LEU2 cells grow at extreme temperatures in the presence of multiple U1 and U4 genes. (A–C) Wild-type cells (SMD1 LHP1), smd1–1 lhp1::LEU2 cells and smd1–1 lhp1::LEU2 cells containing either SNR19 (encoding U1 RNA) in the high copy plasmid pRS424, SNR14 (encoding U4 RNA) in the high copy plasmid pRS426, or both plasmids were streaked to single colonies on YPD medium and grown at 37°C (A) and 16°C (B). The position of each strain is shown in (C).
Interestingly, overexpression of no single U RNA was sufficient to restore growth of the smd1–1 lhp1::LEU2 cells at 16°C. However, cells expressing high copy plasmids containing the U1 and U4 genes were able to grow, although more slowly than wild-type cells (Figure 8B). Expression of high copy plasmids containing all combinations of the U1, U2, U4 and U5 genes in the mutant cells did not significantly increase the growth over that seen with the U1 and U4 plasmids alone (data not shown). Thus, as we did not detect changes in the amounts of U1 snRNPs in smd1–1 lhp1::LEU2 cells during growth in the cold, the levels of U1 snRNPs in these cells may be limiting for growth at 16°C. Alternatively, in addition to the defects that we observed in the accumulation of U4/U6 snRNPs at 16°C, smd1–1 lhp1::LEU2 cells may have defects in U1 snRNP function that are not detected in the native gels.
Discussion
We demonstrated that a mutation in Smd1p, a core protein of the spliceosomal U snRNPs, causes yeast cells to require Lhp1p for growth at low temperatures. Precursors to the U1, U2, U4 and U5 RNAs are bound by Lhp1p in both wild-type and mutant cells. When the mutant cells are grown at low temperature, Lhp1p becomes required for the accumulation of U4/U6 snRNPs. As only the pre–U4 RNA, not the mature RNA, is bound by Smd1p in vitro, we propose that Lhp1p facilitates U4/U6 snRNP assembly by stabilizing the most effective substrate for Sm protein binding.
Our results reveal that the role of the yeast La protein is not limited to the biogenesis of RNA polymerase III transcripts. Instead, Lhp1p plays a more general role in small RNA biogenesis. Consistent with the preference of La proteins for RNAs terminating in UUUOH (Stefano, 1984), each of the pre-U RNAs ends in a run of uridylates. While the mechanism by which snRNA 3′ ends are generated in S.cerevisiae is not fully understood, strains defective in the enzyme RNase III exhibit decreased levels of similar U1, U4 and U5 RNA precursors and reduced levels of mature U2 and U5L RNAs (Chanfreau et al., 1997; Abou Elela and Ares, 1998; Allmang et al., 1999; Seipelt et al., 1999). Also, similar pre-U1, pre-U4 and pre-U5 RNAs accumulate in cells containing mutations in several 3′ exonucleases (Allmang et al., 1999). Thus, the pre-U RNAs bound by Lhp1p are most likely to be processing intermediates, generated by RNase III cleavage and subsequent exonuclease digestion.
Our experiments reveal that the binding of Lhp1p to pre-U RNAs has important consequences for snRNP assembly. As only the pre-U4 RNA is an efficient substrate for Smd1p binding in extracts, the major role of Lhp1p in U4/U6 snRNP assembly may be to stabilize this RNA, thus facilitating Sm protein binding. Since cells that contain wild-type SMD1 do not require Lhp1p, Sm protein binding may normally be sufficiently rapid such that prolonged stabilization of the precursor is unnecessary. As addition of Lhp1p to lhp1::LEU2 extracts resulted in a small increase in Smd1p binding, Lhp1p may also directly facilitate assembly of pre-U4 RNAs into snRNPs by assisting RNA folding, stabilizing RNA structure or interacting with snRNP proteins. Moreover, as Lhp1p has a small effect on Smd1p binding in wild-type extracts, other situations that reduce the efficiency of U snRNP assembly could cause cells to require Lhp1p. In any case, our finding that Lhp1p facilitates U4/U6 snRNP biogenesis supports the hypothesis that Lhp1p functions as a molecular chaperone, i.e. a transiently binding protein, not found in the final assembly, that facilitates the correct fate of newly synthesized RNAs in vivo (Pannone et al., 1998).
Interestingly, Lhp1p and Smd1p may function redundantly to stabilize pre-U4 RNA. In the presence of either LHP1 or SMD1, pre-U4 RNAs are discernible, although they are shorter in SMD1 lhp1::LEU2 cells (Figure 2A). As pre-U4 RNA is undetectable in smd1–1 lhp1::LEU2 cells, binding by either Sm proteins or Lhp1p may stabilize the 3′ extension. Thus, the Sm proteins may bind initially to the 3′ end of the RNA, stabilizing the extension. Alternatively, direct binding by Sm proteins to the Sm site of U4 RNA may stabilize pre-U4 RNA indirectly by influencing RNA structure or by recruiting proteins that slow 3′ end maturation. In this scenario, the 3′ extension could facilitate Smd1p binding by influencing the formation of correctly folded U4 RNA, perhaps by base pairing with mature RNA sequences to form a folding intermediate.
A puzzling aspect of our studies is the finding that smd1–1 lhp1::LEU2 extracts contain lower levels of U1 and U5 RNAs, relative to wild-type cells, than cells that are lysed in hot phenol–SDS. Also, when smd1–1 lhp1::LEU2 cells are grown at 16°C, there are similar discrepancies in the levels of U4 RNAs recovered in the extracts (Figure 6B). While an explanation may be that a fraction of the RNAs are degraded during lysis, addition of vanadyl ribonucleosides to the lysis buffer had no effect (our unpublished data). Moreover, when splicing extracts were prepared from these cells, in vitro synthesized RNAs were equally stable in the mutant and wild-type extracts (Figure 7). While we cannot rule out a technical artifact, a fraction of these RNAs may be present in smd1–1 lhp1::LEU2 cells in a form that results in their rapid degradation upon lysis in aqueous buffers. For example, if incorrectly assembled RNA–protein complexes form aggregates similar to those described for misfolded proteins (e.g. Cheng et al., 1989), the aggregated RNAs could be targeted by nucleases that are released from the vacuole or another compartment during lysis.
We do not yet know whether stabilization of U1, U2 and U5 precursors by Lhp1p similarly facilitates the biogenesis of these snRNPs. In extracts, Smd1p binds mature U5L RNA, and Lhp1p has no detectable effect on Smd1p binding. However, the observation that extracts of smd1–1 lhp1::LEU2 cells contain lower levels of stable U1 and U5 snRNPs than smd1–1 LHP1 cells implies that Lhp1p plays a role in the biogenesis of these RNPs. Lhp1p and Smd1p could function redundantly to stabilize U1 and U5 RNA structure and/or recruit other proteins to the RNA. Alternatively, Lhp1p binding may increase the time window for productive interaction of Smd1p with these RNAs in vivo. Future experiments, such as comparison of Smd1p binding to pre- and mature U1 RNAs, will be required to address this question.
Although we only examined precursors to the spliceosomal U RNAs, Lhp1p may also bind processing intermediates of other RNA polymerase II-transcribed small RNAs. Experiments in which we reprobed our Northern blots to detect the small nucleolar U3 RNA, which functions in rRNA processing, have revealed that 3′-extended forms of this RNA are also bound by Lhp1p (our unpublished data). Thus, Lhp1p probably binds to and stabilizes a variety of small RNA precursors ending in uridylates. It will be interesting to examine whether stabilization of these other RNA precursors by Lhp1p facilitates their assembly into functional RNA–protein complexes.
Does stabilization of pre-U RNAs by the La protein facilitate U snRNP assembly in higher cells? In vertebrates, binding by Sm proteins to pre-U RNAs occurs in the cytoplasm, and several snRNAs undergo 3′ end maturation prior to reimport into the nucleus (Mattaj, 1988). As the human La protein binds a cytoplasmic population of U1 RNAs that are longer than mature U1 RNA (Madore et al., 1984b), the vertebrate protein could function in the cytoplasm to facilitate assembly of pre-U1 RNA into snRNPs. However, the mammalian La protein has not been described to bind U2, U4 or U5 RNA precursors, making analogies difficult. Moreover, as a cytoplasmic phase in snRNP assembly has not been demonstrated in S.cerevisiae, U snRNPs could assemble entirely within the nucleus in this yeast. Consistent with nuclear assembly, pre-U4 RNAs [which are confined to the cytoplasm in mammalian cells (Madore et al., 1984a)] assemble into U4/U6⋅U5 tri-snRNPs in yeast (Figure 6D). Interestingly, the SMN protein, which binds Sm core proteins and is required for snRNP assembly in the vertebrate cytoplasm (Liu et al., 1997; Pellizzoni et al., 1998), has not been identified in S.cerevisiae. Thus, binding by Lhp1p to pre–U RNAs in the nucleus of budding yeast may substitute for the cytoplasmic role played by SMN in other organisms.
Materials and methods
Yeast media and strains
Yeast media were prepared according to Sherman et al. (1991). Wild-type and lhp1::LEU2 strains (Yoo and Wolin, 1997) were CY1 (MATα ura3 lys2 ade2 trp1 his3 leu2 LHP1), CY2 (MATα ura3 lys2 ade2 trp1 his3 leu2 lhp1::LEU2) and CY4 (MATa ura3 lys2 ade2 trp1 his3 leu2 lhp1::LEU2). The smd1–1 mutant and control strains are DX1 (MATα smd1–1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 carrying pATL), DX2 (MATa smd1–1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 carrying pATL), DX3 (MATa SMD1 LHP1 ura3 lys2 ade2 trp1 his3 leu2), DX4 (MATa SMD1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2), DX5 (MATα smd1–1 lhp1::LEU2 ura3 lys2 ade2 trp1 his3 leu2) and DX6 (MATα smd1–1 LHP1 ura3 lys2 ade2 trp1 his3 leu2). Strain Z4 (MATa ura3-52 rpb1–1) and control strain Z1 (ura3-52 RPB1) were gifts of R.Young (Whitehead Institute). The hemagglutinin (HA)-tagged SMD1 strain was a gift of A.Seto and T.Cech (University of Colorado).
Synthetic lethal screen and cloning of SMD1
The synthetic lethal screen was performed as described (Pannone et al., 1998) with the following modifications. CY2 cells carrying pATL, a centromeric plasmid containing LHP1, ADE2 and TRP1 (Pannone et al., 1998), were mutagenized with ethylmethane sulfonate to 25% survival. Cells were plated on synthetic complete medium containing limiting amounts of adenine (SCiade) and screened at 25°C for the inability to lose pATL and form colonies with red sectors. Of 165 000 colonies, 192 did not form sectors. These colonies were transformed with plasmid pSLL28 (Yoo and Wolin, 1997), which contains LHP1, URA3 and LYS2, and tested on SCiade lacking uracil for the ability to lose pATL. Sixteen strains formed sectoring colonies. Of the 16, 14 failed to grow on medium containing 1 μg/ml 5-fluoro-orotic acid, indicating they could not lose pSLL28. Backcrossing to CY2 revealed that 10 strains contained a single recessive mutation that caused them to require pATL. Each strain was mated to CY1 to confirm that the mutation was lethal in combination with lhp1::LEU2. Loss of the plasmid from the diploids, followed by sporulation and tetrad dissection, revealed that five strains contained mutations that resulted in slow growth or lethality in combination with lhp1::LEU2. One strain was backcrossed four times to CY4 to yield strains DX1 and DX2. Crossing of DX2 to CY1, followed by loss of pATL, yielded segregants DX3, DX4, DX5 and DX6.
To clone SMD1, a genomic library in YCp50 was introduced into DX1 and the transformants screened for the ability to lose pATL. One transformant formed red sectors. Subcloning revealed that a PCR-generated fragment containing SMD1 with 511 nucleotides of 5′-flanking sequence and 137 nucleotides of 3′-flanking sequence eliminated the requirement for LHP1. To identify the mutation, genomic DNA from DX1 was amplified and sequenced.
Northern analyses and immunoprecipitations
Total RNA was extracted from yeast using hot phenol and SDS (Ausubel et al., 1998), fractionated in 5% polyacrylamide–8.3 M urea gels and transferred to Zetaprobe GT nylon membranes (Bio-Rad) in 0.5× TBE at 150 mA for 16 h. To resolve U4, U5 and U6 RNA, the bromophenol blue dye was run to the bottom. To resolve U1 and U2, the xylene cyanol was run to the bottom. For native gels, cells were lysed and the extracts fractionated as described (Pannone et al., 1998). Probes were:
U1, 5′ GACCAAGGAGTTTGCATC 3′;
U2, 5′ CAGATACTACACTTGATC 3′;
U4, 5′ CGTATTTCCCGTGCATAAGGAT 3′;
U5, 5′ GGTTCTGGTAAAAGGCAAGAACCATGTTCGTTATAAG 3′;
U6, 5′ AAAACGAAATAAATCTCTTTG 3′;
RPR1, 5′ GACGTCCTACGATTGCAC 3′;
Nme1, 5′ ATAGTAAGCTCCATTGGGTTA 3′.
To detect CRY1 mRNA, the second exon was amplified using 5′ GTTCAAGCTCGTGACAATTCCC 3′ and 5′ GGTTCTAGTACCACCGGTAGC 3′ in the presence of [α-32P]dCTP (400 Ci/mmol) using a CRY1 gene as template (a gift of J.Woolford, University of Pittsburgh). Quantitation was performed using a PhosphorImager (Molecular Dynamics).
For immunoprecipitations, CY1 and CY4 were grown in YPD at 30°C to OD600 = 0.500. After washing in H2O, cells were resuspended in 400 μl of NET-2 (40 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% NP–40) and lysed by vortexing in the presence of glass beads. Following sedimentation at 2000 r.p.m. in a microcentrifuge, the supernatant was sedimented at 100 000 g for 20 min in a Beckman TLA100.2 rotor and subjected to immunoprecipitation using anti-Lhp1p (Yoo and Wolin, 1994) and anti-TMG antibodies (Calbiochem).
Temperature shift and cold shift experiments
For the temperature shift experiment, Z1 and Z4 cells were grown in YPD at 30°C to OD600 = 0.3. After shifting to 37°C, cultures were kept in log phase by diluting in YPD. At intervals, aliquots were collected and total RNA extracted. For the cold shift experiment, strains DX1, DX3, DX4, DX5 and DX6 were grown at 30°C to OD600 = 0.4, and then shifted to 16°C for 39 h. Cells were maintained at OD600 between 0.15 and 0.60 by diluting with YPD. At intervals, cells were collected and stored at –80°C.
Oligonucleotide-directed RNase H cleavage
To identify pre-U2 RNA, 10 μg of total RNA were mixed with 2 μg of 5′ CTGGCCTTGAAACA 3′ in 16 μl and heated to 80°C for 3 min. After cooling to 23°C, 2 μl of 10× buffer [200 mM HEPES–KOH pH 8.0, 500 mM KCl, 100 mM MgCl2, 10 mM dithiothreitol (DTT)] and 2 μl of RNase H (Boehringer Mannheim) were added and the reaction incubated for 30 min at 30°C. To measure 3′ ends, 5 μg of RNA were mixed with 2 ng of 2′-O-methyl-RNA–DNA oligonucleotide in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM KCl, 5% sucrose and 25 mM DTT, and heated to 90°C for 3 min. After cooling to 23°C, 1 μl of RNase H (Pharmacia) was added and the reaction incubated at 37°C for 1 h. Chimeric oligonucleotides were: U2, 5′ ACUGdGdCdCdTUGAAACAACAG 3′; U1, 5′ UAAGAUdCdCdAdCCCGUUCCUA 3′; U4, 5′ CGGAdCdGdAdAUCCUCACUGAUA 3′; and U5, 5′ UGGCAAGCdCdCdAdCAGUAA 3′. After cleavage, 3′ fragments were detected by Northern blotting using the probes U1-3′, 5′ GCATGAAACTTTAAAAGTTTCAGTACTTTAAGA 3′; U2-3′, 5′ GAACGACTCCACAAGTGCGAGGGTCGCGACGTCTCTAAC 3′; U4-3′, 5′ AGGTATTCCAAAAATTCCCTAC 3′; and U5-3′, 5′ AAATAAAATAGAAAAGATAAACGCCCTCC 3′.
Construction of high copy plasmids containing U snRNA genes
To overexpress U1, SNR19 was excised from pTC19 using EcoRI and NarI and ligated into the EcoRI–ClaI sites of pRS424 (Christianson et al., 1992). For U2, pES18 was cut with SalI and XbaI to release SNR20. After filling in with T4 DNA polymerase, the DNA was inserted into the SmaI site of pRS422. For U4, a 0.56 kb EcoRI–BamHI fragment containing SNR14 was excised from pBSU4wt and cloned into the EcoRI–BamHI sites of pRS426. For U5, pDF7 was cut with EcoRI and BamHI, and the SNR7 fragment cloned into the EcoRI–BamHI sites of pRS423. Plasmids pTC19, pES18, pBSU4wt and pDF7 were gifts of C.Collins and C.Guthrie (University of California, San Francisco).
In vitro assembly
Pre-U4 sequences were amplified from genomic DNA using Pfu polymerase (Stratagene) and the primers T7U4 (5′ GCGAATTCTAATACGACTCACTATAGGGTCCTTATGCACGGGAAATACGC 3′) and 5′ GCCGGCGGATCCTTTAAAAGAAAAGAAAAATATGGTTGGGC 3′. Mature U4 sequences were amplified using T7U4 and 5′ GCCGGCGGATCCTTTAAAGGTATTCCAAAAATTCCCTACATAGTC 3′. After digestion with EcoRI and BamHI, the DNAs were cloned into pSP64 (Promega) and sequenced. Upon cleavage with DraI, pre-U4 and mature U4 RNAs were transcribed with T7 RNA polymerase in the presence of 1 mM GpppG and 25 μCi of [α-32P]UTP (both Amersham Pharmacia Biotech) as described (Yisraeli and Melton, 1989). Pre-U5 sequences were amplified from genomic DNA using T7U5 (5′ GCGAATTCTA– ATACGACTCACTATAGGGAAGCAGCTTTACAGATCAATGGC 3′) and 5′ AAAATAGAAAAGATAAACGCCCTCC 3′. Mature U5L sequences were amplified using T7U5 and 5′ ACGCCCTCCTTACTCATTGAGAAAAAGG and transcribed with T7 RNA polymerase. Cell extracts were prepared as described (Ansari and Schwer, 1995). To examine Smd1p binding, 50 000 c.p.m. of each RNA were incubated with 4 μl of extract for 10 min at 23°C in a volume of 10 μl under splicing conditions. The volume was raised to 250 μl with NET-2 and subjected to immunoprecipitation. Rabbit antibodies against amino acids 128–146 of Smd1p coupled to keyhole limpet hemocyanin were raised by AnaSpec, Inc. Lhp1p was purified as described (Yoo and Wolin, 1997).
Acknowledgments
Acknowledgements
We thank C.Collins, C.Guthrie and J.Woolford for providing plasmids, and R.Young, A.Seto and T.Cech for providing yeast strains. We thank Y.-T.Yu and B.Schwer for advice, and A.Horwich, M.Solomon and E.Ullu for comments on the manuscript. This work was supported by grant RO1-GM48410 from the National Institutes of Health. S.L.W. is an Associate Investigator of the Howard Hughes Medical Institute.
References
- Abou Elela S., Ares, M., Jr (1998) Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J., 17, 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E. and Tollervey, D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A. and Schwer, B. (1995) SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J., 14, 4001–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY. [Google Scholar]
- Calvo O., Cuesta, R., Anderson, J., Gutierrez, N., Garcia-Barrio, M.T., Hinnebusch, A.G. and Tamame, M. (1999) GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4167–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Elela, S.A., Ares, M., Jr and Guthrie, C. (1997) Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C., Cech, T.R. and Zaug, A.J. (1997) Polyadenylation of telomerase RNA in budding yeast. RNA, 3, 1337–1551. [PMC free article] [PubMed] [Google Scholar]
- Cheng M.Y., Hartl, F.U., Martin, J., Pollock, R.A., Kalousek, F., Neupert, W., Hallberg, E.M., Hallberg, R.L. and Horwich, A.L. (1989) Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature, 16, 620–625. [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski, R.S., Dante, M., Shero, J.H. and Hieter, P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene, 110, 119–122. [DOI] [PubMed] [Google Scholar]
- Dahlberg J.E. and Lund,E. (1988) The genes and transcription of the major small nuclear RNAs. In Birnstiel,M.L. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag, Berlin, Germany, pp. 38–70. [Google Scholar]
- Inoue H., Hayase, Y., Iwai, S. and Ohtsuka, E. (1987) Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett., 215, 327–330. [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Evans, C.F. and Engelke, D.R. (1991) Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc. Natl Acad. Sci. USA, 88, 6986–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fischer, U., Wong, F. and Dreyfuss, G. (1997) The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell, 90, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Madore S.J., Wieben, E.D., Kunkel, G.R. and Pederson, T. (1984a) Precursors of U4 small nuclear RNA. J. Cell Biol., 99, 1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madore S.J., Wieben, E.D. and Pederson, T. (1984b) Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA–protein complexes. J. Biol. Chem., 259, 1929–1933. [PubMed] [Google Scholar]
- Mattaj I.W. (1988) U snRNP assembly and transport. In Birnstiel,M.L. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag, Berlin, Germany, pp. 100–114. [Google Scholar]
- Noble S.M. and Guthrie, C. (1996) Transcriptional pulse–chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J., 15, 4368–4379. [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe, C., Sexton, J. and Young, R. (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol., 7, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B.K., Xue, D. and Wolin, S.L. (1998) A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J., 17, 7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka, N., Charroux, B. and Dreyfuss, G. (1998) A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell, 95, 615–624. [DOI] [PubMed] [Google Scholar]
- Rinke J. and Steitz, J.A. (1982) Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell, 29, 149–159. [DOI] [PubMed] [Google Scholar]
- Rinke J. and Steitz, J.A. (1985) Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res., 13, 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond B.C. (1993) Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc. Natl Acad. Sci. USA, 90, 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.E. and Clayton, D.A. (1992) Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev., 6, 1975–1985. [DOI] [PubMed] [Google Scholar]
- Seipelt R.L., Zheng, B., Asuru, A. and Rymond, B.C. (1999) U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res., 27, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A.G., Zaug, A.J., Sobel, S.G., Wolin, S.L. and Cech, T.R. (1999) Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature, 401, 177–180. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie, C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Stefano J.E. (1984) Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell, 36, 145–154. [DOI] [PubMed] [Google Scholar]
- Yisraeli J.K. and Melton, D.A. (1989) Synthesis of long, capped transcripts in vitro by SP6 and T7 RNA polymerases. Methods Enzymol., 180, 42–50. [DOI] [PubMed] [Google Scholar]
- Yoo C.J. and Wolin, S.L. (1994) La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell. Biol., 14, 5412–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.J. and Wolin, S.L. (1997) The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell, 89, 393–402. [DOI] [PubMed] [Google Scholar]
- Yu Y.-T., Scharl,E.C., Smith,C.M. and Steitz,J.A. (1998) The growing world of small nuclear ribonucleoproteins. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 487–524. [Google Scholar]