Abstract
Post-transcriptional gene silencing (PTGS) is a homology-dependent RNA degradation process that may target RNA exclusively in the cytoplasm. In plants, PTGS functions as a natural defense mechanism against viruses. We reported previously that the 2b protein encoded by cucumber mosaic cucumovirus (CMV) is a virulence determinant and a suppressor of PTGS initiation in transgenic Nicotiana benthamiana. By fusion with the green fluorescent protein, we now show that the CMV 2b protein localizes to the nuclei of tobacco suspension cells and whole plants via an arginine-rich nuclear localization signal, 22KRRRRR27. We further demonstrate that the nuclear targeting of the 2b protein is required for the efficient suppression of PTGS, indicating that PTGS may be blocked in the nucleus. In addition, our data indicate that the PTGS suppressor activity is important, but not sufficient, for virulence determination by the 2b protein.
Keywords: cucumber mosaic virus/nuclear import/post-transcriptional gene silencing/viral silencing suppressor/virus virulence
Introduction
The diversity of species in which eukaryotic gene expression is downregulated by the expression of exogenously introduced homologous sequences suggests that the phenomenon of homology-dependent gene silencing (HdGS) may be part of a universal gene regulatory system. First described in plants (Napoli et al., 1990; Smith et al., 1990; van der Krol et al., 1990), HdGS has since been identified in fungi (Cogoni and Macino, 1999; van West et al., 1999), Drosophila (PalBhadra et al., 1997), Paramecium (Ruiz et al., 1998) and mice (Bahramian and Zarbl, 1999). In addition, gene silencing by double-stranded RNA, or RNA interference (RNAi), which shares many key features with HdGS (Fire, 1999), has been described in an increasing number of organisms, including nematodes (Fire et al., 1998), trypanosomes (Ngo et al., 1998), Drosophila (Kennerdell and Carthew, 1998), hydra (Lohmann et al., 1999), planarians (Sánchez Alvarado and Newmark, 1999) and zebrafish (Wargelius et al., 1999). Although the mechanism(s) remains incompletely elucidated, distinctions have been made between two types of HdGS (Depicker and van Montagu, 1997; Vaucheret et al., 1998). Thus, transcriptional gene silencing (TGS) is identified by an absence of transcription from the silenced gene and occurs where homology exists between the promoter sequences of affected genes. In contrast, post-transcriptionally silenced (PTGS) genes contain homologous sequence within the coding region. Furthermore, although PTGS genes are actively transcribed, only low levels of steady-state mRNA accumulation can be detected.
Both TGS and PTGS are associated with the sequence-specific methylation of nuclear DNA (reviewed in Kooter et al., 1999). Although the occurrence of such methylation is essential for TGS (Scheid et al., 1998; Jeddeloh et al., 1999), its role in PTGS is not well defined (Jones et al., 1999). Thus, whereas the induction of transgene silencing following virus infection or local leaf infiltration with Agrobacterium was associated with de novo methylation of the corresponding transgene, virus-induced silencing of an endogenous gene occurred in the absence of DNA methylation (Jones et al., 1998, 1999). Furthermore, the frequency and stability of PTGS were unaffected in methylation-defective Neurospora mutants (Cogoni et al., 1996). Moreover, gene silencing events have been reported in Drosophila (PalBhadra et al., 1997; Kennerdell and Carthew, 1998), an organism that lacks DNA methylation (Bird, 1992).
Several models propose that PTGS is initiated by the accumulation of more than threshold amounts of a specific RNA species or through ectopic interactions between homologous DNA species, events that may lead to the production of aberrant RNA as the sequence-specific silencing signal (Lindbo et al., 1993; Baulcombe and English, 1996; English et al., 1996; Sijen et al., 1996; Metzlaff et al., 1997). This aberrant RNA may be in the form of antisense or double-stranded RNA species of low molecular weight, perhaps transcribed and/or amplified by RNA-dependent RNA polymerase (RdRp) (Lindbo et al., 1993). Evidence in support of elements of the proposed models has now been reported. A homolog of the tomato RdRp (Schiebel et al., 1998) is essential for the establishment of PTGS in Neurospora (Cogoni and Macino, 1999). Furthermore, the association of a small antisense RNA species with the occurrence of PTGS in several systems suggests that this antisense RNA is either the silencing signal or the specificity determinant for PTGS (Hamilton and Baulcombe, 1999).
There is mounting evidence that PTGS may target RNA exclusively in the cytoplasm. First, plant viruses replicating in the cytoplasm can function as both the initiator and the target of PTGS (Jones et al., 1998; Baulcombe, 1999; Guo et al., 1999). These RNA viruses are able to induce PTGS of nuclear genes in a homology-dependent manner (Lindbo et al., 1993; Kumagai et al., 1995; Guo and Garcia, 1997; Jones et al., 1998; Ruiz et al., 1998), or PTGS-like degradation of the invading viral RNAs, even in the absence of homologous nuclear sequences (Covey et al., 1997; Ratcliff et al., 1997, 1999). Furthermore, Ruiz et al. (1998) have shown that virus-induced gene silencing was targeted against the exons rather than the introns of an endogenous phytoene desaturase gene, thereby indicating that the mRNA precursors that are absent from the cytoplasm are not the target of PTGS. In addition, in Trypanosoma brucei, RNAi does not occur if pre-mRNA (which is not a target of degradation by RNAi) processing is inhibited (Ngo et al., 1998).
It is probable that the process of gene silencing in plants evolved as a response to selection pressure by pathogenic viruses. This is supported by the finding that many plant viruses encode proteins that can suppress PTGS as a counter-defensive strategy (Anandalakshmi et al., 1998; Beclin et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998; Li et al., 1999; Voinnet et al., 1999). Significantly, we have recently demonstrated that one such protein, the 2b protein of tomato aspermy cucumovirus (TAV), is targeted in Nicotiana tabacum by an alternative host defense mechanism (Li et al., 1999), thereby providing a further indication of the complexities in the battle between host and pathogen.
It has been shown that the viral suppressors identified to date target distinct stages of PTGS. HC-Pro, encoded by potyviruses, is able to ‘switch-off’ gene silencing in plants wherever it is expressed (Brigneti et al., 1998). In contrast, the 2b proteins encoded by TAV and cucumber mosaic virus (CMV) have no effect in tissues where PTGS is established, but are able to prevent the initiation of gene silencing in newly emerging tissues (Brigneti et al., 1998; Li et al., 1999). To understand the mechanism(s) by which the CMV 2b protein (Cmv2b) exerts its counter-defensive suppression effect, we examined the intracellular distribution of Cmv2b by fusion with the green fluorescent protein (GFP). We demonstrate that Cmv2b encodes a functional arginine-rich nuclear localization signal (NLS) and that nuclear targeting is required for the efficient suppression of PTGS, suggesting that PTGS, primarily a cytoplasmic event, may be blocked in the nucleus.
Results
Cmv2b is a nuclear protein
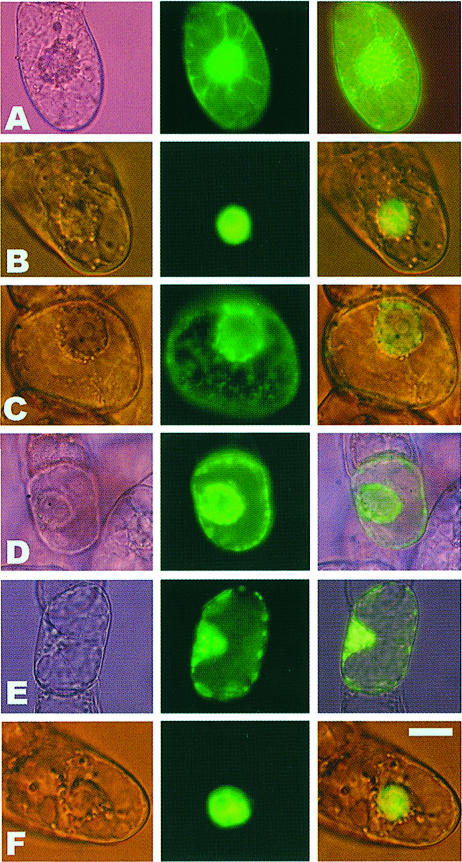
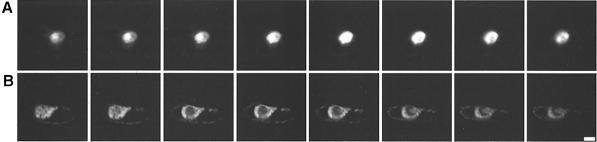
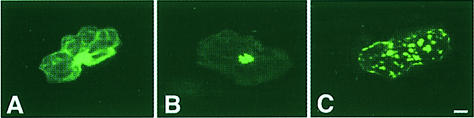
We have used GFP as a visual reporter (Chalfie et al., 1995; Padgett et al., 1996; Grebenok et al., 1997; Fujii et al., 1999) to examine the intracellular distribution of Cmv2b in Bright Yellow–2 (BY2) tobacco suspension cells. Under the transcriptional control of the 35S promoter, Cmv2b was transiently expressed as an N–terminal fusion to GFP (2bGFP). As has been reported previously (Köhler et al., 1997; Zhu et al., 1997; Kost et al., 1998), free GFP expressed in BY2 cells is distributed throughout the cell cytoplasmic and nuclear compartments with only moderate nuclear enrichment (Figure 1A). In contrast, accumulation of 2bGFP was restricted to the nucleus in the majority (60–70%) of those cells showing GFP fluorescence (Figure 1B). Those cells in which fluorescence was seen that did not reflect this pattern commonly showed a distribution indistinguishable from that seen for the free GFP control (Figure 1A). The nuclear distribution of 2bGFP was confirmed under bright field illumination to identify the nuclei (Figure 1B). Further clarification of the extent of the nuclear accumulation of Cmv2b was made using the confocal microscope to obtain optical sections through the nuclei of transfected cells. We resolved that Cmv2b occurred throughout the depth of the nucleus and not only at its periphery (Figure 2A). Investigations of the distribution of Cmv2b in the epidermal cells of intact Nicotiana glutinosa leaves transiently expressing the GFP constructs revealed a punctate distribution for 2bGFP (Figure 3B) in comparison with the more uniform distribution of free GFP (Figure 3A). These discrete, punctate accumulations of 2bGFP could be identified with the nuclei of the transfected leaf cells when viewed under transmitted light microscopy, thereby supporting the nuclear distribution observed earlier in the BY2 cells.
Fig. 1. Transient expression of GFP or GFP fusion proteins in BY2 cells. Free GFP (A), 2bGFP (B), 2b6AGFP (C), 2bQAQGFP (D), 2bQGFP (E) or 2b6A–nlsGFP (F). Micrographs on the left show bright field images of the cells to identify the nuclei. The central images show GFP fluorescence alone. Overlays of the fluorescence and bright field images are shown in the right panel. Bar, 10 μm.
Fig. 2. Optical sections through BY2 cell nuclei. Near-consecutive, serial confocal sections through BY2 cell nuclei transiently expressing 2bGFP (A) or 2b6AGFP (B). Bar, 10 μm; total distance, ∼6 μm.
Fig. 3. Distribution of GFP and GFP fusion proteins in N.glutinosa leaf epidermal cells. Confocal micrographs showing transient expression of free GFP (A), 2bGFP (B) and 2b6AGFP (C). Bar, 10 μm.
Protein sequence analysis (Nakai and Kanehisa, 1992) indicated that Cmv2b contains a monopartite NLS consisting of a lysine followed by five arginines, 22KRRRRR27. In BY2 cells expression of a mutant protein in which all six residues of this predicted NLS were replaced with alanines (2b6AGFP) revealed a punctate cytoplasmic and perinuclear distribution of the fluorescent signal (Figure 1C). The perinuclear distribution of 2b6AGFP was confirmed using confocal microscopy to obtain optical sections through BY2 cells expressing the protein. Whereas Cmv2b accumulated throughout the nucleus (Figure 2A), 2b6AGFP was absent from the nuclear interior and could be seen to form a ‘ring’ encircling the nucleus itself (Figure 2B). This pattern was evident in the majority of the cells transfected with the 2b6AGFP construct (70%). None of the 2b6AGFP-expressing cells demonstrated nuclear enhancement of the fluorescent signal, and those cells that did not reflect the perinuclear/cytoplasmic pattern of accumulation showed a distribution indistinguishable from the free GFP control (data not shown). In the epidermal cells of intact N.glutinosa leaves, 2b6AGFP accumulated in punctate cytoplasmic aggregates throughout the transfected cell, suggesting a non-nuclear distribution (Figure 3C). These findings indicate that the predicted NLS of Cmv2b is essential for the nuclear targeting of the protein.
A smaller three-residue replacement was made within the Cmv2b NLS (23RRR25→QAQ). Transient expression of this construct in BY2 cells yielded the mutant protein, 2bQAQGFP. In a manner reminiscent of 2b6AGFP, 2bQAQGFP was found to accumulate outside of the nuclei in the majority (70%) of the cells transfected (Figure 1D), although the cytoplasmic fluorescence of 2bQAQGFP frequently appeared to be focused both in a perinuclear pattern and at the periphery of the cell (Figure 1D). As for 2b6AGFP (Figure 2B), observation through the confocal microscope confirmed the extra-nuclear distribution of 2bQAQGFP (data not shown). In contrast to 2b6AGFP, however, a low percentage (6%) of the 2bQAQGFP-transfected cells demonstrated a discrete nuclear accumulation akin to that witnessed for 2bGFP. Distinct from the two mutants described above, a single-residue substitution in the NLS (24R→Q) did not abolish the ability of the mutant protein, 2bQGFP, to accumulate in the nucleus of transfected cells (Figure 1E). As with 2bGFP, 60–70% of the cells expressing 2bQGFP showed a strong nuclear enrichment of the fluorescent protein. However, approximately half of those cells showing nuclear enrichment of 2bQGFP also displayed a significant punctate accumulation in the cytoplasm, particularly at the cell periphery (Figure 1E), whereas <10% of the 2bGFP-expressing cells showed a similar accumulation. Discrete accumulations of this type in both the cytoplasm and nucleus differ markedly from the diffuse, combined cytoplasmic and nuclear fluorescence pattern typical of cells expressing GFP alone, the extra-nuclear accumulation of which is particularly evident along the cytoplasmic filaments (Figure 1A).
In order to determine whether the function of the Cmv2b NLS is position dependent, we appended the NLS to the C–terminus of the 2b6A protein, but N–terminal to GFP, to yield 2b6A–nlsGFP. When expressed in BY2 cells, the 2b6A–nlsGFP fusion protein was found to accumulate in the nucleus (Figure 1F). This change in position of the NLS appeared to increase the efficiency of nuclear targeting of the fusion protein as 2b6A–nlsGFP showed significant nuclear enrichment in ∼90% of the transfected BY2 cells compared with 60–70% for 2bGFP. This result illustrates that the identified NLS of Cmv2b is also sufficient to target a cytoplasmic protein to the nucleus and is position independent for that targeting.
Two regions of Cmv2b beyond the NLS are highly conserved in the 2b proteins of other cucumoviruses (Ding et al., 1994). The sequence 39KSPSE43 is invariant, and a second conserved region of 16 residues exists at the C–terminus of Cmv2b, although part of this latter domain is absent in the TAV 2b protein (Ding et al., 1994). 2bΔ5GFP and 2bΔ16GFP each contain a deletion of one of these conserved domains and both retained a nuclear accumulation pattern identical to 2bGFP when transiently expressed in BY2 cells (data not shown). However, when compared with 2bGFP (60–70% of the transfected cells showing nuclear enrichment), a marginally increased proportion of cells transfected with 2bΔ5GFP (∼80%) showed a significant nuclear enrichment of the fluorescent protein (data not shown).
The fusions between Cmv2b or its mutants and GFP described above were also expressed persistently in whole plants from a potato virus X (PVX) vector (pP2C2S; Brigneti et al., 1998). Leaves from Nicotiana benthamiana plants infected with PVX–2bGFP or its derivatives were excised and the intracellular distribution patterns of the fusion proteins examined under the confocal microscope. Similar to our findings using transient expression, 2bGFP again showed fluorescence principally in the nuclei of infected cells when expressed during PVX–2bGFP infections (data not shown). Furthermore, the distribution patterns of the mutant proteins previously observed in transfected BY2 cells were duplicated in whole leaves infected with PVX expressing the respective fusion proteins (data not shown).
Nuclear targeting, silencing suppression and virulence determination
The above experiments demonstrated that Cmv2b encodes an arginine-rich NLS that is essential and sufficient for nuclear targeting in tobacco suspension cells and whole plants. Using an experimental system described previously (Brigneti et al., 1998), we next examined whether the nuclear targeting of Cmv2b is required for its biological functions as a virulence determinant and a suppressor of PTGS.
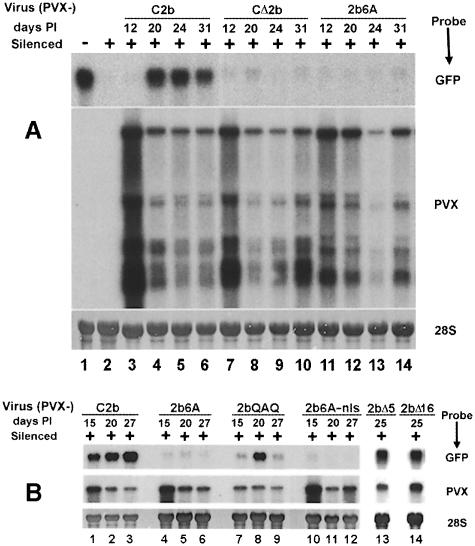
Coding sequences for each of the Cmv2b mutants described in the preceding section (without the GFP coding sequence) were cloned in the PVX vector. As shown previously (Brigneti et al., 1998; Li et al., 1999), on silenced GFP-transgenic N.benthamiana, shoots emerging following systemic infection by PVX expressing Cmv2b (TXMV–2b, herein referred to as PVX–C2b) display green fluorescence under UV illumination. Northern blot analysis of total RNAs extracted from the fluorescing tissues of these plants revealed the presence of significant levels of the transgene RNA from 15 days post-inoculation (d.p.i.) (Figure 4B, lane 1). Both the GFP fluorescence and transgene RNA levels were maintained in these and subsequent leaves throughout the remainder of the plant's growth (Figure 4A, lanes 4–6; Figure 4B, lanes 2 and 3). In contrast, infection by either PVX or a PVX construct containing a non-translatable CMV 2b coding sequence (TXMV-2bΔ, herein referred to as PVX–CΔ2b) resulted in no suppression of silencing (Figure 4A, lanes 7–10) and the plants showed only red chlorophyll fluorescence under UV. Significantly, silenced plants infected with PVX–2b6A showed no GFP fluorescence and the accumulation of GFP mRNA in these plants was as low as that found in PVX–CΔ2b-infected plants (Figure 4A, lanes 11–14; Figure 4B, lanes 4–6). Thus, replacing the NLS of Cmv2b with alanines abolished both the nuclear targeting and the silencing suppressor activity of Cmv2b. Similarly to PVX–2b6A infections, silenced plants inoculated with PVX expressing 2b6A–nls (PVX–2b6A–nls) demonstrated no suppression of PTGS (Figure 4B, lanes 10–12). This indicates that the 2b6A protein is not a functional suppressor of PTGS even when delivered into the nucleus. Therefore, the identified NLS of Cmv2b is required in its original context for the silencing suppression activity of the protein.
Fig. 4. Nuclear targeting and silencing suppression. Completely silenced (+) N.benthamiana plants were infected with in vitro RNA transcripts (A) or sap derived from transcript-inoculated plants (B). Total RNAs were extracted from the new leaves that had emerged after systemic infection was established at the days post-inoculation indicated above each lane, and analyzed by Northern blotting. The specificities of the 32P-labeled riboprobes used are indicated next to the panels. Equivalent loading [5 μg for (A) and 4 μg for (B)] of the total plant RNA for each lane was determined by methylene blue staining and densitometry of the 28S rRNA. (–), RNA extracted from an unsilenced GFP transgenic plant.
The second cytoplasmic/perinuclear-localized mutant of Cmv2b, 2bQAQ, displayed an interesting phenotype when expressed from the PVX vector. GFP fluorescence was not visible at 15 d.p.i. and the level of transgene RNA found (Figure 4B, lane 7) was marginally greater than in silenced plants infected with PVX–2b6A (Figure 4B, lane 4). However, green fluorescence was observed in the petioles, stem and leaf veins of the new leaves emerging after 15 d.p.i., and a significant accumulation of GFP RNA could be detected in these leaves (Figure 4B, lane 8). Intriguingly, additional new leaves emerging after these first green fluorescent leaves appeared showed no GFP fluorescence. Furthermore, transgene RNA accumulation in these later leaves (Figure 4B, lane 9) was as low as in silenced, unsuppressed leaves assayed at 15 d.p.i. (Figure 4B, lane 7). As with each infection, RT–PCR and sequence analysis of extracted RNAs from each time point demonstrated that the 2bQAQ coding sequence cloned into the PVX vector was stably expressed and accurately maintained in the viral progeny. Thus, although 2bQAQ accumulates outside of the nucleus, it retains a delayed, transient and tissue-limited ability to suppress the initiation of gene silencing. In contrast, inoculation of silenced N.benthamiana with a PVX construct expressing 2bQ resulted in a pattern of suppression (as shown by green fluorescence and Northern detection of the transgene RNA) identical to that seen for PVX–C2b (data not shown). The substitutions in both 2bQAQ and 2bQ were made according to the sequence alignment of six different 2b proteins (Ding et al., 1994) and were not expected to disrupt the conserved structure of the protein. The triplet QAQ is encoded by the TAV 2b protein, whereas the 2bQ substitution conforms to the sequence of the 2b protein of the Fny strain of CMV. Both the TAV 2b (Li et al., 1999) and Fny 2b (H.-S.Guo, unpublished data) proteins suppress PTGS more efficiently than the CMV Q–strain 2b protein used herein, suggesting that the PTGS suppression ability of the mutated protein should not be compromised. That the nuclear-targeted 2bQ remains an efficient silencing suppressor, whereas the suppressor activity of the non-nuclear 2bQAQ is severely compromised, further emphasizes the importance of nuclear targeting in PTGS suppression. It is not clear whether the weak suppressor activity of 2bQAQ is caused by a limited diffusion of the protein into the nuclei or as a consequence of the low level of discrete nuclear accumulation seen in 2bQAQ-expressing cells.
Two further constructs that demonstrated nuclear targeting when fused with GFP, 2bΔ5 and 2bΔ16, contained deletions outside of the NLS. Expression of these deletion mutants from the PVX genome resulted in PTGS suppression at least as efficient as the wild-type CMV 2b protein (Figure 4B, lanes 13 and 14). Taken together, the above results provide a strong correlation between the nuclear accumulation of Cmv2b and its efficiency as a suppressor of PTGS.
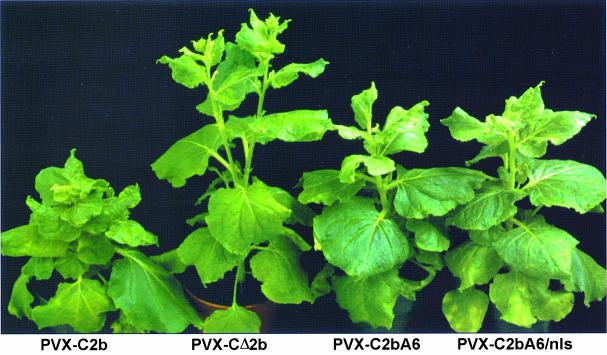
The 2b protein is an important virulence determinant of CMV (Ding et al., 1995). Furthermore, inoculation of PVX–C2b onto N.benthamiana resulted in a much more severe systemic disease than did PVX–CΔ2b infections (Figure 5), indicating that Cmv2b also functions as a determinant of virulence when expressed from the PVX genome. Under the growth conditions set in the Conviron growth chambers (constant 22°C and 75% relative humidity with a 16 h photoperiod), PVX–C2b infection resulted in severe stunting and leaf deformation. In contrast to infections using recombinant PVX expressing the TAV 2b protein (Li et al., 1999), PVX–C2b did not lead to the death of the infected plants under these conditions. No differences in virulence were observed when either wild-type or silenced GFP-transgenic plants were used for the infectivity studies. Using this assay system, it was found that PVX–2bQ (not shown) was as virulent as PVX–C2b (Figure 5). In contrast, PVX–2b6A and PVX–2b6A–nls, both of which were defective in silencing suppression, exhibited significantly reduced stunting and leaf symptoms when compared with PVX–C2b (Figure 5). PVX–2bQAQ, which was also compromised in its ability to suppress PTGS, showed a similar degree of stunting but a slightly more severe leaf mosaic when compared with PVX–2b6A and PVX–2b6A–nls (data not shown). Thus, these data indicate that PTGS suppression plays an important role in virulence determination.
Fig. 5. Silencing suppression and virulence determination. Symptoms on N.benthamiana plants inoculated with in vitro transcripts of, from left to right, pPVX–C2b, pPVX–CΔ2b, pPVX–2b6A or pPVX–2b6A–nls.
Interestingly, although PVX–2b6A and PVX–2b6A–nls were less virulent than PVX–C2b, both were found to be more virulent than PVX–CΔ2b (Figure 5). This indicates that 2b6A and 2b6A–nls retained some ability to exert the virulence determinant function despite being inactive in PTGS suppression. Furthermore, similar to PVX–CΔ2b, N.benthamiana plants systemically infected with PVX–2bΔ5 and PVX–2bΔ16 showed no apparent stunting (data not shown), suggesting that 2bΔ5 and 2bΔ16, both of which retained the wild-type suppressor activity, were not functional virulence determinants.
Discussion
In this report we have identified and demonstrated the functionality of an arginine-rich NLS in the 2b protein encoded by CMV. We have further provided evidence that nuclear targeting is involved in the suppression of PTGS. These findings indicate that Cmv2b may suppress PTGS in the cell nucleus and suggest that the intracellular signaling of PTGS, considered a primarily cytoplasmic process, may include a critical step in the nucleus.
The selective transport of proteins between the nucleus and the cytoplasm occurs through multi-subunit nuclear pore complexes (NPCs), which can accommodate the passive diffusion of molecules up to 9 nm in diameter (Palmeri and Malim, 1999). Thus, the translocation of proteins with masses exceeding 40–60 kDa is dependent upon active mechanisms classically mediated by basic, lysine-rich NLSs on the targeted protein, which act as binding sites for importin α (Görlich and Mattaj, 1996). Importin α in turn interacts with importin β, which mediates docking of the resultant ternary complex to the cytoplasmic face of the NPC. The subsequent translocation of this heterotrimer across the NPC is an energy-dependent process involving the Ran GTPase. Binding of the GTP-bound form of Ran to importin β at the nuclear face of the NPC triggers the disassembly of the heterotrimer and importin α/β are subsequently recycled back to the cytoplasm, while the NLS-bearing protein remains in the nucleus (Görlich and Mattaj, 1996).
Whilst comparable to the NLSs found in the large T–antigen of simian virus 40 (Görlich and Mattaj, 1996), the identified NLS of Cmv2b is a subclass of these monopartite NLSs and is more akin to the newly recognized arginine-rich NLSs found in the Rex protein of human T–cell leukemia virus type 1 (Palmeri and Malim, 1999) and the HIV–1 Tat and Rev proteins (Truant and Cullen, 1999). In contrast to the lysine-rich NLSs, these arginine-rich signals are independent of importin α and have been demonstrated to interact directly with importin β (Palmeri and Malim, 1999; Truant and Cullen, 1999). Using site-directed mutagenesis, we have demonstrated that the predicted arginine-rich NLS of Cmv2b is essential and sufficient for targeting the protein to the nucleus in tobacco suspension cells and whole N.benthamiana plants. However, it remains to be determined whether Cmv2b translocation is independent of importin α.
All of the 2b proteins encoded by the cucumoviruses (Ding et al., 1994; Shi et al., 1997a) contain potential NLSs in the N–terminal region (Figure 6, bold). The NLSs encoded by CMV subgroup IA (Fny and WAII), IB (K) and II (Q) strains are conserved by sequence and position (subgroup designations according to Roosinck et al., 1999). The predicted NLS encoded by the Fny and K 2b proteins, with a glutamine in the third position (Figure 6), is nearly identical to that encoded by 2bQGFP that is functional in vivo, suggesting that they may also accumulate in the cell nucleus. Indeed, cell fractionation and differential sedimentation studies also indicated an association of the Fny 2b protein with the nucleus (Mayers et al., 2000). Furthermore, the 2b proteins encoded by both TAV and peanut stunt virus (PSV) possess multiple NLSs in a combination of monopartite (Figure 6, bold) and bipartite sequences (Figure 6, boxed). Transient expression studies similar to those described have shown that the TAV 2b protein, when fused with GFP, is targeted to the nucleus (A.Lucy, unpublished data). It would therefore appear that all of the 2b proteins encoded by the cucumoviruses are likely to be targeted to the nuclei of invaded cells.
Fig. 6. Nuclear localization signals in the N–terminal region of the cucumoviral 2b proteins. Monopartite NLSs are identified with bold lettering and predicted NESs are in bold italics. Shaded residues indicate potential CKII phosphorylation sites. The TAV and PSV 2b proteins also encode putative bipartite NLS in the N–terminal half of the protein (boxed regions).
Further sequence analyses have identified potential phosphorylation sites for casein kinase II (CKII) (S/TxxD/E) and CDK2 (40SP41) conserved in all of the cucumoviral 2b proteins (Figure 6, shaded). This combination of an NLS, a CKII site and a CDC2-CDK2 site constitutes a CcN motif, which has been shown to be involved in the phosphorylation-regulated nuclear localization of a variety of proteins (Jans, 1995; Jans and Hubner, 1996). In addition, several of the aligned cucumoviral 2b proteins also contain dispersed leucine or isoleucine residues (Figure 6, bold italics) characteristic of the canonical nuclear export signals (NES; Görlich and Mattaj, 1996; Hood and Silver, 1999) of, for example, HIV–1 Rev protein (Fischer et al., 1995) and cyclin B1 (Hagting et al., 1999). Taken together, the existence of the putative CcN and NES motifs in Cmv2b suggests that the protein may shuttle between the nucleus and the cytoplasm, an effect perhaps controlled by the phosphorylation state of the protein. In this regard, it is of interest to note that deletion of the putative phosphorylation domain (40SPSE43; Figure 6) in 2bΔ5GFP was shown to have an enhancing effect on the nuclear accumulation of the protein. It is not clear whether the 30–40% of cells expressing 2bGFP that show a free GFP pattern of fluorescence are due to the activity of the NES and shuttling, nor, indeed, whether the enhanced nuclear accumulation of 2bΔ5GFP is due to an absence of regulation by phosphorylation.
It is interesting that Cmv2b, a suppressor of PTGS, targets to the cell nucleus. Replacing the predicted NLS of six residues with alanines (2b6A) led to the non-nuclear accumulation of the mutant 2b protein and abolished its activity in PTGS suppression. Substitution of part of this NLS with residues conserved in the aligned positions of related 2b proteins (Figure 6) further supports the link between nuclear targeting and PTGS suppression. A single-residue substitution (2bQ) that did not prevent the nuclear targeting of the 2b protein also had no detectable effect on its activity in PTGS suppression. In contrast, a three-residue substitution that prevented nuclear accumulation had a profound effect on PTGS suppression. The resultant mutant protein, 2bQAQ, exhibited a delayed, weak and transient suppressor activity. In view of the low molecular weight of Cmv2b (11.3 kDa; Ding et al., 1994), it cannot be ruled out that the observed activity of 2bQAQ resulted from low levels of the mutant protein entering the nucleus by diffusion. 2b6A may also diffuse into the nucleus, but was not an active silencing suppressor, perhaps because it contains non-conserved substitutions in contrast to 2bQ and 2bQAQ. Furthermore, although 2b6A–nlsGFP was efficiently targeted to the nuclei, 2b6A–nls was unable to suppress PTGS; thus, not only nuclear targeting, but also the original context and sequence of the NLS are required for active silencing suppression. Taken together, our data indicate that Cmv2b may suppress PTGS of the GFP transgene in the cell nucleus of N.benthamiana. This suggests that the establishment of PTGS in a new cell, which may eventually lead to RNA degradation in the cytoplasm, includes an indispensable step in the nucleus. This suggestion is supported by the observed de novo cytosine methylation of nuclear DNA associated with viral RNA replication in the cytoplasm (Jones et al., 1998) and the recent demonstration that the Caenorhabditis elegans lir–1/lin–26 pre-mRNA, which accumulates in the nucleus, is an effective target of RNAi (Bosher et al., 1999).
The precise relationship between the nuclear accumulation of Cmv2b and its silencing suppressor activity remains elusive, although it is conceivable that the viral protein functions as a competitor for nuclear trafficking, thereby effecting the nuclear accumulation of a protein factor(s) critical for PTGS initiation. Alternatively, it may be that Cmv2b functions in the nucleus as a general transcriptional activator or to enhance the transcription of a negative regulator of PTGS. In support of such a proposal, Jones et al. (1999) have recently demonstrated that transgene methylation is maintained in silenced plants infected with CMV, suggesting that the 2b protein does not suppress PTGS by a direct effect on the silenced gene.
Available data suggest that the silencing suppression activity encoded by plant viruses is closely related to virulence determination. The viral silencing suppressors identified to date (Anandalakshmi et al., 1998; Brigneti et al., 1998; Kaschau and Carrington, 1998; Voinnet et al., 1999) are all virulence determinants of their respective viruses and include the 2b proteins encoded by CMV and TAV (Ding et al., 1995, 1996), the potyviral HC–Pro (Cronin et al., 1995), the tomato bushy stunt tombusvirus 19k protein (Scholthof et al., 1995), the rice yellow mottle sobemo virus P1 protein (Bonneau et al., 1998) and the African cassava mosaic begomo virus AC2 protein (Hong et al., 1997). We have further shown here that Cmv2b mutants (2b6A, 2b6A–nls, 2bQAQ) that are defective in silencing suppression are also much less virulent than the wild type when expressed from the PVX vector, demonstrating for the first time a direct correlation between the suppression of gene silencing and the development of virus disease. Interestingly, we have also found that PVX–2bΔ5 and PVX–2bΔ16 were only as virulent as PVX–CΔ2b in N.benthamiana, even though both 2bΔ5 and 2bΔ16 retained the wild-type silencing suppression activity. Indeed, both of these mutations similarly revealed reduced virulence when expressed from the parental CMV background (Ding et al., 1995). Our data thus suggest that virulence determination by plant viruses is more complex than anticipated and requires other activities in addition to the suppression of PTGS. Nevertheless, 2bΔ5 and 2bΔ16, as efficient suppressors of PTGS but lacking the adverse disease-inducing characteristics of the parental 2b protein, may prove useful in plant biotechnology applications as tools to suppress PTGS of introduced transgenes.
Materials and methods
Plasmid constructs
DNA manipulations and cloning were carried out using standard procedures (Sambrook et al., 1989) unless stated otherwise. Mutant proteins are described using the numerical coordinates of the first and last substituted amino acids, given in single-letter code. DNA inserts in all constructs were sequenced in two orientations prior to use to ensure the fidelity of the PCRs.
Constructs for transient expression. Transient gene expression was controlled by the cauliflower mosaic virus 35S promoter and terminator as provided by plasmid pCass2 (Shi et al., 1997b). An EcoRV–SacI fragment containing the red-wavelength-shifted, plant-adapted GFP coding sequence (Haseloff and Amos, 1995; Heim et al., 1995) downstream of the translational enhancer of tobacco mosaic virus was obtained from pE6113–GFP (Zheng et al., 1997). This fragment was ligated into similarly digested pCass2 to give pGFP. XbaI and NcoI restriction enzyme sites were introduced by PCR onto the 5′ and 3′ ends, respectively, of the CMV 2b open reading frame (ORF) (Q strain) as encoded by pSK2b (Ding et al., 1994) and the resultant fragment ligated into similarly digested pGFP to yield p2bGFP. The introduction of an NcoI site at the 3′ end of the CMV 2b fragment replaced the termination codon of ORF 2b with a serine codon to produce an in-frame N–terminal translational fusion with GFP. A serine residue is thus present between the 2b protein and GFP in all of the GFP-expressing constructs examined herein. Site-directed mutagenesis of ORF 2b was performed using the three-step PCR protocol described by Ding et al. (1995). PCR fragments for each mutant contained XbaI and NcoI sites at the 5′ and 3′ ends, respectively, for ligation into pGFP as for p2bGFP to create p2b6AGFP, p2bQAQGFP, p2bQGFP, p2bΔ5GFP and p2bΔ16GFP. Sequence encoding the NLS from Cmv2b (22KRRRRR27) was appended onto the 3′ end of the 2b6A sequence by PCR and the product restriction digested as before and ligated into similarly digested pGFP to give p2b6A–nlsGFP. Flanking proline and valine residues surrounding the 2b6A–nls motif (pKRRRRRvs) were provided to simulate the context of the SV40 large T–antigen NLS (Görlich and Mattaj, 1996), which when translationally fused to the C–terminus of 2b6AGFP directed the protein into the nucleus (A.Lucy, unpublished data).
PVX constructs. Coding sequences for GFP and 2bGFP or its mutants were obtained as XbaI–EcoRI fragments from the transient expression constructs described above, end-filled and ligated into the (end-filled) SalI site of the PVX vector (pP2C2S; Brigneti et al., 1998) to give pPVX–GFP, pPVX–2bGFP, pPVX–2b6AGFP, pPVX–2bQAQGFP, pPVX–2bQGFP, pPVX–2bΔ5GFP, pPVX–2bΔ16GFP and pPVX–2b6A–nlsGFP. pPVX–C2b (pTXMV–2b) and pPVX–CΔ2b (pTXMV–2bΔ) have been described previously (Brigneti et al., 1998). ORFs encoding the mutant 2b proteins described above (without the GFP coding sequence) were PCR amplified from the transient expression constructs, with the introduction of a 3′ stop codon to replace the serine residue (NcoI site) and ligated into pP2C2S at the (end-filled) SalI site to give pPVX–2b6A, pPVX–2bQAQ, pPVX–2bQ, pPVX–2bΔ5, pPVX–2bΔ16 and pPVX–2b6A–nls.
Transient expression and subcellular localization in tobacco suspension cells
GFP fusion proteins were transiently expressed in N.tabacum BY2 tobacco suspension cells maintained as described by Banjoko and Trelease (1995). Cells were subcultured every 7 days or harvested after 3 days in culture for biolistic bombardment.
All constructs used for transient expression were purified using the Plasmid Maxi-Kit (Qiagen) and coated onto 1–μm-diameter gold beads (Bio-Rad) following Sanford et al. (1993). The Bio-Rad Model PDS-1000/He Biolistic Particle Delivery System set at a firing pressure of 1100 p.s.i. (firing distance 6 cm) was used to introduce the DNA into the target cells biolistically. The protocol for bombardment was largely according to Ratnayaka and Oard (1995), except that the cells were adhered onto agar plates rather than filter paper. Between bombardment and observation, cells were incubated at 20°C in total darkness. Mature, fully expanded leaves from healthy N.glutinosa plants were excised and placed on damp filter paper for the whole-leaf bombardments under conditions similar to the BY2 cells, except that following bombardment the leaves were incubated at 20°C with a 16 h photoperiod. Transient expression was examined under blue-light excitation (489 nm) from 16 h post-transfection and observed for the following 56 h. Cells showing GFP fluorescence were also examined under bright field illumination to identify the nuclei. Several hundred cells were examined for each transfection in repeat experiments. The distribution of the GFP fluorescence was scored objectively as follows: strong nuclear enrichment, cytoplasmic without nuclear exclusion and cytoplasmic with nuclear exclusion. Cytoplasmic distributions could be further classified as diffuse or punctate and any intra-nuclear compartmentalization was noted where clearly defined. Light micrographs were taken on Kodak Elitechrome ISO 200 slide film using a Nikon E800M microscope. Slides were then professionally scanned to disc at high resolution (Kodak). Stored images were collated in Adobe Photoshop for figure preparation. Additional observations and optical sections were made using the Bio–Rad MRC–1024 Confocal Imaging System attached to a Zeiss Axioplan microscope. Selected images were collated and printed from the Adobe Photoshop program without enhancement.
In vitro transcription, plant infection and Northern blot analysis
In vitro transcription reactions to produce infectious recombinant PVX RNAs, plant infections, induction of PTGS by Agrobacterium infiltration, GFP imaging and Northern blot analysis were as described previously (Li et al., 1999).
Virus progeny RNA analysis
Viral progeny RNAs were extracted from plants infected with each of the PVX constructs described at the time points indicated in the text and analyzed by RT–PCR and DNA sequencing (Li et al., 1999). Briefly, the cucumoviral 2b coding sequences were first amplified by RT–PCR using a pair of primers flanking the SalI site of pP2C2S. The amplified fragments were then purified from agarose gels and either sequenced directly using the same pair of primers or cloned into a plasmid vector before sequencing.
Acknowledgments
Acknowledgements
We thank Lee Zhang for providing pE6113–GFP, Hong–Sien Kwee for the BY2 cell starter culture and colleagues in our group for useful discussions. This work was financially supported by the National Science and Technology Board, Singapore.
References
- Anandalakshmi R., Pruss, G.J., Ge, X., Marathe, R., Smith, T.H. and Vance, V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl Acad. Sci. USA, 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahramian M.B. and Zarbl, H. (1999) Transcriptional and posttranscriptional silencing of rodent α1(I) collagen by a homologous transcriptionally self-silenced transgene. Mol. Cell. Biol., 19, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjoko A. and Trelease, R.N. (1995) Development and application of an in vivo plant peroxisome import system. Plant Physiol., 107, 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D.C. (1999) Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol., 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. and English, J.J. (1996) Ectopic pairing of homologous DNA and post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol., 32, 79–88. [DOI] [PubMed] [Google Scholar]
- Beclin C., Berthome, R., Palauqui, J.C., Tepfer, M. and Vaucheret, H. (1998) Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral transgenes. Virology, 252, 313–317. [DOI] [PubMed] [Google Scholar]
- Bird A. (1992) The essentials of DNA methylation. Cell, 70, 5–8. [DOI] [PubMed] [Google Scholar]
- Bonneau C., Brugidou, C., Chen, L., Beachy, R.N. and Fauquet, C. (1998) Expression of the rice yellow mottle virus P1 protein in vitro and in vivo and its involvement in virus spread. Virology, 244, 79–86. [DOI] [PubMed] [Google Scholar]
- Bosher J.M., Dufourcq, P., Sookhareea, S. and Labouesse, M. (1999) RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics, 153, 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J., 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chalfie M., Tu, Y., Euskirchen, G., Ward, W.W. and Prasher, D.C. (1995) Green fluorescent protein as a marker for gene expression. Science, 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Cogoni C. and Macino, G. (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature, 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni C., Irelan, J.T., Schumacher, M., Schmidhauser, T.J., Selker, E.U. and Macino, G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Covey S.N., Al–Kaff, N.S., Langara, A. and Turner, D.S. (1997) Plants combat infection by gene silencing. Nature, 385, 781–782. [Google Scholar]
- Cronin S., Verchot, J., Haldeman–Cahill, R., Schaad, M.C. and Carrington, J.C. (1995) Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell, 7, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker A., van Montagu, M. (1997) Post-transcriptional gene silencing in plants. Curr. Opin. Cell Biol., 9, 373–382. [DOI] [PubMed] [Google Scholar]
- Ding S.W., Anderson, B.J., Haase, H.R. and Symons, R.H. (1994) New overlapping gene encoded by the cucumber mosaic virus genome. Virology, 198, 593–601. [DOI] [PubMed] [Google Scholar]
- Ding S.W., Li, W.X. and Symons, R.H. (1995) A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J., 14, 5762–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.W., Shi, B.J., Li, W.X. and Symons, R.H. (1996) An interspecies hybrid RNA virus is significantly more virulent than either parental virus. Proc. Natl Acad. Sci. USA, 93, 7470–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J.J., Mueller, E. and Baulcombe, D.C. (1996) Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell, 8, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. (1999) RNA-triggered gene silencing. Trends Genet., 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber, J., Boelens, W.C., Mattaj, I.W. and Luhrmann, R. (1995) The HIV–1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fujii G., Tsuchiya, R., Ezoe, E. and Hirohashi, S. (1999) Analysis of nuclear localization signals using a green fluorescent protein-fusion protein library. Exp. Cell Res., 251, 299–306. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Mattaj, I.W. (1996) Nucleocytoplasmic transport. Science, 271, 1513–1518. [DOI] [PubMed] [Google Scholar]
- Grebenok R.J., Pierson, E., Lambert, G.M., Gong, F.C., Afonso, C.L., Haldeman–Cahill, R., Carrington, J.C. and Galbraith, D.W. (1997) Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J., 11, 573–586. [DOI] [PubMed] [Google Scholar]
- Guo H.-S. and Garcia, J.A. (1997) Delayed resistance to plum pox potyvirus mediated by a mutated RNA replicase gene: involvement of a gene silencing mechanism. Mol. Plant Microbe Interact., 10, 160–170. [Google Scholar]
- Guo H.-S., Lopez-Moya, J.J. and Garcia, J.A. (1999) Mitotic stability of infection-induced resistance to plum pox potyvirus associated with transgene silencing and DNA methylation. Mol. Plant Microbe Interact., 12, 103–111. [DOI] [PubMed] [Google Scholar]
- Hagting A., Jackman, M., Simpson, K. and Pines, J. (1999) Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol., 9, 680–689. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. (1999) A species of small antisense RNA in post-transcriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Haseloff J. and Amos, B. (1995) GFP in plants. Trends Genet., 11, 328–329. [DOI] [PubMed] [Google Scholar]
- Heim R., Cubitt, A.B. and Tsien, R.Y. (1995) Improved green fluorescence. Nature, 373, 663–664. [DOI] [PubMed] [Google Scholar]
- Hong Y., Saunders, K. and Stanley, J. (1997) Transactivation of dianthin transgene expression by African cassava mosaic virus AC2. Virology, 228, 383–387. [DOI] [PubMed] [Google Scholar]
- Hood J. and Silver, P.A. (1999) In or out? Regulating nuclear transport. Curr. Opin. Cell Biol., 11, 241–247. [DOI] [PubMed] [Google Scholar]
- Jans D.A. (1995) The regulation of protein transport to the nucleus by phosphorylation. Biochem. J., 311, 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans D.A. and Hubner, S. (1996) Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev., 76, 651–685. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes, T.L. and Richards, E.J. (1999) Maintenance of genomic methylation requires a SW12/SNF2-like protein. Nature Genet., 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jones A.L., Thomas,C.L. and Maule,A.J. (1998) De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J., 17, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J. and Baulcombe, D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D. and Carrington, J.C. (1998) A counterdefensive strategy of plant viruses: suppression of post-transcriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Kennerdell J.R. and Carthew, R.W. (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- Köhler R.H., Zipfel, W.R., Webb, W.W. and Hanson, M.R. (1997) The green fluorescent protein as a marker to visualize plant mitochondria in vivo. Plant J., 11, 613–621. [DOI] [PubMed] [Google Scholar]
- Kooter J.M., Matzke, M.A. and Meyer, P. (1999) Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci., 4, 340–347. [DOI] [PubMed] [Google Scholar]
- Kost B., Spielhofer, P. and Chua, N.H. (1998) A GFP–mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J., 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Kumagai M.H., Donson, J., Della–Cioppa, G., Harvey, D., Hanley, K. and Grille, L.K. (1995) Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl Acad. Sci. USA, 92, 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.W., Lucy, A.P., Guo, H.–S., Li, W.X., Ji, L.H., Wong, S.M. and Ding, S.W. (1999) Strong host resistance targeted against a viral suppressor of the plant gene silencing defense mechanism. EMBO J., 18, 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo J.A., Silva–Rosales, L., Proebsting, W.M. and Dougherty, W.G. (1993) Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell, 5, 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann J.U., Endl, I. and Bosch, T.C. (1999) Silencing of developmental genes of Hydra. Dev. Biol., 214, 211–214. [DOI] [PubMed] [Google Scholar]
- Mayers C.N., Palukaitis, P. and Carr, J.P. (2000) Subcellular distribution analysis of the cucumber mosaic virus 2b protein. J. Gen. Virol., 81, 219–226. [DOI] [PubMed] [Google Scholar]
- Metzlaff M., O'Dell, M., Cluster, P.D. and Flavell, R.B. (1997) RNA-mediated degradation and chalcone synthase A silencing in petunia. Cell, 88, 845–854. [DOI] [PubMed] [Google Scholar]
- Nakai K. and Kanehisa, M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. http://psort.nibb.ac.jp:8800/form.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., Lemieux, C. and Jorgensen, R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H., Tschudi, C., Gull, K. and Ullu, E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett H.S., Epel, B.L., Kahn, T.W., Heinlein, M., Watanabe, Y. and Beachy, R.N. (1996) Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J., 10, 1079–1088. [DOI] [PubMed] [Google Scholar]
- PalBhadra M., Bhadra, U. and Birchler, J.A. (1997) Cosuppression in Drosophila: gene silencing of alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell, 90, 479–490. [DOI] [PubMed] [Google Scholar]
- Palmeri D. and Malim, M.H. (1999) Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol. Cell. Biol., 19, 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F.G., Harrison, B.D. and Baulcombe, D.C. (1997) A similarity between viral defense and gene silencing in plants. Science, 276, 1558–1560. [DOI] [PubMed] [Google Scholar]
- Ratcliff F.G., MacFarlane, S.A. and Baulcombe, D.C. (1999) Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell, 11, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayaka I.J.S and Oard, J.H. (1995) A rapid method to monitor DNA precipitation onto microcarriers before particle bombardment. Plant Cell Rep., 14, 794–798. [DOI] [PubMed] [Google Scholar]
- Roosinck M.J., Zhang, L. and Hellwald, K.H. (1999) Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol., 73, 6752–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F., Vayassie, L., Klotz, C., Sperling, L. and Madeddu, L. (1998) Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell, 9, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sánchez Alvarado A. and Newmark, P.A. (1999) Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl Acad. Sci. USA, 96, 5049–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.C., Smith, F.D. and Russell, J.A. (1993) Optimizing the biolistic process for different biological applications. Methods Enzymol., 217, 483–509. [DOI] [PubMed] [Google Scholar]
- Scheid O.M., Afsar, K. and Paszkowski, J. (1998) Release of epigenetic gene silencing by trans-acting mutations in Arabidopsis. Proc. Natl Acad. Sci. USA, 95, 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel W., Pelissier, T., Riedel, L., Thalmier, S., Schiebel, R., Kempe, D., Lottspeich, F., Sanger, H.L. and Wassenegger, M. (1998) Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell, 10, 2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof H.B., Scholthof, K.B. and Jackson, A.O. (1995) Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell, 7, 1157–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B.J., Ding, S.W. and Symons, R.H. (1997a) In vivo expression of an overlapping gene encoded by the cucumoviruses. J. Gen. Virol., 78, 237–241. [DOI] [PubMed] [Google Scholar]
- Shi B.J., Ding, S.W. and Symons, R.H. (1997b) Plasmid vector for cloning infectious cDNAs from plant RNA viruses: high infectivity of cDNA clones of tomato aspermy cucumovirus. J. Gen. Virol., 78, 1181–1185. [DOI] [PubMed] [Google Scholar]
- Sijen T., Wellink, J., Hiriart, J.B., van Kammen, A. (1996) RNA-mediated virus resistance: role of repeated transgenes and delineation of targeted regions. Plant Cell, 8, 2277–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.J., Watson, C.F., Bird, C.P., Ray, J., Schuch, W. and Grierson, D. (1990) Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol. Gen. Genet., 224, 477–481. [DOI] [PubMed] [Google Scholar]
- Truant R. and Cullen, B.R. (1999) The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β–dependent nuclear localization signals. Mol. Cell. Biol., 19, 1210–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Krol A.R., Mur, L.A., Beld, M., Mol, J.N.M. and Stuitje, A.R. (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van West P., Kamoun, S., van't Klooster, J.W. and Govers, F. (1999) Internuclear gene silencing in Phytophthora infestans. Mol. Cell, 3, 339–348. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Mourrain, P., Palauqui, J.C. and Vernhettes, S. (1998) Transgene-induced gene silencing in plants. Plant J., 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto, Y.M. and Baulcombe, D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl Acad. Sci. USA, 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargelius A., Ellingsen, S. and Fjose, A. (1999) Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun., 263, 156–161. [DOI] [PubMed] [Google Scholar]
- Zheng H., Wang, G. and Zhang, L. (1997) Alfalfa mosaic virus movement protein induces tubules in plant protoplasts. Mol. Plant Microbe Interact., 10, 1010–1014. [Google Scholar]
- Zhu X.F., Suzuki, K., Saito, T., Okada, K., Tanaka, K., Nakagawa, T., Matsuda, H. and Kawamukai, M. (1997) Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Mol. Biol., 35, 331–341. [DOI] [PubMed] [Google Scholar]